Abstract
Genome editing based on CRISPR (clustered regularly interspaced short palindromic repeats)-associated nuclease (Cas9) has been successfully applied in dozens of diverse plant and animal species, including the nematode Caenorhabditis elegans. The rapid life cycle and easy access to the ovary by micro-injection make C. elegans an ideal organism both for applying CRISPR-Cas9 genome editing technology and for optimizing genome-editing protocols. Here we report efficient and straightforward CRISPR-Cas9 genome-editing methods for C. elegans, including a Co-CRISPR strategy that facilitates detection of genome-editing events. We describe methods for detecting homologous recombination (HR) events, including direct screening methods as well as new selection/counterselection strategies. Our findings reveal a surprisingly high frequency of HR-mediated gene conversion, making it possible to rapidly and precisely edit the C. elegans genome both with and without the use of co-inserted marker genes.
Keywords: CRISPR-Cas9 system, Co-CRISPR, CRISPR-Cas9-mediated HR, CRISPR-Cas9-induced indels, blasticidin selection
SEQUENCE-specific immunity mechanisms such as RNA interference (Voinnet 2001; Zamore 2001; Grishok and Mello 2002; Hannon 2002) and CRISPR (clustered regularly interspaced short palindromic repeats)-Cas9 (Horvath and Barrangou 2010; Bhaya et al. 2011; Terns and Terns 2011; Wiedenheft et al. 2012) provide sophisticated cellular defense against invasive nucleic acids. Understanding how these defense systems work has enabled researchers to redirect them at cellular targets, providing powerful tools for manipulating both gene expression and the cellular genome itself. The CRISPR-Cas9 system is a bacterial antiviral mechanism that captures fragments of viral DNA in specialized genomic regions for reexpression as small-guide RNAs (sgRNAs) (Bhaya et al. 2011; Terns and Terns 2011; Wiedenheft et al. 2012). In bacterial cells Cas9–sgRNA complexes provide acquired immunity against viral pathogens (Bhaya et al. 2011; Terns and Terns 2011; Wiedenheft et al. 2012). When coexpressed along with an artificial sgRNA designed to target a cellular gene, the Cas9 nuclease has been shown to efficiently direct the formation of double-strand breaks at the corresponding target locus (Jinek et al. 2012). Though bacterial in origin, this mechanism works efficiently even within the context of eukaryotic chromatin (Gilbert et al. 2013). Genome editing using CRISPR-Cas9 has recently been demonstrated in numerous organisms, providing a powerful new tool with rapidly growing—if not infinite—potential for diverse biological applications (Bassett et al. 2013; Chang et al. 2013; Cho et al. 2013a; Cong et al. 2013; Dicarlo et al. 2013; Dickinson et al. 2013; Feng et al. 2013, 2014; Friedland et al. 2013; Gratz et al. 2013; Jiang et al. 2013; Mali et al. 2013b; Wang et al. 2013; Ma et al. 2014; Yu et al. 2014; Zhou et al. 2014).
The CRISPR-Cas9 system has also been successfully applied to Caenorhabditis elegans. Methods that have been used to express Cas9 include mRNA injection and transgene-driven expression from a constitutive or an inducible promoter (Chen et al. 2013; Chiu et al. 2013; Cho et al. 2013b; Dickinson et al. 2013; Friedland et al. 2013; Katic and Grosshans 2013; Lo et al. 2013; Tzur et al. 2013; Waaijers et al. 2013; Zhao et al. 2014). The U6 promoter has been used to drive sgRNA expression (Chiu et al. 2013; Dickinson et al. 2013; Friedland et al. 2013; Katic and Grosshans 2013; Waaijers et al. 2013). The system has been used widely to produce small insertions and deletions (indels) that shift the reading frame of the target gene, often resulting in premature termination of translation and loss-of-function phenotypes (Chiu et al. 2013; Cho et al. 2013b; Friedland et al. 2013; Lo et al. 2013; Waaijers et al. 2013). In addition, single-strand oligonucleotides have been used as donor molecules to precisely alter a target gene through homologous recombination (HR) (Zhao et al. 2014), and a selection scheme has been developed that allows the HR-mediated insertion of large sequence tags such as GFP (Chen et al. 2013; Dickinson et al. 2013; Tzur et al. 2013).
Despite these important advances, current CRISPR protocols for inducing indels and HR events in C. elegans could benefit from refinement. For example, different sgRNAs targeting the same gene can result in substantially variable DNA cleavage efficiencies (Bassett et al. 2013; Chen et al. 2013; Wang et al. 2014); thus, identifying active sgRNAs can be time consuming and costly.
In this study, we investigate several strategies with which to streamline CRISPR-Cas9-mediated genome editing in C. elegans. We describe a Co-CRISPR strategy that can facilitate the identification of functional sgRNAs and can enrich for transgenic animals carrying an HR event. We show that HR events can be identified without the need for selection at a rate of ∼1% to as high as 10% of F1 transgenic animals scored. This high frequency allows HR events to be identified by directly scoring for GFP expression or by PCR screening to detect HR-induced DNA polymorphisms. Direct screening allows precise genome alterations that minimize the footprint of DNA alterations, such as inserted selectable markers, at the target locus. However in some cases, such as whole-gene deletion assays that may induce lethality, selection can be useful for both identifying and maintaining HR events. We therefore describe a straightforward selection/counterselection protocol that facilitates recovery of HR events where having a marker inserted at the target site might be tolerated or useful. Together the findings presented here take much of the guesswork out of using the CRISPR-Cas9 system in C. elegans, and the Co-CRISPR strategy employed here may also prove useful in other organisms.
Materials and Methods
Genetics
All strains in this study were derived in the Bristol N2 background and maintained on nematode growth medium (NGM) plates seeded with OP50 (Brenner 1974).
Selection of sgRNA target sequences
We manually searched for target sequences consisting of G(N)19NGG (Wiedenheft et al. 2012; Friedland et al. 2013) near the desired mutation site. For HR-mediated repair experiments such as gfp knock-in and introduction of point mutation, we selected the target sequences where it was possible to introduce a silent mutation in the PAM site. Target sequences are listed in Supporting Information, Table S1.
Preparation of sgRNA constructs
We replaced the unc-119 target sequence in pU6::unc-119 sgRNA vector (Friedland et al. 2013) with the desired target sequence using overlap extension PCR. The pU6::unc-119 sgRNA vector was diluted to 2 ng/µl and used as a template to generate two overlapping fragments. The first was amplified using the primers CMo16428 and sgRNA R, resulting in the U6 promoter fused to the GN19 target sequence (U6p::GN19). The second was amplified using the primers CMo16429 and sgRNA F, resulting in the GN19 target sequence fused to the sgRNA scaffold and U6 3′-UTR. These two PCR products were mixed together, diluted 1:50, and used as a template for a PCR reaction with primers CMo16428 and CMo16429. The resulting pU6::target sequence::sgRNA scaffold::U6 3′-UTR fusion products were gel purified and inserted into the pCR-Blunt II-TOPO vector (Invitrogen, no. K2800-20). We used iProof high-fidelity DNA polymerase (Bio-Rad, no. 172-5300) in all PCR reactions above to minimize errors of PCR amplification, and all the constructs were confirmed by DNA sequencing. Primers sequences are listed in Table S2.
Preparation of HR donor vectors
pie-1 donor plasmids (point mutations and gfp and flag fusions):
pie-1 genomic sequence (LGIII:12,425,767-12,428,049) was amplified using the primers C_PIE-1 PF and C-PIE-1 PR and the resulting PCR product was inserted into the pCR-Blunt II-TOPO vector (Invitrogen, no. K2800-20).
The K68A and K68R mutations were introduced by PCR sewing (or overlap extension PCR). The pie-1 plasmid described above was used as a template to generate overlapping PCR products with the corresponding site-specific mutations. The overlapping PCR products were mixed together (1:1), diluted 50-fold with water, and used as a template in the PCR-sewing step with an external primer pair. The fused PCR products were gel purified and cloned into the pCR-Blunt II-TOPO vector.
For building gfp::pie-1 donor constructs, an NheI restriction site was inserted immediately after and in frame with the start codon of pie-1 by PCR sewing. A plasmid containing wild-type or mutant pie-1 sequence was used as a template to generate a left-arm PCR product flanked by BsiWI and NheI restriction sites and a right-arm PCR product flanked by NheI and NgoMIV restriction sites. The products were digested with NheI, purified using a PCR cleanup kit, and ligated together. The ligated products were cloned into the pCR-Blunt II-TOPO vector, and plasmids containing the appropriately ligated fragments were identified. A BsiWI and NgoMIV fragment, containing the in frame NheI site immediately after the start codon, was released and ligated to similarly digested pie-1 constructs. The GFP coding region amplified from pPD95.75 (Addgene) was inserted into the NheI site.
For pie-1::gfp or pie-1::flag, a 1.6-kb fragment (LGIII 12,428,172–12,429,798) was amplified from genomic DNA and inserted into the pCR-Blunt II-TOPO vector (Invitrogen, no. K2800-20). Overlap extension PCR was used to introduce an NheI site immediately before the stop codon in this fragment of pie-1. A 3×flag sequence (gattacaaagaccatgatggtgactataaggatcatgatattgactataaagacgatgacgataag) was inserted into the NheI site.
Finally, we used PCR sewing to introduce silent mutations that disrupt the PAM site (NGG to NTG) in each HR donor. The above plasmids were used as templates to generate the initial PCR products for PCR sewing. The final products were cloned into the pCR-Blunt II-TOPO vector.
We used iProof high-fidelity DNA polymerase (Bio-Rad, no. 172-5300) in all PCR reactions above. Primers are listed in Table S3.
mCherry::vet-2 and flag::vet-2 donor construct:
A 2411-bp DNA fragment of the vet-2 gene, including 1249 bp of sequence upstream and 1162 bp downstream of the vet-2 start codon (corresponding to the genomic sequence LGI:10,845,543-10,847,953), was amplified from genomic DNA and inserted into pBluescript KS (+) vector (Addgene). An XmaI site was introduced by PCR immediately after the vet-2 start codon. The mcherry coding sequence amplified from pCFJ90 (Addgene) or 3×flag sequence was inserted into the XmaI site.
smo-1::flag donor plasmid:
smo-1 genomic sequence (LGI: 1,340,243–1,341,558) was amplified from N2 genomic DNA and inserted into the pCR-Blunt II-TOPO vector (Invitrogen, no. K2800-20). Overlap extension PCR was used to introduce an NheI site immediately before the stop codon in this fragment of smo-1. The resulting PCR product was cloned into the pCR-Blunt II-TOPO vector. A 3×flag fragment with NheI overhangs was generated by annealing two overlapping oligonucleotides and ligated into the smo-1 donor plasmid. We mutated the PAM site (Figure S3C) as described for the pie-1 donors above.
gfp::pie-1 for MosSCI:
A 3744-bp fragment (ScaI–NotI) containing the pie-1 promoter was excised from pID3.01B (Addgene) and inserted into a modified MosSCI LGII vector (B1496) in which a NotI site was added to pCFJ151 (Frokjaer-Jensen et al. 2008; Shirayama et al. 2012). A 2631-bp PCR fragment containing the pie-1 open reading frame (ORF) and 3′-UTR was then inserted into the resulting plasmid to make a gfp::pie-1 plasmid for MosSCI. The plasmid was injected into the strain EG4322 at a concentration of 10 ng/μl by direct injection method to insert a single-copy gfp::pie-1 transgene on chromosome II at position 8,420,159.
BSD-fusion to pie-1:
The nucleotide sequence of the Blasticidin S resistance gene (BSD) from Aspergillus terreus was codon optimized for C. elegans and an artificial intron (gtaagagattttttaaaaatttattttttacactgttttttctcag) was inserted into the middle of the BSD ORF: the entire gene was de novo synthesized by GenScript. The BSD fragment containing the BSD ORF (439 bp), rpl-28 promoter (568 bp), and rpl-28 3′-UTR (568 bp) was inserted into pBluescript KS (+) vector (Addgene). The complete sequence of BSD marker is available upon request. A 1077-bp fragment of pie-1 left homology was inserted into the XbaI site before the rpl-28 promoter and a 1017-bp fragment of pie-1 right homology was inserted into the SalI–ApaI site after the rpl-28 3′-UTR. Blasticidin S (AG scientific, no. B-1247) was used to select animals transformed with the BSD gene.
cb-unc-119(+) donor plasmid:
Cbr-unc-119(+) (2216 bp) was amplified from pCFJ151 (Frokjaer-Jensen et al. 2008; Shirayama et al. 2012) using primers tailed on the 5′ end and the 3′ end with the loxP (ataacttcgtataatgtatgctatacgaagttat) sequence (Dickinson et al. 2013). This loxP::Cbr-unc-119(+)::loxP fragment was inserted into pBluescript-KS(+) vector (Addgene) linearized with XhoI. A 1006-bp fragment of the sequence upstream of the oma-1(tm1396) deletion (LGIV: 8,884,663-8,891,662) was inserted into the SpeI site on one side of the loxP::Cbr-unc-119(+)::loxP cassette, and a 1000-bp fragment of the sequence downstream of the oma-1(tm1396) deletion (LGIV:8,887,927-8,894,926) was inserted into the ApaI site after the loxP site.
Preparation of heat-shock-Cas9 plasmid
The Mos1 transposase ORF in pJL44 (Addgene) was replaced with Cas9 from Peft3::Cas9 vector (Friedland et al. 2013) to generate hs::Cas9 (pWU34) construct.
Micro-injection
DNA mixtures were micro-injected into the gonads of young adult worms. Plasmids for injection were prepared using a midiprep plasmid purification kit (Qiagen, no. 12143). For Co-CRISPR, we injected 50 ng/µl each vectors [Cas9 vector, unc-22 sgRNA vector (Co-CRISPR), two untested-sgRNAs, and pRF4::rol-6(su1006)] (Figure 2A). Micro-injection mixtures for HR contained 50 ng/μl each Cas9 vector, sgRNA vector, pRF4::rol-6(su1006), and HR donor vector. The final concentration of DNA in the injection mix did not exceed 200 ng/µl. For injection mixes with five different plasmids, 40 ng/µl of each plasmid was added. For HR experiments, we injected 40–60 worms and for disruptions, 20–30 worms. After recovering from injection, each worm was placed onto an individual plate.
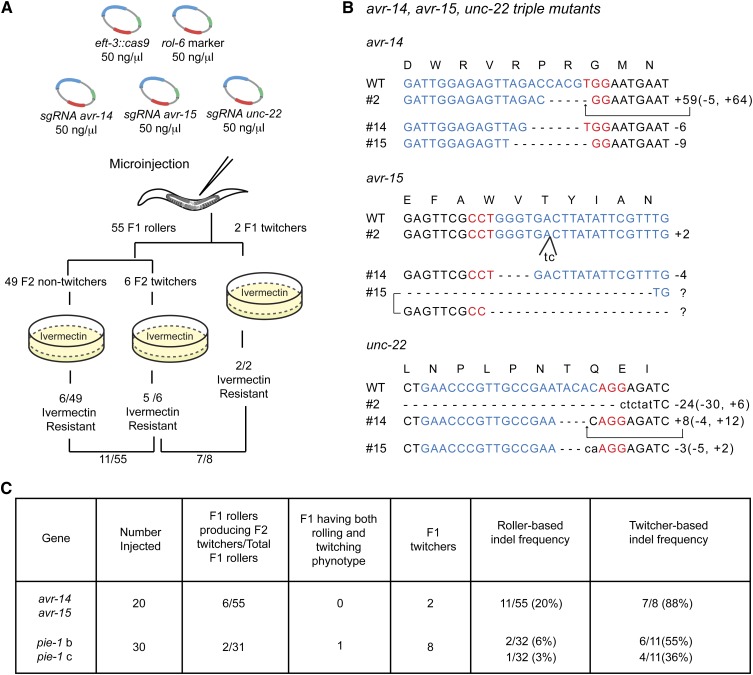
Figure 2.
“unc-22” Co-CRISPR as a marker to indicate actively expressed Cas9. (A) Schematic of Co-CRISPR strategy to identify functional sgRNAs targeting avr genes. sgRNAs targeting avr-14 and avr-15 were co-injected with a functional unc-22 sgRNA, the Cas9 expression vector, and the rol-6 transformation marker. F1 rollers or twitchers were transferred to individual plates. The plates were allowed to starve, and then they were copied to plates containing 2 ng/ml ivermectin to identify CRISPR-Cas9-induced avr-14; avr-15 double mutants. (B) Indel sequences in avr-14; unc-22; avr-15 triple mutants. avr-15 isolate 15 carried different indels on each allele. Sequences labeled with a question mark could not be precisely determined. (C) Comparison of twitcher-based indel frequency and roller-based indel frequency.
Screening for indels using Co-CRISPR
To validate untested sgRNAs we injected mixtures containing the unc-22 sgRNA and up to several untested sgRNAs (as described above). Three days after injection, F1 rollers and F1 twitchers were picked individually to plates and allowed to produce F2 progeny for 2–3 days. F1 twitchers and F1 rollers with twitching F2 progeny were then transferred to 20 µl lysis buffer for PCR, PAGE (see below), and/or DNA sequencing analysis. Total time from injection to indel detection was ∼6–7 days.
Screening for HR events
Direct detection of GFP:
This procedure works for donor vectors that cannot drive GFP without first integrating into the genomic target site. For GFP::PIE-1 it was necessary to mount gravid F1 rollers three at a time under coverslips on 2% agarose pads for screening at 40× magnification using a Zeiss Axioplan2 microscope. For bright GFP constructs, it should be possible to screen using a fluorescence dissecting scope. GFP-positive animals were recovered by carefully removing the coverslip and transferring to individual plates. After laying eggs for 1 day they were individually lysed in 20 μl lysis buffer for PCR confirmation of the GFP insertion. GFP-positive F2 homozygotes were then identified and correct insertion of GFP was confirmed by DNA sequencing. Total time from injection to recovery of heterozygotes was 3–4 days.
PCR detection:
F1 rollers were picked individually to plates and allowed to lay eggs for 1 day. For the Co-CRISPR assay, F1 rollers were allowed to produce F2 progeny for 2–3 days so that F2 twitcher progeny could be identified. (Note that F1 roller animals that segregate twitching progeny should be selected as these animals exhibit the highest HR frequency, while nonrolling F1 twitchers should be avoided; see Results and Discussion). F1 animals were then transferred into lysis buffer in indexed PCR tubes and were screened using primers outside the homology arms followed by restriction enzyme digestion to detect the insertion. In some experiments, 1 µl of the initial PCR reaction was used as a template for a second PCR reaction with primers within the donor sequences. Although useful, this latter procedure gave several false positives in our hands. Total time from injection to recovery of heterozygotes was 4 days. For the Co-CRISPR strategy, 3 more days were required to recover heterozygotes.
Selection/counterselection method:
Four days after injection, gravid F1 rolling adults were placed in groups of 10–15 animals per plate onto media containing ivermectin and blasticidin (Figure 4B). After 3–4 more days, the plates were scored for viable, fertile progeny. Insertion of BSD at the target locus was then confirmed by PCR and DNA sequencing (as described above). The total time from injection to recovery of HR events was 7–10 days. Although slightly longer in duration this procedure required ∼10 times less labor as only the relatively rare viable populations were subject to PCR and DNA sequence analysis. For donor molecules containing the Cbr-unc-119(+) selection, the procedure was essentially the same; however, blasticidin was omitted from the selective media and the recipient strain was both Cbr-unc-119 mutant and ivermectin resistant. Primers for screening HR events are listed in Table S3.
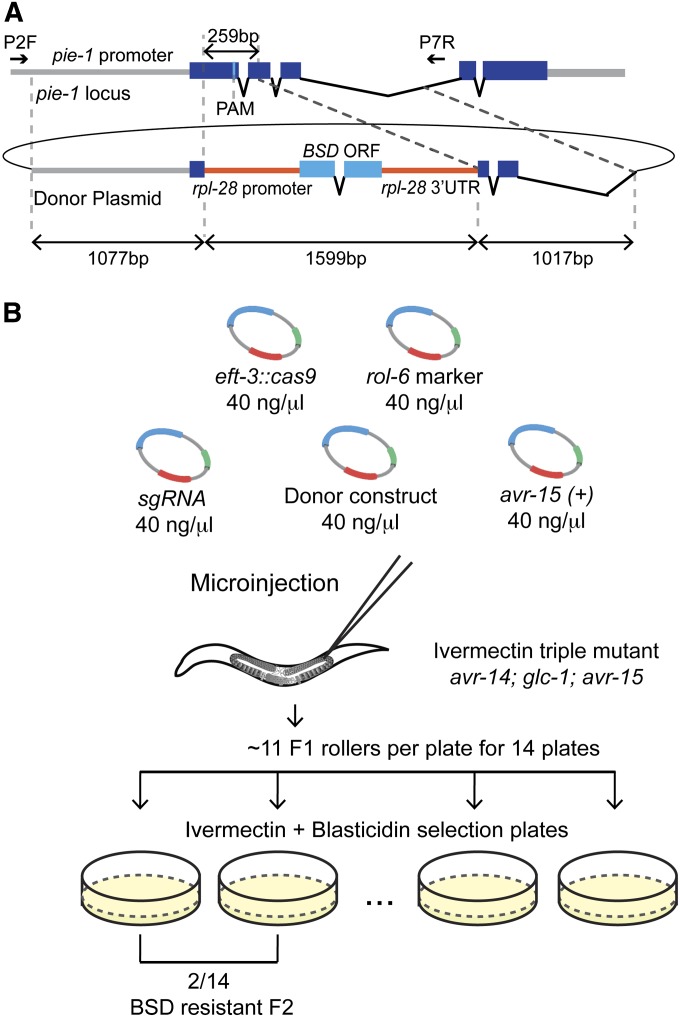
Figure 4.
A blasticidin-resistance marker to select pie-1 knockout mutants. (A) Schematic of the Cas9/sgRNA target sequence and an HR donor plasmid in which a heterologous blasticidin-resistance (BSD) gene replaces a region of pie-1 and is flanked by 1-kb homology arms. The BSD gene is under the control of the rpl-28 promoter (568 bp) and 3′-UTR (568 bp). (B) Schematic of the blasticidin selection strategy to precisely delete the pie-1 gene. pie-1a sgRNA was co-injected with the Cas9 expression vector, the rol-6 transformation marker, the pie-1∆::BSD donor construct, and the pCCM416::Pmyo-2::avr-15(+) counterselection vector. The indicated number of F1 rollers was transferred to the plates containing 2 ng/ml ivermectin to select against the extrachromosomal array and 100 μg/ml blasticidin to identify BSD knock-in lines. We identified two plates with resistant, fertile adults among 14 plates, 3–4 days after transferring animals.
Imaging
Images were captured with an ORCA-ER digital camera (Hamamatsu) and AxioVision (Zeiss) software.
Screening for small indels by PCR and PAGE
We designed primers to amplify (∼30 cycles) PCR products of 60–65 bp encompassing the CRISPR-Cas9 target site. PCR products were resolved on 15% polyacrylamide gels to distinguish dsDNA molecules that differ by as little as 1 bp. We found that we could screen for indels even in HR experiments, but it required two PCR steps. In the first PCR reaction (∼20 cycles), primers outside of the homology arms were used to avoid amplifying the donor sequence. In the second reaction (∼15 cycles), 1 µl of the first PCR reaction was used as a template to generate the 60- to 65-bp PCR product encompassing the CRISPR-Cas9 target site. TaKaRa Ex Taq (Takara, no. RR001) was used for the PCR reactions above. Primer sequences are listed in Table S3.
Immunoblotting
One hundred adult worms were lysed in 80 µl of 1× sample buffer (25 µl of M9 containing 100 worms, 25 µl of 2× lysis buffer, 20 µl of 4× NuPAGE LDS Sample buffer (Invitrogen, no. NP0008), and 10 µl of β-mercaptoethanol by boiling for 20 min, freezing, and boiling again for 10 min. The worm lysate proteins were separated on 4–12% NuPAGE Tris–acetate Mini Gels (Invitrogen, no. NP0335BOX). Proteins were transferred to Immun-Blot PVDF Membrane (Bio-Rad, no.162-0177) at 100 V for 1 hr at 4°. Mouse monoclonal anti-PIE-1 antibody (P4G5) (Mello et al. 1996) and rabbit polyclonal anti-PGL-1 antibody was used at 1:50 and 1:500, respectively.
Results and Discussion
Using a visible cotransformation marker enriches for genome-editing events
While conducting CRISPR-Cas9 experiments to induce mutations in the pie-1 gene, we used the dominant co-injection marker rol-6 to monitor injection quality. From 60 injected animals, we obtained 93 fertile F1 rollers. Remarkably, we noted that several of these F1 rollers (5/93) produced 100% dead embryos exhibiting the distinctive pie-1 maternal-effect embryonic lethal phenotype (Mello et al. 1996) (Figure 1A). Genomic sequencing of these F1 adults identified lesions in the pie-1 gene consistent with Cas9-directed cleavage (Figure 1B). In some cases the maternal and paternal alleles exhibited distinct lesions, while in others, the same lesion was found in both alleles (Figure 1B). Since the DNA was delivered into the ovary of an adult, after the switch from sperm to oocyte development, the paternal allele must have been targeted in the F1 zygote soon after fertilization. The fact that both alleles exhibit identical lesions in some animals suggests that a chromosome previously cut by Cas9 and repaired by non-homologous end joining (NHEJ) was used as a donor molecule to copy the lesion into the homolog.
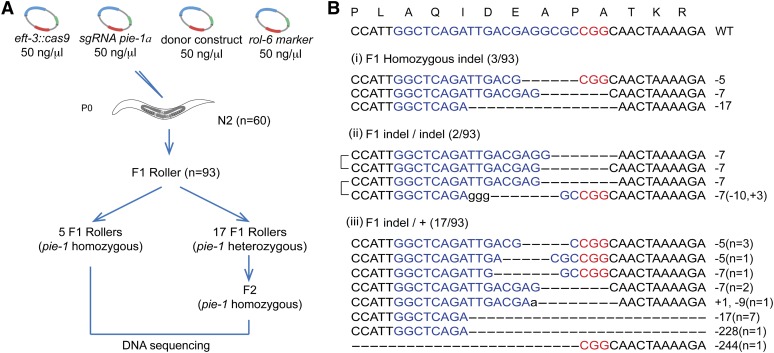
Figure 1.
Efficient CRISPR-Cas9-mediated gene disruption in transgenic animals. (A) Schematic of screen for CRISPR-Cas9 genome editing events. The dominant transformation marker rol-6 was co-injected with Cas9, pie-1a sgRNA, and donor plasmids. F1 rollers were screened for NHEJ-mediated indels by DNA sequencing. Among 93 F1 rollers, 22 indels were obtained. (B) Sequences of the wild-type pie-1 target site (top) and CRISPR-Cas9-mediated indels among F1 animals: (i) pie-1 homozygotes carrying the same indel on both alleles; (ii) pie-1 homozygotes carrying a different indel on each allele; and (iii) pie-1 heterozygotes. Lowercase letters indicate insertions, and dashes indicate deletions. The PAM is marked in red, and target sequences are marked in blue. The number of deleted (−) or inserted (+) bases is indicated to the right of each indel. The numbers in parentheses in (iii) represent the number of animals with the indels shown.
If the activation of Cas9 in the germline is broadly or nonspecifically mutagenic, then some injected animals would be expected to segregate novel mutants, including mutants with non-pie-1 dead-embryo phenotypes. To look for evidence of off-target mutagenesis, we screened among the progeny of F1 rollers for animals producing dead embryos, or other visible phenotypes. A careful examination of F2 and F3 populations revealed 17 populations from 93 F1 rollers that segregated numerous dead embryos (Figure 1A). However, examination of these dead embryos by Nomarski microscopy revealed the distinctive pie-1 mutant phenotype and no other phenotypes. Each of these 17 strains segregated pie-1 homozygotes at the expected Mendelian frequency, indicating that the original F1 rollers were heterozygous for pie-1 loss-of-function mutations. Sequencing of these strains revealed indels in the region of the pie-1 gene targeted by CRISPR-Cas9 (Figure 1B). In some cases, to avoid the delay and added cost of DNA sequencing, genomic DNA prepared from lysates of each candidate was amplified as ∼60-bp PCR products that were then analyzed on a 15% PAGE gel. This analysis easily detected lesions as small as 5 nt (Figure 1B and Figure S1A).
In addition to F1 rollers, we randomly selected F1 nonroller sibling progeny that were produced during the same time window as the F1 rollers. Among 100 nonroller siblings, we failed to find animals segregating dead embryos. Thus, using the dominant visible rol-6 marker to identify F1 transgenic animals (rollers) also identified animals in which Cas9 was active. It is important to note, however, that very few of the animals with pie-1 mutations continued to exhibit the roller phenotype in subsequent generations, suggesting that the rol-6 transgene expression was transient and present only in the F1 generation.
A Co-CRISPR strategy to detect genome-editing events
In practice, we have found that about half of sgRNAs tested are not effective. Thus, while the rol-6 marker was clearly useful for finding animals with CRISPR-Cas9-induced lesions, we nevertheless frequently had to screen through dozens or even hundreds of F1 rollers by PCR or sequencing only to conclude that CRISPR-Cas9 was not active in the injection. We therefore reasoned that co-injecting a proven sgRNA (one that works well and results in an easily recognized visible phenotype) would allow us to more directly identify animals in which Cas9 is active. We tested this strategy using an sgRNA targeting the muscle structural gene unc-22 (Moerman and Baillie 1979; Benian et al. 1993). We chose this sgRNA both because unc-22 loss of function causes a distinctive recessive paralyzed twitching phenotype that is easy to score and because this sgRNA works moderately well compared to other sgRNAs (Table S1). Thus, F1 and F2 unc-22 twitchers should arise from animals exposed to the greatest levels of Cas9 activity and should therefore also have active Cas9 loaded with the co-injected sgRNAs.
To test the Co-CRISPR strategy, we co-injected the unc-22 sgRNA with two previously validated sgRNAs targeting avr-14 and avr-15 (Figure 2A), two genes whose wild-type activities redundantly confer sensitivity to the potent nematicide ivermectin (Dent et al. 2000). The rol-6 marker was also included in these injections to facilitate the identification of twitchers that arise in the F2 among the progeny of F1 roller animals. We then measured, among 55 F1 rollers, the frequency of ivermectin-resistant strains (20%, n = 11) and twitcher strains (11%, n = 6) (Figure 2A). Strikingly, selecting for the twitching phenotype dramatically enriched for animals exhibiting ivermectin resistance. For example, among 8 F1 animals that were either twitching themselves or produced twitcher progeny, 7 (88%) also produced progeny resistant to ivermectin (Figure 2, A and C). We confirmed Co-CRISPR activity by sequencing the lesions in several of these strains (Figure 2B).
Similar results were obtained in several additional Co-CRISPR experiments (Figure 2C and data not shown). For example, we used this approach to test two uncharacterized sgRNAs targeting the 3′ end of pie-1 (Figure 2C and Figure S1B). Among 11 twitcher lines identified in the F1 or F2, we identified three indels by PCR and PAGE for one of the two sgRNAs (Figure S1B) and a single indel for the other sgRNA (Figure S1C). Sequence analysis confirmed these indels, which included a 6-nt deletion, a 24-nt insertion, and an 11-nt deletion for one sgRNA and a 16-nt insertion for the other. However, the PAGE detection method clearly underestimated the frequency of indels. Sequence analysis identified three heterozygous deletion mutations of 42, 43, and 603 nt that deleted primer binding sites and were thus too large to be detected by our PCR and PAGE analysis (Figure S1B). These unusually large deletions may reflect simultaneous cutting induced by the two adjacent sgRNAs whose targets are separated by 61 bp in this experiment (Figure S1B). In conclusion, these findings suggest that PAGE analysis of 10–20 Co-CRISPR lines should be sufficient to determine if an uncharacterized sgRNA is active. It should be noted that since many F1 rollers analyzed were heterozygous for unc-22 lesions, it was usually possible to find non-Unc segregants with an indel in the co-targeted gene using the unc-22 Co-CRISPR assay. However, if unc-22 is inconvenient for a particular experiment, our findings suggest that alternative Co-CRISPR sgRNAs targeting, for example, genes that when mutated confer resistance to ivermectin or benomyl or other genes with visible mutant phenotypes may be substituted (Table S1).
The observation that using nearby sgRNAs can induce deletions that remove the intervening sequence is consistent with previous findings in which large deletions were produced in this way (Horii et al. 2013; Ran et al. 2013; Ren et al. 2013; Wang et al. 2013; Zhou et al. 2014). Thus the Co-CRISPR strategy should facilitate the identification of deletions that remove the interval between two sgRNAs. However, further experimentation will be required to determine how large an interval can readily be eliminated. For the purpose of validating sgRNAs, our findings suggest that large deletions produced by testing multiple nearby sgRNAs simultaneously may confound the analysis of which sgRNAs are active. On the other hand, pooling sgRNAs targeting a number of different genes that are distant from one another in the genome should, in principle, allow several sgRNAs to be validated in a single Co-CRISPR micro-injection experiment.
Identification of HR events without a coselectable marker
We next sought to use CRISPR-induced double-strand breaks to drive HR. Several types of editing are possible, ranging from changing a single amino acid to inserting a protein tag such as GFP, or even deleting the entire target gene. In designing donor molecules to introduce point mutations or epitope fusions, we found it necessary to alter the sgRNA target sequence in the donor by mutating the protospacer adjacent motif (PAM) site or by introducing mismatches within the seed region (Jinek et al. 2012; Cong et al. 2013; Jiang et al. 2013; Sternberg et al. 2014). In our experience, failure to take this step frequently results in HR events containing CRISPR-Cas9-induced indels or a very low frequency of HR events, sometimes 0% (data not shown).
Previous studies successfully used single-strand oligonucleotide donor molecules (Zhao et al. 2014) or double-strand plasmid donor molecules (Dickinson et al. 2013) to induce HR events in C. elegans. However these studies relied on screening for a selectable phenotype introduced by the HR event. Given the high frequencies of NHEJ events detected in the studies above, we wondered if CRISPR-Cas9-mediated HR events could be recovered directly without the need for selection.
To test this idea, we decided to use CRISPR-Cas9-mediated HR to introduce the gfp coding sequence immediately downstream of the start codon in the endogenous pie-1 locus (Figure 3A). The donor plasmid in this experiment contained NheI restriction sites flanking the gfp coding sequence, 1-kb homology arms, and a silent mutation that disrupts the PAM sequence at the sgRNA target site (Figure 3A). We generated three different donor constructs: gfp::pie-1(WT), gfp::pie-1(K68A), and gfp::pie-1(K68R). Each donor molecule was co-injected with vectors to express the sgRNA, Cas9, and rol-6 marker. We then directly examined the resulting F1 rolling animals for GFP::PIE-1 expression in the germline and embryos using epifluorescence microscopy (Figure 3B, see Materials and Methods). Using this approach, we obtained 9 independent gfp::pie-1(K68A) lines from 92 F1 rollers, 1 gfp::pie-1(K68R) line from 69 F1 rollers, and 1 gfp::pie-1(WT) line from 72 F1 rollers. Subsequent analyses revealed that each of these F1 animals was heterozygous for gfp::pie-1, and each strain incorporated both the gfp coding sequence and the PAM site mutation, as well as the linked K68A and K68R missense mutations. For unknown reasons, we found that one of the nine gfp::pie-1(K68A) lines could not be maintained.
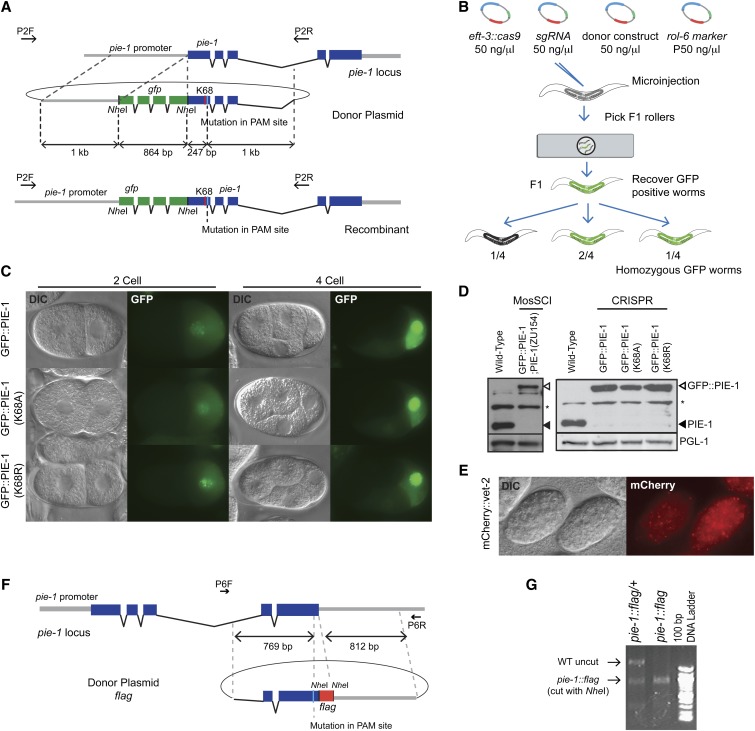
Figure 3.
HR-mediated knock-in to generate fusion genes at endogenous loci. (A) Schematic of the Cas9/sgRNA target site and the donor plasmid for gfp::pie-1 knock-ins. The donor plasmid contains the gfp coding sequence inserted immediately after the start codon of pie-1, 1 kb of homology flanking the CRISPR-Cas9 cleavage site, and a silent mutation in the PAM site. (B) Strategy to screen for gfp knock-in lines. We placed three F1 rollers at a time on a 2% agar pad and screened for GFP expression using epifluorescence microscopy. GFP-expressing worms were individually recovered and allowed to make F2 progeny for 1 day before being lysed for PCR and DNA sequence analysis. We confirmed Mendelian inheritance of gfp knock-in alleles among F2 progeny. (C) GFP::PIE-1 expression in the germline of two- to four-cell embryos of gfp::pie-1 knock-in strains. (D) Immunoblot analysis showing PIE-1 expression levels in wild-type animals, MosSCI-mediated gfp::pie-1 knock-in animals, and CRISPR-Cas9-mediated gfp knock-in animals. A MosSCI strain of gfp::pie-1; pie-1(zu154) was obtained by crossing gfp::pie-1 (LGII) with the pie-1(zu154) (LGIII) null mutant. (E) mCherry expression in late embryos of the mCherry::vet-2 knock-in strain. (F) Schematic of Cas9/sgRNA target sequence, PAM site, and donor plasmid for pie-1::flag knock-in. The PAM is located in the last exon of pie-1. The donor plasmid includes flag coding sequence immediately before the pie-1 stop codon and ∼800-bp homology arms flanking the target site. (G) PCR and restriction analysis of an HR event. PCR products were generated using the primers indicated in F, and the products were digested with NheI. The pie-1::flag gene conversion introduces an NheI RFLP that is observed in F1 heterozygous and F2 homozygous pie-1::flag animals.
The high rates of HR observed in the above study suggested that it should also be possible to recover HR events by screening DNA isolated from F1 rollers using PCR. To test this idea, we designed donor molecules to insert the pie-1 lysine 68 lesions without tag sequences (Figure S2A) or to insert sequences encoding the FLAG epitope immediately before the stop codon of the pie-1 gene (Figure 3F). For these HR experiments, we used 300 bp (no tag) and 800 bp (flag tag) flanking homology arms along with previously tested sgRNAs (Figure 3F, Figure S1B, and Figure S2A). We then used PCR to amplify the genomic DNA sequence from F1 rollers and restriction analysis to identify F1 heterozygotes carrying the desired insertion (Figure 3G, Figure S2, B and C). These studies identified two K68A HR events among 93 F1 rollers (60 injected worms) and three flag HR events among 84 F1 rollers (40 injected worms) (data not shown). A similar PCR-detection strategy was used to introduce mcherry into the gene vet-2. In this experiment, mCherry expression was not visible in adult F1 rollers, but was easily detected among the F2 embryos produced by PCR-positive animals (Figure 3E). Taken together these findings show that CRISPR-Cas9-mediated HR occurs at a remarkably high frequency in C. elegans.
We compared the expression and localization of GFP::PIE-1 protein in our newly generated gfp::pie-1 knock-in strains to strains in which gfp::pie-1 was inserted at a heterologous site in the genome by MosSCI (Frokjaer-Jensen et al. 2008; Shirayama et al. 2012). The knock-in strains showed the expected localization of PIE-1 in two- to four-cell embryos (Figure 3C). Strikingly, immunoblot analysis using the PIE-1 monoclonal antibody (P4G5) revealed that GFP::PIE-1 protein was expressed at a much higher level in the CRISPR-Cas9-induced knock-in strains, similar to the expression level of endogenous PIE-1 protein (Figure 3D). These results are consistent with a previous study (Dickinson et al. 2013) and indicate, perhaps not surprisingly, that insertion of GFP into the endogenous locus can achieve near optimal expression levels of the tagged protein.
Co-CRISPR for identifying HR events
Most of the HR work described above was performed before we realized the utility of Co-CRISPR markers for validating sgRNAs. To determine if the Co-CRISPR strategy could facilitate recovery of HR events, we co-injected unc-22 sgRNAs along with CRISPR HR injection mixes targeting vet-2, pie-1, and smo-1 genes (Figure S3). The findings from these studies suggest that using a Co-CRISPR marker can increase the frequency of HR events in the range of approximately two- to fourfold over those observed by first selecting F1 roller animals (Table 1). Interestingly, however, these studies required a modification to the Co-CRISPR screening strategy. For testing sgRNAs using Co-CRISPR, we found that F1 and F2 twitchers were equally likely to exhibit co-induction of indels with the second sgRNA. However, our data suggest that HR events were not enriched and might be depleted among nonrolling, F1 twitcher animals. One possible explanation for this surprising finding is that Cas9–sgRNA complexes may assemble in the germline cytoplasm and then segregate into maturing oocytes independently of the co-injected DNA (including both the roller DNA and of course the donor DNA plasmids). Zygotes inheriting programmed Cas9 could undergo NHEJ, but HR-directed repair would not be possible without the donor vector. Consistent with this reasoning, we found that in most cases HR events were enriched only among F1 animals that were both rolling, and thus had inherited the injected DNA, and also segregated twitching progeny, indicating that Cas9 was active (Table 1). For example among 145 F1 rollers, we found seven animals heterozygous for a 3′ insertion of gfp into the pie-1 locus. Among the F1 twitchers that were nonrolling, zero were GFP positive, while among the 4 F1 rollers that segregated twitching progeny, 2 (50%) were GFP positive. One convenient aspect of searching for HR events among F1 rollers heterozygous for unc-22 twitchers was that the unlinked twitcher phenotype could easily be segregated away in subsequent generations. These findings suggest that Co-CRISPR screening can enhance the detection of HR events. Indeed, we always found at least one HR event among the F1 rollers with twitcher progeny (3/29, 2/4, and 1/12). However, in most cases additional HR events were also recovered by scoring all the F1 rollers (Table 1).
Table 1. Co-CRISPR strategy for HR events.
| HR donor/targeted gene | No. injected | F1 rollers producing F2 twitchers/total F1 rollers | F1 having both rolling and twitching phenotype | F1 twitchers | F1 Twitcher-based HR frequency | Roller-based HR frequency | Roller producing F2 twitchers-based HR frequency |
|---|---|---|---|---|---|---|---|
| flag::vet-2/vet-2 | 40 | 29/65 | 0 | 62 | 2/62 (3%) | 4/65 (6%) | 3/29 (10%) |
| pie-1::gfp/pie-1 | 40 | 4/145 | 0 | 7 | 0/7 (0%) | 7/145 (5%) | 2/4 (50%) |
| smo-1::flag/smo-1 | 40 | 12/55 | 10 | 22 | 1/22 (5%) | 1/55 (2%) | 1/12 (8%) |
A selection/counterselection strategy for recovering HR events
The above findings demonstrate that selections are not necessary for identifying and recovering HR events using the CRISPR-Cas9 system. However, for some experiments a dominant selection could save considerable time and expense, especially where insertion of heterologous DNA is likely to be tolerated, for example, when generating a null allele of a gene or when one wishes to precisely delete noncoding genes or regulatory elements. The inserted marker also has the potential benefit of providing a selection for maintaining strains that may not be homozygous viable. Previous studies have described several selection strategies, including unc-119, NeomycinR, PuromycinR, and HygromycinR (Frokjaer-Jensen et al. 2008; Giordano-Santini et al. 2010; Semple et al. 2010; Frokjaer-Jensen et al. 2012; Radman et al. 2013). To test a selection/counterselection scheme for CRISPR-induced HR, we decided to employ the unc-119(+) marker as well as a new worm antibiotic-resistance marker expressing the bacterial BSD gene as selectable markers and the avr-15 gene as a counterselectable marker. We have previously shown that introducing an avr-15(+) plasmid into extrachromosomal arrays and balancer chromosomes can be used to facilitate their counterselection by growth on ivermectin (Duchaine et al. 2006; Shirayama et al. 2012). This counterselection can be thus used to remove Cbr-unc-119(+) or BSD(+) extrachromosomal transgenes, thereby facilitating the recovery of animals bearing an HR-induced insertion of the selectable marker. This counterselection approach requires a starting strain resistant to ivermectin, which is conferred by lesions in both the avr-14 and avr-15 genes. Ivermectin-resistant double-mutant strains are essentially wild type in appearance, and Cbr-unc-119 ivermectin-resistant strains are available, or as noted above, new strains can readily be rendered ivermectin resistant by simply co-injecting sgRNAs targeting avr-14 and avr-15 (Figure 2).
To test this selection/counterselection strategy we first designed a donor plasmid containing the BSD gene flanked with 1-kb pie-1 homology arms (Figure 4A). We injected 58 ivermectin-resistant animals with a mix containing this donor plasmid along with a validated pie-1 sgRNA vector, the Peft-3::Cas9 vector, the rol-6 vector, and the avr-15(+) vector. Gravid F1 rollers were then placed ∼11 per plate directly onto plates containing both blasticidin and ivermectin (Figure 4B; see Materials and Methods). After 3–4 days we found that 3 of 14 plates produced blasticidin-resistant, fertile animals (Figure 4B). In a second experiment, we injected the same injection mixture into 40 animals and obtained 103 F1 rollers, from which we identified four blasticidin-resistant strains. In each case, the desired HR events were confirmed both by phenotype and DNA sequence analysis (data not shown).
We also tested Cbr-unc-119(+) as a selectable marker in this selection/counterselection assay. For this experiment we sought to enlarge an oma-1 deletion (tm1396), replacing the oma-1 promoter, coding region, and 3′UTR with the cb-unc-119(+) gene flanked by loxP sites (Dickinson et al. 2013) (Figure S4). We injected 15 ivermectin-resistant Cbr-unc-119; oma-1(tm1396) animals with a mix containing oma-1 sgRNA, Cbr-unc-119(+) oma-1 gene replacement donor vector, a heat-shock Cas9 vector, the rol-6 vector, and the avr-15(+) vector. Injected animals were allowed to recover for 2 hr after injection and were then heat shocked at 34° for 1.5 hr (Waaijers et al. 2013). Gravid non-Unc F1 rollers were collected 15 per plate on six plates, and each population was subjected to ivermectin counterselection. After 4 days, one of the six plates produced healthy ivermectin resistant non-Unc-119 nonrolling animals. Insertion of the Cbr-unc-119 vector and deletion of the oma-1 sequences was confirmed by PCR (data not shown).
Optimizing sgRNA and donor molecule selection
There is much work still to do to optimize CRISPR methodology for C. elegans. For example, it remains unclear at this point why upward of half of the sgRNAs tested fail to induce events. The sgRNAs that we have tested and the activities observed are summarized in Table S1, and all of the active sgRNA vectors will be made available (through Addgene). Another area requiring more study is how best to optimize HR donors. All of the HR donor molecules used in the experiments described here were circular plasmids with at least 300-bp homology arms (Figure S2A). For GFP insertion we used either 800-bp or 1-kb homology arms and observed roughly equal frequencies of HR in both cases (Figure 3A and Figure S3B). Future studies should explore shorter homology arms and other types of donor molecules including linear dsDNA donor molecules produced, for example, by PCR, as well as chemically synthesized ssDNA. It will also be important to explore the optimal distance between the cut site and the homology arm. Increasing this distance requires longer gene-conversion tracts and in other organisms is correlated with reduced frequency of the desired homologous event (Paques and Haber 1999). Our findings suggest that gene-conversion tracts of ∼250 bp are common in C. elegans. Optimizing HR conditions for each type of donor molecule will likely require extensive experimentation to generate statistically significant findings on relative efficiencies. Although there is still much work to do, the efficiencies reported here are already remarkably high. For example, indels were frequently identified in >10% of F1 rollers, and the ratio of HR events to the total number of CRISPR-Cas9-induced-repair events was consistently ∼10% in experiments where both HR and indels were monitored.
Our findings provide a versatile framework for using CRISPR-Cas9 genome editing in C. elegans, and the Co-CRISPR strategy we employ is likely to be of value for CRISPR-Cas9 studies in other organisms. The tools described here, however, are likely to be just the beginning of what will be possible in the near future. For example, the use of catalytically inactive Cas9 fusion proteins to tether regulators to DNA targets has not been described yet in C. elegans, but it is already finding many exciting applications in other systems (Cheng et al. 2013; Larson et al. 2013; Mali et al. 2013a; Qi et al. 2013; Kearns et al. 2014). CRISPR-Cas9 technology should also dramatically facilitate the use of other nematode models, including species distantly related to Caenorhaditis elegans and perhaps parasitic nematodes. The ability to efficiently engineer genomes will only enhance the utility of model organisms where gene variants can now be generated and analyzed rapidly and cost effectively, facilitating the production of new animals models for human disease-associated alleles. Moreover, CRISPR-Cas9-engineered strains with special alleles of important genes can be used as starting strains in forward genetic screens, including suppressor and enhancer screens, which are extremely powerful in C. elegans. It is now easier than ever for researchers to use C. elegans to explore the function of conserved genes of interest. Indeed, the CRISPR-Cas9 technology lowers the barrier to move from one system to another, effectively making all organisms one, when exploring conserved cellular mechanisms.
Supplementary Material
Acknowledgments
We thank John Calarco for sharing information and materials prior to publication and Wen Tang for building Phsp::Cas9 plasmid. We are grateful to members of the Mello and Ambros labs for input and discussion. M. Seth is a Howard Hughes Medical Institute International Student Research Fellow. This work was supported by a National Institutes of Health grant (GM058800) to C.C.M. C.C.M. is a Howard Hughes Medical Institute Investigator.
Footnotes
Communicating editor: O. Hobert
Literature Cited
- Bassett A. R., Tibbit C., Ponting C. P., Liu J. L., 2013. Highly efficient targeted mutagenesis of Drosophila with the CRISPR/Cas9 system. Cell Rep. 4: 220–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benian G. M., L’Hernault S. W., Morris M. E., 1993. Additional sequence complexity in the muscle gene, unc-22, and its encoded protein, twitchin, of Caenorhabditis elegans. Genetics 134: 1097–1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhaya D., Davison M., Barrangou R., 2011. CRISPR-Cas systems in bacteria and archaea: versatile small RNAs for adaptive defense and regulation. Annu. Rev. Genet. 45: 273–297. [DOI] [PubMed] [Google Scholar]
- Brenner S., 1974. The genetics of Caenorhabditis elegans. Genetics 77: 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang N., Sun C., Gao L., Zhu D., Xu X., et al. , 2013. Genome editing with RNA-guided Cas9 nuclease in zebrafish embryos. Cell Res. 23: 465–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C., Fenk L. A., de Bono M., 2013. Efficient genome editing in Caenorhabditis elegans by CRISPR-targeted homologous recombination. Nucleic Acids Res. 41: e193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng A. W., Wang H., Yang H., Shi L., Katz Y., et al. , 2013. Multiplexed activation of endogenous genes by CRISPR-on, an RNA-guided transcriptional activator system. Cell Res. 23: 1163–1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu H., Schwartz H. T., Antoshechkin I., Sternberg P. W., 2013. Transgene-free genome editing in Caenorhabditis elegans using CRISPR-Cas. Genetics 195: 1167–1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho S. W., Kim S., Kim J. M., Kim J. S., 2013a Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat. Biotechnol. 31: 230–232. [DOI] [PubMed] [Google Scholar]
- Cho S. W., Lee J., Carroll D., Kim J. S., Lee J., 2013b Heritable gene knockout in Caenorhabditis elegans by direct injection of Cas9-sgRNA ribonucleoproteins. Genetics 195: 1177–1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong L., Ran F. A., Cox D., Lin S., Barretto R., et al. , 2013. Multiplex genome engineering using CRISPR/Cas systems. Science 339: 819–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dent J. A., Smith M. M., Vassilatis D. K., Avery L., 2000. The genetics of ivermectin resistance in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 97: 2674–2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dicarlo J. E., Conley A. J., Penttila M., Jantti J., Wang H. H., et al. , 2013. Yeast Oligo-Mediated Genome Engineering (YOGE). ACS Synth Biol 2: 741–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson D. J., Ward J. D., Reiner D. J., Goldstein B., 2013. Engineering the Caenorhabditis elegans genome using Cas9-triggered homologous recombination. Nat. Methods 10: 1028–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duchaine T. F., Wohlschlegel J. A., Kennedy S., Bei Y., Conte D., Jr, et al. , 2006. Functional proteomics reveals the biochemical niche of C. elegans DCR-1 in multiple small-RNA-mediated pathways. Cell 124: 343–354. [DOI] [PubMed] [Google Scholar]
- Feng Z., Mao Y., Xu N., Zhang B., Wei P., et al. , 2014. Multigeneration analysis reveals the inheritance, specificity, and patterns of CRISPR/Cas-induced gene modifications in Arabidopsis. Proc. Natl. Acad. Sci. USA 111: 4632–4637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Z., Zhang B., Ding W., Liu X., Yang D. L., et al. , 2013. Efficient genome editing in plants using a CRISPR/Cas system. Cell Res. 23: 1229–1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedland A. E., Tzur Y. B., Esvelt K. M., Colaiacovo M. P., Church G. M., et al. , 2013. Heritable genome editing in C. elegans via a CRISPR-Cas9 system. Nat. Methods 10: 741–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frokjaer-Jensen C., Davis M. W., Hopkins C. E., Newman B. J., Thummel J. M., et al. , 2008. Single-copy insertion of transgenes in Caenorhabditis elegans. Nat. Genet. 40: 1375–1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frokjaer-Jensen C., Davis M. W., Ailion M., Jorgensen E. M., 2012. Improved Mos1-mediated transgenesis in C. elegans. Nat. Methods 9: 117–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert L. A., Larson M. H., Morsut L., Liu Z., Brar G. A., et al. , 2013. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell 154: 442–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giordano-Santini R., Milstein S., Svrzikapa N., Tu D., Johnsen R., et al. , 2010. An antibiotic selection marker for nematode transgenesis. Nat. Methods 7: 721–723. [DOI] [PubMed] [Google Scholar]
- Gratz S. J., Cummings A. M., Nguyen J. N., Hamm D. C., Donohue L. K., et al. , 2013. Genome engineering of Drosophila with the CRISPR RNA-guided Cas9 nuclease. Genetics 194: 1029–1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grishok A., Mello C. C., 2002. RNAi (Nematodes: Caenorhabditis elegans). Adv. Genet. 46: 339–360. [DOI] [PubMed] [Google Scholar]
- Hannon G. J., 2002. RNA interference. Nature 418: 244–251. [DOI] [PubMed] [Google Scholar]
- Horii T., Morita S., Kimura M., Kobayashi R., Tamura D., et al. , 2013. Genome engineering of mammalian haploid embryonic stem cells using the Cas9/RNA system. Peer J. 1: e230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath P., Barrangou R., 2010. CRISPR/Cas, the immune system of bacteria and archaea. Science 327: 167–170. [DOI] [PubMed] [Google Scholar]
- Jiang W., Bikard D., Cox D., Zhang F., Marraffini L. A., 2013. RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nat. Biotechnol. 31: 233–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J. A., et al. , 2012. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337: 816–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katic I., Grosshans H., 2013. Targeted heritable mutation and gene conversion by Cas9-CRISPR in Caenorhabditis elegans. Genetics 195: 1173–1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearns N. A., Genga R. M., Enuameh M. S., Garber M., Wolfe S. A., et al. , 2014. Cas9 effector-mediated regulation of transcription and differentiation in human pluripotent stem cells. Development 141: 219–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson M. H., Gilbert L. A., Wang X., Lim W. A., Weissman J. S., et al. , 2013. CRISPR interference (CRISPRi) for sequence-specific control of gene expression. Nat. Protoc. 8: 2180–2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo T. W., Pickle C. S., Lin S., Ralston E. J., Gurling M., et al. , 2013. Precise and heritable genome editing in evolutionarily diverse nematodes using TALENs and CRISPR/Cas9 to engineer insertions and deletions. Genetics 195: 331–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y., Shen B., Zhang X., Lu Y., Chen W., et al. , 2014. Heritable multiplex genetic engineering in rats using CRISPR/Cas9. PLoS ONE 9: e89413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P., Aach J., Stranges P. B., Esvelt K. M., Moosburner M., et al. , 2013a CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat. Biotechnol. 31: 833–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P., Yang L., Esvelt K. M., Aach J., Guell M., et al. , 2013b RNA-guided human genome engineering via Cas9. Science 339: 823–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mello C. C., Schubert C., Draper B., Zhang W., Lobel R., et al. , 1996. The PIE-1 protein and germline specification in C. elegans embryos. Nature 382: 710–712. [DOI] [PubMed] [Google Scholar]
- Moerman D. G., Baillie D. L., 1979. Genetic organization in Caenorhabditis elegans: fine-structure analysis of the unc-22 gene. Genetics 91: 95–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paques F., Haber J. E., 1999. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 63: 349–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi L. S., Larson M. H., Gilbert L. A., Doudna J. A., Weissman J. S., et al. , 2013. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell 152: 1173–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radman I., Greiss S., Chin J. W., 2013. Efficient and rapid C. elegans transgenesis by bombardment and hygromycin B selection. PLoS ONE 8: e76019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran F. A., Hsu P. D., Lin C. Y., Gootenberg J. S., Konermann S., et al. , 2013. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell 154: 1380–1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren X., Sun J., Housden B. E., Hu Y., Roesel C., et al. , 2013. Optimized gene editing technology for Drosophila melanogaster using germ line-specific Cas9. Proc. Natl. Acad. Sci. USA 110: 19012–19017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semple J. I., Garcia-Verdugo R., Lehner B., 2010. Rapid selection of transgenic C. elegans using antibiotic resistance. Nat. Methods 7: 725–727. [DOI] [PubMed] [Google Scholar]
- Shirayama M., Seth M., Lee H. C., Gu W., Ishidate T., et al. , 2012. piRNAs initiate an epigenetic memory of nonself RNA in the C. elegans germline. Cell 150: 65–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sternberg S. H., Redding S., Jinek M., Greene E. C., Doudna J. A., 2014. DNA interrogation by the CRISPR RNA-guided endonuclease Cas9. Nature 507: 62–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terns M. P., Terns R. M., 2011. CRISPR-based adaptive immune systems. Curr. Opin. Microbiol. 14: 321–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzur Y. B., Friedland A. E., Nadarajan S., Church G. M., Calarco J. A., et al. , 2013. Heritable custom genomic modifications in Caenorhabditis elegans via a CRISPR-Cas9 system. Genetics 195: 1181–1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voinnet O., 2001. RNA silencing as a plant immune system against viruses. Trends Genet. 17: 449–459. [DOI] [PubMed] [Google Scholar]
- Waaijers S., Portegijs V., Kerver J., Lemmens B. B., Tijsterman M., et al. , 2013. CRISPR/Cas9-targeted mutagenesis in Caenorhabditis elegans. Genetics 195: 1187–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Yang H., Shivalila C. S., Dawlaty M. M., Cheng A. W., et al. , 2013. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell 153: 910–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T., Wei J. J., Sabatini D. M., Lander E. S., 2014. Genetic screens in human cells using the CRISPR-Cas9 system. Science 343: 80–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiedenheft B., Sternberg S. H., Doudna J. A., 2012. RNA-guided genetic silencing systems in bacteria and archaea. Nature 482: 331–338. [DOI] [PubMed] [Google Scholar]
- Yu, Z., H. Chen, J. Liu, H. Zhang, Y. Yan et al., 2014 Various applications of TALEN- and CRISPR/Cas9-mediated homologous recombination to modify the Drosophila genome. Biol Open 3: 271–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamore P. D., 2001. RNA interference: listening to the sound of silence. Nat. Struct. Biol. 8: 746–750. [DOI] [PubMed] [Google Scholar]
- Zhao P., Zhang Z., Ke H., Yue Y., Xue D., 2014. Oligonucleotide-based targeted gene editing in C. elegans via the CRISPR/Cas9 system. Cell Res. 24: 247–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J., Shen B., Zhang W., Wang J., Yang J., et al. , 2014. One-step generation of different immunodeficient mice with multiple gene modifications by CRISPR/Cas9 mediated genome engineering. Int. J. Biochem. Cell Biol. 46: 49–55. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.