Abstract
The Ras/MAPK-signaling pathway plays pivotal roles during development of metazoans by controlling cell proliferation and cell differentiation elicited, in several instances, by receptor tyrosine kinases (RTKs). While the internal mechanism of RTK-driven Ras/MAPK signaling is well understood, far less is known regarding its interplay with other corequired signaling events involved in developmental decisions. In a genetic screen designed to identify new regulators of RTK/Ras/MAPK signaling during Drosophila eye development, we identified the small GTPase Rap1, PDZ-GEF, and Canoe as components contributing to Ras/MAPK-mediated R7 cell differentiation. Rap1 signaling has recently been found to participate in assembling cadherin-based adherens junctions in various fly epithelial tissues. Here, we show that Rap1 activity is required for the integrity of the apical domains of developing photoreceptor cells and that reduced Rap1 signaling hampers the apical accumulation of the Sevenless RTK in presumptive R7 cells. It thus appears that, in addition to its role in cell–cell adhesion, Rap1 signaling controls the partitioning of the epithelial cell membrane, which in turn influences signaling events that rely on apico-basal cell polarity.
Keywords: receptor tyrosine kinase, Rap1, Ras, MAPK, Drosophila eye development
EPITHELIAL tissues rely on the complex interplay of diverse signaling events to adopt their appropriate shape, size, and function. Development of the Drosophila compound eye is a well-documented example of epithelial morphogenesis. Since its inception as an experimental system over 3 decades ago (Ready et al. 1976), its development has been intensively studied (Cagan 2009; Roignant and Treisman 2009). Over the years, this system has produced insights on various signaling pathways and cellular behaviors, and several of these have been found to lead to human diseases or syndromes when dysregulated.
The Drosophila eye is composed of hundreds of identical units called ommatidia, each comprising an equal complement of 26 cells [eight photoreceptor neurons (R1–R8), four cone cells, eleven pigments cells, and three mechanosensory bristle cells] (Kumar 2012). Mosaic analysis during eye development has revealed the absence of a clonal relationship between differentiating cells (Ready et al. 1976). Instead, cell fate specification in each ommatidial cluster occurs in a stereotyped manner by inductive cues from neighboring cells that entail adhesive contacts and an elaborate network of signaling pathways, among which signaling through the EGFR/Ras/MAPK pathway remains one of the best understood events (Roignant and Treisman 2009). Except for the founding R8 photoreceptor cell present in each ommatidium, EGFR activity is required for the differentiation of all other retinal cells. Yet a second receptor tyrosine kinase (RTK), known as Sevenless (SEV), also participates in these developmental decisions, but only in the presumptive R7 cells (Freeman 1997).
In addition to the core members of the Ras/MAPK pathway (e.g., Ras, RAF, MEK, and MAPK), genetic screens based on modulation of the R7 cell specification led to the identification of novel components that act as general regulators of RTK/Ras/MAPK signaling. Kinase Suppressor of Ras (KSR) is one of these and corresponds to a pseudokinase related to RAF family kinases (Therrien et al. 1995). KSR has the ability to bridge RAF and MEK together as well as to drive the catalytic activation of RAF through heterodimerization of their respective kinase domain (Udell et al. 2011). Another component identified by genetic means and functionally related to RAF and KSR is Connector eNhancer of KSR (CNK) (Therrien et al. 1998). CNK is an evolutionarily conserved protein that comprises multiple protein- and lipid-interaction domains whose function in Ras/MAPK-mediated processes is also conserved (Claperon and Therrien 2007). Previous work has demonstrated the ability of Drosophila CNK to bind separately to RAF and KSR/MEK, thereby enabling the formation of a RAF/KSR/MEK complex (Douziech et al. 2006). In addition, CNK appears to have a dual role in the Ras/MAPK pathway by keeping RAF inhibited in the absence of an upstream RTK signal and by promoting RAF activation upon RTK signaling (Douziech et al. 2003). The mechanism governing these functions of CNK has been partly solved and requires the action of both Ras and the Src family kinase Src42A (Laberge et al. 2005). More recently, CNK has been shown to form a complex with Steppke, the fly cytohesin homolog, and collaborate in EGFR-induced MAPK activation by a mechanism that has yet to be characterized (Hahn et al. 2013). Whether CNK plays other roles outside the Ras/MAPK pathway in flies is currently not clear, although mammalian CNKs have been found to participate in signaling events regulated by other small GTPases, such as Rho, Rac, Ral, and Arf6 (Lanigan et al. 2003; Venkateswarlu 2003; Jaffe et al. 2005; Lim et al. 2010).
To identify functional partners of CNK that may reveal new RTK/MAPK pathway components or unveil other roles for this scaffolding protein, we conducted a genetic screen in the Drosophila eye to isolate dominant modifiers of a CNK-dependent rough eye phenotype. The screen led to the isolation of 24 complementation groups of enhancers including, as expected, several genes encoding bona fide components of RTK/Ras/MAPK pathways. Nine novel loci have also been identified. Interestingly, three of those (Roughened, PDZ-GEF, and canoe) encode Rap1-signaling components, which together suggest a functional relationship between CNK and Rap1 signaling. Previous work reported that Rap1 activity is required for adherens junction formation and adhesive contacts between developing photoreceptors (O’Keefe et al. 2009). This phenomenon appeared to be critical for maintaining EGFR signaling between cells, although Rap1 activity did not seem autonomously required for the commitment of any cell type. A variation of this model was recently proposed when a detailed genetic analysis of Rap1 requirement demonstrated its essential cell-autonomous role in presumptive R7 cells (Mavromatakis and Tomlinson 2012). However, the mechanism by which Rap1 promotes R7 cell differentiation has not been determined.
Here, we show that Rap1 activity induces and/or maintains the formation of the apical domain of developing photoreceptor cells and that this event correlates with the apical accumulation and activity of SEV in presumptive R7 cells. These results suggest that Rap1 signaling controls cell polarity and thereby influences signaling events that rely on plasma membrane partitioning.
Materials and Methods
CNK C-terminal-dependent screen, mapping, and sequencing
A Kpn1-Not1 PCR fragment encompassing amino acids 382–1557 [CNK C-Terminal, (CCT)] was cloned into the psE vector (Dickson et al. 1992) and introduced into the w1118 background by P-element-mediated germline transformation as described previously (Rubin and Spradling 1982). For the mutagenesis, w1118 males isogenic for the second and the third chromosomes were fed overnight with a 1% sucrose solution containing 25 mM ethyl methanesulfonate (EMS). Mutagenized males were then crossed with either CyO, P[sE-CCT]/Adv or TM3, P[sE-CCT]/e, ftz, ry virgin females. Approximately 75,000 F1 progeny were scored for enhancement of the sE-CCT rough eye phenotype using a Leica MZ8 stereomicroscope. Allelism was assessed by complementation tests based on recessive lethality. Complementation groups corresponding to loci encoding components previously linked to the RTK/Ras/MAPK pathway were determined by conducting complementation tests with the following alleles: rlE-1171, SE-1602, cnkE-1088, EgfrE-1063, SosP566, kisE-1679, ksrE-1572, pntE-1265, Ras85De1b, dos1.46, and maskEP3498. The remaining groups were mapped using meiotic, deletion, and P-element mapping. When possible, genetic identification for these groups was determined by lethal complementation tests using the following alleles: RCD3, RrvB1, PDZ-GEFEP388, PDZ-GEFk13720, cno2, Btk29Ak00206, swm37Dh-1, swmF14, swmF15, Prp19v22146, Prp19v22147, Dl1, Dl12, Bre101640, Nsf2A6, and Nsf2A15. Exon sequencing for a subset of the new loci was performed to ascertain their molecular identity as previously described (Therrien et al. 1998).
Genetics, fly stocks, and microscopy
Fly maintenance and genetic interaction studies were conducted according to standard procedures. The following stocks were used: b1, pr1, c1, px1, sp1 (second chromosome mapping stock); ru1, h1, th1, st1, cu1, sr1, e1, ca1 (third chromosome mapping stock); hypomorphic rl1 and phl12 mutant alleles; several deficiencies including Df(2L)BSC5/SM6a, P-element insertions (Bloomington Stock Center), UAS-CNKIR, UAS-Rap1IR, UAS-PDZ-GEFIR, UAS-EcadIR, UAS-EgfrIR, UAS-sevIR (VDRC), and hs-flp122; Act > CD2 > Gal4; UAS-GFP (Neufeld et al. 1998); GFP-Rap1 driven by the endogenous Rap1 promoter (Knox and Brown 2002); sev-lacZ (Basler et al. 1989); and GMR-sev (Tomlinson et al. 2011).
UAS construct-expressing FLP-out Gal4 clones were induced 72 hr after egg deposition by a 10- to 15-min heat shock at 38°. Third instar larval eye discs were dissected 96 hr after heat shocks. Pupal eye discs were dissected 45 hr after puparium formation.
Scanning electron microscopy and sectioning of adult fly eyes were performed as described by Kimmel et al. (1990) and Tomlinson and Ready (1987), respectively. The UAS-Rap1V12 line was generated by P-element-mediated germline transformation as previously described (Rubin and Spradling 1982).
Immunostaining
Eye-antennal imaginal discs from third instar larvae were dissected in plain Schneider medium (Invitrogen), fixed in 1× phosphate-buffered saline (1× PBS); 4% paraformaldehyde for 15 min at room temperature and washed three times with PBT (1× PBS; 0.2% Triton X-100). Primary antibodies were incubated in PBT + 2.5% fat-free dry milk overnight at 4° with gentle rocking. Primary antibodies and dilutions were rat anti-Elav [1/20; Developmental Studies Hybridoma Bank (DSHB)], mouse anti-Prospero (1/20; DSHB), mouse anti-dpMAPK (1/1000; Sigma), rat anti-E-Cadherin (1/20; DSHB), mouse anti-armadillo (1/20; DSHB), rabbit anti-aPKC (1/200; Santa Cruz Biotechnology), mouse anti-Dlg (1/20; DSHB), mouse anti-Sevenless (1/20; kindly provided by G. M. Rubin), rat anti-EGFR (1/1000; kindly provided by B.-Z. Shilo), mouse anti-Cut (1/100; DSHB), mouse anti-pTyr (1/20, 4G10), mouse anti-β-Gal (1/2000; Promega), rabbit anti-Runt (kindly provided by A. Brand), mouse anti-CNK (1/200; Douziech et al. 2003), rabbit anti-Bazooka (1:1000; kindly provided by A. Wodarz), rabbit anti-Patj (1:1000; kindly provided by E. Knust). Imaginal discs were then washed three times with PBT and incubated with appropriate species-specific Alexa 555-, Alexa 647- (1/1000; Life Technologies), or Cy3-conjugated secondary antibodies (1/1000; Jackson Immuno-Research Laboratories) for 2 hr at room temperature in PBT. Tissues were again washed three times with PBT and then mounted in Mowiol 4-88 (Sigma-Aldrich) and imaged using a Zeiss LSM 510 confocal microscope.
Cell transfection and plasmid constructs
S2 cells were grown in Schneider (Life Technologies) medium supplemented with 10% fetal bovine serum and 0.1% penicillin/streptomycin. A total of 7 × 106 (for total lysates) or 18 × 106 (for immunoprecipitations) cells were transfected using Effectene (Qiagen). Protein expression was induced by adding CuSO4 (0.7 mm) to the medium 24 hr post-transfection. Thirty-six hours post-induction, cells were lysed in NP-40 lysis buffer (20 mM Tris at pH 8.0, 137 mM NaCl, 10% glycerol, 1% Igepal CA-630, 1 mM EDTA, 0.15 units/ml aprotinin, 20 μM leupeptin, and 1× Sigma phosphatase inhibitor cocktail) for 15 min at 4°. Sources and dilutions for antibodies are the following: α-Flag M2 mAb (1:5000; Sigma); α-Pyo mAb (1:2000; Douziech et al. 2003); a-HA mAb (1:2000; Douziech et al. 2003); and α-pMAPK (1:2000; Sigma).
HA-RasV12, HA-MAPK, Pyo-RAF, and Flag-CNK have been previously described in (Douziech et al. 2003). Standard cloning procedures were used to generate HA-Rap1V12 and Pyo-PDZ-GEFΔNT constructs. The HA-Rap1V12 or Pyo-Rap1V12 constructs encompass the entire Drosophila Rap1 open reading frame, harbor a G12V change, and encode a hemagglutinin (HA) or polyoma (Pyo) epitope tag at their N terminus. The Pyo-PDZ-GEFΔNT encodes an N-terminal truncation of Drosophila PDZ-GEF (lacks the first 234 amino acid residues) fused to a Pyo epitope tag at its N terminus. All S2 cell-expressing constructs use the pMet backbone (Therrien et al. 1998) and are thus copper-inducible. The Rap1V12 cDNA was also inserted in the pUAST (Brand and Perrimon 1993) P-element vector.
Results
A screen for modifiers of a CNK-dependent rough eye phenotype
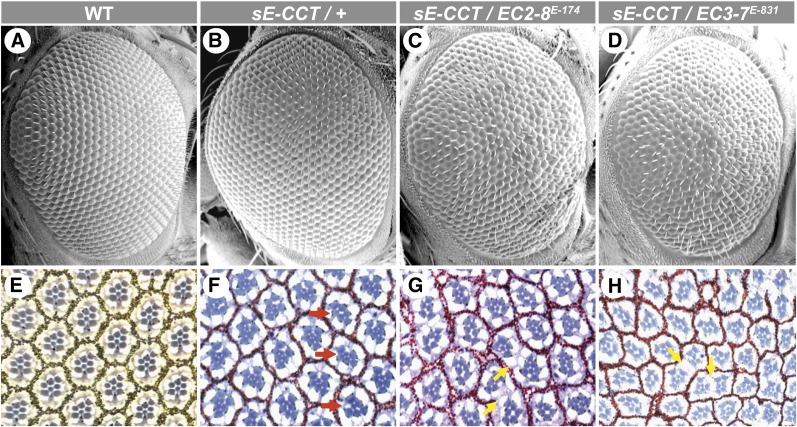
We previously showed that expression of a CCT (amino acids 382–1557) in vivo or in Drosophila S2 cells exhibited dominant-negative activity with respect to the Ras/MAPK pathway owing to its ability to bind and inhibit the RAF Ser/Thr kinase (Therrien et al. 1999; Douziech et al. 2003). Consistent with this, expression of CCT during eye development impeded Ras/MAPK-dependent differentiation events as exemplified by the loss of photoreceptor cells, which in turn caused a mild roughening of the adult eye surface (Figure 1, A, B, E, and F; Table 1).
Figure 1.
EC2-8 and EC3-7 enhance the sE-CCT rough eye phenotype. (A–D) Scanning electron micrographs and (E–H) apical sections of adult fly retinae of the indicated genotypes. (A and E) WT. (B and F) sE-CCT/+. (C and G) sE-CCT/EC2-8E-174. (D and H) sE-CCT/EC3-7E-831. The apical section of a WT eye (E) shows the typical trapezoid arrangement of rhabdomeres marking the six outer photoreceptor cells as well as the R7 photoreceptor cell (smaller centrally located rhabdomere). Granules produced by pigment cells can also be seen surrounding individual ommatidia. In F, ommatidia missing photoreceptor cells are indicated with a red arrow. EC2-8 and EC3-7 alleles not only enhance the loss of photoreceptor cells induced by CCT (G and H), but also lead to the disappearance of pigment cells (yellow arrows). Anterior is to the right.
Table 1. Summary of photoreceptor cell counts.
| Genotype | % of ommatidia missing R7 cell | % of ommatidia missing at least one outer photoreceptor cell | No. of ommatidia analyzed |
|---|---|---|---|
| sE-CCT/+ | 3 | 7 | 617 |
| sE-CCT/PDZ-GEFE-174 | 25 | 25 | 490 |
| sE-CCT/ PDZ-GEFE-696 | 13 | 25 | 686 |
| sE-CCT/+ ; Rap1E-722/+ | 9 | 10 | 509 |
| sE-CCT/+ ; Rap1E-831/+ | 63 | 58 | 631 |
| PDZ-GEFE-174 | 40 | 10 | 505 |
| PDZ-GEFE-174/Df(2L)BSC5 | 54 | 11 | 612 |
| PDZ-GEFE-696/Df(2L)BSC5 | 66 | 62 | 253 |
| aRap1E-814 | 21 | 20 | 321 |
Only recognizable ommatidia/rhabdomeres were scored.
To identify novel components that functionally work together with CNK, we took advantage of the CCT rough eye phenotype and conducted a genetic screen to isolate heterozygous mutations that acted as dominant enhancers. As CCT expression cripples Ras/MAPK signaling, loss-of-function mutations in genes that normally operate in concert with CNK might worsen the phenotype and thereby are expected to emerge as enhancers. Eye roughness was relatively weak, and for this reason suppressors were not sought after. Approximately 75,000 F1 progeny from EMS-mutagenized parent males were scored for enhanced rough eyes. A total of 407 Enhancers of CCT (EC) were isolated, of which 276 fell into one of 24 recessive lethal complementation groups (Table 2). Representative examples of rough eye enhancements are shown in Figure 1.
Table 2. Groups of dominant enhancers of sE-CCT on second and third chromosomes.
| Groups | Genes | Cytological position | No. of alleles |
|---|---|---|---|
| Chromosome II | |||
| EC2-1 | rl | h41 | 10 |
| EC2-2 | S | 21E4 | 21 |
| EC2-3 | cnk | 54B7 | 9 |
| EC2-4 | Egfr | 57E9-F1 | 35 |
| EC2-5 | Sos | 34D1 | 44 |
| EC2-7 | kis | 21B4-B5 | 4 |
| EC2-8 | Gef26/PDZ-GEFa | 26C2-C3 | 6 |
| EC2-9 | Btk29Aa | 29A1-A3 | 2 |
| EC2-10 | swma | 37E4 | 4 |
| EC2-11 | Prp19a | 55C9-D1 | 3 |
| EC2-12 | ND | 43A4-C1 | 3 |
| Chromosome III | |||
| EC3-1 | ksr | 83A5 | 13 |
| EC3-2 | pnt | 94E10-E13 | 33 |
| EC3-3 | Ras85D | 85D19 | 25 |
| EC3-4 | dos | 62E7 | 9 |
| EC3-5 | Dl | 92A1-A2 | 27 |
| EC3-6 | mask | 95F3-F5 | 6 |
| EC3-7 | R/Rap1a | 62B7 | 5 |
| EC3-8 | Bre1a | 64E8 | 2 |
| EC3-9 | ND | 68A7-B1 | 2 |
| EC3-10 | cnoa | 82F4-F6 | 3 |
| EC3-11 | Nsf2a | 87F15 | 5 |
| EC3-12 | ND | ND | 3 |
| EC3-13 | ND | 98B6-E5 | 2 |
Mutations reported in Figure 2.
ND, not determined.
Meiotic-, deletion-, and/or transposon-based mapping were used to position the various groups to specific loci. Consistent with the role of CNK in RTK/Ras/MAPK signaling, 11 groups correspond to genes that had been identified in previous genetic screens targeting this pathway in Drosophila (Simon et al. 1991; Karim et al. 1996; Raabe et al. 1996; Therrien et al. 2000; Smith et al. 2002). These include rl, S, cnk, Egfr, Sos, and kis on the second chromosome and ksr, pnt, Ras85D, dos, and mask on the third chromosome (Table 2). Moreover, we isolated mutations in three genes encoding members of the Rap1-signaling pathway: PDZ-GEF, a Rap1-specific guanine nucleotide exchange factor also known as Dizzy or Gef26 (Lee et al. 2002); Roughened (R), a Rap1 GTPase homolog (Hariharan et al. 1991); and Canoe (CNO), a Rap1 effector known as AF6 in mammals (Miyamoto et al. 1995; Boettner et al. 2003). Finally, we isolated mutations in other loci not previously identified in RTK/Ras/MAPK-dependent modifier screens. They correspond to Btk29A, swm, Prp19, Dl, Bre1, and Nsf2 (Table 2). Additional information on these loci is provided in the Discussion.
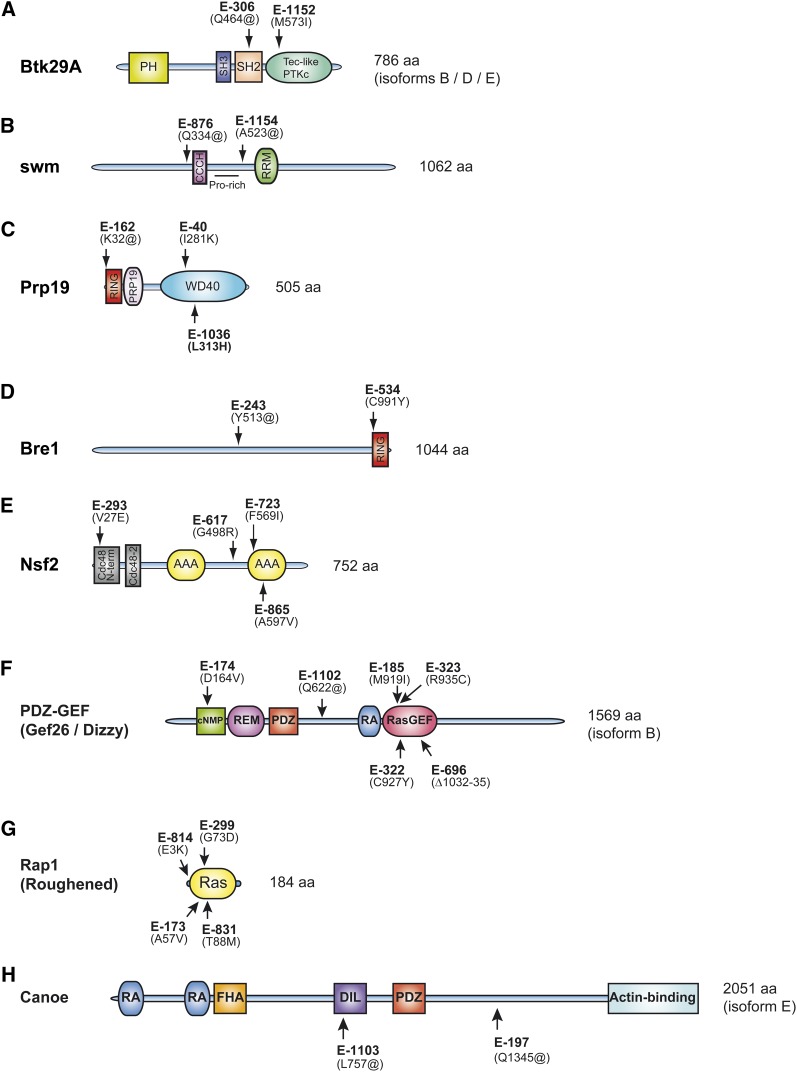
To confirm that the novel groups were allelic to the aforementioned loci, we isolated genomic DNA from flies heterozygous to those groups noted by table footnote a in Table 2 and sequenced their exons. In most cases, specific point mutations introducing either a STOP codon or an amino acid change in conserved residues were found (Figure 2), thus confirming allelism and suggesting that several of the identified alleles are loss-of-functions. One exception to this rule is the Rap1E-722 allele as no mutation in the Rap1 open reading frame was detected.
Figure 2.
Identification of the molecular lesions of eight Enhancers of CCT. (A–H) Protein domain composition (according to the Conserved Domains tool at the National Center for Biotechnology Information) of proteins encoded by loci not previously identified in RTK/RAS-dependent modifier screens is shown. To confirm allelism between the complementation groups and the assigned loci, exon sequences for at least two heterozygous mutations for each group were determined. Mutations in the expected locus were found for each group. The position of the resulting amino acid lesion for each sequenced allele is shown on the protein schematics. “@” denotes STOP codons.
Rap1 signaling is required for RTK-dependent cell differentiation during eye development
The fact that three Rap1 pathway components were identified in the screen strongly suggested a functional relationship between Rap1 signaling and CCT activity. We thus decided to further characterize the role of these components in eye development.
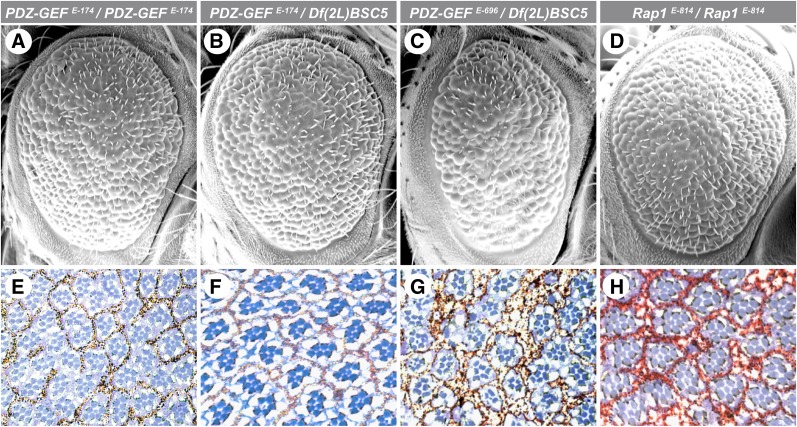
To confirm that mutations in Rap1 pathway components genuinely exacerbated the dominant negative activity of CCT, we examined whether the EC2-8 and EC3-7 alleles (hereafter referred to as PDZ-GEF and Rap1, respectively) also increased the loss of photoreceptor cells caused by CCT expression. As shown in Figure 1, F–H, and quantified in Table 1, heterozygous mutations for both genes considerably augmented CCT-mediated photoreceptor loss. The ability of mutations in Rap1 pathway components to enhance CCT activity suggests that Rap1 signaling positively influences Ras/MAPK signaling. Consistent with this interpretation, we found that both PDZ-GEF and Rap1 mutant alleles genetically interact with a Raf hypomorphic allele during eye development (Supporting Information, Figure S1). Moreover, hypomorphic alleles of PDZ-GEF and Rap1 that show variable degrees of viability as homozygotes have severe rough eye phenotypes accompanied with a large proportion of ommatidia missing photoreceptor cells (Figure 3, A–H, and Table 1), which phenocopied hypomorphic conditions of Ras/MAPK pathway components such as CNK (Therrien et al. 1998). The mutant PDZ-GEF and Rap1 alleles also led to a loss of pigment granules and thereby suggest a failure in the pigment cell differentiation program (Figure 1, F–H). Although this phenotype has not been quantified and its potential link to the loss of photoreceptor cells has not been investigated, it remains consistent with a defect in RTK/Ras/MAPK signaling (Freeman 1997).
Figure 3.
PDZ-GEF and Rap1 are required for photoreceptor cell differentiation. (A–D) Scanning electron micrographs and (E–H) apical sections of adult fly retinae of the indicated genotypes. (A and E) PDZ-GEFE-174/PDZ-GEFE-174. (B and F) PDZ-GEFE-174/Df(2L)BSC5. (C and G) PDZ-GEFE-696/Df(2L)BSC5. (D and H) Rap1E-814/Rap1E-814. Anterior is to the right.
A number of studies have reported a link between Rap1 and RTK signaling during Drosophila development, but the underlying molecular connection is unclear. In two of these, Rap1 is suggested to serve as a direct RAF activator, which would explain how it influences RTK-dependent MAPK signaling (Lee et al. 2002; Mishra et al. 2005). In contrast, a third study failed to detect such a role downstream of EGFR during eye and wing development (O’Keefe et al. 2009). Instead, the authors reported that Rap1 activity is required for cadherin-based adherens junction formation and that elimination of Rap1 impedes EGFR signaling as a consequence of reduced adhesive contacts between differentiating cells.
Given that RAF has been proposed to work as a direct effector of Rap1 in Drosophila (Lee et al. 2002; Mishra et al. 2005), we verified whether ectopic expression of a constitutively activated form of Rap1 (Rap1V12) during eye development stimulated RAF activity by assessing the levels of the dually phosphorylated (active) form of MAPK (pMAPK). Surprisingly, unlike RasV12, clonal Rap1V12 expression in third instar eye discs had no significant effect on pMAPK levels (Figure S2) despite the fact that it considerably altered eye development and produced a few extra R7 cells (Figure S3, A–C) as reported previously (Mirey et al. 2003; Mavromatakis and Tomlinson 2012) as well as extra cone cells and pigment cells (Figure S3D). Likewise, no activation of MAPK was detected when Rap1V12 was overexpressed in S2 cells (Figure S4A). Furthermore, in contrast to Ras, depletion of Rap1 by RNA interference (RNAi) in S2 cells did not compromise EGFR- or SEV-induced MAPK activation (Figures S4, B and C). Interestingly, Rap1V12 associated with RAF, but significantly more Rap1V12 was required compared to RasV12 (Figure S4D). Together, these results suggest that RAF is not a direct effector of Rap1 in these biological settings. However, considering the genetic data supporting a role for Rap1 in MAPK signaling (Lee et al. 2002; Mishra et al. 2005; Mavromatakis and Tomlinson 2012), it is possible that the mechanism is indirect.
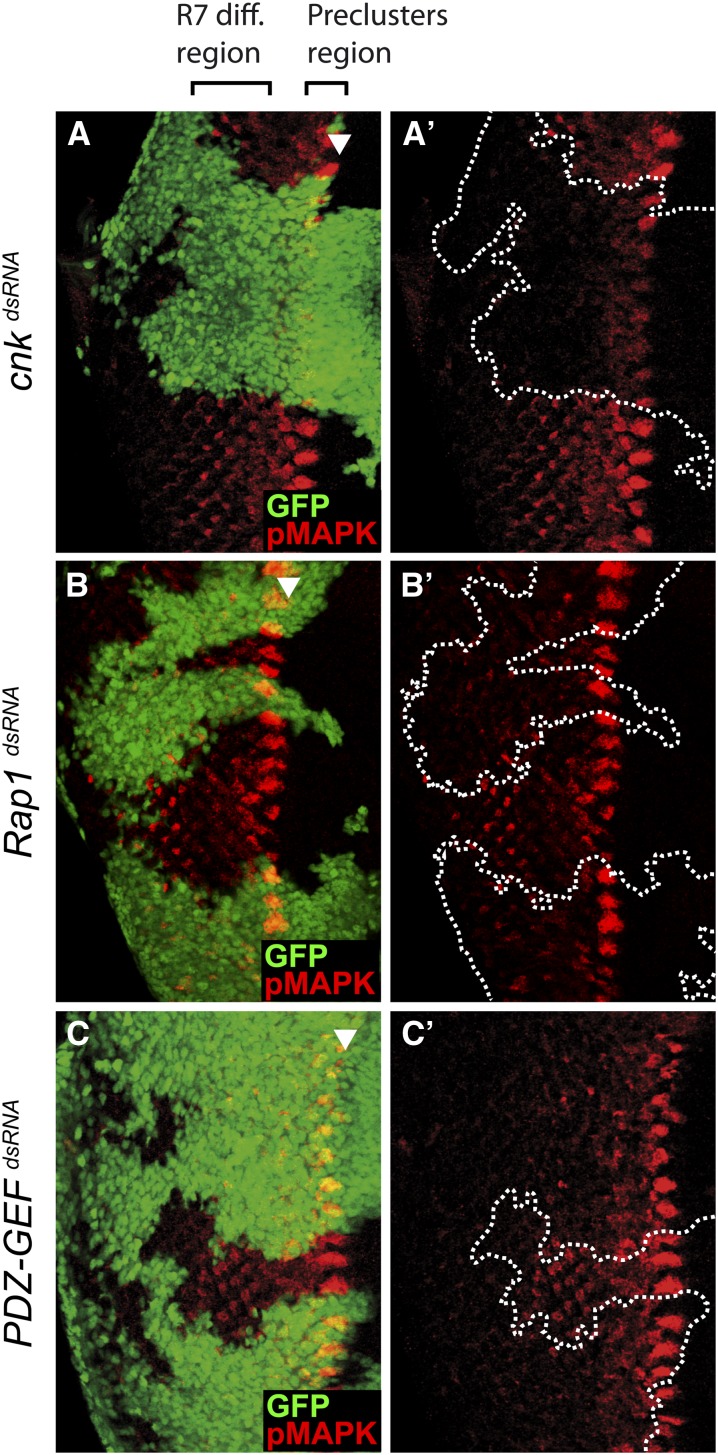
In third instar eye discs, activation of MAPK primarily depends on EGFR activity, and the resulting pMAPK levels are initially detected in groups of cells within the morphogenetic furrow that give rise posteriorly to clusters of maturing photoreceptors (Gabay et al. 1997) (Figure S5). Although less prominent, pMAPK staining is also detected in developing photoreceptor cells posterior to the furrow. The signal is detectable up to 12 rows behind the furrow and encompasses the R7 cell differentiation region, which requires inputs from both the EGFR and the SEV RTKs. As expected, depletion of EGFR by clonally expressing an Egfr dsRNA during eye development abrogated pMAPK levels in the two zones described above (Figure S5A). In contrast, depletion of SEV affected only pMAPK levels in the R7 differentiation region (Figure S5B). We used the same experimental paradigm to compare the effect of depleting CNK, Rap1, or PDZ-GEF by RNAi on pMAPK levels. Consistent with its role as a Ras/MAPK pathway component, reducing the dose of CNK severely decreased pMAPK levels in the morphogenetic furrow as well as in the R7 differentiation region (Figure 4A). Interestingly, depletion of either Rap1 or PDZ-GEF in the morphogenetic furrow had no impact on pMAPK induction. However, the second wave of pMAPK signal patterning the R7 cell differentiation area was greatly reduced (Figure 4, B and C). Similar observations were made in homozygous Rap1 or PDZ-GEF mutant clones (Figure S6 and Figure S7). These findings suggest that Rap1 signaling is not generally required for RTK-mediated MAPK induction, but can influence it in a region-specific manner.
Figure 4.
Knockdown of Rap1 or PDZ-GEF attenuates pMAPK levels in the R7 cell differentiation region of third instar larval eye discs. (A–A′ to C–C′) Random clones (GFP-positive cells) expressing the indicated dsRNAs in third instar eye discs were induced using the FLP-out/Gal4 system. pMAPK levels were monitored by immunofluorescence. The arrowhead denotes the position of the morphogenetic furrow. Anterior is to the right.
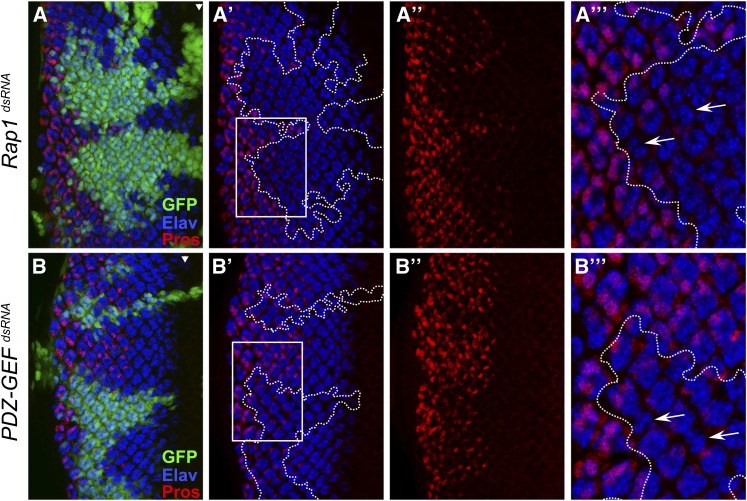
Given the critical importance of MAPK activation for cell differentiation during eye development, the ability of Rap1 depletion to affect MAPK activation specifically in the R7 differentiation zone should have detrimental consequences on R7 photoreceptors. To verify this, we stained the third instar eye disc using an anti-Elav antibody to highlight differentiated photoreceptor neurons in combination with an anti-Prospero (Pros) antibody, which specifically marks non-neuronal cone cells and R7 cells (Kauffmann et al. 1996). In agreement with their effect on MAPK activity, Rap1 and PDZ-GEF RNAi severely impeded the number of Elav/Pros double-positive cells, thus indicating a failure of R7 cells to differentiate (Figure 5, A and B). Except for the general lack of R7 cells, only a few ommatidia appear to be missing several photoreceptor cells (Figure 5). This latter observation is consistent with previous work reporting that elimination of Rap1 activity has partial effects on photoreceptor cell differentiation (O’Keefe et al. 2009). Together, while these findings indicate that Rap1 signaling plays a role during photoreceptor cell differentiation, this role is particularly critical for setting up the R7 cell differentiation program.
Figure 5.
Knockdown of Rap1 or PDZ-GEF impedes R7 cell fate commitment. (A–A′′′ to B–B′′′) Random clones (GFP-positive cells) expressing the indicated dsRNAs in third instar eye discs were induced using the FLP-out/Gal4 system. The Elav and Prospero (Pros) markers were then revealed by immunofluorescence. Enlarged areas (A′′′ and B′′′) correspond to the boxed regions shown in A′ and B′ and highlight (arrows) the less frequent examples of ommatidia lacking several Elav-positive cells in Rap1 or PDZ-GEF dsRNA-expressing areas. Arrowheads denote the position of the morphogenetic furrow. Anterior is to the right.
Compromised Rap1 signaling perturbs apical domain formation of developing photoreceptor cells
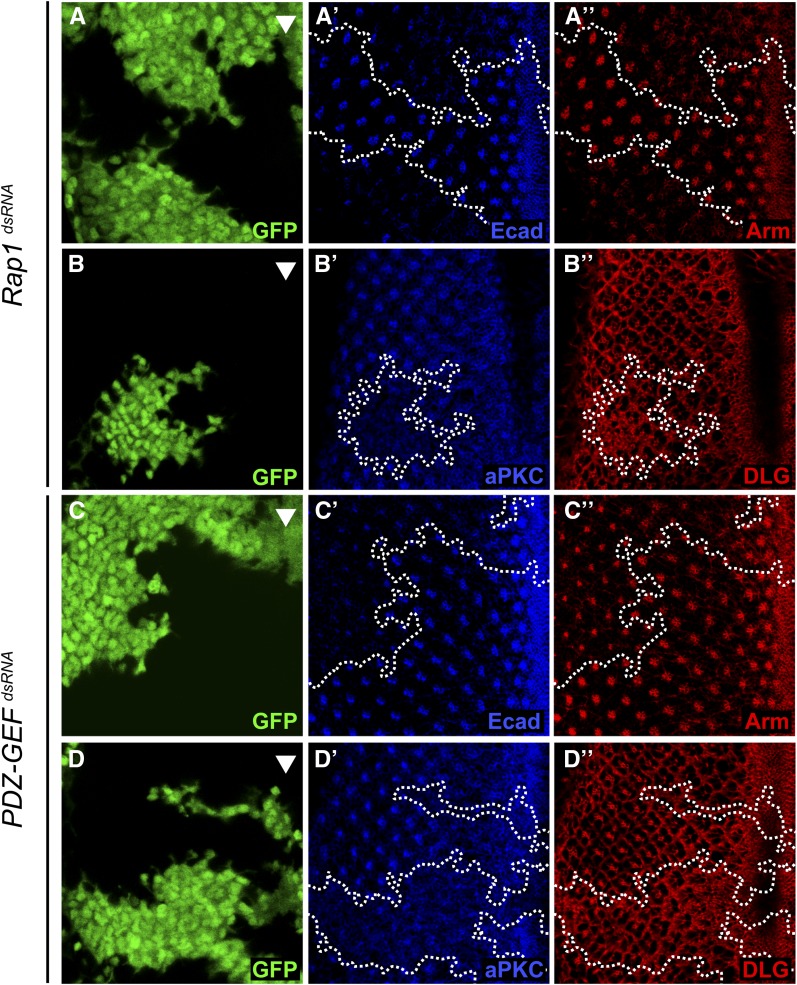
The results presented above suggest that Rap1 does not influence MAPK signaling through a direct action on RAF. We therefore wanted to address the mechanism by which Rap1 impinged on R7 cell differentiation. Rap1 activity is now well recognized for its role in cadherin-based cell–cell adhesion, and various Rap1 pathway components, including Rap1 itself, have been found to localize to adherens junctions (Knox and Brown 2002) (Figure S8). Consistent with this, depletion of either Rap1 or PDZ-GEF by RNAi significantly reduced the epithelial cadherin (Ecad) and Armadillo (Arm; fly β-catenin) levels that normally define the adherens junctions of developing ommatidia (Figure 6, A and C). Despite the clear impact on adherens junctions, no major cell adhesion defects could be observed in these RNAi-based knockdown conditions (GFP-positive cells do not significantly intermingle with non-GFP-positive cells), which is consistent with the fact that most outer photoreceptor cells differentiated normally. The reason for this could be that there are still sufficient levels of adherens junctions to support a relatively normal cell–cell adhesion. Another proposed role for adherens junctions is the establishment/maintenance of apicobasal polarity in the fly embryo, whereby adherens junctions participate in the maintenance of the apical domain and may also serve to physically separate antagonistically acting apical determinants from basolateral determinants (Bilder et al. 2003; Tanentzapf and Tepass 2003; Kaplan et al. 2009). To verify this possibility, we examined the effect of disrupting adherens junctions on the polarity of developing photoreceptor cells. For this, Ecad dsRNA-expressing clones in third instar eye discs were stained with anti-aPKC and anti-DLG, which, respectively, mark the apical and basolateral membranes (Woods et al. 1996; Wodarz et al. 2000). Ecad RNAi clearly reduced aPKC levels, but did not affect DLG levels (Figure S9A). This observation is consistent with an impaired apical compartment. We next verified whether reducing Rap1 activity would have similar consequences. As shown in Figure 6, both Rap1 and PDZ-GEF RNAi diminished aPKC staining in neuronal clusters, but did not reduce DLG levels and, if anything, appeared to slightly elevate DLG levels (Figure 6, B and D). A disrupted apical accumulation of aPKC was also seen in PDZ-GEF mutant clones (Figure S7). The impact of reduced Rap1 signaling was not restricted to aPKC as it also impeded the accumulation of two other apical markers, namely, Bazooka (BAZ) and PATJ (Figure S10), which further supports the idea of a perturbed apical domain. Together, these observations suggest that adherens junctions play a role in apical domain formation/maintenance, as observed in fly embryos (Bilder et al. 2003), and that Rap1 signaling also participates in this event.
Figure 6.
Rap1 and PDZ-GEF dsRNAs impair the expression of adherens junction and apical markers in third instar eye discs. (A–A′′ to D–D′′) Random clones (GFP-positive cells) expressing the indicated dsRNAs in third instar larval eye discs were induced using the FLP-out/Gal4 system. Eye discs were then immunostained with (A′ and C′) anti-Ecad;, (A′′ and C′′) anti-Arm, (B′ and D′) anti-aPKC, and (B′′ and D′′) anti-DLG antibodies. The arrowhead denotes the position of the morphogenetic furrow. Anterior is to the right.
Given the functional link between CNK and Rap1 pathway components, we examined whether CNK also played a role in adherens junction and/or apical domain formation in differentiating photoreceptor cells. As shown in Figure S11, CNK is expressed within the morphogenetic furrow as well as a few rows posterior to it, but its expression rapidly declines more posteriorly. Unlike for Rap1 or PDZ-GEF, knockdown of CNK by RNAi had no impact on the levels of adherens junction (Ecad) or apical domain (aPKC) markers within recruited photoreceptor cells (Figure S11, A–C). Nonetheless, owing to its role as a Ras/MAPK pathway component (Therrien et al. 1998), the loss of CNK impacted cell differentiation and thereby reduced the size of ommatidial clusters (Figure S11B).
Rap1 signaling controls the levels of apically localized tyrosine-phosphorylated proteins of developing photoreceptor cells
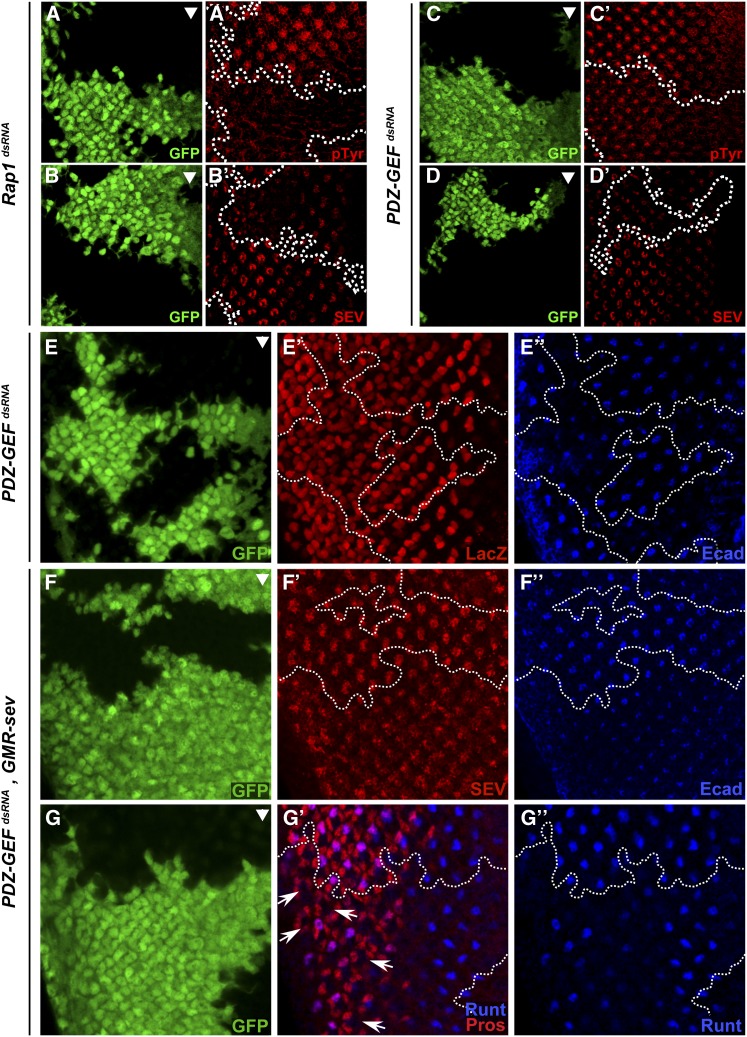
A number of signaling proteins mediating RTK signals, such as DRK and DOS, have been found to localize to the apical region of differentiating eye cells (Olivier et al. 1993; Raabe et al. 1996). Since several of these proteins, including the RTKs, are tyrosine-phosphorylated in response to EGFR and SEV activation, we stained third instar eye discs with an anti-pTyr antibody as a readout for RTK signaling. As shown in Figure 7, the knockdown of either Rap1 or PDZ-GEF greatly reduced pTyr levels that are normally associated with the apical surface of developing ommatidia (Figure 7, A and C). These findings demonstrate the ability of Rap1 signaling to control RTK signaling in developing photoreceptor cells by possibly influencing the formation of the apical domain.
Figure 7.
Rap1 signaling controls SEV levels through a post-transcriptional mechanism. (A–G) Random clones (GFP-positive cells) expressing the indicated dsRNAs in third instar eye discs were induced using the FLP-out/Gal4 system and immunostained with (A′ and C′) anti-pTyr, (B′, D′ and F′) anti-SEV, (E′) anti-β-Gal, (E′′–F′′) anti-Ecad, and (G′ and G′′) anti-Runt and anti-Prospero (Pros) antibodies. (A′–D′) Rap1 and PDZ-GEF dsRNAs impede SEV and pTyr levels in developing photoreceptor cells. However, reducing Rap1 activity, as achieved using PDZ-GEF dsRNA (E–E′′), does not prevent sev-LacZ expression. On the other hand, SEV overexpression using a pGMR-sev transgene (F and G) does not restore SEV levels to the apical domain (F′), nor does it rescue R7 cell differentiation as assessed by the loss of Runt/Pros double-positive cells (G′; arrows point to several examples) in the PDZ-GEF dsRNA-expressing area. (F′′–G′′) Anti-Ecad staining was performed as a control to demonstrate the activity of the PDZ-GEF dsRNA. Arrowheads mark the position of the morphogenetic furrow. Anterior is to the right.
Next, we examined whether Rap1 signaling was also involved in the proper localization of EGFR, which has been found to localize to both apical and basolateral membranes of epithelial cells (Dumstrei et al. 2002). EGFR expression is the highest within the morphogenetic furrow and in developing clusters of photoreceptors two to three rows behind the furrow and then sharply declines, but localizes throughout the cell membranes (Figure S12). However, neither its expression nor its subcellular distribution was affected upon depleting Rap1 or PDZ-GEF activity (Figure S12).
Rap1 signaling is required for the apical accumulation of SEV
The SEV RTK localizes to the apical domain of presumptive R7 cells, and this subcellular distribution is likely critical for interacting with the ligand BOSS presented at the apical surface of the apposing R8 cells (Banerjee et al. 1987; Tomlinson and Ready 1987; Kramer et al. 1991). Because R7 cells are particularly sensitive to Rap1 depletion, we wondered whether this could be due to a failure to properly localize SEV to the apical membranes. Even though SEV is required only for R7 cell differentiation, it is detected on the apical surface of several cells within each developing ommatidium where it is first expressed in differentiating R3/R4 photoreceptor cells (Banerjee et al. 1987; Tomlinson et al. 1987) (Figure 7). Strikingly, reducing the dose of either Rap1 or PDZ-GEF by RNAi or eliminating their presence in homozygous mutant clones greatly affected the accumulation of SEV at the apical surface of the cells (Figure 7, B and D; Figure S6; and Figure S7). Interestingly, a similar observation was made upon depleting Ecad by RNAi (Figure S9B). This suggests that the effect of Rap1 signaling on SEV apical accumulation is related to its impact on adherens junction formation. We verified whether this was accompanied by a relocalization of SEV to basolateral membranes, but did not find any evidence for this (Figure S9, C–E), which indicated that the overall levels of SEV were reduced.
Given that Rap1 modulates MAPK signaling in the R7 cell differentiation area, we verified whether SEV levels were controlled by a MAPK-dependent positive feedback mechanism. To verify this, we compared SEV levels in third instar eye discs between WT and a hypomorphic allele of mapk (rl1), which produces a mild rough eye phenotype (Figure S13) owing to moderate cell differentiation failures (Biggs et al. 1994). In this context, no difference in overall SEV levels could be detected (Figure S13), thus suggesting that SEV levels are not influenced by MAPK-mediated signaling.
Next, we examined whether Rap1 signaling controlled sev gene expression. To this end, we took advantage of an enhancer trap line carrying a LacZ reporter construct inserted in the sev locus that recapitulates a normal sev expression pattern during eye development (Basler et al. 1989). The impairment of Rap1 signaling had no impact on sev-lacZ expression and therefore indicated that Rap1 influences SEV levels post-transcriptionally (Figure 7E). Further supporting this model, overexpressed SEV using a pGMR-sev transgene also failed to localize to the apical domain of developing ommatidial cell clusters depleted of Rap1 activity and, accordingly, did not rescue the loss of R7 cells (Figure 7, F and G). Finally, we tested whether Rap1V12 overexpression modulated SEV levels, but did not find any evidence for this (Figure S14), which also supports the conclusion that Rap1 does not directly control SEV expression. Interestingly, however, Rap1V12 expression elevated adherens junction formation in cells surrounding nascent ommatidia next to the morphogenetic furrow and thereby raised the apparent number of cells per cluster (Figure S14). Whether this phenomenon explains the supernumerary eye-specific cells seen in more mature Rap1V12 clones (Figure S3) will require further investigation.
Discussion
In this report, we describe a genetic screen in Drosophila for dominant modifiers of a CNK-dependent rough eye phenotype. We then characterize two of those modifiers, Rap1 and PDZ-GEF, to further shed light on the mechanism by which Rap1-mediated events influence photoreceptor cell development.
Modifiers of dominant-negative CNK
Given the role CNK plays in RTK-elicited Ras/MAPK signaling, mutations in loci encoding general components of this pathway in flies were recovered in the CCT screen. They correspond to Star [S; trafficking factor for the EGFR ligand Spitz (Mayer and Nusslein-Volhard 1988; Lee et al. 2001; Tsruya et al. 2002)], Egfr, daughter of sevenless (dos; Gab2 homolog; Herbst et al. 1996; Raabe et al. 1996), Son of sevenless (Sos; RasGEF; Simon et al. 1991), Ras85D, rl/mapk, ksr, and pointed (pnt; ETS domain transcription factor mediating MAPK activity; Brunner et al. 1994; O’Neill et al. 1994) (Table 2). Mutations in two genes previously identified in RTK-dependent screens, but of unclear function, were also isolated. These are kismet [kis; chromatin remodeler (Therrien et al. 2000)] and multiple ankyrin repeats single KH domain [mask; putative RNA-binding protein (Smith et al. 2002)].
Mutant alleles not identified in classical RTK/MAPK-dependent genetic screens, but for which a functional link to RTK signaling in flies or in other organisms had been established were also recovered. These include Btk family kinase at 29A [Btk29A; the single representative of Tec family kinases (Readinger et al. 2009)]; Delta (Dl; Baker and Rubin 1992); and three Rap1 pathway loci, PDZ-GEF, Rap1, and canoe (cno) (Li et al. 1997; Lee et al. 2002; Gaengel and Mlodzik 2003; O’Keefe et al. 2009). Finally, mutations were isolated in four additional loci not previously reported to influence RTK/MAPK signaling: Pre-mRNA-processing factor 19 (Prp19), BREFeldin A sensitivity 1 (Bre1), NEM sensitive factor 2 (Nsf2), and second mitotic wave missing (swm).
The Prp19 locus encodes a core spliceosome component (Chan et al. 2003). We recently unveiled a specific role for these proteins in the Ras/MAPK pathway. Indeed, we identified several splicing factors, including Prp19 and Prp8, in a genome-wide RNAi screen in S2 cells for modulators of Ras-induced MAPK activation (Ashton-Beaucage et al. 2014). Incidentally, a single mutant allele of Prp8 was also recovered in the CCT screen (reported in Ashton-Beaucage et al. 2014). Characterization of their implication in the pathway revealed that they specifically regulate MAPK protein levels by controlling the alternative splicing of selected introns of the mapk pre-mRNAs. It is thus likely that Prp19 alleles were recovered in the CCT screen because of their impact on endogenous MAPK levels during eye development.
The association that the last three genes (Bre1, Nsf2, and swm) might have with respect to CCT activity is less clear. Bre1 encodes a RING finger-containing E3 ligase mediating histone H2B monoubiquitination (Bray et al. 2005). This modification contributes to specific histone epigenetic changes such as histone H3K4 and H3K79 methylation that correlate with transcriptional activation (Wright et al. 2011). A role for Bre1 in Notch- and Wingless-dependent gene expression has been reported (Bray et al. 2005; Mohan et al. 2010), but whether it acts similarly downstream of RTK signaling is not known. Given the concerted, yet distinct role Notch and EGFR signaling play in morphogenetic furrow progression and thereby in eye development (Doroquez and Rebay 2006), it could well be that, as for Dl, Bre1 was recovered primarily for its function in Notch signaling.
Nsf2 encodes an AAA ATPase involved in vesicular trafficking and synaptic vesicle release (Boulianne and Trimble 1995; Pallanck et al. 1995). Previous genetic studies have also associated this gene with Notch and Wingless signaling (Stewart et al. 2001), and thus this could be the basis for the isolation of Nsf2 alleles. Alternatively, Nsf2 activity could be required in trafficking events directly involved in RTK signaling.
The swm gene [aka Su(Rux)2B (Dong et al. 1997)] encodes a novel protein that comprises a CCCH zinc finger and a RNA recognition motif (Casso et al. 2008). SWM localizes to the nucleus and was found to play multiple roles during Drosophila development (Casso et al. 2008), although its precise molecular function is not known. During eye development, Swm regulates the proliferation of undifferentiated cells by controlling their G1/S transition (Dong et al. 1997). In particular, third instar eye discs deprived of SWM activity are reduced in size and, as epitomized by the gene name, they lack the second mitotic wave, which corresponds to a row of cells located at a few cells distance posterior to the morphogenetic furrow that undergo a unique and synchronous round of cell division (Baker 2001). This event increases the pool of uncommitted cells used for completing ommatidial assembly. Both Notch and EGFR signaling are essential for cell cycle progression of the uncommitted cells in the second mitotic wave, but act at distinct steps (Baonza et al. 2002; Firth and Baker 2005). Whether the swm alleles were recovered because of their impact on the second mitotic wave or for another role of SWM in differentiating cells remains to be investigated.
Rap1 signaling is required upstream of SEV for MAPK activation in the R7 cell precursors
The ability of the CCT screen to identify mutations in three loci linked to Rap1 signaling strongly suggests a functional relationship between CNK and Rap1 activity. Yet, we did not find evidence for physical association between CNK and Rap1 or PDZ-GEF (Figure S4E), and thus the molecular underpinning of this relationship is currently not known. One possibility for their genetic interactions could be through their separate roles in RTK-mediated events. Rap1 signaling promotes adherens junction formation in differentiating photoreceptor cells, which contributes to their clustering (O’Keefe et al. 2009). This phenomenon, in turn, is thought to enable the cells to respond to extracellular cues promoting differentiation. By lowering Rap1 activity, cohesive contacts between differentiating cells would be suboptimal and thereby would impede ommatidial assembly to some degree. In this scenario, the impact of CCT expression on photoreceptor cell differentiation would be exacerbated by heterozygous mutations in Rap1-signaling components as these would reduce the sensitivity of developing cells to differentiation cues.
Interestingly, it has been noted that loss of Rap1 activity does not prevent EGFR-induced MAPK activation per se, and thus Rap1 does not appear to work like Ras as a direct RAF activator (O’Keefe et al. 2009). However, the data for this conclusion were based on small Rap1 mutant clones that were close or within the morphogenetic furrow. We extended this work by producing larger clones depleted in Rap1 or PDZ-GEF activity. These clones covered the zone where R7 cell commitment normally occurs. Markedly, we found that reduced Rap1 signaling in this area considerably decreased MAPK activity as well as global pTyr levels (Figure 4 and Figure 6) and thereby mimicked the loss of RTK activity. Consistent with this, a strong impairment in R7 cell fate specification was observed (Figure 5).
In agreement with our findings, Mavromatakis and Tomlinson (2012) recently showed by genetic means that R7 cell fate specification had an absolute requirement in Rap1 activity. According to their model, R7 cell precursors sense higher Notch signaling owing to their position in the developing ommatidium, which at this stage is antagonistic to Ras/MAPK-mediated neuronal differentiation. To counteract Notch signaling, Mavromatakis and Tomlinson (2012) proposed that presumptive R7 cells turn on two RTKs (EGFR and SEV) to produce higher MAPK activity. Intriguingly, their work suggested that Rap1 was required downstream of SEV, although they could not distinguish whether Rap1 acted through the canonical MAPK pathway or parallel to it. We investigated this and found that Rap1 does not seem to work directly through the MAPK pathway since ectopic expression of Rap1V12 during eye development or in cultured S2 cells did not promote MAPK phosphorylation (Figure S2 and Figure S4). Moreover, depletion of Rap1 or PDZ-GEF by RNAi in S2 cells had no consequence on MAPK activation induced by SEV or EGFR (Figure S4). Although the precise mechanism by which Rap1 influences signaling downstream of SEV remains to be delineated, our combined data suggest that Rap1 works at two distinct levels in SEV-mediated signaling, that is, upstream of SEV by modulating the apical localization of SEV and downstream of SEV by a mechanism that has yet to be characterized.
Rap1 signaling, adherens junction, and apico-basal polarity
Adherens junctions form a belt-like microdomain that encircles epithelial cells apically and that play a major role in cell–cell adhesion, motility, and polarity (Etienne-Manneville 2011). One of the core structural components of adherens junctions is Ecad, which is a transmembrane glycoprotein that forms Ca2+-dependent homophilic interactions between adjacent cells. The intracellular portion of Ecad is complexed to the catenins that, in turn, mediate linkage to the actomyosin cytoskeleton (Yonemura 2011). Studies conducted over the past 10 years in both vertebrate and invertebrate organisms demonstrate the critical role that Rap1 signaling plays in modulating the connections of adherens junctions to the actomyosin network, which then influence cell–cell adhesion, cell shape, and cell migration (Frische and Zwartkruis 2010).
Although our data are consistent with this view, they also hint at a new role for Rap1 signaling that is to control apical domain formation in developing photoreceptor cells. Given that adherens junctions may act as physical barriers between apical and basolateral membrane compartments, the influence of Rap1 on adherens junction dynamics could represent the mechanism by which Rap1 exerts its effect on the apical domain compartment. A more exciting alternative would be that Rap1 activity directly controls the formation of the apical domain. Work conducted in fly embryos by Peifer and colleagues recently provided evidence supporting this model (Choi et al. 2013). Indeed, not only did they find that Rap1 activity is essential for establishing the apico-basal polarity of cellularizing embryos, but their data also suggest that it has a direct impact on the apical localization of Bazooka, a member of the Par complex, which then orchestrates apical domain assembly. Whether Rap1 signaling has a direct influence on cell polarity during eye development is still unclear. Nonetheless, further characterization of the impact that Rap1 signaling has on apical domain formation/maintenance should reveal novel aspects by which cell compartmentalization is brought about and regulated as well as how it connects to downstream signaling events in epithelial cells.
Supplementary Material
Acknowledgments
We thank the Bloomington and Vienna Drosophila RNAi Collection stock centers; the Developmental Studies Hybridoma Bank; and A. Brand, N. Brown, B. Edgar, E. Knust, J. McDonald, G. Rubin, B.-Z. Shilo, A. Tomlinson, and A. Wodarz for fly lines and reagents used in this study. Special thanks also to D. Ashton-Beaucage, F. Bergeron-Labrecque, and G. Gavory for critical reading of the manuscript. We thank C. Charbonneau and R. Lambert, respectively, of the Institute for Research in Immunology and Cancer Bioimaging and Genomics facilities for assistance with microscopy and DNA sequencing. H.K. is recipient of a postdoctoral fellowship from the Research Council of Norway. This work was supported by a grant (MOP-15375) from the Canadian Institutes for Health Research (to M.T.).
Footnotes
Communicating editor: L. Cooley
These authors contributed equally to this work.
Literature Cited
- Ashton-Beaucage D., Udell C. M., Gendron P., Sahmi M., Lefrançois M., et al. , 2014. A functional screen reveals an extensive layer of transcriptional and splicing control underlying RAS/MAPK signaling in Drosophila. PLoS Biol. 12: e1001809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker N. E., 2001. Cell proliferation, survival, and death in the Drosophila eye. Semin. Cell Dev. Biol. 12: 499–507. [DOI] [PubMed] [Google Scholar]
- Baker N. E., Rubin G. M., 1992. Ellipse mutations in the Drosophila homologue of the EGF receptor affect pattern formation, cell division, and cell death in eye imaginal discs. Dev. Biol. 150: 381–396. [DOI] [PubMed] [Google Scholar]
- Banerjee U., Renfranz P. J., Hinton D. R., Rabin B. A., Benzer S., 1987. The sevenless+ protein is expressed apically in cell membranes of developing Drosophila retina; it is not restricted to cell R7. Cell 51: 151–158. [DOI] [PubMed] [Google Scholar]
- Baonza A., Murawsky C. M., Travers A. A., Freeman M., 2002. Pointed and Tramtrack69 establish an EGFR-dependent transcriptional switch to regulate mitosis. Nat. Cell Biol. 4: 976–980. [DOI] [PubMed] [Google Scholar]
- Basler K., Siegrist P., Hafen E., 1989. The spatial and temporal expression pattern of sevenless is exclusively controlled by gene-internal elements. EMBO J. 8: 2381–2386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biggs W. H., Zavitz K. H., Dickson B., van der Straten A., Brunner D., et al. , 1994. The Drosophila rolled locus encodes a MAP kinase required in the sevenless signal transduction pathway. EMBO J. 13: 1628–1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilder D., Schober M., Perrimon N., 2003. Integrated activity of PDZ protein complexes regulates epithelial polarity. Nat. Cell Biol. 5: 53–58. [DOI] [PubMed] [Google Scholar]
- Boettner B., Harjes P., Ishimaru S., Heke M., Fan H. Q., et al. , 2003. The AF-6 homolog canoe acts as a Rap1 effector during dorsal closure of the Drosophila embryo. Genetics 165: 159–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulianne G. L., Trimble W. S., 1995. Identification of a second homolog of N-ethylmaleimide-sensitive fusion protein that is expressed in the nervous system and secretory tissues of Drosophila. Proc. Natl. Acad. Sci. USA 92: 7095–7099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand A. H., Perrimon N., 1993. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118: 401–415. [DOI] [PubMed] [Google Scholar]
- Bray S., Musisi H., Bienz M., 2005. Bre1 is required for Notch signaling and histone modification. Dev. Cell 8: 279–286. [DOI] [PubMed] [Google Scholar]
- Brunner D., Ducker K., Oellers N., Hafen E., Scholz H., et al. , 1994. The ETS domain protein pointed-P2 is a target of MAP kinase in the sevenless signal transduction pathway. Nature 370: 386–389. [DOI] [PubMed] [Google Scholar]
- Cagan R., 2009. Principles of Drosophila eye differentiation. Curr. Top. Dev. Biol. 89: 115–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casso D. J., Liu S., Iwaki D. D., Ogden S. K., Kornberg T. B., 2008. A screen for modifiers of hedgehog signaling in Drosophila melanogaster identifies swm and mts. Genetics 178: 1399–1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan S. P., Kao D. I., Tsai W. Y., Cheng S. C., 2003. The Prp19p-associated complex in spliceosome activation. Science 302: 279–282. [DOI] [PubMed] [Google Scholar]
- Choi W., Harris N. J., Sumigray K. D., Peifer M., 2013. Rap1 and Canoe/afadin are essential for establishment of apical-basal polarity in the Drosophila embryo. Mol. Biol. Cell 24: 945–963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claperon A., Therrien M., 2007. KSR and CNK: two scaffolds regulating RAS-mediated RAF activation. Oncogene 26: 3143–3158. [DOI] [PubMed] [Google Scholar]
- Dickson B., Sprenger F., Morrison D., Hafen E., 1992. Raf functions downstream of Ras1 in the Sevenless signal transduction pathway. Nature 360: 600–603. [DOI] [PubMed] [Google Scholar]
- Dong X., Zavitz K. H., Thomas B. J., Lin M., Campbell S., et al. , 1997. Control of G1 in the developing Drosophila eye: rca1 regulates Cyclin A. Genes Dev. 11: 94–105. [DOI] [PubMed] [Google Scholar]
- Doroquez D. B., Rebay I., 2006. Signal integration during development: mechanisms of EGFR and Notch pathway function and cross-talk. Crit. Rev. Biochem. Mol. Biol. 41: 339–385. [DOI] [PubMed] [Google Scholar]
- Douziech M., Roy F., Laberge G., Lefrancois M., Armengod A. V., et al. , 2003. Bimodal regulation of RAF by CNK in Drosophila. EMBO J. 22: 5068–5078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douziech M., Sahmi M., Laberge G., Therrien M., 2006. A KSR/CNK complex mediated by HYP, a novel SAM domain-containing protein, regulates RAS-dependent RAF activation in Drosophila. Genes Dev. 20: 807–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumstrei K., Wang F., Shy D., Tepass U., Hartenstein V., 2002. Interaction between EGFR signaling and DE-cadherin during nervous system morphogenesis. Development 129: 3983–3994. [DOI] [PubMed] [Google Scholar]
- Etienne-Manneville S., 2011. Control of polarized cell morphology and motility by adherens junctions. Semin. Cell Dev. Biol. 22: 850–857. [DOI] [PubMed] [Google Scholar]
- Firth L. C., Baker N. E., 2005. Extracellular signals responsible for spatially regulated proliferation in the differentiating Drosophila eye. Dev. Cell 8: 541–551. [DOI] [PubMed] [Google Scholar]
- Freeman M., 1997. Cell determination strategies in the Drosophila eye. Development 124: 261–270. [DOI] [PubMed] [Google Scholar]
- Frische E. W., Zwartkruis F. J., 2010. Rap1, a mercenary among the Ras-like GTPases. Dev. Biol. 340: 1–9. [DOI] [PubMed] [Google Scholar]
- Gabay L., Seger R., Shilo B. Z., 1997. In situ activation pattern of Drosophila EGF receptor pathway during development. Science 277: 1103–1106. [DOI] [PubMed] [Google Scholar]
- Gaengel K., Mlodzik M., 2003. Egfr signaling regulates ommatidial rotation and cell motility in the Drosophila eye via MAPK/Pnt signaling and the Ras effector Canoe/AF6. Development 130: 5413–5423. [DOI] [PubMed] [Google Scholar]
- Hahn I., Fuss B., Peters A., Werner T., Sieberg A., et al. , 2013. The Drosophila Arf GEF Steppke controls MAPK activation in EGFR signaling. J. Cell Sci. 126: 2470–2479. [DOI] [PubMed] [Google Scholar]
- Hariharan I. K., Carthew R. W., Rubin G. M., 1991. The Drosophila roughened mutation: activation of a rap homolog disrupts eye development and interferes with cell determination. Cell 67: 717–722. [DOI] [PubMed] [Google Scholar]
- Herbst R., Carroll P. M., Allard J. D., Schilling J., Raabe T., et al. , 1996. Daughter of sevenless is a substrate of the phosphotyrosine phosphatase Corkscrew and functions during sevenless signaling. Cell 85: 899–909. [DOI] [PubMed] [Google Scholar]
- Jaffe A. B., Hall A., Schmidt A., 2005. Association of CNK1 with Rho guanine nucleotide exchange factors controls signaling specificity downstream of Rho. Curr. Biol. 15: 405–412. [DOI] [PubMed] [Google Scholar]
- Kaplan N. A., Liu X., Tolwinski N. S., 2009. Epithelial polarity: interactions between junctions and apical-basal machinery. Genetics 183: 897–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karim F. D., Chang H. C., Therrien M., Wassarman D. A., Laverty T., et al. , 1996. A screen for genes that function downstream of Ras1 during Drosophila eye development. Genetics 143: 315–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauffmann R. C., Li S., Gallagher P. A., Zhang J., Carthew R. W., 1996. Ras1 signaling and transcriptional competence in the R7 cell of Drosophila. Genes Dev. 10: 2167–2178. [DOI] [PubMed] [Google Scholar]
- Kimmel B. E., Heberlein U., Rubin G. M., 1990. The homeo domain protein rough is expressed in a subset of cells in the developing Drosophila eye where it can specify photoreceptor cell subtype. Genes Dev. 4: 712–727. [DOI] [PubMed] [Google Scholar]
- Knox A. L., Brown N. H., 2002. Rap1 GTPase regulation of adherens junction positioning and cell adhesion. Science 295: 1285–1288. [DOI] [PubMed] [Google Scholar]
- Kramer H., Cagan R. L., Zipursky S. L., 1991. Interaction of bride of sevenless membrane-bound ligand and the sevenless tyrosine-kinase receptor. Nature 352: 207–212. [DOI] [PubMed] [Google Scholar]
- Kumar J. P., 2012. Building an ommatidium one cell at a time. Dev. Dyn. 241: 136–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laberge G., Douziech M., Therrien M., 2005. Src42 binding activity regulates Drosophila RAF by a novel CNK-dependent derepression mechanism. EMBO J. 24: 487–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanigan T. M., Liu A., Huang Y. Z., Mei L., Margolis B., et al. , 2003. Human homologue of Drosophila CNK interacts with Ras effector proteins Raf and Rlf. FASEB J. 17: 2048–2060. [DOI] [PubMed] [Google Scholar]
- Lee J. H., Cho K. S., Lee J., Kim D., Lee S. B., et al. , 2002. Drosophila PDZ-GEF, a guanine nucleotide exchange factor for Rap1 GTPase, reveals a novel upstream regulatory mechanism in the mitogen-activated protein kinase signaling pathway. Mol. Cell. Biol. 22: 7658–7666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. R., Urban S., Garvey C. F., Freeman M., 2001. Regulated intracellular ligand transport and proteolysis control EGF signal activation in Drosophila. Cell 107: 161–171. [DOI] [PubMed] [Google Scholar]
- Li Q., Hariharan I. K., Chen F., Huang Y., Fischer J. A., 1997. Genetic interactions with Rap1 and Ras1 reveal a second function for the fat facets deubiquitinating enzyme in Drosophila eye development. Proc. Natl. Acad. Sci. USA 94: 12515–12520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim J., Zhou M., Veenstra T. D., Morrison D. K., 2010. The CNK1 scaffold binds cytohesins and promotes insulin pathway signaling. Genes Dev. 24: 1496–1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mavromatakis Y. E., Tomlinson A., 2012. The role of the small GTPase Rap in Drosophila R7 photoreceptor specification. Proc. Natl. Acad. Sci. USA 109: 3844–3849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer U., Nusslein-Volhard C., 1988. A group of genes required for pattern formation in the ventral ectoderm of the Drosophila embryo. Genes Dev. 2: 1496–1511. [DOI] [PubMed] [Google Scholar]
- Mirey G., Balakireva M., L’Hoste C., Rossé C., Voegeling S., et al. , 2003. A Ral guanine exchange factor-Ral pathway is conserved in Drosophila melanogaster and sheds new light on the connectivity of the Ral, Ras, and Rap pathways. Mol. Cell. Biol. 23: 1112–1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra S., Smolik S. M., Forte M. A., Stork P. J., 2005. Ras-independent activation of ERK signaling via the torso receptor tyrosine kinase is mediated by Rap1. Curr. Biol. 15: 366–370. [DOI] [PubMed] [Google Scholar]
- Miyamoto H., Nihonmatsu I., Kondo S., Ueda R., Togashi S., et al. , 1995. canoe encodes a novel protein containing a GLGF/DHR motif and functions with Notch and scabrous in common developmental pathways in Drosophila. Genes Dev. 9: 612–625. [DOI] [PubMed] [Google Scholar]
- Mohan M., Herz H. M., Takahashi Y. H., Lin C., Lai K. C., et al. , 2010. Linking H3K79 trimethylation to Wnt signaling through a novel Dot1-containing complex (DotCom). Genes Dev. 24: 574–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neufeld T. P., de la Cruz A. F., Johnston L. A., Edgar B. A., 1998. Coordination of growth and cell division in the Drosophila wing. Cell 93: 1183–1193. [DOI] [PubMed] [Google Scholar]
- O’Keefe D. D., Gonzalez-Nino E., Burnett M., Dylla L., Lambeth S. M., et al. , 2009. Rap1 maintains adhesion between cells to affect Egfr signaling and planar cell polarity in Drosophila. Dev. Biol. 333: 143–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivier J. P., Raabe T., Henkemeyer M., Dickson B., Mbamalu G., et al. , 1993. A Drosophila SH2–SH3 adaptor protein implicated in coupling the sevenless tyrosine kinase to an activator of Ras guanine nucleotide exchange, Sos. Cell 73: 179–191. [DOI] [PubMed] [Google Scholar]
- O’Neill E. M., Rebay I., Tjian R., Rubin G. M., 1994. The activities of two Ets-related transcription factors required for Drosophila eye development are modulated by the Ras/MAPK pathway. Cell 78: 137–147. [DOI] [PubMed] [Google Scholar]
- Pallanck L., Ordway R. W., Ramaswami M., Chi W. Y., Krishnan K. S., et al. , 1995. Distinct roles for N-ethylmaleimide-sensitive fusion protein (NSF) suggested by the identification of a second Drosophila NSF homolog. J. Biol. Chem. 270: 18742–18744. [DOI] [PubMed] [Google Scholar]
- Raabe T., Riesgo-Escovar J., Liu X., Bausenwein B. S., Deak P., et al. , 1996. DOS, a novel pleckstrin homology domain-containing protein required for signal transduction between sevenless and Ras1 in Drosophila. Cell 85: 911–920. [DOI] [PubMed] [Google Scholar]
- Readinger J. A., Mueller K. L., Venegas A. M., Horai R., Schwartzberg P. L., 2009. Tec kinases regulate T-lymphocyte development and function: new insights into the roles of Itk and Rlk/Txk. Immunol. Rev. 228: 93–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ready D. F., Hanson T. E., Benzer S., 1976. Development of the Drosophila retina, a neurocrystalline lattice. Dev. Biol. 53: 217–240. [DOI] [PubMed] [Google Scholar]
- Roignant J. Y., Treisman J. E., 2009. Pattern formation in the Drosophila eye disc. Int. J. Dev. Biol. 53: 795–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin G. M., Spradling A. C., 1982. Genetic transformation of Drosophila with transposable element vectors. Science 218: 348–353. [DOI] [PubMed] [Google Scholar]
- Simon M. A., Bowtell D. D., Dodson G. S., Laverty T. R., Rubin G. M., 1991. Ras1 and a putative guanine nucleotide exchange factor perform crucial steps in signaling by the sevenless protein tyrosine kinase. Cell 67: 701–716. [DOI] [PubMed] [Google Scholar]
- Smith R. K., Carroll P. M., Allard J. D., Simon M. A., 2002. MASK, a large ankyrin repeat and KH domain-containing protein involved in Drosophila receptor tyrosine kinase signaling. Development 129: 71–82. [DOI] [PubMed] [Google Scholar]
- Stewart B. A., Mohtashami M., Zhou L., Trimble W. S., Boulianne G. L., 2001. SNARE-dependent signaling at the Drosophila wing margin. Dev. Biol. 234: 13–23. [DOI] [PubMed] [Google Scholar]
- Tanentzapf G., Tepass U., 2003. Interactions between the crumbs, lethal giant larvae and bazooka pathways in epithelial polarization. Nat. Cell Biol. 5: 46–52. [DOI] [PubMed] [Google Scholar]
- Therrien M., Chang H. C., Solomon N. M., Karim F. D., Wassarman D. A., et al. , 1995. KSR, a novel protein kinase required for RAS signal transduction. Cell 83: 879–888. [DOI] [PubMed] [Google Scholar]
- Therrien M., Wong A. M., Rubin G. M., 1998. CNK, a RAF-binding multidomain protein required for RAS signaling. Cell 95: 343–353. [DOI] [PubMed] [Google Scholar]
- Therrien M., Wong A. M., Kwan E., Rubin G. M., 1999. Functional analysis of CNK in RAS signaling. Proc. Natl. Acad. Sci. USA 96: 13259–13263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Therrien M., Morrison D. K., Wong A. M., Rubin G. M., 2000. A genetic screen for modifiers of a kinase suppressor of Ras-dependent rough eye phenotype in Drosophila. Genetics 156: 1231–1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomlinson A., Ready D. F., 1987. Cell fate in the Drosophila ommatidium. Dev. Biol. 123: 264–275. [DOI] [PubMed] [Google Scholar]
- Tomlinson A., Bowtell D. D., Hafen E., Rubin G. M., 1987. Localization of the sevenless protein, a putative receptor for positional information, in the eye imaginal disc of Drosophila. Cell 51: 143–150. [DOI] [PubMed] [Google Scholar]
- Tomlinson A., Mavromatakis Y. E., Struhl G., 2011. Three distinct roles for notch in Drosophila R7 photoreceptor specification. PLoS Biol. 9: e1001132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsruya R., Schlesinger A., Reich A., Gabay L., Sapir A., et al. , 2002. Intracellular trafficking by Star regulates cleavage of the Drosophila EGF receptor ligand Spitz. Genes Dev. 16: 222–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udell C. M., Rajakulendran T., Sicheri F., Therrien M., 2011. Mechanistic principles of RAF kinase signaling. Cell. Mol. Life Sci. 68: 553–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkateswarlu K., 2003. Interaction protein for cytohesin exchange factors 1 (IPCEF1) binds cytohesin 2 and modifies its activity. J. Biol. Chem. 278: 43460–43469. [DOI] [PubMed] [Google Scholar]
- Wodarz A., Ramrath A., Grimm A., Knust E., 2000. Drosophila atypical protein kinase C associates with Bazooka and controls polarity of epithelia and neuroblasts. J. Cell Biol. 150: 1361–1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods D. F., Hough C., Peel D., Callaini G., Bryant P. J., 1996. Dlg protein is required for junction structure, cell polarity, and proliferation control in Drosophila epithelia. J. Cell Biol. 134: 1469–1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright D. E., Wang C. Y., Kao C. F., 2011. Flickin’ the ubiquitin switch: the role of H2B ubiquitylation in development. Epigenetics 6: 1165–1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonemura S., 2011. Cadherin-actin interactions at adherens junctions. Curr. Opin. Cell Biol. 23: 515–522. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.