Abstract
Phosphorylated tyrosine hydroxylase (TH) can form complexes with 14-3-3 proteins, resulting in enzyme activation and stabilization. Although TH was among the first binding partners identified for these ubiquitous regulatory proteins, the binding stoichiometry and the activation mechanism remain unknown. To address this, we performed native mass spectrometry analyses of human TH (nonphosphorylated or phosphorylated on Ser19 (TH-pS19), Ser40 (TH-pS40), or Ser19 and Ser40 (TH-pS19pS40)) alone and together with 14-3-3γ. Tetrameric TH-pS19 (224 kDa) bound 14-3-3γ (58.3 kDa) with high affinity (Kd = 3.2 nM), generating complexes containing either one (282.4 kDa) or two (340.8 kDa) dimers of 14-3-3. Electron microscopy also revealed one major population of an asymmetric complex, consistent with one TH tetramer and one 14-3-3 dimer, and a minor population of a symmetric complex of one TH tetramer with two 14-3-3 dimers. Lower phosphorylation stoichiometries (0.15–0.54 phosphate/monomer) produced moderate changes in binding kinetics, but native MS detected much less of the symmetric TH:14-3-3γ complex. Interestingly, dephosphorylation of [32P]-TH-pS19 was mono-exponential for low phosphorylation stoichiometries (0.18–0.52), and addition of phosphatase accelerated the dissociation of the TH-pS19:14-3-3γ complex 3- to 4-fold. All together this is consistent with a model in which the pS19 residues in the TH tetramer contribute differently in the association to 14-3-3γ. Complex formation between TH-pS40 and 14-3-3γ was not detected via native MS, and surface plasmon resonance showed that the interaction was very weak. Furthermore, TH-pS19pS40 behaved similarly to TH-pS19 in terms of binding stoichiometry and affinity (Kd = 2.1 nM). However, we found that 14-3-3γ inhibited the phosphorylation rate of TH-pS19 by PKA (3.5-fold) on Ser40. We therefore conclude that Ser40 does not significantly contribute to the binding of 14-3-3γ, and rather has reduced accessibility in the TH:14-3-3γ complex. This adds to our understanding of the fine-tuned physiological regulation of TH, including hierarchical phosphorylation at multiple sites.
Tyrosine hydroxylase (TH)1 is a tetrahydrobiopterin-dependent enzyme that catalyzes the conversion of l-tyrosine to l-DOPA. This is the rate-limiting reaction in the synthesis of dopamine and other catecholamine neurotransmitters. For human TH (hTH), alternative splicing of transcripts mainly occurs at the N-terminal segment, adjacent to the regulatory domain, generating isoforms 1 to 4 (hTH1–hTH4) (1–3). Here we have studied hTH1, which is very similar to rat and mouse TH in length and sequence, is the major isoform in brain and peripheral tissues, and is the most studied human isoform in situ and in vitro.
The synthesis of catecholamines is tightly controlled through the regulation of TH activity, localization, and stability (3–5). TH protein abundance is subject to regulatory mechanisms at the transcriptional and translational levels (3, 6, 7). Other mechanisms include posttranslational modifications, notably phosphorylation at different N-terminal residues, as well as other short-term effects such as binding to 14-3-3 proteins, feedback inhibition by catecholamines, and substrate inhibition (8–12). Phosphorylation of the enzyme occurs at different N-terminal Ser/Thr residues (Thr8, Ser19, Ser31, and Ser40) (see Refs. 13 and 14 for reviews on the specific kinases involved and the main functional effects of phosphorylation at these sites). Little is known about the possible effects of phosphorylation at Ser/Thr8. However, it is well established that phosphorylation at Ser31 and Ser40 leads to an increase of TH activity for the purified protein and in catecholamine-producing cells (15–17). In particular, strong activation is observed upon Ser40 phosphorylation, as it increases the maximal activity of TH and its affinity for the cofactor tetrahydrobiopterin, whereas it decreases the binding affinity for inhibitory catecholamines by about 200-fold (18, 19).
In contrast, the regulatory functional role of Ser19-phosphorylation of TH is not yet clear. In vitro experiments using the highly Ser19-specific p38-regulated/activated protein kinase (PRAK) show no evidence of Ser19-phosphorylation-induced activation of TH unless 14-3-3 proteins are also added (20). Other Ser19-directed kinases such as Ca2+/calmodulin-dependent protein kinase II and mitogen-activated protein kinase–activated protein kinase-2 also phosphorylate TH on Ser40. For these less specific kinases, the activation observed is proportional to the phosphorylation stoichiometry at Ser40 (17). Similarly, cell experiments on potassium-stimulated PC12 cells show overlapping temporal increases in TH activity with phosphorylation of Ser40, but not of Ser19 (21). Ser19 phosphorylation may directly exert only modest changes in TH activity, but it appears to modulate the phosphorylation rate of the activity regulatory site Ser40 (20, 22). It has also been reported that Ser19 phosphorylation regulates the degradation of TH through the ubiquitin-proteasome pathway (23). Thus, much of the uncertainty regarding the functional importance of Ser19 phosphorylation of TH seems to result from an unresolved understanding of how this phosphorylation regulates protein binding, in particular to 14-3-3 proteins (10, 24).
14-3-3 constitutes a family of ubiquitous proteins involved in many cellular functions, mostly via subcellular sequestration and scaffolding of other proteins to which they bind in a serine/threonine-phosphorylation-dependent manner (25–27). In humans there are seven 14-3-3 isoforms (β, γ, ε, η, ζ, σ, and τ/θ) with high sequence identity. Based on the specific localization of the different 14-3-3 isoforms (28–30), the regulation of TH function upon 14-3-3 binding is also expected to modulate its subcellular distribution. Although all 14-3-3 isoforms are soluble cytoplasmic proteins, 14-3-3γ and -ε have been shown to have an increased propensity to interact peripherally with membranes and could increase membrane binding of their cargo proteins (29–31).
Recently, a combination of methods has provided structural information on the conformation adopted by peptides corresponding to residues 1–43 of hTH1 in its nonphosphorylated (TH-(1–43)) and Ser19-phosphorylated (THp-(1–43)) states (32). This N-terminal region of TH represents an extension to the regulatory ACT domain. The x-ray structure of 14-3-3γ complexed with THp-(1–43) provided structural information only on central residues around pSer19 but showed that each 14-3-3γ dimer binds two peptides, one in each adjacent subunit, and that the bound peptides adopt a more extended conformation around pSer19 than when free in solution (32) (PDB 4J6S; supplemental Fig. S1A). However, this information cannot be extended to the structural understanding in terms of stoichiometry and architecture for the complex formed between Ser19-phosphorylated, full-length TH (TH-pS19) and 14-3-3. So far, attempts to obtain the crystal structure of the complex have been unsuccessful, and biophysical and conformational studies have only verified the high-affinity binding of several 14-3-3 isoforms to TH-pS19 (33, 34). The dimeric 14-3-3 proteins have two phosphopeptide concave binding grooves, and for several target proteins the two sites must be occupied in order for high-affinity binding to occur. However, in many cases one site contributes the most to binding (gatekeeper site), and another site, that can be more degenerate from the typical 14-3-3 recognition sequence, helps to strengthen the binding further (26, 35, 36).
Thus, a tetramer of TH-pS19 (phosphorylated in all four subunits) could in theory be able to form TH-pS19:14-3-3 complexes that might include one to four 14-3-3 dimers, and this stoichiometry might depend on, among other things, structural limitations. Furthermore, although Ser40-phosphorylated TH (TH-pS40) does not seem to bind to mammalian 14-3-3 proteins, this issue is not totally clear, as a direct association of TH-pS40 to the yeast 14-3-3 proteins BMH1 and BMH2 has been reported (23). Thus, an even higher degree of variability of binding modes and stoichiometries might arise for TH phosphorylated at both Ser19 and Ser40 (TH-pS19pS40) in all four subunits. Studies revealing that the phosphorylation at Ser40 is stimulated upon phosphorylation at Ser19 (22, 37) suggest that a sequential phosphorylation might be followed by a conformational change in the N terminus, allowing the binding of each phosphorylated site to adjacent 14-3-3 monomers in each dimer. Such binding of 14-3-3 to dissimilar sites in the same protein chain has been reported for several target proteins such as PKCε, Foxo4, and Raf-1 (38–40) (see also Ref. 41 for review).
Elucidating the structural details of the TH-pS19:14-3-3γ and TH-pS19pS40:14-3-3γ complexes appears crucial to understand the regulatory effects of complex formation on catecholamine synthesis. In this work we investigated possible binding stoichiometries of the protein complexes. To give a satisfactory answer to this question, full stoichiometry of phosphorylation was needed on Ser19 and Ser40, including the double-phosphorylated TH, as achieved in this work. Furthermore, native mass spectrometry, with the gentleness of the electrospray ionization technique that retains protein tertiary and quaternary structure (42, 43), represents a suitable method for solving the stoichiometry of this complex. We also visualized complex formation by electron microscopy (EM) and performed additional analyses utilizing surface plasmon resonance (SPR). Our results provide a frame within which to interpret both the available results on multisite phosphorylation in TH and the observations on stoichiometry and functional effects of Ser19 and Ser40 phosphorylation.
EXPERIMENTAL PROCEDURES
Protein Expression and Purification
The construct (His)6-ZZ-hTH1 was prepared by cloning the human TH1 gene in the pET-ZZ-1a vector (44).2 The construct, which codes for a fusion protein, with a Tobacco etch virus protease-cutting site between the N-terminal His-ZZ fusion partner and hTH1, was expressed in Escherichia coli (BL21 Codon Plus(DE3), Stratagene, La Jolla, CA) in auto-induction media at 37 °C overnight (45). Bacteria were lysed by French press in 50 mM sodium phosphate, pH 7.0, 300 mM NaCl, 0.5 mg/ml Lysozyme, 1 U/ml Benzonase, Roche protease inhibitor mixture, 10 mM benzamidine, 1 mM phenylmethyl-sulfonyl-fluoride. The fusion protein was purified using TALON® metal affinity resin (Clontech, Mountain View, CA). The fusion tag was removed via proteolytic cleavage using Tobacco etch virus protease (1:25 (mg) Tobacco etch virus protease:TH) in 15 mM Hepes, pH 7.4, 150 mM NaCl, 1 mM DTT, 5% glycerol for 4 h on ice before centrifugation (13,000 × g for 10 min) and gel filtration (Superdex 200 10/300 GL, GE Healthcare, UK) in the same buffer without DTT. The homogeneity of the preparation and verification of an intact N-terminal of TH was confirmed using SDS-PAGE and mass spectrometry.
The 14-3-3γ was expressed in E. coli (BL21 Codon Plus (DE3), Stratagene) using the pGEX-2T expression vector (kindly provided by Prof. A. Aitken, Edinburgh, Scotland, UK) via induction (1 mM isopropyl 1-thio-β-D-galactopyranoside) for 4 h at 30 °C. Bacteria were lysed by French press, and GST–14-3-3 fusion proteins were purified on glutathione Sepharose 4B (GE Healthcare) as described previously (30).
Protein Phosphorylation and Dephosphorylation
For optimal phosphorylation of TH (2 mg/ml) by active PRAK (7.5 U/ml (Division of Signal Transduction Therapy, University of Dundee, Scotland, UK)) on Ser19 or by the catalytic subunit of PKA (100 nM, kindly provided by Prof. S. O. Døskeland, University of Bergen (46)) on Ser40, a 50 mM Na-β-glycerophosphate buffer, pH 7.5, containing 5% glycerol, 1 mM DTT, 0.1 mM EGTA, 0.5 mM ATP, 5 mM MgCl2 was used. The phosphorylation was performed at 25 °C for 45 min (PRAK) or 25 min (PKA). To generate comparable samples of TH in different phosphorylation states, we first performed ± phosphorylation of Ser40 using PKA, with a control sample (without kinase) treated the same way. After 25 min the reaction was stopped by the addition of the PKA inhibitor H-89 (5 μM, Sigma-Aldrich), and each of the two samples was split in two before (±) Ser19 phosphorylation was performed using PRAK. Finally, we obtained nonphosphorylated TH (control), Ser19-phosphorylated TH (TH-pS19), Ser40-phosphorylated TH (TH-pS40), and TH phosphorylated on both Ser19 and Ser40 (TH-pS19pS40). We added 50 mM NaF after phosphorylation, and the samples were gel-filtrated on a Superdex 200 column equilibrated with 15 mM Hepes, pH 7.4, 150 mM NaCl. The stoichiometry of all phosphorylations was measured using [32P]-γ-ATP labeling of equivalent parallel samples by spotting on phospho-cellulose paper (p81), washing in 75 mM ortho-phosphoric acid, and employing scintillation counting.
Phosphorylation of TH for dephosphorylation studies was performed at the following conditions: TH (2 mg/ml), PRAK (7 U/ml), 25 mM Hepes, pH 7.2, 130 mM KCl, 0.1 mM EGTA, 0.1 mM ATP, 0.01 mCi [γ-32P]ATP (PerkinElmer), 5 mM MgCl2, 10% glycerol, 1 mM DTT, 25 °C. Aliquots were taken at different times (5–50 min) and mixed with the PRAK inhibitor epigallocatechin gallate (EGCG) (100 μM) before dephosphorylation assays. TH (5 μM) was then preincubated for 10 min on ice with or without 14-3-3γ (7.5 μM) prior to 5-fold dilution in dephosphorylation assay using shrimp alkaline phosphatase (SAP) (0.14 U/μl, Thermo Fisher Scientific), 25 mM Hepes, pH 7.2, 130 mM KCl, 2 mM MgCl2, 1 mM DTT, at 15 °C or 25 °C. Aliquots were taken at different time points, spotted on phospho-cellulose paper, washed, and counted to measure remaining labeled protein.
For measurement of the rate of phosphorylation of TH in the presence or absence of 14-3-3γ by PRAK or PKA, we used the following conditions: TH (2.5 μM), PRAK (5 U/ml), PKA (1 nM catalytic subunit), 14-3-3γ (10 μM), 25 mM Hepes, pH 7.2, 130 mM KCl, 0.1 mM EGTA, 0.1 mM ATP, 0.01 mCi [γ-32P]ATP (PerkinElmer), 5 mM MgCl2, 10% glycerol, 1 mM DTT, 0.5 mg/ml BSA, 0.1 mg/ml soya bean trypsin inhibitor, 25 °C. Peptide containing Ser19 (RRAVS19ELQDTK) was from the University of Dundee (UK), and the PKA substrate peptide Kemptide was obtained from Sigma-Aldrich (St. Louis, MO).
Mapping of the Phosphorylation Sites via Mass Spectrometry
Label-free quantitative proteomics-type experiments were carried out to assess the phosphorylation level in samples of TH, TH-pSer19, TH-pSer40, and TH-pSer19pSer40. The purified samples were diluted to a concentration of 0.2 μM with 5 M urea and subsequently subjected to reduction and alkylation using 45 mM DTT (Sigma) and 100 mM iodoacetamide (Sigma). Digestion was performed by incubating the samples overnight at 37 °C with trypsin at a 1:50 enzyme:protein ratio. Peptides were diluted with 10% formic acid and subjected to nano-LC-MS/MS analysis using an Orbitrap Q-Exactive instrument (Thermo Fisher Scientific). Raw data were analyzed using Proteome Discoverer 1.3 (Thermo Fisher Scientific), and Ser phosphorylation was assessed in a label-free fashion based on peptide intensity.
SPR Measurements
To measure the binding affinity for the interaction of TH and 14-3-3, we used a BIAcore 3000 instrument (GE Healthcare). All measurements and procedures were performed using HBS-P buffer obtained from the manufacturer (10 mM Hepes, pH 7.4, 150 mM NaCl, and 0.005% polysorbate 20) at 25 °C. 14-3-3γ was immobilized on a CM5 sensor chip (GE Healthcare) by means of amine coupling in 10 mM acetic acid (pH 5.0) as described previously (30, 33). Immobilization of 600 to 1000 response units was achieved, and different concentrations of TH (nonphosphorylated, TH-pS19, TH-pS40, and TH-pS19pS40) were injected (30 μl/min) after a stable baseline was observed. We also used immobilization of 14-3-3 using a GST-capture kit (GE Healthcare), where a monoclonal antibody against GST was immobilized using amine coupling as described by the manufacturer. GST-14-3-3γ was then immobilized by binding to the antibody (using GST as a control), and different forms and concentrations of TH were then injected. This procedure readily allowed regeneration of the chip. The sensorgrams were analyzed using BIAevaluation software, version 3.2 (GE Healthcare Life Sciences), and the dissociation and association rate constants were obtained by fitting the data using a heterogeneous model (Eq. 1) for the dissociation phase (the first 220 s) and through Langmuir association models for the association phase. The dissociation rate constant, representing the highest population of complexes (>80%), was used to fit the association rate constants.
where Robs is the observed response starting from time t0 and R1 and R2 are the responses contributed by complex types 1 and 2, characterized by the dissociation rate constants kd1 and kd2, respectively.
Native Electrophoresis and Immunodetection
Tetrameric TH and dimeric 14-3-3γ were mixed at a 1:3 subunit molar ratio (TH:14-3-3γ) by incubating 10 μM of purified TH (nonphosphorylated TH, TH-pS19, and TH-pS19pS40) with 30 μM 14-3-3γ for 5 min at room temperature. Samples were mixed with 4× loading buffer (0.2% bromphenol blue; 50% glycerol in 20 mM Na-Hepes, pH 7.0, 200 mM NaCl) and separated at 250 V for 90 min on a continuous-gradient 4–16% NativePAGE pre-casted gel (LifeTechnologies, Carlsbad, CA) using as running buffer 50 mM BisTris, 50 mM Tricine, pH 7.5. Gels were stained with Coomassie 5% and distained with 10% methanol, 10% acetic acid, with gentle shaking until a clear background was obtained. Alternatively, after electrophoretic separation, gels were incubated in 12 mM Tris, 96 mM glycine, pH 8.3, 0.1% SDS for 20 min at room temperature before being transferred to a nitrocellulose membrane using the TurboBlot transfer system from Bio-Rad. Membranes were incubated with specific antibodies for rabbit-anti-TH (1:1000; Thermo Scientific) and rabbit-anti-14-3-3γ (1:1000; Novus Biologicals, Littleton, CO), and with goat-anti-rabbit (1:2000; Santa Cruz, Dallas, TX) as a secondary antibody. Membranes were developed using the ECL system and imaged using a ChemiDoc instrument (Bio-Rad).
Native Mass Spectrometry
Purified samples (14-3-3γ, nonphosphorylated TH, TH-pSer19, and TH-pSer19pSer40) were exchanged into 150 mM ammonium acetate, pH 7.5 or pH 6.1, using 10-kDa molecular weight cut-off spin-filter columns (Amicon Ultra-0.5 Centrifugal Filter Unit, Millipore, Billerica, MA). 2 μl of each sample were sprayed at a concentration of 5 μM on an electrospray ionization TOF mass spectrometer (LCT, Waters, Manchester, UK). In particular, for the analysis of the complexes, TH (nonphosphorylated TH, TH-pS19, or TH-pS19pS40) and 14-3-3γ samples were prepared at both 1:0.5 and 1:3 molar subunit mixing ratios. Gold-coated borosilicate capillaries, made in-house for nano-electrospray (using a Sutter P-97 puller (Sutter Instruments Co., Novato, CA) and an Edwards Scancoat Six sputter-coater (Edwards Laboratories, Milpitas, CA)), were used to directly infuse the samples into the instrument. Source backing pressure was increased to 6.5 mbar. Mass calibration was performed using 25 mg/ml CsI. MassLynx V4.1 (Waters) was used for data analysis and therefore for experimental mass determination.
Electron Microscopy
The TH-pS19:14-3-3γ complex was formed by mixing TH-pS19 (5.9 μM) with 14-3-3γ (17.8 μM) (1:3 TH-pS19:14-3-3γ molar ratio) in binding buffer (30 mM Hepes, pH 7.4, 200 mM NaCl, 2% glycerol) for 20 min at 25 °C. Complex purification was performed by GraFix (47) in a glycerol (10% to 30%)/glutaraldehyde (0% to 0.15%) gradient at 32,000 rpm by a SW 55Ti rotor (Beckman Coulter) for 16 h at 4 °C. Mobility controls of individual proteins and complex without glutaraldehyde were also performed under the same conditions. Gradients were fractionated in 200-μl aliquots and analyzed via SDS-PAGE and Coomassie and silver staining.
Aliquots of either TH-pS19 or the TH-pS19:14-3-3γ complex were applied to glow-discharged carbon grids and stained with 2% (w/v) uranyl acetate. Micrographs were recorded in a JEOL JEM-1010 electron microscope, operated at 80 kV, with a TemCam F416 camera (TVIPS GmbH, Gauting, Germany) at 52,000 nominal magnification to a final 2.97 Å pixel−1 resolution. Individual particles in each micrograph were selected automatically and extracted with the XMIPP software package (48, 49). Image classification was performed using free-pattern maximum-likelihood procedures (50). Characteristic and homogeneous particles of both specimens were selected after the two-dimensional classification procedure and aligned for a final two-dimensional image using the same software package.
RESULTS
TH Purification and Phosphorylation
The TH tetramer is structurally organized as a dimer of dimers (supplemental Fig. S1B), an organization that may allow for a number of possible complexes between phosphorylated TH and 14-3-3. To minimize possible interference in our results from inhomogeneity of the N-terminal structure, we used a His-ZZ-fusion expression and purification strategy with a Tobacco etch virus protease cleavage site, which provided highly homogeneous TH preparation as judged by results from SDS-PAGE, peptide fingerprinting, N-terminal sequencing, size exclusion chromatography, and dynamic light scattering.2 We have previously described the phosphorylation of TH by PRAK, which has the highest selectivity for Ser19 among the currently known TH kinases (20). Using PRAK and/or PKA, we were able to obtain TH phosphorylated to full stoichiometry on Ser19 (TH-pS19), Ser40 (TH-pS40), and both Ser19 and Ser40 (TH-pS19pS40) as judged by 32P-incorporation. We verified the phosphorylation of Ser19 using peptide fingerprinting with quantitative mass spectrometry. From this analysis and the masses obtained using native MS (see below), we conclude that our preparations of phosphorylated TH were primarily what we expected (supplemental Figs. S2–S4).
Assessing the Binding Kinetics of 14-3-3γ with Different Phosphorylated Forms of TH
The dimeric 14-3-3 proteins can bind two phospho-Ser/Thr residues. For some target proteins this is necessary for optimal high-affinity binding, but there can be an unequal contribution to the binding affinity between the two phosphorylation sites (39, 40). Thus, one site can be crucial for complex formation, whereas the second site, which can be more degenerate from the optimal 14-3-3 binding sequence, contributes by further increasing the binding affinity of the complex (38). As there is evidence of the involvement of Ser40 phosphorylation of TH in binding to yeast and some mammalian 14-3-3 proteins (33), we wanted to investigate whether the interaction between 14-3-3γ and TH was different for TH-pS19 and TH-pS19pS40. For TH-pS19pS40, there are in fact four possible configurations of phospho-Ser recognition and binding to one 14-3-3 dimer (supplemental Fig. S1C, i–iv).
Using SPR, we measured the binding affinity of TH-pS19 and TH-pS19pS40 to 14-3-3γ immobilized by amine coupling (Figs. 1A and 1B). We did not observe any binding to 14-3-3γ when we used nonphosphorylated TH, and we noted very low binding with TH-pS40 (Fig. 1C). Injections of different concentrations of TH (1–100 nM) were performed for at least two different immobilizations, and the resulting sensorgrams were analyzed to obtain the dissociation and association rate constants (Table I). As described previously, we consistently observed heterogeneity in the dissociation rate constant, possibly arising from two different populations, where the larger population (83% to 92%) had a slow dissociation rate constant (Table I) and the remainder had a rate constant of 0.018 ± 0.004 s−1. Using the dissociation rate constant from the larger population, we estimated association rate constants (ka) and the resulting equilibrium dissociation constant (Kd) for the protein interactions (Table I). Thus, we estimated a Kd value of ∼3.2 nM or ∼2.1 nM for the interaction between 14-3-3γ and TH-pS19 or TH-pS19pS40, respectively. The rate constants were also very similar for the two types of complexes, suggesting that these phospho-species of TH associate with similar strength and kinetics to 14-3-3γ. It therefore seemed unlikely that 14-3-3γ engaged in different modes of binding for TH phosphorylated on only Ser19 than for TH with both Ser19 and Ser40 phosphorylated (Table I). The phospho-Ser recognition configurations noted in parts iii and iv of supplemental Fig. S1C thus appear unlikely.
Fig. 1.
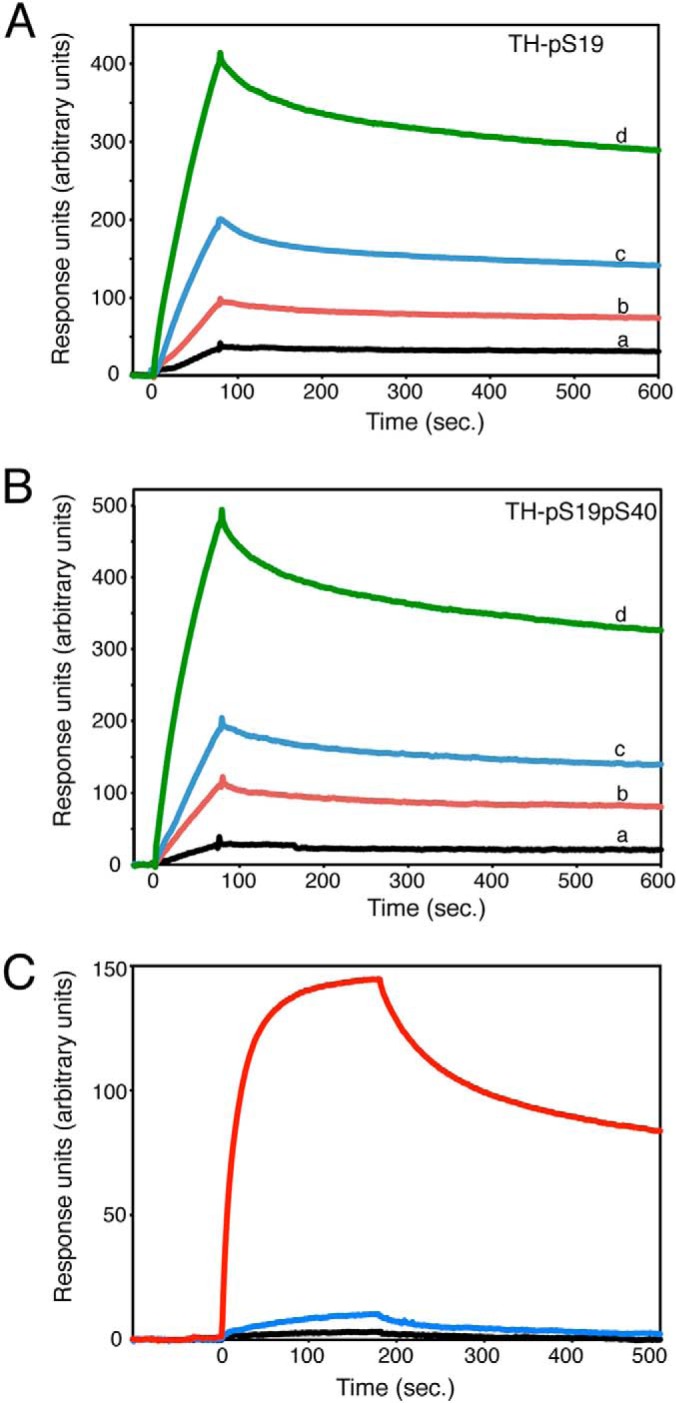
Binding of different phosphorylated forms of TH to 14-3-3γ by SPR. 14-3-3γ was immobilized via amine coupling as described under “Experimental Procedures.” TH-pS19 (A) and TH-pS19pS40 (B) were injected at different concentrations (10 nM (black, a), 25 nM (red, b), 50 nM (blue, c), and 100 nM (green, d)), and the association to 14-3-3γ was monitored in terms of response units. C, GST-14-3-3γ was immobilized to a CM5 chip (GE Healthcare) using a GST-immobilization kit (GE Healthcare) according to recommendations by the manufacturer. Sensorgrams were recorded at a flow rate of 30 μl/min, and the figure compares injected (90 μl) nonphosphorylated TH (100 nM, black), TH-pS40 (100 nM, blue), and TH-pS19 (100 nM, red).
Table I. Binding kinetics of the complex between 14-3-3 and phosphorylated TH. We measured the association and dissociation rate constants for binding of two different phosphorylated forms of TH (TH-pS19 and TH-pS19pS40) to 14-3-3γ using SPR. TH was phosphorylated on Ser19 using PRAK and on both Ser19 and Ser40 using PRAK and PKA (see “Experimental Procedures” for details). Two to four separate immobilizations of 14-3-3 were used, and different concentrations of TH were injected for each of them. Values of rate constants are shown ± S.E.M. (n = 4 to 7). The quality of the model fitting is reflected in the average squared residual (ASR), which corresponded to R2 values > 0.99987. Other reported values are shown for comparison.
| Phosphosite (TH-) | 14-3-3 isoform | ka (105 m−1 s−1)a | ASR of fit | kd (10−3 s−1) | ASR of fit | kd (nM) | Reference |
|---|---|---|---|---|---|---|---|
| pS19 | 14-3-3γ | 1.44 ± 0.38 | 1.74 | 0.46 ± 0.03 | 0.286 | 3.2 ± 0.9 | This work |
| pS19pS40 | 14-3-3γ | 1.59 ± 0.54 | 0.233 | 0.36 ± 0.06 | 0.155 | 2.1 ± 0.6 | This work |
| pS19b | 14-3-3γ | 7.8 ± 1.7 | 0.514 | 0.45 ± 0.04 | 0.047 | 0.57 ± 0.16 | This work |
| pS19pS40c | 14-3-3η | 5.4 | – | 1.6 | – | 3.0 | (24) |
| pS19pS40d | 14-3-3ζ | 0.2 | – | 0.26 | – | 10 | (33) |
| pS19pS40d | BMH1 | 5.1 | – | 2.7/0.09 | – | 5/0.2 | (33) |
| pS19 | 14-3-3ζ | – | – | 2.6 | (20) | ||
| pS19 | BMH1 | – | – | 1.4 | (20) |
a Fitted with a Langmuir association model to binding curves of all experiments simultaneously using TH subunit concentration. S.E.M. was estimated from values obtained by fitting each experiment.
b TH-pS19 phosphorylated to a stoichiometry of 0.17. The ka values were fitted using the concentration of phosphorylated TH subunits.
c Phosphorylated in vitro with Ca2+/calmodulin-dependent protein kinase II, which phosphorylates Ser19 and Ser40 in about a 2:1 ratio.
d Phosphorylated in vitro by mitogen-activated protein kinase–activated protein kinase-2, which phosphorylates Ser19 and Ser40 in about a 1:2 ratio.
Native PAGE and Immunodetection
In order to further investigate complex formation between the different forms of TH and 14-3-3γ, we performed native PAGE. Native electrophoresis using gradient native gels and BisTris and Tricine running buffers gave the best resolution, although band positions in the gel could not be assigned to precise molecular weights. The proteins were transferred from the gels to nitrocellulose membranes and stained with Ponceau Red before specific immunodetection of TH or 14-3-3γ. TH, as well as its two phosphorylated forms, showed an intense upper band and a lower, more diffuse band (Fig. 2) that could be associated with tetrameric TH and a minor dimeric form of the enzyme, respectively. The acidic 14-3-3γ ran closer to the front of the gel and displayed a double band pattern. The incubation of TH-pS19 or TH-pS19pS40 with 14-3-3γ at a TH:14-3-3γ subunit ratio of 1:3 led to a similar upward shift of both bands of TH, and the lower band became sharper and more intense. The incubation of nonphosphorylated TH with 14-3-3γ did not render any band differences indicating that complex formation requires TH phosphorylation.
Fig. 2.

Native PAGE and immunodetection of 14-3-3γ, TH, and phosphorylated forms of TH, alone and in complex. TH (TH, TH-pS19, or TH-pS19pS40) and 14-3-3γ complexes, formed by incubation of TH:14-3-3 at a mixing ratio of 1:3, were separated via native PAGE and transferred to nitrocellulose membranes. Ponceau Red staining was used to visualize all protein forms, and specific antibodies anti-TH and anti-14-3-3γ were used for TH and 14-3-3γ detection.
Elucidating the Configuration of the TH:14-3-3 Complex Using Native Mass Spectrometry
Although native PAGE corroborated the formation of a stable complex between 14-3-3γ and both TH-pS19 and TH-pS19pS40, the exact stoichiometry of the complex could be assessed only through accurate determination of the masses of the complexes. To this end, we selected native MS as the method of choice for these measurements.
To confirm the oligomeric state of both TH and 14-3-3γ, we first analyzed the spectra of the isolated proteins. The native MS spectrum of 14-3-3γ indicated a very homogeneous preparation consisting mostly of dimers (2 × 29.2 = 58.4 Da) (supplemental Fig. S3A), in agreement with earlier nanoelectrospray ionization MS analysis of this isoform (51). With regard to TH, native MS analysis confirmed the tetrameric oligomerization state for TH, TH-pS19, TH-pS40, and TH-pS19pS40 (223.8–224.3 kDa) (supplemental Figs. S3 and S4). The native mass spectrum of TH-pS19 showed two clearly distinct charge-state envelopes. This difference in charge uptake suggests two different conformations for TH. Most probably, the highly charged conformation was caused by the partial unfolding of a small region of the protein. The same behavior was observed for TH, TH-pS19pS40, and TH-pS40 (supplemental Figs. S3 and S4).
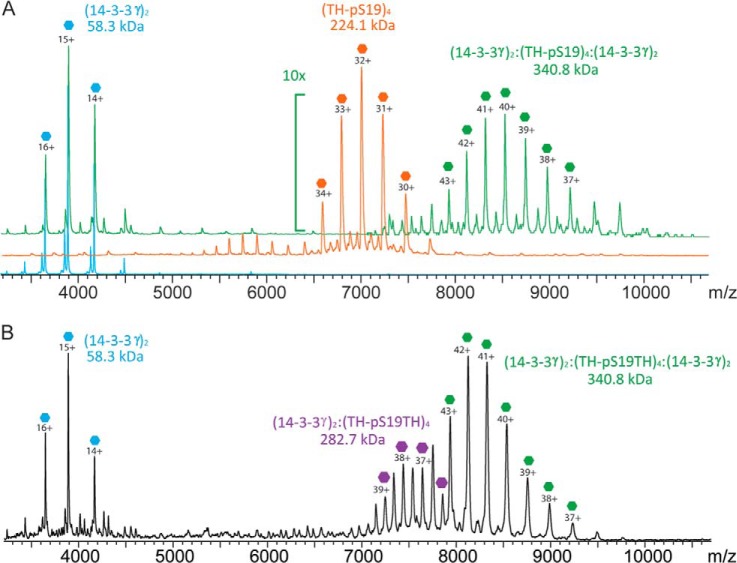
Mixing TH-pS19 with 14-3-3γ at a 1:3 ratio (TH-pS19:14-3-3, subunit ratios) induced the formation of a 340.8-kDa complex. This mass corresponds to the tetrameric TH-pS19 bound to two dimers of 14-3-3γ (Fig. 3A). The formation of the complex (14-3-3γ)2:(TH)4:(14-3-3γ)2 was observed also for TH-pSer19pSer40 (341.0 kDa). In contrast, no complex could be observed when nonphosphorylated TH or TH-pS40 was mixed with 14-3-3γ (supplemental Fig. S4). When the amount of 14-3-3γ was decreased, an additional charge-state distribution was detected corresponding to a (14-3-3γ)2:(TH-pS19)4 complex (282.7 kDa) (Fig. 3B), but no evidence was found of TH bound to a single subunit of 14-3-3γ. The observed masses and related stoichiometries of the investigated proteins and complexes are summarized in Table II.
Fig. 3.
Native MS of TH-pS19 in complex with 14-3-3γ. Native MS experiments were performed on purified TH-pS19 and 14-3-3γ and on their complexes. A, overlay of native mass spectra of the 14-3-3γ dimer (blue trace), the TH-pS19 tetramer (orange trace), and the (14-3-3γ)2:(TH)4:(14-3-3γ)2 complex (green trace) formed upon mixing of TH-pS19 tetramer with 14-3-3γ at a subunit mixing ratio of 1:3. B, native mass spectrum of the TH-pS19 tetramer mixed with 14-3-3γ at a subunit mixing ratio of 1:1. When we reduced the amount of 14-3-3γ, both the (14-3-3γ)2:(TH)4 complex and the (14-3-3γ)2:(TH)4:(14-3-3γ)2 complex were detected together with the free 14-3-3γ dimer.
Table II. Overview of the results obtained via native MS. Observed masses and related stoichiometries of the single components and TH:14-3-3γ complexes as determined via native MS. Expected masses and mass errors are also shown.
| Proteins | Stoichiometry of the protein complex | Measured mass (kDa) | Expected mass (kDa) | Error (%) |
|---|---|---|---|---|
| 14-3-3γ | (14-3-3γ)2 | 58.3 | 58.3 | 0.0 |
| TH | (TH)4 | 223.8 | 222.6 | 0.5 |
| TH-pS19 | (TH-pS19)4 | 224.1 | 222.9 | 0.5 |
| TH-pS40 | (TH-pS40)4 | 224.2 | 222.9 | 0.6 |
| TH-pS19pS40 | (TH-pS19pS40)4 | 224.3 | 223.2 | 0.5 |
| TH and 14-3-3γ | No complex detected between TH and 14-3-3γ | |||
| TH-pS19 and 14-3-3γ | (14-3-3γ)2:(TH-pS19)4:(14-3-3γ)2 | 340.8 | 339.5 | 0.4 |
| (14-3-3γ)2:(TH-pS19)4 | 282.7 | 281.2 | 0.5 | |
| TH-pS40 and 14-3-3γ | No complex detected between TH-pS40 and 14-3-3γ | |||
| TH-pS19pS40 and 14-3-3γ | (14-3-3γ)2:(TH-pS19p40)4:(14-3-3γ)2 | 341.0 | 339.8 | 0.4 |
The phospho-Ser19-TH:14-3-3γ complex was further analyzed in MS/MS experiments. Dissociation of (14-3-3γ)2:(TH)4:(14-3-3γ)2 upon collision-induced dissociation revealed that only a single monomer of 14-3-3γ fell off the complex, stripping the majority of the charges, in line with what normally is observed in this type of experiment (supplemental Fig. S5) (52, 53).
Finally, in order to investigate the complex formation under low stoichiometry of phosphorylation, TH was phosphorylated at Ser19 using PRAK to 0.24, 0.38, and 0.54 mol phosphate/TH subunit. Native MS spectra of these TH preparations with 14-3-3γ showed much more prominent formation of the complex with only one 14-3-3γ dimer bound (supplemental Fig. S6). Only for the phosphorylation stoichiometry of 0.54 could a reasonable amount of the (14-3-3γ)2:(TH)4:(14-3-3γ)2 complex be detected. Still, also at this phosphorylation stoichiometry, a considerable amount of TH was detected not bound to 14-3-3 (supplemental Fig. S6). Notably, if random Ser19 phosphorylation is assumed, a binomial distribution of Ser19-phosphorylated subunits in the TH tetramer predicts less completely unphosphorylated TH (4.5%, 15%, and 33% at stoichiometries of 0.54, 0.38, and 0.24, respectively).
EM
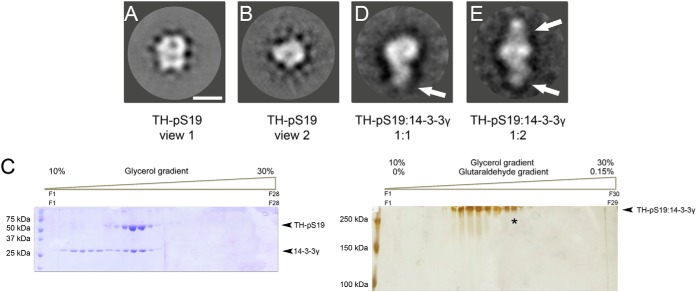
The TH-pS19:14-3-3γ complex was further investigated using EM. First, aliquots of purified TH-pS19 were negatively stained and observed at the electron microscope. A total of 6347 particles were selected and processed, and the classification procedure revealed a major population whose average image (Fig. 4A) showed a particle of approximately 115 Å in length and 95 Å in width, in agreement with the hydrodynamic diameter of TH (about 12 nm) obtained via dynamic light scattering (data not shown). The size of the particle, together with the presence of four stain excluding masses, pointed to tetrameric TH-pS19 (represented in supplemental Fig. S1B). Another minor population represented a particle with approximate dimensions of 85-Å length and 95-Å width (Fig. 4B), which we believe to be the orthogonal view of the major population.
Fig. 4.
Electron microscopy analysis of TH-pS19 and the TH-pS19:14-3-3 complexes. A, two-dimensional average image of TH-pS19 representing the largest population of particles. Bar at bottom right = 100 Å. B, two-dimensional average image of a minority population of particles of TH-pS19. C, glycerol gradients in the absence (left) or presence (right) of cross-linker. The asterisk points to the fraction used for the EM analysis of the TH-pS19:14-3-3 complexes. D, two-dimensional average image of the largest population of TH-pS19:14-3-3 complexes, with one 14-3-3 dimer (signaled by the white arrow) bound to the TH-pS19. E, two-dimensional average image of the minority population of TH-pS19:14-3-3 complexes, with presumably two 14-3-3 dimers bound on opposite sides of TH-pS19.
Subsequently, TH-pS19 was mixed with 14-3-3γ in a 1:3 molar ratio. The putative complex between the two proteins (TH-pS19:14-3-3γ) was purified via gel filtration and visualized by means of EM, but only a complex consistent with (14-3-3γ)2:(TH-pS19)4 (1:1 TH:14-3-3 oligomer stoichiometry) was found (data not shown). Because of the presumably transient character of the (14-3-3γ)2:(TH)4:(14-3-3γ)2 complex (1:2 TH:14-3-3 oligomer stoichiometry) found by MS, we decided to carry out a GraFix purification (Fig. 4C; see also supplemental Fig. S7). SDS-PAGE of the complex isolated via glycerol/glutaraldehyde gradient showed higher bands, presumably the TH-pS19:14-3-3γ complex, that were analyzed via EM. All fractions contained complexes with a 1:1 oligomer stoichiometry, and only the last fractions (those marked with asterisks in Fig. 4C) revealed a lower percentage of complexes with an apparent 1:2 oligomer stoichiometry. A total of 12,396 particles were selected and processed as described “Experimental Procedures.” The classification procedure revealed a dominant population whose average image is shown in Fig. 4D. The image clearly shows two stain excluding masses: a large one very similar to the one obtained for control TH-pS19 (Fig. 4B) that can be therefore assigned to the tetrameric TH-pS19, and a smaller mass (pointed out by the white arrow in Fig. 4D) located at one side of the larger one that we assign to a 14-3-3γ dimer. The classification procedures also found a small population (∼8%) of a particle with a third stain excluding mass of a similar mass and in an opposite position to the previously assigned 14-3-3γ dimer (Fig. 4E). We assigned this new particle to a TH-pS19:14-3-3γ symmetric complex (1:2 oligomer stoichiometry).
The Kinetics of Complex Formation between 14-3-3γ and TH-pS19 at Different Phosphorylation Stoichiometries
The majority of the interactions between 14-3-3γ and TH mentioned so far in this work were studied at high phosphorylation stoichiometry. However, at lower phosphorylation stoichiometries native MS detected little of the (14-3-3γ)2:(TH)4:(14-3-3γ)2 complex (supplemental Fig. S6), which might suggest a decreased binding affinity. A low degree of Ser19 phosphorylation might affect the binding strength between TH and the 14-3-3γ dimer, as well as the protein complex composition (supplemental Fig. S1C, configurations i and ii, respectively). We expected that only one pSer19 in each tetramer would be available for binding to 14-3-3 at low phosphorylation stoichiometries of TH. We therefore performed SPR measurements of TH-pS19 with low phosphorylation stoichiometry (<0.2, TH-pS19L), where the PRAK phosphorylation had been stopped by addition of the inhibitor epigallocatechin gallate prior to preparation of the sample for SPR.
The dissociation rate of the TH-pS19:14-3-3 complexes for TH-pS19L was very similar to that observed for full stoichiometry (TH-pS19H, Table I, Fig. 5A). This similarity was also observed for the smaller population of rapid dissociating complex, which had a similar size and rate constant as found for high phosphorylation levels (82% to 91%, 0.019 s−1). To fit the association rate constant, we used the concentration of phosphorylated TH subunits, which assumes that the rate-limiting step of complex formation is concentration dependent. We then obtained a higher ka value for TH-pS19L than for TH-pS19H, leading to a lower apparent Kd value for the TH-pS19L:14-3-3γ complex (Table I). The ratios of the ka values for TH-pS19L and TH-pS19H corresponded roughly to the concentration ratio obtained when correcting for the phosphorylation stoichiometry. Similarly as noted by native MS at lower phosphorylation stoichiometries, a binomial distribution predicted only 48% of the TH tetramers as completely nonphosphorylated and unable to bind 14-3-3 at 17% Ser19-phosphorylation, whereas 39% of the tetramers would contain one phosphorylated subunit.
Fig. 5.

Effect of 14-3-3γ on TH phosphorylation and dephosphorylation. A, binding of TH phosphorylated on Ser19 by PRAK to immobilized 14-3-3γ. TH was phosphorylated to a stoichiometry of 0.17 (red) or 1.0 (blue) before preparation for injections at subunit concentrations of 5, 25, and 50 nM. Sensorgrams were scaled for illustration of kinetics. B, TH was [32P]-labeled on Ser19 to different stoichiometries using PRAK before incubation with the PRAK inhibitor epigallocatechin gallate and 14-3-3γ (7.5 μM, TH-pS19:14-3-3 mixing ratio of 1:1.5). The complex was then diluted 1/10 in buffer containing high levels of shrimp alkaline phosphatase (SAP) (145 U/ml), and the temporal decay of 32P-Ser19 was monitored as described under “Experimental Procedures.” Controls without 14-3-3 and without SAP were also measured (dotted lines). Exponential decay functions were fitted to each curve, and the corresponding rate constants were 0.089, 0.125, and 0.157 min−1 for TH-pS19 phosphorylated to 51% (○), 33% (▵), and 18.5% (□), respectively. Insignificant change in phosphorylation stoichiometry was observed in the absence of SAP (horizontal dotted lines), and a high rate of dephosphorylation was measured in the absence of 14-3-3γ (lower dotted lines). C, the phosphorylation of TH-pS19 (2.5 μM, pre-phosphorylated on Ser19 by PRAK) by PKA in the absence (○) or presence (●) of 14-3-3γ (10 μM).
To corroborate these results, we measured the kinetics of binding and dissociation of the complex at a range of phosphorylation stoichiometries (of TH-pS19, 0.15–0.54), using GST-14-3-3 capture for immobilization, which also allowed more rapid regeneration of the chip. However, we observed only minor differences in the dissociation rate (4.5–5.1 ± 0.7 10−4 s−1) when we used direct amine coupling of 14-3-3 and slightly higher, but also similar (6.3–6.9 ± 0.3 10−4 s−1) using GST-14-3-3 capture. Using the latter immobilization procedure, we found a slight increase in complex formation as a function of increasing phosphorylation that was more pronounced at injections of lower concentrations (<25 nM). When fitting each injection separately, we also observed higher estimates of ka for injections of lower TH concentrations. However, there were minimal differences between the average ka values for the different phosphorylation stoichiometries (2.4, 1.8, and 1.2 ± 0.7 μM−1s−1 for stoichiometries of 0.15, 0.24, and 0.38, respectively). All taken together, this could suggest that the kinetics of complex formation included multiple steps that showed stronger concentration dependence at lower TH concentrations.
As an alternative measure of the configuration of the complex formed between TH-pS19 and 14-3-3γ, we performed dephosphorylation rate experiments of the TH-pS19:14-3-3γ complex (formed at a molar mixing ratio of 1.0:1.5), where TH was labeled on Ser19 using [32P]-γ-ATP. Using high levels of the nonspecific phosphatase SAP in the incubation buffer, we performed dephosphorylation measurements as indicators of temporal reorganizations and dissociation events of the TH-pS19:14-3-3γ complex. TH phosphorylation was stopped with epigallocatechin gallate at different times, the stoichiometry was measured prior to incubation with 14-3-3γ, and dephosphorylation was monitored as remaining phosphorylated TH (%) at 15 °C or 25 °C. Insignificant changes in the phosphorylation stoichiometry were observed for incubations without added SAP, and in the absence of 14-3-3γ we observed a highly efficient dephosphorylation rate of TH-pS19. The dephosphorylation measurements were fitted to exponential decay functions. When we compared the rate constants for decay of TH-pS19 at 25 °C, the stoichiometries of 0.18, 0.33, and 0.52 showed only modest decreases in the rate of pSer19 decay by 2.7, 2.2, and 1.5 10−3 s−1, respectively (Fig. 5B). Similarly, at 15 °C we found little difference (0.028–0.045 min−1) among the dephosphorylation rates for TH with different phosphorylation stoichiometries (supplemental Fig. S8A).
The dephosphorylation experiments did, however, estimate a more rapid dissociation of the TH-pS19:14-3-3γ complex than measured via SPR. We therefore performed an SPR experiment in which SAP was injected just after formation of the TH-pS19:14-3-3γ complex to see whether SAP affected the dissociation process. We found that injection of high levels of SAP increased the dissociation rate 3- to 4-fold (supplemental Fig. S8B). With this taken into consideration, the two methods now agreed on the dissociation rate of the complex. This also suggests that the protein complex might be dynamic or that allosteric interactions take place in the protein complex.
The Complex between 14-3-3γ and TH Shows Altered Accessibility of Ser40
The sequential order of structural rearrangements that occur during binding of 14-3-3 to phosphorylated TH is still unknown. When 14-3-3 proteins are present during phosphorylation, as is expected to be the situation in cells, the sequential phosphorylation of tetrameric TH may be affected by the binding of 14-3-3 proteins. As a measure of the geometry of the protein complex, we wanted to test whether 14-3-3, when present during phosphorylation of Ser19, influences the rate or accessibility to the remaining Ser19 sites. We did not observe any significant effect of 14-3-3γ (10 μM subunit, 5-fold higher than the concentration of the TH subunit) on the rate of TH phosphorylation with PRAK (not shown). Furthermore, the presence of 14-3-3γ at similar or higher concentrations did not affect the PRAK phosphorylation of Ser19 peptides of TH.
In the absence of 14-3-3, phosphorylation of Ser19 has been shown to make TH a better substrate for PKA and Ca2+/calmodulin-dependent protein kinase II, probably by increasing the accessibility to Ser40 through a conformational change (20, 22). Because Ser40 phosphorylation had only moderate effects on the binding affinity of TH-pS19 to 14-3-3, we questioned whether Ser40 would be as accessible for phosphorylation in the TH-pS19:14-3-3 complex as in free TH. Therefore, we performed TH Ser40 phosphorylation experiments (with PKA) using TH previously phosphorylated to full stoichiometry on Ser19 (TH-pS19) in the presence and absence of 14-3-3γ. An excess of 14-3-3γ (1:3 TH-pS19:14-3-3γ subunits) was used in these experiments to ensure that TH was fully bound to 14-3-3. Control experiments using a peptide substrate of PKA, kemptide, as well as nonphosphorylated TH, showed that the presence of 14-3-3γ did not by itself alter the activity of PKA, nor did 14-3-3γ have any effect on the rate of Ser40 phosphorylation for nonphosphorylated TH. However, we found that the pre-incubation of 14-3-3γ with TH-pS19 reduced the initial rate of Ser40 phosphorylation of TH by PKA by about 3.5-fold (Fig. 5C), suggesting that there is steric interference at this site in the 14-3-3γ:TH-pS19 complex.
DISCUSSION
The significance of TH regulation by 14-3-3 proteins still remains enigmatic after its discovery several decades ago. Knowledge about the underlying structural requirements for complex formation between both proteins can increase our understanding of the relevance of this protein complex in dopamine-synthesizing cells. We therefore set out to investigate how TH phosphorylation stoichiometry and multiphosphorylation affect the complex formation and possible complex configurations. In particular, we believed that the use of native MS, with its mild ionization protocol and high mass accuracy, could overcome the challenges of unequivocal protomer assignment. More than 300 phosphorylated partners of 14-3-3 have been identified so far (54). Nonetheless, this is to our knowledge the first report on the use of native MS to study the complex between 14-3-3 and a target protein, and the first to microscopically visualize such a complex.
Structural and Kinetic Considerations of the TH:14-3-3 Protein Complexes
Previous structural investigations of 14-3-3 protein complexes have revealed that both symmetric and highly asymmetric complexes can form. Thus, the crystal structure of 14-3-3ζ with aromatic amino acid N-acetyl transferase revealed a symmetric protein complex of 1:1 stoichiometry in which the phosphorylated N-terminals of aromatic amino acid N-acetyl transferase extend into the 14-3-3 phospho-Ser/Thr recognition motif and where aromatic amino acid N-acetyl transferase also interacts extensively with distant areas outside the 14-3-3 binding groove (55). In a more recent postulated complex structure between 14-3-3 and regulator of G-protein signaling 3, a similarly extended structure was proposed for the sequence composing the phosphorylation site, whereas extensive interactions were reported between regulator of G-protein signaling 3 and 14-3-3 outside the binding groove of one of the 14-3-3 subunits (56, 57). Consistently, we also expect that regions of TH outside its phosphosites at the N-terminal region make a considerable contribution to the binding interface with 14-3-3 proteins. In fact, much lower affinities (Kd ∼ 0.5 μM) are measured for binding of phospho-Ser19-TH peptides THp-(1–43) (30) than for the full-length protein, although a difference in entropy loss can explain some of this (Table I). Furthermore, we have previously shown that heparin, which activates TH through interaction with its N-terminal domain (residues 70–90) (58), competitively inhibits binding of TH-pS19 to 14-3-3 (33). X-ray crystallography and molecular dynamics simulations have revealed the extended structure of the 14-3-3γ binding region around pSer19 (32), similar to the 14-3-3 interacting phosphorylated regions in 14-3-3-bound aromatic amino acid N-acetyl transferase (55) and regulator of G-protein signaling 3 (56). However, it is difficult to elucidate interactions outside the Ser/Thr recognition motif of 14-3-3 because the available crystal structure of the complex (supplemental Fig. S1A) unfortunately does not include the complete regulatory N-terminal domain.
The structure of truncated TH (59) and that of the corresponding composite model of full-length TH including the recent NMR structure of the regulatory domain (see Ref. 60 and supplemental Fig. S1B) are symmetric, in agreement with the EM image obtained for the full-length enzyme (Figs. 4A and 4B). The N-terminal regulatory domains of TH thus seem to place themselves two and two on opposite sides of the plane of four catalytic domains, preserving the symmetry in the structure, and Ser19 phosphorylation does not seem to alter the symmetry of this structure (Fig. 4A).
With respect to the putative binding configurations that could occur between TH-pS19pS40 and both binding sites in one dimer of 14-3-3, four likely complexes (as illustrated in supplemental Fig. S1C) were initially envisioned in this work. Results from SPR, showing that phosphorylation of Ser40 did not have a large effect on the binding affinity for 14-3-3γ, indicate that configurations iii and iv based on direct binding of pSer40 to the phospho-Ser recognition site of 14-3-3 appear unlikely. Furthermore, results from SPR analysis using TH-pS19 with high and a range of low phosphorylation stoichiometries suggest that only one pSer19 residue, simultaneously and unrestrained, binds to the phospho-Ser recognition motifs of the same 14-3-3γ dimer (configuration i, supplemental Fig. S1C). MS experiments also supported a certain asymmetry within the bound 14-3-3 dimer, as only a single monomer of 14-3-3 fell off the (14-3-3γ)2:(TH)4:(14-3-3γ)2 complex upon collision-induced dissociation (supplemental Fig. S5).
Still, PRAK may preferentially phosphorylate neighboring N-terminals of the TH tetramer, even at low stoichiometries. For this reason, we cannot exclude the possibility that binding of two phospho-Ser19 residues to the two phospho-Ser recognition motifs in the 14-3-3γ dimer (comprising mainly Arg57, Arg132, and Tyr133) predominates for all measurements. The observation that TH:14-3-3 complexes at moderate phosphorylation stoichiometries (supplemental Fig. S6) were dominated by the low molecular complex ((14-3-3γ)2:(TH)4) and showed greater amounts of uncomplexed TH than expected could suggest that phosphorylation of Ser19 is not binomially distributed between the tetramers.
Another possibility is that binding of the two 14-3-3γ dimers occurs with different affinities. Using SPR where 14-3-3 is immobilized, it is likely that only one type of interaction will be measured. Still, for both types of immobilization we observed two populations, with the minor population (<20% for direct amine coupling, <30% for GST-capture) having a higher dissociation rate constant (∼20-fold). However, SPR experiments where 14-3-3γ was injected after TH:14-3-3 complex formation did not reveal any additional binding (data not shown). Under the conditions used for native MS, the (14-3-3γ)2:(TH-pS19)4:(14-3-3γ)2 complex dominated, whereas the (14-3-3γ)2:(TH-pS19)4 complex was more abundant when we used EM. As the native MS experiments showed little formation of the (14-3-3γ)2:(TH)4:(14-3-3γ)2 complex at about 50% phosphorylation, we are uncertain whether the mono-exponential decay observed during dephosphorylation is relevant for the conditions in which two 14-3-3 proteins bind. The accelerated dissociation of the complex by SAP as measured via SPR suggests a somewhat dynamic association between the two proteins in which one of the pSer19 residues can become exposed and dephosphorylated. The ∼4-fold increase in kd seen with SAP treatment could suggest that the remaining pSer19 residue contributes with a Kd of about 12 nM to the complex formation. However, it could also be a measure of the rate constant for restructuring (breathing) of the protein complex that becomes rate-limiting relative to complex dissociation upon the first dephosphorylation. More detailed experiments are needed to provide definite answers to these questions.
We did not pursue a more extensive model fitting to our SPR experiments, and the association rate constant was fitted assuming that the rate was dependent on the concentration of phosphorylated subunits. However, as Ser19 phosphorylation was necessary for binding of TH, it is likely that an initial step of complex formation involves phospho-Ser recognition similar to that of peptide binding to 14-3-3. For this initial step the rate would depend on the concentration of TH-pS19, which could be different for the various phospho-forms of the TH tetramer. Still, subsequent rearrangements of the proteins into a more stable complex may dampen the concentration dependence of the initial complex formation. Subsequent transitions that become rate-limiting could explain why higher ka values were fitted for lower protein concentrations.
The TH:14-3-3 Complex in Relation to Observed in Vivo TH Phosphorylation Levels
The phosphorylation status of TH has been investigated in several cell types and brain regions under both resting and stimulated conditions. Through the use of phospho-specific antibodies, the phosphorylation stoichiometry of TH has been estimated on Ser19 and Ser40 (12–14). In different brain regions of rat, Ser19 was found to be 10% to 35% phosphorylated, whereas Ser40 was less phosphorylated (5% to 12%) (12). Also, in isolated cells particularly Ser19 phosphorylation has been observed at substantial levels (70%) (21), close to the phosphorylation stoichiometry used in most experiments here. Native MS experiments at low phosphorylation stoichiometries suggest that the complex (14-3-3γ)2:(TH)4, with only one 14-3-3 dimer bound per TH tetramer, would be more abundant at conditions typically reported in cells and different brain regions. The high-molecular-weight complex (14-3-3γ)2:(TH)4:(14-3-3γ)2 corresponding to two 14-3-3 dimers bound to the TH tetramer may only exist at conditions of high levels of stimulation and may be functionally related to such conditions.
Control of TH Phosphorylation by 14-3-3 Binding
We have previously reported that binding of 14-3-3 to phosphorylated TH inhibits dephosphorylation of the protein (33). For binding of 14-3-3 to TH-pS19, it is expected that this will delay dephosphorylation of the residues that are directly involved in binding. Based on dephosphorylation experiments involving 32P-labeled TH-pS19, binding of 14-3-3 to a Ser19-phosphorylated subunit seems to affect the dephosphorylation rate of pSer19 of the neighboring N terminus. Binding of 14-3-3 therefore seems to promote the same phosphorylation status of both TH subunits. As we did not observe a similar reduction in the phosphorylation rate, this should lead to a cooperative phosphorylation response of TH on Ser19 in the presence of 14-3-3 proteins. The physiological relevance of such cooperativity remains to be elucidated, in particular because the functional role of 14-3-3 proteins on TH is still unresolved.
Phosphorylation of TH on Ser19, by either PRAK or Ca2+/calmodulin-dependent protein kinase II, has been found to increase the rate of Ser40 phosphorylation by PKA 2- to 3-fold, referred to as hierarchical phosphorylation (20, 22). It was therefore surprising that binding of 14-3-3 proteins had an inhibitory effect on Ser40 phosphorylation. This would counteract the hierarchical phosphorylation observed in the absence of 14-3-3 proteins. The negative effect of 14-3-3 binding therefore seems to even out the phosphorylation hierarchy between these two sites. Observations of the hierarchical phosphorylation in cells have been somewhat contradictory. In bovine adrenal chromaffin cells, pathways that increase Ser19 phosphorylation were also found to synergistically increase Ser40 phosphorylation (37). However, in striatal slices such a relationship was not observed (61). Thus, the conditions for hierarchical phosphorylation between Ser19 and Ser40 in cells might therefore be more complicated and not depend only on phosphorylation itself.
The TH:14-3-3 Complex and TH Functionality
The 14-3-3 proteins are generally considered as activators of TH, though this notion has been challenged (17, 21). Our results suggest that the functionality of the TH:14-3-3γ complexes is not confined to the regulation of TH activity only. There are several reasons for this. An optimal complex formation for the activation of TH would not inhibit Ser40 phosphorylation, which is the site that gives the strongest activation of the enzyme (5, 14). Also, a symmetric complex that allows a similar favorable conformational change in all four subunits would be expected to be more optimal for activation. We therefore propose that the TH:14-3-3 complexes described here are not only optimized for controlling TH activity. This is also in concordance with the moderate activation found for 14-3-3γ on TH-pS19 (30). We have previously described the membrane binding properties of 14-3-3γ and TH that may actively locate TH to different subcellular locations (30). Recently, the interaction of TH with components of the downstream biosynthesis pathway has been described (4), which, together with reports of other binding partners (62, 63), suggests that 14-3-3 proteins could also function to modulate the interaction of TH with other proteins.
In conclusion, based on the reported TH phosphorylation status on Ser19, we expect that the physiologically dominating TH:14-3-3 complex will consist of one 14-3-3 dimer bound to one TH tetramer. Interestingly, this is expected to leave the opposite face of TH unaffected by 14-3-3 in terms of interactions with other proteins. This should allow unrestricted and hierarchical phosphorylation of the N-terminal Ser40 residues situated at the side free of 14-3-3. Although Ser40 phosphorylation had little impact on the interaction with 14-3-3γ under the conditions used here, we cannot exclude situations where this would be different, such as for other 14-3-3 and TH isoforms. These speculations have to be tested experimentally; however, it might suggest that the tetramer of TH operates in several modes, which may be controlled by its interaction with 14-3-3 proteins.
Supplementary Material
Footnotes
Author contributions: R.K., S.R., J.V., A.J.H., and A.M. designed research; R.K., S.R., A.J., S.A., and S.G. performed research; A.J., S.A., and J.H. contributed new reagents or analytic tools; R.K., S.R., A.J., S.A., S.G., J.H., J.V., A.J.H., and A.M. analyzed data; R.K., J.V., A.J.H., and A.M. wrote the paper.
* We thank The Norwegian Research Council, The Kristian Gerhard Jebsen Foundation, the Western Norway Health Authorities, and the Meltzer Fund for financial support. A.J.F. was also supported by grants from Fundación Ramón Areces and the Marie Curie IEF program. J.M.V. was supported by the Spanish Ministry of Science and Innovation (Grant No. BFU2013-44202). This work was in part supported by the PRIME-XS project (Grant No. 262067), funded by the European Union 7th Framework Programme. S.R. and A.J.R.H. acknowledge support by STW (Project 10805) and The Netherlands Proteomics Center, embedded in The Netherlands Genomics Initiative.
 This article contains supplemental material.
This article contains supplemental material.
2 Bezem, M. T., Baumann, A., Meyer, R., Martinez, A., and Flydal, M. I., manuscript in preparation.
1 The abbreviations used are:
- TH
- tyrosine hydroxylase
- hTH
- human tyrosine hydroxylase
- EM
- electron microscopy
- PKA
- cAMP-dependent protein kinase
- PRAK
- p38-regulated/activated protein kinase
- SAP
- shrimp alkaline phosphatase
- SPR
- surface plasmon resonance
- TH-pS19
- tyrosine hydroxylase phosphorylated at Ser19
- TH-pS40
- tyrosine hydroxylase phosphorylated at Ser40
- TH-pS19pS40
- tyrosine hydroxylase phosphorylated at both Ser19 and Ser40.
REFERENCES
- 1. Grima B., Lamouroux A., Boni C., Julien J. F., Javoy-Agid F., Mallet J. (1987) A single human gene encoding multiple tyrosine hydroxylases with different predicted functional characteristics. Nature 326, 707–711 [DOI] [PubMed] [Google Scholar]
- 2. Kaneda N., Kobayashi K., Ichinose H., Kishi F., Nakazawa A., Kurosawa Y., Fujita K., Nagatsu T. (1987) Isolation of a novel cDNA clone for human tyrosine hydroxylase: alternative RNA splicing produces four kinds of mRNA from a single gene. Biochem. Biophys. Res. Commun. 146, 971–975 [DOI] [PubMed] [Google Scholar]
- 3. Nagatsu T. (1995) Tyrosine hydroxylase: human isoforms, structure and regulation in physiology and pathology. Essays Biochem. 30, 15–35 [PubMed] [Google Scholar]
- 4. Cartier E. A., Parra L. A., Baust T. B., Quiroz M., Salazar G., Faundez V., Egana L., Torres G. E. (2010) A biochemical and functional protein complex involving dopamine synthesis and transport into synaptic vesicles. J. Biol. Chem. 285, 1957–1966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Daubner S. C., Le T., Wang S. (2011) Tyrosine hydroxylase and regulation of dopamine synthesis. Arch. Biochem. Biophys. 508, 1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chen X., Xu L., Radcliffe P., Sun B., Tank A. W. (2008) Activation of tyrosine hydroxylase mRNA translation by cAMP in midbrain dopaminergic neurons. Mol. Pharmacol. 73, 1816–1828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lewis E. J., Harrington C. A., Chikaraishi D. M. (1987) Transcriptional regulation of the tyrosine hydroxylase gene by glucocorticoid and cyclic AMP. Proc. Natl. Acad. Sci. U.S.A. 84, 3550–3554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Andersson K. K., Cox D. D., Que L., Jr., Flatmark T., Haavik J. (1988) Resonance Raman studies on the blue-green-colored bovine adrenal tyrosine 3-monooxygenase (tyrosine hydroxylase). Evidence that the feedback inhibitors adrenaline and noradrenaline are coordinated to iron. J. Biol. Chem. 263, 18621–18626 [PubMed] [Google Scholar]
- 9. Haycock J. W., Bennett W. F., George R. J., Waymire J. C. (1982) Multiple site phosphorylation of tyrosine hydroxylase. Differential regulation in situ by a 8-bromo-cAMP and acetylcholine. J. Biol. Chem. 257, 13699–13703 [PubMed] [Google Scholar]
- 10. Ichimura T., Isobe T., Okuyama T., Takahashi N., Araki K., Kuwano R., Takahashi Y. (1988) Molecular cloning of cDNA coding for brain-specific 14-3-3 protein, a protein kinase-dependent activator of tyrosine and tryptophan hydroxylases. Proc. Natl. Acad. Sci. U.S.A. 85, 7084–7088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Nagatsu T., Ichinose H. (1999) Regulation of pteridine-requiring enzymes by the cofactor tetrahydrobiopterin. Mol. Neurobiol. 19, 79–96 [DOI] [PubMed] [Google Scholar]
- 12. Salvatore M. F., Pruett B. S. (2012) Dichotomy of tyrosine hydroxylase and dopamine regulation between somatodendritic and terminal field areas of nigrostriatal and mesoaccumbens pathways. PLoS One 7, e29867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Daubner S. C., Le T., Wang S. (2011) Tyrosine hydroxylase and regulation of dopamine synthesis. Arch. Biochem. Biophys. 508, 1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Dunkley P. R., Bobrovskaya L., Graham M. E., von Nagy-Felsobuki E. I., Dickson P. W. (2004) Tyrosine hydroxylase phosphorylation: regulation and consequences. J. Neurochem. 91, 1025–1043 [DOI] [PubMed] [Google Scholar]
- 15. Haycock J. W., Wakade A. R. (1992) Activation and multiple-site phosphorylation of tyrosine hydroxylase in perfused rat adrenal glands. J. Neurochem. 58, 57–64 [DOI] [PubMed] [Google Scholar]
- 16. Andersson K. K., Vassort C., Brennan B. A., Que L., Jr., Haavik J., Flatmark T., Gros F., Thibault J. (1992) Purification and characterization of the blue-green rat phaeochromocytoma (PC12) tyrosine hydroxylase with a dopamine-Fe(III) complex. Reversal of the endogenous feedback inhibition by phosphorylation of serine-40. Biochem. J. 284 (Pt 3), 687–695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sutherland C., Alterio J., Campbell D. G., Le Bourdelles B., Mallet J., Haavik J., Cohen P. (1993) Phosphorylation and activation of human tyrosine hydroxylase in vitro by mitogen-activated protein (MAP) kinase and MAP-kinase-activated kinases 1 and 2. Eur. J. Biochem. 217, 715–722 [DOI] [PubMed] [Google Scholar]
- 18. Sura G. R., Daubner S. C., Fitzpatrick P. F. (2004) Effects of phosphorylation by protein kinase A on binding of catecholamines to the human tyrosine hydroxylase isoforms. J. Neurochem. 90, 970–978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Almas B., Le Bourdelles B., Flatmark T., Mallet J., Haavik J. (1992) Regulation of recombinant human tyrosine hydroxylase isozymes by catecholamine binding and phosphorylation. Structure/activity studies and mechanistic implications. Eur. J. Biochem. 209, 249–255 [DOI] [PubMed] [Google Scholar]
- 20. Toska K., Kleppe R., Armstrong C. G., Morrice N. A., Cohen P., Haavik J. (2002) Regulation of tyrosine hydroxylase by stress-activated protein kinases. J. Neurochem. 83, 775–783 [DOI] [PubMed] [Google Scholar]
- 21. Haycock J. W., Lew J. Y., Garcia-Espana A., Lee K. Y., Harada K., Meller E., Goldstein M. (1998) Role of serine-19 phosphorylation in regulating tyrosine hydroxylase studied with site- and phosphospecific antibodies and site-directed mutagenesis. J. Neurochem. 71, 1670–1675 [DOI] [PubMed] [Google Scholar]
- 22. Bevilaqua L. R., Graham M. E., Dunkley P. R., von Nagy-Felsobuki E. I., Dickson P. W. (2001) Phosphorylation of Ser(19) alters the conformation of tyrosine hydroxylase to increase the rate of phosphorylation of Ser(40). J. Biol. Chem. 276, 40411–40416 [DOI] [PubMed] [Google Scholar]
- 23. Nakashima A., Mori K., Kaneko Y. S., Hayashi N., Nagatsu T., Ota A. (2011) Phosphorylation of the N-terminal portion of tyrosine hydroxylase triggers proteasomal digestion of the enzyme. Biochem. Biophys. Res. Commun. 407, 343–347 [DOI] [PubMed] [Google Scholar]
- 24. Itagaki C., Isobe T., Taoka M., Natsume T., Nomura N., Horigome T., Omata S., Ichinose H., Nagatsu T., Greene L. A., Ichimura T. (1999) Stimulus-coupled interaction of tyrosine hydroxylase with 14-3-3 proteins. Biochemistry 38, 15673–15680 [DOI] [PubMed] [Google Scholar]
- 25. Kleppe R., Martinez A., Doskeland S. O., Haavik J. (2011) The 14-3-3 proteins in regulation of cellular metabolism. Semin. Cell Dev. Biol. 22, 713–719 [DOI] [PubMed] [Google Scholar]
- 26. Obsil T., Obsilova V. (2011) Structural basis of 14-3-3 protein functions. Semin. Cell Dev. Biol. 22, 663–672 [DOI] [PubMed] [Google Scholar]
- 27. Aitken A. (2011) Post-translational modification of 14-3-3 isoforms and regulation of cellular function. Semin. Cell Dev. Biol. 22, 673–680 [DOI] [PubMed] [Google Scholar]
- 28. Roth D., Morgan A., Martin H., Jones D., Martens G. J., Aitken A., Burgoyne R. D. (1994) Characterization of 14-3-3 proteins in adrenal chromaffin cells and demonstration of isoform-specific phospholipid binding. Biochem. J. 301, 305–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Martin H., Rostas J., Patel Y., Aitken A. (1994) Subcellular localisation of 14-3-3 isoforms in rat brain using specific antibodies. J. Neurochem. 63, 2259–2265 [DOI] [PubMed] [Google Scholar]
- 30. Halskau O., Ying M., Baumann A., Kleppe R., Rodriguez-Larrea D., Almas B., Haavik J., Martinez A. (2009) Three-way interaction between 14-3-3 proteins, the N-terminal region of tyrosine hydroxylase, and negatively charged membranes. J. Biol. Chem. 284, 32758–32769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bustad H. J., Skjaerven L., Ying M., Halskau O., Baumann A., Rodriguez-Larrea D., Costas M., Underhaug J., Sanchez-Ruiz J. M., Martinez A. (2012) The peripheral binding of 14-3-3gamma to membranes involves isoform-specific histidine residues. PLoS ONE 7, e49671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Skjevik A. A., Mileni M., Baumann A., Halskau O., Teigen K., Stevens R. C., Martinez A. (2014) The N-terminal sequence of tyrosine hydroxylase is a conformationally versatile motif that binds 14-3-3 proteins and membranes. J. Mol. Biol. 426, 150–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kleppe R., Toska K., Haavik J. (2001) Interaction of phosphorylated tyrosine hydroxylase with 14-3-3 proteins: evidence for a phosphoserine 40-dependent association. J. Neurochem. 77, 1097–1107 [DOI] [PubMed] [Google Scholar]
- 34. Obsilova V., Nedbalkova E., Silhan J., Boura E., Herman P., Vecer J., Sulc M., Teisinger J., Dyda F., Obsil T. (2008) The 14-3-3 protein affects the conformation of the regulatory domain of human tyrosine hydroxylase. Biochemistry 47, 1768–1777 [DOI] [PubMed] [Google Scholar]
- 35. Yaffe M. B. (2002) How do 14-3-3 proteins work? Gatekeeper phosphorylation and the molecular anvil hypothesis. FEBS Lett. 513, 53–57 [DOI] [PubMed] [Google Scholar]
- 36. Gardino A. K., Smerdon S. J., Yaffe M. B. (2006) Structural determinants of 14-3-3 binding specificities and regulation of subcellular localization of 14-3-3-ligand complexes: a comparison of the x-ray crystal structures of all human 14-3-3 isoforms. Semin. Cancer Biol. 16, 173–182 [DOI] [PubMed] [Google Scholar]
- 37. Bobrovskaya L., Dunkley P. R., Dickson P. W. (2004) Phosphorylation of Ser19 increases both Ser40 phosphorylation and enzyme activity of tyrosine hydroxylase in intact cells. J. Neurochem. 90, 857–864 [DOI] [PubMed] [Google Scholar]
- 38. Kostelecky B., Saurin A. T., Purkiss A., Parker P. J., McDonald N. Q. (2009) Recognition of an intra-chain tandem 14-3-3 binding site within PKCepsilon. EMBO Rep. 10, 983–989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Obsil T., Ghirlando R., Anderson D. E., Hickman A. B., Dyda F. (2003) Two 14-3-3 binding motifs are required for stable association of Forkhead transcription factor FOXO4 with 14-3-3 proteins and inhibition of DNA binding. Biochemistry 42, 15264–15272 [DOI] [PubMed] [Google Scholar]
- 40. Molzan M., Ottmann C. (2012) Synergistic binding of the phosphorylated S233- and S259-binding sites of C-RAF to one 14-3-3zeta dimer. J. Mol. Biol. 423, 486–495 [DOI] [PubMed] [Google Scholar]
- 41. Johnson C., Crowther S., Stafford M. J., Campbell D. G., Toth R., MacKintosh C. (2010) Bioinformatic and experimental survey of 14-3-3-binding sites. Biochem. J. 427, 69–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Sharon M., Robinson C. V. (2007) The role of mass spectrometry in structure elucidation of dynamic protein complexes. Annu. Rev. Biochem. 76, 167–193 [DOI] [PubMed] [Google Scholar]
- 43. Heck A. J. (2008) Native mass spectrometry: a bridge between interactomics and structural biology. Nat. Methods 5, 927–933 [DOI] [PubMed] [Google Scholar]
- 44. Bogomolovas J., Simon B., Sattler M., Stier G. (2009) Screening of fusion partners for high yield expression and purification of bioactive viscotoxins. Protein Expr. Purif. 64, 16–23 [DOI] [PubMed] [Google Scholar]
- 45. Studier F. W. (2005) Protein production by auto-induction in high density shaking cultures. Protein Expr. Purif. 41, 207–234 [DOI] [PubMed] [Google Scholar]
- 46. Kopperud R., Christensen A. E., Kjarland E., Viste K., Kleivdal H., Doskeland S. O. (2002) Formation of inactive cAMP-saturated holoenzyme of cAMP-dependent protein kinase under physiological conditions. J. Biol. Chem. 277, 13443–13448 [DOI] [PubMed] [Google Scholar]
- 47. Kastner B., Fischer N., Golas M. M., Sander B., Dube P., Boehringer D., Hartmuth K., Deckert J., Hauer F., Wolf E., Uchtenhagen H., Urlaub H., Herzog F., Peters J. M., Poerschke D., Luhrmann R., Stark H. (2008) GraFix: sample preparation for single-particle electron cryomicroscopy. Nat. Methods 5, 53–55 [DOI] [PubMed] [Google Scholar]
- 48. de la Rosa-Trevin J. M., Oton J., Marabini R., Zaldivar A., Vargas J., Carazo J. M., Sorzano C. O. (2013) Xmipp 3.0: an improved software suite for image processing in electron microscopy. J. Struct. Biol. 184, 321–328 [DOI] [PubMed] [Google Scholar]
- 49. Scheres S. H., Nunez-Ramirez R., Sorzano C. O., Carazo J. M., Marabini R. (2008) Image processing for electron microscopy single-particle analysis using XMIPP. Nat. Protoc. 3, 977–990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Scheres S. H., Valle M., Nunez R., Sorzano C. O., Marabini R., Herman G. T., Carazo J. M. (2005) Maximum-likelihood multi-reference refinement for electron microscopy images. J. Mol. Biol. 348, 139–149 [DOI] [PubMed] [Google Scholar]
- 51. Yang X., Lee W. H., Sobott F., Papagrigoriou E., Robinson C. V., Grossmann J. G., Sundstrom M., Doyle D. A., Elkins J. M. (2006) Structural basis for protein-protein interactions in the 14-3-3 protein family. Proc. Natl. Acad. Sci. U.S.A. 103, 17237–17242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Benesch J. L., Aquilina J. A., Ruotolo B. T., Sobott F., Robinson C. V. (2006) Tandem mass spectrometry reveals the quaternary organization of macromolecular assemblies. Chem. Biol. 13, 597–605 [DOI] [PubMed] [Google Scholar]
- 53. van Duijn E., Barbu I. M., Barendregt A., Jore M. M., Wiedenheft B., Lundgren M., Westra E. R., Brouns S. J., Doudna J. A., van der Oost J., Heck A. J. (2012) Native tandem and ion mobility mass spectrometry highlight structural and modular similarities in clustered-regularly-interspaced shot-palindromic-repeats (CRISPR)-associated protein complexes from Escherichia coli and Pseudomonas aeruginosa. Mol. Cell. Proteomics 11, 1430–1441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Johnson C., Tinti M., Wood N. T., Campbell D. G., Toth R., Dubois F., Geraghty K. M., Wong B. H., Brown L. J., Tyler J., Gernez A., Chen S., Synowsky S., MacKintosh C. (2011) Visualization and biochemical analyses of the emerging mammalian 14-3-3-phosphoproteome. Mol. Cell. Proteomics 10, M110.005751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Obsil T., Ghirlando R., Klein D. C., Ganguly S., Dyda F. (2001) Crystal structure of the 14-3-3zeta:serotonin N-acetyltransferase complex. A role for scaffolding in enzyme regulation. Cell 105, 257–267 [DOI] [PubMed] [Google Scholar]
- 56. Rezabkova L., Man P., Novak P., Herman P., Vecer J., Obsilova V., Obsil T. (2011) Structural basis for the 14-3-3 protein-dependent inhibition of the regulator of G protein signaling 3 (RGS3) function. J. Biol. Chem. 286, 43527–43536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Rezabkova L., Boura E., Herman P., Vecer J., Bourova L., Sulc M., Svoboda P., Obsilova V., Obsil T. (2010) 14-3-3 protein interacts with and affects the structure of RGS domain of regulator of G protein signaling 3 (RGS3). J. Struct. Biol. 170, 451–461 [DOI] [PubMed] [Google Scholar]
- 58. Daubner S. C., Piper M. M. (1995) Deletion mutants of tyrosine hydroxylase identify a region critical for heparin binding. Protein Sci. 4, 538–541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Goodwill K. E., Sabatier C., Marks C., Raag R., Fitzpatrick P. F., Stevens R. C. (1997) Crystal structure of tyrosine hydroxylase at 2.3 A and its implications for inherited neurodegenerative diseases. Nat. Struct. Biol. 4, 578–585 [DOI] [PubMed] [Google Scholar]
- 60. Zhang S., Huang T., Ilangovan U., Hinck A. P., Fitzpatrick P. F. (2014) The solution structure of the regulatory domain of tyrosine hydroxylase. J. Mol. Biol. 426, 1483–1497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Lindgren N., Xu Z. Q., Lindskog M., Herrera-Marschitz M., Goiny M., Haycock J., Goldstein M., Hokfelt T., Fisone G. (2000) Regulation of tyrosine hydroxylase activity and phosphorylation at Ser(19) and Ser(40) via activation of glutamate NMDA receptors in rat striatum. J. Neurochem. 74, 2470–2477 [DOI] [PubMed] [Google Scholar]
- 62. Lou H., Montoya S. E., Alerte T. N., Wang J., Wu J., Peng X., Hong C. S., Friedrich E. E., Mader S. A., Pedersen C. J., Marcus B. S., McCormack A. L., Di Monte D. A., Daubner S. C., Perez R. G. (2010) Serine 129 phosphorylation reduces the ability of alpha-synuclein to regulate tyrosine hydroxylase and protein phosphatase 2A in vitro and in vivo. J. Biol. Chem. 285, 17648–17661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Sachs N. A., Vaillancourt R. R. (2004) Cyclin-dependent kinase 11p110 and casein kinase 2 (CK2) inhibit the interaction between tyrosine hydroxylase and 14-3-3. J. Neurochem. 88, 51–62 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.