Abstract
Herpesviral capsids are assembled in the host cell nucleus before being translocated into the cytoplasm for further maturation. The crossing of the nuclear envelope represents a major event that requires the formation of the nuclear egress complex (NEC). Previous studies demonstrated that human cytomegalovirus (HCMV) proteins pUL50 and pUL53, as well as their homologs in all members of Herpesviridae, interact with each other at the nuclear envelope and form the heterodimeric core of the NEC. In order to characterize further the viral and cellular protein content of the multimeric NEC, the native complex was isolated from HCMV-infected human primary fibroblasts at various time points and analyzed using quantitative proteomics. Previously postulated components of the HCMV-specific NEC, as well as novel potential NEC-associated proteins such as emerin, were identified. In this regard, interaction and colocalization between emerin and pUL50 were confirmed by coimmunoprecipitation and confocal microscopy analyses, respectively. A functional validation of viral and cellular NEC constituents was achieved through siRNA-mediated knockdown experiments. The important role of emerin in NEC functionality was demonstrated by a reduction of viral replication when emerin expression was down-regulated. Moreover, under such conditions, reduced production of viral proteins and deregulation of viral late cytoplasmic maturation were observed. Combined, these data prove the functional importance of emerin as an NEC component, associated with pUL50, pUL53, pUL97, p32/gC1qR, and further regulatory proteins. Summarized, our findings provide the first proteomics-based characterization and functional validation of the HCMV-specific multimeric NEC.
Viruses are tightly linked to the regulatory processes governing the metabolic state of their host cells. This regulatory linkage is reflected by viral activation or silencing of gene expression and productive replication in response to cellular changes in signaling, cell cycle, apoptosis, differentiation, and other parameters. Viruses also tend to exert a strong influence on regulatory cellular pathways and the developmental fate of virus-infected tissues (1, 2). These examples of virus-cell interregulation have been studied in detail, but in many cases the essential molecular mechanisms are still poorly understood. In the field of herpesviruses, profound efforts in molecular research have been undertaken to characterize those direct protein–protein interactions that regulate cross-talk between the virus and its host. Multi-protein complexes composed of both viral and cellular constituents were identified in several stages of herpesviral lytic replication. In particular, detailed studies on the replication of human cytomegalovirus (HCMV)1 in primary fibroblasts and other permissive cell types have provided very interesting insights into the nature of chimeric multi-protein complexes. These examples were described for viral entry, viral response to intrinsic immunity, intracellular transport of viral products, nucleocytoplasmic egress of viral capsids, and other processes (3–6). In classical approaches, protein–protein interaction was studied by means of approved methods including yeast two-hybrid, coimmunoprecipitation (CoIP), and pulldown analyses with purified proteins. More recently, very sensitive methods have been introduced into this field, such as proteomic analysis using tandem mass spectrometry (MS/MS), confocal imaging techniques, surface plasmon resonance analysis, and others.
During HCMV replication, the translocation of genome-containing viral capsids from the nucleus to the cytoplasm (nuclear egress) is one of the most crucial steps. In this process, the nuclear envelope represents a barrier consisting of three distinct elements: nuclear membranes, nuclear pores, and the proteinaceous network of the nuclear lamina. The viral capsids traverse the nuclear envelope by budding through nuclear membranes. Importantly, HCMV capsids access the inner nuclear membrane by overcoming the proteinaceous network of the nuclear lamina. To regulate the serial steps in this procedure, a multimeric protein complex is formed, termed the nuclear egress complex (NEC) (4, 7). One of the main tasks of the NEC is the distortion of the nuclear lamina. Our recent studies identified the formation of lamina-depleted areas that result from the recruitment of sophisticated enzymatic activities to these specific sites at the lamina (8). Viral and cellular effectors, such as protein kinases, a proline cis/trans isomerase, and possibly further regulatory proteins, are involved in this process (4). It is commonly accepted that the core NEC is composed of two viral proteins, namely, pUL50 and pUL53 (9–13). Moreover, the association of pUL50–pUL53 with a number of viral and cellular proteins supports the concept of a multimeric NEC that may include the viral protein kinase pUL97, multi-ligand binding protein p32/gC1qR, lamin B receptor, and protein kinase C (PKC) (14).
In this work, we first confirmed the major role played by pUL50 and pUL53 in NEC formation. The pUL50–pUL53 core NEC was then used as bait for the identification of other NEC components at different time points post-infection. Quantitative MS-based proteomics confirmed known members of the multimeric NEC and also identified the cellular inner nuclear membrane protein emerin as a novel NEC constituent. Importantly, colocalization of emerin with the HCMV-specific NEC and its interaction with pUL50 were demonstrated for the first time. Knockdown experiments provided functional validation of the importance of emerin and other NEC proteins for HCMV replication. Together, these data provide an extended mechanistic model for the composition and function of the HCMV-specific NEC.
EXPERIMENTAL PROCEDURES
Immunoprecipitation of HCMV Native NEC
Primary human foreskin fibroblasts (HFFs) were cultivated and infected with recombinant HCMV AD169-GFP UL53-F or AD169-GFP UL50-HA in 550-ml cell culture flasks at a density of 5 × 106 to 6 × 106 cells as described previously (15). Cells were harvested at time points between 1 and 10 days post-infection (dpi) (supplemental Table S2), and the native NEC was immunoprecipitated from total cell lysates as previously described (14). Briefly, infected cells were lysed in 500 μl of CoIP buffer (50 mm Tris/HCl, pH 8.0, 150 mm NaCl, 5 mm EDTA, 0.5% Nonidet P-40, 1 mm PMSF, 2 mg of aprotinin ml−1, 2 mg of leupeptin ml−1, 2 mg of pepstatin ml−1) and used for immunoprecipitation with tag-specific antibodies against pUL53-F (1.2 μl of mouse monoclonal antibody mAb-FLAG (M2; Sigma)) or pUL50-HA (3 μl of mouse mAb-HA (12CA5; Roche Applied Science)) coupled to protein A–Sepharose beads (GE Healthcare Life Sciences) for 1.5 to 3 h at 4 °C under rotation. Mouse mAb-GFP (3 μl of clones 7.1/13.1; Roche Applied Science) or a mouse Fab fragment (1.2 μl; Jackson ImmunoResearch Laboratories, West Grove, PA) was used as a control for specificity.
Quality Control of Purified Samples
To ensure high quality of the immunoprecipitated protein complexes, samples were analyzed both qualitatively and quantitatively. First, samples were analyzed to ensure successful CoIP of the core NEC proteins via standard Western blot (Wb) detection using antibodies against hemagglutinin (HA)-tagged or untagged pUL50 (mouse mAb-HA (12CA5) and rabbit pAb-UL50 (10)) and FLAG-tagged or untagged pUL53 (mouse mAb-FLAG (M2) and mouse pAb-UL53 (16)). Second, proteins were separated on 12.5% SDS-PAGE gels and Coomassie stained with InstantBlue™ (Expedeon, San Diego, CA) for 1 h at room temperature under continuous gentle agitation. Thereafter, the gels were subjected to silver staining procedures (all steps at room temperature). For fixation, gels were incubated for 2 h in fixative A (30% methanol, 7.5% acetic acid), washed three times in 10% ethanol for 5 min, and incubated for 30 min in fixative B (1% glutaraldehyde). After five additional washing steps with 10% ethanol for 5 min, fixed gels were incubated with silver nitrate solution (0.2% AgNO3, 0.2% NaOH, 0.28% NH4OH) for 30 min and washed again four times in 10% ethanol. Finally, protein bands were visualized by incubation with staining solution (0.005% citric acid, 0.02% formaldehyde) for 30 to 45 min.
Proteomic Analyses
Preparation and In-gel Digestion of NEC Proteins
Immunoprecipitated complexes were solubilized in Laemmli buffer before stacking of proteins in the top layer of a 4–12% NuPAGE gel (Invitrogen) for separation followed by R-250 Coomassie Blue staining. The gel band was manually excised and cut into pieces before being washed by six successive incubations in 25 mm NH4HCO3 for 15 min each, followed by six incubations in 25 mm NH4HCO3 containing 50% (v/v) acetonitrile. Gel pieces were then dehydrated with 100% acetonitrile and incubated with 10 mm DTT in 25 mm NH4HCO3 for 45 min at 53 °C and with 55 mm iodoacetamide in 25 mm NH4HCO3 for 35 min in the dark. Alkylation was stopped by the addition of 10 mm DTT in 25 mm NH4HCO3 (10-min incubation). Gel pieces were then washed again by incubation in 25 mm NH4HCO3 followed by dehydration with 100% acetonitrile. Modified trypsin (sequencing grade, Promega, Madison, WI) in 25 mm NH4HCO3 was added to the dehydrated gel pieces for incubation at 37 °C overnight. Peptides were extracted from gel pieces in three sequential extraction steps (each 15 min) in 30 μl of 50% acetonitrile, 30 μl of 5% formic acid, and finally 30 μl of 100% acetonitrile. The pooled supernatants were dried under vacuum.
Nano-LC-MS/MS Analyses
The dried extracted peptides were resuspended in 5% acetonitrile and 0.1% trifluoroacetic acid and analyzed via online nano-LC-MS/MS (Ultimate 3000, Dionex, Germering, Germany; LTQ-Orbitrap Velos Pro, Thermo Fisher Scientific). Peptides were applied onto a 300 μm × 5 mm PepMap C18 precolumn and separated on a 75 μm × 250 mm C18 column (PepMap, Dionex). The nano-LC method consisted of a 120-min gradient at a flow rate of 300 nl/min, ranging from 5% to 37% acetronitrile in 0.1% formic acid for 114 min before reaching 72% acetronitrile in 0.1% formic acid for the last 6 min. MS and MS/MS data were acquired using Xcalibur (Thermo Fisher Scientific). The spray voltage was set at 1.4 kV, and the heated capillary was adjusted to 200 °C. Survey full-scan MS spectra (m/z = 400–1600) were obtained in the Orbitrap with a resolution of 60,000 after the accumulation of 106 ions (maximum filling time: 500 ms). The 20 most intense ions from the preview survey scan delivered by the Orbitrap were fragmented via collision-induced dissociation (collision energy: 35%) in the LTQ after the accumulation of 104 ions (maximum filling time: 100 ms).
Data Analyses
Data were processed automatically using Mascot Daemon software (version 2.3.2, Matrix Science, London, United Kingdom). Concomitant searches of the UniProt protein data bank (Homo sapiens and HCMV strain AD169, 125,420 sequences), classical contaminant protein sequence databases (260 sequences, homemade), and the corresponding reversed databases were performed using Mascot (version 2.4). An ESI-TRAP was chosen as the instrument, trypsin/P was selected as the enzyme, and two missed cleavages were allowed. Precursor and fragment mass error tolerances were set at 10 ppm and 0.6 Da. The following peptide modifications were allowed during the search: carbamidomethyl (C, fixes), acetyl (N-ter, variable), oxidation (M, variable), and deamidation (NQ, variable). The IRMa software (version 1.31.1) (17) was used to filter the results by conservation of rank 1 peptides, peptide identification with a false discovery rate of <1% (as calculated from peptide scores by employing the reverse database strategy), and a minimum of one specific peptide per identified protein group. Filtered results were uploaded into a relational mass spectrometry identification database (MSIdb) before compilation, grouping, and comparison of the protein groups from the different samples using a homemade tool (hEIDI). The algorithm compares each protein group of the union reference with each protein group of the individual identification and computes a similarity index for each protein group based on the Dice coefficient (18).
Statistical Analyses
Differential analysis of control and NEC samples was performed using extracted specific spectral counts (SSCs) and a β-binomial test considering the within-sample and between-sample variations in a single statistical model (19). The threshold for statistical significance was set at 0.05. To select for robust candidates in NEC samples, only proteins exhibiting a mean SSC of at least 4 were retained.
Transient Transfection and Eukaryotic Expression Plasmids
293T cells were cultivated and transfected with expression plasmids coding for HA- or FLAG-tagged cytomegaloviral proteins by the use of Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocol. Expression plasmids pcDNA-UL50-HA, pcDNA-UL53-F, pcDNA-UL97-F, pcDNA-UL44-F, pcDNA-UL69-F, pcDNA-UL84-F, pcDNA-UL26-F, and pcDNA-IE2-F and plasmids encoding the N-terminal deletion (i.e. encoded amino acids 5–397, 10–397, 15–397, 40–397, or 100–397) and C-terminal deletion mutants of pUL50 (i.e. encoded amino acids 1–358, 1–340, 1–310, or 1–280) have been previously described (9, 12, 14, 20, 21). As a vector control, pcDNA3.1 (Invitrogen) was used in CoIP assays.
CoIP Assay
Transfected 293T cells or HCMV-infected HFFs were used for protein–protein interaction experiments utilizing CoIP. Immunoprecipitation was performed 2 days post-transfection (PEI transfection method) or 3 dpi under previously described conditions (14) using 2 μl of mouse mAb-FLAG (M2), pAb-HA (HA.11), or mouse mAb-emerin (H-12). CoIP samples and expression controls taken prior to the addition of CoIP antibody were subjected to standard Wb analysis using mouse mAbs and rabbit pAbs as follows: mAb-FLAG (M2), pAb-FLAG (F7425), mAb-HA (12CA5), mAb-emerin (H-12), and mouse mAb-UL50 (UL50.01).
Generation and Reconstitution of Recombinant HCMVs
To generate recombinant HCMVs harboring point mutants of pUL50 (i.e. single mutants E56A, Q13A, and L116A; double mutant E56A/Y57A) fused to a C-terminal HA-tag, traceless bacterial artificial chromosome (BAC) mutagenesis (22) of the BAC clone AD169-GFP UL50-HA (15) was performed in Escherichia coli strain GS1783 (23). A detailed description of the generation and verification of recombinant viral genomes, as well as virus reconstitution, can be found in Ref. 15. Oligonucleotide primers used for the generation of PCR products are depicted in supplemental Table S1.
Indirect Immunofluorescence Assay and Confocal Laser-scanning Microscopy
HFF cells were cultivated and grown on coverslips for infection with HCMV laboratory strain AD169 or recombinant viruses at multiplicities of infection (MOIs) between 0.1 and 0.5. At 3 dpi, cells were fixed, permeabilized, and used for indirect immunofluorescence staining as previously described (14) with pAbs and mAbs indicated in the respective experiments. Secondary antibodies used for double staining were Alexa Fluor® 488-conjugated goat anti-rabbit IgG (H+L) and Alexa Fluor® 555-conjugated goat anti-mouse IgG (H+L; New England Biolabs GmbH, Ipswitch, MA). Images were acquired using a TCS SP5 confocal laser-scanning microscope equipped with a ×63 HCX PL APO CS oil immersion objective lens (Leica Microsystems, Wetzlar, Germany). For three-dimensional images, z-series of 100 confocal sections were recorded along 9.78 μm (z-axis) with a pinhole of 0.5 airy units. Images and z-series were analyzed using LAS AF software (Leica Microsystems).
Transient Knockdown of Emerin, pUL50, pUL97, and p32/gC1qR
Knockdown of Emerin, pUL50, and pUL97
HFFs were cultivated in 12-well or 24-well plates and grown to ∼60% confluence before transient transfection with synthetic siRNAs (50 to 60 nm) targeting the cellular EMD gene (coding for emerin; sc-35296, Santa Cruz Biotechnology Inc., Dallas, TX) or the viral UL50 or UL97 genes (see supplemental Table S1; Lipofectamine 2000, Invitrogen). A scrambled sequence (sc-44238, Santa Cruz Biotechnology Inc.) was used as a negative control in siRNA experiments. Cells were infected 24 h post-transfection with recombinant HCMV AD169-GFP, HCMV strain AD169, or recombinant HCMV TB40 UL32-GFP (24) at an MOI of 0.2 to 0.3 GFP/IE1-fu/cell. Cells were harvested at various time points post-infection and analyzed with regard to the influence of the specific knockdown on different aspects of HCMV replication: viral protein expression via Wb detection, HCMV replication efficiency via automated GFP fluorometry (15), and formation of the cytoplasmic viral assembly compartment (cVAC) via confocal microscopy. For the effect of emerin knockdown on HCMV replication, in addition to siRNA transfection followed by HCMV infection, a second set of samples was tested with an initial HCMV infection and subsequent transfection of the siRNAs at 1 dpi. In both cases, cells were harvested at 7 dpi, and HCMV replication efficiency was determined via automated GFP fluorometry.
Knockdown of Cellular p32/gC1qR
Knockdown of endogenous p32/gC1qR expression was achieved via the transduction of a siRNA-expressing adenovirus construct into HFFs according to an optimized procedure as described previously (25). Briefly, HFFs were transduced with 5 × 105 adenoviral knockdown particles per cell coding for a siRNA targeting the cellular p32/gC1qR gene or a scrambled sequence used as a negative control. Transduction of both types of knockdown particles was done at time points 1 d before and 2 d after HCMV infection. HCMV infection was performed with the reporter virus AD169-GFP (MOI: 0.2 GFP-fu/cell).
Quantitative Analyses of Viral Protein Expression Levels in Emerin Knockdown Cells
HFFs transiently transfected with siRNAs and infected with HCMV were lysed at 7 dpi. Viral protein expression was subsequently analyzed via Wb staining using protein-specific antibodies. Expression levels of immediate early, early, and late marker proteins of HCMV infection (IE1p72, pUL44, and major capsid protein (MCP)) were determined by AIDA Image Analyzer version 4.22 (Raytest Isotopenmessgeraete GmbH, Straubenhardt, Germany). Consecutive exposure times of the same Wb staining of a certain protein were evaluated via densitometry and compared with the corresponding staining of the loading control β-actin. Mean percentages of the expression levels of the emerin knockdown relative to the negative control are given in the respective experiments.
Polyclonal Antisera and Monoclonal Antibodies
Mouse mAbs against pUL50 (UL50.01 from clone 1A11) and pUL53 (UL53.01 from clone 7H8) were produced in the laboratory of Stipan Jonjic and Tihana Lenac (University of Rijeka, Croatia) by the use of bacterially expressed fragments of pUL50 (amino acids 1–181) and pUL53 (50–292). Further antibodies were provided as follows: rabbit pAb-UL97 (26), mouse pAb-UL53 (16), rabbit pAb-p32/gC1qR (27), rabbit pAb-FLAG (F7425; Sigma), rabbit pAb-HA (HA.11; Covance Inc., Princeton, NJ), mouse mAb-UL97 (28), rabbit mAb-lamin A/C (EPR4100; Abcam, Cambridge, United Kingdom), mouse mAb-emerin (H-12; Santa Cruz Biotechnology Inc.), mouse mAb-PKC (A-3; Santa Cruz Biotechnology Inc.), mAb-β-actin (AC-15; Sigma), mAb-FLAG (M2; Sigma), and mAb-HA (12CA5; Roche Applied Science). mAbs against viral IE1p72, MCP, and pp28 were kindly provided by William Britt (University of Alabama). mAb against pUL44 was kindly provided by Bodo Plachter (University of Mainz, Germany), and pAb against pUL50 was kindly provided by James Alwine (University of Pennsylvania).
RESULTS
Interaction between pUL50 and pUL53 Is a Requirement for NEC Formation
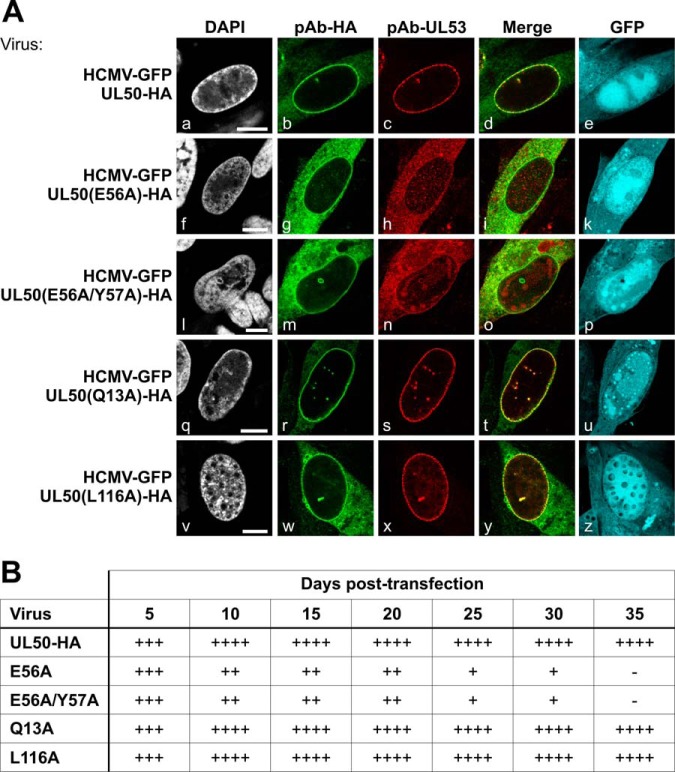
The core of the HCMV-specific NEC comprises the nuclear egress proteins pUL50 and pUL53. For studying individual functions of the HCMV's core NEC proteins, BACmid-derived viruses carrying tagged versions of the two proteins (HCMV-GFP UL50-HA and HCMV-GFP UL53-FLAG) were generated recently (15). These recombinant HCMVs have been characterized on a molecular basis, and findings indicate regular core NEC formation at the nuclear envelope (Figs. 1A and 2A) (15). As reported in 2012, site-specific UL50 replacement mutation (E56 and E57) in transiently expressed proteins abrogates pUL50–pUL53 interaction (9). In order to validate pUL50 and pUL53 as promising baits for purification of a multimeric NEC, we generated additional recombinant HCMVs carrying mutant pUL50 (E56A and E56A/Y57A as defective mutants; Q13A and L116A as potentially unimpaired controls). For virus reconstitution, BACmid DNA was transfected into human MRC-5 fibroblasts to monitor the expression of viral proteins. The progress of reconstitution was monitored via fluorescence microscopy. In essence, the production of infectious virus could be proven for samples derived from the parental BACmid (HCMV-GFP UL50-HA) and from mutants Q13A and L116A by the occurrence of viral cytopathic effect, observation of GFP-positive cells, and production of viral stocks from harvested media 35 days post-transfection. In contrast, no production of infectious virus was obtained with mutants E56A and E56A/Y57A (Fig. 1B). Furthermore, a confocal microscopy study visualized the production of BACmid-derived pUL50-HA and pUL53 and the recruitment of the two proteins to the nuclear envelope of transfected primary HFFs (Fig. 1A). For the interaction-incompetent pUL50 (i.e. mutants E56A and E56A/Y57A), we could not detect regular core NEC formation. This was indicated by mislocalization of pUL50-HA and pUL53 in the cytoplasm and the lack of distinct colocalization at the nuclear rim of BACmid-transfected cells (Fig. 1A, panels f–p). In contrast, control mutants Q13A and L116A (Fig. 1A, panels q–z) behaved very similarly to the parental virus (HCMV-GFP UL50-HA), with a clear localization of pUL50-HA and pUL53 at the nuclear rim (Fig. 1A, panels a–e). Combined, these results clearly point to the essential role of a correct mode of pUL50–pUL53 interaction, as well as nuclear rim localization, for core NEC formation and, consequently, for further steps in virus production.
Fig. 1.
Essential role of the E56 and Y57 residues of pUL50 in HCMV core NEC assembly. HFFs were transfected with recombinant HCMV BACmid DNA carrying the mutations indicated and expressing GFP as a marker of virus-positive cells. A, 10 to 15 days post-transfection, cells were fixed and used for confocal immunofluorescence analysis to detect the recruitment of core NEC pUL50–pUL53 to the nuclear rim (pAb-HA for the detection of pUL50-HA, mAb-UL53 for pUL53). Note the lack of both rim recruitment and colocalization of pUL50 and pUL53 with pUL50 mutations E56A and E56A/Y57A. DAPI, 4′,6-diamidino-2-phenylindole; scale bars, 10 μm. B, number of GFP-positive cells per well of a six-well plate transfected with mutant BACmids at different days post-transfection. -, none; +, >10; ++, 10–100; +++, 100–1000; ++++, >1000.
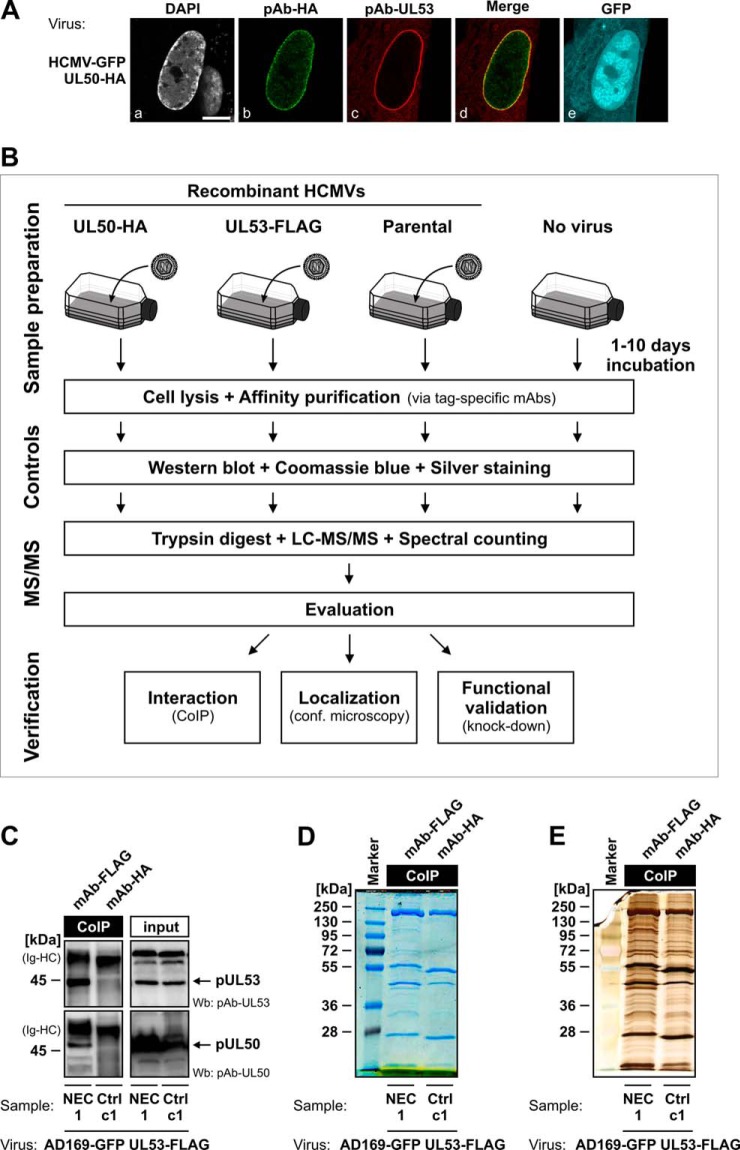
Fig. 2.
Schematic depiction of the purification and proteomic analysis of the multimeric NEC from HCMV-infected HFFs. A, core NEC formation in HCMV-infected HFFs. HFFs were infected with HCMV-GFP UL50-HA, fixed at 7 dpi, and stained with pAb-HA and pAb-UL53 as indicated. Scale bar, 10 μm. B, proteomics workflow for purification and analysis of the native NEC using recombinant HCMVs carrying tagged versions of pUL50 and pUL53. C–E, quality controls. CoIP samples of the pUL50–pUL53 core NEC were analyzed via Wb analysis using detection antibodies pAb-UL53 and pAb-UL50 (C). Differences in the precipitated proteomes of specific NEC samples relative to controls were examined by means of Coomassie Blue (D) and silver staining (E) of SDS-PAGE gels. Quality control samples are only depicted for NEC sample 1 and its direct control c1, but Wb analysis and Coomassie Blue and silver stainings were conducted similarly for all other NEC and control samples.
Purification of the Native NEC from HCMV-infected Human Fibroblasts
The recombinant viruses HCMV UL50-HA and HCMV UL53-FLAG represent tools of choice for the purification of the native HCMV-specific NEC (Fig. 2B). For this purpose, HFFs were infected at various MOIs with these viruses and harvested at time points between 1 and 10 dpi. CoIP of total cell lysates was performed by the use of tag-specific antibodies to purify the multimeric NEC from HCMV-infected cells. Protein complexes were then subjected to several steps of quality control, including proteomic pattern analysis using Wb and Coomassie Blue and silver staining of proteins in SDS-PAGE gels (Figs. 2C–2E). As expected, pUL50 and pUL53 were detected only in NEC-specific samples, and not in control samples (Fig. 2C). Several protein bands specifically visible in NEC samples (Figs. 2D and 2E) were considered as putative NEC components to be identified. Sample preparation conditions were varied during four independent MS/MS experiments with alternate use of viruses, CoIP antibodies, and controls, as well as varying incubation periods of infection (supplemental Table S2).
Identification of Postulated Components of the HCMV-specific NEC and Discovery of Novel Candidates via Proteomic Analysis
MS-based quantitative proteomic analysis was used to identify the set of proteins coimmunoprecipitating with native NEC. Interestingly, a general difference in the protein contents of precipitated complexes was detected between the samples taken from different time points post-infection. In samples from days 1 and 2 post-infection, no or only very low numbers of pUL50 or pUL53 peptides were recovered (supplemental Tables S3 and S4; data have been deposited in the ProteomeXchange repository (www.proteomexchange.org) with accession number PXD000536). This could be explained by relatively low levels of tagged pUL50 or pUL53 expressed in cells infected with these recombinant viruses at this initial stage of infection (15). At later time points, pUL50 and pUL53 were consistently identified. In order to highlight valuable candidates of the NEC, we performed a statistical analysis comparing SSC values of the proteins identified in control and NEC samples at early (i.e. samples taken at 3 and 4 dpi) versus late time points of analysis (i.e. 7 dpi and later). Cut-off criteria were as follows: (i) statistical p value of enrichment < 0.05, (ii) at least 4-fold enrichment in NEC samples relative to control samples, and (iii) a minimum SSC of 4. Only proteins positive for all three criteria were considered as significantly enriched in NEC samples (supplemental Table S3). Notably, immunoprecipitation of either pUL50 or pUL53 resulted in comparable results. However, it should be noted that pUL50-specific immunoprecipitations generally yielded lower SSC values (supplemental Table S3), most likely due to slightly delayed replication kinetics of recombinant HCMV UL50-HA (15). As a main outcome of this proteomic analysis, we were able to confirm previously postulated constituents of the HCMV-specific NEC, and we also identified novel candidates (Table I). In samples harvested at 3 to 4 dpi, only two viral proteins, pUL45 and pUL83 (pp65), known to be associated with virions, were found enriched with the pUL50–pUL53 core NEC. In contrast, at later time points (7 dpi and later), the viral protein kinase pUL97 and the cellular proteins p32/gC1qR, importin subunit β-1, and emerin were revealed as associated with pUL50–pUL53. Thus, these results suggest a change in NEC composition during the course of infection, particularly characterized by an enrichment of cellular proteins at late time points (Table I).
Table I. Functional description of proteins associated with the pUL50–pUL53 core NEC during the time course of infection.
| Protein name | UniProt I.D. | Mean SSC | Mean SSC ratioa (NEC/control) | p value | Subcellular localization | Known functionb |
|---|---|---|---|---|---|---|
| Transient composition of the NEC (samples taken at 3 and 4 dpi (i.e. samples NEC 7, 8, 10, and 11)) | ||||||
| HCMV proteins | ||||||
| pUL45 | P16782 | 6.3 | 5.0 | 3.55E-02 | Vi | Function unknown |
| pUL50 | P16791 | 12.5 | NEC only | 2.75E-05 | INM | Nuclear egress; NEC formation |
| pUL53 | P16794 | 26.5 | NEC only | 3.91E-05 | INM | Nuclear egress; NEC formation |
| pUL83 (pp65) | P06725 | 96.5 | 4.5 | 2.03E-02 | C, N, Vi | Counteracts host antiviral immune response |
| Final composition of the NEC (samples taken at 7 dpi and later (i.e. samples NEC 1, 2, 3, and 4)) | ||||||
| HCMV proteins | ||||||
| pUL50 | P16791 | 48.5 | 26.9 | 4.68E-04 | INM | Nuclear egress; NEC formation |
| pUL53 | P16794 | 57.8 | 144.4 | 5.51E-06 | INM | Nuclear egress; NEC formation |
| pUL97 | P16788 | 5.3 | 6.6 | 4.28E-02 | N, Vi | S/T-protein kinase; multifunctional (including phosphorylation of nuclear lamins during nuclear egress) |
| Cellular proteins | ||||||
| Emerin | P50402 | 5.0 | 25.0 | 3.96E-03 | INM | Localization of non-farnesylated prelamin A/C; stabilization and formation of a nuclear actin cortical network |
| Importin subunit β-1 | Q14974 | 6.0 | 10.0 | 1.30E-02 | C, N | Protein transport |
| LRRF-interacting protein 2 | Q9Y608 | 4.0 | 5.0 | 2.23E-02 | C | Potential activator of canonical Wnt signaling pathway |
| p32/gC1qR | Q07021 | 7.8 | NEC only | 2.39E-04 | C, Mt, N | Multifunctional protein; potential involvement in inflammation and infection processes, ribosome biogenesis, regulation of apoptosis, transcriptional regulation, and pre-mRNA splicing |
| Ribophorin I | P04843 | 4.8 | NEC only | 1.34E-03 | ERM | Protein glycosylation |
| SERCA2 | P16615 | 8.3 | NEC only | 5.43E-04 | ERM, SRM | Calcium transport |
Notes: ERM/SRM, endoplasmic/sarcoplasmic reticulum membrane; C, cytoplasm; INM, inner nuclear membrane; Mt, mitochondrion; N, nucleus; Vi, virion.
a Ratio between mean SSC of the NEC samples and mean SSC of the corresponding control samples (see supplemental Table S2).
b Information taken from Protein Knowledgebase (UniProtKB) and Ref. 2.
We classified proteins according to their functional relevance for nuclear egress. Therefore, we categorized the enriched proteins in terms of localization and specifically focused on proteins that either localize in the nucleus of infected cells or are associated with HCMV particles (Table I). Our results confirmed some proteins of the previously postulated multimeric NEC, namely, pUL50, pUL53, pUL97, and p32/gC1qR (14). For several of these proteins, including pUL53 and pUL97, functional nuclear localization signals have been identified recently (15, 29). In this context, it is interesting that importin subunit β-1 was identified in NEC samples. Importins are considered responsible for the nuclear localization of nuclear localization signal–carrying NEC proteins, but they are probably not actively involved in the process of HCMV nuclear egress. It was intriguing, however, to identify the inner nuclear membrane protein emerin among potential HCMV-specific NEC components, as its involvement in the nuclear egress of herpes simplex virus type 1 (HSV-1) was reported by others (30–32). These findings suggest that emerin may play a conserved role in the nuclear egress of HCMV and other herpesviruses.
Interaction of Emerin with pUL50
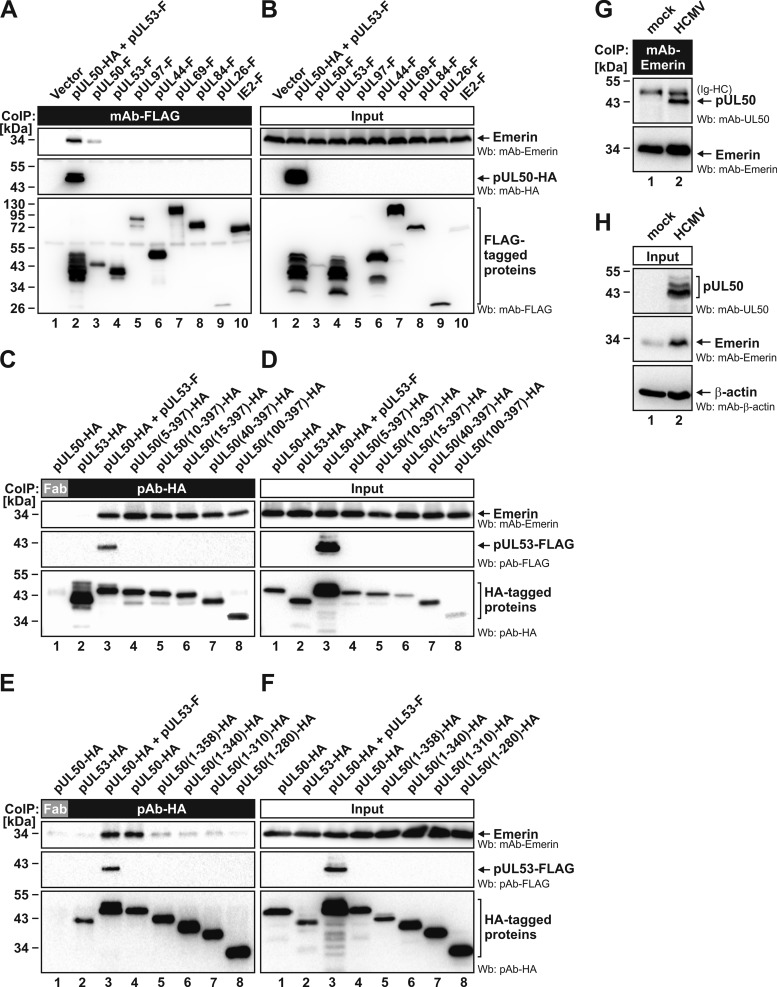
To verify results obtained via our quantitative proteomic approach, identified proteins were characterized in further protein–protein interaction analyses. For the previously postulated NEC proteins, former studies revealed an interaction network including pUL50, pUL53, pUL97, and p32/gC1qR (summarized in Table II) (9, 12, 14, 33). To address the question of whether endogenous emerin interacts with pUL50, pUL53, or both, CoIP experiments were performed with proteins from transiently transfected and HCMV-infected cells (Fig. 3). First, FLAG-tagged versions of pUL50, pUL53, or viral control proteins were transiently expressed in 293T cells and immunoprecipitated with mAb-FLAG. CoIP of endogenous emerin was tested via Wb. Our results indicated that emerin specifically interacts with pUL50 (as well as with coexpressed pUL50–pUL53), but not with pUL53 alone (Fig. 3A). CoIP of emerin with pUL50 alone was clearly detectable, although the pUL50 expression level remained much lower than that of pUL50–pUL53 coexpression (Fig. 3B; explained by the previous finding that coexpression stabilizes the pUL50–pUL53 complex) (9). All other viral proteins tested (Fig. 3A, lanes 4–10; including the NEC-associated kinase pUL97) did not show interaction with emerin. Second, a mapping experiment was performed to identify the region responsible for emerin interaction. For this purpose, a series of N- and C-terminal deletion mutants of pUL50 were analyzed in CoIP assays (Figs. 3C–3F). As a striking result, amino acids 1 to 99 proved to be dispensable for interaction (Figs. 3C and 3D, lane 8), and C-terminal truncations prevented CoIP of emerin (Figs. 3E and 3F). Thus, region 100 to 397 of pUL50 is competent and sufficient for emerin interaction. Finally, a further experiment using material from HCMV-infected HFFs and mAb-emerin for CoIP (reverse setting compared with Fig. 3A and Table I) likewise revealed pUL50 interaction with emerin (Figs. 3G and 3H). Together, these results confirmed the association of emerin with the HCMV-specific NEC, probably mediated via a direct emerin–pUL50 interaction.
Table II. Overview of proteins associated with the pUL50–pUL53 core NEC.
| NEC constituent | NEC purification |
Test for specific interaction |
pUL50–pUL53 colocalization |
|||
|---|---|---|---|---|---|---|
| MS/MSa |
CoIP (8, 12, 14, 15) |
Y2H (12, 14) |
Confocal microscopy (8, 12, 14, 15) |
|||
| HCMV infection | Transient transfection | Transient transfection | HCMV infection | Transient transfection | ||
| pUL50-HA-/pUL53-FLAG-associated | With pUL50 | With pUL53 | With pUL50 | |||
| pUL50 | + | − | + | − | + | + |
| pUL53 | + | + | − | + | + | + |
| pUL97 | + | − | − | − | −/+ | + |
| p32/gC1qR | + | + | −/+b | + | −/+ | +c |
| Emerin | + | + | − | n.d. | + | n.d. |
| PKC | − | +d | − | +e | − | +d |
Notes: n.d., not determined.
a See Table I for a complete list of proteins.
b Not consistent with a negative yeast two-hybrid result.
c Partial colocalization with pUL50 (data not shown).
d PKC isoform α.
e PKC isoforms ϵ and ζ.
Fig. 3.
Specific interaction between endogenous emerin and pUL50 in transfected and HCMV-infected cells. A–F, FLAG-/HA-tagged HCMV proteins, pUL50 deletion mutants, or pcDNA3.1 (vector) were transiently expressed in 293T cells as indicated. Cells were lysed at 2 days post-transfection, and this was followed by immunoprecipitation of the FLAG-tagged viral proteins and the HA-tagged pUL50 deletion mutants using mouse mAb-FLAG (A) and rabbit pAb-HA (C and E), respectively. A mouse Fab fragment was used as a control for specificity (C and E, lane 1). Coimmunoprecipitates and expression control samples were subjected to Wb analysis using tag-specific antibodies or mAb-emerin. G, H, HFFs were infected with HCMV strain AD169 at an MOI of 0.1 or remained uninfected (mock). At 3 dpi, cells were lysed and used for CoIP analysis with mAb-emerin. Detection of coimmunoprecipitates and of expression controls was performed by means of Western blotting using mAb-UL50, mAb-emerin, and mAb-β-actin. Ig-HC, cross-reactive band for immunoglobulin heavy chain.
Localization Study of NEC Components
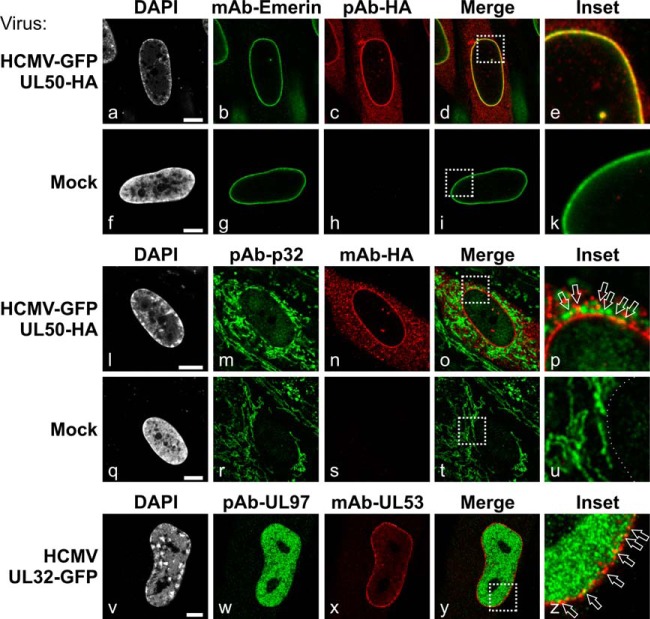
An extended confocal microscopy study was performed to investigate the intracellular localization of previously postulated and novel NEC components in HCMV-infected versus mock-infected fibroblasts (Fig. 4 and supplemental Fig. S1). For this purpose, HFFs were infected with HCMV (strain AD169) or recombinant virus HCMV-GFP UL50-HA (15) or HCMV UL32-GFP (24). Notably, pUL50 was found to colocalize with emerin at the nuclear envelope of HCMV-infected cells (Fig. 4, panels a–e). This result confirms the data obtained via proteomic and CoIP analyses. Concerning p32/gC1qR, our results demonstrated a relocalization in HCMV-infected cells (Fig. 4, panels l–p; supplemental Fig. S1B). The predominant mitochondrial protein p32/gC1qR is able to partly accumulate in the nucleus under specific stimuli including phosphorylation (34, 35). Phosphorylation of p32/gC1qR by the viral kinase pUL97 was demonstrated previously (33). In the present work, p32/gC1qR localization changed from a tubular cytoplasmic localization (mitochondria-associated in mock-infected cells) (36) to a punctate, granular-shaped cytoplasmic localization with some faint staining in the nucleus of infected cells (Fig. 4, panels l–p). Importantly, p32/gC1qR partially colocalized with pUL50 in dot-like structures at the nuclear rim (Fig. 4, panel p). In addition, viral pUL97 showed a partial colocalization with pUL50 (data not shown) and pUL53 (Fig. 4, panels v–z) restricted to areas of the nuclear rim, as similarly reported in other studies (29, 37). Finally, PKC, which interacts and colocalizes with pUL50 in transfected cells (12), was found relocalized in HCMV-infected cells (supplemental Fig. S1A). PKC showed translocation into the nucleus (compared with cytoplasmic localization in mock-infected cells) and partially colocalized with pUL97 in dot-like structures inside the nucleoplasm with a slight accumulation at the nuclear rim. This finding is particularly interesting, considering that an association of PKC with the cytomegalovirus-specific NEC is presently the topic of controversial discussion (8, 12, 37, 38) and our MS/MS data did not provide a final resolution to this controversy. In conclusion, the identified NEC proteins exhibited marked colocalization with pUL50 or pUL53 in HCMV-infected fibroblasts and/or profound intracellular relocalization.
Fig. 4.
Intracellular localization of NEC proteins in HCMV-infected primary fibroblasts. HFFs were infected with recombinant HCMV-GFP pUL50-HA (panels a–e and l–p) or recombinant HCMV UL32-GFP (panels v–z) or remained uninfected (mock; panels f–k and q–u). At 7 dpi (panels a–u) or 3 dpi (panels v–z), cells were fixed and coimmunostained with the indicated antibodies. Arrows point to yellow dots of merged signals of antibody staining indicating a partial colocalization of the respective proteins. Scale bars, 7.5 μm.
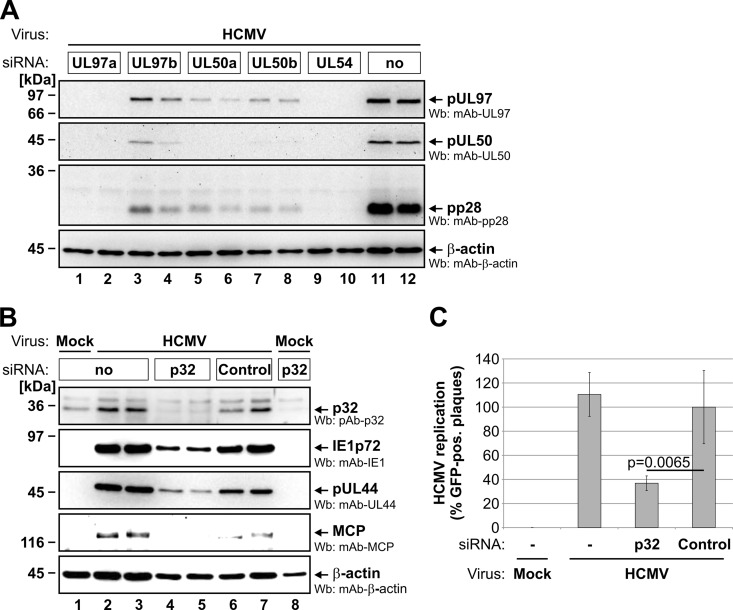
Influence of Knockdown of Specific NEC Proteins on HCMV Replication
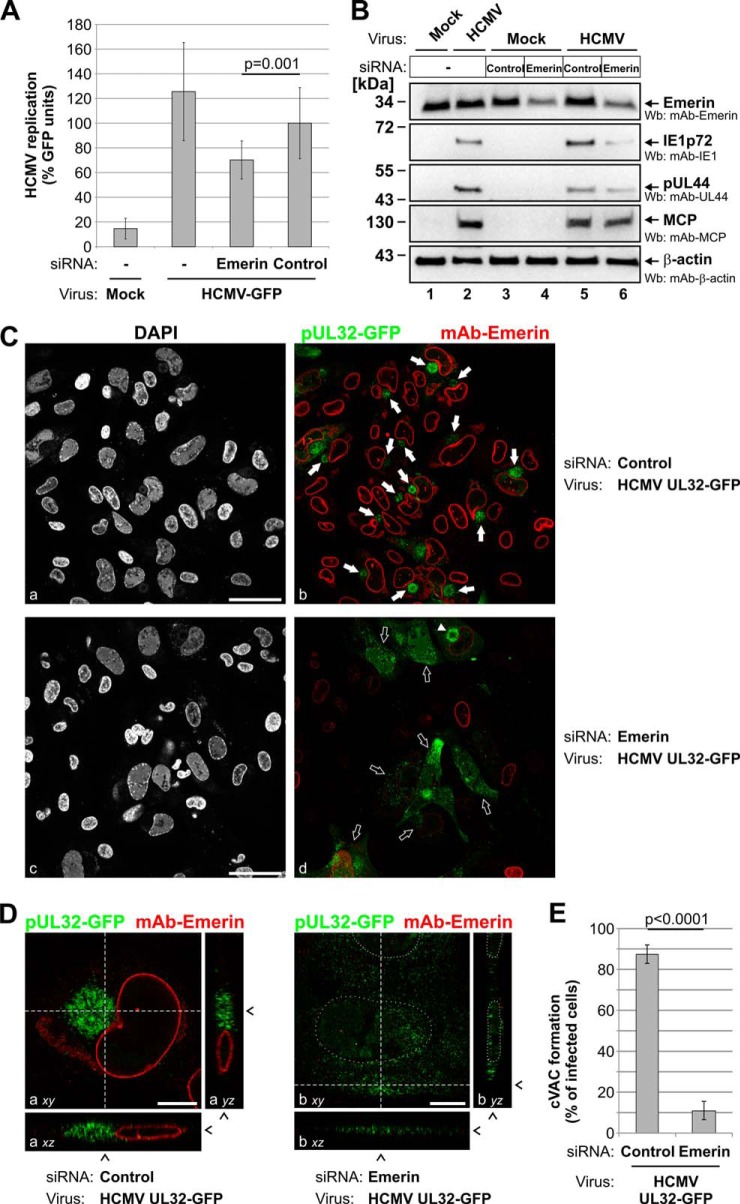
A functional validation of identified NEC proteins was performed through knockdown experiments. For the established NEC components pUL50, pUL97, and p32/gC1qR, knockdown was performed by means of transfection of synthetic siRNAs (specifically directed to the expression of viral pUL50 and pUL97; supplemental Table S1) or transduction of an adenoviral siRNA construct (previously established for knockdown of human p32/gC1qR) (25). In either case, knockdown of these NEC proteins resulted in reduced expression levels of essential viral proteins (Figs. 5A and 5B) and in significant inhibition of HCMV replication (Fig. 5C), as expected. Next, the effect of a siRNA-mediated knockdown of the novel NEC constituent emerin on HCMV infection was analyzed in detail (Fig. 6). First, when the endogenous expression of emerin was reduced by the transfection of a pool of three target-specific siRNAs (supplemental Fig. S2A), HCMV infection was markedly inhibited in these emerin-silenced HFFs relative to control cells, as demonstrated by a GFP-based HCMV replication assay (Fig. 6A). Second, knockdown of emerin effected a reduction of the expression levels of marker proteins for different phases of HCMV replication (Fig. 6B): immediate early marker IE1p72, early marker pUL44, and late marker MCP. Densitometric analyses of Wb stainings revealed that emerin-specific siRNAs reduced levels of emerin to 50% (Fig. 6B, lane 6) relative to an unspecific control siRNA (lane 5). Whereas levels of IE1p72 and pUL44 were markedly reduced upon emerin knockdown (reduction of IE1p72 levels to 23% and of pUL44 to 54%), MCP levels were only slightly affected (reduced to 75%). Finally, confocal microscopy was used to determine whether emerin knockdown interferes with late maturation of HCMV particles (Figs. 6C–6E). Notably, a characteristic feature of HCMV-infected cells is an enlarged, kidney-shaped nucleus bent around a perinuclear body, the cVAC (40, 41). Within the cVAC, viral particles undergo essential steps of late maturation such as the attachment of remaining tegument proteins and acquisition of the final envelope (6). In order to analyze the effect of emerin knockdown on late maturation events, we infected HFFs with recombinant HCMV UL32-GFP, expressing a fusion between pUL32 (pp150) and GFP as a reporter protein (24). The reporter protein pUL32-GFP associates with viral capsids in the nucleus, allowing visualization of viral capsids at various stages throughout the HCMV replication cycle (8, 24). At 5 dpi, most of the HCMV-infected control cells showed a cytoplasmic accumulation of GFP-labeled viral capsids (Fig. 6C, panels a and b, solid arrows). Three-dimensional images from z-series of confocal images representing only one infected cell (Fig. 6D, panel a) further suggested that this accumulation of viral capsids resembles the discoid structure of the cVAC (41, 42). In HFFs transfected with emerin-specific siRNAs (which resulted in the reduction of endogenous emerin levels in ∼80% of transfected cells; supplemental Fig. S2B), infection with HCMV UL32-GFP still led to massive viral capsid production in the nuclei and the cytoplasm of infected cells (Fig. 6C, panels c and d). However, in cells with considerably reduced levels of emerin, GFP-labeled capsids did not accumulate in viral cVACs, but were generally found throughout the whole cytoplasm (Fig. 6D, panel b, open arrows). In cells with only slightly reduced levels of emerin, cVAC formation was still observed in a few cells even under knockdown conditions (Fig. 6C, panel d, solid triangle). These findings indicated that late maturation in terms of cVAC formation was significantly deregulated with the knockdown of emerin (Fig. 6E). Thus, emerin and the NEC proteins pUL50, pUL97, and p32/gC1qR exert a strong influence on the efficiency of HCMV replication.
Fig. 5.
Knockdown analysis with HCMV-infected primary fibroblasts to verify the functional role of the established NEC proteins pUL50, pUL97, and p32/gC1qR. HFFs were transfected with synthetic siRNAs directed to the viral UL97 or UL50 genes (A) or transduced with adenoviral knockdown particles expressing siRNA directed to the cellular p32/gC1qR or a scrambled sequence as a control (B, C). In each case, transfected/transduced cells were additionally infected with the reporter virus HCMV-GFP. At 7 days (A) or 6 days (B, C) post-HCMV infection, the production of viral proteins was monitored by means of Wb analysis (A, B) and the formation of GFP-positive viral plaques was determined via wide-field microscopy (C). For A, transfection of siRNA targeting HCMV pUL54 strictly inhibiting HCMV replication was used as a positive control (39). For C, Student's t test was used to calculate statistical significance (a mean of four values with standard deviations is depicted for each sample).
Fig. 6.
Effect of emerin knockdown on HCMV replication, viral protein expression, and viral late maturation. HFFs were transiently transfected with emerin-specific siRNAs or with a scrambled siRNA as a control. In addition, transfected cells were infected with the reporter virus HCMV-GFP (A), HCMV strain AD169 (B), or recombinant HCMV UL32-GFP (C–E) as indicated. At 7 dpi, the production of viral proteins was monitored by means of Wb analysis (A), and HCMV replication efficiency was determined via automated GFP fluorometry (B). For A, Student's t test was used to calculate statistical significance (samples were performed in quadruplicate, and GFP signals were measured in duplicate; a mean of eight values and standard deviations are depicted). C–E, at 5 dpi, cells were fixed and immunostained with mAb-emerin. Representative confocal images (C) or examples of corresponding z-series with higher magnification (D) illustrate emerin immunostaining (red) and the distribution of GFP-labeled viral capsids (green) under emerin knockdown conditions or in control cells as indicated. Solid arrows, distribution of viral capsids in a cVAC-like fashion in emerin-expressing cells; open arrows, distribution of viral capsids throughout the cytoplasm of cells with no detectable emerin staining; solid triangle, distribution of viral capsids in a cVAC-like fashion in cells with slightly reduced levels of emerin staining; dashed lines/arrowheads, optical sections through the z stack (xy) or the focal plane (xz and yz); dotted line, nuclear margin deduced from the z-series of the corresponding DAPI images. E, quantitation of cVAC formation in emerin knockdown cells relative to control cells. The percentage of infected cells, showing GFP-labeled viral capsids in cVAC-like fashion, was determined by scoring 15 microscopic fields (>330 cells) for each setting. Student's t test was used to calculate statistical significance.
DISCUSSION
In this study, we used a combination of CoIP and MS-based quantitative proteomic analysis to explore the composition of the HCMV-specific multimeric NEC. The main findings are (i) that the native NEC can be isolated from HCMV-infected human fibroblasts by the use of pUL50-/pUL53-tagged recombinant viruses; (ii) that viral and cellular constituents, at least in part, could be confirmed as previously postulated; (iii) that emerin was validated as a novel NEC constituent; and (iv) that specific knockdown of the individual NEC constituents pUL50, pUL97, p32/gC1qR, and emerin interfered with efficient HCMV replication.
Constituents of the HCMV-specific Multimeric NEC
The current findings on the multi-component nature of the HCMV-specific NEC implicate a refined view of its mechanistic basis. Previously, we suggested a dynamic sequence of regulatory and enzymatic activities contributing to the NEC function. The most important players were categorized into functional groups: viral core NEC proteins (pUL50–pUL53), cellular targeting and adaptor proteins (LBR, lamins A/C, p32/gC1qR), regulatory protein kinases phosphorylating nuclear lamins (pUL97, PKC) and enzymatic effectors producing lamina-depleted areas for viral capsid egress (isomerase Pin1) (8). In this work, we were able to identify pUL50, pUL53, pUL97, p32/gC1qR, and several more NEC candidates. In this regard, we proved the major role played by p32/gC1qR in HCMV replication in primary fibroblasts. However, not all of the postulated members of the NEC, although individually detected via classical protein–protein interaction methods (e.g. yeast two-hybrid and CoIP analyses) or colocalization studies, were detected in our proteomic analysis of HCMV-specific NEC purified from primary fibroblasts (Table II). In part, this may be explained by the low-affinity binding and highly dynamic association-dissociation of regulatory enzymes, such as Pin1 and PKC. In addition, methodological limitations of static, two-dimensional measurements of protein interaction have to be taken into account (i.e. interactions will never all happen at the same time in the same place) when addressing the challenging task of characterizing complete interactomes (43). Another postulated NEC component, lamin B receptor, is tightly integrated into cellular membrane entities through transmembrane domains and may be lost in NEC preparation using CoIP. Interestingly, our results from the present and previous analyses suggest that the association of the viral core NEC with functional components of the entire multi-component NEC involves pUL50 as the major interactor scaffold. Interaction with pUL50 could be demonstrated for at least four NEC components, such as p32/gC1qR, PKC, emerin, and pUL53. Noticeably, pUL53 is a high-affinity interactor of pUL50, but interaction with further NEC components has not been conclusively demonstrated (11, 14, 37).
Our proteomic approach identified the inner nuclear membrane protein emerin as a novel constituent associated with the core NEC. Notably, a knockdown of emerin expression resulted in a strong impairment of HCMV replication. Emerin does not harbor enzymatic activities and may be considered a novel docking molecule for guiding the entire HCMV-specific NEC to preferred sites in the nuclear lamina. A role for emerin in nuclear replication and egress of other human herpesviruses (HSV-1 and Kaposi's sarcoma–associated herpesvirus) has recently been proposed by others (31, 32, 44). In this work, we obtained evidence of its interaction with pUL50. Interestingly, emerin is a member of the LAP2-emerin-MAN1 family of nuclear proteins, and thus this protein exhibits the LAP2-emerin-MAN1 domain as a common structural feature that mediates interaction with the barrier-to-autointegration factor (45–47). This factor is required for the assembly of emerin at the nuclear envelope (48). Despite the presence of a common protein interaction domain in emerin, it is tempting to speculate that the LAP2-emerin-MAN1 domain is also involved in interaction with pUL50. The structure of the emerin–barrier-to-autointegration factor complex revealed that the LAP2-emerin-MAN1 domain is bound by a deep concave cleft formed by the barrier-to-autointegration factor dimer (45). Our domain mapping analysis of pUL50 suggests that the C-terminal portion is responsible for emerin interaction.
Timely Coordinated Assembly of the Entire Multimeric NEC
We were also interested in exploring whether the NEC's composition generally varies along the time course of the HCMV lytic replication cycle. To answer this question, we performed MS/MS analyses of NEC purified from HCMV-infected cells at consecutive time points. A major finding was that the complexes isolated at early and late time points differed strongly in their composition. From our data, we can speculate that, first, the heteromer pUL50–pUL53 constitutes the viral core NEC at the nuclear envelope. Then this core recruits further proteins to form a multi-component NEC, possibly in the varieties of a transient and a final composition (Table I), as seen at later stages of viral replication. Here, the incorporation of the viral protein kinase pUL97 is of special importance. The enzymatic activity of pUL97 is responsible for the local disassembly of the nuclear lamina during the late phase of HCMV infection (8, 33, 49). Experiments with recombinant HCMVs (expressing mutant pUL50) pointed to the essential role of the pUL50–pUL53 interaction and the fundamental role of E56 and Y57 of pUL50 in the formation of the NEC. In addition, assembly of the entire multimeric NEC might be regulated by another mechanism. It is tempting to speculate that specific protein phosphorylation is essential for coordinated formation of the NEC. Major components of the NEC, such as pUL50, pUL53, and p32/gC1qR, are phosphorylated by pUL97 or cellular PKC (12, 33). PKC is additionally important for phosphorylation of emerin in HSV-1-infected cells (30), and pUL97 is capable of autophosphorylation (50–52). Thus, the coordinated fashion of NEC assembly might be regulated by phosphorylation of its constituents.
Another aspect of NEC assembly is the connection between different replicative multiprotein complexes formed in the HCMV-infected host nucleus. It appears plausible that herpesviral complexes involved in DNA replication, DNA encapsidation, capsid assembly, and nuclear egress might evolve from each other on the basis of their temporal association and dissociation (6, 53). Some of our results were in line with this hypothesis, notably the identification of virion-associated proteins pUL45 and pUL83 (Table I) only at early time points. Interestingly, the viral proteins pUL89 (MCP) and pUL85 were also enriched in these samples but were not considered significant according to the chosen cut-off limits of our quantitative proteomic analysis (3.2-fold enrichment of both proteins in NEC samples relative to controls; supplemental Table S3). The two proteins are constituents of viral capsids and therefore may provide a link between nuclear viral particles and the NEC as succeeding steps.
Differential NEC Assembly Comparing Human and Murine CMVs
The properties of NEC formation and function appear to be a conserved feature within the family Herpesviridae. All herpesviruses analyzed so far utilize a core NEC of pUL50–pUL53 homologs as a scaffold to assemble further NEC regulators and docking and adaptor proteins (7). However, a comparison between the findings of the present study on human CMV and a related study on murine CMV (54) revealed that the findings were only partly consistent. The central NEC components pUL50/pM50, pUL53/pM53, and p32/gC1qR were identified accordingly, but a huge number of NEC-associated proteins differed between the two systems. This may indicate that higher conservation can be assumed among human herpesviruses, whereas differences are seen between human and murine CMVs. The differences between the individual protein compositions of herpesviral NECs may indicate an analogy, rather than a homology, of proteins fulfilling NEC functions. In summary, our approach describes for the first time the composition of the native NEC isolated from HCMV-infected primary fibroblasts. This refined knowledge about the HCMV-specific NEC will support future studies on virus–host interaction, HCMV pathogenesis, and novel NEC-based antiviral strategies.
Supplementary Material
Acknowledgments
We thank Stipan Jonjic (University of Rijeka, Croatia), James Alwine (Department of Cancer Biology, University of Pennsylvania), Paola Dal Monte (University of Bologna, Italy), Donald M. Coen (Harvard Medical School, Boston, MA), William C. Russel (University of St Andrews, UK), William Britt, and Mark N. Prichard (University of Alabama) for providing antibodies against viral pUL53, pUL50, pUL97, and cellular p32/gC1qR. We are grateful to Eva Borst and Martin Messerle (Institute for Virology, Hannover Medical School, Germany), Karsten Tischer (Institute for Virology, Free University of Berlin, Germany), and Gregory Smith (Department of Microbiology-Immunology, Northwestern University of Chicago) for supplying us with materials and methods for BACmid recombination. We are grateful to Anna Greco (Université Lyon, France) and Thomas Stamminger for scientific support and to Hanife Bahsi and Isabel Zeitträger for excellent technical assistance. We thank Victoria Jackiw, Language Center of the University of Erlangen-Nuremberg, for reading the manuscript.
Footnotes
Author contributions: J.M., W.D.R., Y.C., and M.M. designed research; J.M., A.K., C.H., E.S., C.S., S.W., T.L., S.T.H., Y.C., and M.M. performed research; C.C. and S.P. contributed new reagents or analytic tools; J.M., A.K., C.H., M.F., H.S., Y.C., and M.M. analyzed data; J.M., H.S., Y.C., and M.M. wrote the paper; C.H. provided scientific support.
* This work has been supported by the PRIME-XS project (Grant No. 262067, funded by the European Union 7th Framework Programme), Research Foundation Medicine of Univ. Erlangen-Nuremberg (JM-2010), ELAN program of University of Erlangen-Nuremberg (Vi-12-10-29-1), Wilhelm Sander-Stiftung (2011.085.1), Bayerische Forschungsstiftung (ForBiMed/I1), and Deutsche Forschungsgemeinschaft (DFG MA 1289/7–1 and SFB 796 C3/A2).
 This article contains supplemental material.
This article contains supplemental material.
1 The abbreviations used are:
- HCMV
- human cytomegalovirus
- BAC
- bacterial artificial chromosome
- CoIP
- coimmunoprecipitation
- dpi
- days post-infection
- cVAC
- cytoplasmic virus assembly compartment
- HFF
- primary human foreskin fibroblast
- mAb
- monoclonal antibody
- MCP
- major capsid protein
- MOI
- multiplicity of infection
- NEC
- nuclear egress complex
- pAb
- polyclonal antiserum
- PKC
- protein kinase C
- SSC
- specific spectral count
- Wb
- Western blot.
REFERENCES
- 1. Pe'ery T., Mathews M. B. (2007) Viral Conquest of the Host Cell. In Fields Virology (Knipe D. M., Howley P. M., eds), pp. 169–208, Lippincott Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 2. Mocarski E. S., Shenk T., Pass R. F. (2007) Cytomegaloviruses. In Fields Virology (Knipe D. M., Howley P. M., eds), pp. 2701–2772, Lippincott Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 3. Mettenleiter T. C., Müller F., Granzow H., Klupp B. G. (2013) The way out: what we know and do not know about herpesvirus nuclear egress. Cell. Microbiol. 15, 170–178 [DOI] [PubMed] [Google Scholar]
- 4. Marschall M., Feichtinger S., Milbradt J. (2011) Regulatory roles of protein kinases in cytomegalovirus replication. Adv. Virus Res. 80, 69–101 [DOI] [PubMed] [Google Scholar]
- 5. Johnson D. C., Baines J. D. (2011) Herpesviruses remodel host membranes for virus egress. Nat. Rev. Microbiol. 9, 382–394 [DOI] [PubMed] [Google Scholar]
- 6. Tandon R., Mocarski E. S. (2012) Viral and host control of cytomegalovirus maturation. Trends Microbiol. 20, 392–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lee C. P., Chen M. R. (2010) Escape of herpesviruses from the nucleus. Rev. Med. Virol. 20, 214–230 [DOI] [PubMed] [Google Scholar]
- 8. Milbradt J., Webel R., Auerochs S., Sticht H., Marschall M. (2010) Novel mode of phosphorylation-triggered reorganization of the nuclear lamina during nuclear egress of human cytomegalovirus. J. Biol. Chem. 285, 13979–13989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Milbradt J., Auerochs S., Sevvana M., Muller Y. A., Sticht H., Marschall M. (2012) Specific residues of a conserved domain in the N terminus of the human cytomegalovirus pUL50 protein determine its intranuclear interaction with pUL53. J. Biol. Chem. 287, 24004–24016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Buchkovich N. J., Maguire T. G., Alwine J. C. (2010) Role of the endoplasmic reticulum chaperone BiP, SUN domain proteins, and dynein in altering nuclear morphology during human cytomegalovirus infection. J. Virol. 84, 7005–7017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sam M. D., Evans B. T., Coen D. M., Hogle J. M. (2009) Biochemical, biophysical, and mutational analyses of subunit interactions of the human cytomegalovirus nuclear egress complex. J. Virol. 83, 2996–3006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Milbradt J., Auerochs S., Marschall M. (2007) Cytomegaloviral proteins pUL50 and pUL53 are associated with the nuclear lamina and interact with cellular protein kinase C. J. Gen. Virol. 88, 2642–2650 [DOI] [PubMed] [Google Scholar]
- 13. Schnee M., Ruzsics Z., Bubeck A., Koszinowski U. H. (2006) Common and specific properties of herpesvirus UL34/UL31 protein family members revealed by protein complementation assay. J. Virol. 80, 11658–11666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Milbradt J., Auerochs S., Sticht H., Marschall M. (2009) Cytomegaloviral proteins that associate with the nuclear lamina: components of a postulated nuclear egress complex. J. Gen. Virol. 90, 579–590 [DOI] [PubMed] [Google Scholar]
- 15. Schmeiser C., Borst E., Sticht H., Marschall M., Milbradt J. (2013) The cytomegalovirus egress proteins pUL50 and pUL53 are translocated to the nuclear envelope through two distinct modes of nuclear import. J. Gen. Virol. 94, 2056–2069 [DOI] [PubMed] [Google Scholar]
- 16. Dal Monte P., Pignatelli S., Zini N., Maraldi N. M., Perret E., Prevost M. C., Landini M. P. (2002) Analysis of intracellular and intraviral localization of the human cytomegalovirus UL53 protein. J. Gen. Virol. 83, 1005–1012 [DOI] [PubMed] [Google Scholar]
- 17. Dupierris V., Masselon C., Court M., Kieffer-Jaquinod S., Bruley C. (2009) A toolbox for validation of mass spectrometry peptides identification and generation of database: IRMa. Bioinformatics 25, 1980–1981 [DOI] [PubMed] [Google Scholar]
- 18. Casabona M. G., Vandenbrouck Y., Attree I., Couté Y. (2013) Proteomic characterization of Pseudomonas aeruginosa PAO1 inner membrane. Proteomics 13, 2419–2423 [DOI] [PubMed] [Google Scholar]
- 19. Pham T. V., Piersma S. R., Warmoes M., Jimenez C. R. (2010) On the beta-binomial model for analysis of spectral count data in label-free tandem mass spectrometry-based proteomics. Bioinformatics 26, 363–369 [DOI] [PubMed] [Google Scholar]
- 20. Thomas M., Rechter S., Milbradt J., Auerochs S., Müller R., Stamminger T., Marschall M. (2009) Cytomegaloviral protein kinase pUL97 interacts with the nuclear mRNA export factor pUL69 to modulate its intranuclear localization and activity. J. Gen. Virol. 90, 567–578 [DOI] [PubMed] [Google Scholar]
- 21. Marschall M., Freitag M., Suchy P., Romaker D., Kupfer R., Hanke M., Stamminger T. (2003) The protein kinase pUL97 of human cytomegalovirus interacts with and phosphorylates the DNA polymerase processivity factor pUL44. Virology 311, 60–71 [DOI] [PubMed] [Google Scholar]
- 22. Tischer B. K., von Einem J., Kaufer B., Osterrieder N. (2006) Two-step red-mediated recombination for versatile high-efficiency markerless DNA manipulation in Escherichia coli. BioTechniques 40, 191–197 [DOI] [PubMed] [Google Scholar]
- 23. Tischer B. K., Smith G. A., Osterrieder N. (2010) En passant mutagenesis: a two step markerless red recombination system. Methods Mol. Biol. 634, 421–430 [DOI] [PubMed] [Google Scholar]
- 24. Sampaio K. L., Cavignac Y., Stierhof Y. D., Sinzger C. (2005) Human cytomegalovirus labeled with green fluorescent protein for live analysis of intracellular particle movements. J. Virol. 79, 2754–2767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Claus C., Chey S., Heinrich S., Reins M., Richardt B., Pinkert S., Fechner H., Gaunitz F., Schäfer I., Seibel P., Liebert U. G. (2011) Involvement of p32 and microtubules in alteration of mitochondrial functions by rubella virus. J. Virol. 85, 3881–3892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kamil J. P., Coen D. M. (2007) Human cytomegalovirus protein kinase UL97 forms a complex with the tegument phosphoprotein pp65. J. Virol. 81, 10659–10668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Matthews D. A., Russell W. C. (1998) Adenovirus core protein V interacts with p32—a protein which is associated with both the mitochondria and the nucleus. J. Gen. Virol. 79, 1677–1685 [DOI] [PubMed] [Google Scholar]
- 28. Gill R. B., James S. H., Prichard M. N. (2012) Human cytomegalovirus UL97 kinase alters the accumulation of CDK1. J. Gen. Virol. 93, 1743–1755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Webel R., Solbak S. M., Held C., Milbradt J., Groβ A., Eichler J., Wittenberg T., Jardin C., Sticht H., Fossen T., Marschall M. (2012) Nuclear import of isoforms of the cytomegalovirus kinase pUL97 is mediated by differential activity of NLS1 and NLS2 both acting through classical importin-α binding. J. Gen. Virol. 93, 1756–1768 [DOI] [PubMed] [Google Scholar]
- 30. Leach N. R., Roller R. J. (2010) Significance of host cell kinases in herpes simplex virus type 1 egress and lamin-associated protein disassembly from the nuclear lamina. Virology 406, 127–137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Leach N., Bjerke S. L., Christensen D. K., Bouchard J. M., Mou F., Park R., Baines J., Haraguchi T., Roller R. J. (2007) Emerin is hyperphosphorylated and redistributed in herpes simplex virus type 1-infected cells in a manner dependent on both UL34 and US3. J. Virol. 81, 10792–10803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Morris J. B., Hofemeister H., O'Hare P. (2007) Herpes simplex virus infection induces phosphorylation and delocalization of emerin, a key inner nuclear membrane protein. J. Virol. 81, 4429–4437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Marschall M., Marzi A., aus dem Siepen P., Jochmann R., Kalmer M., Auerochs S., Lischka P., Leis M., Stamminger T. (2005) Cellular p32 recruits cytomegalovirus kinase pUL97 to redistribute the nuclear lamina. J. Biol. Chem. 280, 33357–33367 [DOI] [PubMed] [Google Scholar]
- 34. Brokstad K. A., Kalland K. H., Russell W. C., Matthews D. A. (2001) Mitochondrial protein p32 can accumulate in the nucleus. Biochem. Biophys. Res. Commun. 281, 1161–1169 [DOI] [PubMed] [Google Scholar]
- 35. van Leeuwen H. C., O'Hare P. (2001) Retargeting of the mitochondrial protein p32/gC1Qr to a cytoplasmic compartment and the cell surface. J. Cell Sci. 114, 2115–2123 [DOI] [PubMed] [Google Scholar]
- 36. Yagi M., Uchiumi T., Takazaki S., Okuno B., Nomura M., Yoshida S., Kanki T., Kang D. (2012) p32/gC1qR is indispensable for fetal development and mitochondrial translation: importance of its RNA-binding ability. Nucleic Acids Res. 40, 9717–9737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sharma M., Kamil J. P., Coughlin M., Reim N. I., Coen D. M. (2014) Human cytomegalovirus UL50 and UL53 recruit viral protein kinase UL97, not protein kinase C, for disruption of nuclear lamina and nuclear egress in infected cells. J. Virol. 88, 249–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Muranyi W., Haas J., Wagner M., Krohne G., Koszinowski U. H. (2002) Cytomegalovirus recruitment of cellular kinases to dissolve the nuclear lamina. Science 297, 854–857 [DOI] [PubMed] [Google Scholar]
- 39. Hamilton S. T., Milbradt J., Marschall M., Rawlinson W. D. (2014) Human cytomegalovirus replication is strictly inhibited by siRNAs targeting UL54, UL97 or UL122/123 gene transcripts. PLoS One 9, e97231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Alwine J. C. (2012) The human cytomegalovirus assembly compartment: a masterpiece of viral manipulation of cellular processes that facilitates assembly and egress. PLoS Pathog. 8, e1002878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Das S., Pellett P. E. (2011) Spatial relationships between markers for secretory and endosomal machinery in human cytomegalovirus-infected cells versus those in uninfected cells. J. Virol. 85, 5864–5879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Das S., Vasanji A., Pellett P. E. (2007) Three-dimensional structure of the human cytomegalovirus cytoplasmic virion assembly complex includes a reoriented secretory apparatus. J. Virol. 81, 11861–11869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Jensen L. J., Bork P. (2008) Biochemistry. Not comparable, but complementary. Science 322, 56–57 [DOI] [PubMed] [Google Scholar]
- 44. Farina A., Santarelli R., Bloise R., Gonnella R., Granato M., Bei R., Modesti A., Cirone M., Bengtsson L., Angeloni A., Faggioni A. (2013) KSHV ORF67 encoded lytic protein localizes on the nuclear membrane and alters emerin distribution. Virus Res. 175, 143–150 [DOI] [PubMed] [Google Scholar]
- 45. Cai M., Huang Y., Ghirlando R., Wilson K. L., Craigie R., Clore G. M. (2001) Solution structure of the constant region of nuclear envelope protein LAP2 reveals two LEM-domain structures: one binds BAF and the other binds DNA. EMBO J. 20, 4399–4407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Cai M., Huang Y., Suh J. Y., Louis J. M., Ghirlando R., Craigie R., Clore G. M. (2007) Solution NMR structure of the barrier-to-autointegration factor-emerin complex. J. Biol. Chem. 282, 14525–14535 [DOI] [PubMed] [Google Scholar]
- 47. Laguri C., Gilquin B., Wolff N., Romi-Lebrun R., Courchay K., Callebaut I., Worman H. J., Zinn-Justin S. (2001) Structural characterization of the LEM motif common to three human inner nuclear membrane proteins. Structure 9, 503–511 [DOI] [PubMed] [Google Scholar]
- 48. Haraguchi T., Koujin T., Segura-Totten M., Lee K. K., Matsuoka Y., Yoneda Y., Wilson K. L., Hiraoka Y. (2001) BAF is required for emerin assembly into the reforming nuclear envelope. J. Cell Sci. 114, 4575–4585 [DOI] [PubMed] [Google Scholar]
- 49. Hamirally S., Kamil J. P., Ndassa-Colday Y. M., Lin A. J., Jahng W. J., Baek M. C., Noton S., Silva L. A., Simpson-Holley M., Knipe D. M., Golan D. E., Marto J. A., Coen D. M. (2009) Viral mimicry of Cdc2/cyclin-dependent kinase 1 mediates disruption of nuclear lamina during human cytomegalovirus nuclear egress. PLoS Pathog. 5, e1000275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Michel D., Kramer S., Höhn S., Schaarschmidt P., Wunderlich K., Mertens T. (1999) Amino acids of conserved kinase motifs of cytomegalovirus protein UL97 are essential for autophosphorylation. J. Virol. 73, 8898–8901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Baek M. C., Krosky P. M., Coen D. M. (2002) Relationship between autophosphorylation and phosphorylation of exogenous substrates by the human cytomegalovirus UL97 protein kinase. J. Virol. 76, 11943–11952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Marschall M., Stein-Gerlach M., Freitag M., Kupfer R., van den Bogaard M., Stamminger T. (2002) Direct targeting of human cytomegalovirus protein kinase pUL97 by kinase inhibitors is a novel principle for antiviral therapy. J. Gen. Virol. 83, 1013–1023 [DOI] [PubMed] [Google Scholar]
- 53. Vizoso Pinto M. G., Pothineni V. R., Haase R., Woidy M., Lotz-Havla A. S., Gersting S. W., Muntau A. C., Haas J., Sommer M., Arvin A. M., Baiker A. (2011) Varicella zoster virus ORF25 gene product: an essential hub protein linking encapsidation proteins and the nuclear egress complex. J. Proteome Res. 10, 5374–5382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Lemnitzer F., Raschbichler V., Kolodziejczak D., Israel L., Imhof A., Bailer S. M., Koszinowski U., Ruzsics Z. (2013) Mouse cytomegalovirus egress protein pM50 interacts with cellular endophilin-A2. Cell. Microbiol. 15, 335–351 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.