Abstract
In 2010, a large outbreak of rotavirus gastroenteritis occurred in the Alice Springs region of the Northern Territory, Australia. The outbreak occurred 43 months after the introduction of the G1P[8] rotavirus vaccine Rotarix®. Forty-three infants were hospitalized during the outbreak and analysis of fecal samples from each infant revealed a G1P[8] rotavirus strain. The outbreak strain was adapted to cell culture and neutralization assays were performed using VP7 and VP4 neutralizing monoclonal antibodies. The outbreak strain exhibited a distinct neutralization resistance pattern compared to the Rotarix® vaccine strain. Whole genome sequencing of the 2010 outbreak virus strain demonstrated numerous amino acid differences compared to the Rotarix® vaccine strain in the characterized neutralization epitopes of the VP7 and VP4 proteins. Phylogenetic analysis of the outbreak strain revealed a close genetic relationship to global strains, in particular RVA/Human-wt/BEL/BE0098/2009/G1P[8] and RVA/Human-wt/BEL/BE00038/2008/G1P[8] for numerous genes. The 2010 outbreak strain was likely introduced from a globally circulating population of strains rather than evolving from an endemic Australian strain. The outbreak strain possessed antigenic differences in the VP7 and VP4 proteins compared to the Rotarix® vaccine strain. The outbreak was associated with moderate vaccine coverage and possibly low vaccine take in the population.
Keywords: Australia; diarrheal outbreak; G1P[8]; full-genome analysis; rotavirus, Rotarix®
INTRODUCTION
Rotavirus is the predominant cause of acute gastroenteritis in young children worldwide.1 A high burden of rotavirus disease is present in both developing and developed countries resulting in an estimated 453 000 annual deaths, principally in Asia and sub-Saharan Africa.1,2 Rotavirus belongs to the Reoviridae virus family and is a non-enveloped, icosahedral virus and the 11 segment double-stranded RNA (dsRNA) genome encodes six structural viral proteins (VP1–4, VP6, VP7) and six non-structural proteins (NSP1–5/6).3 Rotavirus strains can be classified into eight groups (groups A–H) based on the genetic characteristics of the inner capsid protein (VP6); group A strains are the most common cause of rotavirus disease in humans.4
Rotavirus strains can be further classified based on the two outer capsid proteins into G (glycoprotein, VP7) and P (protease-sensitive, VP4) genotypes respectively; these proteins also elicit type-specific and cross-reactive neutralizing antibody responses.3 A genotyping classification system based on the open reading frame of each gene has been adopted; Gx–P[x]–Ix–Rx–Cx–Mx–Ax–Nx–Tx–Ex–Hx.5 To date, 27 G (VP7), 37 P (VP4), 17 I (VP6), 9 R (VP1), 9 C (VP2), 8 M (VP3), 18 A (NSP1), 10 N (NSP2), 12 T (NSP3), 15 E (NSP4) and 11 H (NSP5) genotypes have been described.5,6,7,8,9
Two live-oral rotavirus vaccines; Rotarix® (GlaxoSmithKline Vaccines, Rixensart, Belgium) and RotaTeq® (Merck and Co. Inc., Whitehouse Station, NJ, USA) have been demonstrated to be efficacious in large clinical trials and are included in vaccination programs of numerous countries worldwide.10,11,12 Rotarix® and RotaTeq® have been highly efficacious in decreasing the burden of rotavirus gastroenteritis in several countries worldwide including Brazil, Belgium, the United States, Nicaragua, Austria and Mexico.13,14,15,16,17 Rotarix® is a live-attenuated monovalent vaccine comprised of a single G1P[8] strain and is administered in a two-dose schedule at 2 and 4 months of age.18 RotaTeq® is a live-attenuated, pentavalent, human-bovine reassortant vaccine administered in a three-dose schedule at 2, 4 and 6 months of age.19
Rotavirus vaccines were introduced into the Australian National Immunization Program in July 2007. Rotarix® was introduced in the Northern Territory in October 2006 due to the high burden of rotavirus disease experienced in the region, particularly in Indigenous infants.20 Vaccine introduction has decreased the burden of rotavirus disease in several locations around Australia.20 The sole exception is Central Australia a large region of the Northern Territory encompassing Alice Springs and surrounding communities with a high Indigenous Australian population.21 In Central Australia there has been no clear decline in rotavirus notification rates despite the introduction of routine infant vaccination with Rotarix®.21,22 Outbreaks of rotavirus gastroenteritis have continued to occur in Central Australia in the vaccine era, particularly affecting the town of Alice Springs and surrounding communities which place a high demand on health-care facilities.21
During May and June 2010, a large G1P[8] rotavirus outbreak occurred in Alice Springs that resulted in a considerable number of children hospitalized with severe gastroenteritis. The aim of this study was to perform a genetic and antigenic characterization of the G1P[8] rotavirus strain responsible for the outbreak. A comparison of the outbreak strain to other Australian and international G1P[8] strains was performed to establish its context within the global rotavirus population.
MATERIALS AND METHODS
Stool specimens
A total of 43 fecal samples were collected from infants (≤38 months of age) presenting to hospital with severe gastroenteritis during a rotavirus outbreak in the Alice Springs region of the Northern Territory between 10 May and 15 June 2010. Patient information including date of birth, date of sample collection and gender was routinely collected. The length of hospitalization, immunization status with regard to the Rotarix® rotavirus vaccine was obtained where possible. Fecal samples were frozen, stored at −70°C and forwarded to the Australian Rotavirus Reference Centre in Melbourne, Victoria. G1P[8] samples collected in 2010 were analyzed from Darwin (n=20), Gove (n=12), Katherine (n=26), Tennant Creek (n=16) and remote regions of the Northern Territory and Western Australia (n=28). Additional G1P[8] samples circulating in neighbouring states South Australia (n=7), Queensland (n=2) and Western Australia (n=5) in 2010 were also analyzed.
Nucleic acid extraction
Rotavirus dsRNA was extracted from clarified 20% (w/v) fecal suspensions using a RNA extraction kit (QIAamp® Viral RNA mini kit (spin protocol), Qiagen, Inc., Hilden, Germany) in accordance with the manufacturer's instructions.
Polyacrylamide gel electrophoresis
The 11 segments of dsRNA were separated on 10% (w/v) polyacrylamide gel with 3% (w/v) polyacrylamide stacking gel at 25 mA for 16 h. The genome migration patterns (electropherotypes) were visualized by silver staining according to the established protocol.23,24
Viruses and adaptation of strains to MA104 culture
The 2010 G1P[8] outbreak strain was adapted to culture in MA104 cells. Filtered 20% (w/v) fecal extracts were activated with 10 µg/mL porcine trypsin (Sigma, St Louis, MO, USA) at 37°C for 30 min and inoculated onto 2.5×106 MA104 cells in DMEM supplemented with 1 µg/mL porcine trypsin (Sigma). Cultures were incubated at 37°C with 5% CO2 and maintained in suspension culture using a rotary mixer. At 24 h post inoculation, an additional 1×106 MA104 cells were added to the suspension culture. At 96 h post inoculation, the virus was released by three cycles of freeze–thaw at −80°C and clarified by centrifugation at 4000g for 10 min. The viruses underwent serial passage in suspension for six passages and were adapted to stationary phase by three passages in MA104 cells. The electropherotypes of the adapted strains were compared to the pattern derived from the original stool sample. The VP7 and VP4 genes of the adapted virus was sequenced as previously described to ensure conservation of protein sequence following adaptation.25 Virus titer was monitored during passage using indirect immunofluorescence in MA104 cells.26 The standard human rotavirus strains RV4 (G1P[8]), F45 (G9P[8]) and D (G1P[8]), as well as Rotarix®, were propagated in MA104 cells in the presence of trypsin.
Neutralization of strains with monoclonal antibody and polyclonal antisera
Adapted 2010 G1P[8] outbreak virus and control viruses were studied in a fluorescent focus reduction neutralization assay with neutralizing monoclonal antibodies (N-MAbs) and polyclonal sera, as described previously.26 Four rotavirus N-MAbs RV4:1, RV4:2, RV4:3 and RV4:5, reactive with the VP7 antigen of human G1P[8] rotavirus RV4, were used.27 The VP4-specific N-MAb F45:4 reactive to P[8] antigen of F45 was also used.28,29 Rabbit hyperimmune antisera raised to RV4 was also used.27 The criteria for resistance to neutralization was defined as a reduction in neutralization titer of at least 1 log, when compared to the homologous virus titer.
Amplification and nucleotide sequencing of the rotavirus genome segments
The extracted dsRNAs of five representative G1P[8] 2010 outbreak samples were sent to the J Craig Venter Institute (Rockville, MD, USA) for high-throughput reverse transcription-polymerase chain reaction and Sanger sequencing as previously described.30 Briefly, reverse transcription-polymerase chain reaction primers were designed at 600 bp intervals along the sense and antisense RNA strand of each gene to ensure high coverage by reverse transcription-polymerase chain reaction.
Phylogenetic analysis
Nucleotide similarity searches were conducted using the Basic Local Alignment Search Tool server on the GenBank database at the National Center for Biotechnology Information, Bethesda, MD, USA (http://www.ncbi.nlm.nih.gov). The nucleotide (nt) and deduced amino acid (aa) sequences of each gene were compared with sequences available in the GenBank database that possessed the entire open reading frame and multiple alignments constructed using the MUSCLE algorithm in the MEGA5.20 program.31,32 The optimal evolutionary model was selected for each gene based upon the Akaike information criterion (corrected) ranking implemented in jModelTest.33,34 Maximum likelihood phylogenetic trees using the models of nucleotide substitution GTR+I+GG4 (VP1, VP2, VP3, NSP1 and NSP3), GTR+GG4 (VP4 and VP6), TrN+GG4 (VP7, NSP2 and NSP4) and HKY+GG4 (NSP5) were generated using MEGA5.20.31 The robustness of branches was assessed by bootstrap analysis with 1000 pseudoreplicate runs. Nucleotide and amino acid distance matrixes were calculated using the P-distance algorithm in MEGA5.20.31 Structural analysis of the VP7 protein (PDB ID: 3FMG) was performed using PyMOL.35
Assignment of genotypes
The genotypes of each of the 11 genome segments of the 2010 G1P[8] outbreak strains were determined using the online rotavirus genotyping tool RotaC v2.0 (http://rotac.regatools.be).36
Accession numbers
The nucleotide sequences for genes described in this study have been deposited in GenBank under the accession numbers RVA/Human-wt/AUS/CK00096/2010/G1P[8] (JX027934–JX027944), RVA/Human-wt/AUS/CK00097/2010/G1P[8] (JX027945–JX027955), RVA/Human-wt/AUS/CK00099/2010/G1P[8] (JX027956–JX027966) and RVA/Human-wt/AUS/CK00100/2010/G1P[8] (JX027967–JX027977). For simplicity, the strains will be referred to by their common name CK00096, CK00097, CK00098, CK00099 and CK00100.
RESULTS
Sample characterization
G1P[8] (n=43) samples were collected from patients hospitalized with severe gastroenteritis during the 2010 Alice Springs rotavirus outbreak. The average age of infants was 5.9 months (1–38 months), and 53.3% of patients were male. Based on age, 42/43 infants were eligible to be vaccinated with at least the primary dose of Rotarix®. A single patient was too young to be vaccinated. Vaccination status was available for 29 patients, seven patients had received the first dose of Rotarix® and ten patients had received both doses. Five patients did not receive the second dose despite being eligible. Seven patients had not been vaccinated despite being of an eligible age. The remaining 13 patients had no vaccination history recorded and were assumed to not have been vaccinated.
Polyacrylamide gel electrophoresis
All Alice Springs samples analyzed during the outbreak had an identical electropherotype and strains possessing the outbreak electropherotype circulated in the region until mid-August, 2010. Strains analyzed from other regions of the Northern Territory (excluding Gove) and remote Western Australia exclusively possessed the outbreak electrotype detected between May and July 2010. The outbreak electropherotype was also observed in a single South Australian, a single Queensland and three Western Australian samples circulating in June/July 2010.
Neutralization of 2010 G1P[8] outbreak samples
Fluorescent focus reduction neutralization assay was used to identify potential antigenic differences between a culture adapted isolate (V474) from the 2010 G1P[8] outbreak, Rotarix® and control viruses. A panel of VP7 N-MAbs and polyclonal sera derived against RV4 (G1P[8]) and N-MAb derived to VP4 of F45 (G9P[8]) were used to determine neutralization profiles (Table 1). The 2010 G1P[8] outbreak virus was neutralized with the N-MAbs RV4:1, RV4:2, F45:4; however, it was resistant to neutralization by the VP7 MAbs RV4:3 and RV4:5, and rabbit anti-RV4 sera. In contrast, Rotarix® was neutralized by the VP7 N-MAbs RV4:1, RV4:2, RV4:3, RV4:5 and the rabbit RV4 polyclonal sera, but was resistant to neutralization by the VP4 N-MAb F45:4. The distinct neutralization patterns of the 2010 G1P[8] outbreak virus and Rotarix® demonstrate differences in the antigenic profile of these viruses, suggesting alterations in VP7 and VP4 proteins.
Table 1. Fluorescent focus reduction neutralization assay.
| Strain | RV4:1 | RV4:2 | RV4:3 | RV4:5 | F45:4 | RV4 (poly) |
|---|---|---|---|---|---|---|
| RV4 | 720 000 | 125 000 | 210 000 | 125 000 | 12 000 | 78 000 |
| D | 41 000 | 3783 | 330 000 | <100 | ND | 59 000 |
| F45 | ND | ND | ND | ND | 1800 | ND |
| V474 | 137 000 | 29 333 | <100 | 327 | 30 000 | 230 |
| ROTARIX | 295 000 | 4867 | 203 667 | 2417 | 593 | 40 667 |
Abbreviation: ND, not done.
Neutralization titers are reported as the reciprocal dilution, which results in a 50% fluorescent focus reduction. Titers are representative of three independent assays.
Comparison of the Alice Springs 2010 G1P[8] outbreak strain to Rotarix® vaccine VP7 and VP4 genes
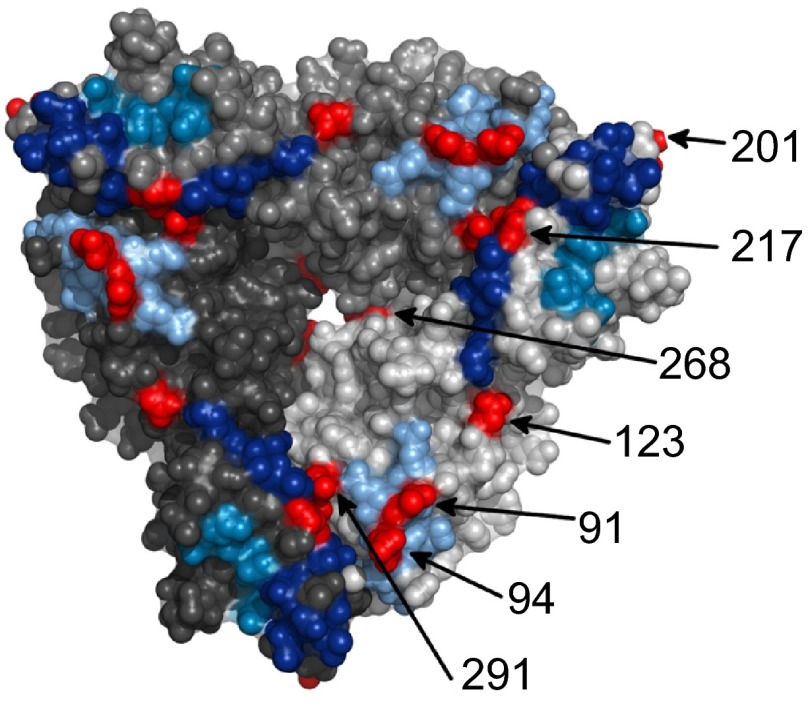
The VP7 gene of the 2010 outbreak samples possessed 94.2% nt and 95.7% aa identity the VP7 gene of Rotarix®. The amino acid differences between the outbreak strains and Rotarix® were analyzed and mapped to the VP7 trimer (Figure 1), identifying several changes in regions of biological function (Figure 2). Several changes in the VP7 protein were located within antigenic regions, T91N and N94S in antigenic region A and M217T in antigenic region C. An additional change was identified at position K291R previously identified in neutralization escape mutants.27 The N94S change identified in the outbreak virus correlated with the loss of neutralization by N-MAb RV4:3 and RV4 polyclonal sera.27 Similarly, the outbreak virus was resistant to neutralization with RV4:5 when compared to Rotarix®. The N-MAb RV4:5 requires the sequence asparagine-lysine at position 147–148 in antigenic region B for neutralization, plus additional unidentified amino acids.27 Resistance to RV4:5 by strain D correlates with the N147S change in antigenic region B (Figure 2). The outbreak strain is resistant to RV4:5, despite a conserved amino acid sequence in antigenic region B when compared to RV4 and Rotarix®. This suggests that additional surface exposed amino acids, potentially the S123N change, are responsible neutralization resistance.
Figure 1.
A surface representation of the VP7 trimer (PDB ID: 3FMG). Each trimer is colored a different shade of gray and the antigenic regions A, C and F are colored light blue, dark blue and mid blue, respectively. The surface exposed amino acid residues that differ between the 2010 G1P[8] Alice Springs outbreak strain and Rotarix® vaccine strain are shown in red.
Figure 2.

Alignment of VP7 gene of the prototype G1P[8] strains RV4, D, the 2010 G1P[8] Alice Springs outbreak strain and Rotarix® vaccine strain. Amino acid differences between these strains are shaded. Residues comprising the antigenic regions are defined within in brackets. The sites shown to escape neutralization with monoclonal antibodies include RV4:1 (sites 147 and 148, filled circle), RV4:2 (sites 213, filled square), RV4:3 (site 94, filled triangle) and RV4:5: (sites 147, 148 and other undefined sites, filled right pointing triangle).
The VP4 genes of the 2010 outbreak samples possessed 90.2%–90.3% nt and 94.5%–94.6% aa identity to the Rotarix® vaccine strain. The VP4 protein undoes proteolytic cleavage by trypsin into two subunits; VP8* (aa 1–247) and VP5* (aa 248–776) which enhances viral infectivity.3 The Y385D change that differentiated Rotarix® and the outbreak virus was identified within the hydrophobic apex of VP5*, a conformationally dependent antigenic region.37,38 The change at position 385 correlates with neutralization resistance of Rotarix® with N-MAb F45:4, which selects neutralization escape mutants at position 392 within this antigenic region of VP5*.29
Whole genome analysis of the Alice Springs 2010 G1P[8] outbreak samples
Five representative strains collected during the outbreak were selected for whole genome sequence analysis and possessed the archetypal Wa-like genome constellation G1-P[8]–I1–R1–C1–M1–A1–N1–T1–E1–H1. A high-quality sequence read could not be achieved for CK00098 and the genome was not included in the phylogenetic analysis. CK00096, CK00097, CK00099 and CK00100 shared 100% nt and aa identity for all genes except, NSP1 (99.9% nt and 99.8% aa), NSP2 and VP4 (99.9% nt and 99.7% aa).
Phylogenetic analysis of the Alice Springs 2010 G1P[8] outbreak samples
Phylogenetic analysis of each of the 11 gene segments was conducted to investigate the genetic relationship of the outbreak strain to Australian and global strains. In each tree, the 2010 G1P[8] outbreak strains clustered within large, diverse clades that were comprised of contemporary global isolates (Supplementary Figures S1, S2 and S3).
The VP7 gene of the outbreak strain shared the highest genetic identity (99.9% nt and 99.7% aa) to numerous strains circulating in Asia between 2004 and 2010 and RVA/Human-wt/BEL/BE00098/2009/G1P[8]. The outbreak strains shared the highest genetic similarity to RVA/Human-wt/BEL/BE00098/2009/G1P[8] for the VP4 (99.7% nt and aa), VP2 (99.9% nt and aa), VP6 (99.7% nt and 100% aa), NSP1 (99.7% nt and 99.8% aa) and NSP3 (99.9% nt and 100% aa) genes.
The outbreak strain shared the highest genetic similarity to RVA/Human-wt/BEL/BE00038/2008/G1P[8] for VP1 (99.8% nt and 99.9% aa) and VP3 (99.8% nt and 99.8% aa). The NSP2 gene shared 99.7% nt and aa similarity to RVA/Human-wt/BEL/BE00098/2009/G1P[8], RVA/Human-wt/USA/VU-06-07-21/2006/G3P[8] and RVA/Human-wt/USA/2007719674/2007/G1P[8], while the NSP4 gene shared 99.6% nt and 100% aa similarity to RVA/Human-wt/USA/VU-08-09-24/2008/G3P[8], RVA/Human-wt/USA/2007719674/2007/G1P[8] and RVA/Human-wt/RUS/Nov08-3281/2008/G3P[8]. The NSP5 gene shared 99.8% nt and 99.5% aa similarity to RVA/Human-wt/THA/CU875-BK/2010/G1P[8], RVA/Human-wt/USA/2009726997/2009/G3P[8] and RVA/Human-wt/USA/VU08-09-7/2008/G3P[8].
DISCUSSION
The inclusion of rotavirus vaccines onto the Australian National Immunization Program has decreased rotavirus associated hospitalizations, emergency room visits and episodes of gastroenteritis in several regions.20 In contrast, vaccination has not been associated with decreased rotavirus notifications in Central Australia; a large region of the Northern Territory encompassing Alice Springs and surrounding communities with a high Indigenous Australian population.21 We report the characterization of a G1P[8] rotavirus outbreak that occurred in Alice Springs between May and June 2010, which resulted in the hospitalization of 43 patients. This is the first G1P[8] outbreak in the Northern Territory following the introduction of the homotypic Rotarix® vaccine. Electropherotype and sequence analysis identified that a single G1P[8] strain was responsible for the outbreak. The strain was also observed to be circulating in high numbers in other regions of the Northern Territory (Darwin, Katherine and Tennant Creek); 500–1500 km from Alice Springs. The 2010 G1P[8] outbreak strain was also circulating in low numbers in Queensland, South Australia and Western Australia in the months following the outbreak. However, routine surveillance in the 2010–2011 period revealed that G1P[8] strains accounted for 26.5% of strains identified Australia-wide and no state (regardless of vaccine used) experienced a significant detection of G1P[8] strains except the Northern Territory, due to the continued circulation of the outbreak strain in the region.39
Both the VP7 and VP4 proteins are involved in eliciting protective immunity and the production of neutralizing antibodies.3 Antigenic differences were identified between the outbreak G1P[8] strain and Rotarix® in both VP7 and VP4 using N-MAbs, correlating with amino acid changes in known neutralization epitopes. Similar changes in antigenic regions of VP7 and VP4 have been previously reported in other contemporary G1P[8] strains circulating in Belgium and globally.40 Most neutralizing antibodies to the parental strain of Rotarix® (89-12) are to the VP4 protein; five VP4 amino acid changes (G51D, L167F, S331F, D385Y and N695I) identified during vaccine attenuation are thought to significantly reduce serum neutralization titers following Rotarix® vaccination when compared to infection with the parental 89-12 strain.41 One VP4 amino acid change identified during 89-12 attenuation is D385Y, a site selected in neutralization escape mutants of the human rotavirus KU (G1P[8]).41,42 The Y385D change which differentiates Rotarix® and the outbreak G1P[8] strain suggests similar antigenic changes in VP4 as a potential mechanism for neutralization escape. However, preliminary data using sera from children who seroconverted to Rotarix® indicate that it is able to neutralize the G1P[8] outbreak strain (unpublished observations). In addition, multiple other viral antigens including VP2, VP6, NSP2 and NSP4 are thought to be important in rotavirus immunity.43
Phylogenetic analysis of the 2010 outbreak samples identified the strain was similar to globally circulating G1P[8] strains, forming large diverse clusters with contemporary G1P[8] strains isolated from numerous countries. The outbreak strain shared the highest degree of genetic identity to G1P[8] strains circulating in Belgium in 2008 and 2009 for several genes. Individual genes also shared a high degree of genetic identity to G3P[8] strains isolated in America (2006–2009), Russia (2008) and a G1P[8] strain isolated in Thailand in 2010. Interestingly, the VP7 gene shared the highest genetic relatedness to a widely circulating gene identified in Hong Kong, Japan and China between 2004, 2006–2008 and 2010, as well as Belgium. The finding that the 2010 G1P[8] outbreak strain shared a higher degree of genetic relatedness to global strains than to previously characterized Australian G1P[8] strains suggests that this strain was recently introduced from a global population rather than evolving from an endemic Australian strain.
Rotarix® vaccine effectiveness (VE) has varied in Alice Springs during previous rotavirus outbreaks. During the 2009 G9P[8] outbreak, VE for two doses against all hospitalizations for gastroenteritis was 77.7% (95% confidence interval, 40.2%–91.7%).44 A lower two-dose VE of 19% (95% confidence interval: −105%–68%) was observed during a 2009 G2P[4] outbreak.45 While the VE in the current outbreak has not been determined, the identification of a homotypic G1P[8] strain among fully vaccinated individuals suggests a possibly reduced VE in this period. In 2010, the reported two-dose Rotarix® vaccine coverage in the Northern Territory was 78.4%, considerably lower than other Australian states (83.2%–88.0%), with only 71.1% of Indigenous infants vaccinated compared to 85.1% of non-Indigenous infants.21,46 Vaccine coverage of children hospitalized with rotavirus gastroenteritis during the outbreak was low, with 50% of eligible infants vaccinated.
The overall efficacy of Rotarix® has been lower in South Africa and Malawi compared to European and Latin American studies, correlating with lower anti-rotavirus seroconversion and IgA titer following vaccination.47,48 In addition, the homotypic and heterotypic immunity acquired following natural rotavirus infections is less in settings with a high disease burden and other comorbidities.49 Several factors have been implicated in lower natural immunity and VE in these settings, including host characteristics such as poor nutritional status, underlying environmental enteropathy, and high maternal anti-rotavirus antibodies which might neutralize vaccine viruses. Among Indigenous infants in the Northern Territory, the burden of diarrheal disease is high, with admissions coded for enteric infections occurring at a rate 10-fold higher than among non-Indigenous infants.50 It is possible that comorbidities present in the Northern Territory Indigenous population contributes to reduced immunological response following Rotarix® vaccination. Assessing rotavirus sera responses and IgA titer following vaccination may provide important insights into the effectiveness of Rotarix® in this population.
In conclusion, we characterized a G1P[8] rotavirus outbreak in Central Australia in the vaccine era. The G1P[8] outbreak strain exhibited high genetic relatedness to contemporary global strains and exhibited antigenic differences to the VP4 and VP7 proteins of Rotarix®. The outbreak was more likely the result of one or more factors including suboptimal vaccine uptake, low primary immune responses or waning immunity in this population rather than the circulation of a strain associated with increased pathogenicity or vaccine escape. This study emphasizes the need for continued surveillance of rotavirus strains to help guide current and future vaccination strategies.
Acknowledgments
We gratefully acknowledge Dr Nicole Donker (Enteric Virus Group, Murdoch Childrens Research Institute, Royal Children's Hospital, Melbourne, Australia) for assistance with structural analysis of the VP7 protein and critical review of the manuscript. We thank Timothy Stockwell, Natalie Fedorova and Susmita Shrivastava (The J Craig Venter Institute, Rockville, MD, USA) for assistance with assembly, closure and annotation of viral segment sequences. Carl D Kirkwood is director of Australian Rotavirus Surveillance Program, which is supported by research grants from vaccine manufacturers CSL, GSK and Merck. This work was supported by grants from the National Health and Medical Research Council (NHMRC) of Australia (1031473); Australian Commonwealth Department of Health; GSK Biologicals (Melbourne, Australia); and CSL (Melbourne, Australia). Carl D Kirkwood is supported by a Career Development Award from the NHMRC of Australia (607347). Thomas L Snelling is supported by a NHMRC Frank Fenner Early Career Research Fellowship (1036229) and by a Fiona Stanley Investigator Award. This study was supported by the Victorian Government's Operational Infrastructure Support Program. This project was funded in part with federal funds from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services under contract number HHSN272200900007C.
Footnotes
Note: Supplementary Information for this article can be found on Emerging Microbes and Infections' website (http://www.nature.com/emi/).
Supplementary Information
References
- Glass RI, Parashar UD, Bresee JS, et al. Rotavirus vaccines: current prospects and future challenges. Lancet. 2006;368:323–332. doi: 10.1016/S0140-6736(06)68815-6. [DOI] [PubMed] [Google Scholar]
- Tate JE, Burton AH, Boschi-Pinto C, Steele AD, Duque J, Parashar UD. 2008 estimate of worldwide rotavirus-associated mortality in children younger than 5 years before the introduction of universal rotavirus vaccination programmes: a systematic review and meta-analysis. Lancet Infect Dis. 2012;12:136–141. doi: 10.1016/S1473-3099(11)70253-5. [DOI] [PubMed] [Google Scholar]
- Estes MK, Kapikian AZ.Fields' virology5th ed. Vol. 2. Philadelphia, PA; Lippincott Williams & Wilkins; 2007 [Google Scholar]
- Matthijnssens J, Otto PH, Ciarlet M, Desselberger U, van Ranst M, Johne R. VP6-sequence-based cutoff values as a criterion for rotavirus species demarcation. Arch Virol. 2012;157:1177–1182. doi: 10.1007/s00705-012-1273-3. [DOI] [PubMed] [Google Scholar]
- Matthijnssens J, Ciarlet M, McDonald SM, et al. Uniformity of rotavirus strain nomenclature proposed by the Rotavirus Classification Working Group (RCWG) Arch Virol. 2011;156:1397–1413. doi: 10.1007/s00705-011-1006-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo D, Liu J, Lu Y, et al. Full genomic analysis of rabbit rotavirus G3P[14] strain N5 in China: identification of a novel VP6 genotype. Infect Genet Evol. 2012;12:1567–1576. doi: 10.1016/j.meegid.2012.06.010. [DOI] [PubMed] [Google Scholar]
- Papp H, Al-Mutairi LZ, Chehadeh W, et al. Novel NSP4 genotype in a camel G10P[15] rotavirus strain. Acta Microbiol Immunol Hung. 2012;59:411–421. doi: 10.1556/AMicr.59.2012.3.11. [DOI] [PubMed] [Google Scholar]
- Trojnar E, Sachsenroder J, Twardziok S, Reetz J, Otto PH, Johne R. Identification of an avian group A rotavirus containing a novel VP4 gene with a close relationship to those of mammalian rotaviruses. J Gen Virol. 2013;94:136–142. doi: 10.1099/vir.0.047381-0. [DOI] [PubMed] [Google Scholar]
- Jere KC, Esona MD, Ali YH, et al. Novel NSP1 genotype characterised in an African camel G8P[11] rotavirus strain. Infect Genet Evol. 2014;21:58–66. doi: 10.1016/j.meegid.2013.10.002. [DOI] [PubMed] [Google Scholar]
- Dennehy PH. Effects of vaccine on rotavirus disease in the pediatric population. Curr Opin Pediatr. 2012;24:76–84. doi: 10.1097/MOP.0b013e32834ee594. [DOI] [PubMed] [Google Scholar]
- Vesikari T, Matson DO, Dennehy P, et al. Safety and efficacy of a pentavalent human-bovine (WC3) reassortant rotavirus vaccine. N Engl J Med. 2006;354:23–33. doi: 10.1056/NEJMoa052664. [DOI] [PubMed] [Google Scholar]
- Ruiz-Palacios GM, Perez-Schael I, Velazquez FR, et al. Safety and efficacy of an attenuated vaccine against severe rotavirus gastroenteritis. N Engl J Med. 2006;354:11–22. doi: 10.1056/NEJMoa052434. [DOI] [PubMed] [Google Scholar]
- Patel M, Pedreira C, de Oliveira LH, et al. Duration of protection of pentavalent rotavirus vaccination in Nicaragua. Pediatrics. 2012;130:e365–e372. doi: 10.1542/peds.2011-3478. [DOI] [PubMed] [Google Scholar]
- Paulke-Korinek M, Kundi M, Rendi-Wagner P, et al. Herd immunity after two years of the universal mass vaccination program against rotavirus gastroenteritis in Austria. Vaccine. 2011;29:2791–2796. doi: 10.1016/j.vaccine.2011.01.104. [DOI] [PubMed] [Google Scholar]
- Hanquet G, Ducoffre G, Vergison A, et al. Impact of rotavirus vaccination on laboratory confirmed cases in Belgium. Vaccine. 2011;29:4698–4703. doi: 10.1016/j.vaccine.2011.04.098. [DOI] [PubMed] [Google Scholar]
- Ichihara MY, Rodrigues LC, Teles Santos CA, et al. Effectiveness of rotavirus vaccine against hospitalized rotavirus diarrhea: a case–control study. Vaccine. 2014;32:2740–2747. doi: 10.1016/j.vaccine.2014.01.007. [DOI] [PubMed] [Google Scholar]
- Quintanar-Solares M, Yen C, Richardson V, Esparza-Aguilar M, Parashar UD, Patel MM. Impact of rotavirus vaccination on diarrhea-related hospitalizations among children <5 years of age in Mexico. Pediatr Infect Dis J. 2011;30 1 Suppl:S11–S15. doi: 10.1097/INF.0b013e3181fefb32. [DOI] [PubMed] [Google Scholar]
- Vesikari T, Karvonen A, Prymula R, et al. Efficacy of human rotavirus vaccine against rotavirus gastroenteritis during the first 2 years of life in European infants: randomised, double-blind controlled study. Lancet. 2007;370:1757–1763. doi: 10.1016/S0140-6736(07)61744-9. [DOI] [PubMed] [Google Scholar]
- Heaton PM, Goveia MG, Miller JM.Development of a pentavalent rotavirus vaccine against prevalent serotypes of rotavirus gastroenteritis J Infect Dis 2005192Suppl 1): S17–S21. [DOI] [PubMed] [Google Scholar]
- Buttery JP, Lambert SB, Grimwood K, et al. Reduction in rotavirus-associated acute gastroenteritis following introduction of rotavirus vaccine into Australia's National Childhood vaccine schedule Pediatr Infect Dis J 201130(1 Suppl): S25–29. [DOI] [PubMed] [Google Scholar]
- Snelling T, Markey P, Carapetis J, Andrews R. Rotavirus in the Northern Territory before and after vaccination. Microbiol Aust. 2012;33:61–63. [Google Scholar]
- Macartney KK, Porwal M, Dalton D, et al. Decline in rotavirus hospitalisations following introduction of Australia's national rotavirus immunisation programme. J Paediatr Child Health. 2011;47:266–270. doi: 10.1111/j.1440-1754.2010.01953.x. [DOI] [PubMed] [Google Scholar]
- Dyall-Smith ML, Holmes IH. Sequence homology between human and animal rotavirus serotype-specific glycoproteins. Nucleic Acids Res. 1984;12:3973–3982. doi: 10.1093/nar/12.9.3973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herring AJ, Inglis NF, Ojeh CK, Snodgrass DR, Menzies JD. Rapid diagnosis of rotavirus infection by direct detection of viral nucleic acid in silver-stained polyacrylamide gels. J Clin Microbiol. 1982;16:473–477. doi: 10.1128/jcm.16.3.473-477.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowley D, Donato CM, Roczo-Farkas S, Kirkwood CD. Novel G10P[14] rotavirus strain, Northern Territory, Australia. Emerg Infect Dis. 2013;19:1324–1327. doi: 10.3201/eid.1908.121653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulson BS, Fowler KJ, Bishop RF, Cotton RG. Neutralizing monoclonal antibodies to human rotavirus and indications of antigenic drift among strains from neonates. J Virol. 1985;54:14–20. doi: 10.1128/jvi.54.1.14-20.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulson BS, Kirkwood C. Relation of VP7 amino acid sequence to monoclonal antibody neutralization of rotavirus and rotavirus monotype. J Virol. 1991;65:5968–5974. doi: 10.1128/jvi.65.11.5968-5974.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulson BS. Typing of human rotavirus VP4 by an enzyme immunoassay using monoclonal antibodies. J Clin Microbiol. 1993;31:1–8. doi: 10.1128/jcm.31.1.1-8.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkwood CD, Bishop RF, Coulson BS. Human rotavirus VP4 contains strain-specific, serotype-specific and cross-reactive neutralization sites. Arch Virol. 1996;141:587–600. doi: 10.1007/BF01718319. [DOI] [PubMed] [Google Scholar]
- McDonald SM, Matthijnssens J, McAllen JK, et al. Evolutionary dynamics of human rotaviruses: balancing reassortment with preferred genome constellations. PLoS Pathog. 2009;5:e1000634. doi: 10.1371/journal.ppat.1000634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darriba D, Taboada GL, Doallo R, Posada D. jModelTest 2: more models, new heuristics and parallel computing. Nat Meth. 2012;9:772. doi: 10.1038/nmeth.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- DeLano WL. Unraveling hot spots in binding interfaces: progress and challenges. Curr Opin Struct Biol. 2002;12:14–20. doi: 10.1016/s0959-440x(02)00283-x. [DOI] [PubMed] [Google Scholar]
- Maes P, Matthijnssens J, Rahman M, van Ranst M. RotaC: a web-based tool for the complete genome classification of group A rotaviruses. BMC Microbiol. 2009;9:238. doi: 10.1186/1471-2180-9-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trask SD, Kim IS, Harrison SC, Dormitzer PR. A rotavirus spike protein conformational intermediate binds lipid bilayers. J Virol. 2010;84:1764–1770. doi: 10.1128/JVI.01682-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackow ER, Shaw RD, Matsui SM, Vo PT, Dang MN, Greenberg HB. The rhesus rotavirus gene encoding protein VP3: location of amino acids involved in homologous and heterologous rotavirus neutralization and identification of a putative fusion region. Proc Natl Acad Sci USA. 1988;85:645–649. doi: 10.1073/pnas.85.3.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkwood CD, Roczo S, Boniface K, Bishop RF, Barnes GL, Australian Rotavirus Surveillance Group Australian Rotavirus Surveillance Program annual report, 2010/11. Commun Dis Intell Q Rep. 2011;35:281–287. [PubMed] [Google Scholar]
- Zeller M, Patton JT, Heylen E, et al. Genetic analyses reveal differences in the VP7 and VP4 antigenic epitopes between human rotaviruses circulating in Belgium and rotaviruses in Rotarix and RotaTeq. J Clin Microbiol. 2012;50:966–976. doi: 10.1128/JCM.05590-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward RL, Kirkwood CD, Sander DS, et al. Reductions in cross-neutralizing antibody responses in infants after attenuation of the human rotavirus vaccine candidate 89-12. J Infect Dis. 2006;194:1729–1736. doi: 10.1086/509623. [DOI] [PubMed] [Google Scholar]
- Kobayashi N, Taniguchi K, Urasawa S. Identification of operationally overlapping and independent cross-reactive neutralization regions on human rotavirus VP4. J Gen Virol. 1990;71:2615–2623. doi: 10.1099/0022-1317-71-11-2615. [DOI] [PubMed] [Google Scholar]
- Desselberger U, Huppertz HI. Immune responses to rotavirus infection and vaccination and associated correlates of protection. J Infect Dis. 2011;203:188–195. doi: 10.1093/infdis/jiq031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snelling TL, Schultz R, Graham J, et al. Rotavirus and the indigenous children of the Australian outback: monovalent vaccine effective in a high-burden setting. Clin Infect Dis. 2009;49:428–431. doi: 10.1086/600395. [DOI] [PubMed] [Google Scholar]
- Snelling TL, Andrews RM, Kirkwood CD, Culvenor S, Carapetis JR. Case–control evaluation of the effectiveness of the G1P[8] human rotavirus vaccine during an outbreak of rotavirus G2P[4] infection in central Australia. Clin Infect Dis. 2011;52:191–199. doi: 10.1093/cid/ciq101. [DOI] [PubMed] [Google Scholar]
- Hull B, Dey A, Menzies R, McIntyre P. Annual immunisation coverage report, 2010. Commun Dis Intell Q Rep. 2013;37:E21–E39. [PubMed] [Google Scholar]
- Patel M, Glass RI, Jiang B, Santosham M, Lopman B, Parashar U. A systematic review of anti-rotavirus serum IgA antibody titer as a potential correlate of rotavirus vaccine efficacy. J Infect Dis. 2013;208:284–294. doi: 10.1093/infdis/jit166. [DOI] [PubMed] [Google Scholar]
- Madhi SA, Cunliffe NA, Steele D, et al. Effect of human rotavirus vaccine on severe diarrhea in African infants. N Engl J Med. 2010;362:289–298. doi: 10.1056/NEJMoa0904797. [DOI] [PubMed] [Google Scholar]
- Gladstone BP, Ramani S, Mukhopadhya I, et al. Protective effect of natural rotavirus infection in an Indian birth cohort. N Engl J Med. 2011;365:337–346. doi: 10.1056/NEJMoa1006261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li QS, Guthridge S, d'Espaignet ET, Paterson B.From infancy to young adulthood: health status in the Northern Territory, 2006. Darwin: Department of Health and Community Services; 2006. Available at: http://digitallibrary.health.nt.gov.au/dspace/bitstream/10137/84/1/infancy_to_young_adulthood_2006.pdf (accessed 13 June 2013). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.