Abstract
Interactions between environmental contaminants can lead to non-additive effects that may affect the toxicity and risk assessment of a mixture. Comprehensive time course and dose-response studies with 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD), non-dioxin-like 2,2′,4,4′,5,5′-hexachlorobiphenyl (PCB153) and their mixture were performed in immature, ovariectomized C57BL/6 mice. Mice were gavaged once with 30 μg/kg TCDD, 300 mg/kg PCB153, a mixture of 30 μg/kg TCDD with 300 mg/kg PCB153 (MIX) or sesame oil vehicle for 4,12, 24,72 or 168 h. In the 24 h dose-response study, animals were gavaged with TCDD (0.3,1, 3, 6, 10, 15, 30, 45 μg/kg), PCB153 (3,10, 30, 60, 100, 150, 300, 450 mg/kg), MIX (0.3+3, 1+10, 3+30, 6+60, 10+100, 15+150, 30+300, 45 μg/kg TCDD+450 mg/kg PCB153, respectively) or vehicle. All three treatments significantly increased relative liver weights (RLW), with MIX eliciting significantly greater increases compared to TCDD and PCB153 alone. Histologically, MIX induced hepatocellular hypertrophy, vacuolization, inflammation, hyperplasia and necrosis, a combination of TCDD and PCB153 responses. Complementary lipid analyses identified significant increases in hepatic triglycerides in MIX and TCDD samples, while PCB153 had no effect on lipids. Hepatic PCB153 levels were also significantly increased with TCDD co-treatment. Microarray analysis identified 167 TCDD, 185 PCB153 and 388 MIX unique differentially expressed genes. Statistical modeling of quantitative real-time PCR analysis of Pla2g12a, Serpinb6a, Nqo1, Srxn1, and Dysf verified non-additive expression following MIX treatment compared to TCDD and PCB153 alone. In summary, TCDD and PCB153 co-treatment elicited specific non-additive gene expression effects that are consistent with RLW increases, histopathology, and hepatic lipid accumulation.
Keywords: TCDD, PCB153, dose-response, mixture, non-additive interaction, liver, gene expression
INTRODUCTION
Polyhalogenated aromatic hydrocarbons (PHAHs), including polychlorinated biphenyls (PCBs) and polychlorinated dibenzo-p-dioxins (PCDDs) are persistent environmental contaminants that elicit species- and tissue-specific toxic effects. 2,3,7,8-Tetrachlorodibenzo-p-dioxin (TCDD) and other PCDDs are by-products of various activities including municipal solid waste and sewage sludge incineration, herbicide production, pulp and paper bleaching, backyard barrel burning, and other processes (Poland and Glover 1973; Safe et al. 1982; Mason and Safe 1986; Commoner et al. 1987). Despite banning the production and use of PCBs in many industrial applications in the late 1970s (Safe et al. 1982; Mullin et al. 1984; Safe 1990), they are still present in the environment and pose a potential health concern. The chemical stability of PCBs and PCDDs coupled with their lipophilic nature has led to bioaccumulation and biomagnification in the food chain, particularly in fatty tissues (Kimbrough 1995; Safe 1997).
The broad spectrum of effects elicited by PCBs and PCDDs include hepatotoxicity, immune suppression, reproductive toxicity, endocrine disruption, developmental toxicity, and carcinogenicity (Poland and Knutson 1982; Knerr and Schrenk 2006). Overall toxicity is determined by the chemical s structural similarity to TCDD, and ability to bind and activate the aryl hydrocarbon receptor (AhR) (Van den Berg et al. 2006). PCB congeners containing meta and para chlorines on the biphenyl ring assume coplanar conformation, resembling dioxin (dioxin-like PCBs), and are more toxic than non-coplanar PCBs possessing ortho chlorine substituents which have reduced AhR binding affinity (Safe 1990; Safe 1994).
The effects of dioxin and dioxin-like PCBs are mediated through the activation of AhR, a cytosolic basic helix-loop-helix periodicity-AhR nuclear translocator (ARNT)-simple-minded domain containing transcription factor (Poland and Knutson 1982; Denison and Heath-Pagliuso 1998; Safe 2001; Denison et al. 2002). Following ligand binding, chaperone proteins dissociate from the AhR, allowing translocation to the nucleus and heterodimerization with ARNT. AhR:ARNT heterodimers interact with dioxin response elements (DREs) in the regulatory regions of target genes, leading to changes in gene expression (Denison and Heath-Pagliuso 1998; Nebert et al. 2000).
The non-dioxin-like, non-coplanar 2,2′,4,4′,5,5′-hexachlorobiphenyl (PCB153) is the PCB congener at the highest concentrations in human tissue on molar basis, and is found in high concentrations in environmental and other biological samples (Schecter et al. 1994; NTP 2006a). PCB153 does not bind or activate the AhR, but elicits constitutive androstane receptor (CAR)/pregnane X receptor (PXR)-mediated responses (Craft et al. 2002; Honkakoski et al. 2003; Tabb et al. 2004; Kopec et al. 2010b). Following ligand activation, CAR/PXR translocates to the nucleus to heterodimerize with the retinoid X receptor and binds to CAR and PXR response elements (CAREs and PXREs) in the regulatory regions of target genes to elicit changes in gene expression (Blumberg and Evans 1998; Masahiko and Honkakoski 2000; Lemaire et al. 2007).
Traditionally, environmental risk assessments have focused on single congener exposures. However, PCBs and dioxins exist as complex mixtures, and their interactions can influence the toxicity of a mixture. For example, interactions could include additive effects, or non-additive responses (i.e., synergistic or antagonistic) (Staal et al. 2007). Previous studies have reported non-additive interactions between PCB153 and dioxin-like chemicals including the inhibition of TCDD- and PCB126-induced cleft palate and immunotoxicity, and suppression of ethoxyresorufin-O-deethylase (EROD) (Biegel et al. 1989; Zhao et al. 1997; Suh et al. 2003). In contrast, others reported synergistic induction in mouse hepatic EROD and aryl hydrocarbon hydroxylase activities compared to TCDD treatment alone (Bannister and Safe 1987). Co-treatment has also been shown to elicit synergistic effects on porphyrin accumulation in Sprague-Dawley rats (van Birgelen et al. 1996a). However, no study has examined the non-additive effects of PCB153 and TCDD co-treatment on gene expression.
In this report, time course and dose-dependent hepatic gene expression studies with complementary computational response element searches, histopathology, lipid profiling and tissue analyses of PCB153 and TCDD levels were performed to evaluate the hepatic effects elicited by a mixture of TCDD and PCB153 in immature, ovariectomized C57BL/6 mice. This study complements our previous report (Kopec et al. 2010b) by examining the dose-dependent mixture effects elicited by PCB153 and TCDD. More specifically, dose-response non-linear logistic modeling identified non-additive gene expression interactions that are consistent with the non-additive phenotypic responses elicited by TCDD and PCB153 co-treatment.
MATERIALS AND METHODS
Animal Husbandry
Female C57BL/6 mice, ovariectomized by the supplier on postnatal day (PND) 20, with body weights (BW) within 10% of the average, were obtained from Charles Rivers Laboratories (Portage, MI) on PND 25. Animals were housed in polycarbonate cages containing cellulose fiber chips (Aspen Chip Laboratory Bedding, Northeastern Products, Warrensberg, NY) with 30–40% humidity and a 12 h light/dark cycle (07:00 h – 19:00 h), had free access to deionized water and were fed ad libitum with Harlan Teklad 22/5 Rodent Diet 8640 (Madison, WI). Animals were acclimatized prior to dosing on PND 28. Immature ovariectomized mice were used to facilitate comparisons with other data sets obtained using the same model, study design and analysis methods (Boverhof et al. 2005; Kopec et al. 2008; Kopec et al. 2010a). Immature animals are more responsive to AhR ligands, and ovariectomy negates potential interactions with estrogens produced by the maturing ovaries. All procedures were carried out with the approval of the Michigan State University All-University Committee on Animal Use and Care.
Time Course and Dose-Response Studies
A stock solution of PCB153 (99.9% purity, AccuStandard, New Haven, CT) was first dissolved in acetone (J.T. Baker), followed by dilution with sesame oil (Sigma, St. Louis, MO), and evaporation of acetone under nitrogen gas. A stock solution of TCDD was a gift from the Dow Chemical Company (Midland, MI). PCB153 and TCDD stock solutions were diluted using sesame oil to achieve the desired dose. Animals were orally gavaged using 1.5 inch feeding needles with 2.25 mm ball ends (Cadence Science, Lake Success, NY). For the time course study, mice (n=5 per group) were administered 0.1 ml of a single dose of 30 μg/kg TCDD, 300 mg/kg PCB153, a mixture (MIX) of 1:10,000 ratio of TCDD:PCB153 (30 μg/kg TCDD with 300 mg/kg PCB153) or sesame oil vehicle and sacrificed after 4, 12, 24, 72 or 168 h. Individual TCDD and PCB153 treatments have been published (Kopec et al. 2010b). The time points and doses used were selected to facilitate comparisons with previous studies (Boverhof et al. 2005; Boverhof et al. 2006; Kopec et al. 2008; Kopec et al. 2010a). 30 μg/kg TCDD was selected because it elicited maximum induction of Cyp1a1 mRNA levels without inducing significant changes in body weight gain (Boverhof et al. 2005). 300 mg/kg PCB153 was selected based on a previous dose-response study (Kopec et al. 2010b), eliciting maximum induction of Cyp2b10 mRNA levels and having no negative effect on body weight gain. The mixture (30 μg/kg TCDD+300 mg/kg PCB153) was selected based on the characterized responses of the individual components. The ratio of 1:10,000 TCDD:PCB153 approaches reports of the relative concentrations of TCDD and PCB153 in human samples (Schecter et al. 1994).
In the dose-response study, mice (n=5 per group) were gavaged with TCDD (0.3, 1, 3, 6, 10, 15, 30, 45 μg/kg), PCB153 (3, 10, 30, 60, 100, 150, 300, 450 mg/kg), MIX (0.3+3, 1+10, 3+30, 6+60, 10+100, 15+150, 30+300, 45 μg/kg TCDD + 450 mg/kg PCB153, respectively) or vehicle, and sacrificed 24 h post dose by cervical dislocation. Livers were removed, sections were weighed, flash frozen in liquid nitrogen, and stored at −80°C. A section of the right liver lobe was fixed in 10% neutral buffered formalin (Sigma) for histological analysis. For lipid staining, the remaining right lobe of the liver was frozen in Tissue-Tek O.C.T. compound (Sakura, Torrance, CA).
Histological Analysis
Fixed liver tissues were sectioned and processed in ethanol, xylene, and paraffin using a Thermo Electron Excelsior tissue processor (Waltham, MA). Tissues were embedded in paraffin with Miles Tissue Tek II embedding center, after which paraffin blocks were sectioned at 5 μm with a rotary microtome. Liver sections were placed on glass microscope slides, washed twice in xylene for 5 min, followed by four quick washes in ethanol and rinsing in water. Slides were placed in Gill 2 hematoxylin (Thermo Fisher Scientific, Waltham, MA) for 1.5 min followed by 2–3 quick dips in 1% glacial acetic acid water and rinsed with running water for 2–3 min. Slides were then rinsed in ethanol and counterstained with 1% eosin Y-phloxine B solution (Sigma) followed by multiple rinses in ethanol and xylene.
For lipid staining, liver samples were sectioned at 6 μm, fixed in 10% neutral buffered formalin for 5 min, rinsed with water and immersed in 100% propylene glycol for 5 min. Slides were stained with Oil Red O solution (Sigma) for 8 min at 60°C, placed in 80% propylene glycol for 5 min and rinsed in water for 15 min. Slides were counterstained with Gill 2 hematoxylin for 30 sec and washed with water for 30 min. Coverslips were attached using aqueous mounting media. All the histological processing was performed at Michigan State University Investigative Histopathology Laboratory, Division of Human Pathology using a modified version of previously published procedures (Sheehan and Hrapchak 1980).
Lipid Analysis by Gas Chromatography-Mass Spectrometry (GC-MS)
TCDD, PCB153, MIX and vehicle control liver samples (~100 mg) from the time course (24, 72, 168 h) and dose-response (24 h) studies were homogenized (Polytron PT2100, Kinematica AG, Luzern, CH) in 40% methanol and acidified with concentrated HCl. Lipids were extracted with chloroform:methanol (2:1) containing 1 mM 2,6-di-tert-butyl-4-methylphenol (B HT; Sigma) and extraction efficiency controls (19:1n9 FFA [free fatty acid] and 19:0 TAG [triacylglycerol]) were added (Nu-Chek Prep, Elysian, MN). Protein and aqueous phases were re-extracted with chloroform and the organic phases were pooled. A derivatization standard (19:2n6 FFA; Nu-Chek Prep) was added and samples were dried under nitrogen, resuspended in 2% non-aqueous methanolic HCl and incubated at 60°C overnight. Samples were cooled to room temperature and 0.9% (w/v) NaCl and hexane were added. The organic phase was collected and a loading control (17:1n9 fatty acid methyl ester [FAME]; Nu-Chek Prep) was added. Samples were dried under nitrogen, resuspended in equal volumes of hexane and separated on Agilent 6890N gas chromatograph interfaced to Agilent 5973 mass spectrometer with DB23 column (30 m length, 0.25 mm internal diameter, 0.25 μm film thickness). Samples were run on the following temperature program: 50°C to 150°C at 40°C/min, to 185°C at 5°C/min, to 235°C at 3°C/min, to 250°C at 10°C/min. Peak areas were integrated with MassLynx software (Waters, Milford, MA) and data were normalized to the starting amount of liver and loading controls. Principal component analysis of temporal lipid profiles (represented as the average fold change induction of the 21 identified fatty acids) was performed in R 2.6.0. Data were extracted and used to generate PCA plots in GraphPad Prism 5.0 (La Jolla, CA).
Hepatic Triglyceride Measurement
Frozen liver samples (~100 mg) were homogenized (Polytron PT2100, Kinematica) in 1 ml of 1.15% KCl. Triglycerides were extracted from 200 μl of hepatic homogenate with 800 μl of isopropyl alcohol by vortex-mixing for 10 min. The samples were centrifuged for 5 min at 800 x g at room temperature and supernatant was collected into separate vials. The concentration of hepatic triglycerides was determined using a commercial L-Type Triglyceride M kit (Wako Diagnostics, Richmond, VA) with Multi-Calibrator Lipids as a standard (Wako Diagnostics). The measurements were performed according to the manufacturer’s protocol with 20 μl of the triglyceride extract incubated with 150 μl of Reagent 1 followed by incubation with 50 μl of Reagent 2. Final results were normalized to the starting amount of liver (Supplementary Fig. 5).
Quantification of Hepatic PCB153 and TCDD Levels
Liver samples were processed in parallel with laboratory blanks and a reference or background sample at Wellington Laboratories Inc. (Guelph, ON, Canada). Samples (100 to 500 mg) were transferred to a tared screw cap culture tube and weights were recorded. Samples were spiked with 13C12-PCB153 and 13C12-2,3,7,8-TCDD surrogates and digested with hydrochloric acid. Each digested sample was then split between two screw cap tubes and 3–4 ml hexane was added to each tube followed by vigorous mixing. Tubes were centrifuged, and the organic layer was removed. Hexane extraction was repeated three times per screw cap tube and the six hexane fractions were combined. The hexane fraction was then split evenly prior to clean-up and one fraction was archived. The other fraction was cleaned-up using a small multi-layer (acid/base/neutral) silica gel column eluted with 20–25 ml of hexane. The eluate was concentrated on a rotary evaporator and then transferred to a conical microvial with pentane and dichloromethane rinses and allowed to dry. Immediately prior to injection on the high-resolution gas chromatograph/high-resolution mass spectrometer (HRGC-HRMS) system 13C12-PCB111 and 13C12-1,2,3,4-TCDD injection standards were added to the conical microvial. The identification and quantification of PCB153 and TCDD was performed using an Agilent (Santa Clara, CA, USA) 6890 series HRGC with direct capillary interface to a Waters Autospec Ultima HRMS. Chromatographic separations were carried out on a 60 m DB5 (0.25 mm ID, 0.25 μm film thickness) column in constant flow mode (Helium, 1 ml/min). All injections were 1 μl and a splitless injection was used. The mass spectrometer was operated in EI+ selective ion recording mode at a mass resolving power of 10,000 or greater. The % recovery ranged between 78–111% for 13C12-PCB153 and between 80–107% for 13C12-2,3,7,8-TCDD. The detection limits were 5.5 pg/g (ppt) for TCDD and 44.5 pg/g (ppt) for PCB153.
RNA Isolation
Liver samples (~100 mg stored at −80°C) were immediately transferred to 1 ml TRIzol (Invitrogen, Carlsbad, CA) and homogenized using a Mixer Mill 300 tissue homogenizer (Retsch, Germany). Total RNA was isolated according to the manufacturer s protocol with an additional acid phenol:chloroform extraction. Isolated RNA was resuspended in an RNase-free RNA storage solution (Ambion Inc., Austin, TX) that has a low pH and contains sodium citrate to minimize RNA hydrolysis. RNA was quantified (A260), and quality assessed by determining the A260/A280 ratio and by visual inspection of 2 μg on a denaturing gel.
Microarray Experimental Design
TCDD, PCB153, MIX and vehicle RNA samples from the time course study were individually hybridized to 4 × 44 K Agilent microarrays (version 1; Agilent Technologies, Inc., Santa Clara, CA). Three biological replicates were performed using one-color labeling (Cy3) for each time point and treatment, according to the manufacturer s protocol (Agilent Manual: G4140-90040 v. 5.7). Microarray slides were scanned at 532 nm (Cy3) on a GenePix 4000B scanner (Molecular Devices, Union City, CA). Images were analyzed for feature and background intensities using GenePix Pro 6.0 (Molecular Devices). All data were managed in TIMS dbZach data management system (Burgoon and Zacharewski 2007).
Microarray Analysis
All microarray data passed our laboratory quality assurance protocol (Burgoon et al. 2005). Microarray data were normalized using a semi-parametric approach (Eckel et al. 2005) and the posterior probabilities were calculated using an empirical Bayes method based on a per gene and dose basis using model-based t values (Eckel et al. 2004). Gene expression data were ranked and prioritized using P1(t) values >0.999 and |fold change|>1.5 to identify differentially expressed genes. Individual chemical (TCDD and PCB153) temporal microarray datasets have been published (Kopec et al. 2010b). Data obtained from the time course microarray were used as a screen to identify putative non-additive candidates for further verification using QRTPCR in the 24 h dose-response study. Complete temporal microarray datasets (TCDD, PCB153 and MIX) with corresponding statistics comparing TCDD vs. PCB153, TCDD vs. MIX and MIX vs. PCB153 at each time point are available as Supplementary Table 2.
Quantitative Real-Time PCR (QRTPCR)
QRTPCR verification of microarray responses was performed as described previously (Boverhof et al. 2005). Briefly, 1 μg of total RNA was reverse transcribed by SuperScript II (Invitrogen) using an anchored oligo-dT primer as described by the manufacturer. The cDNA (1.0 μl) was used as a template in a 30 μl PCR reaction containing 0.1 μM of forward and reverse gene-specific primers, 3 mM MgCl2, 1 mM dNTPs, 0.025 IU AmpliTaq Gold, and 1 × SYBR Green PCR buffer (Applied Biosystems, Foster City, CA). Supplementary Table 3 provides the names, abbreviations, accession numbers, forward and reverse primer sequences, and amplicon sizes. PCR amplification was conducted on an Applied Biosystems PRISM 7500 Sequence Detection System. cDNAs were quantified using a standard curve approach and the copy number of each sample was standardized to 3 housekeeping genes (Actb, Gapdh, Hprt) to control for the differences in RNA loading, quality, and cDNA synthesis (Vandesompele et al. 2002). For graphing purposes, the relative expression levels were scaled such that the expression level of the time-matched control group was equal to one.
Computational Putative DNA Response Element Identification
Putative dioxin response elements (DREs) (Sun et al. 2004; Dere et al. 2011a), constitutive androstane receptor response elements (CAREs) (Phillips et al. 2009) and pregnane X receptor response elements (PXREs) (Kopec et al. 2010b) were computationally identified using Position Weight Matrices (PWM) specific to each element. Genomic regions (−10,000 bp relative to the transcriptional start site [TSS] together with 5′-untranslated region [UTR]) were obtained from the University of California, Santa Cruz, Genome Browser for mouse (build 37), computationally searched, and each DRE, CARE and PXRE was scored using a specific PWM. The genomic region 10,000 bp upstream of the TSS and the 5′ UTR was examined to include more distal putative response elements which have recently been identified and may regulate gene expression. These regions were also selected to facilitate comparisons to other studies examing similar regions (Boverhof et al. 2006; Kopec et al. 2010b; Dere et al. 2011a; Dere et al. 2011b). Matrix similarity scores (MSS) >0.80 are considered to be putative response elements.
Dose-Response Non-linear Logistic Modeling
Gene-specific non-linear models were fit to investigate the dose-response relationship for Dysf, Pla2g12a, Serpinb6a, Srxn1, Nqo1, Got1, Elovl5, Dexi, Akr1c20, and Srebf1 identified in the microarray study as putative candidates exhibiting non-additive gene expression and further verified using QRTPCR in the 24 h dose-response study. Specifically, for each gene a non-linear logistic model for the mean was used to estimate the dose-response relationship for TCDD and PCB153, according to:
where β0 is the unknown intercept parameter, βi, i=1,2, is the unknown slope parameter associated with the ith single chemical, xi is the dose associated with the ith chemical, γ is the maximum-effect parameter, and μ add is the expected value of the response under the additive model. Simultaneously, the MIX data were fit with a mixture model:
where t is the total dose associated with MIX data. For a fixed-ratio of TCDD and PCB153 given by (α1, α2, such that α1 + α2 =1), the slope for the mixture under the assumption of additivity is θadd= β1α1+ β2α2. Based on preliminary analyses, using the quasi-likelihood estimation criterion, the assumption is made that variance is a function of the mean: Var(y)= τμ, where τ is the parameter associated with the functional form of the variance and has to be estimated. The quasi-likelihood criterion has assumptions only concerning the first and second moments, and some regularity conditions related to the regression equation: E(Y)= μ(θ) and Var(Y)= τV(μ). It is of interest to maximize the quasi-likelihood function given by:
Gene-specific plots of the mean versus the variance for TCDD, PCB153 and MIX confirmed the relationship between the mean and the variance. In the study, the additive dose-response model for the fixed-ratio mixture based on TCDD and PCB153 data alone was compared to the mixture model obtained using MIX data. Therefore, the null hypothesis tested was θmix= θadd, where θmix and θadd are the parameter estimates assuming the mixture and the additive models, respectively. The modeling (PROC NLIN) was performed with SAS 9.2 (SAS Institute, Cary, NC).
Functional Gene Annotation and Statistical Analysis
Annotation and functional categorization of the differentially regulated genes was performed using Database for Annotation, Visualization and Integrated Discovery (DAVID) (Dennis et al. 2003) and Ingenuity Pathway Analysis (Ingenuity Systems, Redwood City, CA). All statistical analyses were performed with SAS 9.2. Unless stated otherwise, all data were analyzed by analysis of variance (ANOVA) followed by Dunnett’s or Tukey’s post hoc test. Differences between treatment groups were considered significant when p<0.05.
RESULTS
Organ and Body Weights
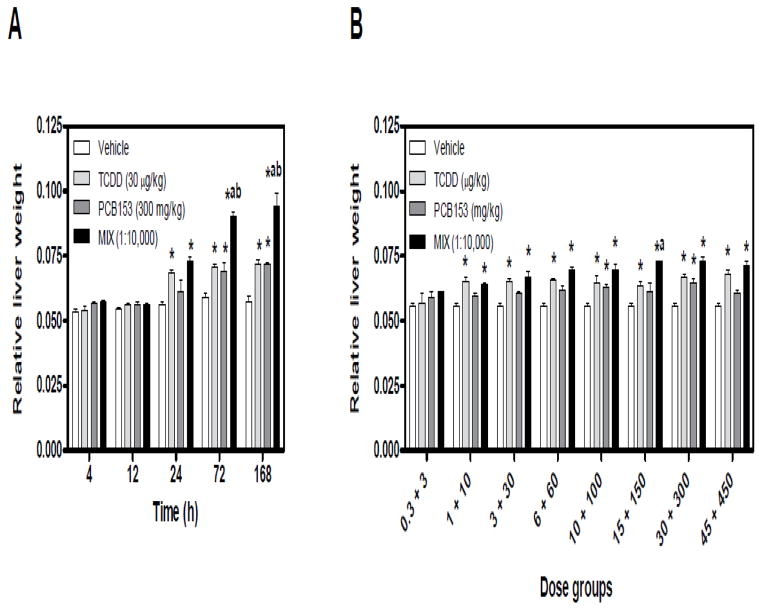
Exposure to dioxins and PCBs elicits temporal and dose-dependent increases in liver weight in rodents (Denison and Heath-Pagliuso 1998; Craft et al. 2002; Boverhof et al. 2006; NTP 2006a; NTP 2006b; Kopec et al. 2010b). In the time course study, significant increases in relative liver weight (RLW) were observed with 300 mg/kg PCB153 (72, 168 h), 30 μg/kg TCDD and MIX (24, 72, 168 h), relative to time-matched vehicle controls (Fig. 1A). TCDD and PCB153 treatment resulted in comparable increases in RLW, while MIX induced significantly greater increases in RLW compared to TCDD and PCB153 alone at 72 and 168 h (Fig. 1A). These results are consistent with reported increases in RLW in C57BL/6 and B6C3F1 mice 3, 7 and 13 days after a single gavage of TCDD and PCB153 mixture (De Jongh et al. 1995; van Birgelen et al. 1996b).
Figure 1.
Temporal and dose-dependent changes in the relative liver weight following exposure to TCDD, PCB153, and MIX. (A) In the time course study, 30 μg/kg TCDD + 300 mg/kg PCB153 were co-administered (MIX, 1:10,000 TCDD to PCB153). (B) In the 24 h dose-response, MIX (μg/kg TCDD + mg/kg PCB153) was administered at 1:10,000 ratio of TCDD to PCB153, respectively. Results are displayed as mean ± standard error (SE) for 5 independent replicates. Data were analyzed by analysis of variance (ANOVA) followed by Tukey’s post hoc test: *p<0.05 vs. vehicle, ap<0.05 vs. PCB153 and bp<0.05 vs. TCDD. Time course increases in relative liver weight induced by individual TCDD and PCB153 treatments are from Kopec et al. (2010b).
In the 24 h dose-response study, TCDD and MIX elicited comparable increases in RLW in all but the lowest dose groups (Fig. 1B). RLW increases at 15+150 (μg/kg TCDD + mg/kg PCB153, respectively) were also greater compared to 150 mg/kg PCB153 (Fig. 1B). In contrast, PCB153 increased RLW only at 100 and 300 mg/kg.
Minimal (≤ 10%) decreases in body weight gain were observed with PCB153 (300 and 450 mg/kg) and MIX (three highest dose group combinations) at 24 h, but are not indicative of systemic toxicity (FDA 2008). Supplementary Tables 1A–B provide time course and dose-response data for terminal body weight, body weight gain, liver weight and relative liver weights.
Hepatic Tissue Level Quantification
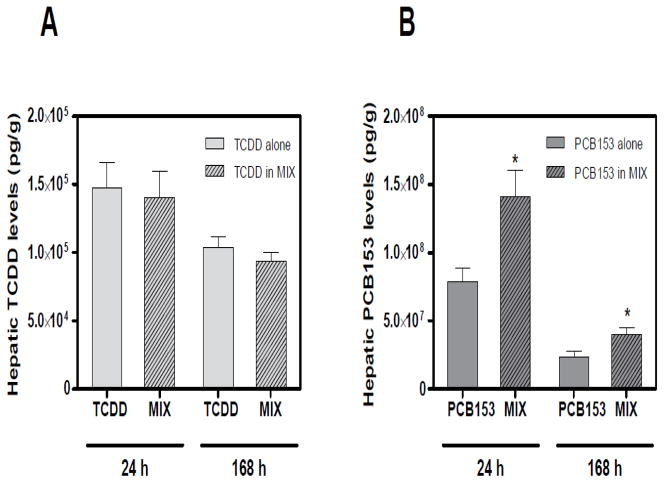
HRGC-HRMS analysis of liver samples at 24 and 168 h indicate that hepatic PCB153 accumulation was potentiated following TCDD co-treatment compared to PCB153 alone (Fig. 2B). No changes were detected in the hepatic TCDD accumulation following PCB153 co-administration (Fig. 2A). In addition, PCB153 levels dramatically decreased at 168 h, compared to more modest reductions in TCDD levels after seven days (Kopec et al. 2010b).
Figure 2.
Hepatic (A) TCDD and (B) PCB153 tissue levels per g liver wet weight measured by HRGC-HRMS (Kopec et al. 2010b). PCB153 levels were potentiated following co-administration with TCDD (in MIX) at 24 and 168 h. However no changes in TCDD hepatic levels were detected following co-treatment with PCB153 (in MIX). Results are displayed as mean ± SE for 3 independent replicates. Data were analyzed by ANOVA followed by Tukey’s post hoc test: *p<0.05 vs. time-matched PCB153 levels alone.
Histopathology
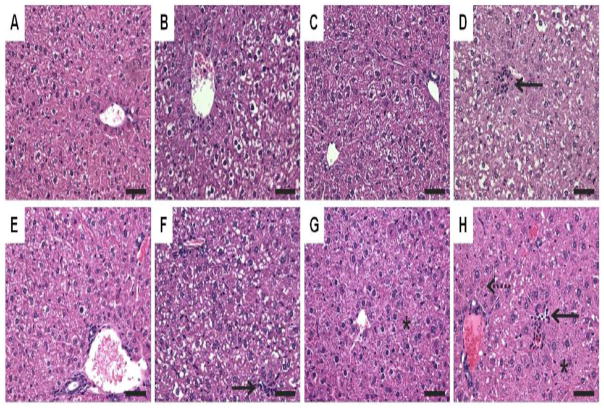
In the time course study, TCDD elicited moderate multifocal inflammation and marked hepatocellular vacuolization localized in periportal regions, extending to midzonal and centrilobular regions at later time points. Instances of vacuolization corresponded with Oil Red O (ORO) staining indicative of neutral lipid accumulation (Fig. 3B,F, Supplementary Fig. 1B,F). These changes are consistent with the reported hepatic effects of TCDD in mice (Boverhof et al. 2005; Boverhof et al. 2006).
Figure 3.
Standard hematoxylin and eosin staining of liver sections at 24 (top panel: A–D) and 168 h (bottom panel: E–H). (A&E) Sesame oil elicited minimal vacuolization. (B&F) 30 μg/kg TCDD elicited vacuolization and inflammation (solid arrow). (C&G) 300 mg/kg PCB153 resulted in minimal vacuolization and hypertrophy (asterisk). (D&H) MIX resulted in vacuolization, inflammation (solid arrow), hypertrophy (asterisk) and necrosis (dashed arrow). Bars = 50 μm.
PCB153 elicited vacuolization comparable to levels observed in vehicles, as well as mild to moderate centrilobular hypertrophy with cytoplasmic granularity (Fig. 3C,G) (Kopec et al. 2010b). In contrast, PCB153 did not elicit inflammation, while MIX elicited a combination of TCDD and PCB153 responses. Vacuolization and ORO staining in MIX were the most severe at 24 h, decreasing to TCDD-exposed levels at 168 h. MIX-treated livers also had moderate multifocal inflammation and a temporal increase in cellular hypertrophy that was comparable to time-matched TCDD and PCB153-exposed livers, respectively. Furthermore, MIX elicited minimal necrosis and instances of mitotic figures indicative of hyperplasia between 72 and 168 h (Fig. 3D,H).
Liver microscopic changes were also dose-responsive (Supplementary Fig. 2). TCDD elicited moderate periportal hepatocellular vacuolization and minimal multifocal inflammation (15, 30, 45 μg/kg), while PCB153 elicited mild to moderate centrilobular hypertrophy (300 and 450 mg/kg) and minimal to mild hepatocellular vacuolization. MIX induced moderate to marked periportal hepatocellular vacuolization and centrilobular hypertrophy and minimal mixed cell infiltration (15+15, 30+300, 45 μg/kg TCDD + 450 mg/kg PCB153, respectively).
Lipid Profiling
Total lipids extracted from TCDD, PCB153, MIX and control livers from the time course (24, 72 and 168 h) and dose-response studies were derivatized to fatty acid methyl esters. Twenty one fatty acid species were measured using GC-MS. Principal component (PC) analysis (Supplementary Fig. 3) identified a time- and treatment-dependent separation of fatty acid (FA) profiles (as fold changes) around PC1 and PC2, respectively. Treatment and time-dependent induction in individual fatty acids is represented in Supplementary Fig 4. TCDD induced the most pronounced changes in total FAs (Table 1), consistent with the ORO staining. MIX-elicited increases in total FAs were comparable across time, with significant changes at 24 and 168 h (Table 1).
Table 1.
GC-MS analysis of fatty acid composition.
| Time (h) | Treatment | SFA | MUFA | n3 PUFA | n6 PUFA | Total FA |
|---|---|---|---|---|---|---|
| 24 | Vehicle | 322.09 ± 38.38 | 170.21 ± 41.37 | 103.94 ± 9.56 | 349.80 ± 42.12 | 946.04 ± 119.47 |
| TCDD | 327.67 ± 14.47 | 197.54 ± 54.99 | 106.95 ± 10.60 | 363.20 ± 32.40 | 995.37 ± 99.90 | |
| PCB153 | 304.38 ± 35.47 | 134.35 ± 31.72 | 103.70 ± 11.67 | 331.29 ± 26.02 | 873.73 ± 80.71 | |
| MIX | 350.49 ± 14.14 | 264.58 ± 27.06*a | 114.20 ± 6.99 | 447.42 ± 29.96*ab | 1176.69 ± 73.34*a | |
| 72 | Vehicle | 311.76 ± 32.18 | 184.24 ± 33.04 | 93.26 ± 10.15 | 317.51 ± 22.72 | 906.77 ± 60.97 |
| TCDD | 353.26 ± 29.51 | 394.28 ± 63.50*a | 109.14 ± 2.92 | 432.07 ± 40.07* | 1288.74 ± 126.72*a | |
| PCB153 | 326.72 ± 45.42 | 213.53 ± 100.64 | 98.27 ± 15.94 | 326.28 ± 64.06 | 964.8 ± 191.49 | |
| MIX | 327.58 ± 22.21 | 257.33 ± 67.52 | 104.47 ± 5.89 | 394.45 ± 23.05 | 1083.84 ± 115.65 | |
| 168 | Vehicle | 297.26 ± 6.39 | 178.44 ± 24.29 | 97.07 ± 7.28 | 333.55 ± 14.33 | 906.32 ± 33.60 |
| TCDD | 384.92 ± 44.64* | 432.65 ± 75.20*a | 130.39 ± 6.76*a | 526.26 ± 46.53*a | 1474.21 ± 160.53*a | |
| PCB153 | 329.79 ± 46.76 | 184.20 ± 19.16 | 104.39 ± 8.46 | 342.23 ± 26.61 | 960.61 ± 95.75 | |
| MIX | 383.33 ± 14.70* | 272.31 ± 62.84*ab | 121.32 ± 11.21*a | 461.61 ± 41.32*a | 1238.56 ± 107.18*ab |
Hepatic fatty acids were measured by GC-MS. Peak areas were integrated using MassLynx software. The results are displayed in arbitrary units and reported as mean ± SD of 5 independent replicates. Data were analyzed by ANOVA followed by Tukey’s post hoc test: *p<0.05 vs. vehicle, ap<0.05 vs. PCB153 and bp<0.05 vs. TCDD.
FA – fatty acids; SFA – saturated fatty acids; MUFA – monounsaturated fatty acids; PUFA – polyunsaturated fatty acids. TCDD and PCB153 data are from Kopec et al. (2010b).
TCDD significantly induced saturated (SFA), monounsaturated (MUFA) and n3 and n6 polyunsaturated fatty acids (PUFA) in a time-dependent fashion. In contrast, PCB153 did not change total FAs or any specific FA (Table 1), consistent with the lack of ORO staining, and CAR-mediated inhibition of lipogenesis (Zhai et al. 2010). However, MIX elicited differences in SFA, MUFA, and PUFA levels when compared to individual treatments (Table 1). For example, MIX significantly decreased MUFA and total FA levels (168 h), but increased n6 PUFA (24 h) levels compared to TCDD (Table 1). These effects are consistent with the induction of triglyceride levels by MIX (Supplementary Fig. 5) and ORO staining (Supplementary Fig. 1). GC-MS analysis of the dose-response study also revealed higher FA accumulation in MIX-treated livers compared to TCDD groups, although the differences did not achieve statistical significance (Supplementary Fig. 6).
Temporal Microarray Gene Expression
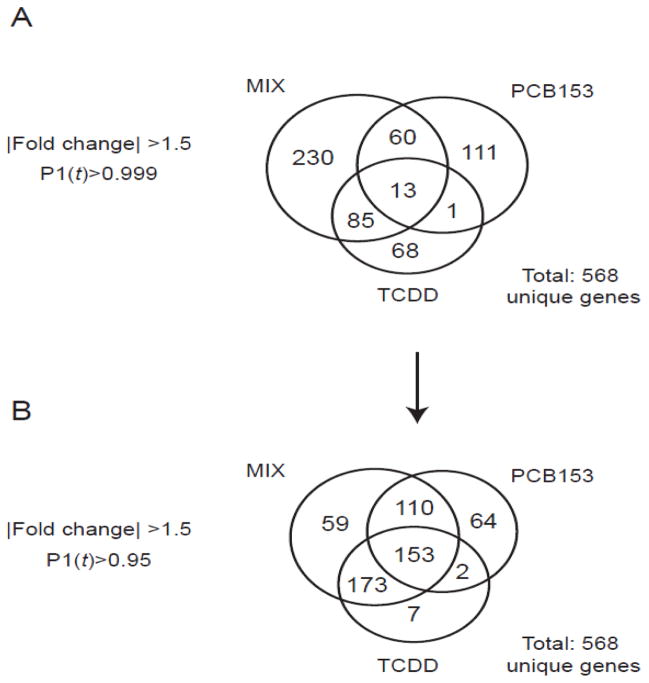
Hepatic gene expression was evaluated using whole-genome 4×44K Agilent oligonucleotide microarrays containing ~21,000 unique annotated genes. Statistical analysis of the time course data identified 568 unique gene expression changes, with 167, 185 and 388 genes differentially regulated in response to TCDD, PCB153, and MIX, respectively (|fold change|>1.5, P1(t)>0.999) at one or more time points. MIX elicited the highest number of differentially expressed genes at each time point. Even with more stringent fold change criteria (|fold change|>2.0), MIX consistently elicited the highest number of differentially expressed genes (339 unique genes) relative to TCDD (151 unique genes) and PCB153 (121 unique genes).
Comparative analysis identified 68 TCDD-, 111 PCB153-, and 230 MIX-specific responses (|fold change|>1.5, P1(t)>0.999). Only 13 genes were differentially expressed by all three treatments (Fig. 4A). When the statistical criteria were relaxed (|fold change|>1.5, P1(t)>0.95) (Fig. 4B), the number of overlapping genes increased. A complementary Venn diagram (|fold change|>2.0) is included (Supplementary Fig. 7A–B).
Figure 4.
Temporal microarray data sets for 30 μg/kg TCDD, 300 mg/kg PCB153 (Kopec et al. 2010b), and MIX (1:10,000 TCDD:PCB153) groups were compared using (A) stringent (|fold change|>1.5, P1(t)>0.999) and (B) relaxed (|fold change|>1.5, P1(t)>0.95) selection criteria. Numbers in the Venn diagram represent unique genes.
Functional annotation of the 568 differentially regulated genes was associated with xenobiotic metabolism, oxidoreduction/oxidative stress, lipid metabolism, immune response, cell division and differentiation, cell death, and transport. Many of these over-represented functions (Table 2) could be anchored to the changes in RLW, histopathology, and lipid profiles. For example, the differential expression of genes involved in lipid metabolism and transport was consistent with the hepatic fatty accumulation, increased hepatic triglyceride levels, and changes in specific FAs. TCDD and MIX induced significant fatty vacuolization between 24–168 h consistent with Cd36 (~6 fold) and Fabp12 (~54–34 fold) induction, which were not affected by PCB153. Down-regulation of Elovl5 (~2 fold) and up-regulation of Insig2 (~6 fold) by PCB153, in contrast to TCDD, was also consistent the fatty acid profile indicating a lack of lipid accumulation in PCB153-exposed livers.
Table 2.
Functional categorization and potential response element regulation of select hepatic genes differentially expressed by 30 μg/kg TCDD, 300 mg/kg PCB153, and MIX.
| Functional category | Gene ID | Gene name | Gene symbol | TCDD* | PCB153* | MIX* | DREsa | PXREsa | CAREsa |
|---|---|---|---|---|---|---|---|---|---|
| 11761 | Aldehyde oxidase 1 | Aox1 | 2.2 | 2.9 | 4.8 | yes | yes | yes | |
| 11997 | Aldo-keto reductase family 1, member B7 | Akr1b7 | NC | 22.1 | 16.8 | yes | yes | yes | |
| 432720 | Aldo-keto reductase family 1, member C19 | Akr1c19 | NC | 1.7 | 2.0 | yes | yes | no | |
| 116852 | Aldo-keto reductase family 1, member C20 | Akr1c20 | −1.7 | −1.6 | −3.0 | no | yes | no | |
| Xenobiotic metabolism/Oxidoreductase activity/Oxidative stress | 13076 | Cytochrome P450, family 1, subfamily a, polypeptide 1 | Cyp1a1 | 348.1 | NC | 348.2 | yes | yes | yes |
| 13077 | Cytochrome P450, family 1, subfamily a, polypeptide 2 | Cyp1a2 | 2.4 | 1.7 | 2.4 | yes | yes | yes | |
| 13088 | Cytochrome P450, family 2, subfamily b, polypeptide 10 | Cyp2b10 | NC | 16.1 | 24.2 | yes | yes | yes | |
| 13089 | Cytochrome P450, family 2, subfamily b, polypeptide 13 | Cyp2b13 | NC | 1.9 | 1.9 | yes | yes | yes | |
| 13094 | Cytochrome P450, family 2, subfamily b, polypeptide 9 | Cyp2b9 | NC | 7.6 | 6.2 | no | yes | yes | |
| 72082 | Cytochrome P450, family 2, subfamily c, polypeptide 55 | Cyp2c55 | NC | 48.4 | 48.4 | yes | yes | yes | |
| 13112 | Cytochrome P450, family 3, subfamily a, polypeptide 11 | Cyp3a11b | −1.9 | 4.0 | 3.1 | yes | yes | yes | |
| 13113 | Cytochrome P450, family 3, subfamily a, polypeptide 13 | Cyp3a13 | −1.9 | 2.5 | NC | no | yes | no | |
| 56388 | Cytochrome P450, family 3, subfamily a, polypeptide 25 | Cyp3a25 | NC | 3.8 | 3.0 | no | yes | yes | |
| 56050 | Cytochrome P450, family 39, subfamily a, polypeptide 1 | Cyp39a1 | NC | 2.5 | 2.0 | yes | yes | no | |
| 14776 | Glutathione peroxidase 2 | Gpx2 | NC | 4.3 | 8.9 | yes | yes | yes | |
| 14858 | Glutathione S-transferase, alpha 2 (Yc2) | Gsta2 | 5.8 | 4.1 | 11.3 | yes | yes | yes | |
| 14863 | Glutathione S-transferase, mu 2 | Gstm2 | 2.0 | 2.0 | 2.2 | yes | yes | yes | |
| 14865 | Glutathione S-transferase, mu 4 | Gstm4 | 2.4 | 4.0 | 5.6 | yes | yes | yes | |
| 14872 | Glutathione S-transferase, theta 2 | Gstt2 | 2.0 | 2.0 | 2.2 | yes | yes | yes | |
| 18104 | NAD(P)H dehydrogenase, quinone 1 | Nqo1s | 4.6 | 2.8 | 12.6 | yes | yes | yes | |
| 76650 | Sulfiredoxin 1 homolog (S. cerevisiae) | Srxn1s | 2.6 | 2.3 | 3.9 | yes | yes | yes | |
| 394435 | UDP glucuronosyltransferase 1 family, polypeptide A6B | Ugt1a6b | 5.6 | NC | 6.4 | yes | yes | no | |
| 22235 | UDP-glucose dehydrogenase | Ugdh | 2.7 | 2.5 | 2.7 | yes | yes | yes | |
| 12491 | CD36 antigen | Cd36 | 5.7 | NC | 6.1 | yes | yes | yes | |
| 26903 | Dysferlin | Dysfs | 3.3 | 1.5 | 14.9 | yes | yes | yes | |
| 170439 | ELOVL family member 6, elongation of long chain fatty acids (yeast) | Elovl6 | −4.3 | −3.8 | −5.2 | yes | yes | yes | |
| Lipid and glucose metabolism | 75497 | Fatty acid binding protein 12 | Fabp12 | 53.7 | −1.5 | 34.4 | yes | yes | yes |
| 14104 | Fatty acid synthase | Fasn | −4.3 | −3.9 | −6.4 | yes | yes | yes | |
| 14718 | Glutamate oxaloacetate transaminase 1, soluble | Got1 | −2.6 | NC | −4.3 | yes | yes | yes | |
| 14571 | Glycerol phosphate dehydrogenase 2, mitochondrial | Gpd2 | −3.0 | 1.5 | −3.3 | yes | yes | yes | |
| 72999 | Insulin induced gene 2 | Insig2 | NC | 5.9 | 4.0 | yes | yes | yes | |
| 53357 | Phospholipase A2, group VI | Pla2g6 | NC | 2.5 | 1.8 | yes | yes | yes | |
| 66350 | Phospholipase A2, group XIIA | Pla2g12as | 3.7 | 2.7 | 7.7 | yes | yes | yes | |
| 228026 | Pyruvate dehydrogenase kinase, isoenzyme 1 | Pdk1 | −1.6 | NC | 1.6 | yes | yes | no | |
| 20250 | Stearoyl-Coenzyme A desaturase 2 | Scd2 | 2.0 | NC | NC | yes | yes | yes | |
| 20788 | Sterol regulatory element binding factor 2 | Srebf2 | −1.7 | −2.1 | −1.6 | yes | yes | yes | |
| 20787 | Sterol regulatory element binding transcription factor 1 | Srebf1 | −2.3 | −2.4 | −4.6 | yes | yes | yes | |
| 12125 | BCL2-like 11 (apoptosis facilitator) | Bcl2l11 | 1.9 | NC | 4.2 | yes | yes | no | |
| Cell cycle/Cell death | 13197 | Growth arrest and DNA-damage-inducible 45 alpha | Gadd45a | −3.6 | 9.4 | 3.1 | yes | no | yes |
| 17873 | Growth arrest and DNA-damage-inducible 45 beta | Gadd45b | 5.7 | 10.8 | 29.2 | yes | yes | yes | |
| 53415 | HIV-1 tat interactive protein 2, homolog (human) | Htatip2 | 3.5 | NC | 4.4 | yes | yes | no | |
| 15937 | Immediate early response 3 | Ier3 | 3.5 | NC | 3.8 | yes | yes | yes | |
| 17869 | Myelocytomatosis oncogene | Myc | 3.0 | 4.2 | 7.2 | yes | yes | yes | |
| 211323 | Neuregulin 1 | Nrg1 | 2.8 | NC | 6.7 | yes | yes | yes | |
| 78688 | Nucleolar protein 3 (apoptosis repressor with CARD domain) | Nol3 | NC | 4.8 | 4.7 | yes | yes | no | |
| 22033 | Tnf receptor-associated factor 5 | Traf5 | 2.1 | NC | 2.3 | yes | yes | yes | |
| 21813 | Transforming growth factor, beta receptor II | Tgfbr2 | 1.6 | 1.8 | 2.4 | yes | yes | yes | |
| 29820 | Tumor necrosis factor receptor superfamily, member 19 | Tnfrsf19 | 3.9 | 2.6 | 7.7 | yes | yes | yes | |
| 21928 | Tumor necrosis factor, alpha-induced protein 2 | Tnfaip2 | 6.7 | NC | 6.9 | yes | yes | yes | |
| 66443 | Tumor necrosis factor, alpha-induced protein 8-like 1 | Tnfaip8l1 | 3.0 | NC | 4.1 | yes | yes | yes | |
| Immune response | 12774 | Chemokine (C-C motif) receptor 5 | Ccr5 | 2.4 | −1.6 | NC | no | yes | yes |
| 56644 | C-type lectin domain family 7, member a | Clec7a | 3.9 | NC | 2.9 | yes | yes | yes | |
| 58239 | Dexamethasone-induced transcript | Dexi | −1.9 | −1.9 | −4.3 | yes | yes | no | |
| 14129 | Fc receptor, IgG, high affinity I | Fcgr1 | 2.4 | NC | 1.9 | yes | yes | no | |
| 14969 | Histocompatibility 2, class II antigen E beta | H2-Eb1 | 7.8 | NC | 5.0 | yes | yes | yes | |
| 14998 | Histocompatibility 2, class II, locus DMa | H2-DMa | 4.0 | NC | 2.7 | yes | yes | yes | |
| 14999 | Histocompatibility 2, class II, locus Mb1 | H2-DMb1 | 4.9 | NC | 3.2 | yes | yes | yes | |
| 15900 | Interferon regulatory factor 8 | Irf8 | 3.1 | NC | 2.0 | yes | yes | yes | |
| 16994 | Lymphotoxin B | Ltb | 2.9 | NC | 2.3 | yes | yes | yes | |
| 17304 | Milk fat globule-EGF factor 8 protein | Mfge8 | 4.6 | NC | 5.0 | yes | yes | yes | |
| 20719 | Serine (or cysteine) peptidase inhibitor, clade B, member 6a | Serpinb6as | 5.1 | NC | 11.0 | yes | yes | yes | |
| 20208 | Serum amyloid A 1 | Saa1 | 3.4 | NC | 3.2 | yes | yes | no | |
| 20209 | Serum amyloid A 2 | Saa2 | 3.2 | NC | 3.2 | yes | yes | yes | |
| 20210 | Serum amyloid A 3 | Saa3 | 7.8 | NC | 3.5 | yes | yes | no |
NC - no change.
Maximum fold change on the microarray (|fold change| >1.5, P1(t) > 0.90).
Putative DNA response elements identified by computational searches (see Materials and Methods).
QRTPCR results. The Agilent microarray Cyp3a11 probe maps the 60-mer region between positions 1105 and 1164 of the Cyp3a11 gene, which is 2053 bp long, while Cyp3a11 primers (listed in a Supplementary Table 3) amplify the 119 bp fragment between 1701 and 1819 bp suggesting that both approaches amplify non-overlapping regions of the gene.
Synergistic response verified by non-linear logistic modeling of QRTPCR dose-response data.
Histopathology also revealed alterations in cell size (hypertrophy), as well as cell division and cell death (hyperplasia and necrosis) (Fig. 3 and Supplementary Fig. 2). TCDD and MIX induced genes that promote cell death including Bcl2l11 (~2–4 fold), Tnfaip2 (~7 fold), Tnfaip8l1 (~3–4 fold), Traf5 (~2 fold), and Htatip2 (~4 fold), and were not regulated by PCB153, which did not elicit cell death.
Moreover, TCDD induced immune response genes (~2–8 fold; Clec7a, Fcgr1, H2-Eb1, H2-DMa, H2-DMb1, Irf8, Ltb, Mfg28 and Saa1-3), consistent with the marked multifocal inflammation at later time points. In contrast, PCB153 did not alter the expression of these genes, while MIX elicited significant inflammation, reflected by the induction of the same immune related genes as TCDD treatment alone.
Genes involved in xenobiotic metabolism and oxidoreductase activities exhibited the highest induction by all three treatments. This included cytochrome P450s (e.g. Cyp1a1, Cyp1a2, Cyp2b10, Cyp2b13, Cyp2b9, Cyp2c55, Cyp23a25, Cyp39a1) and glutathione S-transferases (e.g. Gsta2, Gstm2, Gstt2, Gstm4), as well as Nqo1, Ugdh, and Aox1. For example, Cyp1a1 was induced ~348 fold by TCDD and MIX. Unlike treatment-specific induction of Cyp1a (TCDD) and Cyp2b/2c/23a/39a (PCB153) families, MIX elicited the differential expression of all cytochrome P450 subfamilies, suggesting the dual activation of AhR and CAR/PXR pathways. Computational analysis identified putative DREs, CAREs and PXREs in the promoter regions (−10,000 bp relative to the TSS together with the 5′ UTR) of differentially regulated genes (Table 2) (Kopec et al. 2010b; Dere et al. 2011a).
QRTPCR Verification of Microarray Responses
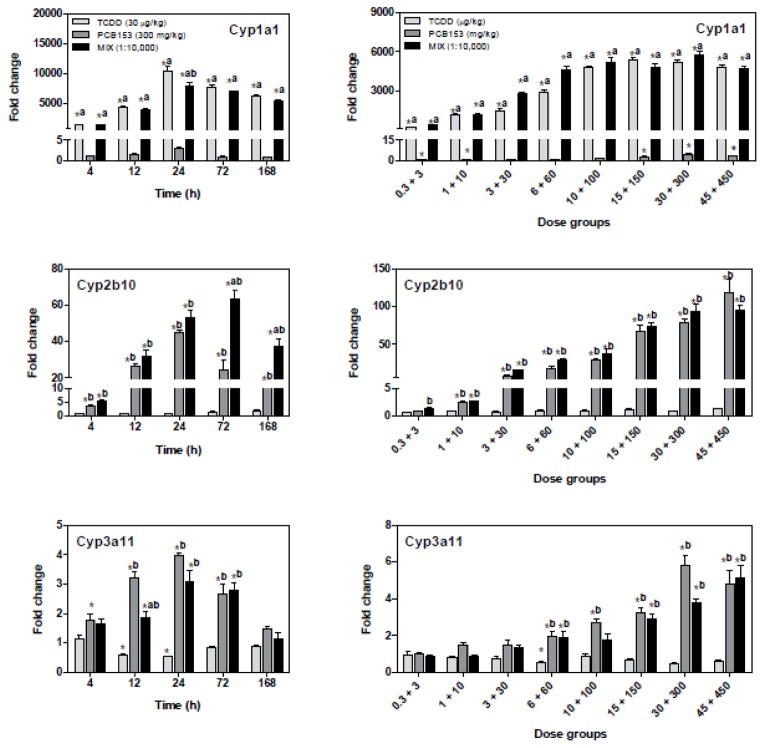
In addition to functional categorization of the gene expression responses, the temporal microarray dataset with corresponding statistics (Supplementary Table 2) were used to identify putative non-additive gene expression in MIX treatment compared to TCDD and PCB153 alone. QRTPCR verified the temporal and dose-dependent expression of 13 genes. Cyp1a1, Cyp2b10, and Cyp3a11 differential expression (Fig. 5) was confirmed as positive controls for AhR, CAR and PXR-mediated responses, respectively (Nebert et al. 1990; Maglich et al. 2002; Honkakoski et al. 2003). Although microarray data suggested that Cyp3a11 was not differentially expressed, it was verified by QRTPCR, resulting in amplification of a different region of the 2053 bp long Cyp3a11 gene relative to the microarray. More specifically, the microarray probe maps the 60-mer region between positions 1105 and 1164, while Cyp3a11 primers amplified the 119 bp fragment between 1701 and 1819 bp suggesting that both techniques amplified non-overlapping regions of the Cyp3a11 gene.
Figure 5.
QRTPCR verification of AhR, CAR, and PXR-regulated genes. TCDD induced Cyp1a1 (AhR), while PCB153 induced Cyp2b10 and Cyp3a11 (CAR/PXR), and MIX induced all three genes. All fold changes were calculated relative to time-matched vehicle controls. The genes are represented by their official gene symbols. Results are displayed as mean ± SE for 5 independent replicates. Data were analyzed by ANOVA followed by Tukey’s post hoc test: *p<0.05 vs. time-matched vehicle, ap<0.05 vs. PCB153 and bp<0.05 vs. TCDD. TCDD and PCB153 temporal data are from Kopec et al. (2010b).
TCDD significantly induced Cyp1a1, while PCB153 induced Cyp2b10 and Cyp3a11. The induction of Cyp1a1, Cyp2b10, and Cyp3a11 by MIX was comparable to TCDD and PCB153 alone, and therefore was not considered for non-additive evaluation.
Statistical Verification of Non-Additive Gene Expression
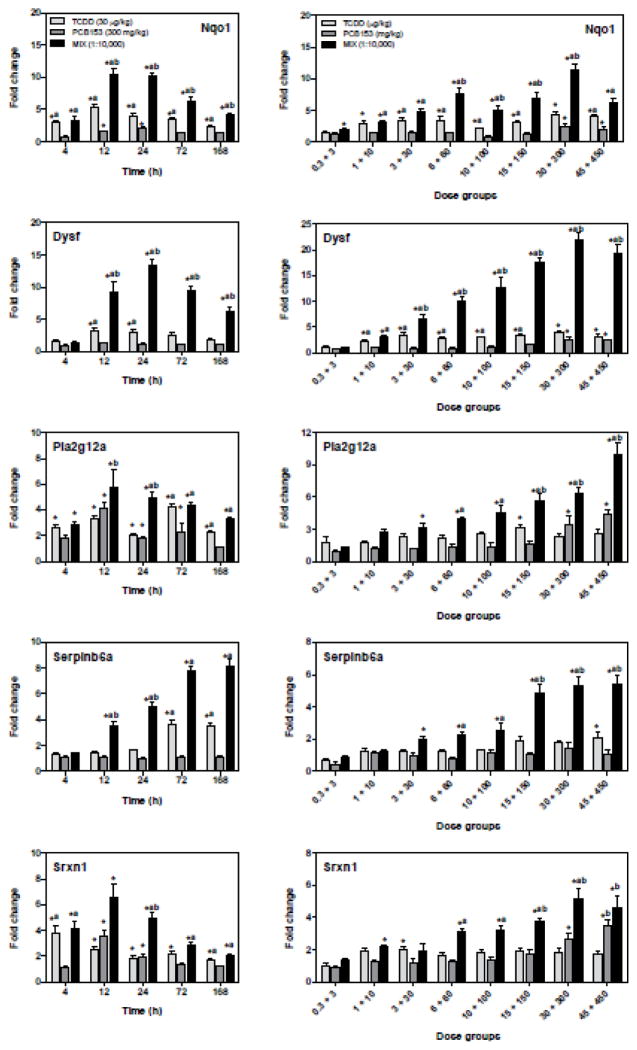
Analysis of gene expression time course data suggested that Nqo1, Dysf, Pla2g12a, Serpinb6a, Srxn1 (Fig. 6) and Elovl5, Srebf1, Dexi, Got1, Akr1c20 (Supplementary Fig. 8) exhibited potential non-additive interactions. The time course and dose-response profiles elicited by TCDD, PCB153, and MIX were verified using QRTPCR and further examined using gene-specific non-linear models to statistically determine synergistic, antagonistic, or additive interactions. Initial analysis confirmed that the variance is proportional to the mean: Var(Y)= τV(μ). Linear regression provided parameter estimates used as initial values for βi’s and maximum fold expression for each gene was used as the initial value for γ. The final model fit for the expected value for each gene was:
where β0 is the unknown intercept, β1 is the unknown slope parameter associated with TCDD doses, β2 is the unknown parameter associated with PCB153 doses, θmix is the parameter associated with doses of the mixture TCDD+PCB153, and Ii are indicator functions for the chemical groups, where:
where i=1, i=2, and i=3 represents TCDD, PCB153, and MIX, respectively. The intercept parameter α is assumed to be zero. Plots of the residuals against the predicted values were assessed for goodness of fit of the models, and no abnormalities were found.
Figure 6.
Temporal and dose-dependent QRTPCR verification of putative non-additive genes. Genes exhibiting putative non-additive interactions initially identified in the time course study were verified by QRTPCR in the time course study and 24 h dose-response study. The genes are represented by their official gene symbols. Results are displayed as mean ± SE for 5 independent replicates. Data were analyzed by ANOVA followed by Tukey’s post hoc test: *p<0.05 vs. time-matched vehicle, ap<0.05 vs. PCB153 and bp<0.05 vs. TCDD.
To determine statistically significant deviation from additivity, the quasi-likelihood ratio test was used to compare the empirical mixture model with the reduced additivity model based on the F-distribution (Gennings et al. 2002):
where θadd= β1α1+ β2α2, where α1 and α are the ‘mix ratios’ for TCDD and PCB153 respectively. The ‘mix ratios’ were obtained by dividing the dose by the total dose. For example, when the TCDD dose is 1 μg/kg, the PCB153 dose is 10000 μg/kg (10 mg/kg), so the ‘mix ratios’ are and such that Σiαi = 1.
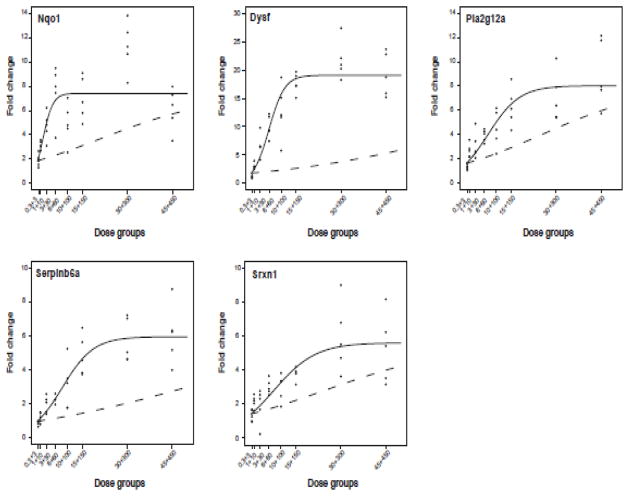
Each graph in Fig. 7 illustrates the overlay between the additive model generated using TCDD and PCB153 data alone and the mixture model fitted using MIX data for each gene. The quasi-likelihood ratio test determined that the mixture model was significantly (p<0.0001) different from the additive model for Nqo1, Dysf, Pla2g12a, Serpinb6a and Srxn1, indicating co-administration of TCDD plus PCB153 elicited synergistic differential gene expression.
Figure 7.
Non-linear logistic modeling of QRTPCR dose-response data. Graphs depict the additive model (dashed line) generated using TCDD and PCB153 data and the mixture model (solid line) fitted using MIX data (black dots). MIX data for Nqo1, Dysf, Pla2g12a, Serpinb6a, and Srxn1 fit the mixture model better than the additivity model, indicating a statistically significant (p<0.0001) synergistic interaction.
Srebf1, Akr1c20 and Dexi approached significant non-additive expression (p=0.0619, p=0.0615, p=0.0806, respectively), while Elovl5 (p=0.5164) and Got1 (p=0.9181) was not significantly different from additivity (Supplementary Fig. 9). Supplementary Table 4 contains summary statistics, parameter values for each gene and results of the quasi-likelihood ratio test.
DISCUSSION
Non-additive interactions confound mixture risk assessment. Therefore special considerations are required, including adequate dose-response characterization for each mixture component, applying a “no-interaction” hypothesis to determine synergy or antagonism, testing mixture components across a sufficient range, using appropriate models to statistically determine departure from additivity, and assessing interactions at relevant levels of biological organization to assure meaningful interpretation of the results (Borgert et al. 2001; Borgert 2007).
This study complements our previous report evaluating the temporal effects of single chemical toxicity (Kopec et al. 2010b), by identifying non-additive dose-dependent effects elicited by a mixture of TCDD and PCB153. The non-additive hepatic differential gene expression was phenotypically anchored and compared to effects elicited by individual TCDD and PCB153 treatments. Statistical modeling confirmed non-additive effects in a subset of differentially expressed genes that could be functionally associated with changes in gross physiology, histopathology, and hepatic lipid composition.
Mixtures of dioxin and non-dioxin-like chemicals increase hepatic disposition compared to single chemical administration (De Jongh et al. 1995). In the current study, hepatic PCB153 levels were significantly increased following co-administration with TCDD at 24 and 168 h. These results are consistent with rat studies that also reported TCDD co-treatment increased PCB153 levels (Van der Kolk et al. 1992). Co-administration of PCB153 with dioxin-like PCB156 also led to significant hepatic retention of PCB153 after 7 and 14 days in C57BL/6J mice (De Jongh et al. 1992). In B6C3F1 mice, TCDD-elicited increases in PCB153 levels were specific to the liver and not observed in other tissues (van Birgelen et al. 1996b). The NTP co-treatment study (PCB153+PCB126) in Sprague-Dawley rats reported that at high mixture dose combinations, the levels of PCB126 were lower compared to hepatic concentrations of the PCB126 group alone, suggesting that PCB153 antagonizes the accumulation of PCB126 in the liver (NTP 2006d; NTP 2006c). In contrast, TCDD levels did not change with PCB153 co-treatment, as previously reported (De Jongh et al. 1995; van Birgelen et al. 1996b).
Hepatic sequestration of TCDD and other dioxin-like PCBs is mediated by binding to Cyp1a2 protein (DeVito et al. 1998). Cyp1a2 null mice exhibit 10-fold lower hepatic TCDD accumulation compared to wild types (Diliberto et al. 1999). PCB153 has a higher affinity for adipose compared to hepatic tissue (van Birgelen et al. 1996b) and unlike TCDD and dioxin-like PCBs it is not sequestered by Cyp1a2 protein. However, TCDD induces fatty accumulation in the liver (Boverhof et al. 2005), increasing hepatic lipophilicity, which may contribute to hepatic PCB153 accumulation following co-administration with TCDD (Van der Kolk et al. 1992; van Birgelen et al. 1996b).
MIX induced xenobiotic metabolism gene expression, comparable to levels elicited by individual treatments. Cytochrome P450 induction was the highest among all functional categories, and included prototypical AhR (Cyp1a1, Cyp1a2) and CAR/PXR (Cyp2b9, Cyp2b10, Cyp2b13, Cyp23a25, Cyp3a11) regulated genes (Nebert et al. 1990; Maglich et al. 2002; Honkakoski et al. 2003). Nqo1, a known AhR- and nuclear factor erythroid 2-related factor 2 (Nrf2)- inducible gene (Yeager et al. 2009), also displayed non-additive induction following MIX treatment, which may serve cytoprotective roles (Ma et al. 2004; Klaassen and Reisman 2010). In addition, MIX synergistically induced Srxn1, sulfiredoxin 1 homolog, an Nrf2-dependent oxidoreductase, which protects lung tissue from smoke-induced oxidative injury (Park et al. 2009; Singh et al. 2009). Synergistic induction of Nqo1 and Srxn1 may provide additional defense from the xenobiotic stress induced by MIX. Enzyme induction also leads to endoplasmic reticulum proliferation increasing hepatocyte size, which may be a factor in increased RLW induced by MIX compared to TCDD and PCB153 alone.
Among oxidoreductase and xenobiotic metabolism genes, aldo-keto reductase (AKR) superfamily members were also differentially expressed including Akr1b7 and Akr1c19 induction and the repression of Akr1c20. AKRs are involved in NAD(P)H-dependent oxidoreductions of a variety of natural and foreign substrates and have been implicated in alleviating toxicity associated with lipid peroxidation (Liu et al. 2009). Expression of Akr1b7 is abolished in CAR and PXR null mice (Liu et al. 2009), consistent with PCB153 and MIX-mediated Akr1b7 induction. Moreover, putative CAREs and PXREs were identified in the upstream regulatory regions. Statistical modeling approached significant (p=0.0615) synergistic repression of Akr1c20 following MIX treatment. Akr1c20 expression is liver-specific, but has unknown function (Matsumoto et al. 2006). Based on structural similarity to 17β-hydroxysteroid dehydrogenase type 5 and comparable enzymatic properties, Akr1c20 may be involved in steroid metabolism and reduction of non-steroidal α-dicarbonyl compounds (Matsumoto et al. 2006).
Integrating histopathology, ORO staining, GC-MS fatty acid and triglyceride analyses suggests that TCDD- and MIX-elicited hepatic lipid accumulation also contribute to increased RLW. The AhR-mediated fatty accumulation (steatosis) can be associated with the differential expression of lipid transport and metabolism genes. TCDD induces the expression of the fatty acid transporter, Cd36 (Lee et al. 2010), which was also induced by MIX. Cd36 facilitates hepatic uptake of free fatty acids and is up-regulated in non-alcoholic fatty liver disease patients (Greco et al. 2008). TCDD and MIX also induced fatty acid binding protein 12 (Fabp12), which also facilitates hepatic uptake of free fatty acids (Tijet et al. 2006). In contrast, PCB153 treatment alone did not alter Cd36 or Fabp12 expression, consistent with lack of hepatic fat accumulation.
Furthermore, PCB153 down-regulated a number of lipid metabolism genes, including Elovl5, which was induced by TCDD, suggesting divergent regulation. Elovl5 null mice experience steatosis, which has been attributed to activation of SREBP-1c protein (Moon et al. 2008). MIX approached significant (p=0.0619) synergistic repression of Srebf1 consistent with less fat accumulation at 168 h compared to TCDD, which did not affect Srebf1 expression. In contrast, PCB153 repressed Srebf1 expression, consistent with the lack of lipid accumulation. Repression of SREBP-mediated lipogenesis in Zucker diabetic fatty rats is also associated with CAR/PXR-mediated over-expression of insulin induced genes (Insig1/2) (Takaishi et al. 2004; Roth et al. 2008). PCB153 and MIX induced Insig2, while TCDD down-regulated Insig2 at 4 h, consistent with the observed hepatic phenotypes.
Pla2g12a, a novel secretory phospholipase, which is involved in the digestion of dietary phospholipids, and the production of molecules that induce inflammatory responses (Gelb et al. 2000) was also synergistically induced by MIX. Although Pla2g12a induction may be linked to elevated free fatty acids, reports suggest it has weak catalytic activity and contributes little to increase cellular fatty acid release (Gelb et al. 2000; Murakami et al. 2003).
High doses of TCDD and dioxin-like chemicals cause feed refusal, body weight loss, and exhaustion of energy sources, collectively referred to as “wasting syndrome” (Viluksela et al. 1995). PCB153 is linked to increased glucose consumption in MCF-10A human non-tumorigenic mammary epithelial cells (Venkatesha et al. 2010). TCDD and MIX repression of the glucose metabolism genes, including glutamate oxaloacetate soluble 1, (Got1), glycerol-phosphate dehydrogenase 2, mitochondrial (Gpd2), and pyruvate dehydrogenase kinase, isoenzyme 1 (Pdk1), are consistent with the TCDD repression of gluconeogenesis in chick embryo hepatocytes and dioxin-mediated reversal of hyperglycemia in a diabetes rat model (Diani-Moore et al. 2010; Fried et al. 2010). MIX also repressed Got1 expression, but modeling did not confirm non-additive expression.
The lack of hepatocellular necrosis in PCB153-treated mice compared to TCDD and MIX, is consistent with reports of decreases in PCB153-mediated cell death in mouse hepatocytes (Tharappel et al. 2002; Lu et al. 2004). In contrast, TCDD and MIX induced Tnfaip2 and Tnfaip8l1, members of the tumor necrosis factor (TNF)-signaling pathway, which mediate cell death and inflammation (Tracey and Cerami 1994). In addition, TCDD and MIX, but not PCB153, induced pro-apoptotic Bcl2l11and Htatip2 (Shi et al. 2008; Seitz et al. 2010).
TCPOBOP, a potent CAR ligand, also increased hepatic proliferation by 30–50% when co-treated with TCDD (Mitchell et al. 2010), suggesting TCDD synergizes with TCPOBOP to induce hyperplasia (Mitchell et al. 2010). MIX-induced hyperplasia is consistent with the induction of Myc, a transcription factor associated with cell proliferation, growth and metabolism (Dang 1999). Early response 3 (Ier3), which is anti-apoptotic and associated with cell proliferation (Wu 2003), is regulated by Myc, and was induced by TCDD and MIX, but not PCB153.
Histopathology also revealed PCB153 did not elicit inflammatory cell infiltration, unlike TCDD and MIX. The number of immune related genes increased following TCDD and MIX treatment, including major histocompatibility complex molecules H2-Eb1, H2-DMa, and H2-DMb1 (Gottstein et al. 2010). However, increases in the levels of these transcripts are not due to AhR-mediated expression but rather the coincident infiltration of immune cells.
MIX synergistically induced Serpinb6a, a serine protease inhibitor associated with inflammation and cell death (Scott et al. 2007). Serpinb6a is highly expressed by mast cells in all organs and may regulate endogenous β-tryptase in the cytoplasm, a biomarker of mast cell activation and mastocytosis immune response (Strik et al. 2004). Although PCB153 did not affect Serpinb6a expression, its induction by TCDD at later time points and synergistic induction by MIX was paralleled by the increased incidence of immune cell infiltration.
MIX also suppressed Dexi, a dexamethasone induced transcript that is regulated by PXR (Huss and Kasper 2000; Edgar et al. 2001) between 24 and 168 h. In contrast, Dexi was only transiently repressed by TCDD or PCB153 alone. Glucocorticoids, like dexamethasone, are linked to the suppression of inflammatory responses, partially by regulated expression of TNF (Noti et al. 2010). MIX approached significant (p=0.0806) synergistic suppression of Dexi, concomitant with histopathological findings indicating significant immune cell infiltration.
Dysferlin (Dysf), a transmembrane protein implicated in calcium-dependent sarcolemmal membrane repair, exhibited the most dramatic synergistic response following MIX treatment. Dysf-deficient skeletal muscles induce and activate key inflammasome adaptor components, including NACHT, LRR and PYD-containing proteins (Rawat et al. 2010). The C2 domains of dysferlin have lipid binding specificity, and facilitate interactions with lipid bilayer components including phosphatidylserine (PtdS), phosphatidylinositol 4-phosphate (PtdIns[4]P) and phosphatidylinositol 4,5-bisphosphate (PtdIns[4,5]P2) in a calcium-dependent fashion (Boesze-Battaglia and Schimmel 1997; Di Paolo and De Camilli 2006; Therrien et al. 2009). Interestingly, deficiencies in phosphatidylcholine synthesis, another major membrane component, are linked to muscular dystrophy (Li et al. 2005). MIX elicited the most significant changes in hepatic architecture including cell size and number that may be linked to cellular membrane stress. Dysf induction by MIX during membrane injury may lead to calcium-dependent interactions with PtdS and PtdIns to patch and repair membrane damage (Therrien et al. 2009).
In summary, we have used a “no-interaction” hypothesis to statistically determine departure from additivity in a dose-response study linking gene expression changes to phenotypic endpoints. PCB153 co-administration with TCDD elicited significant synergistic gene expression responses associated with increases in RLW, hepatic cell size and number, immune response and lipid accumulation in ovariectomized C57BL/6 mice at 24 h. Complementary dose-response studies at other time points are likely to identify additional non-additive interactions that can be phenotypically anchored to apical endpoints. Interestingly, our analysis did not identify examples of significant gene expression antagonism, likely due to insufficient power and modest levels of repression. Moreover, non-additive changes in protein, enzymatic activity and metabolite levels may not be reflected in microarray data (Borgert 2007). Consequently, the further development of predictive models that integrate pharmacokinetic and pharmacodynamic components from disparate data sets (e.g., microarray, proteomic, metabolomic) are required in order to further elucidate the effects of mixture composition on elicited toxic effects.
Supplementary Material
Highlights.
MIX (TCDD:PCB153 at 1:10,000 ratio) exposure leads to non-additive gene expression.
MIX-induced liver weights are significantly greater relative to single chemicals.
MIX exposure leads to potentiation of hepatic PCB153 levels compared to TCDD.
MIX synergistically induces expression of Nqo1, Dysf, Pla2g12a, Serpinb6a, and Srxn1.
Non-additive gene expression supports putative non-additive phenotypic responses.
Acknowledgments
FUNDING: This work was supported by the National Institute of Environmental Health Sciences Superfund Basic Research Program (P42ES04911).
The authors thank Agnes Forgacs, Michelle Angrish and Suntae Kim for critically reviewing this manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bannister R, Safe S. Synergistic interactions of 2,3,7,8-TCDD and 2,2′,4,4′,5,5′-hexachlorobiphenyl in C57BL/6J and DBA/2J mice: role of the Ah receptor. Toxicology. 1987;44:159–169. doi: 10.1016/0300-483x(87)90146-6. [DOI] [PubMed] [Google Scholar]
- Biegel L, Harris M, Davis D, Rosengren R, Safe L, Safe S. 2,2′,4,4′,5,5′-hexachlorobiphenyl as a 2,3,7,8-tetrachlorodibenzo-p-dioxin antagonist in C57BL/6J mice. Toxicol Appl Pharmacol. 1989;97:561–571. doi: 10.1016/0041-008x(89)90261-5. [DOI] [PubMed] [Google Scholar]
- Blumberg B, Evans RM. Orphan nuclear receptors--new ligands and new possibilities. Genes & development. 1998;12:3149–3155. doi: 10.1101/gad.12.20.3149. [DOI] [PubMed] [Google Scholar]
- Boesze-Battaglia K, Schimmel R. Cell membrane lipid composition and distribution: implications for cell function and lessons learned from photoreceptors and platelets. J Exp Biol. 1997;200:2927–2936. doi: 10.1242/jeb.200.23.2927. [DOI] [PubMed] [Google Scholar]
- Borgert CJ. Predicting interactions from mechanistic information: can omic data validate theories? Toxicol Appl Pharmacol. 2007;223:114–120. doi: 10.1016/j.taap.2007.01.002. [DOI] [PubMed] [Google Scholar]
- Borgert CJ, Price B, Wells CS, Simon GS. Evaluating chemical interaction studies for mixture risk assessment. Human and Ecological Risk Assessment. 2001;7:259–305. [Google Scholar]
- Boverhof DR, Burgoon LD, Tashiro C, Chittim B, Harkema JR, Jump DB, Zacharewski TR. Temporal and dose-dependent hepatic gene expression patterns in mice provide new insights into TCDD-Mediated hepatotoxicity. Toxicol Sci. 2005;85:1048–1063. doi: 10.1093/toxsci/kfi162. [DOI] [PubMed] [Google Scholar]
- Boverhof DR, Burgoon LD, Tashiro C, Sharratt B, Chittim B, Harkema JR, Mendrick DL, Zacharewski TR. Comparative toxicogenomic analysis of the hepatotoxic effects of TCDD in Sprague Dawley rats and C57BL/6 mice. Toxicol Sci. 2006;94:398–416. doi: 10.1093/toxsci/kfl100. [DOI] [PubMed] [Google Scholar]
- Burgoon LD, Eckel-Passow JE, Gennings C, Boverhof DR, Burt JW, Fong CJ, Zacharewski TR. Protocols for the assurance of microarray data quality and process control. Nucleic Acids Res. 2005;33:e172. doi: 10.1093/nar/gni167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgoon LD, Zacharewski TR. dbZach toxicogenomic information management system. Pharmacogenomics. 2007;8:287–291. doi: 10.2217/14622416.8.3.287. [DOI] [PubMed] [Google Scholar]
- Commoner B, Shapiro K, Webster T. The Origin and Health Risks of PCDD and PCDF. Waste Management & Research. 1987;5:327–346. [Google Scholar]
- Craft ES, DeVito MJ, Crofton KM. Comparative responsiveness of hypothyroxinemia and hepatic enzyme induction in Long-Evans rats versus C57BL/6J mice exposed to TCDD-like and phenobarbital-like polychlorinated biphenyl congeners. Toxicol Sci. 2002;68:372–380. doi: 10.1093/toxsci/68.2.372. [DOI] [PubMed] [Google Scholar]
- Dang CV. c-Myc target genes involved in cell growth, apoptosis, and metabolism. Molecular and cellular biology. 1999;19:1–11. doi: 10.1128/mcb.19.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Jongh J, DeVito M, Diliberto J, Van den Berg M, Birnbaum L. The effects of 2,2′,4,4′,5,5′-hexachlorobiphenyl cotreatment on the disposition of 2,3,7,8-tetrachlorodibenzo-p-dioxin in mice. Toxicol Lett. 1995;80:131–137. doi: 10.1016/0378-4274(95)03387-z. [DOI] [PubMed] [Google Scholar]
- De Jongh J, Wondergem F, Seinen W, Van den Berg M. Toxicokinetic interactions in the liver of the C57BL/6J mouse after administration of a single oral dose of a binary mixture of some PCBs. Chemosphere. 1992;25:1165–1170. [Google Scholar]
- Denison MS, Heath-Pagliuso S. The Ah receptor: a regulator of the biochemical and toxicological actions of structurally diverse chemicals. Bull Environ Contam Toxicol. 1998;61:557–568. doi: 10.1007/pl00002973. [DOI] [PubMed] [Google Scholar]
- Denison MS, Pandini A, Nagy SR, Baldwin EP, Bonati L. Ligand binding and activation of the Ah receptor. Chemico-biological interactions. 2002;141:3–24. doi: 10.1016/s0009-2797(02)00063-7. [DOI] [PubMed] [Google Scholar]
- Dennis G, Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome biology. 2003;4:P3. [PubMed] [Google Scholar]
- Dere E, Forgacs AL, Zacharewski TR, Burgoon LD. Genome-wide computational analysis of dioxin response element location and distribution in the human, mouse and rat genomes. Chem Res Toxicol. 2011a;24:494–504. doi: 10.1021/tx100328r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dere E, Lee AW, Burgoon LD, Zacharewski TR. Differences in TCDD-elicited gene expression profiles in human HepG2, mouse Hepa1c1c7 and rat H4IIE hepatoma cells. BMC Genomics. 2011b;12:193. doi: 10.1186/1471-2164-12-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeVito MJ, Ross DG, Dupuy AE, Jr, Ferrario J, McDaniel D, Birnbaum LS. Dose-response relationships for disposition and hepatic sequestration of polyhalogenated dibenzo-p-dioxins, dibenzofurans, and biphenyls following subchronic treatment in mice. Toxicol Sci. 1998;46:223–234. doi: 10.1006/toxs.1998.2530. [DOI] [PubMed] [Google Scholar]
- Di Paolo G, De Camilli P. Phosphoinositides in cell regulation and membrane dynamics. Nature. 2006;443:651–657. doi: 10.1038/nature05185. [DOI] [PubMed] [Google Scholar]
- Diani-Moore S, Ram P, Li X, Mondal P, Youn DY, Sauve AA, Rifkind AB. Identification of the aryl hydrocarbon receptor target gene Tiparp as a mediator of suppression of hepatic gluconeogenesis by 2,3,7,8-tetrachlorodibenzo-p-dioxin and of nicotinamide as a corrective agent for this effect. J Biol Chem. 2010;285:38801–38810. doi: 10.1074/jbc.M110.131573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diliberto JJ, Burgin DE, Birnbaum LS. Effects of CYP1A2 on disposition of 2,3,7, 8-tetrachlorodibenzo-p-dioxin, 2,3,4,7,8-pentachlorodibenzofuran, and 2,2′,4,4′,5,5′-hexachlorobiphenyl in CYP1A2 knockout and parental (C57BL/6N and 129/Sv) strains of mice. Toxicol Appl Pharmacol. 1999;159:52–64. doi: 10.1006/taap.1999.8720. [DOI] [PubMed] [Google Scholar]
- Eckel JE, Gennings C, Chinchilli VM, Burgoon LD, Zacharewski TR. Empirical bayes gene screening tool for time-course or dose-response microarray data. J Biopharm Stat. 2004;14:647–670. doi: 10.1081/BIP-200025656. [DOI] [PubMed] [Google Scholar]
- Eckel JE, Gennings C, Therneau TM, Burgoon LD, Boverhof DR, Zacharewski TR. Normalization of two-channel microarray experiments: a semiparametric approach. Bioinformatics. 2005;21:1078–1083. doi: 10.1093/bioinformatics/bti105. [DOI] [PubMed] [Google Scholar]
- Edgar AJ, Birks EJ, Yacoub MH, Polak JM. Cloning of dexamethasone-induced transcript: a novel glucocorticoid-induced gene that is upregulated in emphysema. Am J Respir Cell Mol Biol. 2001;25:119–124. doi: 10.1165/ajrcmb.25.1.4417. [DOI] [PubMed] [Google Scholar]
- FDA. Dose Selection for Carcinogenicity Studies. 2008:1–10. [Google Scholar]
- Fried KW, Guo GL, Esterly N, Kong B, Rozman KK. 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) reverses hyperglycemia in a type II diabetes mellitus rat model by a mechanism unrelated to PPAR gamma. Drug Chem Toxicol. 2010;33:261–268. doi: 10.3109/01480540903390026. [DOI] [PubMed] [Google Scholar]
- Gelb MH, Valentin E, Ghomashchi F, Lazdunski M, Lambeau G. Cloning and recombinant expression of a structurally novel human secreted phospholipase A2. J Biol Chem. 2000;275:39823–39826. doi: 10.1074/jbc.C000671200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gennings C, Carter WH, Campain JA, Bae DS, Yang RSH. Statistical analysis of interactive cytotoxicity in human epidermal keratinocytes following exposure to a mixture of four metals. J Agric Biol Environ Stat. 2002;7:58–73. [Google Scholar]
- Gottstein B, Wittwer M, Schild M, Merli M, Leib SL, Muller N, Muller J, Jaggi R. Hepatic gene expression profile in mice perorally infected with Echinococcus multilocularis eggs. PLoS One. 2010;5:e9779. doi: 10.1371/journal.pone.0009779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greco D, Kotronen A, Westerbacka J, Puig O, Arkkila P, Kiviluoto T, Laitinen S, Kolak M, Fisher RM, Hamsten A, Auvinen P, Yki-Jarvinen H. Gene expression in human NAFLD. Am J Physiol Gastrointest Liver Physiol. 2008;294:G1281–1287. doi: 10.1152/ajpgi.00074.2008. [DOI] [PubMed] [Google Scholar]
- Honkakoski P, Sueyoshi T, Negishi M. Drug-activated nuclear receptors CAR and PXR. Annals of medicine. 2003;35:172–182. doi: 10.1080/07853890310008224. [DOI] [PubMed] [Google Scholar]
- Huss JM, Kasper CB. Two-stage glucocorticoid induction of CYP3A23 through both the glucocorticoid and pregnane X receptors. Mol Pharmacol. 2000;58:48–57. doi: 10.1124/mol.58.1.48. [DOI] [PubMed] [Google Scholar]
- Kimbrough RD. Polychlorinated biphenyls (PCBs) and human health: an update. Crit Rev Toxicol. 1995;25:133–163. doi: 10.3109/10408449509021611. [DOI] [PubMed] [Google Scholar]
- Klaassen CD, Reisman SA. Nrf2 the rescue: effects of the antioxidative/electrophilic response on the liver. Toxicol Appl Pharmacol. 2010;244:57–65. doi: 10.1016/j.taap.2010.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knerr S, Schrenk D. Carcinogenicity of “non-dioxinlike” polychlorinated biphenyls. Crit Rev Toxicol. 2006;36:663–694. doi: 10.1080/10408440600845304. [DOI] [PubMed] [Google Scholar]
- Kopec AK, Boverhof DR, Burgoon LD, Ibrahim-Aibo D, Harkema JR, Tashiro C, Chittim B, Zacharewski TR. Comparative Toxicogenomic Examination of the Hepatic Effects of PCB126 and TCDD in Immature, Ovariectomized C57BL/6 Mice. Toxicol Sci. 2008;102:61–75. doi: 10.1093/toxsci/kfm289. [DOI] [PubMed] [Google Scholar]
- Kopec AK, Burgoon LD, Ibrahim-Aibo D, Burg AR, Lee AW, Tashiro C, Potter D, Sharratt B, Harkema JR, Rowlands JC, Budinsky RA, Zacharewski TR. Automated Dose-Response Analysis and Comparative Toxicogenomic Evaluation of the Hepatic Effects Elicited by TCDD, TCDF and PCB126 in C57BL/6 Mice. Toxicol Sci. 2010a;118:286–297. doi: 10.1093/toxsci/kfq236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopec AK, Burgoon LD, Ibrahim-Aibo D, Mets BD, Tashiro C, Potter D, Sharratt B, Harkema JR, Zacharewski TR. PCB153-elicited hepatic responses in the immature, ovariectomized C57BL/6 mice: Comparative toxicogenomic effects of dioxin and non-dioxin-like ligands. Toxicol Appl Pharmacol. 2010b;243:359–371. doi: 10.1016/j.taap.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Wada T, Febbraio M, He J, Matsubara T, Lee MJ, Gonzalez FJ, Xie W. A Novel Role for the Dioxin Receptor in Fatty Acid Metabolism and Hepatic Steatosis. Gastroenterology. 2010;139:653–663. doi: 10.1053/j.gastro.2010.03.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemaire G, Benod C, Nahoum V, Pillon A, Boussioux AM, Guichou JF, Subra G, Pascussi JM, Bourguet W, Chavanieu A, Balaguer P. Discovery of a highly active ligand of human pregnane x receptor: a case study from pharmacophore modeling and virtual screening to “in vivo” biological activity. Mol Pharmacol. 2007;72:572–581. doi: 10.1124/mol.106.033415. [DOI] [PubMed] [Google Scholar]
- Li Z, Agellon LB, Vance DE. Phosphatidylcholine homeostasis and liver failure. J Biol Chem. 2005;280:37798–37802. doi: 10.1074/jbc.M508575200. [DOI] [PubMed] [Google Scholar]
- Liu MJ, Takahashi Y, Wada T, He J, Gao J, Tian Y, Li S, Xie W. The aldo-keto reductase Akr1b7 gene is a common transcriptional target of xenobiotic receptors pregnane X receptor and constitutive androstane receptor. Mol Pharmacol. 2009;76:604–611. doi: 10.1124/mol.109.057455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Z, Lee EY, Robertson LW, Glauert HP, Spear BT. Effect of 2,2′,4,4′,5,5′-hexachlorobiphenyl (PCB-153) on hepatocyte proliferation and apoptosis in mice deficient in the p50 subunit of the transcription factor NF-kappaB. Toxicol Sci. 2004;81:35–42. doi: 10.1093/toxsci/kfh193. [DOI] [PubMed] [Google Scholar]
- Ma Q, Kinneer K, Bi Y, Chan JY, Kan YW. Induction of murine NAD(P)H:quinone oxidoreductase by 2,3,7,8-tetrachlorodibenzo-p-dioxin requires the CNC (cap ‘n’ collar) basic leucine zipper transcription factor Nrf2 (nuclear factor erythroid 2-related factor 2): cross-interaction between AhR (aryl hydrocarbon receptor) and Nrf2 signal transduction. The Biochemical journal. 2004;377:205–213. doi: 10.1042/BJ20031123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maglich JM, Stoltz CM, Goodwin B, Hawkins-Brown D, Moore JT, Kliewer SA. Nuclear pregnane x receptor and constitutive androstane receptor regulate overlapping but distinct sets of genes involved in xenobiotic detoxification. Mol Pharmacol. 2002;62:638–646. doi: 10.1124/mol.62.3.638. [DOI] [PubMed] [Google Scholar]
- Masahiko N, Honkakoski P. Induction of drug metabolism by nuclear receptor CAR: molecular mechanisms and implications for drug research. Eur J Pharm Sci. 2000;11:259–264. doi: 10.1016/s0928-0987(00)00112-3. [DOI] [PubMed] [Google Scholar]
- Mason G, Safe S. Synthesis, biologic and toxic effects of the major 2,3,7,8-tetrachlorodibenzo-p-dioxin metabolites in the rat. Toxicology. 1986;41:153–159. doi: 10.1016/0300-483x(86)90196-4. [DOI] [PubMed] [Google Scholar]
- Matsumoto K, Endo S, Ishikura S, Matsunaga T, Tajima K, El-Kabbani O, Hara A. Enzymatic properties of a member (AKR1C20) of the aldo-keto reductase family. Biological & pharmaceutical bulletin. 2006;29:539–542. doi: 10.1248/bpb.29.539. [DOI] [PubMed] [Google Scholar]
- Mitchell KA, Wilson SR, Elferink CJ. The activated aryl hydrocarbon receptor synergizes mitogen-induced murine liver hyperplasia. Toxicology. 2010;276:103–109. doi: 10.1016/j.tox.2010.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon YA, Hammer RE, Horton JD. Deletion of ELOVL5 leads to fatty liver through activation of SREBP-1c in mice. Journal of lipid research. 2008;50:412–423. doi: 10.1194/jlr.M800383-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullin MD, Pochini CM, McCrindle S, Romkes M, Safe SH, Safe LM. High-resolution PCB analysis: synthesis and chromatographic properties of all 209 PCB congeners. Environ Set Technol. 1984;18:468–476. doi: 10.1021/es00124a014. [DOI] [PubMed] [Google Scholar]
- Murakami M, Masuda S, Shimbara S, Bezzine S, Lazdunski M, Lambeau G, Gelb MH, Matsukura S, Kokubu F, Adachi M, Kudo I. Cellular arachidonate-releasing function of novel classes of secretory phospholipase A2s (groups III and XII) J Biol Chem. 2003;278:10657–10667. doi: 10.1074/jbc.M211325200. [DOI] [PubMed] [Google Scholar]
- Nebert DW, Petersen DD, Fornace AJ., Jr Cellular responses to oxidative stress: the [Ah] gene battery as a paradigm. Environ Health Perspect. 1990;88:13–25. doi: 10.1289/ehp.908813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nebert DW, Roe AL, Dieter MZ, Solis WA, Yang Y, Dalton TP. Role of the aromatic hydrocarbon receptor and [Ah] gene battery in the oxidative stress response, cell cycle control, and apoptosis. Biochem Pharmacol. 2000;59:65–85. doi: 10.1016/s0006-2952(99)00310-x. [DOI] [PubMed] [Google Scholar]
- Noti M, Corazza N, Mueller C, Berger B, Brunner T. TNF suppresses acute intestinal inflammation by inducing local glucocorticoid synthesis. The Journal of experimental medicine. 2010;207:1057–1066. doi: 10.1084/jem.20090849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NTP. NTP technical report on the toxicology and carcinogenesis studies of 2, 2 ′,4,4′,5,5′-hexachlorobiphenyl (PCB 153) (CAS No. 35065-27-1) in female Harlan Sprague-Dawley rats (Gavage studies) Natl Toxicol Program Tech Rep Ser 4–168. 2006a [PubMed] [Google Scholar]
- NTP. NTP technical report on the toxicology and carcinogenesis studies of 2, 3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) (CAS No. 1746-01-6) in female Harlan Sprague-Dawley rats (Gavage Studies) Natl Toxicol Program Tech Rep Ser 4–232. 2006b [PubMed] [Google Scholar]
- NTP. NTP Toxicology and Carcinogenesis Studies of 3, 3′,4,4′,5-pentachlorobiphenyl (PCB 126) (CAS No. 57465-28-8) in Female Harlan Sprague-Dawley Rats (Gavage Studies) National Toxicology Program technical report series 4–246. 2006c [PubMed] [Google Scholar]
- NTP. NTP Toxicology and Carcinogenesis Studies of a Binary Mixture of 3, 3′,4,4′,5-Pentachlorobiphenyl (PCB 126) (CAS No. 57465-28-8) and 2,2′,4,4′,5,5′-Hexachlorobiphenyl (PCB 153) (CAS No. 35065-27-1) in Female Harlan Sprague-Dawley Rats (Gavage Studies) National Toxicology Program technical report series 1–258. 2006d [PubMed] [Google Scholar]
- Park JW, Mieyal JJ, Rhee SG, Chock PB. Deglutathionylation of 2-Cys peroxiredoxin is specifically catalyzed by sulfiredoxin. J Biol Chem. 2009;284:23364–23374. doi: 10.1074/jbc.M109.021394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips JM, Burgoon LD, Goodman JI. Phenobarbital (PB) Elicits Unique, Early Changes in the Expression of Hepatic Genes That Affect Critical Pathways in Tumor-Prone B6C3F1 Mice. Toxicol Sci. 2009;109:193–205. doi: 10.1093/toxsci/kfp050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poland A, Glover E. Studies on the mechanism of toxicity of the chlorinated dibenzo-p-dioxins. Environ Health Perspect. 1973;5:245–251. doi: 10.1289/ehp.7305245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poland A, Knutson JC. 2,3,7,8-tetrachlorodibenzo-p-dioxin and related halogenated aromatic hydrocarbons: examination of the mechanism of toxicity. Annu Rev Pharmacol Toxicol. 1982;22:517–554. doi: 10.1146/annurev.pa.22.040182.002505. [DOI] [PubMed] [Google Scholar]
- Rawat R, Cohen TV, Ampong B, Francia D, Henriques-Pons A, Hoffman EP, Nagaraju K. Inflammasome up-regulation and activation in dysferlin-deficient skeletal muscle. Am J Pathol. 2010;176:2891–2900. doi: 10.2353/ajpath.2010.090058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth A, Looser R, Kaufmann M, Blattler SM, Rencurel F, Huang W, Moore DD, Meyer UA. Regulatory cross-talk between drug metabolism and lipid homeostasis: constitutive androstane receptor and pregnane X receptor increase Insig-1 expression. Mol Pharmacol. 2008;73:1282–1289. doi: 10.1124/mol.107.041012. [DOI] [PubMed] [Google Scholar]
- Safe S. Polychlorinated biphenyls (PCBs), dibenzo-p-dioxins (PCDDs), dibenzofurans (PCDFs), and related compounds: environmental and mechanistic considerations which support the development of toxic equivalency factors (TEFs) Crit Rev Toxicol. 1990;21:51–88. doi: 10.3109/10408449009089873. [DOI] [PubMed] [Google Scholar]
- Safe S. Limitations of the toxic equivalency factor approach for risk assessment of TCDD and related compounds. Teratog Carcinog Mutagen. 1997;17:285–304. [PubMed] [Google Scholar]
- Safe S. Molecular biology of the Ah receptor and its role in carcinogenesis. Toxicol Lett. 2001;120:1–7. doi: 10.1016/s0378-4274(01)00301-0. [DOI] [PubMed] [Google Scholar]
- Safe S, Robertson LW, Safe L, Parkinson A, Bandiera S, Sawyer T, Campbell MA. Halogenated biphenyls: molecular toxicology. Can J Physiol Pharmacol. 1982;60:1057–1064. doi: 10.1139/y82-151. [DOI] [PubMed] [Google Scholar]
- Safe SH. Polychlorinated biphenyls (PCBs): environmental impact, biochemical and toxic responses, and implications for risk assessment. Crit Rev Toxicol. 1994;24:87–149. doi: 10.3109/10408449409049308. [DOI] [PubMed] [Google Scholar]
- Schecter A, Stanley J, Boggess K, Masuda Y, Mes J, Wolff M, Furst P, Furst C, Wilson-Yang K, Chisholm B. Polychlorinated biphenyl levels in the tissues of exposed and nonexposed humans. Environ Health Perspect. 1994;102(Suppl 1):149–158. doi: 10.1289/ehp.94102s1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott FL, Sun J, Whisstock JC, Kato K, Bird PI. SerpinB6 is an inhibitor of kallikrein-8 in keratinocytes. Journal of biochemistry. 2007;142:435–442. doi: 10.1093/jb/mvm156. [DOI] [PubMed] [Google Scholar]
- Seitz SJ, Schleithoff ES, Koch A, Schuster A, Teufel A, Staib F, Stremmel W, Melino G, Krammer PH, Schilling T, Muller M. Chemotherapy-induced apoptosis in hepatocellular carcinoma involves the p53 family and is mediated via the extrinsic and the intrinsic pathway. Int J Cancer. 2010;126:2049–2066. doi: 10.1002/ijc.24861. [DOI] [PubMed] [Google Scholar]
- Sheehan DC, Hrapchak BB. Theory and Practice of Histotechnology. Mosby; St. Louis: 1980. [Google Scholar]
- Shi M, Yan SG, Xie ST, Wang HN. Tip30-induced apoptosis requires translocation of Bax and involves mitochondrial release of cytochrome c and Smac/DIABLO in hepatocellular carcinoma cells. Biochimica et biophysica acta. 2008;1783:263–274. doi: 10.1016/j.bbamcr.2007.10.006. [DOI] [PubMed] [Google Scholar]
- Singh A, Ling G, Suhasini AN, Zhang P, Yamamoto M, Navas-Acien A, Cosgrove G, Tuder RM, Kensler TW, Watson WH, Biswal S. Nrf2-dependent sulfiredoxin-1 expression protects against cigarette smoke-induced oxidative stress in lungs. Free radical biology & medicine. 2009;46:376–386. doi: 10.1016/j.freeradbiomed.2008.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staal YC, Hebels DG, van Herwijnen MH, Gottschalk RW, van Schooten FJ, van Delft JH. Binary PAH mixtures cause additive or antagonistic effects on gene expression but synergistic effects on DNA adduct formation. Carcinogenesis. 2007;28:2632–2640. doi: 10.1093/carcin/bgm182. [DOI] [PubMed] [Google Scholar]
- Strik MC, Wolbink A, Wouters D, Bladergroen BA, Verlaan AR, van Houdt IS, Hijlkema S, Hack CE, Kummer JA. Intracellular serpin SERPINB6 (PI6) is abundantly expressed by human mast cells and forms complexes with beta-tryptase monomers. Blood. 2004;103:2710–2717. doi: 10.1182/blood-2003-08-2981. [DOI] [PubMed] [Google Scholar]
- Suh J, Kang JS, Yang KH, Kaminski NE. Antagonism of aryl hydrocarbon receptor-dependent induction of CYP1A1 and inhibition of IgM expression by di-ortho-substituted polychlorinated biphenyls. Toxicol Appl Pharmacol. 2003;187:11–21. doi: 10.1016/s0041-008x(02)00040-6. [DOI] [PubMed] [Google Scholar]
- Sun YV, Boverhof DR, Burgoon LD, Fielden MR, Zacharewski TR. Comparative analysis of dioxin response elements in human, mouse and rat genomic sequences. Nucleic Acids Res. 2004;32:4512–4523. doi: 10.1093/nar/gkh782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabb MM, Kholodovych V, Grun F, Zhou C, Welsh WJ, Blumberg B. Highly chlorinated PCBs inhibit the human xenobiotic response mediated by the steroid and xenobiotic receptor (SXR) Environ Health Perspect. 2004;112:163–169. doi: 10.1289/ehp.6560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takaishi K, Duplomb L, Wang MY, Li J, Unger RH. Hepatic insig-1 or -2 overexpression reduces lipogenesis in obese Zucker diabetic fatty rats and in fasted/refed normal rats. Proc Natl Acad Sci U S A. 2004;101:7106–7111. doi: 10.1073/pnas.0401715101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tharappel JC, Lee EY, Robertson LW, Spear BT, Glauert HP. Regulation of cell proliferation, apoptosis, and transcription factor activities during the promotion of liver carcinogenesis by polychlorinated biphenyls. Toxicol Appl Pharmacol. 2002;179:172–184. doi: 10.1006/taap.2001.9360. [DOI] [PubMed] [Google Scholar]
- Therrien C, Di Fulvio S, Pickles S, Sinnreich M. Characterization of lipid binding specificities of dysferlin C2 domains reveals novel interactions with phosphoinositides. Biochemistry. 2009;48:2377–2384. doi: 10.1021/bi802242r. [DOI] [PubMed] [Google Scholar]
- Tijet N, Boutros PC, Moffat ID, Okey AB, Tuomisto J, Pohjanvirta R. Aryl hydrocarbon receptor regulates distinct dioxin-dependent and dioxin-independent gene batteries. Mol Pharmacol. 2006;69:140–153. doi: 10.1124/mol.105.018705. [DOI] [PubMed] [Google Scholar]
- Tracey KJ, Cerami A. Tumor necrosis factor: a pleiotropic cytokine and therapeutic target. Annu Rev Med. 1994;45:491–503. doi: 10.1146/annurev.med.45.1.491. [DOI] [PubMed] [Google Scholar]
- van Birgelen AP, Fase KM, van der Kolk J, Poiger H, Brouwer A, Seinen W, van den Berg M. Synergistic effect of 2,2′,4,4′,5,5′-hexachlorobiphenyl and 2,3,7,8-tetrachlorodibenzo-p-dioxin on hepatic porphyrin levels in the rat. Environ Health Perspect. 1996a;104:550–557. doi: 10.1289/ehp.96104550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Birgelen AP, Ross DG, DeVito MJ, Birnbaum LS. Interactive effects between 2,3,7,8-tetrachlorodibenzo-p-dioxin and 2,2′,4,4′,5,5′-hexachlorobiphenyl in female B6C3F1 mice: tissue distribution and tissue-specific enzyme induction. Fundam Appl Toxicol. 1996b;34:118–131. doi: 10.1006/faat.1996.0182. [DOI] [PubMed] [Google Scholar]
- Van den Berg M, Birnbaum LS, Denison M, De Vito M, Farland W, Feeley M, Fiedler H, Hakansson H, Hanberg A, Haws L, Rose M, Safe S, Schrenk D, Tohyama C, Tritscher A, Tuomisto J, Tysklind M, Walker N, Peterson RE. The 2005 World Health Organization reevaluation of human and Mammalian toxic equivalency factors for dioxins and dioxin-like compounds. Toxicol Sci. 2006;93:223–241. doi: 10.1093/toxsci/kfl055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Kolk J, van Birgelen AP, Poiger H, Schlatter C. Interactions of 2,2′,4,4′,5,5′-hexachlorobiphenyl and 2,3,7,8-tetrachlorodibenzo-p-dioxin in a subchronic feeding study in the rat. Chemosphere. 1992;25:2023–2027. [Google Scholar]
- Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome biology. 2002;3:RESEARCH0034. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatesha VA, Kalen AL, Sarsour EH, Goswami PC. PCB-153 exposure coordinates cell cycle progression and cellular metabolism in human mammary epithelial cells. Toxicol Lett. 2010;196:110–116. doi: 10.1016/j.toxlet.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viluksela M, Stahl BU, Rozman KK. Tissue-specific effects of 2,3,7,8-Tetrachlorodibenzo-p-dioxin (TCDD) on the activity of phosphoenolpyruvate carboxykinase (PEPCK) in rats. Toxicol Appl Pharmacol. 1995;135:308–315. doi: 10.1006/taap.1995.1237. [DOI] [PubMed] [Google Scholar]
- Wu MX. Roles of the stress-induced gene IEX-1 in regulation of cell death and oncogenesis. Apoptosis. 2003;8:11–18. doi: 10.1023/a:1021688600370. [DOI] [PubMed] [Google Scholar]
- Yeager RL, Reisman SA, Aleksunes LM, Klaassen CD. Introducing the “TCDD-inducible AhR-Nrf2 gene battery”. Toxicol Sci. 2009;111:238–246. doi: 10.1093/toxsci/kfp115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai Y, Wada T, Zhang B, Khadem S, Ren S, Kuruba R, Li S, Xie W. A functional cross-talk between liver X receptor-alpha and constitutive androstane receptor links lipogenesis and xenobiotic responses. Mol Pharmacol. 2010;78:666–674. doi: 10.1124/mol.110.064618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao F, Mayura K, Harper N, Safe SH, Phillips TD. Inhibition of 3,3′,4,4′,5-pentachlorobiphenyl-induced fetal cleft palate and immunotoxicity in C57BL/6 mice by 2,2′,4,4′,5,5′-hexachlorobiphenyl. Chemosphere. 1997;34:1605–1613. doi: 10.1016/s0045-6535(97)00456-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.