Abstract
Although oxidative tissue injury often accompanies viral infection, there is little understanding of how it influences virus replication. We show that multiple hepatitis C virus (HCV) genotypes are exquisitely sensitive to oxidative membrane damage, a property distinguishing them from other pathogenic RNA viruses. Lipid peroxidation, regulated in part through sphingosine kinase 2, severely restricts HCV replication in Huh-7 cells and primary human hepatoblasts. Endogenous oxidative membrane damage lowers the 50% effective concentration of direct-acting antivirals, suggesting critical regulation of the conformation of the NS3/4A protease and NS5B polymerase, membrane-bound HCV replicase components. Resistance to lipid peroxidation maps genetically to trans-membrane and membrane-proximal residues within these proteins, and is essential for robust replication in cell culture, as exemplified by the atypical JFH1 strain. Thus, the typical, wild-type HCV replicase is uniquely regulated by lipid peroxidation, providing a novel mechanism for attenuating replication in stressed tissue and possibly facilitating long-term viral persistence.
Reactive oxygen species (ROS) are an unavoidable by-product of aerobic metabolism and a double-edged sword for complex cellular systems1. While central to many disease states, ROS also function as second messengers during embryonic development and, in macrophages, contribute to host defense against infection2,3. Viral infections frequently induce ROS generation, either by stimulating host immune responses or by direct tissue injury4. Hepatitis C virus (HCV), an hepatotropic RNA virus with a unique capacity for persistence5, induces substantial intrahepatic oxidative stress, thereby promoting liver injury6,7. Limited data suggest lipid peroxidation restricts HCV replication8, but how it impairs viral replicative machinery is unknown.
Although HCV is a leading cause of cirrhosis and liver cancer5, many details of its replication remain obscure since most HCV strains replicate poorly in cell culture. A notable exception is JFH1, a genotype 2a virus recovered from a patient with fulminant hepatitis9. JFH1 recapitulates the entire virus lifecycle and replicates efficiently in Huh-7 hepatoma cells9–11. In recent years, it has become a laboratory standard, used in most studies of HCV replication. However, there is very limited understanding of the robust replication phenotype that sets it apart from other HCVs12,13.
Like all positive-strand RNA viruses, the HCV genome is synthesized by a multi-protein replicase complex that assembles in association with intracellular membranes. Known as the ‘membranous web’ in HCV-infected cells14,15, this specialized cytoplasmic compartment provides a platform for viral RNA synthesis. Its membranes are enriched in cholesterol, sphingolipids, and phosphatidylinositol-4-phosphate16,17. Assembly of the membranous web involves recruitment of phosphatidylinositol-4-phosphate-3 kinase and annexin A217–19, and possibly also direct membrane remodeling by nonstructural HCV proteins20. While lipid metabolism also plays key roles in later steps in the virus lifecycle21, viral RNA synthesis is thus closely linked to modifications of intracellular membranes.
Sphingolipids are increased in abundance within the replicase membranes, and are important factors in HCV replication22–25. Sphingomyelin (SM) interacts with and in some genotypes stimulates NS5B, the viral RNA-dependent RNA polymerase23,26. While studying these virus-host interactions in cell culture, we discovered that JFH1 differs from other HCV strains in its response to inhibitors of sphingolipid converting enzymes. These initial observations led to experiments that demonstrate the HCV replicase to be exquisitely sensitive to endogenous lipid peroxidation, a feature lacking in the atypical JFH1 strain and other pathogenic RNA viruses. Our findings suggest that HCV possesses a unique capacity to sense lipid peroxides induced by infection, and to respond to their presence by restricting viral RNA synthesis, thereby limiting virus replication and possibly facilitating virus persistence.
RESULTS
Sphingosine kinase 2 regulates HCV replication
We determined how inhibitors of sphingolipid converting enzymes influence replication of two cell culture-adapted HCVs: H77S.3/GLuc, a genotype 1a virus, and HJ3-5/GLuc, an inter-genotypic chimera expressing the genotype 2a JFH1 replicase (Fig. 1a). To assess replication, we monitored Gaussia princeps luciferase (GLuc) produced from in-frame insertions in each viral genome after transfecting Huh-7.5 cells with synthetic RNA27. Surprisingly, these viral RNAs demonstrated contrary responses to many inhibitors, including most notably SKI, a sphingosine kinase (SPHK) inhibitor (Fig. 1b and Supplementary Fig. 1a,b). We also observed contrasting responses to sphingolipid supplementation (Supplementary Fig. 1c). SKI (1 μM) enhanced replication of H77S.3/GLuc as well as N.2/GLuc, a cell culture-adapted genotype 1b virus (Fig. 1a), by 3–6 fold, while suppressing replication of HJ3-5 (Fig. 1b,c). These effects were evident within 48 h of exposure. SKI also enhanced H77S.3 protein expression 10-fold, while slightly suppressing HJ3-5 protein expression (Fig. 1d). Thus, changes in the cellular environment induced by SKI favor H77S.3/GLuc and N.2/GLuc replication, while inhibiting HJ3-5/GLuc. These effects are not due to altered cell proliferation or viral RNA translation, and were also observed with autonomously replicating, subgenomic HCV RNAs (“replicons”) in multiple cell types (Supplementary Fig. 2).
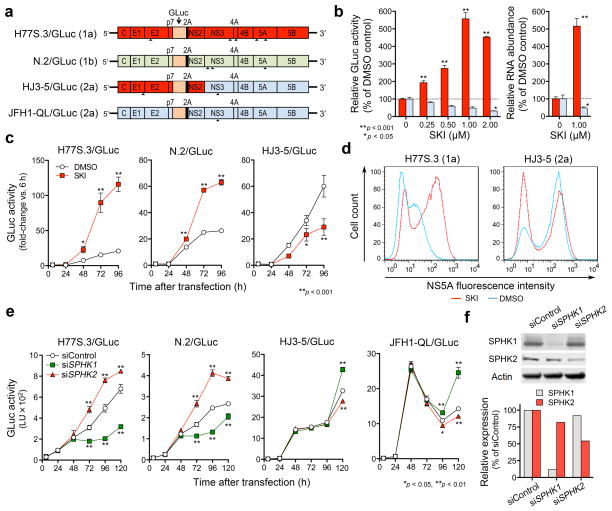
Figure 1.
SKI enhances genotype 1 HCV replication while suppressing JFH1-based viruses by inhibiting type 2 sphingosine kinase (SPHK2). (a) HCV RNA genomes that express Gaussia Luciferase (GLuc) fused to foot-and-mouth disease virus 2A autoprotease as part of the HCV polyprotein. Arrowheads indicate cell culture-adaptive mutations. (b) (left) Dose-response effects of SKI on replication of H77S.3/GLuc (red) or HJ3-5/GLuc (blue) RNAs in Huh-7.5 cells. (right) Effect of 1 μM SKI on replication of H77S.3 (red) or HJ3-5 (blue) RNAs. Data shown represent relative amounts of GLuc secreted between 48–72 h (left) or intracellular RNA levels at 72 h (right). (c) Effect of 1 μM SKI on GLuc activities of the indicated viruses are presented as fold-change from baseline (6 h). (d) Flow cytometric analysis of NS5A expression in Huh-7.5 cells electroporated with H77S.3 or HJ3-5 RNA and treated with 1 μM SKI or DMSO. (e–f) Effect of siRNAs targeting SPHK isoforms or non-targeting control siRNA on replication of different HCV RNAs (e) and abundance of each SPHK isoform (f). Results represent the mean ± s.e.m. from two independent (b,c,d) or triplicate (e) experiments. *P < 0.05, **P < 0.01.
SPHK is expressed as two isoforms28, which we individually silenced by transfecting cells with gene-specific siRNAs. Partial SPHK2 depletion enhanced replication of H77S.3/GLuc and N.2/GLuc, while SPHK1 depletion inhibited both viruses (Fig. 1e,f). In contrast, replication of HJ3-5/GLuc and cell culture-adapted JFH1 (JFH-QL/GLuc) virus was increased following SPHK1 depletion, and decreased after SPHK2 knockdown. Neither knockdown significantly affected cell proliferation (Supplementary Fig. 2e). Thus, SKI enhances replication of H77S.3/GLuc and N.2/GLuc by inhibiting SPHK2. Consistent with this, SKI preferentially inhibited SPHK2 in cell-free assays (Supplementary Fig. 2f), demonstrating no activity against endogenous SPHK1 at low concentrations (≤ 2 μM) (Supplementary Fig. 2g).
Lipid peroxidation is a key factor in SKI regulation of HCV
Polyunsaturated fatty acids (PUFAs) inhibit replication of genotype 1b HCV replicons by inducing lipid peroxidation8,29. Surprisingly, although PUFAs such as arachidonic acid (ARA), docosahexaenoic acid (DHA) or linoleic acid (LA) potently suppressed H77S.3/GLuc replication without affecting cell viability, HJ3-5/GLuc was highly resistant (Fig. 2a,b and Supplementary Fig. 5a,b). Thus PUFAs appear to phenocopy the effect of SPHK2 on HCV replication, suggesting that SPHK2 promotes lipid peroxidation. Consistent with this hypothesis, SKI completely abolished the inhibitory effects of PUFAs on H77S.3/GLuc and N.2/GLuc (Fig. 2c). SKI also lowered the abundance of malondialdehyde (MDA), a secondary product of peroxidative degradation (Fig. 2d). SKI was as effective as the lipophilic antioxidants vitamin E (VE) and coenzyme Q10 (CoQ10) in reducing MDA abundance in HCV-infected cells, and like VE and CoQ10 prevented large increases in lipid peroxidation induced by PUFAs (Fig. 2d,e). SKI also reduced both endogenous and PUFA-induced 8-isoprostane, an alternative biomarker of lipid peroxidation (Fig. 2e). Conversely, RNAi-mediated depletion of SPHK1 increased MDA abundance, while SPHK2 knockdown, like SKI, reduced it (Fig. 2f). Thus, the contrasting effects of SPHK1 and SPHK2 on HCV replication can be explained by opposing actions on peroxidation of endogenous PUFA. These results identify SPHK2 as an important mediator of lipid peroxidation.
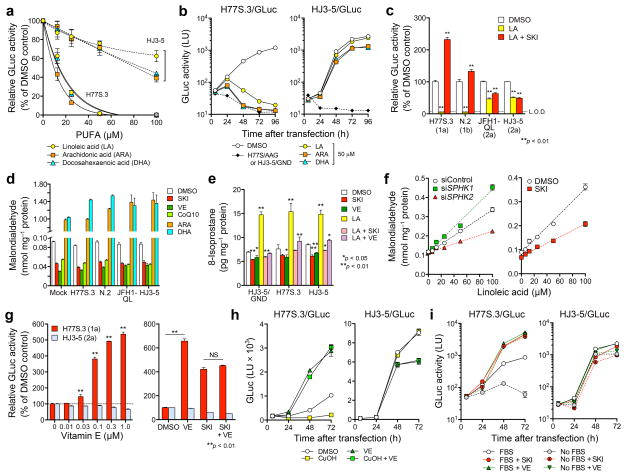
Figure 2.
Differential regulation of HCV strains by SPHK2-mediated lipid peroxidation. (a) Dose-dependent effects of PUFAs on H77S.3/GLuc and HJ3-5/GLuc RNAs in Huh-7.5 cells. Data shown represent percent GLuc activity secreted between 48–72 h relative to DMSO control. (b) Growth kinetics of H77S.3/GLuc and HJ3-5/GLuc RNAs in the presence of 50 μM PUFAs. Data shown are mean ± s.e.m. of GLuc activity in supernatant fluids of two replicate cultures. (c) Cells transfected with HCV RNAs encoding GLuc were treated with DMSO, 100 μM LA or 100 μM LA plus 1 μM SKI. Data shown represent percent GLuc activity secreted between 48–72 h relative to DMSO control. L.O.D. = limit of detection. (d) Effect of 1 μM SKI, 1 μM VE, 100 μM CoQ10 or 50 μM ARA or DHA, on intracellular malondialdehyde (MDA) abundance in cells transfected with the indicated HCV/GLuc RNAs at 72 h. MDA was significantly increased by PUFAs and reduced by SKI or lipophilic antioxidants (P < 0.01). (e) Analysis of 8-isoprostane abundance in cells electroporated with the indicated HCV RNAs and grown in the presence of 1 μM SKI or VE, or 50 μM LA with or without 1 μM SKI and VE for 48 h. (f) Effect of siRNA targeting SPHK isoforms (see Fig. 1f) on MDA accumulation after treatment with increasing concentrations of LA (6.25, 12.5, 25, 50, 100 μM) for 24 h (left panel). MDA levels in Huh-7.5 cells treated with increasing concentrations of LA in the presence of DMSO or 1 μM SKI (right panel). (g) Effects of increasing concentrations of VE (left), 1 μM VE alone, or 1 μM VE plus 1 μM SKI (right) on replication of H77S.3/GLuc and HJ3-5/GLuc RNAs. Data shown represent GLuc secreted between 48–72 h relative to DMSO control. (h) GLuc secretion from Huh-7.5 cells transfected as in a and treated with 10 μM CuOH with or without 10 μM VE. (i) Influence of SKI or VE (each 1 μM) on replication of H77S.3 and HJ3-5 viruses expressing GLuc in cells cultured in the presence or absence of 10% FBS. Medium containing 10% FBS was replaced with FBS-free or 10% FBS media containing SKI, VE or DMSO 6 h after RNA transfection. Data shown represent mean GLuc activity ± s.e.m. from two (a–f,i) or three (g,h) independent experiments. *P < 0.05, **P < 0.01.
Lipid-soluble antioxidants, including VE (α-, rac-β- and γ-tocopherols), CoQ10, and butylated hydroxytoluene (BHT), enhanced H77S.3 replication in a concentration-dependent fashion, as described for a genotype 1b replicon8, but suppressed HJ3-5 replication (Fig. 2g and Supplementary Fig. 5c–e). Importantly, the effects of VE and SKI on H77S.3/GLuc or N.2/GLuc replication were not additive (Fig. 2g and Supplementary Fig. 5f), suggesting that both act via a common antioxidant mechanism. Antioxidants that are not soluble in lipids, such as ebselen and N-acetyl cysteine, and the NADPH oxidase inhibitor diphenyleneiodonium had no effect on H77S.3/GLuc replication (Supplementary Fig. 5g), while cumene hydroperoxide (CuOH), a lipophilic oxidant, suppressed H77S.3/GLuc replication in a VE-reversible fashion (Fig. 2h). Fetal bovine serum (FBS) contains substantial quantities of lipophilic antioxidants8. H77S.3/GLuc replication was 20-fold lower in FBS-free cultures, but enhanced 100-fold with either SKI or VE (Fig. 2i). HJ3-5/GLuc replication was relatively unimpaired in FBS-free medium.
Collectively, these data provide evidence that endogenous lipid peroxidation restricts H77S.3/GLuc and N.2/GLuc replication, while the JFH1 replicase is resistant to both basal and chemically-induced lipid peroxidation.
Endogenous lipid peroxidation restricts infectious HCV yield
Both SKI and VE induced a 10-fold increase in the yield of infectious virus released by H77S.3 RNA-transfected cells, reaching ~20,000 focus-forming units (FFU) ml−1 (Fig. 3a and Supplementary Fig. 6a). Infectious N.2 virus yields were increased up to 100-fold (Fig. 3a). SKI also enhanced virus spread when H77S.3 RNA-transfected cells were co-cultured with non-transfected CFSE-labeled cells (Fig. 3b). In contrast, neither SKI nor VE enhanced infectious yields of JFH1-QL or HJ3-5 virus (Fig. 3a), while SKI reduced the spread of HJ3-5 virus by >40% (Fig. 3b).
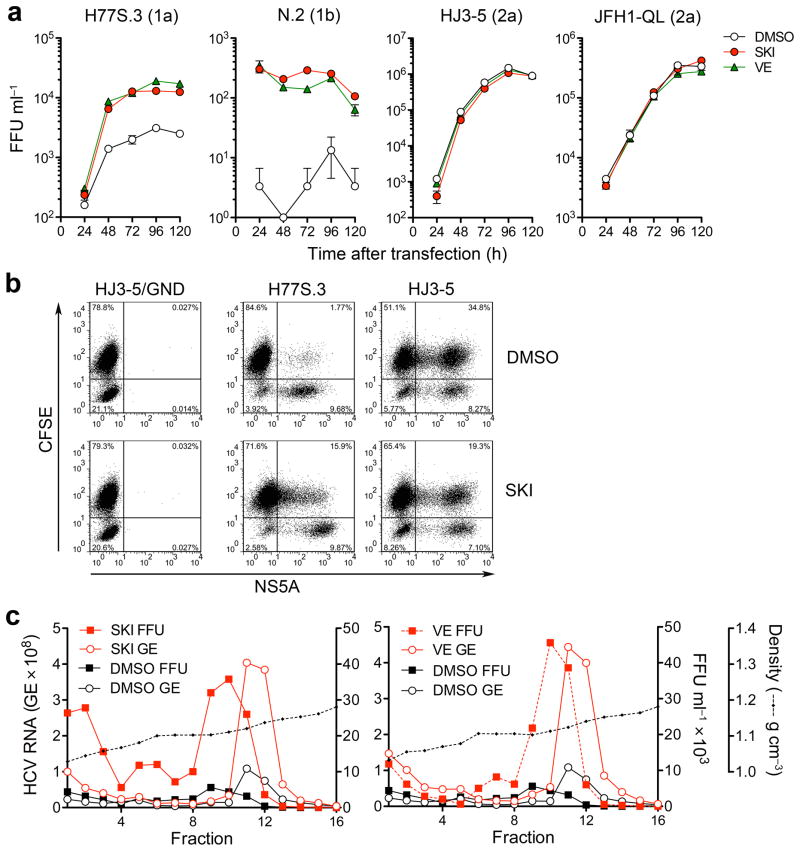
Figure 3.
Inhibition of lipid peroxidation by SKI or VE promotes production and spread of infectious genotype 1 HCV. (a) Infectious virus yields from Huh-7.5 cells transfected with the indicated viral RNAs and grown in the presence of 1 μM SKI, 1 μM VE or DMSO vehicle. Culture supernatant fluids were harvested and replaced with fresh media containing compounds every 24 h. Infectivity titers are expressed as focus forming units (FFU) ml−1. Data shown are mean ± s.e.m. from three replicate cultures. (b) SKI promotes spread of H77S.3 but inhibits HJ3-5 virus. Cells electroporated with H77S.3 or HJ3-5 RNAs were mixed with carboxyfluorescein succinimidyl ester (CFSE)-labeled naïve Huh-7.5 cells; CFSE/NS5A double-positive cells (upper right quadrant) are indicative of virus spread. (c) Buoyant density of H77S.3 virus particles released from H77S.3 RNA-transfected Huh-7.5 cells grown in 1 μM SKI (left) or 1 μM VE (right) vs. DMSO control. Fractions from isopycnic iodixanol gradients were assayed for infectious virus (FFU) or HCV RNA (GE = genome equivalents). The data shown are representative of two independent experiments.
HCV particles produced in cell culture have heterogeneous buoyant densities30. However most H77S.3 particles produced in the absence of SKI or VE banded between 1.12–1.14 g cm−3 in isopycnic gradients, with peak infectivity banding between 1.10–1.11 g cm−3 (Fig. 3c). Neither SKI nor VE altered the distribution of RNA-containing or infectious particles in gradients, but they substantially increased the abundance of both (Fig. 3c). SKI and VE also caused modest increases the specific infectivity of the most infectious particles (Supplementary Fig. 6b).
Lipid peroxidation restricts HCV in primary hepatocytes
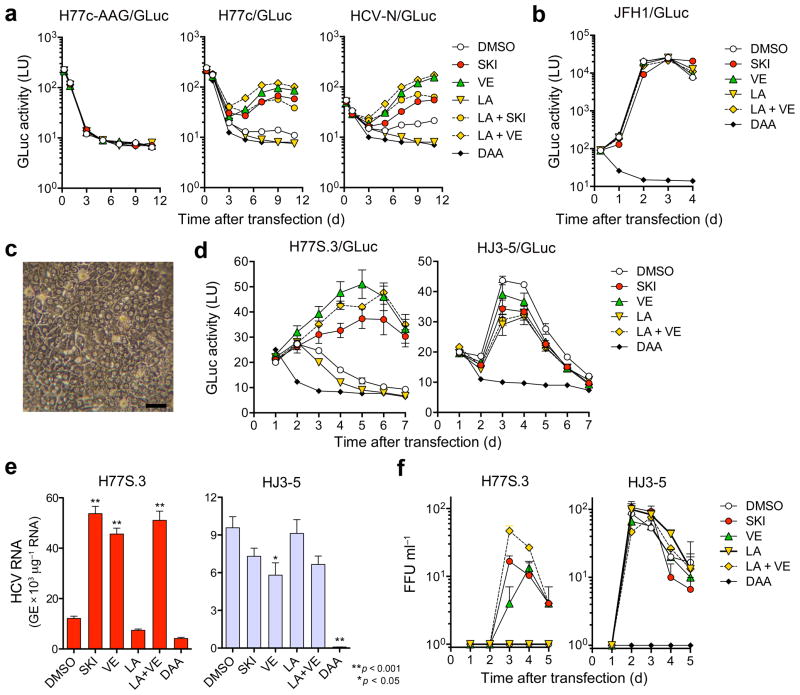
To assess replication of wild-type HCV genomes possessing no cell culture-adaptive mutations, we inserted GLuc sequence into infectious molecular clones of H77c and N31,32. Both H77c/GLuc and N/GLuc RNAs produced more GLuc in electroporated cells than RNA with a lethal mutation in NS5B, H77c/GLuc-AAG (Fig. 4a). This was eliminated by direct-acting antivirals (DAA) targeting NS5B, confirming it represents replication. In contrast, treatment with either SKI or VE markedly increased GLuc, while LA reduced it to background (Fig. 4a). Moreover, the inhibitory effect of LA was reversed by co-treatment with SKI or VE. In contrast, the wild-type JFH1/GLuc was not affected by SKI, VE, or LA (Fig. 4b). Thus, endogenous lipid peroxidation is a critical restriction factor for H77c and N viruses, but not wild-type JFH1.
Figure 4.
Lipid peroxidation regulates wild-type HCV replication and represses cell culture-adapted virus in primary human liver cultures. (a) Effects of SKI, VE (each 1 μM), LA (20 μM), LA + SKI, LA + VE, or a DAA (MK-0608, 10 μM) on replication of wild-type H77c/GLuc or HCV-N/GLuc RNAs, or a replication-defective control (H77c/GLuc-AAG) in Huh-7.5 cells. (b) Wild-type JFH1/GLuc RNA was electroporated and treated with drugs as in (a) with PSI-6130 (10 μM) as the DAA control. (c) Phase contrast microscopy of fetal hepatoblasts at 3 d. Scale bar, 50 μm. (d) Human fetal hepatoblasts (HFH) were infected with H77S.3/GLuc or HJ3-5/GLuc viruses in HFH media containing SKI or VE (each 1 μM), LA (50 μM), LA + VE, or a DAA, MK-0608 or PSI-6130 (each 10 μM) and assayed for GLuc. Results represent mean ± s.e.m. from three replicate cultures with cells from two donors. (e) HFH were infected with H77S.3 or HJ3-5 (MOI = 0.01) and treated as in d. Cell-associated viral RNA was quantified by qRT-PCR at 5 d (*P < 0.05, **P < 0.01). (f) Infectious virus released from H77S.3- or HJ3-5 virus-inoculated HFH (MOI = 0.01). Virus was quantified by FFU assay. Results represent mean ± s.e.m. from three replicate cultures.
SKI and VE also promoted replication of the H77S.3/GLuc reporter virus in primary human fetal hepatoblasts (HFH), sustaining GLuc expression for 7 d after low multiplicity infection (Fig. 4c,d). In contrast, LA suppressed H77S.3 infection in HFH in a VE-reversible manner (Fig. 4d, left). H77S.3/GLuc replicated efficiently only in the presence of VE or SKI, while HJ3-5/GLuc was inhibited by both (Fig. 4d, right). We observed similar results in cells infected with H77S.3 or HJ3-5 virus (Fig. 4e). Production of infectious H77S.3 virus was detected in HFH cultures only in the presence of VE or SKI, while infectious HJ3-5 yields were not enhanced by either (Fig. 4f). While disruption of innate immune responses also promotes HCV replication in HFH33, neither SKI nor VE reduced Sendai virus activation of the interferon-β promoter (Supplementary Fig. 7).
Lipid peroxidation reduces the EC50 of direct antivirals
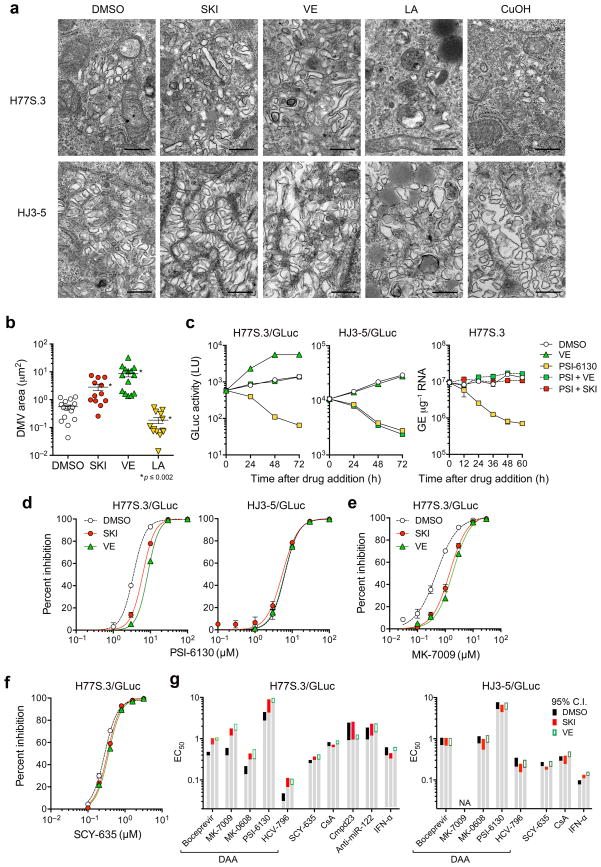
Both SKI and VE treatment increased the area occupied by the membranous web in H77S.3-infected cells (Fig. 5a,b), without altering the morphology of double-membrane vesicles which are the likely site of genome replication14. Increased lipid peroxidation had the opposite effect, while the HJ3-5 membranous web was insensitive to changes in lipid peroxidation.
Figure 5.
Lipid peroxidation reduces HCV-induced membranous web abundance and alters the EC50 of DAAs. (a) Transmission electron microscopic images of the membranous web in Huh-7.5 cells electroporated with H77S.3 or HJ3-5 RNA and treated with DMSO, SKI (1 μM), VE (1 μM), LA (50 μM), or CuOH (10 μM). Scale bar = 500 nm. (b) Quantitation of area occupied by double-membrane vesicles (DMV) within individual cells infected with H77S.3 virus and treated with SKI, VE, or LA as in a. *P ≤ 0.002 vs. DMSO by two-sided Mann-Whitney test. (c) SKI and VE mask the antiviral effect of PSI-6130 against H77S.3/GLuc replication. (left) Huh-7.5 cells electroporated with H77S.3/GLuc or HJ3-5/GLuc RNA were cultured for 7 d, then treated with DMSO, 10 μM PSI-6130, 1 μM VE or both PSI-6130 and VE. Culture supernatant fluids were replaced every 24 h and assayed for GLuc activity. (right) Huh-7.5 cells were electroporated with H77S.3 RNA, cultured for 5 d and then treated with DMSO, PSI-6130, SKI or VE, or PSI-6130 plus SKI or VE. Cell-associated HCV RNA was quantified by qRT-PCR. Results represent mean ± s.e.m from two (left) or three (right) replicate cultures. (d) Inhibition of H77S.3 (left) and HJ3-5 (right) replication by the NS5B inhibitor PSI-6130 in the presence of SKI or VE (each 1 μM) or DMSO vehicle. Inhibition was assessed by quantifying GLuc secreted 48–72 h after drug addition. Results represent mean ± s.e.m. of two replicate cultures. (e,f) Inhibition of H77S.3 replication by (e) MK-7009, an NS3/4A inhibitor, and (f) SCY-635, a host-targeting cyclophilin inhibitor. (g) EC50 values of representative direct- versus indirect-acting antivirals against H77S.3 (left) and HJ3-5 (right) viruses in the presence of SKI or VE (each 1 μM). Assays were carried out as in panels d–f. Colored bars represent limits of the 95% c.i. of EC50 values calculated from Hill plots. ‘NA’ = not measureable due to poor antiviral activity. See Supplementary Fig. 9 for additional details.
Reverse molecular genetic studies involving exchanges between the H77S.3 and JFH1 genomes suggested that the peroxidation resistance phenotype of JFH1 involves multiple nonstructural proteins within the replicase (Supplementary Results and Supplementary Fig. 8). Consistent with this, we observed a surprising increase in the 50% effective concentration (EC50) of DAAs targeting the NS3/4A protease and NS5B polymerase after suppressing endogenous lipid peroxidation in H77S.3/GLuc infected cells. Both SKI and VE masked the antiviral effects of PSI-6130, a potent NS5B inhibitor (Fig. 5c), due in part to an increase in its EC50 from 3.49 μM (95% c.i. 2.48–4.89) to 6.22 μM (4.43–8.72) and 8.46 μM (7.60–9.44), respectively (Fig. 5d). SKI and VE also increased the EC50 of MK-7009, an inhibitor of the NS3/4A protease, from 0.488 nM (95% c.i. 0.411–0.578) to 1.45 nM (1.23–1.72) and 1.90 nM (1.64–2.22) (Fig. 5e). The EC50 of other inhibitors targeting NS3/4A (boceprevir), and NS5B (HCV-796 and MK-0608) were similarly increased against H77S.3/GLuc by SKI and VE, but neither significantly altered the EC50 of SCY-635, cyclosporine A, compound 23, or anti-miR-122, inhibitors targeting essential HCV host factors, or interferon-α (Fig. 5f,g and Supplementary Fig. 9). In contrast, SKI and VE caused no change in the EC50 of any antiviral against HJ3-5/GLuc (Fig. 5d,g and Supplementary Fig. 10), indicating that VE and SKI do not impair cellular uptake or metabolism of DAAs. Changes in the EC50 against H77S.3/GLuc thus likely reflect altered affinity of the DAAs for NS3/4A and NS5B, suggesting that peroxidation modulates conformation of these replicase proteins.
Resistance to lipid peroxidation maps to NS4A and NS5B
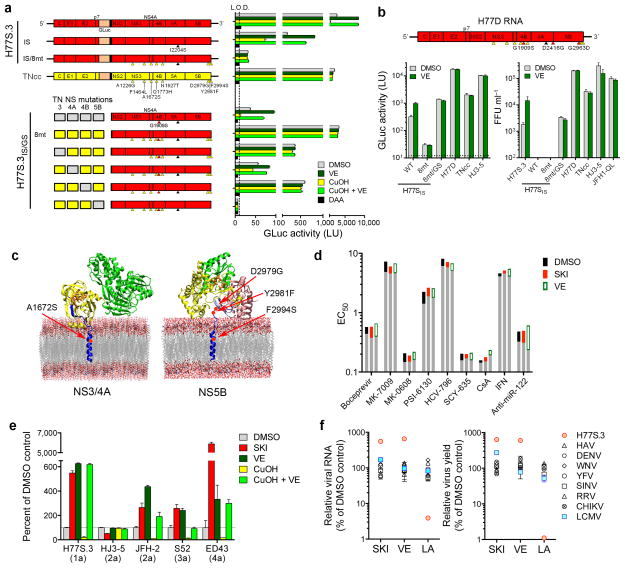
TNcc is a recently described genotype 1a virus with 8 cell culture-adaptive mutations that replicates almost as well as JFH1 in Huh-7.5 cells34. Surprisingly, we found it completely resistant to lipid peroxidation (Fig. 6a and Supplementary Fig. 10a). We introduced all 8 TNcc mutations into H77S.3 to determine whether they would confer peroxidation resistance. This RNA (H77S.3/GLuc8mt) failed to replicate, but removal of a key H77S adaptive mutation (S2204I in NS5A)12 restored low-level replication, and this virus, H77S.3/GLucIS/8mt, was resistant to lipid peroxidation (Fig. 6a and Supplementary Fig. 10b). Continued passage of cells infected with H77S.3IS/8mt resulted in additional mutations in NS4B (G1909S), NS5A (D2416G) and NS5B (G2963D) that together enhanced replication by 850-fold (see Supplementary Results and Supplementary Fig. 11). NS4B G1909S compensates for negative effects of TNcc mutations placed into the H77S.3 background, but does not confer peroxidation resistance (Fig. 6a). When all 3 mutations were introduced into H77S.3/GLucIS/8mt (designated H77D), infectious virus yields were comparable to HJ3-5 or JFH1-QL, and not increased by VE supplementation (Fig. 6b). Importantly, the EC50 of DAAs against H77D virus was not altered by SKI or VE (Fig. 6d and Supplementary Fig. 9h).
Figure 6.
Resistance to lipid peroxidation is tightly linked to robust replication in cell culture. (a) (upper panel) Cell culture-adaptive mutations in TNcc34 (yellow arrowheads) confer resistance to lipid peroxidation when introduced into H77S.3/GLucIS (H77S.3/GLuc in which the adaptive mutation S2204I has been removed (black arrowhead), see Supplementary Fig. 10b for details). Huh-7.5 cells were treated with DMSO, 1 μM SKI, 1 μM VE, 10 μM CuOH, CuOH plus VE, or 30 μM sofosbuvir (DAA) beginning 6 h following RNA electroporation. GLuc secretion was measured between 48–72 h. (lower panel) TNcc mutations in NS3 (helicase) and NS4B are not required for lipid peroxidation resistance. Combinations of TNcc substitutions were introduced into H77S.3/GLucIS/GS (NS proteins shown only) that contains the compensatory mutation G1909S (GS) in NS4B (red arrowhead, see Supplementary Fig. 11). Data shown represent mean GLuc activity ± s.e.m. from two independent experiments. L.O.D. = limit of detection. (b) (top) H77D genome containing the I2204S substitution (black), 8 TNcc-derived mutations (yellow) and 3 novel compensatory mutations (red) in the H77S.3 background. (bottom) Huh-7.5 cells transfected with the indicated RNAs encoding GLuc (left) or lacking GLuc (right) were treated with DMSO or 1 μM VE and secreted GLuc activity (left) or released infectious virus (right) measured between 48–72 h. Data shown represent means ± s.d. from triplicate cultures in a representative experiment. (c) Structural models of (left) NS4A and (right) NS5B membrane interactions showing key residues that determine sensitivity to lipid peroxidation. (d) EC50 of direct- vs. indirect-acting antivirals against H77D in the presence of SKI or VE (each 1 μM). Assays were carried out as in Fig. 5g. (e) Huh-7.5 cells transfected with H77S.3/GLuc or HJ3-5/GLuc RNA, genome-length JFH-2 RNA, or subgenomic RNAs encoding FLuc (S52 and ED43) were treated with DMSO, 1 μM SKI or VE, 10 uM CuOH or CuOH plus VE. Data shown represent percent GLuc (H77S.3 and HJ3-5), RNA copies (JFH-2) or FLuc activities (S52 and ED43) at 72 h relative to DMSO controls. Data shown represent mean ± s.e.m. from three independent experiments. (f) The HCV replicase is uniquely regulated by lipid peroxidation. The impact of SKI or VE (each 1 μM) or LA (50 μM) on the abundance of viral RNA (left) or yields of infectious virus (right) was determined for a panel of RNA viruses following infection of Huh-7.5 cells. In addition to H77S.3, viruses studied included the hepatotropic picornavirus, hepatitis A virus (HAV), representative flaviviruses (dengue virus, DENV; West Nile virus, WNV; yellow fever virus, YFV), alphaviruses (Sindbis virus, SINV; Ross River virus, RRV; Chikungunya virus, CHIKV), and lymphocytic choriomeningitis virus (LCMV). Data shown are mean ± s.e.m. from three replicate cultures. See Supplementary Fig. 12 for additional details.
Introducing the compensatory G1909S mutation into H77S.3/GLucIS (H77S.3/GLucIS/GS) allowed us to identify A1672S (NS4A) as essential for peroxidation resistance, while mutations in NS3 and NS4B are not required (Fig. 6a). Mutations in NS5B were essential for replication of peroxidation resistant virus, but neither these nor the NS4A mutation alone conferred peroxidation resistance (Supplementary Fig. 10c). Thus, mutations in both NS4A and NS5B are required for genotype 1a resistance. These mutations are within or in close proximity to the transmembrane domains of these proteins (Fig. 6c), consistent with direct involvement of these residues in resistance to lipid peroxidation.
Regulation by lipid peroxidation is unique to HCV
In addition to genotypes 1a (H77S.3) and 1b (HCV-N.2) (Figs. 2c and 3a), replication of genotype 2a (JFH2), 3a (S52), and 4a (ED43) HCVs was enhanced by treatment with SKI or VE, and inhibited by CuOH-induced lipid peroxidation (Fig. 6e). JFH1 is thus unique among wild-type HCV in its resistance to lipid peroxidation.
As with HCV, the genomes of other positive-strand RNA viruses are synthesized by replicase complexes that assemble in association with cytoplasmic membranes and are thus at risk for damage due to lipid peroxidation. Yet, like JFH1, the replication of other pathogenic flaviviruses, picornaviruses, and alphaviruses is neither enhanced by SKI or VE, nor suppressed by LA (Fig. 6f and Supplementary Fig. 12a–c). This is also true for clone 13 lymphocytic choriomeningitis virus, an ambisense RNA virus that establishes persistent infections (Fig. 6f and Supplementary Fig. 13d). Thus, most viral RNA replicases have evolved in ways that prevent or mitigate the potentially negative effects of lipid peroxidation. HCV is a clear exception, suggesting that its sensitivity to lipid peroxidation provides a distinct survival advantage.
DISCUSSION
Lipid peroxides are formed on polyunsaturated fatty acid chains within membranes by reactive intermediates produced during oxidative stress. They alter membrane fluidity and permeability and potentially contribute to a variety of disease states35. The degradation products of these lipid peroxides include reactive aldehydes, such as acrolein, 4-hydroxy-2-nonenal, and MDA, that add to this damage by forming adducts with membrane proteins, thereby modulating their biological activities35,36. The opposing effects of SPHK1 and SPHK2 on lipid peroxidation (Fig. 2f) have not been noted previously and remain unexplained. SPHK1 is predominantly cytosolic and translocates to the plasma membrane upon activation37, while SPHK2 is more likely to be associated with intracellular membranes28. Sphingosine-1-phosphate produced by SPHK functions as a messenger in several signaling pathways, and is a cofactor for enzymes involved in signal transduction and transcriptional regulation38,39. However, it has no effect on HCV replication in cell cultures (Supplementary Fig. 1c), and only minimally increases MDA abundance (Supplementary Fig. 1d).
During HCV infection, oxidative stress is caused by both host inflammatory responses and direct interactions of viral proteins with mitochondria7,40. Our results show that the wild-type H77S and N.2 replicases are highly sensitive to both endogenous and PUFA-induced lipid peroxidation. Con1 and OR6, other genotype 1 HCVs, are also inhibited by PUFA-induced peroxidation8,41. Since lipid peroxidation also inhibits multiple other HCV genotypes (Fig. 6e), it is a common feature of HCV.
In genotype 1 virus, resistance to lipid peroxidation maps to residues within or near the transmembrane domains of NS4A and NS5B (Fig. 6c), key components of the replicase complex. Reactive aldehydes derived from degraded lipid peroxides could form adducts with residues within these transmembrane domains, impairing their capacity for essential interactions and thereby degrading replicase activity. The A1672S mutation promotes oligodimerization of the NS4A transmembrane domain, an interaction necessary for efficient replicase function42. Thus, A1672S might confer resistance to peroxidation by restoring NS4A dimerization impaired by adduct formation. Adduct formation could similarly affect the NS5B transmembrane domain. While hypothetical, such effects could explain changes in the EC50 of DAAs targeting NS3/4A and NS5B (Fig. 5d,e,g). Alternatively, proper membrane localization and assembly of nonstructural proteins within the replicase may require lipids esterified with non-oxidized fatty acid chains. Indeed, monounsaturated fatty acid supplements such as oleic acid stimulate H77S.3/GLuc and N.2/GLuc replication but not JFH1 (Supplementary Fig. 5h). Yet a third possibility is that peroxidation could induce changes in membrane fluidity that alter replicase conformation35.
We are drawn to the possibility that HCV exploits lipid peroxidation as a means of autoregulating its replication, and that lipid peroxides act as a brake, down-regulating the efficiency of genome amplification when reaching a threshold abundance. Such a model suggests that HCV possesses a conserved peroxidation ‘sensor’, mapping in part to the transmembrane domains of NS4A and NS5B, that governs replication efficiency, thereby limiting tissue damage, reducing viral exposure to the immune system, and facilitating viral persistence. While hepatotoxicity associated with diets deficient in lipophilic antioxidants presents a technical barrier to testing this hypothesis in murine models of hepatitis C43, HCV replication is regulated by lipid peroxidation in primary human hepatocytes (Fig. 4c–f). Related RNA viruses that are capable of establishing persistent infection appear to auto-restrict replication via alternative mechanisms. For example, bovine viral diarrhea virus, a pestivirus, down-regulates replicase formation by limiting cleavage of its NS2-3 protein, thereby arresting RNA synthesis and enabling a noncytolytic phenotype44. Thus, reducing the efficiency of the replicase may be a common theme for RNA viruses that establish persistent infection.
Like most other positive-strand RNA viruses (Fig. 6f), the genotype 2a JFH1 virus is highly resistant to lipid peroxidation (Fig. 2a,b,h). This suggests JFH1 may be a loss-of-function mutant that no longer senses lipid peroxides and auto-restricts its replicase activity. Only one of the 4 amino acid substitutions conferring peroxidation resistance in H77S.3 (NS5B F2981) exists in JFH1, and the molecular basis of its peroxidation resistance remains to be determined.
ONLINE METHODS
Cells and reagents
Huh-7.5 cells were grown in Dulbecco’s modified Eagle’s medium (DMEM), High Glucose supplemented with 10% fetal bovine serum (FBS), 1×Penicillin-Streptomycin, 1×GlutaMAX and 1×MEM Non-Essential Amino Acids Solution (Gibco). BD-BioCoat™ collagen-I coated plates were purchased from BD Biosciences. SKI [2-(p-Hydroxyanilino)-4-(p-chlorophenyl) thiazole] was obtained from Merck Millipore. Myriocin, fumonisin B1, N-[2-hydroxy-1-(4-morpholinylmethyl)-2-phenylethyl]-decanamide (PDMP), D-erythro-2-tetradecanoylamino-1-phenyl-1-propanol (D-MAPP), dihydrosphingosine, C-2 and C-8 ceramides, sphingosine, sphingosine 1-phosphate, lovastatin, ebselen, arachidonic acid, docosahexaenoic acid, and linoleic acid were from Cayman Chemical. Vitamin E (α-, rac-β-, and γ-tocopherols), 4-deoxypyridoxine hydrochloride (DOP), coenzyme Q10, butylated hydroxytoluene, N-acetyl-L-cysteine, diphenyleneiodonium chloride, oleic acid, and cyclosporine A were from Sigma-Aldrich. nSMase spiroepoxide and cumene hydroperoxide were from Santa Cruz Biotechnology, D609 was from Enzo Life Sciences, and sofosbuvir (PSI-7977) was from Chemscene. Locked nucleic acid anti-miR-122 was synthesized by Exiqon. A selective PI4KIIIα inhibitor, compound 2345, was provided by Raffaele De Francesco. Relative cell numbers were assessed using the WST-1 reagent (Millipore) or determination of protein content. Protein concentrations in samples were determined using the Protein Assay kit (Bio-Rad) with bovine serum albumin as a standard.
Fetal liver cells
Tissue samples were supplied by the accredited agency, Advanced Biological Resources, and came from fetuses between 15–20 weeks gestation obtained during elective terminations of pregnancy. Tissue was processed as described elsewhere46–48, and isolated hepatoblasts seeded at density of 5.2×105 cm−2 on 12- or 24-well plates and in regular Kubota’s Medium49 supplemented with 5% FBS. Following an overnight incubation, the medium was changed to a variation of Kubota’s Medium (HFH medium) comprised of DMEM supplemented with 25 mM HEPES, 1 nM selenium, 0.1% BSA, 4.5 mM niacinamide, 0.1 nM zinc sulfate heptahydrate, 10 nM hydrocortisone, 5 μg ml−1 transferrin/Fe, 5 μg ml−1 insulin, 2 mM L-glutamine, antibiotics, and 2% FBS. The use of commercially procured fetal liver cells was reviewed by the UNC Office of Human Research Ethics and was determined not to require approval by the UNC Institutional Review Board.
Plasmids
pHJ3-550, pHJ3-5/GLuc2A (referred to here as pHJ3-5/GLuc), pHJ3-5/GND, pH77c51, pH77S.3, pH77S.3/GLuc2A (referred to here as pH77S.3/GLuc), and pH77S/GLuc2A-AAG12,27,52 have been described. pHCV-N.2 is a modified version of HCV3-9b32 that contains cell culture-adaptive mutations in NS3 and NS5A (A1099T, E1203G and S2204I in the polyprotein). Mutations in the SM-binding domains, 5′UTR, 3′UTR, and nonstructural protein regions were generated by site-directed mutagenesis. The Gaussia princeps luciferase (GLuc)-coding sequence followed by the foot-and-mouth disease virus 2A protease-coding sequence was inserted between p7 and NS2 in pH77c, pHCV-N.2, pJFH1 (wild-type), and pJFH1-QL (containing the cell culture-adaptive mutation Q221L in the NS3 helicase) using a strategy applied previously to pH77S27. JFH1-QL was used for experiments unless otherwise indicated. Other cell culture-adapted genotype 1a (TNcc), 2a (JFH-2/AS/mtT3), 3a (S52/SG-Feo(SH)) and 4a (ED43/SG-Feo(K)) HCV strains and RNA replicons have been described34,53,54.
Luciferase assays
For the Gaussia luciferase (GLuc) assay, cells transfected with HCV RNA encoding GLuc were treated with drugs at 6 h after transfection, and the culture media were harvested, refed with fresh media containing drugs, and assayed for GLuc at 24 h intervals. Secreted GLuc activity was measured as described52. For the Firefly luciferase (FLuc) assay, cell monolayers were washed with PBS and lysed in Passive Lysis Buffer (Promega), and the lysates analyzed with the Luciferase Assay System (Promega) according to the manufacturer’s instructions. Each individual experiment used duplicate or triplicate cell cultures. Results shown represent the mean ± s.e.m. from multiple independent experiments.
RNA transcription
RNA transcripts were synthesized in vitro as described previously27.
HCV RNA transfection
At 24 h before transfection, 7.5×104 Huh-7.5 cells were seeded onto a 24-well plate. One day later, media were replaced with fresh media, and the cells transfected with 0.25 μg (per well) HCV RNA encoding GLuc using the TransIT mRNA transfection kit (Mirus) according to the manufacturer’s protocol. After 6 h incubation at 37°C, supernatant fluids were removed for GLuc assay and replaced with fresh media containing compound. Alternatively, 10 μg of HCV RNA was mixed with 5×106 Huh-7.5 cells and electroporated into cells using a Gene Pulser Xcell Total System (Bio-Rad) as described previously52. Transfection of wild-type HCV RNA was performed by electroporating 5 μg HCV RNA in 2.5×106 Huh-7.5 cells and seeded into collagen-coated plates (BD Biosciences). Cells were grown in DMEM supplemented with 25 mM HEPES, 7 ng ml−1 glucagon, 100 nM hydrocortisone, 5 μg ml−1 insulin, 2 mM GlutaMAX, antibiotics, and 2% FBS. Culture supernatants were replaced with the media supplemented with drugs at 6 h and every 48 h thereafter and assayed for GLuc activity.
HCV production
For virus production, subconfluent Huh-7.5 cells in a 100 mm diameter dish were transfected with 5 μg HCV RNA using the TransIT mRNA transfection kit as above and split at 1:2 ratio at 6 h after transfection. Cells were then fed with media supplemented with 50 mM HEPES (Cellgro) and the supernatants harvested and replaced with fresh media every 24 h. Cells were passaged at a 1:2 ratio again 3 d after transfection. Media containing HCV were supplemented with an additional 50 mM HEPES and stored at 4°C until assayed for infectivity. Gaussia luciferase H77S.3/GLuc reporter virus was produced by electroporating 5 μg H77S.3/GLuc RNA into 2.5×106 Huh-7.5 cells. Cells were fed with medium containing 25 mM HEPES and 10 μM VE at 3 h and grown for 3 d until subconfluent. Cells were then split 1:3 into medium containing 25 mM HEPES and the supernatant harvested on the next day and stored at 4°C until use. HJ3-5/GLuc virus was produced in medium lacking VE and stored in –80°C until use. Infectious titers were determined by TCID50 using GLuc activity produced at 72 h after inoculation.
HCV infectivity assays
Huh-7.5 cells were seeded at 5×104 cells per well into 48-well plates 24 h before inoculation with 100 μl of culture medium. Cells were fed with media containing 1 μM VE 24 h later to facilitate visualization of core protein expression, fixed with methanol-acetone (1:1) at –20°C for 10 min 72 h after inoculation (48 h for JFH1-QL and HJ3-5), and stained for intracellular core antigen with a mouse monoclonal antibody C7–50 (Thermo SCIENTIFIC, 1:300 dilution). Clusters of infected cells identified by staining for core antigen were considered to constitute a single infectious focus, and the data expressed as focus-forming units (FFU) ml−1.
HCV infection in fetal hepatoblasts
Cells were inoculated with HCV encoding GLuc (MOI = 0.001) for 6 h. After washing 5 times with HFH medium, cells were incubated for an additional 18 h to determine baseline GLuc secretion. Culture supernatant fluids were replaced at 24 h intervals with HFH media containing drugs and assayed for GLuc.
Flow cytometry
Huh-7.5 cells electroporated with H77S.3 or HJ3-5 RNA were treated with 1 μM SKI or DMSO beginning at 24 h, and analyzed for NS5A expression by flow cytometry at 96 h. Virus spread assays were adapted from a previously described method55. Briefly, Huh-7.5 cells were electroporated with H77S.3, HJ3-5 or HJ3-5/GND RNAs and cultured for 24 h. The electroporated (producer) cells were then co-cultured at a 1:4 ratio with naïve Huh-7.5 (recipient) cells pre-labeled with 5 μM carboxyfluorescein diacetate succinimidyl ester (CellTrace™ CFSE Cell Proliferation Kit, Invitrogen) in the presence of different compounds (see Figure legends) for 48 h. Cells were stained for NS5A protein and analyzed by flow cytometry as described previously56.
Equilibrium ultracentrifugation
Filtered supernatant fluids collected from transfected cell cultures were concentrated 50-fold using Centricon Plus-70 Centrifugal Filter Units (100-kDa exclusion) (Millipore), then layered on top of a pre-formed continuous 10–40% iodixanol (OptiPrep, Sigma-Aldrich) gradient in Hanks’ balanced salt solution (HBSS, Invitrogen). Gradients were centrifuged in a SureSpin 630 Swinging Bucket Rotor (Thermo SCIENTIFIC) at 30,000 rpm for 24 h at 4°C, and fractions were collected from the top of the tube. The density of each fraction was calculated from the refractive index measured with a refractometer (ATAGO). RNA was isolated from each fraction using QIAamp Viral RNA kit (Qiagen) and the viral amount quantified by qRT-PCR as described below. Infectious virus titers in each fraction were determined as described above.
qRT-PCR
One-step qRT-PCR analysis of HCV RNA in Huh-7.5 cells was carried out as described52. HCV RNA in primary human fetal hepatoblast cultures was detected by means of a two-step qRT-PCR procedure using SuperScript III First-Strand Synthesis SuperMix for qRT-PCR (Invitrogen), followed by TaqMan qPCR analysis with primer pairs and a probe targeting a conserved 221-base sequence within the 5′UTR of the genome and iQ Supermix (Bio-Rad)52.
RNA interference
Validated siRNA targeting human SPHK1 (SI02660455)57 was purchased from Qiagen. siRNA targeting human SPHK2 (5′-CGUCACGGUUAAAGAGAAA-3′)39 and control siRNA (#2) were from Dharmacon. siRNA (20 nM) was transfected into cells using siLentfect Lipid Reagent (Bio-Rad) according to the manufacturer’s protocol.
Immunoblots
Immunoblotting was carried out using standard methods with the following antibodies: mouse monoclonal antibodies to β-actin (AC-74, Sigma, 1:10,000), HCV NS3 (ab65407, abcam, 1:500); and rabbit polyclonal antibodies to SPHK1 (A302-177A, Bethyl Laboratories, 1:2000), SPHK2 (ab37977, abcam, 1:500). Protein bands were visualized and quantified with an Odyssey Infrared Imaging System (Li-Cor Biosciences).
Sphingosine kinase assay
Sphingosine kinase activity was determined as described previously28. Recombinant human SPHK1 and SPHK2 proteins were obtained from BPS Bioscience. SPHK1 activity was determined in the presence of 0.25% Triton X-100, which inhibits SPHK228. The labeled S1P was separated by TLC on Silica Gel G-60 (Whatman) with 1-butanol/ethanol/acetic acid/water (80:20:10:20, v/v) and visualized and quantified by phosphorimager (Bio-Rad).
Quantification of cholesterol and triglyceride levels
Cells were scraped and lysed in PBS containing 1% Triton X-100 and complete protease inhibitor cocktail (Roche). Cell lysates were clarified by centrifugation at 15,000 rpm at 4°C for 10 min. Cholesterol contents were determined using the Amplex Red Cholesterol Assay Kit (Invitrogen), according to the manufacturer’s protocol. Triglyceride levels in cells grown on 96-well plates were determined using Triglyceride Assay Kit (Zen-Bio) as per manufacturer’s instruction. The values were normalized to the total protein content.
Lipid peroxidation assays
Malondialdehyde (MDA), a product of lipid peroxidation, was quantified by the thiobarbituric acid reactive substances (TBARS) Assay Kit (Cayman Chemical). Cells transfected with HCV RNAs were grown in the presence of different drugs, and analyzed for intracellular malondialdehyde (MDA) abundance at 48–72 h as indicated in legends. Cells scraped into PBS containing complete protease inhibitor cocktail (Roche) were homogenized by sonication on ice using Sonic Dismembrator (FB-120, Fisher Scientific). The amount of MDA in 100 μl of cell homogenates was analyzed by a fluorescent method as described by the manufacturer. Lipid peroxidation levels were expressed as the amount of MDA normalized to the amount of total protein. Alternatively, the lipid peroxidation product, 8-isoprostane, was quantified using the 8-Isoprostane EIA kit (Cayman Chemical) according to the manufacturer’s recommended procedures.
Innate immune response reporter assays
IFN-β, IRF-3, and NFκB-dependent promoter activities were assayed using firefly luciferase reporters, pIFN-β-Luc, p4xIRF3-Luc, or pPRDII-Luc as described previously58. Cells were co-transfected with the reporter plasmid pRL-CMV, and the firefly luciferase results normalized to Renilla luciferase activity in order to control for potential differences in transfection efficiency. The luminescence was measured on a Synergy 2 (Bio-Tek) Multi-Mode Microplate Reader.
Mass spectrometry of sphingolipids and metabolites
Cells were washed extensively with PBS and scraped into tubes. An aliquot of cells was taken for protein and total lipid phosphate measurements. After addition of a sphingolipid internal standard cocktail (Avanti Polar Lipids), the lipids were extracted and individual sphingolipid species were quantified by liquid chromatography, electrospray ionization-tandem mass spectrometry as described previously59,60.
Electron microscopy
Huh-7.5 cells (5×106 cells) electroporated with 5 μg HCV RNA were seeded into a 6-well plate and media containing compounds added 24 h later. At 48 h after transfection, cells were fixed with 3% glutaraldehyde in 0.15M sodium phosphate buffer, pH 7.4, for 1 h at room temperature and stored at 4°C until processed. Following three rinses with 0.15M sodium phosphate buffer, pH 7.4, monolayers were post-fixed with 1% osmium tetroxide for 1 h, washed in deionized water, and stained en bloc with 2% aqueous uranyl acetate for 20 min. The cells were dehydrated using increasing concentrations of ethanol (30%, 50%, 75%, 100%, 100%, 10 min each) and embedded in Polybed 812 epoxy resin (Polysciences, Inc.). Cell layers were sectioned en face to the substrate at 70 nm using a diamond knife and a Leica Ultracut UCT microtome (Leica Microsystems). Ultrathin sections were mounted on 200 mesh copper grids and stained with 4% aqueous uranyl acetate and Reynolds’ lead citrate61. The grids were observed at 80 kV using a LEO EM910 transmission electron microscope (Carl Zeiss SMT, LLC). Digital images were taken using a Gatan Orius SC 1000 CCD Camera with DigitalMicrograph 3.11.0 software (Gatan, Inc.).
Quantitation of hepatitis A virus, flavivirus, alphavirus, and lymphocytic choriomeningitis virus RNA
The abundance of HAV, RRV, SINV, and LCMV RNA was determined using iScript One-Step RT-PCR kit with SYBR Green (Bio-Rad) and primer pairs as follows: HAV, forward 5′-GGTAGGCTACGGGTGAAAC-3′ and reverse 5′-AACAACTCACCAATATCCGC-3′, RRV, forward 5′-AGAGTGCGGAAGACCCAGAG-3′ and reverse 5′-CCGTGATCTTACCGGACACA-3′, SINV, forward 5′-GAGGTAGTAGCACAGCAGG-3′, and reverse, 5′-CGGAAAACATTCTACGAGC-3′, LCMV, forward 5′-CATTCACCTGGACTTTGTCAGACTC-3′, and reverse, 5′-GCAACTGCTGTGTTCCCGAAAC-3′. WNV and YFV RNA levels were quantified using a previously described method62 with primers as follows: WNV, forward 5′-TCAGCGATCTCTCCACCAAAG-3′ and reverse 5′-GGGTCAGCACGTTTGTCATTG-3′, YFV, forward 5′-CTGTCCCAATCTCAGTCC-3′ and reverse 5′-AATGCTCCCTTTCCCAAATA-3′. DENV RNA was quantified on a 7900 HT Real-Time PCR System (ABI) using primers and probe as described63.
The infrared fluorescent immunofocus assay for infectious hepatitis A virus (HAV) was done using FRhK-4 cells as previously described64. Titration of infectious alphaviruses was performed in duplicate by visualization of plaques on Vero cells seeded in 12-well plates. Plates were incubated with inoculum at 37°C with 5% CO2 for 1 h with periodic agitation. Inocula were then removed and plates overlaid with 1.25% carboxymethylcellulose in MEM supplemented with 3% FBS, 1×Penicillin-Streptomycin, 2 mM L-glutamine, and HEPES. All of these reagents were identical to those used for parallel studies of HCV. At 48 h after infection, plates were fixed with 4% paraformaldehyde, and visualized with at 0.25% crystal violet solution. Infectious titers of WNV and YFN were determined by plaque assay on confluent BHK cell monolayers in 6-well plates. Cells were incubated with virus inoclula for 2 h at 37°C then washed prior to addition of a 1% methylcellulose media overlay and further incubated for 3 d. Plaques were visualized with Giemsa staining. DENV titers were determined by plaque assay on Vero 76 cell monolayers in 96-well plates. Cells were incubated with virus inocula for 2 h at 37°C then washed prior to addition of a 0.8% methylcellulose media overlay and further incubated for 3 d. Following fixation with ice-cold acetone methanol solution (50:50 v/v). Cells were immunostained using DENV E-specific monoclonal antibody 4G2 (UNC Antibody Core Facility) followed by HRP-conjugated anti-mouse IgG secondary antibody (KPL). Infectious foci were visualized using Vector VIP reagent (Vector Labs). LCMV titers were determined by plaque assay on confluent Vero cell monolayers in 6-well plates65. Cells were incubated with virus inocula for 80 min at 37°C prior to addition of a 1:1 mixture of 1% agarose and EMEM medium containing 10% FCS, 2 mM L-glutamine, 2% penicillin, and 2% streptomycin. Cells were further incubated for 5 d at 37°C. Plaques were visualized with crystal violet.
Modeling of HCV nonstructural protein-membrane interactions
Membrane topologies of HCV nonstructural proteins were modeled as suggested by Bartenschlager et al.66 using the Protein Data Bank (PDB) structure 4A92 (http://www.rcsb.org/pdb/home/home.do) for the NS3/4A protease-helicase (Fig. 6c, left) and PDB 1GX6 for the NS5B RNA-dependent RNA polymerase (Fig. 6c, right). Secondary structure predictions were generated on the Jpred3 server (http://www.compbio.dundee.ac.uk/www-jpred/). Visual Molecular Dynamics (VMD) with the plugin “Membrane” was applied for visualization of protein-membrane interactions67.
Statistical methods
Unless noted otherwise, all between-group comparisons were carried out by 2-way ANOVA using Prism 6.0 software (GraphPad Software Inc.). For determination of EC50 concentrations of DAAs, data were fit to a four-parameter dose response curve with variable slope using Prism 5.0c for Mac OS X software (GraphPad Software, Inc.). Results are reported as the estimated EC50 ± 95 confidence interval (Fig. 5g).
Supplementary Material
Acknowledgments
We thank L.F. Ping and W. Lovell for expert technical assistance; R. Purcell (National Institute of Allergy and Infectious Diseases) and J. Bukh (Copenhagen University Hospital, Denmark) for pCV-H77C and pTNcc plasmids; C.M. Rice and M. Saeed (The Rockefeller University) for Huh-7.5 cells and S52/SG-Feo and ED43/SG-Feo plasmids; T. Wakita (National Institute of Infectious Diseases, Japan) for pJFH1 and pJFH-2 plasmids; M.J. Otto (Pharmasset, Inc.), A.Y. Howe (Merck Research Laboratory), A. Sluder (SCYNEXIS, Inc.) and R. De Francesco (Istituto Nazionale di Genetica Molecolare, Italy) for antiviral compounds; and, Z. Feng (University of North Carolina) for HAV stocks. We also thank S.A. Weinman for critical reading of the manuscript, and R.A. Coleman and T. Masaki for helpful discussions. This work was supported by grants from the National Institutes of Health: RO1-AI095690, RO1-CA164029, U19-AI109965 (S.M.L.), R01-AI075090 (M.Y.), RO1-AI073335 (C.C.K.), RO1-DE018304 (D.P.D.), U54-GM069338 (S.B.), an NCI Center Core Support Grant to the Lineberger Comprehensive Cancer Center (P30-CA016086), the University Cancer Research Fund, and by funding from Vesta Therapeutics and the Dow Chemical Company (L.M.R.). C.W. was supported by the Deutsche Forschungsgemeinschaft (WE 4388/3-1 and WE 4388/6-1). I.A. was supported by the CIPSM excellence cluster.
Footnotes
Author contributions. D.Y. and S.M.L conceived the study and wrote the paper; D.Y., D.R.M., E.W., V.J.M., Y.W., P.E.C., C.E.M., D.G.W., and I.M. conducted experiments; C.W. and I.A. modeled membrane interactions of proteins; S.B. and A.H.M. carried out mass spectrometry analysis of sphingolipids; M.Y., S.K., T.S., T.O., S.M.P. and L.M.R. provided research materials; all authors discussed the results and commented on the manuscript.
References
- 1.Kalyanaraman B. Teaching the basics of redox biology to medical and graduate students: Oxidants, antioxidants and disease mechanisms. Redox Biol. 2013;1:244–257. doi: 10.1016/j.redox.2013.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nathan C, Cunningham-Bussel A. Beyond oxidative stress: an immunologist’s guide to reactive oxygen species. Nat Rev Immunol. 2013;13:349–361. doi: 10.1038/nri3423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dennery PA. Effects of oxidative stress on embryonic development. Birth Defects Res C Embryo Today. 2007;81:155–162. doi: 10.1002/bdrc.20098. [DOI] [PubMed] [Google Scholar]
- 4.Valyi-Nagy T, Dermody TS. Role of oxidative damage in the pathogenesis of viral infections of the nervous system. Histol Histopathol. 2005;20:957–967. doi: 10.14670/HH-20.957. [DOI] [PubMed] [Google Scholar]
- 5.Thomas DL. Global control of hepatitis C: where challenge meets opportunity. Nat Med. 2013;19:850–858. doi: 10.1038/nm.3184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Choi J. Oxidative stress, endogenous antioxidants, alcohol, and hepatitis C: pathogenic interactions and therapeutic considerations. Free Radic Biol Med. 2012;52:1135–1150. doi: 10.1016/j.freeradbiomed.2012.01.008. [DOI] [PubMed] [Google Scholar]
- 7.Okuda M, et al. Mitochondrial injury, oxidative stress, and antioxidant gene expression are induced by hepatitis C virus core protein. Gastroenterology. 2002;122:366–375. doi: 10.1053/gast.2002.30983. [DOI] [PubMed] [Google Scholar]
- 8.Huang H, Chen Y, Ye J. Inhibition of hepatitis C virus replication by peroxidation of arachidonate and restoration by vitamin E. Proc Natl Acad Sci USA. 2007;104:18666–18670. doi: 10.1073/pnas.0708423104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wakita T, et al. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat Med. 2005;11:791–796. doi: 10.1038/nm1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lindenbach BD, et al. Complete replication of hepatitis C virus in cell culture. Science. 2005;309:623–626. doi: 10.1126/science.1114016. [DOI] [PubMed] [Google Scholar]
- 11.Zhong J, et al. Robust hepatitis C virus infection in vitro. Proc Natl Acad Sci USA. 2005;102:9294–9299. doi: 10.1073/pnas.0503596102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yi M, Villanueva RA, Thomas DL, Wakita T, Lemon SM. Production of infectious genotype 1a hepatitis C virus (Hutchinson strain) in cultured human hepatoma cells. Proc Natl Acad Sci USA. 2006;103:2310–2315. doi: 10.1073/pnas.0510727103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pietschmann T, et al. Production of infectious genotype 1b virus particles in cell culture and impairment by replication enhancing mutations. PLoS Pathog. 2009;5:e1000475. doi: 10.1371/journal.ppat.1000475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Paul D, Hoppe S, Saher G, Krijnse-Locker J, Bartenschlager R. Morphological and biochemical characterization of the membranous hepatitis C virus replication compartment. J Virol. 2013;87:10612–10627. doi: 10.1128/JVI.01370-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gosert R, et al. Identification of the hepatitis C virus RNA replication complex in Huh-7 cells harboring subgenomic replicons. J Virol. 2003;77:5487–5492. doi: 10.1128/JVI.77.9.5487-5492.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shi ST, Lee KJ, Aizaki H, Hwang SB, Lai MM. Hepatitis C virus RNA replication occurs on a detergent-resistant membrane that cofractionates with caveolin-2. J Virol. 2003;77:4160–4168. doi: 10.1128/JVI.77.7.4160-4168.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hsu NY, et al. Viral reorganization of the secretory pathway generates distinct organelles for RNA replication. Cell. 2010;141:799–811. doi: 10.1016/j.cell.2010.03.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reiss S, et al. Recruitment and activation of a lipid kinase by hepatitis C virus NS5A is essential for integrity of the membranous replication compartment. Cell Host Microbe. 2011;9:32–45. doi: 10.1016/j.chom.2010.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Saxena V, Lai CK, Chao TC, Jeng KS, Lai MM. Annexin A2 is involved in the formation of hepatitis C virus replication complex on the lipid raft. J Virol. 2012;86:4139–4150. doi: 10.1128/JVI.06327-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Romero-Brey I, et al. Three-dimensional architecture and biogenesis of membrane structures associated with hepatitis C virus replication. PLoS Pathog. 2012;8:e1003056. doi: 10.1371/journal.ppat.1003056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miyanari Y, et al. The lipid droplet is an important organelle for hepatitis C virus production. Nat Cell Biol. 2007;9:1089–1097. doi: 10.1038/ncb1631. [DOI] [PubMed] [Google Scholar]
- 22.Aizaki H, et al. Critical role of virion-associated cholesterol and sphingolipid in hepatitis C virus infection. J Virol. 2008;82:5715–5724. doi: 10.1128/JVI.02530-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sakamoto H, et al. Host sphingolipid biosynthesis as a target for hepatitis C virus therapy. Nat Chem Biol. 2005;1:333–337. doi: 10.1038/nchembio742. [DOI] [PubMed] [Google Scholar]
- 24.Umehara T, et al. Serine palmitoyltransferase inhibitor suppresses HCV replication in a mouse model. Biochem Biophys Res Commun. 2006;346:67–73. doi: 10.1016/j.bbrc.2006.05.085. [DOI] [PubMed] [Google Scholar]
- 25.Hirata Y, et al. Self-enhancement of hepatitis C virus replication by promotion of specific sphingolipid biosynthesis. PLoS Pathog. 2012;8:e1002860. doi: 10.1371/journal.ppat.1002860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Weng L, et al. Sphingomyelin activates hepatitis C virus RNA polymerase in a genotype-specific manner. J Virol. 2010;84:11761–11770. doi: 10.1128/JVI.00638-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shimakami T, et al. Protease inhibitor-resistant hepatitis C virus mutants with reduced fitness from impaired production of infectious virus. Gastroenterology. 2011;140:667–675. doi: 10.1053/j.gastro.2010.10.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu H, et al. Molecular cloning and functional characterization of a novel mammalian sphingosine kinase type 2 isoform. J Biol Chem. 2000;275:19513–19520. doi: 10.1074/jbc.M002759200. [DOI] [PubMed] [Google Scholar]
- 29.Kapadia SB, Chisari FV. Hepatitis C virus RNA replication is regulated by host geranylgeranylation and fatty acids. Proc Natl Acad Sci USA. 2005;102:2561–2566. doi: 10.1073/pnas.0409834102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lindenbach BD, et al. Cell culture-grown hepatitis C virus is infectious in vivo and can be recultured in vitro. Proc Natl Acad Sci USA. 2006;103:3805–3809. doi: 10.1073/pnas.0511218103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yanagi M, Purcell RH, Emerson SU, Bukh J. Transcripts from a single full-length cDNA clone of hepatitis C virus are infectious when directly transfected into liver of a chimpanzee. Proc Natl Acad Sci USA. 1997;97:8738–8743. doi: 10.1073/pnas.94.16.8738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Beard MR, et al. An infectious molecular clone of a Japanese genotype 1b hepatitis C virus. Hepatology. 1999;30:316–324. doi: 10.1002/hep.510300137. [DOI] [PubMed] [Google Scholar]
- 33.Andrus L, et al. Expression of paramyxovirus V proteins promotes replication and spread of hepatitis C virus in cultures of primary human fetal liver cells. Hepatology. 2011;54:1901–1912. doi: 10.1002/hep.24557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li YP, et al. Highly efficient full-length hepatitis C virus genotype 1 (strain TN) infectious culture system. Proc Natl Acad Sci USA. 2012;109:19757–19762. doi: 10.1073/pnas.1218260109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bochkov VN, et al. Generation and biological activities of oxidized phospholipids. Antioxid Redox Signal. 2010;12:1009–1059. doi: 10.1089/ars.2009.2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pizzimenti S, et al. Interaction of aldehydes derived from lipid peroxidation and membrane proteins. Front Physiol. 2013;4:242. doi: 10.3389/fphys.2013.00242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pitson SM, et al. Activation of sphingosine kinase 1 by ERK1/2-mediated phosphorylation. EMBO J. 2003;22:5491–5500. doi: 10.1093/emboj/cdg540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Alvarez SE, et al. Sphingosine-1-phosphate is a missing cofactor for the E3 ubiquitin ligase TRAF2. Nature. 2010;465:1084–1088. doi: 10.1038/nature09128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hait NC, et al. Regulation of histone acetylation in the nucleus by sphingosine-1-phosphate. Science. 2009;325:1254–1257. doi: 10.1126/science.1176709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jain SK, et al. Oxidative stress in chronic hepatitis C: not just a feature of late stage disease. J Hepatol. 2002;36:805–811. doi: 10.1016/s0168-8278(02)00060-0. [DOI] [PubMed] [Google Scholar]
- 41.Yano M, et al. Comprehensive analysis of the effects of ordinary nutrients on hepatitis C virus RNA replication in cell culture. Antimicrob Agents Chemother. 2007;51:2016–2027. doi: 10.1128/AAC.01426-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kohlway A, et al. Hepatitis C virus RNA replication and virus particle assembly require specific dimerization of the NS4A protein transmembrane domain. J Virol. 2014;88:628–642. doi: 10.1128/JVI.02052-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ibrahim W, et al. Oxidative stress and antioxidant status in mouse liver: effects of dietary lipid, vitamin E and iron. J Nutr. 1997;127:1401–1406. doi: 10.1093/jn/127.7.1401. [DOI] [PubMed] [Google Scholar]
- 44.Lackner T, Muller A, Konig M, Thiel HJ, Tautz N. Persistence of bovine viral diarrhea virus is determined by a cellular cofactor of a viral autoprotease. J Virol. 2005;79:9746–9755. doi: 10.1128/JVI.79.15.9746-9755.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Leivers AL, et al. Discovery of Selective Small Molecule Type III Phosphatidylinositol 4-Kinase Alpha (PI4KIIIalpha) Inhibitors as Anti Hepatitis C (HCV) Agents. J Med Chem. 2014;57:2091–2106. doi: 10.1021/jm400781h. [DOI] [PubMed] [Google Scholar]
- 46.Oikawa T, et al. Sal-like protein 4 (SALL4), a stem cell biomarker in liver cancers. Hepatology. 2013;57:1469–1483. doi: 10.1002/hep.26159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schmelzer E, et al. Human hepatic stem cells from fetal and postnatal donors. J Exp Med. 2007;204:1973–1987. doi: 10.1084/jem.20061603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang Y, et al. Paracrine signals from mesenchymal cell populations govern the expansion and differentiation of human hepatic stem cells to adult liver fates. Hepatology. 2010;52:1443–1454. doi: 10.1002/hep.23829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kubota H, Reid LM. Clonogenic hepatoblasts, common precursors for hepatocytic and biliary lineages, are lacking classical major histocompatibility complex class I antigen. Proc Natl Acad Sci USA. 2000;97:12132–12137. doi: 10.1073/pnas.97.22.12132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ma Y, Yates J, Liang Y, Lemon SM, Yi M. NS3 helicase domains involved in infectious intracellular hepatitis C virus particle assembly. J Virol. 2008;82:7624–7639. doi: 10.1128/JVI.00724-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yanagi M, Purcell RH, Emerson SU, Bukh J. Transcripts from a single full-length cDNA clone of hepatitis C virus are infectious when directly transfected into the liver of a chimpanzee. Proc Natl Acad Sci USA. 1997;94:8738–8743. doi: 10.1073/pnas.94.16.8738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shimakami T, et al. Stabilization of hepatitis C virus RNA by an Ago2-miR-122 complex. Proc Natl Acad Sci USA. 2012;109:941–946. doi: 10.1073/pnas.1112263109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Date T, et al. Novel cell culture-adapted genotype 2a hepatitis C virus infectious clone. J Virol. 2012;86:10805–10820. doi: 10.1128/JVI.07235-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Saeed M, et al. Efficient replication of genotype 3a and 4a hepatitis C virus replicons in human hepatoma cells. Antimicrob Agents Chemother. 2012;56:5365–5373. doi: 10.1128/AAC.01256-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Timpe JM, et al. Hepatitis C virus cell-cell transmission in hepatoma cells in the presence of neutralizing antibodies. Hepatology. 2008;47:17–24. doi: 10.1002/hep.21959. [DOI] [PubMed] [Google Scholar]
- 56.Kannan RP, Hensley LL, Evers LE, Lemon SM, McGivern DR. Hepatitis C virus infection causes cell cycle arrest at the level of initiation of mitosis. J Virol. 2011;85:7989–8001. doi: 10.1128/JVI.00280-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Puneet P, et al. SphK1 regulates proinflammatory responses associated with endotoxin and polymicrobial sepsis. Science. 2010;328:1290–1294. doi: 10.1126/science.1188635. [DOI] [PubMed] [Google Scholar]
- 58.Dansako H, et al. Class A scavenger receptor 1 (MSR1) restricts hepatitis C virus replication by mediating toll-like receptor 3 recognition of viral RNAs produced in neighboring cells. PLoS Pathog. 2013;9:e1003345. doi: 10.1371/journal.ppat.1003345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shaner RL, et al. Quantitative analysis of sphingolipids for lipidomics using triple quadrupole and quadrupole linear ion trap mass spectrometers. J Lipid Res. 2009;50:1692–1707. doi: 10.1194/jlr.D800051-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sullards MC, Liu Y, Chen Y, Merrill AH., Jr Analysis of mammalian sphingolipids by liquid chromatography tandem mass spectrometry (LC-MS/MS) and tissue imaging mass spectrometry (TIMS) Biochim Biophys Acta. 2011;1811:838–853. doi: 10.1016/j.bbalip.2011.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Reynolds ES. The use of lead citrate at high pH as an electron-opaque stain in electron microscopy. J Cell Biol. 1963;17:208–212. doi: 10.1083/jcb.17.1.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Papin JF, Vahrson W, Dittmer DP. SYBR green-based real-time quantitative PCR assay for detection of West Nile Virus circumvents false-negative results due to strain variability. J Clin Microbiol. 2004;42:1511–1518. doi: 10.1128/JCM.42.4.1511-1518.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gurukumar KR, et al. Development of real time PCR for detection and quantitation of Dengue Viruses. Virol J. 2009;6:10. doi: 10.1186/1743-422X-6-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Qu L, et al. Disruption of TLR3 signaling due to cleavage of TRIF by the hepatitis A virus protease-polymerase processing intermediate, 3CD. PLoS Pathog. 2011;7:e1002169. doi: 10.1371/journal.ppat.1002169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ahmed R, Salmi A, Butler LD, Chiller JM, Oldstone MB. Selection of genetic variants of lymphocytic choriomeningitis virus in spleens of persistently infected mice. Role in suppression of cytotoxic T lymphocyte response and viral persistence. J Exp Med. 1984;160:521–540. doi: 10.1084/jem.160.2.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bartenschlager R, Lohmann V, Penin F. The molecular and structural basis of advanced antiviral therapy for hepatitis C virus infection. Nat Rev Microbiol. 2013;11:482–496. doi: 10.1038/nrmicro3046. [DOI] [PubMed] [Google Scholar]
- 67.Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. J Mol Graph. 1996;14:33–38. 27–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.