Abstract
Nanopore sequencing of DNA is a single-molecule technique that may achieve long reads, low cost and high speed with minimal sample preparation and instrumentation. Here, we build on recent progress with respect to nanopore resolution and DNA control to interpret the procession of ion current levels observed during the translocation of DNA through the pore MspA. As approximately four nucleotides affect the ion current of each level, we measured the ion current corresponding to all 256 four-nucleotide combinations (quadromers). This quadromer map is highly predictive of ion current levels of previously unmeasured sequences derived from the bacteriophage phi X 174 genome. Furthermore, we show nanopore sequencing reads of phi X 174 up to 4,500 bases in length that can be unambiguously aligned to the phi X 174 reference genome, and demonstrate proof-of-concept utility with respect to hybrid genome assembly and polymorphism detection. This work provides the foundation for nanopore sequencing of long, complex, natural DNA strands.
DNA sequencing is stimulating biomedical and other life sciences research through its expanding scope1 and has a rapidly growing presence in clinical medicine2. These developments are driven in part by the successful completion of the Human Genome Project3 and in part by the introduction of new sequencing technologies that have dramatically reduced the cost of DNA sequencing4. Although such `next-generation' sequencing technologies have matured considerably since early proof-of-concepts5–7, nearly all remain limited to short sequence reads (with the exception of real-time sequencing from elongating polymerases8) and rely on complex, expensive instrumentation. Most platforms are also limited with respect to speed and require extensive sample preparation steps prior to sequencing.
Nanopore sequencing, independently proposed by Church and Deamer in the mid-1990s, has tremendous potential to overcome these limitations and achieve long reads, low cost and high speed while requiring minimal sample preparation and instrumentation9–12. However, this promise has faced substantial technical challenges, such that despite nearly 20 years of effort, nanopore-derived sequence reads that align to complex, natural DNA sequences have yet to be demonstrated.
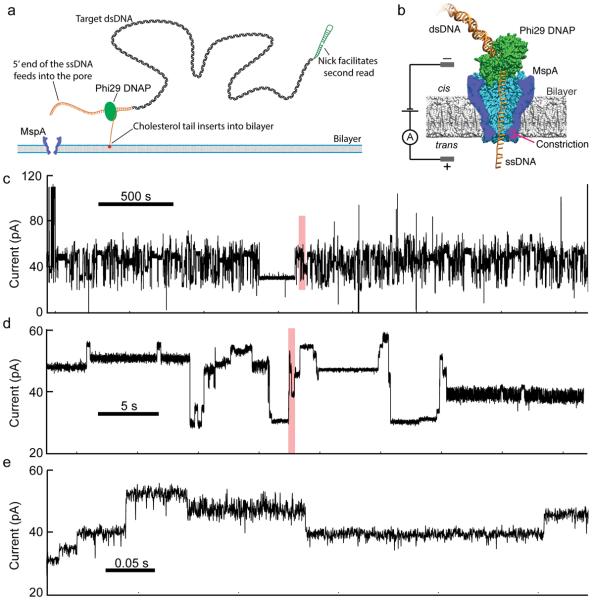
In nanopore devices directed at DNA sequencing, a salt solution is divided into cis and trans wells by a thin membrane. A single nanometer-scale pore in the membrane connects the cis and trans wells electrically. When a voltage is applied across this membrane, ion current flows through the pore; this current provides the primary signal. DNA is negatively charged and is electrophoretically attracted into the pore. When single-stranded (ss) DNA enters the pore, it blocks some fraction of the ion current. The fraction of the ion current that is blocked depends on the identity of nucleotides within the pore13–15. Key challenges of this technique are single-nucleotide resolution and control of the DNA translocation. Single-nucleotide resolution was recently enabled through the development of MspA, a protein pore with a short and narrow constriction11, 13, 15, 16. DNA translocation control was also recently enabled through the use of molecular motors such as phi29 DNA Polymerase (DNAP)11, 17 (Fig. 1).
Figure 1.
Experimental schematic and raw data. (a) Method of adapting dsDNA for nanopore sequencing. The first adaptor (orange) includes a cholesterol tail which inserts into the membrane, increasing DNA capture rates32 while, the long 5' single stranded overhang facilitates insertion into the pore. A second adaptor (green) enables re-reading of the pore using the DNAP's synthesis mode11, 17. (b) The protein nanopore MspA is shown in blue, phi 29 DNAP in green and DNA in orange. An applied voltage across the bilayer drives an ion current through the pore and an amplifier measures the current. DNA bases within the constriction determine the ion current. Phi 29 DNAP steps DNA through the pore in single-nucleotide steps. (c–e) Raw data for a representative 3000-second time window. Ion current changes as DNA is fed through the pore in single-nucleotide steps. Panels d and e each show a 1% section of the preceding panel's data shaded in red.
We have found that the currents in the MspA pore are determined by about four nucleotides at any given time11, 15. Each four-nucleotide combination (i.e. quadromer) has its own unique current value and in a few cases, nucleotides outside of a quadromer can have a small additional influence on the current. This prompted us to measure the ion current associated with each of the 256 possible quadromers.
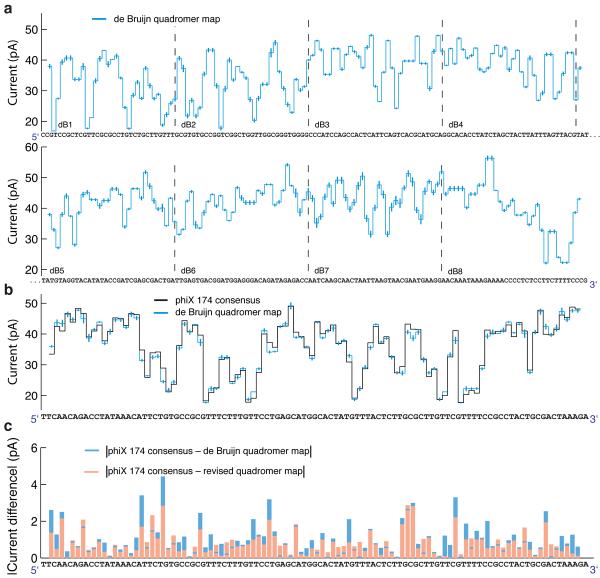
We constructed a 256 nucleotide-long cyclical de Bruijn sequence18 containing all possible combinations of four nucleotides (Supplementary Table 1). We divided the de Bruijn sequence into eight separate strands (Supplementary Table 1) and synthesized these with appropriate modifications to facilitate insertion into the pore, to initiate proper polymerase function and to allow for ion current calibration11, 17, 19 (Fig. 1a). Phi29 DNAP–based control of translocation results in two reads from each DNA molecule: (i) `unzipping` wherein one strand of the DNA moves 5' to 3' through the pore as the polymerase is forced to unzip the complimentary strand, and (ii) `synthesis' wherein the DNA moves 3' to 5' after the primer enters the DNAP's active site and the DNAP begins synthesizing a second complimentary strand11. As the polymerase moves along the strand, one nucleotide at a time, the identity of the quadromer within the MspA pore shifts in lock-step (Fig 1b), resulting in discrete changes in the measured ion current (Fig. 1c). We performed nanopore sequencing of all eight strands, averaging signals observed across multiple molecules of each strand to estimate the current level of the 256 quadromers, i.e. a `quadromer map' (Fig. 2a, Supplementary Figs. 2–4, Supplementary Table 2).
Figure 2.
A quadromer map predicts current levels for previously unmeasured DNA. (a) Current levels observed for all possible 4-nucleotide sequences (quadromers) measured in eight segments of a 256-nucleotide de Bruijn sequence. (b) The black trace shows a consensus based on 22 reads of phi X 174 DNA. This is compared to predicted current levels based on the de Bruijn quadromer values. Error bars are the variance of the measured quadromer values. We use a consensus to correct for insertion/deletion errors caused by the stochastic motion of the phi29 DNAP11. (c) Absolute current difference between quadromer map and measured consensus for the ~100 level sequence shown in panel b using the de Bruijn quadromer map (blue) and the revised quadromer map (red). In most instances, the revised map improves the predictive ability of our map. The correlation coefficient between measured values and the de Bruijn quadromer values is 0.9905 (95% confidence bounds [0.9859–0.9936]). The correlation coefficient between measured values and the revised quadromer values is 0.9938 (95% confidence bounds [0.9908–0.9958]).
We next sought to evaluate whether the quadromer map constructed by nanopore sequencing of the de Bruijn sequence was predictive of current levels for previously unmeasured, natural DNA sequences. To assess this, we constructed and nanopore sequenced a genomic DNA sequencing library from the bacteriophage phi X 174. Specifically, we attached asymmetric adaptors (a nicked hairpin adaptor and a cholesterol tailed adaptor; green and orange respectively in Fig. 1a) to the ends of linearized phi X 174 dsDNA, and used phi29 DNAP to draw ssDNA through a mutant MspA pore in single-nucleotide steps11 (Fig. 1b,c). The measured current level sequences were then compared to predicted current levels based on the quadromer map. Figure 2b shows quadromer map–based predicted current levels versus a consensus of 22 nanopore reads for a representative ~100 nucleotide (nt) region of the phi X 174 genome.
Overall, the predicted current levels from the de Bruijn sequence-based quadromer map strongly match the observed current levels from nanopore sequencing of the phi X 174 DNA (r = 0.9905, 95% confidence bounds [0.9859–0.9936]). However, differences between prediction and measurement are not due to statistical fluctuation (For the 100 levels shown, χ2 = 150, corresponding to a p-value < 0.001, indicating quadromers alone are not sufficient to describe all the variation). Close analysis suggests that this error is dominated by shifts in the positioning of the DNA within the pore's constriction owing either to DNA secondary structure within the vestibule or DNA interactions with the pore vestibule or constriction. In some instances, nucleotides outside of the quadromer also have a small effect. However, as independent reads of the same sequence yield extremely reproducible current values11, 19, we conclude that the small differences between prediction and observation are systematic and likely due to sequence context outside of the quadromer itself. The source of these systematics is a subject of our ongoing research.
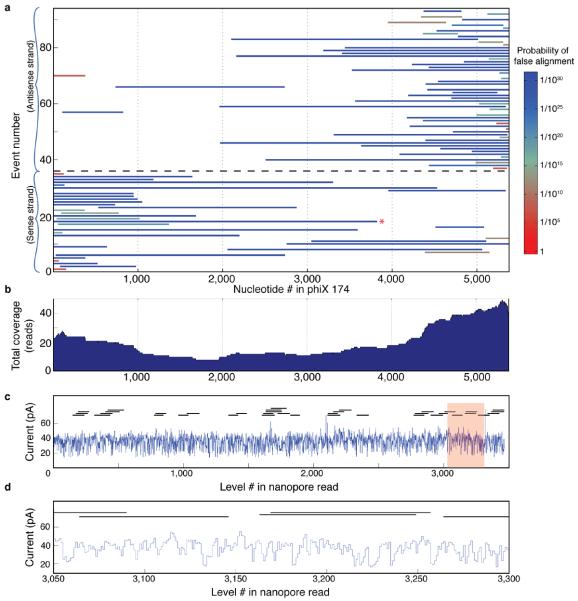
The strong homology between quadromer-based current predictions and nanopore sequencing reads can be used to perform alignments to reference genomes and sequence databases with high confidence. As a first assessment, we subjected three PCR amplicons derived from phi X 174 to nanopore sequencing in a blinded fashion, i.e. the individuals performing sequencing and analysis were not aware of the genomic positions of the amplicons. After extracting the current levels from nanopore reads using a custom algorithm (Supplementary Discussion), we aligned the observed current levels from each read to predicted current levels obtained by applying the quadromer map to the known phi X 174 genome sequence (Fig. 3a,b). Our alignment algorithm is similar to Needleman-Wunsch alignment20, 21 but allows for backsteps in the series of levels (Fig. 3c and Supplementary Figs. 5 and 6). We assessed the confidence of these alignments by comparing alignment scores with those obtained against random sequences (Supplementary Fig. 7). The vast majority (30 out of 31) of nanopore sequencing reads with a probability of false alignment below 1 × 10−4 aligned to one of three regions; un-blinding confirmed that these corresponded to the locations along the phi X 174 genome from which the three PCR amplicons were derived (Supplementary Fig. 8).
Figure 3.
Raw data to alignment. (a) Raw data are processed using a level-finding algorithm (Supplementary Discussion) to identify transitions between levels in the current trace. A subsequent filter removes most repeated levels, which likely result from polymerase backsteps (indicated by `*'). (b) Extract the sequence of median current values of each level. (c) Align the current values to predicted values from the reference sequence using the quadromer map (Fig. 2a). Alignment is performed with a dynamic programming alignment algorithm similar to Needleman-Wunch alignment20 (Supplementary Discussion). In some locations, levels are skipped in the nanopore read either owing to motions of the DNAP or errors made by the level finding algorithm. In other places, backsteps result in multiple reads of the same level. We determine read boundaries from the first and last matched levels in the reference sequence. Read boundaries are indicated by the blue lines. The above alignment had an estimated 6.4× 10−15 probability of false alignment.
We next assessed whether we could achieve long nanopore sequencing reads. We constructed a genomic DNA sequencing library by ligating asymmetric adaptors to the linearized, full-length phi X 174 genome as described above, and this library was nanopore sequenced. We generated 106 long (>200 base-pair) ion current recordings corresponding to single molecules within this library. We aligned these reads to ion current levels predicted with the quadromer map; 92 of these reads aligned with high confidence to the phi X 174 genome (Fig. 4a) with a misalignment probability estimated at < 1×10−10. Within this set of aligned reads, ~60% were >1,000 bp, ~20% were >2,000 bp and ~10% were >3,000 bp. This is in contrast to the length distribution of our library (Supplementary Fig. 9), which contains far longer strands, implying that DNAP dissociation from the strand is the primary cause of event termination. As expected, the 5' end of most reads aligns to the cut site of the restriction enzyme used to linearize the genome, and these reads are split approximately equally between the sense and antisense strands (Fig. 4a). The 92 reads comprise a sum total of 118 kilobases (kb) with mean 21.9-fold coverage of the phi X 174 reference genome (range: 10-fold to 44-fold) (Fig. 4b).
Figure 4.
Alignments to reference sequence and hybrid reconstruction. (a) Coverage plot for 91 nanopore sequencing reads of bacteriophage phi X 174 genomic DNA. Left and right alignment bounds are indicated by the extent of the line for each read. Random attachment of the asymmetric adaptors results in reads of both sense and antisense strands. Reads below the black dashed line (events 1–38) are sense strands while reads above (events 40–92) are antisense strands. Most reads begin near the 5' end of the linearization cut site and proceed towards the 3' end as the phi29 DNAP unzips the double stranded DNA. (b) Sum total coverage for each region within the phi X 174 genome. This graph indicates the number of reads that cover any given section of the genome using the sense and antisense strands. (c) Hybrid assembly of Illumina sequencing reads using a single nanopore read (Supplementary Discussion). Thirty-eight Illumina reads (horizontal black lines) are aligned to a single 3,819 nt long nanopore read (blue trace; indicated by the red * in panel a). (d) Detail of shaded region in panel c. Six 100 bp Illumina reads are shown where they align to the nanopore read.
The 10,772 bases contained within the phi X 174 sense and antisense strands include on average 35 instances of nearly all quadromers (255 out of 256) in diverse sequence contexts (Supplementary Fig. 10), and are likely to yield a more reliable quadromer map than the de Bruijn sequence alone. We therefore used these independent measurements of each quadromer to generate an improved quadromer map (Supplementary Fig. 11, Supplementary Table 2). The new quadromer map is a closer match to measured levels (Fig. 2c; r = 0.9936, 95% confidence bounds [0.9908–0.9958] as compared to r = 0.9905 with bounds [0.9859–0.9936]).
We then explored the potential of nanopore reads to facilitate hybrid assembly22–24, by aligning short Illumina sequencing reads directly to the nanopore ion current measurements using the afore described alignment software. Specifically, we took 11,000 single-end 100 bp Illumina MiSeq shotgun reads from phi X 174 and aligned these directly to a single 3,800 bp nanopore sequencing read (Supplementary Fig 12). Figure 4c shows the alignment locations of a representative 38 Illumina reads within the nanopore read. As nanopore sequencing develops longer reads and higher throughput, such alignments may facilitate rapid and accurate sorting of short sequence reads into their proper order for de novo genome assembly requiring far lower coverage.
To assess whether long nanopore sequencing reads could be accurately aligned against a large database of naturally occurring DNA sequences, we took one 250-level sub-region of ion currents from three individual long nanopore reads and individually aligned these 250-level regions to a 156 Mb database containing 5,287 viral genomes, including phi X 174. The highest scoring alignment for all nanopore sequencing reads was to the phi X 174 genome, each with high confidence (>99.9996%, Supplemental Fig. 13), implying that nanopore read quality is sufficient for unambiguous species identification. These 250-level alignment `seeds' could then be extended in both directions to the full nanopore sequencing read, yielding alignments to phi X 174 identical to the targeted alignments shown in Figure 4a with high confidence.
Finally, we assessed our ability to detect single nucleotide polymorphisms (SNPs). SNPs can be detected by comparing nanopore reads to a previously measured nanopore consensus19 (comparison to a `reference consensus' minimizes the impact of the systematic, context-dependent deviations from the quadromer map predictions discussed above). To systematically assess our power to detect single-base substitutions, we iteratively inserted a total of 1,044 `mock SNPs' to the reference genome of phi X 174, i.e. introducing quadromer map values corresponding to these SNPs at the appropriate locations in the phi X 174 reference consensus map25. We then aligned 33 of the nanopore sequencing reads from phi X 174 to the modified reference consensus. We successfully called 77.4% of the mock SNPs (Supplementary Discussion). These data and methods provide a starting point for the further development of variant calling algorithms for nanopore sequencing data.
This work demonstrates nanopore sequencing of long, complex, natural DNA strands. By measuring the ion current signal associated with all 256 possible 4-mers as they translocate through the constriction of the MspA nanopore, we report, to our knowledge, the first nanopore `quadromer map.' This map is highly predictive of ion current levels of previously unmeasured, complex, natural DNA sequences. We exploit this reproducible behavior of MspA on quadromers to develop both a level-finding algorithm as well as a dynamic programming alignment algorithm for nanopore sequencing reads. We apply these tools to unambiguously align long nanopore sequencing reads generated from phi X 174 to the corresponding reference genome sequence, including reads that span up to 4,500 bases in length. We then show proof-of-concept utility with respect to organismal identification as well as for hybrid de novo assembly. Lastly, we demonstrate algorithmic approaches for successful SNP detection using nanopore reads. Much of this analysis will be applicable to other nanopores with a short constriction. MspA's ~four nucleotide long recognition zone is in fact advantageous in that it is short enough to have high current contrast yet it is long enough to distinguish homopolymer sections of two or three bases.
A limitation of our present system is that the amplitude of ion current levels alone does not provide enough information for direct de novo sequencing, i.e. conversion of ion currents to accurate sequences in the absence of a reference for alignment and comparison. However, additional information is contained in the variance, the duration and the voltage dependence of each current level that may enable de novo sequencing with improved algorithms. Furthermore, much of the variance in current levels associated with the nanopore sequencing system described here results from the erratic and stochastic motion of the phi29 polymerase as it feeds the DNA through the pore. Switching to a different enzyme, e.g. a helicase, that translates along DNA monotonically and with reduced stochasticity is expected to sharply improve performance.
Notably, the nanopore sequencing performed here was implemented on a low-cost experimental device in a small lab, with essentially real-time results from a single MspA pore. Full realization of nanopore sequencing's potential will require additional progress in areas including nanopore parallelization26, channel setup27–30 and microfluidics31.
Despite the remaining hurdles, our demonstration of a highly predictive quadromer map and of 4,500 bp interpretable nanopore reads, —corresponding to natural DNA sequences and generated in real-time with a low-cost device—represents a major milestone in the nearly 20 year history of this technological paradigm. All experimental methods, raw data and algorithms are made fully available to the research community to facilitate the further maturation of nanopore sequencing.
Online Methods
Pore establishment
A single MspA pore was established in a bilayer as previously described11, 13, 15, 19. Quadromer DNA was ordered from PAN labs at Stanford. DNA oligos were mixed and prepared as previously described11, 19.
Data acquisition
Data was acquired with a sampling rate of 500 kHz on Axopatch 200B or Axopatch 1B amplifiers filtered at 100 kHz. Data was downsampled to 5 kHz by averaging every 100 datapoints. DNA interaction events were detected using a thresholding algorithm as previously described11 and good events were selected automatically using characteristics such as duration, mean, and standard deviation. Levels within good events were then selected either by hand (for the initial quadromer data) or with an automated level-finding algorithm (for all other data). The level finding algorithm is described in detail below (see Supplmentary Fig. 1 for data reduction flowchart).
De Bruijn sequence design
Just as the circular eight letter sequence …AAABABBB… contains all eight three-letter combinations of A and B-AAA, AAB, ABA, BAB, ABB, BBB, BBA, BAA-one can construct a cyclical 256 letter sequence that contains all 256 four-letter combinations of A, C, G, and T18.
The 256 nt long de Bruijn sequence was divided up into eight separate strands. This was to ensure DNA accuracy because high purity custom oligos longer than ~100 were not readily available. Each strand contained part of the quadromer map sequence but also contained an ion current calibration sequence that allowed us to correct for buffer evaporation and voltage offsets. In order to use the phi 29 DNAP control method11, 17, these strands also had a portion of sequence conjugated to a hairpin primer (see table S1 for strand construction and sequences). We made several measurements of each of the eight strands and aligned the extracted current levels to the known DNA sequence (see Supplmentary Figs. 2–4). Oligos were mixed together and annealed as previously described11, 19.
Adaptor design
Adaptors were designed to each contain ½ of a NotI restriction endonuclease site to allow digestion of adaptor-dimers that were produced during the shotgun ligation. Both the fantail (FT-½NotI) and hairpin adaptors (HP-½NotI) were comprised of top and bottom oligos ordered from IDT (FT-½NotI-top: 5'- (Phosphate) (3 Carbon Spacer) AAA AAA ACC TTC C (3 Carbon Spacer) CCT TCC CAT CAT CAT CAG ATC TCA CGC GG -3', FT-½NotI-bot: 5'-(Phosphate) GGC GCA CTC TAG ACT TTT TAA ATT TGG GTT T (3 Carbon Spacer) (Cholesterol) -3', HP-½NotI-top: 5'- (Phosphate) CGC CTA CGG TTT TTC CGT AGG CGT ACG C (Uracil) TAC TTG TAC TTG GCG G -3', HP-½NotI-bot: 5'- (Phosphate) CCG CCA AGT ACA AGT AAG CGT A -3'). The cholesterol tag at the end of the fantail adaptor causes the DNA to bind to the bilayer thereby increasing the DNA concentration near the pore and increasing DNA-pore interactions32.
Oligos were resuspended to 100 μM in 10 mM Tris, and annealed by combining 20 μL of the top and bottom oligos with the addition of 60 μL 10 mM Tris followed by heating to 95°C for 2 minutes and gradual cooling to room temperature in a polystyrene casing. The two-oligo scheme for the hairpin adaptor was designed to prevent synthesis of excessively long oligos. The nick present between the oligos ligates during the adaptor ligation process, followed by subsequent introduction of the desired nick by digestion of the uracil base with USER enzyme.
Library construction
Phi X 174 nanopore libraries were constructed using a shotgun-ligation approach. 0.5 to 4 μg of Phi X 174 gDNA (Thermo Scientific) was restriction-digested using 5U of SspI (NEB) in 1× SspI Reaction Buffer for 2 hours at 37°C to linearize DNA and produce blunt ends followed by SPRI bead purification. DNA was resuspended in 42 μL Elution Buffer (EB, QIAGEN) followed by addition of 5 μL 10× T4 DNA Ligase Buffer (NEB), 1 μL of each annealed adaptor (FT-½NotI and HP-½NotI), and 1 μL (400U) of T4 DNA Ligase (NEB) and incubated overnight at 16°C. Ligase was heat-inactivated at 65°C for 15 minutes followed by cooling on ice and addition of 1 μL 10× T4 DNA Ligase Buffer, 1 μL (1U) USER Enzyme (NEB), 1 μL (20U) NotI-HF (NEB), and 2 μL (10U) λ Exonuclease (NEB) and incubation at 37°C for 2 hours. DNA was then purified using 45 μL SPRI beads. A subset of samples were gel-size-selected to remove adaptor-dimer bands on a 1% agarose gel (SeaKem) and purified using the column-based Gel Purification Kit (QIAGEN) and eluted in EB.
Alignment Algorithm
Alignments were performed using a novel dynamic programming algorithm described later in the supporting text. Quality scores for alignments were estimated by comparing the maximal alignment score to the alignment scores obtained from alignments of measured strands to random DNA sequences with the same GC content as that of phi X 174. A data processing flowchart is available below (Suplementary Fig. 1).
Supplementary Material
Acknowledgments
This work was supported by the National Institutes of Health, National Human Genome Research Institutes (NHGRI) $ 1,000 Genome Program Grants R01HG005115, R01HG006321, and R01HG006283 and graduate research fellowship DGE-0718124 from the National Science Foundation (to A.A.)
Footnotes
Author contributions: A.H.L., I.M.D., A.A., J.S., and J.H.G. designed the research. H.B., A.A., I.C.N., J.M.C., J.M.S., R.D., and K.D. performed the research. A.H.L., I.M.D., B.C.R., H.B., I.C.N., J.M.C., K.W.L. analyzed the data. A.H.L., J.H.G., and J.S. wrote the paper.
Competing interests statement: J.H.G. and I.M.D. have a commercial interest with Illumina Inc. The University of Washington has filed a provisional patent on technologies described herein.
References
- 1.Shendure J, Lieberman Aiden E. The expanding scope of DNA sequencing. Nat Biotechnol. 2012;30(11):1084–1094. doi: 10.1038/nbt.2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McCarthy JJ, McLeod HL, Ginsburg GS. Genomic medicine: a decade of successes, challenges, and opportunities. Sci Transl Med. 2013;5(189):189sr184. doi: 10.1126/scitranslmed.3005785. [DOI] [PubMed] [Google Scholar]
- 3.Finishing the euchromatic sequence of the human genome. Nature. 2004;431(7011):931–945. doi: 10.1038/nature03001. Anonymous. [DOI] [PubMed] [Google Scholar]
- 4.Shendure J, Ji H. Next-generation DNA sequencing. Nat Biotechnol. 2008;26(10):1135–1145. doi: 10.1038/nbt1486. [DOI] [PubMed] [Google Scholar]
- 5.Mitra RD, Shendure J, Olejnik J, Edyta Krzymanska O, Church GM. Fluorescent in situ sequencing on polymerase colonies. Anal Biochem. 2003;320(1):55–65. doi: 10.1016/s0003-2697(03)00291-4. [DOI] [PubMed] [Google Scholar]
- 6.Levene MJ, et al. Zero-mode waveguides for single-molecule analysis at high concentrations. Science. 2003;299(5607):682–686. doi: 10.1126/science.1079700. [DOI] [PubMed] [Google Scholar]
- 7.Braslavsky I, Hebert B, Kartalov E, Quake SR. Sequence information can be obtained from single DNA molecules. Proc Natl Acad Sci U S A. 2003;100(7):3960–3964. doi: 10.1073/pnas.0230489100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eid J, et al. Real-time DNA sequencing from single polymerase molecules. Science. 2009;323(5910):133–138. doi: 10.1126/science.1162986. [DOI] [PubMed] [Google Scholar]
- 9.Kasianowicz JJ, Brandin E, Branton D, Deamer DW. Characterization of individual polynucleotide molecules using a membrane channel. Proc Natl Acad Sci U S A. 1996;93(24):13770–13773. doi: 10.1073/pnas.93.24.13770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Branton D, et al. The potential and challenges of nanopore sequencing. Nature Biotechnol. 2008;26(10):1146–1153. doi: 10.1038/nbt.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Manrao EA, et al. Reading DNA at single-nucleotide resolution with a mutant MspA nanopore and phi29 DNA polymerase. Nature Biotechnol. 2012;30(4):349–U174. doi: 10.1038/nbt.2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wanunu M. Nanopores: A journey towards DNA sequencing. Phys Life Rev. 2012;9(2):125–158. doi: 10.1016/j.plrev.2012.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Derrington IM, et al. Nanopore DNA sequencing with MspA. Proc Natl Acad Sci U S A. 2010;107(37):16060–16065. doi: 10.1073/pnas.1001831107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wallace EVB, et al. Identification of epigenetic DNA modifications with a protein nanopore. Chem. Commun. 2010;46:8195–8197. doi: 10.1039/c0cc02864a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Manrao EA, Derrington IM, Pavlenok M, Niederweis M, Gundlach JH. Nucleotide Discrimination with DNA Immobilized in the MspA Nanopore. PLoS ONE. 2011;6(10):e25723. doi: 10.1371/journal.pone.0025723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Butler TZ, Pavlenok M, Derrington IM, Niederweis M, Gundlach JH. Single-molecule DNA detection with an engineered MspA protein nanopore. Proc Natl Acad Sci U S A. 2008;105(52):20647–20652. doi: 10.1073/pnas.0807514106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cherf GM, et al. Automated forward and reverse ratcheting of DNA in a nanopore at 5-angstrom precision. Nature Biotechnol. 2012;30(4):344–348. doi: 10.1038/nbt.2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.de Bruijn NG. A Combinatorial Problem. Koninklijke Netherlandse Akademie v. Wetenschappen. 1946;49:758–764. [Google Scholar]
- 19.Laszlo AH, et al. Detection and mapping of 5-methylcytosine and 5-hydroxymethylcytosine with nanopore MspA. Proc. Natl. Acad. Sci. U.S.A. 2013;110(47):18904–18909. doi: 10.1073/pnas.1310240110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Needleman SB, Wunsch CD. A General Method Applicable to Search for Similarities in Amino Acid Sequence of 2 Proteins. Journal of Molecular Biology. 1970;48(3):443–453. doi: 10.1016/0022-2836(70)90057-4. [DOI] [PubMed] [Google Scholar]
- 21.Durbin R, Eddy S, Krogh A, Mitchison G. Biological sequence analysis. 2006. pp. 92–96. [Google Scholar]
- 22.Bashir A, et al. A hybrid approach for the automated finishing of bacterial genomes. Nat Biotech. 2012;30(7):701–707. doi: 10.1038/nbt.2288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Koren S, et al. Hybrid error correction and de novo assembly of single-molecule sequencing reads. Nat Biotech. 2012;30(7):693–700. doi: 10.1038/nbt.2280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ribeiro FJ, et al. Finished bacterial genomes from shotgun sequence data. Genome Res. 2012;22(11):2270–2277. doi: 10.1101/gr.141515.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shendure J, et al. Accurate multiplex polony sequencing of an evolved bacterial genome. Science. 2005;309(5741):1728–1732. doi: 10.1126/science.1117389. [DOI] [PubMed] [Google Scholar]
- 26.Baaken G, Sondermann M, Schlemmer C, Ruhe J, Behrends JC. Planar microelectrode-cavity array for high-resolution and parallel electrical recording of membrane ionic currents. Lab Chip. 2008;8(6):938–944. doi: 10.1039/b800431e. [DOI] [PubMed] [Google Scholar]
- 27.Malmstadt N, Nash MA, Purnell RF, Schmidt JJ. Automated formation of lipid bilayer membranes in a microfluidic device. Nano Lett. 2006;6(9):1961–1965. doi: 10.1021/nl0611034. [DOI] [PubMed] [Google Scholar]
- 28.Schibel AE, Edwards T, Kawano R, Lan W, White HS. Quartz nanopore membranes for suspended bilayer ion channel recordings. Anal Chem. 2010;82(17):7259–7266. doi: 10.1021/ac101183j. [DOI] [PubMed] [Google Scholar]
- 29.Heitz BA, Jones IW, Hall HK, Jr, Aspinwall CA, Saavedra SS. Fractional polymerization of a suspended planar bilayer creates a fluid, highly stable membrane for ion channel recordings. J Am Chem Soc. 2010;132(20):7086–7093. doi: 10.1021/ja100245d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Heitz BA, et al. Polymerized planar suspended lipid bilayers for single ion channel recordings: comparison of several dienoyl lipids. Langmuir. 2011;27(5):1882–1890. doi: 10.1021/la1025944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jain T, Guerrero RJ, Aguilar CA, Karnik R. Integration of solid-state nanopores in microfluidic networks via transfer printing of suspended membranes. Anal Chem. 2013;85(8):3871–3878. doi: 10.1021/ac302972c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yusko EC, et al. Controlling protein translocation through nanopores with bio-inspired fluid walls. Nat Nano. 2011;6(4):253–260. doi: 10.1038/nnano.2011.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dawson E, et al. A SNP resource for human chromosome 22: extracting dense clusters of SNPs from the genomic sequence. Genome Res. 2001;11(1):170–178. doi: 10.1101/gr.156901. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.