Abstract
Drug screening against novel targets is warranted to generate biochemical probes and new therapeutic drug leads. Tyrosyl-DNA-phosphodiesterases 1 and 2 (TDP1 and TDP2) are two DNA repair enzymes that have yet to be successfully targeted. TDP1 repairs topoisomerase I-, alkylation-, and chain terminator-induced DNA damage, while TDP2 repairs topoisomerase II-induced DNA damage. Here we report the quantitative high-throughput screening (qHTS) of the NIH Molecular Libraries Small Molecule Repository using recombinant human TDP1. We also developed a secondary screening method using a multiple loading gel-based assay where recombinant TDP1 is replaced by whole cell extract (WCE) from genetically engineered DT40 cells. While developing this assay, we determined the importance of buffer conditions for testing TDP1, and most notably the possible interference of phosphate-based buffers. The high specificity of endogenous TDP1 in WCE allowed the evaluation of a large number of hits with up to 600 samples analyzed per gel via multiple loadings. The increased stringency of the WCE assay eliminated a large fraction of the initial hits collected from the qHTS. Finally, inclusion of a TDP2 counter-screening assay allowed the identification of two novel series of selective TDP1 inhibitors.
Keywords: TDP1, TDP2, topoisomerases, drug discovery, combination therapy
Introduction
Topoisomerase I (Top1)-mediated cleavage complexes resulting from the trapping of Top1 by DNA lesions including abasic sites, oxidized bases, carcinogenic adducts (1–3) and anticancer Top1 inhibitors (topotecan, irinotecan and non-camptothecin Top1 inhibitors (4, 5)) are removed by TDP1 [for review see (6, 7)]. TDP1 acts by cleaving the covalent bond between a 3’-DNA phosphate group and the catalytic tyrosine residue of the trapped Top1 (8–10). TDP1 can also remove a broad range of 3’-blocking DNA lesions including 3’-phoshoglycolates (11, 12), 3’-nucleosides (13, 14), and chain-terminating anticancer and antiviral nucleotide analogs (15). TDP1 has also been shown to act as a backup repair pathway for topoisomerase II (Top2) cleavage complexes (16, 17). Both Top1 and Top2 are pharmacological targets for widely used anticancer drugs. Therefore, TDP1 inhibitors are under consideration for combination therapies with existing anticancer treatments. There is currently no reported TDP1 inhibitor exhibiting a synergistic effect when used in combination with a Top1 inhibitor. Yet, the usefulness of a combination therapy with a TDP1 and a Top1 inhibitor in the clinic is supported by genetic evidence. Genetic inactivation of TDP1 confers hypersensitivity to CPT in human cells (18–20), murine cells (21, 22), chicken cells (17, 23), and in yeast (24). In addition, mutation of the catalytic histidine to an arginine residue at position 493 (H493R) results in the accumulation of covalent TDP1-DNA intermediates (13) ultimately leading to the rare autosomal recessive neurodegenerative disease called spinocerebellar ataxia with axonal neuropathy (SCAN1) (25); SCAN1 cells are hypersensitive to CPT (18–21). Because there is yet no available TDP1 inhibitor active in cells, an indirect way to inhibit the TDP1 pathway is actually to block PARP activity. Indeed, we recently showed that PARP1 is a critical cofactor of TDP1 in cells, acting by stabilizing TDP1 and facilitating its recruitment to Top1cc damage sites (26). This mechanism is one of the underlying molecular mechanisms by which PARP inhibitors synergize with Top1 inhibitors (27–29).
The discovery of TDP1 inhibitors has been challenging because previously known inhibitors either lack selectivity or cellular efficiency suitable for drug development (30). We previously reported the development and optimization of a quantitative high-throughput screening assay (qHTS) based on the AlphaScreen technology for the discovery of TDP1 inhibitors (31). In this study, we report the development of novel biochemical assays with increased stringency for the confirmation of chemical hits obtained from our qHTS campaign using libraries at the National Center for Advancing Translational Sciences1, and the use of TDP2 for counterscreening. We also discuss the importance of reaction conditions and counter screening for the characterization of TDP1-selective inhibitors.
Material and Methods
Chemicals
JLT048 (CAS# 664357; 4-(5-[{[1-(2-fluorobenzyl)-2,5-dioxo-4-imidazolidinylidene] methyl]}-2furyl)benzoic acid) was purchased from ChemBridge Corporation. Camptothecin (CPT) and veliparib were obtained from the Drug Synthesis and Chemistry Branch, Developmental Therapeutics Program, DCTD, NCI.
All reactions were performed under argon in oven-dried or flame-dried glassware. All commercially available reagents were purchased from Sigma Aldrich and used as received. All experiments were monitored by analytical thin layer chromatography (TLC) performed on Silicycle silica gel 60 Å glass supported plates with 0.25mm thickness. Yields are not optimized. Low-resolution mass spectra (electrospray ionization) were acquired on an Agilent Technologies 6130 quadrupole spectrometer coupled to an Agilent Technologies 1200 series HPLC. High resolution mass spectrum-electron ionization sprary (HRMS-ESI) were obtained on an Agilent Technologies 1200 series Dual Absorbance Detector HPLC system equipped with a Phenomenex Luna 75×3mm, C18, 3 µm column at 45 °C (UV detection at 220nm, BW 8nm, and 254nm BW 8nm, flow rate: 0.8 mL/min (increasing), Injection volume: 1.0 µL, sample solvent: 100% Methanol, sample conc.: ~0.01 mg/mL, mobile phase A: Water with 0.1% acetic acid, mobile phase B: Acetonitrile with 0.1% acetic acid) coupled to a Agilent 6210 time-of-flight mass spectrometer (ion source: Duel ESI, min range: 115 m/z, max range: 1400 m/z, scan rate: 0.9 seconds, gas temp: 340°C, gas flow: 10 L/min, nebulizer: 50 PSI, ion polarity: positive, VCap: 3500 V, fragmentor: 175 V, skimmer1: 65 V, OctopoleRFPeak: 250 V, ref mass: enabled (Agilent P/N G1969-85001). Data were analyzed using Agilent Masshunter Workstation Data Acquisition (v B.02.00, Patch 1,2,3) and Agilent Masshunter Qualitative Analysis (v B.02.00, Build 2.0.197.7, Patch 3). Abbreviations are as follows: dimethylsulfoxide – DMSO; sodium hydroxide – NaOH; ethanol – EtOH; methanol – MeOH. Preparation steps for NCGC00183974 are described in Supplemental Materials.
Cells
DT40 knockout cells (TDP1−/−) and complemented with human TDP1 (hTDP1) were established and authenticated by Southern blot, RT-PCR and Western blot in 2012 in the Developmental Therapeutics Branch, CCR NCI (17). DT40 cells were cultured with RPMI 1640 medium (GIBCO 11875-093>) supplemented with 10% fetal calf serum (Gemini Bio-Products 100–106), 1% chicken serum (Invitrogen 16110082), and 50 µM α-mercaptoethanol at 37°C. TDP1-deficient (Tdp1−/−) cells, and TDP1−/− cells complemented with human TDP1 (hTDP1) in chicken DT40 B cell line have previously been reported and described here (17).
Quantitative high throughput screening assay
Tdp1 enzyme in the HTS buffer containing 1X PBS, pH 7.4, 80 mM KCl, and 0.01% Tween-20 (31) was dispensed at 3 µl into 1536-well Kalypsys black solid bottom plates (Kalypsys, San Diego, CA). Compounds and controls (23 nl) were transferred via a Pin Tool station (Kalypsys) at 23 nl. The plates were incubated for 15 min at room temperature, and then 1 ul of DNA substrate (Supplemental Figure S1) was added to start the reaction. After 5 min incubation at room temperature, 1 µl of AlphaScreen donor/acceptor bead mix (PerkinElmer Lifesciences, Waltham, MA) was added and the plates were further incubated for 10 min at room temperature. The detection was then performed on a PerkinElmer Envision reader (PerkinElmer Lifesciences). Compounds were first classified as having full titration curves, partial modulation, partial curve (weaker actives), single point activity (at highest concentration only), or inactive (32). For all active compounds, a score range was given for each curve class type given above. Active compounds received a Pubchem Activity_Score between 40 and 100. Inconclusive compounds received Pubchem Activity_Score between 1 and 39. All inactive compounds received a Pubchem Activity_Score of 0.
Whole cell extract TDP1 assay
DT40 hTDP1 were collected, washed, and centrifuged. Cell pellets were then resuspended in 100 µL of CelLytic M cell lysis reagent (Sigma-Aldrich C2978). After 15 min on ice, lysates were centrifuged at 12,000 g for 10 min, and supernatants were transferred to a new tube. Protein concentrations were determined using a Nanodrop spectrophotometer (Invitrogen), and whole cell extracts were stored at −80 °C. This method of protein concentration determination is acceptable in this case because cellular nucleic acids have been precipitated and discarded during the precipitation step described above and because the amount of WCE to be used is determined by the dilution of WCE required to achieve 30–40% of TDP1 cleavage in the assay. A 5′-[32P]-labeled single-stranded DNA oligonucleotide containing a 3′-phosphotyrosine (N14Y) (31) was incubated at 1 nM with 4 µg/mL of whole cell extract (WCE quantity to achieve 30–40% of TDP1 cleavage) in the absence or presence of inhibitor for 15 min at room temperature in the WCE buffer containing 50 mM Tris HCl, pH 7.5, 80 mM KCl, 2 mM EDTA, 1 mM DTT, 40 µg/mL BSA, and 0.01% Tween-20. Reactions were terminated by the addition of 1 volume of gel loading buffer [99.5% (v/v) formamide, 5 mM EDTA, 0.01% (w/v) xylene cyanol, and 0.01% (w/v) bromophenol blue]. Samples were subjected to a 16% denaturing PAGE (Accugel 19:1, National Diagnostics, Atlanta, GA) in 1X TBE with multiple loadings at 12-min intervals. Gels were run at 70 Watts for a total time of 2.5 hours, dried and exposed to a PhosphorImager screen (GE Healthcare). Gel images were scanned using a Typhoon 8600 (GE Healthcare), and densitometry analyses were performed using the ImageQuant software (GE Healthcare).
Recombinant TDP1 and TDP2 assays
The N14Y DNA substrate was incubated at 1 nM with 10 pM recombinant TDP1 in the absence or presence of inhibitor for 15 min at room temperature in WCE buffer (see above and Figure 3A). When indicated, parallel reactions were performed in the HTS assay buffer containing 1X PBS, pH 7.4, 80 mM KCl, and 0.01% Tween-20 (31) (see Figure 3A). Samples were then analyzed similarly to the WCE TDP1 assay (see above).
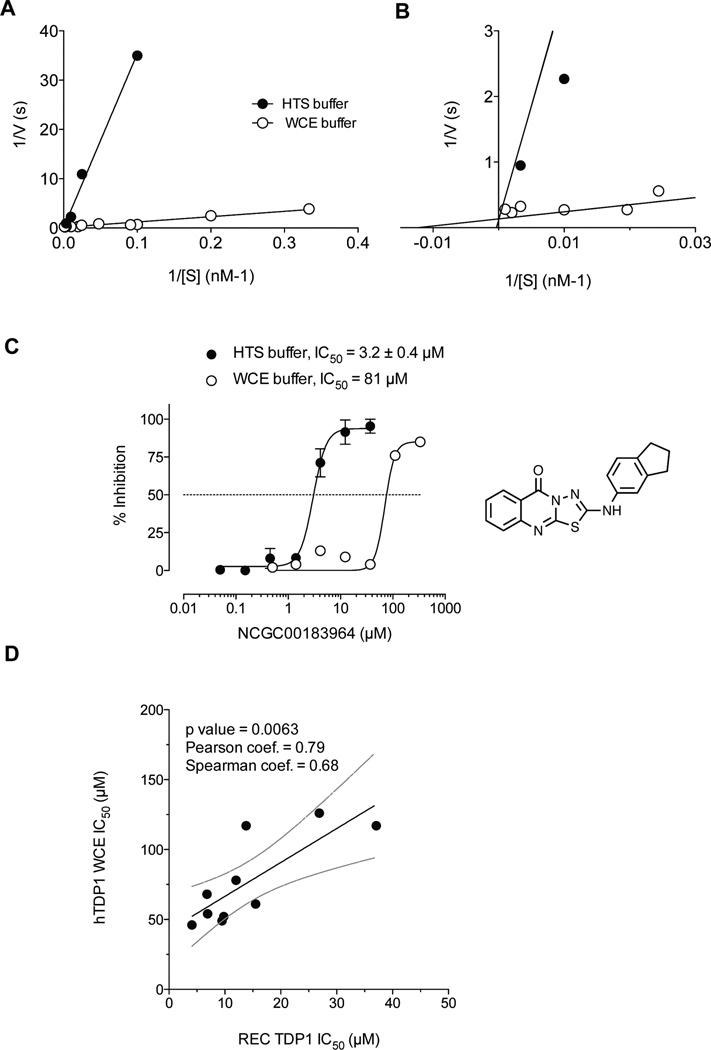
Figure 3.
Differential kinetics of TDP1 reactions in the presence of different buffers. (A) Lineweaver-Burk double reciprocal plot obtained for recombinant TDP1 in the presence HTS buffer or WCE buffer. (B) Intersecting curves in the origin area of the Lineweaver-Burk double-reciprocal plot presented in (A). (C) Concentration response inhibitory curves obtained for NCGC00183964 in HTS and WCE buffers. (D) Correlation between recombinant TDP1 (REC) and endogenous TDP1 (WCE) inhibition by the compounds presented in Figure 1 and Supplementary Table S1. The regression line is represented by a solid line, and dashed lines correspond to 95% confidence interval.
TDP2 reactions were carried out as described previously (33) with the following modifications. The 18-mer single-stranded oligonucleotide DNA substrate (α32P-cordycepin-3’-labeled) was incubated at 1 nM with 25 pM recombinant human TDP2 in the absence or presence of inhibitor for 15 min at room temperature in a buffer containing 50 mM Tris-HCl, pH 7.5, 80 mM KCl, 5 mM MgCl2, 0.1 mM EDTA, 1 mM DTT, 40 µg/mL BSA, and 0.01% Tween 20. Reactions were terminated and treated similarly to WCE and recombinant TDP1 reactions (see above).
Kinetics experiments
To determine the kinetic parameters for the 3’- tyrosyl-DNA phosphodiesterase activity of TDP1, 10 pM and 100 pM of TDP1 was incubated at room temperature with various amount of substrates (N14Y) in excess in WCE and HTS buffer respectively. All reactions were spiked with 1 nM of 32P-labeled N14Y. The extent of reaction progression was followed in a time-dependent manner and terminated at different time by adding 1 volume of gel loading buffer. Samples were analyzed by 16% denaturing PAGE, and the initial portions of the reaction curves were fitted to a linear equation to approximate the pre–steady-state reaction velocities using Prism (Graphpad software). Lineweaver–Burk plot was then generated with the pre–steady-state reaction velocities and the corresponding substrate concentrations.
Surface plasmon resonance analysis
Binding experiments were performed on a Biacore T100 instrument (GE Healthcare, Piscataway NJ). Tdp1 was amine coupled to a CM5 sensor chip (GE Healthcare). Coupling reagents (N-ethyl-N’-(3dimethylaminopropyl)carbodiimide (EDC), N-hydroxysuccinimide (NHS) and ethanolamine were purchased from GE Heathcare,, Neutravidin was obtained from Pierce. In order to protect the amine groups with the active site from modification, TDP1 was bound with a 14 base oligonucleotide before coupling to the surface. Specifically 1 µM Tdp1 was incubated with 2 µM of a 14 base oligonucleotide containing at phosphate group at the 3’ end (GATCTAAAAGACTT) in 10 mM sodium acetate pH 4.5 for 20 min. The CM5 chip surface was activated for 7 min with 0.1 M NHS and 0.4 M EDC at a flow rate of 20 µl/min and TDP1-oligonucleotide mixture was injected until approximately 4000 RU’s was attached. Activated amine groups were quenched with an injection of 1 M solution of ethanolamine pH 8.0 for 7 min. Any bound oligonucleotide was removed by washing the surface with 1 M NaCl. A reference surface was prepared in the same manner without coupling of TDP1. Compound 70 was diluted into running buffer [10 mM Hepes, 150 mM NaCl, 0.01% tween 20 (v/v), 5% DMSO (v/v) pH 7.5] and injected over all flow cells at 30 µl/min at 25°C. Following compound injections, the surface was regenerated with a 30 second injection 1 M NaCl, a 30 second injection of 50% DMSO (v/v) and a 30 second running buffer injection. Each cycle of compound injection was followed by buffer cycle for referencing purposes. A DMSO calibration curve was included to correct for refractive index mismatches between the running buffer and compound dilution series.
Drug combination experiments
Drug cellular sensitivity was measured as previously described (23). Briefly, cells were continuously exposed to various drug concentrations for 72 hours in triplicate. DT40 cells were seeded at 200 cells per well into 384-well white plate (PerkinElmer) in 40 µl of medium. Cell viability was determined at 72 hours by adding 20 µl of ATPlite solution (ATPlite 1-step kit, PerkinElmer). After 5 min incubation, luminescence was measured on an EnVision Plate Reader (PerkinElmer). The ATP level in untreated cells was defined as 100% percent and viability of treated cells was defined as (ATP level of treated cell/ ATP level of untreated cells) ×100.
Results
Quantitative high throughput screening assay
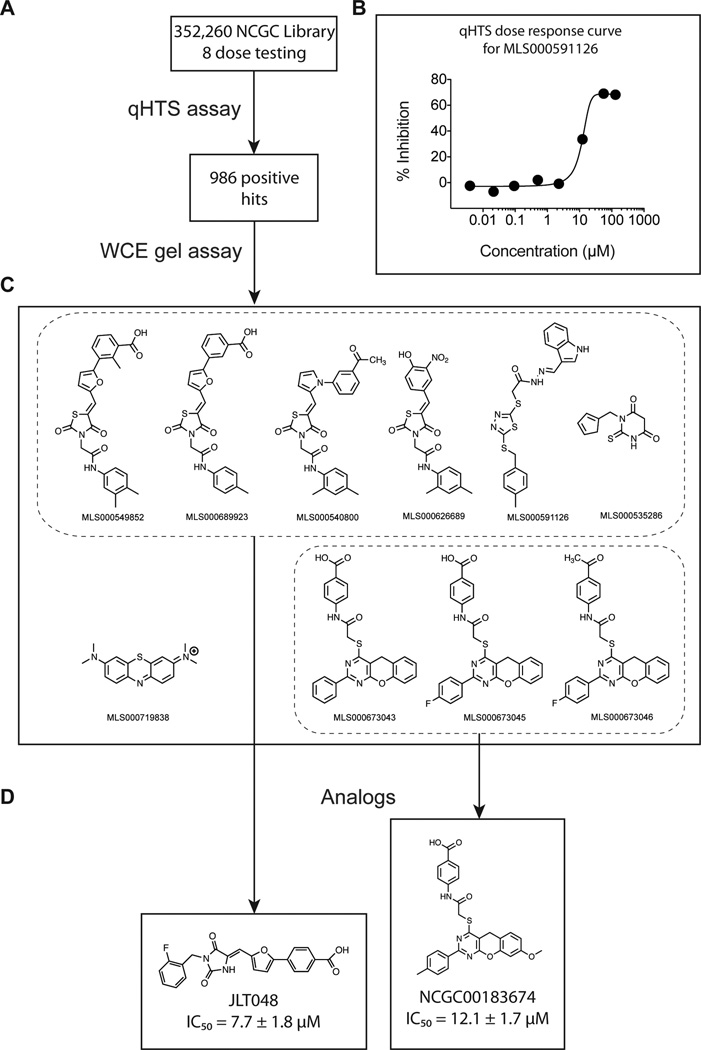
A qHTS was used to screen the 352,260-compound NIH Molecular Libraries Small Molecule Repository (MLSMR) at 8 concentrations against TDP1. This small molecule repository constitutes the NCATS (formerly NCGC) library and therefore screening results can be crossed compared with other assays run by NCATS. The optimization and validation of the TDP1 qHTS assay has previously been reported on the Sigma-Aldrich LOPAC1280 library of 1280 known bioactive small molecules (31). This qHTS, based on the AlphaScreen technology (Supplementary Fig. S1) (31) was run in 1536-well robotic plate format (Supplementary Fig. S1B), and led to the identification of 986 positive hits, which have been deposited into PubChem under the AID# 485290 (http://pubchem.ncbi.nlm.nih.gov). A summary of the assay flowchart and selected chemical structures are included in Figure 1.
Figure 1.
Flow chart summarizing our biochemical screening strategy for TDP1 inhibitors. (A) The 352,260-compound NIH Molecular Libraries Small Molecule Repository (MLSMR) was screened at 8 concentrations against TDP1 by quantitative high throughput screening (qHTS) (31), which led to the identification of 986 positive hits (results accessible online at PubChem AID 485290). (B) Typical qHTS concentration-response for a representative positive compound. (C) The entire set of positives hits was subsequently tested against endogenous human TDP1 from whole cell extracts (WCE) leading to the identification of 10 compounds that can be categorized in 2 chemical groups (dashed rectangles). (D) Two analogs, each representing one chemical group, were selected and further tested.
Novel whole cell extract TDP1 assay
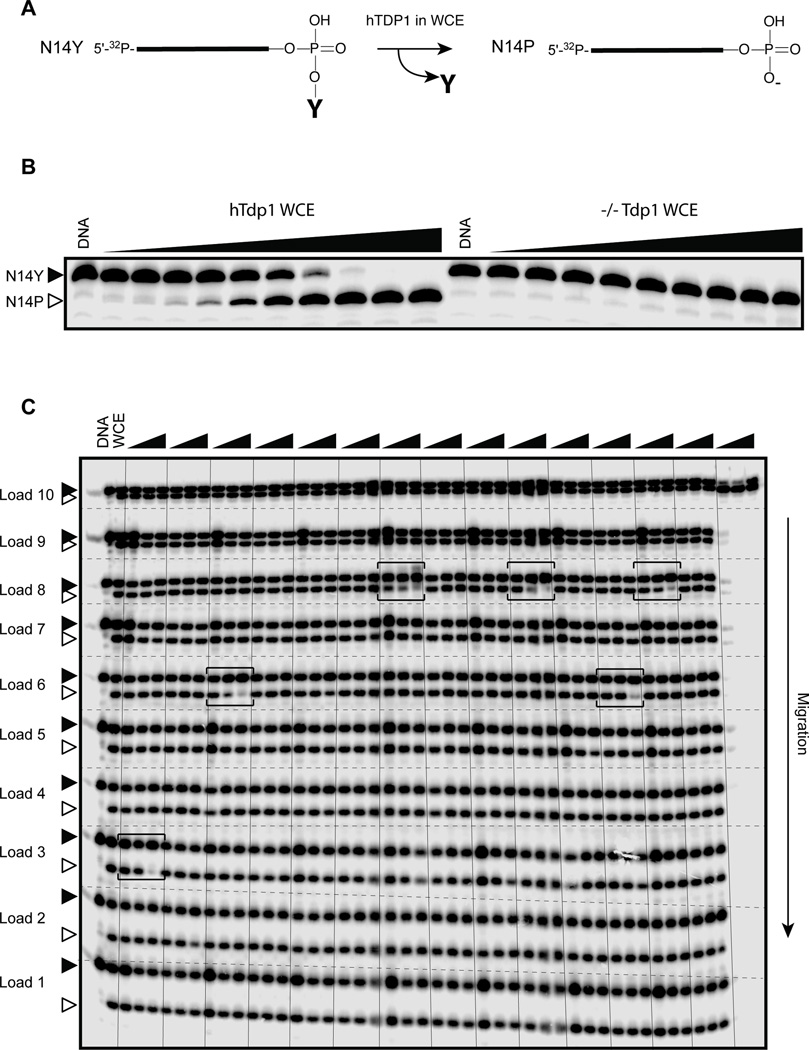
The 986 positive hits from the primary qHTS screen were tested in a novel secondary biochemical assay using whole cell extracts (WCE) in place of recombinant (REC) TDP1. Our recent finding revealed that WCE can selectively process TDP1 substrates (17). WCE were generated from DT40 chicken lymphoma cells that have been genetically modified to express human TDP1 (hTDP1) in a knockout background for the chicken TDP1 gene (TDP1−/−) (17). As shown in Figure 2A–B, endogenous TDP1 from hTDP1 WCE efficiently excised the 3’-phosphotyrosine from its DNA substrate to generate a 3’-phosphate product. This reaction was totally abolished when TDP1−/− WCE was used (Fig. 2B) demonstrating the selectivity of the TDP1 reaction in a cellular extract environment.
Figure 2.
Whole Cell Extract (WCE) TDP1 assay. (A) Schematic representation of the 14-mer single-stranded TDP1 substrate bearing a 3’-phosphotyrosine (N14Y). In the presence of WCE, endogenous TDP1 excises the terminal tyrosine to generate a 14-mer 3’-phosphate DNA product (N14P). (B) Representative gel showing the concentration-dependent appearance of the N14P product in the presence of hTDP1 WCE. This reaction is specific of TDP1 because it is absent with WCE from TDP1 knockout cells (−/−TDP1). WCE concentrations were from 900 µg/ml in 3-fold decrements. (C) Representative gel showing the concentration-dependent inhibition of TDP1 by positive hits (horizontal brackets). Because of the specificity of the TDP1 reaction in WCE, 10 consecutive loadings of 14 compounds tested at 3 concentrations were performed on the same gel.
The secondary WCE screening assay has the advantage to use of native human TDP1 in a physiologically relevant (cellular extract) environment. In contrast to a screen carried out solely with REC-TDP1, a screen carried out with WCE utilizes endogenous native TDP1 with its post-translational modifications and cofactors (18, 34), as well as a vast number of other cellular components that can also affect the drug and the substrate. Additionally, non-specific drug targets present in the WCE may selectively decrease the potency of compounds with a tendency to adsorb on different interfaces. This screening strategy should result in an increased stringency of the assay and allow the elimination of promiscuous inhibitors. The increased reactional complexity in the hTDP1 WCE assay still maintained a high specificity for the TDP1 reaction, as we did not detect non-specific nucleolytic degradation of the DNA substrate even at high concentrations of WCE (Fig. 2B).
Because the phosphotyrosine catalytic excision by TDP1 produces a single product (N14P, see Fig. 2A), we were also able to perform multiple loadings. With 12 min intervals between each loading, up to 600 samples could be analyzed on a single sequencing gel (see representative image in Fig. 2C). WCE screening of the 986 qHTS positive hits led to the confirmation of 10 lead compounds with IC50 values below 111 µM (Fig. 1C and Fig. 2C), indicating that our biochemical assay based on WCE can serve as a robust and efficient secondary screen for the large number of positive hits selected from qHTS assays.
Importance of reaction buffer for TDP1 assays
Our original qHTS assay was run in a buffer required for an optimal signal by the AlphaScreen technology and compatibility with robotic liquid handling (See buffer components in Table 1) (31). On the other hand, WCE conditions could be adapted to more physiological and stringent buffer conditions including the use of serum albumin, metal chelating agents and reducing agents. Table 1 outlines the differences in TDP1 kinetics between these two buffer conditions (the qHTS and WCE buffers). Figure 3A–B shows representative Lineweaver-Burk double-reciprocal plots allowing the determination of KM values of 3936 nM in the HTS buffer versus 80 nM in the WCE buffer (Table 1). This approximately 50-fold difference in KM indicates that TDP1 recognizes its substrate distinctly more efficiently in the WCE buffer than in the HTS buffer. On the other hand, the apparent constants did not vary significantly for the two conditions. TDP1 had a kcat value of 11 and 7 s−1 in the HTS and WCE buffers, respectively (Table 1). The resulting kcat/KM values of 2.8×106 in the HTS buffer and 87.5×106 in the WCE buffer suggest that TDP1 performs approximately 30-fold better in the WCE buffer than in the HTS buffer (Table 1).
Table 1.
Kinetics paramaters
| Buffer |
KM (nM) |
kcat (s−1) |
kcat/KM (M−1s−1) |
|
|---|---|---|---|---|
| HTS buffer | 1× PBS, PH 7.4 | 3936 | 11 | 2.8 × 106 |
| 80 mM KCl | ||||
| 0.01% Tween-20 | ||||
| WCE buffer | 50 mM Tris-HCL, pH 7.5 | 80 | 7 | 87.5 × 106 |
| 80 mM KCl | ||||
| 2 mM EDTA | ||||
| 1 nM DTT | ||||
| 40 µg/ml BSA | ||||
| 0.01% Tween-20 | ||||
The enhanced catalytic activity of TDP1 in the WCE buffer probably explains, at least in part why some compounds tested in the WCE buffer failed to inhibit TDP1 below 100 µM drug concentration, as REC-TDP1 gave a similar difference when it was used under these buffer conditions. This is illustrated in Figure 3C, which shows that compound NCGC00183964 inhibits REC-TDP1 with an IC50 of 3.2 ± 0.4 µM in the HTS buffer, whereas its IC50 was 81 µM in the WCE buffer; a 25-fold reduction in potency. Together, these experiments demonstrate the enhanced stringency of the TDP1 assays in WCE buffer over the HTS buffer.
To compare the WCE and REC-TDP1 assays, IC50 values for the 10 compounds presented in Figure 1 were determined in both assays (Supplementary Table S1). A correlation can be established between the IC50 values determined in the WCE assay and in the REC-TDP1 assay (Fig. 3D; p value = 0.0063 and Pearson & Spearman coefficients = 0.79 and 0.68, respectively). IC50 values determined in the WCE assay were approximately 5-fold higher than those determined in the REC-TDP1 assay (Fig. 3D) reflecting the higher stringency of the hTDP1 WCE assay over the REC-TDP1 assay.
TDP2 Counter-screening assay
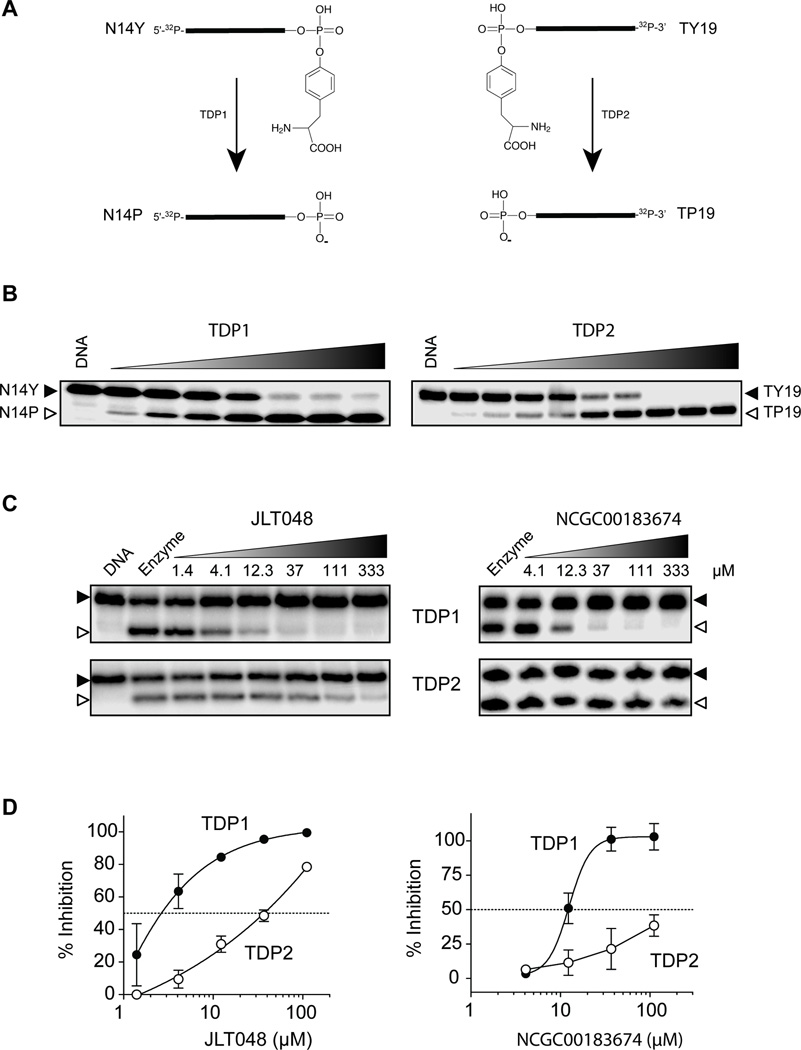
To test the selectivity of TDP1 inhibitors active in the WCE assay, we set up a counter-screening assay with tyrosyl-DNA phosphodiesterase 2 (TDP2). TDP2 (encoded by the TTRAP/TDP2 gene) was recently discovered as a key enzyme involved in the repair of Top2-mediated DNA lesions as it excises the Top2 catalytic tyrosine residue from a trapped Top2-DNA complex (35–39). Similarly to TDP1, TDP2 cleaves a phosphotyrosine bond to generate a phosphate product, but this cleavage occurs preferentially with an opposite polarity compared to TDP1 (Fig. 4A and B) (33, 35, 40, 41). Therefore both enzymes are phosphotyrosine-processing enzymes with opposite preferential polarities (3’-Y for TDP1 and 5’-Y for TDP2; Fig. 4A). In addition, both enzymes preferentially process the same type of single-stranded DNA substrates preferentially (8, 33), which makes TDP2 a relevant counter-screening target for TDP1 inhibitors. Moreover, TDP2 is structurally unrelated to TDP1 (10, 40–42). TDP2 requires magnesium for its catalytic activity (33, 35), which is not the case for TDP1. Therefore TDP2 was chosen as an appropriate counter-screening enzyme for testing the specificity of our TDP1 inhibitors.
Figure 4.
TDP2 counter-screening assay. (A) Schematic representation of the catalytic reaction carried out by recombinant (REC) TDP1 and TDP2. Both enzymes excise a terminal tyrosine residue from single-stranded oligonucleotides but with an opposite polarity: 3’-tyrosine for TDP1 and 5’-tyrosine for TDP2. (B) Representative gel showing enzyme concentration-dependent cleavage reactions for TDP1 and TDP2. REC TDP1 and REC TDP2 concentrations are from 160 pM and 1600 pM in 2-fold decrements, respectively. (C) Representative gels showing concentration-dependent inhibition of recombinant TDP1 (upper gels) and TDP2 (lower gels) by JLT048 and NCGC00183674. (D) Concentration-response curves for JLT048 (left panel) and NCGC00183674 (right panel) with REC TDP1 (solid circles) or REC TDP2 (open circles).
The ten compounds active in the hTDP1 WCE assays can be structurally categorized in two groups (Fig. 1C). Two analogs derived from these two groups, NCGC00183974 (Fig. 1D) and JLT048 (CAS# 664357-58-8, Fig. 1D), both inhibited REC-TDP1 at low micromolar concentrations (Table 2 and Fig. 4C) and their potency was maintained in the WCE assay (Table 2). When tested in parallel against TDP1 and TDP2, JLT048 also inhibited TDP2, albeit with higher IC50 values (Fig. 4C–D, Table 2). NCGC00183974 was more selective for TDP1 with only marginal activity against TDP2 at 111 µM (Fig. 4C–D, Table 2).
Table 2.
IC50 values
| IC50 (µM) | ||||
|---|---|---|---|---|
| TDP1 | TDP2 | |||
| Compound | Structure | REC | WCE | REC |
| NCGC00183674 | 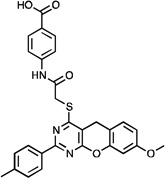 |
12.1 ± 1.7 (n=4) | 51, 59 (n=2) | >111 |
| JLT048 | 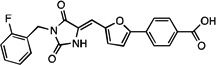 |
7.7 ± 1.8 (n=5) | 115, 148 (n=2) | 32 ±10 (n=3) |
Cellular combination treatment with CPT
To determine whether the two compounds could potentiate the cytotoxic effect of a Top1 inhibitor, NCGC00183974 (Fig. 5A) and JLT048 (Fig. 5B) were tested in combination with CPT in DT40 hTDP1 cells for cytotoxicity. We observed no synergistic effect, in contrast to the PARP inhibitor, veliparib, which showed the expected strong synergism with CPT (26, 27) (Fig. 5C). The two compounds also did not exhibit any cytotoxicity suggesting that they do not enter cells efficiently or/and are inactivated. Therefore, further structural optimization is warranted to improve their cellular profile.
Figure 5.
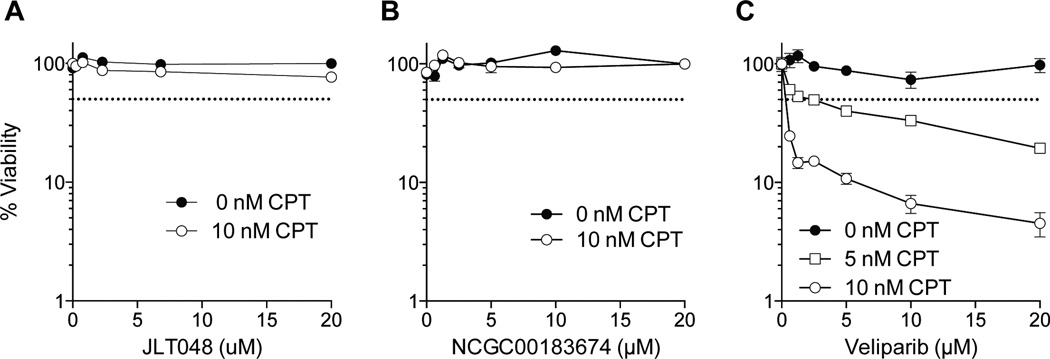
Cellular survival curves in the presence of CPT and various concentrations of JLT048 (A), NCGC00183674 (B) and veliparib (C) in hTDP1 cells.
Discussion
TDP1 and TDP2 are two relatively new DNA repair enzymes, which are rational pharmacological targets (see Introduction). Here we report our screening approach including the development of a novel whole cell extract (WCE) gel-based assay, and counter-screening with TDP2, which led to the identification of two novel TDP1 inhibitors that could serve for further development.
The new WCE assay has the advantage of using native endogenous human TDP1 enzyme in a cellular environment with its cofactors, binding partners (11, 34) and post-translational modifications (43, 44). It is therefore likely to be more biologically relevant than assays based on REC TDP1, as exemplified by the fact that the protein kinase inhibitor, 7-hydroxystaurosporine (UCN-01) was found to inhibit Chk2 purified from cell extract by immunoprecipitation while being ineffective against the recombinant Chk2 enzyme (45). WCE also incorporates a complex cellular mixture, which promotes the adsorption of non-specific small molecules inhibitors to different cellular proteins and components, providing for a more biologically relevant model of inhibitor distribution. The WCE assay is simpler and cheaper than assays using purified recombinant enzymes that require purification steps. In the present study, WCE were generated from DT40 chicken lymphoma cells because these cells have a short doubling-time and can be easily grown in large quantity in suspension. They are frequently used to generate gene knockout cell lines (17, 23) and we previously engineered DT40 cells to express functional human TDP1 in a TDP1 knockout background (17). WCE from human cells can also be used in place of the DT40 WCE (46), which should render the WCE assays applicable to other platforms and reference cell lines.
The WCE gel-based assay is convenient for drug screening because the TDP1 substrate is processed in a single product (see Figs 2 and 4), allowing multiple loading on a single gel (see Fig. 2). The novel WCE assay was run in a more physiologically relevant buffer than the qHTS assay (31, 47, 48). When these two buffers were tested side-by-side, a more efficient TDP1 catalytic activity was observed in the WCE buffer than in the qHTS buffer. We observed a large difference in the Km of TDP1 and only a slight change (within experimental error) in its kcat values. This likely reflects the presence of phosphate salts in the HTS buffer acting as an inhibitor for TDP1. Indeed, we have observed that phosphate likely inhibits TDP1 by competing with its tyrosine-phosphodiester-DNA substrate (10). The other key difference between the two buffer systems is the presence of BSA. After investigating the impact of BSA on the kinetics of TDP1, we found that the removal of BSA from the WCE buffer resulted in a lower kcat but with little impact on the Km value, which may be the result of higher protein adhesion to the tube walls (data not shown). The specific example of TDP1 sheds light on the general importance of reaction buffers when developing screening assays, especially when robotic platforms require specific screening conditions.
TDP2 is TDP1’s counterpart for the repair of Top2-mediated DNA lesions with the cleavage of a 5’-phospho-tyrosine bond. Although both enzymes process single-stranded substrates, they are structurally unrelated and differ in their biochemical mechanisms. TDP1 belongs to the phospholipase D family and its catalytic mechanism involves two HKN motifs and a covalent intermediate (9, 10). On the other hand, TDP2 is a magnesium-dependent phosphodiesterase that hydrolyzes the 5’-phosphotyrosyl bonds without covalent intermediate (33, 40, 41). Dual TDP1-TDP2 inhibitors are therefore likely to be promiscuous (49).
From the 10 TDP1 hits identified by qHTS and confirmed in the WCE assay, two analogs showed selectivity for TDP1 versus TDP2. Surface plasmon resonance experiments showed that the two compounds interacted with TDP1 directly without interacting with the DNA substrate (Supplementary Figure S2). Yet, these inhibitors have some potential liabilities. JLT048 incorporates a methyleneimidazolinedione substructure that gives concerns for potential reactivity as a Michael acceptor (49). NCGC00183974 exhibits a higher selectivity for TDP1 but also inhibit other DNA processing enzymes including DNA polymerase Kappa2. Also, cellular cytotoxicity assays indicate that further studies are warranted to optimize the cellular activity of these series.
In summary, our WCE-based screening approach allowed stringent hit confirmation from qHTS, reducing the number of original hits and markedly enhancing the prospect of discovering selective and relevant inhibitors of TDP1. These results suggest the value of using WCE for the screening of TDP1 inhibitors, and the value of recombinant TDP1 and TDP2 for second line screening assays and mechanism of action studies.
Supplementary Material
Acknowledgments
Gratitude is expressed to Dr. Leyla Celik for computational assistance at Yale.
Financial Information
This study was supported in part by the Intramural Research Program of the Center for Cancer Research (Z01BC006150), National Cancer Institute, NIH, by the NIH R03 Grant MH089814-01 (C. Marchand & Y. Pommier) and by the NIH grant GM32136 (W.L. Jorgensen).
Footnotes
The authors declare no conflict of interest
References
- 1.Pourquier P, Pommier Y. Topoisomerase I-mediated DNA damage. Adv Cancer Res. 2001;80:189–216. doi: 10.1016/s0065-230x(01)80016-6. [DOI] [PubMed] [Google Scholar]
- 2.Dexheimer TS, Kozekova A, Rizzo CJ, Stone MP, Pommier Y. The modulation of topoisomerase I-mediated DNA cleavage and the induction of DNA-topoisomerase I crosslinks by crotonaldehyde-derived DNA adducts. Nucleic Acids Res. 2008;36:4128–4136. doi: 10.1093/nar/gkn334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pommier Y, Kohlhagen G, Pourquier P, Sayer JM, Kroth H, Jerina DM. Benzo[a]pyrene epoxide adducts in DNA are potent inhibitors of a normal topoisomerase I cleavage site and powerful inducers of other topoisomerase I cleavages. Proc Natl Acad Sci USA. 2000;97:2040–2045. doi: 10.1073/pnas.040397497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pommier Y. DNA topoisomerase I inhibitors: chemistry, biology, and interfacial inhibition. Chem Rev. 2009;109:2894–2902. doi: 10.1021/cr900097c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pommier Y. Drugging topoisomerases: lessons and challenges. ACS chemical biology. 2013;8:82–95. doi: 10.1021/cb300648v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dexheimer TS, Antony S, Marchand C, Pommier Y. Tyrosyl-DNA phosphodiesterase as a target for anticancer therapy. Anticancer Agents Med Chem. 2008;8:381–389. doi: 10.2174/187152008784220357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.El-Khamisy SF. To live or to die: a matter of processing damaged DNA termini in neurons. Embo Mol Med. 2011;3:78–88. doi: 10.1002/emmm.201000114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang SW, Burgin AB, Jr, Huizenga BN, Robertson CA, Yao KC, Nash HA. A eukaryotic enzyme that can disjoin dead-end covalent complexes between DNA and type I topoisomerases. Proc Natl Acad Sci U S A. 1996;93:11534–11539. doi: 10.1073/pnas.93.21.11534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Interthal H, Pouliot JJ, Champoux JJ. The tyrosyl-DNA phosphodiesterase Tdp1 is a member of the phospholipase D superfamily. Proc Natl Acad Sci U S A. 2001;98:12009–12014. doi: 10.1073/pnas.211429198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Davies DR, Interthal H, Champoux JJ, Hol WGJ. Crystal structure of a transition state mimic for Tdp1 assembled from vanadate, DNA, and a topoisomerase I-derived peptide. Chem Biol. 2003;10:139–147. doi: 10.1016/s1074-5521(03)00021-8. [DOI] [PubMed] [Google Scholar]
- 11.El-Khamisy SF, Hartsuiker E, Caldecott KW. TDP1 facilitates repair of ionizing radiation-induced DNA single-strand breaks. DNA Repair (Amst) 2007;6:1485–1495. doi: 10.1016/j.dnarep.2007.04.015. [DOI] [PubMed] [Google Scholar]
- 12.Zhou T, Akopiants K, Mohapatra S, Lin PS, Valerie K, Ramsden DA, et al. Tyrosyl-DNA phosphodiesterase and the repair of 3'-phosphoglycolate-terminated DNA double-strand breaks. DNA Repair (Amst) 2009;8:901–911. doi: 10.1016/j.dnarep.2009.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Interthal H, Chen HJ, Champoux JJ. Human Tdp1 cleaves a broad spectrum of substrates, including phosphoamide linkages. J Biol Chem. 2005;280:36518–36528. doi: 10.1074/jbc.M508898200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dexheimer TS, Stephen AG, Fivash MJ, Fisher RJ, Pommier Y. The DNA binding and 3'-end preferential activity of human tyrosyl-DNA phosphodiesterase. Nucleic Acids Res. 2010;38:2444–2452. doi: 10.1093/nar/gkp1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang SY, Murai J, Dalla Rosa I, Dexheimer TS, Naumova A, Gmeiner WH, et al. TDP1 repairs nuclear and mitochondrial DNA damage induced by chain-terminating anticancer and antiviral nucleoside analogs. Nucleic Acids Research. 2013;41:7793–7803. doi: 10.1093/nar/gkt483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nitiss KC, Malik M, He X, White SW, Nitiss JL. Tyrosyl-DNA phosphodiesterase (Tdp1) participates in the repair of Top2-mediated DNA damage. Proc Natl Acad Sci U S A. 2006;103:8953–8958. doi: 10.1073/pnas.0603455103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Murai J, Huang SY, Das BB, Dexheimer TS, Takeda S, Pommier Y. Tyrosyl-DNA phosphodiesterase 1 (TDP1) repairs DNA damage induced by topoisomerases I and II and base alkylation in vertebrate cells. The Journal of biological chemistry. 2012;287:12848–12857. doi: 10.1074/jbc.M111.333963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.El-Khamisy SF, Saifi GM, Weinfeld M, Johansson F, Helleday T, Lupski JR, et al. Defective DNA single-strand break repair in spinocerebellar ataxia with axonal neuropathy-1. Nature. 2005;434:108–113. doi: 10.1038/nature03314. [DOI] [PubMed] [Google Scholar]
- 19.Interthal H, Chen HJ, Kehl-Fie TE, Zotzmann J, Leppard JB, Champoux JJ. SCAN1 mutant Tdp1 accumulates the enzyme--DNA intermediate and causes camptothecin hypersensitivity. EMBO J. 2005;24:2224–2233. doi: 10.1038/sj.emboj.7600694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miao ZH, Agama K, Sordet O, Povirk L, Kohn KW, Pommier Y. Hereditary ataxia SCAN1 cells are defective for the repair of transcription-dependent topoisomerase I cleavage complexes. DNA Repair (Amst) 2006;5:1489–1494. doi: 10.1016/j.dnarep.2006.07.004. [DOI] [PubMed] [Google Scholar]
- 21.Hirano R, Interthal H, Huang C, Nakamura T, Deguchi K, Choi K, et al. Spinocerebellar ataxia with axonal neuropathy: consequence of a Tdp1 recessive neomorphic mutation? EMBO J. 2007;26:4732–4743. doi: 10.1038/sj.emboj.7601885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Katyal S, el-Khamisy SF, Russell HR, Li Y, Ju L, Caldecott KW, et al. TDP1 facilitates chromosomal single-strand break repair in neurons and is neuroprotective in vivo. EMBO J. 2007;26:4720–4731. doi: 10.1038/sj.emboj.7601869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maede Y, Shimizu H, Fukushima T, Kogame T, Nakamura T, Miki T, et al. Differential and common DNA repair pathways for topoisomerase I- and II-targeted drugs in a genetic DT40 repair cell screen panel. Mol Cancer Ther. 2014;13:214–220. doi: 10.1158/1535-7163.MCT-13-0551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pouliot JJ, Robertson CA, Nash HA. Pathways for repair of topoisomerase I covalent complexes in Saccharomyces cerevisiae. Genes Cells. 2001;6:677–687. doi: 10.1046/j.1365-2443.2001.00452.x. [DOI] [PubMed] [Google Scholar]
- 25.Takashima H, Boerkoel CF, John J, Saifi GM, Salih MA, Armstrong D, et al. Mutation of TDP1, encoding a topoisomerase I-dependent DNA damage repair enzyme, in spinocerebellar ataxia with axonal neuropathy. Nat Genet. 2002;32:267–272. doi: 10.1038/ng987. [DOI] [PubMed] [Google Scholar]
- 26.Das BB, Huang SY, Murai J, Rehman I, Ame JC, Sengupta S, et al. PARP1-TDP1 coupling for the repair of topoisomerase I-induced DNA damage. Nucleic Acids Res. 2014 doi: 10.1093/nar/gku088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang YW, Regairaz M, Seiler JA, Agama KK, Doroshow JH, Pommier Y. Poly(ADP-ribose) polymerase and XPF-ERCC1 participate in distinct pathways for the repair of topoisomerase I-induced DNA damage in mammalian cells. Nucleic Acids Res. 2011 doi: 10.1093/nar/gkq1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bowman KJ, Newell DR, Calvert AH, Curtin NJ. Differential effects of the poly (ADP-ribose) polymerase (PARP) inhibitor NU1025 on topoisomerase I and II inhibitor cytotoxicity in L1210 cells in vitro. Br J Cancer. 2001;84:106–112. doi: 10.1054/bjoc.2000.1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Patel AG, Flatten KS, Schneider PA, Dai NT, McDonald JS, Poirier GG, et al. Enhanced killing of cancer cells by poly(ADP-ribose) polymerase inhibitors and topoisomerase I inhibitors reflects poisoning of both enzymes. J Biol Chem. 2012;287:4198–4210. doi: 10.1074/jbc.M111.296475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang SN, Pommier Y, Marchand C. Tyrosyl-DNA Phosphodiesterase 1 (Tdp1) inhibitors. Expert opinion on therapeutic patents. 2011;21:1285–1292. doi: 10.1517/13543776.2011.604314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marchand C, Lea WA, Jadhav A, Dexheimer TS, Austin CP, Inglese J, et al. Identification of phosphotyrosine mimetic inhibitors of human tyrosyl-DNA phosphodiesterase I by a novel AlphaScreen high-throughput assay. Mol Cancer Ther. 2009;8:240–248. doi: 10.1158/1535-7163.MCT-08-0878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Inglese J, Auld DS, Jadhav A, Johnson RL, Simeonov A, Yasgar A, et al. Quantitative high-throughput screening: a titration-based approach that efficiently identifies biological activities in large chemical libraries. Proc Natl Acad Sci U S A. 2006;103:11473–11478. doi: 10.1073/pnas.0604348103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gao R, Huang SY, Marchand C, Pommier Y. Biochemical Characterization of Human Tyrosyl-DNA Phosphodiesterase 2 (TDP2/TTRAP): A Mg2+/Mn2+-DEPENDENT PHOSPHODIESTERASE SPECIFIC FOR THE REPAIR OF TOPOISOMERASE CLEAVAGE COMPLEXES. J Biol Chem. 2012;287:30842–30852. doi: 10.1074/jbc.M112.393983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Plo I, Liao ZY, Barcelo JM, Kohlhagen G, Caldecott KW, Weinfeld M, et al. Association of XRCC1 and tyrosyl DNA phosphodiesterase (Tdp1) for the repair of topoisomerase I-mediated DNA lesions. DNA Repair (Amst) 2003;2:1087–1100. doi: 10.1016/s1568-7864(03)00116-2. [DOI] [PubMed] [Google Scholar]
- 35.Cortes Ledesma F, El Khamisy SF, Zuma MC, Osborn K, Caldecott KW. A human 5'-tyrosyl DNA phosphodiesterase that repairs topoisomerase-mediated DNA damage. Nature. 2009;461:674–678. doi: 10.1038/nature08444. [DOI] [PubMed] [Google Scholar]
- 36.Pommier Y, Leo E, Zhang H, Marchand C. DNA topoisomerases and their poisoning by anticancer and antibacterial drugs. Chem Biol. 2010;17:421–433. doi: 10.1016/j.chembiol.2010.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nitiss JL. Targeting DNA topoisomerase II in cancer chemotherapy. Nat Rev Cancer. 2009;9:338–350. doi: 10.1038/nrc2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nitiss JL. DNA topoisomerase II and its growing repertoire of biological functions. Nat Rev Cancer. 2009;9:327–337. doi: 10.1038/nrc2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fortune JM, Osheroff N. Topoisomerase II as a target for anticancer drugs: when enzymes stop being nice. Prog Nucleic Acid Res Mol Biol. 2000;64:221–253. doi: 10.1016/s0079-6603(00)64006-0. [DOI] [PubMed] [Google Scholar]
- 40.Shi K, Kurahashi K, Gao R, Tsutakawa SE, Tainer JA, Pommier Y, et al. Structural basis for recognition of 5'-phosphotyrosine adducts by Tdp2. Nature structural & molecular biology. 2012;19:1372–1377. doi: 10.1038/nsmb.2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schellenberg MJ, Appel CD, Adhikari S, Robertson PD, Ramsden DA, Williams RS. Mechanism of repair of 5'-topoisomerase II-DNA adducts by mammalian tyrosyl-DNA phosphodiesterase 2. Nature structural & molecular biology. 2012;19:1363–1371. doi: 10.1038/nsmb.2418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Davies DR, Interthal H, Champoux JJ, Hol WGJ. Insights into substrate binding and catalytic mechanism of human tyrosyl-DNA phosphodiesterase (Tdp1) from vanadate and tungstate-inhibited structures. J Mol Biol. 2002;324:917–932. doi: 10.1016/s0022-2836(02)01154-3. [DOI] [PubMed] [Google Scholar]
- 43.Hudson JJ, Chiang SC, Wells OS, Rookyard C, El-Khamisy SF. SUMO modification of the neuroprotective protein TDP1 facilitates chromosomal single-strand break repair. Nature communications. 2012;3:733. doi: 10.1038/ncomms1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Das BB, Antony S, Gupta S, Dexheimer TS, Redon CE, Garfield S, et al. Optimal function of the DNA repair enzyme TDP1 requires its phosphorylation by ATM and/or DNA-PK. The EMBO Journal. 2009;28:3667–3680. doi: 10.1038/emboj.2009.302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yu Q, La Rose J, Zhang H, Takemura H, Kohn KW, Pommier Y. UCN-01 inhibits p53 up-regulation and abrogates gamma-radiation-induced G(2)-M checkpoint independently of p53 by targeting both of the checkpoint kinases, Chk2 and Chk1. Cancer Res. 2002;62:5743–5748. [PubMed] [Google Scholar]
- 46.Gao R, Das B, Chatterjee R, Abaan OD, Agama K, Matuo R, et al. Epigenetic and genetic inactivation of tyrosyl-DNA-phosphodiesterase 1 (TDP1) in human lung cancer cells from the NCI-60 panel. DNA Repair. 2014;13:1–9. doi: 10.1016/j.dnarep.2013.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Antony S, Marchand C, Stephen AG, Thibaut L, Agama KK, Fisher RJ, et al. Novel high-throughput electrochemiluminescent assay for identification of human tyrosyl-DNA phosphodiesterase (Tdp1) inhibitors and characterization of furamidine (NSC 305831) as an inhibitor of Tdp1. Nucleic Acids Res. 2007;35:4474–4484. doi: 10.1093/nar/gkm463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dexheimer TS, Gediya LK, Stephen AG, Weidlich I, Antony S, Marchand C, et al. 4-Pregnen-21-ol-3,20-dione-21-(4-bromobenzenesulfonate) (NSC 88915) and related novel steroid derivatives as tyrosyl-DNA phosphodiesterase (Tdp1) inhibitors. J Med Chem. 2009;52:7122–7131. doi: 10.1021/jm901061s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Baell JB, Holloway GA. New substructure filters for removal of pan assay interference compounds (PAINS) from screening libraries and for their exclusion in bioassays. J Med Chem. 2010;53:2719–2740. doi: 10.1021/jm901137j. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.