Abstract
Hox genes encode homeodomain-containing transcription factors that determine cell and tissue identities in the embryo during development. Hox genes are also expressed in various adult tissues and cancer cells. In Drosophila, expression of cell adhesion molecules, cadherins and integrins, is regulated by Hox proteins operating in hierarchical molecular pathways and plays a crucial role in segment-specific organogenesis. A number of studies using mammalian cultured cells have revealed that cell adhesion molecules responsible for cell-cell and cell-extracellular matrix interactions are downstream targets of Hox proteins. However, whether Hox transcription factors regulate expression of cell adhesion molecules during vertebrate development is still not fully understood. In this review, the potential roles Hox proteins play in cell adhesion and migration during vertebrate body patterning are discussed.
1. Introduction
Homeobox genes (Hox genes) were initially identified in Drosophila through genetic mutations that resulted in transformations of one body segment to another—so-called homeotic transformations [1]. Homeoboxes are 183-bp sequences that encode highly conserved 61-amino-acid homeodomains with helix-turn-helix motifs that are responsible for binding specific DNA sites [2]. Homeodomain proteins are transcription factors that modulate expression levels of their target genes [3, 4]. In amniotes including mammals and birds, 39 Hox genes are arranged in four clusters on different chromosomes. Numerous genetic analyses of loss- and gain-of-function mutations in mice have revealed that Hox genes play pivotal roles in determining the identities of cells and tissues in the developing embryo. In adult animals, Hox expression is required for the proliferation and differentiation of hematopoietic cells [5–7] and renewal of the endometrium [8–10]. Because HOX genes are frequently deregulated in human cancer cells, HOX proteins can be used as both diagnostic markers and therapeutic targets for malignant tumors [11].
Recent studies have converged on identifying downstream Hox target genes. Genome-wide techniques such as microarrays and chromatin immunoprecipitation have been used to identify Hox-regulated genes in Drosophila, mice, and cultured cells. The Hox target genes identified thus far are very diverse with regard to their roles in cellular identity and function. The proteins encoded by the target genes are involved in transcriptional regulation, signal transduction, cell shape, and cell adhesion and migration, as well as in the cell cycle and cell death [12–15]. However, the diverse mechanisms of Hox regulation pose a challenge for elucidating the exact mechanisms by which Hox proteins determine cell identities and where in the molecular cascade they exert their effects. The mechanisms by which Hox transcription factors regulate cellular events are not fully understood.
Since the mouse neural cell adhesion molecule (N-CAM), a mediator of cell adhesion in nervous system tissues during embryonic development, was first identified as a Hox target [16], a number of studies have reported that other cell adhesion molecules, such as cadherins and integrins, are downstream targets of Hox proteins. Cadherins constitute a large superfamily of transmembrane glycoproteins that mediate calcium-dependent intercellular adhesion in most tissues and play important roles in a wide variety of cellular events [17, 18]. Integrins are heterodimers composed of two transmembrane proteins, namely, α and β subunits [19]. The α and β extracellular domains cooperatively bind to extracellular matrix components such as collagen, laminin, and fibronectin. Interactions between integrins and the extracellular matrix modulate essential aspects of cell behavior crucial to the development and maintenance of organisms. In this review, I summarize what is known regarding Hox downstream targets, focusing on molecules mediating cell-cell and cell-extracellular matrix interactions in Drosophila and mammals (Table 1). Furthermore, potential roles for Hox proteins in cell adhesion and migration during vertebrate development will be discussed.
Table 1.
Hox proteins controlling expression of cell adhesion molecules.
| Hox protein | Controlled cell adhesion molecules | Organism | Cell or tissue | Proposed function | References |
|---|---|---|---|---|---|
| Scr | Integrin α subunit (αPS1) (+) |
Drosophila | Salivary gland | Organ formation and migration | [41, 42] |
|
| |||||
| Abd-B | E-Cadherin (+) Nonclassical cadherins (+) |
Drosophila | Posterior spiracle | Organ formation | [40] |
|
| |||||
| Hoxb1 | N-Cadherin (−) Cadherin 6B (−) Cadherin 7 (+) |
Chick | Neural crest cells | EMT | [43] |
|
| |||||
| HOXD1 | β1 Integrin (+) | Human | HUVEC | Increase in cell motility | [44] |
|
| |||||
| Hoxa2 | Cadherin 6B (−) | Chick | Neural crest cells | EMT | [43] |
|
| |||||
| HOXD3 |
β3 Integrin (+) Cadherin 4 (−) |
Human | Erythroleukemia (HEL) |
Increased adhesion to fibronectin | [45, 46] |
|
| |||||
| HOXD3 | β3 Integrin (+) | Human | HUVEC | Conversion to angiogenic phenotype | [47] |
|
| |||||
| HOXD3 |
α3 Integrin (+) β3 Integrin (+) E-Cadherin (−) N-Cadherin (+) |
Human | Lung cancer (A549) |
Increase in cell motility EMT |
[33] |
|
| |||||
| HOXD3 | N-Cadherin (−) | Mouse | Roof plate cells | Expansion in the neural tube | [34] |
|
| |||||
| HOXA4 | β1 Integrin (+) | Human | Ovarian cancer epithelium | Decrease in cell motility | [48] |
|
| |||||
| Hoxc6 | N-CAM (+) | Mouse | NIH 3T3 fibroblast |
Increase in promoter activity | [49] |
|
| |||||
| HOXA7 | E-Cadherin (+) | Human | Ovarian surface epithelium (IOSE-29) |
MET | [50] |
|
| |||||
| HOXB7 | E-Cadherin (−) claudin 1, 4, 7 (−) |
Human | Mammary epithelium (MCF10A) |
EMT | [51] |
|
| |||||
| Hoxb8 | N-CAM (−) | Mouse | NIH 3T3 fibroblast |
Reduction in promoter activity | [16] |
|
| |||||
| Hoxb9 | N-CAM (+) | Mouse | NIH 3T3 fibroblast |
Increase in promoter activity | [16] |
|
| |||||
| HOXB9 | E-Cadherin (−) | Human | Mammary epithelium (MCF10A) |
EMT | [52] |
|
| |||||
| HOXA10 | E-Cadherin (+) | Human | Endometrial carcinoma (SPEC2, KLE) |
MET | [53] |
|
| |||||
| HOXA10 | β3 Integrin (+) | Human | Endometrium | Pathway regulated by sex steroid | [54] |
|
| |||||
| HOXA10 | β3 Integrin (+) | Human | Myeloma (U937) |
Increased adhesion to fibronectin | [55] |
|
| |||||
| HOXA11 | α8 Integrin (+) | Human | Embryonic kidney 239 |
Branching morphogenesis | [56] |
|
| |||||
| Hoxa13 | EphrinA7 (+) | Mouse | Mesenchyme in limb bud | Cell sorting | [57] |
(+): upregulated expression; (−): downregulated expression; HUVEC: human umbilical vein endothelial cells; MET: mesenchymal to epithelial transition; EMT: epithelial to mesenchymal transition.
2. Structural and Functional Organization of Hox Genes in Drosophila and Mammals
In Drosophila, eight Hox genes are clustered in two groups: the Antennapedia complex (ANT-C) and Bithorax complex (BX-C) (Figure 1). The order of genes along the chromosome corresponds to their domains of function along the anterior-posterior axis of the animal. The labial (lab) and Deformed (Dfd) genes specify the head segments, while Sex combs reduced (Scr) and Antennapedia (Antp) are required for the identities of the first and second thoracic segments, respectively. Ultrabithorax (Ubx) is responsible for specifying third thoracic segment identity, and Abdominal A (Abd-A) and Abdominal B (Abd-B) contribute to specifying abdominal segment identities. In homeotic mutants, these specific segmental identities can be changed. For example, a loss-of-function mutation in Ubx gives rise to flies with two sets of wings, due to the transformation of the third thoracic segment into one with second thoracic segment identity. This transformation, referred to as “anteriorization,” is caused by the functional substitution of the more anterior gene Antp for Ubx.
Figure 1.
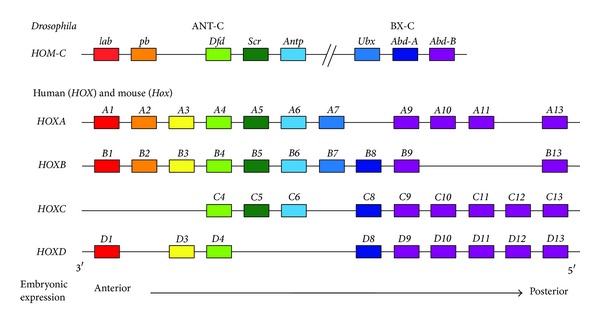
Arrangement of Hox genes in the Drosophila and mammalian genomes. In Drosophila, eight Hox genes clustered on a single chromosome, the homeotic complex (HOM-C), are divided into two groups: the Antennapedia complex (ANT-C) and Bithorax complex (BX-C). ANT-C comprises five Hox genes: labial (lab), proboscipedia (pb), Deformed (Dfd), Sex combs reduced (Scr), and Antennapedia (Antp). The BX-C consists of three Hox genes: Ultrabithorax (Ubx), Abdominal-A (Abd-A), and Abdominal-B (Abd-B). In mammals, 39 Hox genes are divided into four separate clusters (HoxA, HoxB, HoxC, and HoxD) on four different chromosomes. In each cluster, Hox genes are tandem arranged in sequence from 3′ to 5′. Hox genes with the same number are referred to as paralogs. In the embryo, expression of the 3′ paralogs occurs earlier and more anteriorly along the anterior-posterior axis, whereas the 5′ paralogs are expressed later and more posteriorly.
In mammals, 39 Hox genes are organized in four different clusters (HoxA, HoxB, HoxC, and HoxD) found at four distinct chromosomal loci (Figure 1). These clusters are thought to have arisen by two duplication events during the emergence of the vertebrates. Based on the nucleotide sequence similarities between the Hox genes and their Drosophila counterparts, these genes are classified into 13 homology groups, referred to as paralogs [20]. As observed in Drosophila, the order of these paralogs on their respective chromosomes shows collinearity with the spatiotemporal expression pattern of these genes in the embryo [21]. Hox expression can be seen in the neural tube, neural crest, paraxial mesoderm, and surface ectoderm, along the anterior-posterior axis. The 3′ Hox genes are expressed more anteriorly and earlier, while the 5′ Hox genes are expressed more posteriorly and later [22, 23]. Morphological analyses of Hox knockout mice show that the segmental identity of the body along the anterior-posterior axis is primarily determined by the posterior-most Hox gene expressed in the segment [24]. Disruption of all Hox10 paralogs results in the conversion of lumbar vertebrae into thoracic vertebra-like structures with rib projections. Similarly, when all Hox11 paralogs are deleted, sacral vertebrae are transformed into vertebrae with lumbar identity [25]. Thus, homeotic transformations comparable to those in Drosophila occur in mutant mice that are null for all the paralogs belonging to a particular group. To directly investigate how Hox cluster duplications contributed to morphological innovations in vertebrates during evolution, mutant mouse embryos, in which full Hox clusters are deleted, have been generated. Mice lacking all HoxA and HoxD functions in their forelimbs show an early developmental arrest of the limbs and severe truncations of distal elements, suggesting that the evolutionary recruitment of Hox proteins into growing appendages leads to distal extension of tetrapod appendages [26]. Deletion of both HoxA and HoxB clusters results in a heart-looping defect that is recognized as an atavistic phenotype, suggesting that both HoxA and HoxB clusters were necessary for vertebrate heart evolution [27]. In addition, a growing body of recent work highlights the significance of functional organization of Hox gene clusters in vertebrate evolution [28–32].
3. Cell Adhesion Molecules Identified as Hox Realizators during Segment-Specific Organogenesis in Drosophila
In Drosophila, 17 different proteins that contain cadherin domains have been identified. Of these, E-cadherin and two N-cadherins are considered classical types, while the remaining 14 cadherins are regarded as nonclassical cadherins [35]. In addition, Drosophila has 5 integrin α subunits (αPS1–5) and 2 integrin β subunits (βPS and β ν) [36]. These cell adhesion molecules play versatile roles in the development and adult life of Drosophila and interact with cytoplasmic proteins to form adhesion complexes that link their intracellular domains with the cytoskeleton [37].
Posterior spiracles connect the tracheal respiratory systems of Drosophila larvae to the external environment. The Hox gene Abd-B is required to induce the specification and morphogenetic movements required for posterior spiracle formation, as evidenced by the lack of spiracles in Abd-B mutants and formation of ectopic spiracles when Abd-B is ectopically expressed [38, 39]. A study by Lovegrove et al. [40] has provided a framework for understanding how Abd-B controls posterior spiracle formation. Abd-B activates three transcription factors, spalt (sal), empty spiracle (ems), and cut (ct), and a signaling molecule, unpaired (upd, the ligand of the JAK/STAT pathway), the expression of which leads to the activation of realizator molecules controlling cell adhesion. The Abd-B direct target Ct promotes the E-cadherin expression that is responsible for ectodermal cell invagination during the formation of the spiracular chamber, the internal tube connecting the trachea to the exterior of the larva. The expression of four nonclassical cadherins in different spiracle cell domains is controlled by several regulators (Sal, Ems, Ct, and Upd) that partially overlap in expression. E-cadherin and nonclassical cadherins cooperate to control spiracle cell invagination, suggesting that these adhesive molecules, which function in the Abd-B-regulated molecular cascade, play crucial roles in spiracle organogenesis.
The salivary gland is a simple tubular organ composed of two major cell types: secretory and duct cells [58]. The Hox protein Scr, which forms a transcriptional complex with the extradenticle and homothorax homeodomain proteins, is required for salivary gland formation, as evidenced by the complete absence of salivary glands because of Scr loss of function [41]. Although Scr is critical for the specification of salivary gland fates, the protein cannot directly maintain salivary gland cell identity because it disappears early in salivary gland development [58]. Once specified, the salivary gland primordium forms a placode of columnar epithelial cells within the ventral ectoderm [42]. The α PS1 gene, which encodes an integrin α subunit, is expressed in the salivary gland primordium formed within the ventral ectoderm. At later embryonic stages, α PS1 expression is maintained in invaginating and posteriorly migrating secretory cells that keep in contact with the visceral mesoderm substratum. Embryos carrying Scr mutations lack α PS1 expression in the salivary primordium, suggesting that α PS1 is a downstream target of Scr [42]. In α PS1 mutants, the distal tip of invaginating secretory cells reaches the turning point of the visceral mesoderm, but these cells fail to migrate posteriorly [41]. These salivary gland defects, observed when Scr and αPS1 expression is lost, suggest that integrin αPS1 participating in the Scr-directed molecular cascade is essential for the salivary gland to migrate posteriorly along the visceral mesoderm.
4. Hox-Regulated Cell Adhesion Molecule Expression in Cultured Normal and Cancer Cells
The neural cell adhesion molecule (N-CAM), a member of the immunoglobulin superfamily, is involved in cell adhesion, intracellular signaling, and cytoskeleton dynamics [59]. The effects of Hox proteins on N-CAM promoter activity have been investigated by cotransfecting NIH 3T3 mouse embryonic fibroblasts with constitutively active Xenopus Hox constructs and a reporter gene construct containing the mouse N-CAM promoter sequence. Hox2.5 (Hoxb9) greatly increases the transcriptional activity of the reporter gene, while transfection of Hox2.4 (Hoxb8) eliminates its activity [16]. Hoxc6 also stimulates the transcriptional activity driven by the N-CAM promoter [49]. Together, these findings suggest that N-CAM is a downstream target for regulation by Hoxb8, Hoxb9, and Hoxc6.
HOXD3 overexpression in human erythroleukemia HEL cells results in an increase of cell-extracellular matrix adhesiveness, giving rise to elevated β3 integrin expression levels [45, 46]. Human lung carcinoma A549 epithelial cells transfected with HOXD3 exhibit an increase in β3 integrin expression and this modification promotes migratory and invasive behavior [33, 60]. HOXD3 expression elicits phenotypic changes in human umbilical vein endothelial cells (HUVECs), switching them from a resting to angiogenic or invasive state by enhancing αvβ3 integrin expression [47]. HOXD3 directly binds to the β 3 integrin promoter in human microvascular endothelial cells [61]. While HOXD3 causes an increase in β 3 integrin expression in several cell lines, HOXB3, which is paralogous to HOXD3, is not involved in β 3 integrin expression in endothelial cells [62]. Although the HOXA3 paralog is functionally similar to HOXD3 with respect to promotion of cell migration, these transcription factors do not have common downstream target genes [63]. β 3 integrin mRNA levels are increased in endometrial adenocarcinoma cells transfected with a HOXA10 expression vector and are decreased in the cells treated with a HOXA10 antisense construct [54]. HOXA10 directly regulates β 3 integrin expression in endometrial cells, mediating the effects of steroid hormones, estrogen and progesterone, on β 3 integrin expression [54]. The HOXA10 transcription factor interacts with a specific β 3 integrin cis element, activating β 3 integrin transcription during differentiation of U937 cells into a myeloid lineage [55]. Increased adhesion of differentiating U937 cells to fibronectin is dependent upon a HOXA10-induced increase in β3 integrin expression [55]. Thus, expression of β 3 integrin can be controlled by at least two HOX proteins that belong to different paralogous groups, possibly reflecting the redundant functions of the different HOX paralogs.
In addition to β3 integrin, expression of several integrins is reportedly regulated by HOX transcription factors. An approximate 20-fold increase in α 8 integrin expression levels is caused by ectopic Hoxa11 expression in human embryonic kidney 293 cells [56]. During development, α 8 integrin and Hoxa11 are coexpressed in mouse metanephric mesenchyme cells. Mutations in the α 8 integrin gene give rise to a bud branching morphogenesis defect that is very similar to that observed in Hoxa11/Hoxd11 mutant mice. Furthermore, a regional reduction in α 8 integrin expression is found in the developing kidneys of Hoxa11/Hoxd11 mutant mice [56]. These findings suggest that α 8 integrin is a major realizator of Hoxa11/Hoxd11 function in the developing kidney.
In ovarian cancer epithelial cells, HOXA4 suppresses cell motility and spreading through the medium by increasing cell-cell adhesion and β1 integrin protein levels [48]. Loss of HOXD1 expression in HUVECs results in a decrease in cell motility and cell-extracellular matrix adhesiveness, accompanied by decreasing β1 integrin expression levels, suggesting HOXD1 is a positive regulator of cell motility and cell-extracellular matrix adhesiveness in endothelial cells [44]. Thus, it is possible that cell-extracellular matrix interactions mediated by different types of integrin molecules are dependent on the assortment of HOX genes expressed and the amount of protein they produce in nonmalignant and malignant cells.
5. A Role for Hox Proteins in Epithelial to Mesenchymal Transition and Its Reverse Process in Normal and Cancer Cells
Epithelial to mesenchymal transition (EMT) is an event in which adherent epithelial cells are converted into migratory mesenchymal cells that can invade the extracellular matrix. The EMT process is essential for gastrulation and neural crest migration during the development of the early vertebrate embryo. EMT also plays a role in cancer metastasis. Mesenchymal to epithelial transition, the converse of EMT, is observed in many aspects of embryonic development and tumor metastasis, suggesting that epithelial and mesenchymal morphologies are reversible [64].
HOX expression is reported to be closely associated with the transition between epithelial and mesenchymal states. HOXA7 transcripts are absent from normal ovarian surface epithelial cells, but HOXA7 protein is produced in ovarian tumors derived from epithelial cells, which often resemble epithelia composing the Müllerian duct. Ectopic HOXA7 expression in immortalized ovarian surface epithelial (IOSE-29) cells induces E-cadherin expression and downregulates expression of the mesenchymal marker vimentin, enhancing the epithelial phenotype [50]. Hoxa10 is required for proper patterning of the uterus during embryonic development and functional endometrial differentiation in adults [65]. Downregulation of HOXA10 expression in endometrial carcinomas correlates with increased tumor grade and promotes tumor growth and invasive properties [53]. Forced expression of HOXA10 in endometrial carcinoma (SPEC2 and KLE) cells induces E-cadherin expression, suppresses vimentin expression, and inhibits their invasive behavior [53]. The findings described above suggest that HOXA7 and HOXA10 expression promotes mesenchymal to epithelial transition.
In contrast, HOXD3 overexpression in lung cancer A549 cells transforms them from epithelial to mesenchymal morphology (Figure 2) and causes a simultaneous reduction in E-cadherin expression levels and increase in α3 and β3 expression [33]. This was the first study reporting that HOX gene expression enhances the invasive and metastatic properties of human cancer cells. Primary breast carcinomas and distant metastases of various organs exhibit significantly higher HOXB7 expression levels than normal mammary epithelial cells [51]. Overexpression of HOXB7 in MCF10A cells, an immortalized cell line derived from normal human mammary epithelial cells, induces their transformation from cobblestone-like epithelial morphology to spindle-shape mesenchymal morphology, which brings about a dramatic reduction in expression of E-cadherin and tight junction proteins, claudin 1, claudin 4, and claudin 7, as well as an elevation in α-smooth muscle actin expression [51]. Similarly, HOXB9 overexpression in MCF10A cells transforms them from an epithelial phenotype into a mesenchymal phenotype by reducing E-cadherin expression levels and increasing vimentin expression [52]. These findings suggest that HOXD3, HOXB7, and HOXB9 transcription factors serve as EMT inducers in immortalized cells and cancer cells.
Figure 2.
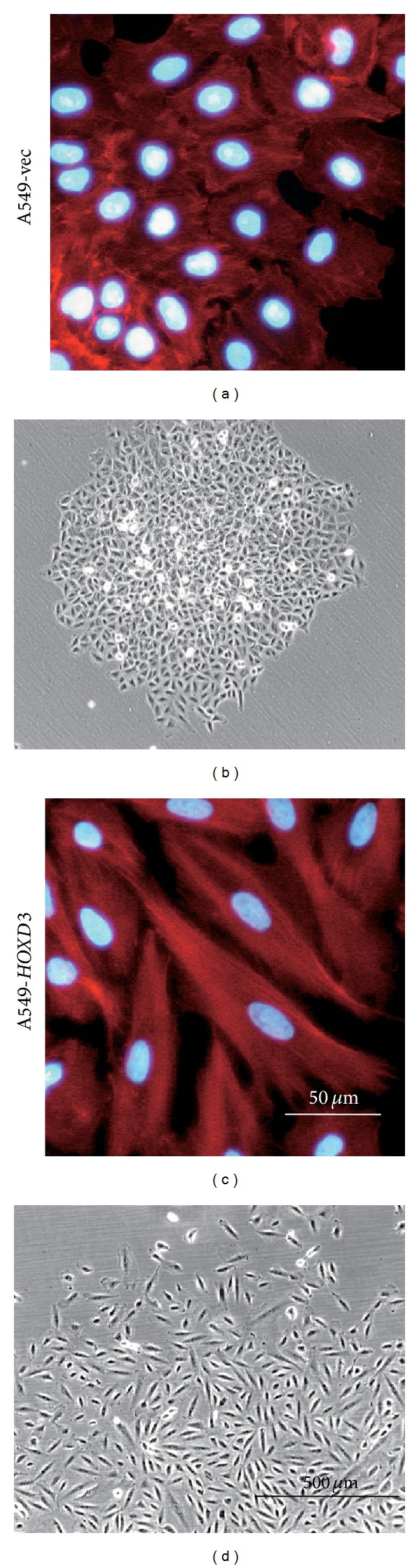
Transition from epithelial to mesenchymal morphology caused by HOXD3 expression in lung cancer A549 cells. A549 cells stably transfected with empty vector (A549-vec) or HOXD3 expression vector (A549-HOXD3) [33] were fixed and stained for nuclei and F-actin by using DAPI and phalloidin-rhodamine, respectively. A549-vec cells have epithelial morphology (a, b), while A549-HOXD3 cells have spindle-shape mesenchymal morphology (c, d). A reduction in E-cadherin expression and an increase in α3 and β3 integrin expression were observed in A549-HOXD3 cells, as compared to A549-vec cells [33].
Whether EMT-inducing HOX proteins have the ability to regulate adhesion molecule gene expression directly or where in the signal transduction pathway HOX proteins exert their effect to induce EMT warrants clarification. HOX proteins have been reported to control expression of some regulatory molecules. HOXA10 inhibits expression of Snail, a zinc-finger transcription factor, in endometrial carcinoma cells [53]. Snail, a key regulator of EMT, downregulates E-cadherin expression, leading to the loss of epithelial morphology in cells undergoing migration during embryonic development as well as tumor progression [66–68]. These results clearly suggest that downregulation of HOXA10 expression induces EMT by elevating Snail expression levels. HOXB9 induces elevated expression of signaling molecules, TGF-β1 and TGF-β2, in MCF10A cells, leading to increased cell motility and acquisition of mesenchymal phenotypes [52]. Members of the TGF-β family play crucial roles in initiating and maintaining EMT during embryonic development and tumor metastasis [69, 70]. These findings indicate that HOXB9 expression induces EMT by activating the TGF-β signaling pathway.
During development of the vertebrate embryo, neural crest cells initially reside within the dorsal neural tube, subsequently undergo EMT to migrate to distant locations, and then differentiate into a wide range of derivatives. When neural crest cells delaminate from the neuroepithelium, N-cadherin and cadherin 6B are downregulated and β1 integrin and cadherin 7 are upregulated [71]. The EMT process is controlled by a hierarchical gene regulatory network in which transcription factors and signaling molecules operate [72]. A recent study [43] has demonstrated that anterior Hox genes interact with components of this network to induce neural crest fates in the chick embryo. Expression of Hoxb1 in the trunk neural tube induces expression of the key transcription factors Snail and Msx1/2, leading to downregulation of N-cadherin and cadherin 6B expression and upregulation of cadherin 7. These changes in cell adhesion molecule expression possibly reflect that Hoxb1 causes neural crest EMT. It is interesting to note that expression of Hox genes participates in EMT events that occur during embryonic morphogenesis as well as tumor progression.
6. Possible Association between Hox Expression and Cell-Cell and Cell-Extracellular Matrix Interactions in the Vertebrate Embryo during Development
When neural crest cells delaminate from the dorsal neural tube by EMT, these cells lose N-cadherin on their surfaces. [73–75]. As mentioned previously, HOXD3 promotes cell motile activity and invasiveness in lung cancer cells [33]. To investigate whether HOXD3 expression regulates cell adhesiveness in dorsal neural tube or roof plate cells in the early mouse embryo, transgenic mouse embryos were generated that overexpress HOXD3 in these cell types under the control of the Wnt1 regulatory element [34]. Dorsal neural tube cells expressing HOXD3 expand ventrally within the neural tube (Figures 3(a), 3(b), 3(e), and 3(f)). This finding raises the possibility that HOXD3-expressing roof plate cells propagate in the dorsal neural tube and then migrate ventrally. Furthermore, in the neural tube ventricular zone, a large number of progenitor cells that do not express N-cadherin protein can be observed in HOXD3-expressing transgenic embryos (Figures 3(c), 3(d), 3(e), and 3(f)). Although HOXD3 expression is localized in the dorsal half of the neural tube and in cells immediately adjacent to the floor plate, progenitor cells that do not express N-cadherin are distributed throughout the ventricular zone. This finding indicates that HOXD3 expression has a non-cell-autonomous effect, negatively affecting N-cadherin expression in cells at a distance from those expressing HOXD3. Therefore, signaling molecules or secreted proteins whose expression is induced by HOXD3 likely reduce N-cadherin expression.
Figure 3.
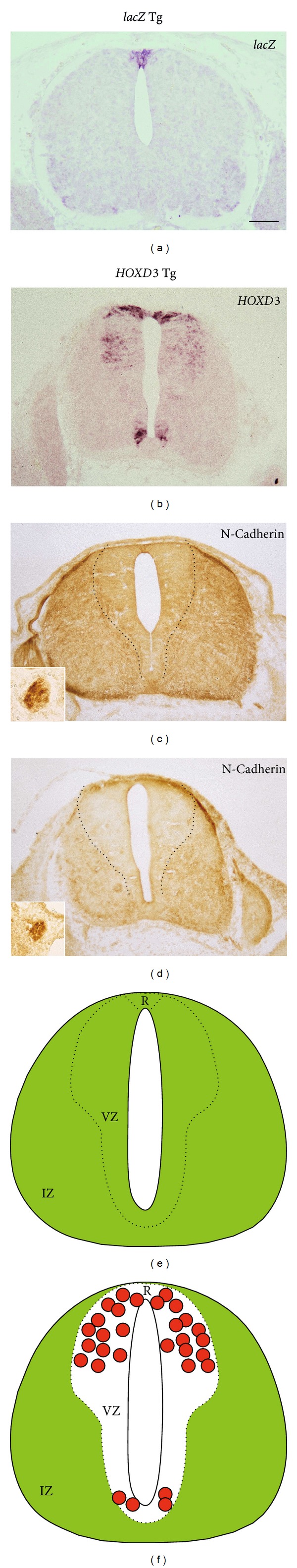
Reduced N-cadherin expression induced by HOXD3 overexpression in the roof plate of the early mouse embryo. (a, b) Expression of lacZ and HOXD3 genes in transverse neural tube sections at the thoracic level of 12.5-day transgenic embryos. Transgenic embryos were generated, in which lacZ and HOXD3 are expressed in the roof plate cells under the control of the Wnt1 regulatory element [34]. These embryos were sectioned and analyzed using in situ hybridization. Expression of lacZ (control) is restricted to roof plate cells within the neural tube, while HOXD3 expression is localized not only in the dorsal neural tube, but also within the ventricular zone and in ventral regions of the neural tube. (c, d) N-Cadherin expression in the thoracic neural tubes of 12.5-day lacZ- and HOXD3-expressing transgenic embryos. Transverse sections were stained using anti-human N-cadherin antibodies [34]. N-Cadherin is strongly expressed in the ventricular zone of lacZ-expressing embryos, whereas the ventricular zone in HOXD3-expressing embryos is composed of a number of progenitor cells that do not express N-cadherin. The ventricular zone is surrounded by dotted lines. Insets show that N-cadherin expression levels in the sympathetic ganglia of lacZ-expressing embryos are similar to those of HOXD3-expressing embryos. (e, f) Summary of the neural tube phenotype in transgenic embryos expressing lacZ and HOXD3. In embryos expressing lacZ, N-cadherin expression (green) is distributed throughout the neural tube. In HOXD3-expressing embryos, roof plate cells expressing HOXD3 (red circles) expand ventrally into the ventricular zone, where almost all N-cadherin-expressing cells are lost. R, roof plate; IZ, intermediate zone; VZ, ventricular zone. Scale bar: 100 μm.
Gastrulation is an essential process in the development of most animals. In amniotes, gastrulation begins with the acquisition of asymmetry in the early embryo. The movements of epiblast cells towards the midline of the embryo form the primitive streak. At the streak, epiblast cells undergo EMT, ingress, and migrate inwardly to their proper positions where they differentiate into mesodermal and endodermal tissues. Consequently, the three definitive germ layers, ectoderm, mesoderm, and endoderm, are organized. The crucial role of FGF signaling in regulating cell migration is highlighted by the effect of altering fibroblast growth factor receptor 1 (FGFR1) expression. In Fgfr1-deficient mouse embryos, epiblast cells fail to undergo EMT, which is required for ingression through the primitive streak [76]. The defect is attributed to a failure in Snail upregulation and E-cadherin downregulation. This finding shows that FGFR1 regulates epiblast cell migration by differentially regulating the intercellular adhesion properties of these cells at the primitive streak. Furthermore, this study suggests that Snail expression downstream of FGFR1 is required for normal downregulation of E-cadherin. In the early chick embryo, PDGF signaling plays a major role in the migration of mesodermal cells during gastrulation [77]. PDGFA expression in the epiblast controls N-cadherin expression and activates PDGFRα, which is required for migration of mesodermal cells away from the primitive streak. The timing of ingression is orchestrated by temporal and spatial collinear activation of Hox genes that starts in the epiblast [78]. Expression of posterior Hox genes can delay the time at which cells ingress from the epiblast into the primitive streak and nascent mesoderm. Within a region of epiblast cells expressing a given Hox gene, a subpopulation of epiblast cells that express the neighboring 5′ Hox gene exists. These cells acquire slightly different migratory properties, and their ingression is slightly delayed. Ingressing cells expressing Hox genes from successive paralogous groups might sort out from each other along the anterior-posterior axis [24, 78]. The target genes of Hox proteins and the mechanism by which they control ingression remain to be elucidated; however, the targets might include genes encoding factors that regulate EMT, such as cell-cell and cell-extracellular matrix adhesion molecules [79].
In the developing mouse embryo, Hox3 paralogs play crucial roles in the formation of neural crest, somatic mesoderm, and endoderm-derived structures in the cervical region, including the pharyngeal arches [80, 81]. Hoxa3 is essential for the development of the thymus, thyroid, parathyroid glands, and ultimobranchial bodies [82]. These organs develop concurrently and they are composed of cells that migrate from their original sites in the pharynx and pharyngeal pouches to their final positions in the cervical and upper thoracic regions. The ultimobranchial bodies fuse with the thyroid; the cells disperse within the thyroid lobes and then differentiate into calcitonin-producing C-cells. Mice doubly mutant for Hoxa3 and Hoxb3 or Hoxa3 and Hoxd3 show that the ultimobranchial bodies fail to migrate to their normal positions in the thyroid, suggesting that expression of Hox3 paralogs is required for the organized movement of primordial organs in the pharyngeal tissues [83]. The thymus and parathyroid glands originate from both the neural crest-derived mesenchymal cells of the pharyngeal arches and the pharyngeal endoderm. Conditional deletion of Hoxa3 alleles from neural crest cells results in the development of ectopic thymus and parathyroid glands [84], raising the possibility that Hoxa3 controls neural crest cell migration in pharyngeal regions. In the chick embryo, knockdown of Hoxa3 function by using antisense morpholino oligonucleotides disrupts the migration of epibranchial placode-derived cells and neural crest cells, indicating that Hoxa3 is required for the migration of these cell types [85]. Although these findings show that Hoxa3 and its paralogs are regulators of cell migration, the target genes for Hox3 proteins are not known. Genes encoding molecules involved in regulating cell-cell and cell-extracellular matrix interactions could be candidate Hox3 paralog targets.
During vertebrate limb development, posterior Hox genes in the HoxA cluster are expressed in a specific spatiotemporal manner along the proximodistal axis. Hoxa13 is expressed in the autopod during normal limb development. In the chick embryo, misexpression of Hoxa13 in the entire limb bud results in a marked size reduction of the zeugopodal cartilage due to homeotic transformation into cartilage of a more distal type [86]. When limb mesenchymal cells are dissociated and cultured in vitro, Hoxa13-expressing cells sort out from Hoxa13-nonexpressing cells. This finding indicates that Hoxa13 expression is involved in modulation of cell-cell adhesiveness. Mice homozygous for a Hoxa13 loss-of-function mutation show major defects in the formation of autopod skeletal elements [87]. Autopod-derived mesenchymal cells in homozygous Hoxa13 mutant embryos fail to form chondrogenic condensations in vitro, and mutant cells in the distal region fail to sort out from wild-type cells in the proximal region [57]. This failure in cell sorting reflects the fact that Hoxa13 expression is involved in determining cell surface properties. Eph proteins, which constitute a large family of receptor tyrosine kinases, interact with cell surface-bound ligands, ephrins [88, 89]. Eph/ephrin juxtacrine signaling modulates cell morphology, motility, and attachment. A marked reduction in EphrinA7 expression prevents mesenchymal cells in the autopod of homozygous Hoxa13 mutant embryos from forming chondrogenic condensations in vivo and in vitro [57]. EphrinA7 has been shown to be a direct downstream target of Hoxa13 and Hoxd13 during limb development [90]. Furthermore, using a CHIP-on-chip approach (chromatin immunoprecipitation with DNA microarray technology), the gene loci of cadherin 12 (also known as Br-cadherin or N-cadherin 2) and protocadherins are identified as direct Hoxd13 binding sites in the developing mouse limb bud [91]. It has been reported that the cadherin 12 protein is exclusively expressed in the developing and adult mouse brain [92, 93]. Cadherin 12 does not seem to function in the limb bud. On the other hand, N-cadherin is abundant in the distal limb bud and increases in the distal region as limb development proceeds [94, 95]. N-Cadherin-positive mesenchymal cells segregate from N-cadherin-negative cells in vitro, suggesting that N-cadherin plays an important role in cell sorting. However, the relation between N-cadherin and expression of Hox genes during limb development is presently unknown.
7. Concluding Remarks
In this review, cell adhesion molecules mediating cell-cell and cell-extracellular matrix interactions, whose expression is directly or indirectly controlled by Hox transcription factors, have been the focus. In Drosophila, cadherins, components of the hierarchical Abd-B-regulated molecular pathway, play an important role in the formation of posterior spiracles during development. Integrin molecules participate in the Scr-directed molecular cascade that is required for salivary gland formation and migration. In cultured normal and malignant mammalian cells, expression of several Hox genes enhances cell-extracellular matrix adhesion and cell motility by activating integrin expression. Several Hox proteins play a role in epithelial-mesenchymal transition and its reverse process by reducing and elevating cadherin expression. Hox proteins likely do not regulate cadherin expression directly; Hox proteins might control cadherin expression by using transcription factors and signaling molecules as intermediaries. To elucidate the exact processes governed by Hox proteins, it is worthwhile to investigate whether cell adhesion molecule expression is directly controlled by Hox proteins or where in the Hox-directed molecular cascade cell adhesion molecules function.
In this review, I have discussed the necessity of Hox expression for neural crest migration, gastrulation, migration of organs in the pharyngeal regions, and limb bud formation in the vertebrate embryo during development. These developmental processesrequire precise regulation of cell adhesion and migration. How Hox proteins are related to expression of cell adhesion molecules during vertebrate body patterning is not fully understood. The highly redundant functions of Hox genes pose a challenge when attempting to clarify the association between Hox transcription factors and expression of a diverse set of cell adhesion molecules. However, as the gaps in the puzzle are filled by future research findings, the precise mechanisms by which Hox proteins govern expression of cell adhesion molecules will be uncovered.
Acknowledgment
The author thanks Dr. Jun-ichi Hamada (Hokkaido University) for providing the photographs.
Conflict of Interests
The author declares that there are no conflicts of interest regarding the publication of this paper.
References
- 1.Lewis EB. A gene complex controlling segmentation in Drosophila . Nature. 1978;276(5688):565–570. doi: 10.1038/276565a0. [DOI] [PubMed] [Google Scholar]
- 2.Qian YQ, Billeter M, Otting G, Müller M, Gehring WJ, Wüthrich K. The structure of the Antennapedia homeodomain determined by NMR spectroscopy in solution: comparison with prokaryotic repressors. Cell. 1989;59(3):573–580. doi: 10.1016/0092-8674(89)90040-8. [DOI] [PubMed] [Google Scholar]
- 3.Levine M, Hoey T. Homeobox proteins as sequence-specific transcription factors. Cell. 1988;55(4):537–540. doi: 10.1016/0092-8674(88)90209-7. [DOI] [PubMed] [Google Scholar]
- 4.McGinnis W, Krumlauf R. Homeobox genes and axial patterning. Cell. 1992;68(2):283–302. doi: 10.1016/0092-8674(92)90471-n. [DOI] [PubMed] [Google Scholar]
- 5.Abramovich C, Pineault N, Ohta H, Humphries RK. Hox genes: from leukemia to hematopoietic stem cell expansion. Annals of the New York Academy of Sciences. 2005;1044:109–116. doi: 10.1196/annals.1349.014. [DOI] [PubMed] [Google Scholar]
- 6.Argiropoulos B, Humphries RK. Hox genes in hematopoiesis and leukemogenesis. Oncogene. 2007;26(47):6766–6776. doi: 10.1038/sj.onc.1210760. [DOI] [PubMed] [Google Scholar]
- 7.Alharbi RA, Pettengell R, Pandha HS, Morgan R. The role of HOX genes in normal hematopoiesis and acute leukemia. Leukemia. 2013;27(5):1000–1008. doi: 10.1038/leu.2012.356. [DOI] [PubMed] [Google Scholar]
- 8.Lim H, Ma L, Ma W, Maas RL, Dey SK. Hoxa-10 regulates uterine stromal cell responsiveness to progesterone during implantation and decidualization in the mouse. Molecular Endocrinology. 1999;13(6):1005–1017. doi: 10.1210/mend.13.6.0284. [DOI] [PubMed] [Google Scholar]
- 9.Cakmak H, Taylor HS. Molecular mechanisms of treatment resistance in endometriosis: the role of progesterone-hox gene interactions. Seminars in Reproductive Medicine. 2010;28(1):69–74. doi: 10.1055/s-0029-1242996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zanatta A, Rocha AM, Carvalho FM, et al. The role of the Hoxa10/HOXA10 gene in the etiology of endometriosis and its related infertility: a review. Journal of Assisted Reproduction and Genetics. 2010;27(12):701–710. doi: 10.1007/s10815-010-9471-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shah N, Sukumar S. The Hox genes and their roles in oncogenesis. Nature Reviews Cancer. 2010;10(5):361–371. doi: 10.1038/nrc2826. [DOI] [PubMed] [Google Scholar]
- 12.Pearson JC, Lemons D, McGinnis W. Modulating Hox gene functions during animal body patterning. Nature Reviews Genetics. 2005;6(12):893–904. doi: 10.1038/nrg1726. [DOI] [PubMed] [Google Scholar]
- 13.Svingen T, Tonissen KF. Hox transcription factors and their elusive mammalian gene targets. Heredity. 2006;97(2):88–96. doi: 10.1038/sj.hdy.6800847. [DOI] [PubMed] [Google Scholar]
- 14.Arderiu G, Cuevas I, Chen A, Carrio M, East L, Boudreau NJ. HoxA5 stabilizes adherens junctions via increased Akt1. Cell Adhesion & Migration. 2007;1(4):185–195. doi: 10.4161/cam.1.4.5448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sánchez-Herrero E. Hox targets and cellular functions. Scientifica. 2013;2013:26 pages. doi: 10.1155/2013/738257.738257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jones FS, Prediger EA, Bittner DA, de Robertis EM, Edelman GM. Cell adhesion molecules as targets for Hox genes: Neural cell adhesion molecule promoter activity is modulated by cotransfection with Hox-2.5 and -2.4. Proceedings of the National Academy of Sciences of the United States of America. 1992;89(6):2086–2090. doi: 10.1073/pnas.89.6.2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Takeichi M. Morphogenetic roles of classic cadherins. Current Opinion in Cell Biology. 1995;7(5):619–627. doi: 10.1016/0955-0674(95)80102-2. [DOI] [PubMed] [Google Scholar]
- 18.Nollet F, Kools P, van Roy F. Phylogenetic analysis of the cadherin superfamily allows identification of six major subfamilies besides several solitary members. Journal of Molecular Biology. 2000;299(3):551–572. doi: 10.1006/jmbi.2000.3777. [DOI] [PubMed] [Google Scholar]
- 19.Hynes RO. Integrins: versatility, modulation, and signaling in cell adhesion. Cell. 1992;69(1):11–25. doi: 10.1016/0092-8674(92)90115-s. [DOI] [PubMed] [Google Scholar]
- 20.Scott MP. Vertebrate homeobox gene nomenclature. Cell. 1992;71(4):551–553. doi: 10.1016/0092-8674(92)90588-4. [DOI] [PubMed] [Google Scholar]
- 21.Duboule D, Dolle P. The structural and functional organization of the murine HOX gene family resembles that of Drosophila homeotic genes. The EMBO Journal. 1989;8(5):1497–1505. doi: 10.1002/j.1460-2075.1989.tb03534.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wilkinson DG, Bhatt S, Cook M, Boncinelli D, Krumlauf R. Segmental expression of Hox-2 homoeobox-containing genes in the developing mouse hindbrain. Nature. 1989;341(6241):405–409. doi: 10.1038/341405a0. [DOI] [PubMed] [Google Scholar]
- 23.Hunt P, Gulisano M, Cook M, et al. A distinct Hox code for the branchial region of the vertebrate head. Nature. 1991;353(6347):861–864. doi: 10.1038/353861a0. [DOI] [PubMed] [Google Scholar]
- 24.Iimura T, Pourquié O. Hox genes in time and space during vertebrate body formation. Development Growth and Differentiation. 2007;49(4):265–275. doi: 10.1111/j.1440-169X.2007.00928.x. [DOI] [PubMed] [Google Scholar]
- 25.Wellik DM, Capecchi MR. Hox10 and Hox11 genes are required to globally pattern the mammalian skeleton. Science. 2003;301(5631):363–367. doi: 10.1126/science.1085672. [DOI] [PubMed] [Google Scholar]
- 26.Kmita M, Tarchini B, Zàkàny J, Logan M, Tabin CJ, Duboule D. Early developmental arrest of mammalian limbs lacking HoxA/HoxD gene function. Nature. 2005;435(7045):1113–1116. doi: 10.1038/nature03648. [DOI] [PubMed] [Google Scholar]
- 27.Soshnikova N, Dewaele R, Janvier P, Krumlauf R, Duboule D. Duplications of hox gene clusters and the emergence of vertebrates. Developmental Biology. 2013;378(2):194–199. doi: 10.1016/j.ydbio.2013.03.004. [DOI] [PubMed] [Google Scholar]
- 28.Di-Poï N, Montoya-Burgos JI, Miller H, Pourquié O, Milinkovitch MC, Duboule D. Changes in Hox genes structure and function during the evolution of the squamate body plan. Nature. 2010;464(7285):99–103. doi: 10.1038/nature08789. [DOI] [PubMed] [Google Scholar]
- 29.Guerreiro I, Nunes A, Woltering JM, et al. Role of a polymorphism in a Hox/Pax-responsive enhancer in the evolution of the vertebrate spine. Proceedings of the National Academy of Sciences of the United States of America. 2013;110(26):10682–10686. doi: 10.1073/pnas.1300592110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vieux-Rochas M, Mascrez B, Krumlauf R, Duboule D. Combined function of HoxA and HoxB clusters in neural crest cells. Developmental Biology. 2013;382(1):293–301. doi: 10.1016/j.ydbio.2013.06.027. [DOI] [PubMed] [Google Scholar]
- 31.Nishimoto S, Minguillon C, Wood S, Logan MPO. A combination of activation and repression by a colinear Hox code controls forelimb-restricted expression of Tbx5 and reveals Hox protein specificity. PLoS Genetics. 2014;10(3) doi: 10.1371/journal.pgen.1004245.e1004245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jung H, Mazzoni EO, Soshnikova N, et al. Evolving Hox activity profiles govern diversity in locomotor systems. Developmental Cell. 2014;29(2):171–187. doi: 10.1016/j.devcel.2014.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hamada J, Omatsu T, Okada F, et al. Overexpression of homeobox gene HOXD3 induces coordinate expression of metastasis-related genes in human lung cancer cells. International Journal of Cancer. 2001;93(4):516–525. doi: 10.1002/ijc.1357. [DOI] [PubMed] [Google Scholar]
- 34.Taniguchi Y, Tanaka O, Sekiguchi M, et al. Enforced expression of the transcription factor HOXD3 under the control of the Wnt1 regulatory element modulates cell adhesion properties in the developing mouse neural tube. Journal of Anatomy. 2011;219(5):589–600. doi: 10.1111/j.1469-7580.2011.01425.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hill E, Broadbent ID, Chothia C, Pettitt J. Cadherin superfamily proteins in Caenorhabditis elegans and Drosophila melanogaster . Journal of Molecular Biology. 2001;305(5):1011–1024. doi: 10.1006/jmbi.2000.4361. [DOI] [PubMed] [Google Scholar]
- 36.Brown NH, Gregory SL, Martin-Bermudo MD. Integrins as mediators of morphogenesis in Drosophila. Developmental Biology. 2000;223(1):1–16. doi: 10.1006/dbio.2000.9711. [DOI] [PubMed] [Google Scholar]
- 37.Bulgakova NA, Klapholz B, Brown NH. Cell adhesion in Drosophila: versatility of cadherin and integrin complexes during development. Current Opinion in Cell Biology. 2012;24(5):702–712. doi: 10.1016/j.ceb.2012.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lamka ML, Boulet AM, Sakonju S. Ectopic expression of UBX and ABD-B proteins during Drosophila embryogenesis: competition, not a functional hierarchy, explains phenotypic suppression. Development. 1992;116(4):841–854. doi: 10.1242/dev.116.4.841. [DOI] [PubMed] [Google Scholar]
- 39.Castelli-Gair J, Greig S, Micklem G, Akam M. Dissecting the temporal requirements for homeotic gene function. Development. 1994;120(7):1983–1995. doi: 10.1242/dev.120.7.1983. [DOI] [PubMed] [Google Scholar]
- 40.Lovegrove B, Simões S, Rivas ML, et al. Coordinated control of cell adhesion, polarity, and cytoskeleton underlies Hox-induced organogenesis in Drosophila. Current Biology. 2006;16(22):2206–2216. doi: 10.1016/j.cub.2006.09.029. [DOI] [PubMed] [Google Scholar]
- 41.Abrams EW, Vining MS, Andrew DJ. Constructing an organ: the Drosophila salivary gland as a model for tube formation. Trends in Cell Biology. 2003;13(5):247–254. doi: 10.1016/s0962-8924(03)00055-2. [DOI] [PubMed] [Google Scholar]
- 42.Bradley PL, Myat MM, Comeaux CA, Andrew DJ. Posterior migration of the salivary gland requires an intact visceral mesoderm and integrin function. Developmental Biology. 2003;257(2):249–262. doi: 10.1016/s0012-1606(03)00103-9. [DOI] [PubMed] [Google Scholar]
- 43.Gouti M, Briscoe J, Gavalas A. Anterior hox genes interact with components of the neural crest specification network to induce neural crest fates. Stem Cells. 2011;29(5):858–870. doi: 10.1002/stem.630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Park H, Choi H, Kim J, et al. Homeobox D1 regulates angiogenic functions of endothelial cells via integrin β1 expression. Biochemical and Biophysical Research Communications. 2011;408(1):186–192. doi: 10.1016/j.bbrc.2011.04.017. [DOI] [PubMed] [Google Scholar]
- 45.Taniguchi Y, Komatsu N, Moriuchi T. Overexpression of the HOX4A (HOXD3) homeobox gene in human erythroleukemia HEL cells results in altered adhesive properties. Blood. 1995;85(10):2786–2794. [PubMed] [Google Scholar]
- 46.Taniguchi Y, Moriuchi T, Inoko H, Kimura M. HOXD3 mediates the switchover of cadherin 4 to β3 integrin gene expression in human erythroleukemia HEL cells. Biomedical Research. 2003;24(3):133–140. [Google Scholar]
- 47.Boudreau N, Andrews C, Srebrow A, Ravanpay A, Cheresh DA. Induction of the angiogenic phenotype by HoxD3. The Journal of Cell Biology. 1997;139(1):257–264. doi: 10.1083/jcb.139.1.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Klausen C, Leung PCK, Auersperg N. Cell motility and spreading are suppressed by HOXA4 in ovarian cancer cells: possible involvement of β1 integrin. Molecular Cancer Research. 2009;7(9):1425–1437. doi: 10.1158/1541-7786.MCR-08-0466. [DOI] [PubMed] [Google Scholar]
- 49.Jones FS, Holst BD, Minowa O, De Robertis EM, Edelman GM. Binding and transcriptional activation of the promoter for the neural cell adhesion molecule by HoxC6 (Hox-3.3) Proceedings of the National Academy of Sciences of the United States of America. 1993;90(14):6557–6561. doi: 10.1073/pnas.90.14.6557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Naora H, Montz FJ, Chai CY, Roden RBS. Aberrant expression of homeobox gene HOXA7 is associated with müllerian-like differentiation of epithelial ovarian tumors and the generation of a specific autologous antibody response. Proceedings of the National Academy of Sciences of the United States of America. 2001;98(26):15209–15214. doi: 10.1073/pnas.011503998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wu X, Chen H, Parker B, et al. HOXB7, a homeodomain protein, is overexpressed in breast cancer and confers epithelial-mesenchymal transition. Cancer Research. 2006;66(19):9527–9534. doi: 10.1158/0008-5472.CAN-05-4470. [DOI] [PubMed] [Google Scholar]
- 52.Hayashida T, Takahashi F, Chiba N, et al. HOXB9, a gene overexpressed in breast cancer, promotes tumorigenicity and lung metastasis. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(3):1100–1105. doi: 10.1073/pnas.0912710107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yoshida H, Broaddus R, Cheng W, Xie S, Naora H. Deregulation of the HOXA10 homeobox gene in endometrial carcinoma: role in epithelial-mesenchymal transition. Cancer Research. 2006;66(2):889–897. doi: 10.1158/0008-5472.CAN-05-2828. [DOI] [PubMed] [Google Scholar]
- 54.Daftary GS, Troy PJ, Bagot CN, Young SL, Taylor HS. Direct regulation of β 3-integrin subunit gene expression by HOXA10 in endometrial cells. Molecular Endocrinology. 2002;16(3):571–579. doi: 10.1210/mend.16.3.0792. [DOI] [PubMed] [Google Scholar]
- 55.Bei L, Lu Y, Bellis SL, Zhou W, Horvath E, Eklund EA. Identification of a HoxA10 activation domain necessary for transcription of the gene encoding β3 integrin during myeloid differentiation. Journal of Biological Chemistry. 2007;282(23):16846–16859. doi: 10.1074/jbc.M609744200. [DOI] [PubMed] [Google Scholar]
- 56.Valerius MT, Patterson LT, Feng Y, Potter SS. Hoxa 11 is upstream of integrin α8 expression in the developing kidney. Proceedings of the National Academy of Sciences of the United States of America. 2002;99(12):8090–8095. doi: 10.1073/pnas.122229199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Stadler HS, Higgins KM, Capecchi MR. Loss of Eph-receptor expression correlates with loss of cell adhesion and chondrogenic capacity in Hoxa13 mutant limbs. Development. 2001;128(21):4177–4188. doi: 10.1242/dev.128.21.4177. [DOI] [PubMed] [Google Scholar]
- 58.Bradley PL, Haberman AS, Andrew DJ. Organ formation in Drosophila: specification and morphogenesis of the salivary gland. BioEssays. 2001;23(10):901–911. doi: 10.1002/bies.1131. [DOI] [PubMed] [Google Scholar]
- 59.Ditlevsen DK, Povlsen GK, Berezin V, Bock E. NCAM-induced intracellular signaling revisited. Journal of Neuroscience Research. 2008;86(4):727–743. doi: 10.1002/jnr.21551. [DOI] [PubMed] [Google Scholar]
- 60.Ohta H, Hamada J, Tada M, et al. HOXD3-overexpression increases integrin αvβ3 expression and deprives E-cadherin while it enhances cell motility in A549 cells. Clinical and Experimental Metastasis. 2006;23(7-8):381–390. doi: 10.1007/s10585-006-9047-5. [DOI] [PubMed] [Google Scholar]
- 61.Boudreau NJ, Varner JA. The homeobox transcription factor Hox D3 promotes integrin α 5 β 1 expression and function during angiogenesis. Journal of Biological Chemistry. 2004;279(6):4862–4868. doi: 10.1074/jbc.M305190200. [DOI] [PubMed] [Google Scholar]
- 62.Myers C, Charboneau A, Boudreau N. Homeobox B3 promotes capillary morphogenesis and angiogenesis. The Journal of Cell Biology. 2000;148(2):343–351. doi: 10.1083/jcb.148.2.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mace KA, Hansen SL, Myers C, Young DM, Boudreau N. HOXA3 induces cell migration in endothelial and epithelial cells promoting angiogenesis and wound repair. Journal of Cell Science. 2005;118(12):2567–2577. doi: 10.1242/jcs.02399. [DOI] [PubMed] [Google Scholar]
- 64.Acloque H, Adams MS, Fishwick K, Bronner-Fraser M, Nieto MA. Epithelial-mesenchymal transitions: the importance of changing cell state in development and disease. The Journal of Clinical Investigation. 2009;119(6):1438–1449. doi: 10.1172/JCI38019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Benson GV, Lim H, Paria BC, Satokata I, Dey SK, Maas RL. Mechanisms of reduced fertility in Hoxa-10 mutant mice: uterine homeosis and loss of maternal Hoxa-10 expression. Development. 1996;122(9):2687–2696. doi: 10.1242/dev.122.9.2687. [DOI] [PubMed] [Google Scholar]
- 66.Cano A, Pérez-Moreno MA, Locascio A, et al. The transcription factor Snail controls epithelial-mesenchymal transitions by repressing E-cadherin expression. Nature Cell Biology. 2000;2(2):76–83. doi: 10.1038/35000025. [DOI] [PubMed] [Google Scholar]
- 67.Batlle E, Sancho E, Franci C, et al. The transcription factor Snail is a repressor of E-cadherin gene expression in epithelial tumour cells. Nature Cell Biology. 2000;2(2):84–89. doi: 10.1038/35000034. [DOI] [PubMed] [Google Scholar]
- 68.Their JP. Epithelial-mesenchymal transitions in tumor progression. Nature Reviews Cancer. 2002;2(6):442–454. doi: 10.1038/nrc822. [DOI] [PubMed] [Google Scholar]
- 69.Moses HL, Serra R. Regulation of differentiation by TGF-β . Current Opinion in Genetics and Development. 1996;6(5):581–586. doi: 10.1016/s0959-437x(96)80087-6. [DOI] [PubMed] [Google Scholar]
- 70.Zavadil J, Böttinger EP. TGF-β and epithelial-to-mesenchymal transitions. Oncogene. 2005;24(37):5764–5774. doi: 10.1038/sj.onc.1208927. [DOI] [PubMed] [Google Scholar]
- 71.Kerosuo L, Bronner-Fraser M. What is bad in cancer is good in the embryo: importance of EMT in neural crest development. Seminars in Cell & Developmental Biology. 2012;23(3):320–332. doi: 10.1016/j.semcdb.2012.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Betancur P, Bronner-Fraser M, Sauka-Spengler T. Assembling neural crest regulatory circuits into a gene regulatory network. Annual Review of Cell and Developmental Biology. 2010;26:581–603. doi: 10.1146/annurev.cellbio.042308.113245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hatta K, Takagi S, Fujisawa H, Takeichi M. Spatial and temporal expression pattern of N-cadherin cell adhesion molecules correlated with morphogenetic processes of chicken embryos. Developmental Biology. 1987;120(1):215–227. doi: 10.1016/0012-1606(87)90119-9. [DOI] [PubMed] [Google Scholar]
- 74.Akitaya T, Bronner-Fraser M. Expression of cell adhesion molecules during initiation and cessation of neural crest cell migration. Developmental Dynamics. 1992;194(1):12–20. doi: 10.1002/aja.1001940103. [DOI] [PubMed] [Google Scholar]
- 75.Monier-Gavelle F, Duband J. Cross talk between adhesion molecules: control of N-cadherin activity by intracellular signals elicited by β1 and β3 integrins in migrating neural crest cells. The Journal of Cell Biology. 1997;137(7):1663–1681. doi: 10.1083/jcb.137.7.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ciruna B, Rossant J. FGF signaling regulates mesoderm cell fate specification and morphogenetic movement at the primitive streak. Developmental Cell. 2001;1(1):37–49. doi: 10.1016/s1534-5807(01)00017-x. [DOI] [PubMed] [Google Scholar]
- 77.Yang X, Chrisman H, Weijer CJ. PDGF signalling controls the migration of mesoderm cells during chick gastrulation by regulating N-cadherin expression. Development. 2008;135(21):3521–3530. doi: 10.1242/dev.023416. [DOI] [PubMed] [Google Scholar]
- 78.Iimura T, Pourquié O. Collinear activation of Hoxb genes during gastrulation is linked to mesoderm cell ingression. Nature. 2006;442(7102):568–571. doi: 10.1038/nature04838. [DOI] [PubMed] [Google Scholar]
- 79.Chuai M, Weijer CJ. Regulation of cell migration during chick gastrulation. Current Opinion in Genetics and Development. 2009;19(4):343–349. doi: 10.1016/j.gde.2009.06.007. [DOI] [PubMed] [Google Scholar]
- 80.Chisaka O, Capecchi MR. Regionally restricted developmental defects resulting from targeted disruption of the mouse homeobox gene hox-1.5. Nature. 1991;350(6318):473–479. doi: 10.1038/350473a0. [DOI] [PubMed] [Google Scholar]
- 81.Manley NR, Capecchi MR. Hox group 3 paralogous genes act synergistically in the formation of somitic and neural crest-derived structures. Developmental Biology. 1997;192(2):274–288. doi: 10.1006/dbio.1997.8765. [DOI] [PubMed] [Google Scholar]
- 82.Manley NR, Capecchi MR. The role of Hoxa-3 in mouse thymus and thyroid development. Development. 1995;121(7):1989–2003. doi: 10.1242/dev.121.7.1989. [DOI] [PubMed] [Google Scholar]
- 83.Manley NR, Capecchi MR. Hox group 3 paralogs regulate the development and migration of the thymus, thyroid, and parathyroid glands. Developmental Biology. 1998;195(1):1–15. doi: 10.1006/dbio.1997.8827. [DOI] [PubMed] [Google Scholar]
- 84.Chen L, Zhao P, Wells L, Amemiya CT, Condie BG, Manley NR. Mouse and zebrafish Hoxa3 orthologues have nonequivalent in vivo protein function. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(23):10555–10560. doi: 10.1073/pnas.1005129107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Watari-Goshima N, Chisaka O. Chicken HOXA3 gene: its expression pattern and role in branchial nerve precursor cell migration. International Journal of Biological Sciences. 2011;7(1):87–101. doi: 10.7150/ijbs.7.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Yokouchi Y, Nakazato S, Yamamoto M, et al. Misexpression of HoxA-13 induces cartilage homeotic transformation and changes cell adhesiveness in chick limb buds. Genes and Development. 1995;9(20):2509–2522. doi: 10.1101/gad.9.20.2509. [DOI] [PubMed] [Google Scholar]
- 87.Fromental-Ramain C, Warot X, Messadecq N, LeMeur M, Dollé P, Chambon P. Hoxa-13 and Hoxd-13 play a crucial role in the patterning of the limb autopod. Development. 1996;122(10):2997–3011. doi: 10.1242/dev.122.10.2997. [DOI] [PubMed] [Google Scholar]
- 88.Klein R. Excitatory Eph receptors and adhesive ephrin ligands. Current Opinion in Cell Biology. 2001;13(2):196–203. doi: 10.1016/s0955-0674(00)00197-6. [DOI] [PubMed] [Google Scholar]
- 89.Klein R. Eph/ephrin signalling during development. Development. 2012;139(22):4105–4109. doi: 10.1242/dev.074997. [DOI] [PubMed] [Google Scholar]
- 90.Salsi V, Zappavigna V. Hoxd13 and Hoxa13 directly control the expression of the EphA7 ephrin tyrosine kinase receptor in developing limbs. Journal of Biological Chemistry. 2006;281(4):1992–1999. doi: 10.1074/jbc.M510900200. [DOI] [PubMed] [Google Scholar]
- 91.Salsi V, Vigano MA, Cocchiarella F, Mantovani R, Zappavigna V. Hoxd13 binds in vivo and regulates the expression of genes acting in key pathways for early limb and skeletal patterning. Developmental Biology. 2008;317(2):497–507. doi: 10.1016/j.ydbio.2008.02.048. [DOI] [PubMed] [Google Scholar]
- 92.Selig S, Lidov HGW, Bruno SA, Segal MM, Kunkel LM. Molecular characterization of Br-cadherin, a developmentally regulated, brain-specific cadherin. Proceedings of the National Academy of Sciences of the United States of America. 1997;94(6):2398–2403. doi: 10.1073/pnas.94.6.2398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Mayer M, Bercsényi K, Géczi K, Szabó G, Lele Z. Expression of two type II cadherins, Cdh12 and Cdh22 in the developing and adult mouse brain. Gene Expression Patterns. 2010;10(7-8):351–360. doi: 10.1016/j.gep.2010.08.002. [DOI] [PubMed] [Google Scholar]
- 94.Yajima H, Hara K, Ide H, Tamura K. Cell adhesiveness and affinity for limb pattern formation. The International Journal of Developmental Biology. 2002;46(7):897–904. [PubMed] [Google Scholar]
- 95.Tamura K, Yonei-Tamura S, Yano T, Yokoyama H, Ide H. The autopod: its formation during limb development. Development Growth and Differentiation. 2008;50(supplement 1):S177–S187. doi: 10.1111/j.1440-169X.2008.01020.x. [DOI] [PubMed] [Google Scholar]


