Abstract
Purpose of Review
This review discusses progress in understanding the impact of immune tolerance on inducing broadly neutralizing antibodies (BnAbs), and how such knowledge can be incorporated into novel immunization approaches.
Recent Findings
Over 120 BnAbs have now been isolated, all of which bear unusual features associated with host tolerance controls, but paradoxically, may also be required for their function. Evidence that poly-/autoreactivity of MPER+ BnAbs can invoke such controls has been demonstrated by knock-in (KI) technology, highlighting its potential for studying the impact of tolerance in the generation of BnAb lineages to distinct Env targets. The requirement for extensive affinity maturation in developing neutralization breadth/potency during infection is being examined, and similar studies in the setting of immunization will be aided by novel vaccine approaches and KI models that either selectively express reverted V(D)J rearrangements, or unrearranged germline segments, from BnAb lineages.
Summary
It is increasingly apparent that immune tolerance, sometimes invoked by self-reactivity that overlaps with BnAb epitope specificity, adds to a formidable set of roadblocks impeding BnAb induction. The path to an effective HIV-1 vaccine may thus benefit from a deeper understanding of host controls, including categorizing which are unique or common at distinct BnAb targets, and ranking those most feasible to overcome by immunization. Ultimately, such emerging information will be critical to incorporate into new vaccine approaches that can be tested in human trials.
Keywords: broadly neutralizing antibodies, polyeractivity, autoreactivity, immune tolerance, somatic hypermutation
Introduction
Despite major advances in treatment and preventative measures, the HIV-1 pandemic has not slowed down significantly, with now over 35 million people infected. Thus, the development of a transmission-preventative HIV-1 vaccine is crucial, but continues to be elusive. Why is this? A correlate of most successful viral vaccines is generating potently neutralizing antibodies (1). HIV-1, however, differs from other viruses for which FDA-approved vaccines have been made, because as an integrating, rapidly mutating retrovirus, it is resistant to immune responses upon establishment of a latently-infected pool of CD4+ T-cells (2). Therefore, it is critical for an HIV vaccine to rapidly induce neutralizing antibodies reactive to a broad spectrum of HIV-1 strains (BnAbs). Studies demonstrating robust protection by passive infusion of BnAbs preceding SHIV challenge in rhesus macaques (3,4) or their transduction in humanized mice prior to HIV-1 infection (5) lend support to this strategy. Furthermore, serum profiling of HIV-1 chronically infected subjects reveal that BnAbs can eventually develop, although only years after transmission, too late to avert disease course, and only in a minority of patients (6–9).
Unfortunately, efforts to elicit significant titers of BnAbs by immunization have failed. Thus, identifying the obstacles impeding BnAb induction by existing immunization regimens and devising strategies capable of overcoming them, is key to HIV-1 vaccine development. Defining the origins and precise characteristics of BnAbs from HIV-1 infected individuals has been facilitated by comprehensive serum profiling assays, improved antigen-specific memory B-cell sorting (10,11) and culture methodologies (12,13), and by high-throughput recombinant Ab cloning techniques (14,15). These efforts led to the discovery of new BnAbs with remarkable breadth/potency, and helped define structural characteristics for four vulnerable areas of the HIV-1 Envelope (Env) targeted by BnAbs: the gp120 CD4-binding site (CD4bs), the gp41 membrane proximal external region (MPER), and two novel peptide-glycan epitope-rich regions in either the gp120 V1/V2 or V3 hypervariable loops gp120 (reviewed in 16••). Furthermore, it is now known that BnAbs targeting more than one region are frequently co-produced in some individuals (8,17,18), passively-administered combinations of two BnAbs to distinct targets can confer near-complete breadth (19,20), and increased selection for escape mutants occur when individual BnAb lineages are produced during infection (21••). These data thus suggest that immunization regimens capable of eliciting more than one class of BnAb may not only be a desirable, but possibly necessary.
Despite this remarkable progress, the field’s best efforts at engineering immunogens capable of presenting Env epitopes remain unsuccessful at inducing BnAbs (16••). Thus, it is becoming clear that “structure-based” approaches (see 22–28) will likely not solve the HIV-1 vaccine problem alone. Attention has now shifted to the host for insight into why BnAbs are difficult to elicit. This review provides a perspective on the potential association of BnAbs with self-reactivity and its role in generating subdominant BnAb responses to vaccines. It is not exhaustive; we will not cover structural considerations, which have been reviewed elsewhere (16••, 29–33). Furthermore, since all BnAbs isolated thus far originate from chronically-infected individuals, wherein secondary B-cell dysregulation/dysfunction effects may have impacted the magnitude/quality of BnAb responses (reviewed in (34•)), we will focus on host factors we predict impact BnAb production not only during infection, but also in vaccination of healthy individuals.
Autoreactivity, polyreactivity, and other unusual BnAb traits associated with tolerance induction
Between 1993–2009, only a handful of BnAbs were identified, of which only three, 2F5, 4E10, and 2G12 were isolated directly from chronically infected patients. In 2005, two of these, 2F5 and 4E10, specific for linear, adjacent epitopes in the MPER, were reported to have elongated, hydrophobic CDRH3 residues, and to exhibit poly-/autoreactivity in vitro (35). Because both sets of traits are potential predictors of negative B-cell selection, based on numerous studies (reviewed in 36), this led to the hypothesis that BnAbs like 2F5 and 4E10 are rarely generated because the B-cells which produce them are subjected to immune tolerance (37). A corollary of this hypothesis, that BnAbs are more readily generated in autoimmune subjects (with defective tolerance) was also indirectly supported by reports of disproportionate infrequency of SLE+ subjects with HIV-1 infection (38–42).
This hypothesis has been independently investigated by three groups, that monitored B-cell development in knock-in (KI) mice expressing the original (mutated) VDJ rearrangements of 2F5 and 4E10 (43,44,45••,46••,47•). 2F5/4E10 (VDJ or VDJ+VJ) KI mice share a striking blockade in immature B-cell generation, a phenotype characteristic of central clonal deletion, and similar to KI mice expressing BCRs with high affinities to well-defined self-antigens (48–50). Furthermore, residual 2F5 and 4E10 KI B-cell populations are under additional tolerance mechanisms including poorly expressing/signaling through, their BCRs (44,45••,46••), thus resembling anergic (unresponsive) B-cells (51) and immature 4E10/2F5 B-cells undergo extensive LC receptor editing that mitigates MPER epitope-associated self-reactivity (44) and apoptotic deletion (44,45••,46••). Thus, these results indicate 2F5 and 4E10 poly-/autoreactivities are indeed sufficient to induce profound negative B-cell selection.
Conserved vertebrate host antigens bound by 2F5 and 4E10 have now been identified: for 2F5, kynureninase (containing a motif identical to the ELDKWA neutralization epitope) and for 4E10, splicing factor-3b subunit-3 (SF3B3) (52••) and type-1 inositol triphosphate (IP3R1) (47•). However, since affinity is only one aspect of an autoantigen’s ability to effect tolerance (53,54), demonstration of these 2F5/4E10 targets as their bona fide self-ligands will require breeding of 2F5/4E10 KI mice to those with targeted disruptions in their putative self-reactive motifs. In terms of relevance to vaccine development, it will be important to determine the extent to which this kind of self-antigen mimicry limits BnAb generation, and the stage in B-cell development when BnAbs normally acquire tolerizing reactivity. Regarding this latter point, the data suggests that it can occur at any of several checkpoints: BnAbs like 2F5 may be tolerized in early BM B-cell development, since KI mice carrying reverted 2F5 BCRs undergo central deletion (Verkoczy L, Haynes BF, unpublished data), whereas others like CH103 and 4E10, whose reverted BCRs lack BnAb and self-specificity in vitro ((21••,55) and Haynes BF, unpublished data) may acquire tolerizing polyreactivity de novo, in the periphery.
Is there a correlation between self-reactivity of BnAbs and their unusual SHM levels?
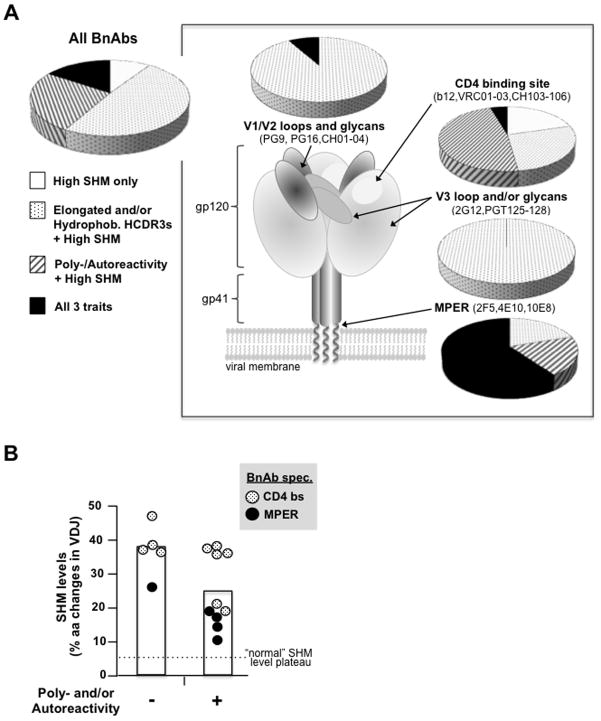
Over 120 BnAbs have now been identified worldwide (10–15, reviewed in 16••,56•). Although these BnAbs often have greater breadth and/or potency than those initially isolated, many share predictive traits of negative selection: elongated HCDR3s, and, as examined by in vitro autoimmune assays, >1/2 exhibit poly- and/or autoreactivity (Fig. 1A). Furthermore, from this representative BnAb dataset emerges an additional feature common to all: an exceptionally high degree of somatic hypermutation (SHM)-mediated aa changes in rearranged immunoglobulins.
Figure 1. Distribution of the three BnAb traits associated with negative selection.
A. The relative distribution of BnAbs with only high SHM (≥10% aa changes in V(D)J rearrangements), or in addition, in vitro poly-/autoreactivity or elongated and/or hydrophobic HCDR3 regions [see (16••) for individual BnAb refs] are shown for either all BnAbs, or broken down into the four general Env regions they target (inset)(16••,29,30); schematic diagram of trimeric Env based on (57••), with representative BnAbs also listed. HCDR3 lengths are reported in Kabat nomenclature and BnAbs with elongated HCDR3s are defined as those ≥2 standard deviations (i.e.≥18aa) above the median length (13.5 aa) in the normal mature human repertoire (58), and unusual hydrophobicity was calculated as previously described (36,58). For all calculations, independently-discovered individual BnAbs and BnAb clonal lineages are equally weighted (and clonal members with different characteristics as fractions therein) and only BnAbs for which all traits have been assessed, including poly-/autoreactivities measured in vitro in common clinical autoimmune screens (14,35,59), are included. B. Correlation between poly-/autoreactivity and SHM levels in BnAbs specific to either the MPER or CD4 binding site. Note the cutoff line labeled “normal SHM level plateau” is defined as the theoretical maximum during typical secondary Ab responses (57••,60,61) and also approximate levels generated in autologous nAb responses. SHM data shown for BnAb lineages are averages of their clonally related BnAb members.
SHM, along with the linked mechanism of BCR affinity-dependent selection, comprise a general “fine-tuning” process that occurs in germinal centers (GC) known as affinity maturation (AM). AM is crucial for generating higher affinity BCRs/secreted Abs and formation of higher-affinity memory B-cells that confer long-term protection against future infections, the hallmark of secondary B-cell responses. During “typical” AM responses, SHM levels increase BCR affinity but can also inadvertently create de novo self-reactivity, and thus normally plateaus at ~5–6%. SHM in excess of that is believed to increase the probability of generating self-reactivity and decrease BCR affinity (62–64), resulting in decreased B-cell survival (57••,60,61). Thus, in addition to elongated CDRH3 regions, and poly-/autoreactivity, exceptionally high SHM levels in BnAb V(D)J rearrangements (15–48%) represents another trait associated with negative selection.
That all BnAbs identified thus far originate from subjects infected with HIV-1 for 2–4 years suggests such remarkable SHM frequencies are products of disfavored and/or highly-convoluted AM pathways (57••). The reasons for why such complex AM pathways are generated in chronic infection were unknown until recent examination of clonal BnAb lineages (21••,65) suggests a component is a co-evolutionary “arms race” between viral and host responses. These studies, and recent findings that most experimentally-reverted unmutated ancestors of BnAbs lack neutralizing specificity (57••,65–66) offer one explanation: naïve BCRs with extensive modification are required to achieve unusual structural requirements for dealing with extensive viral diversification (Fig. 2A). However, the observation that some in vitro poly/autoreactive BnAbs targeting the same general Env regions tend to be less mutated, relative to their less/non-polyreactive counterparts (eg: all MPER BnAbs, relative to 10E8) (Fig. 1B) raises an intriguing alternative hypothesis: negative selection may not only result from excess SHM, but also be its cause. At least two mechanisms address how tolerizing autoreactivity of BnAbs could drive unusually-high SHM (52••,57••,69••) (Figs. 2B, C). One possibility (52••,57••) (Fig. 2B) is that B-cells with germline BCRs that bind self-mimicking BnAb epitopes do not contribute to AM, because they are deleted in early development, necessitating recruitment of weakly cross-reactive, mutated B-cell clones to generate BnAbs via AM. We have proposed a second possibility (69••) (Fig. 2C), that peripheral tolerance drives additional SHM, when BnAb reactivity and self-reactivity are coupled. In this scenario, GC-mutated, self-reactive B-cells with BnAb specificity initially attempt to escape negative selection by acquiring additional mutations that remove the self-reactivity, but that also inadvertently remove BnAb specificity. Such a “tug-of-war” process would likely invoke multiple rounds of mutation/selection to de-couple self-reactivity from affinity, yielding high SHM levels. Evidence for this, previously proposed as a model for the GC reaction (70), is observed in 2F5 KI mice. In these mice self-reactivity encoded by the original 2F5 V(D)J rearrangement involves a high degree of neutralization epitope mimicry (45••,52••) that is difficult to fully eliminate by receptor editing (43,44), resulting in an anergic, mature B-cell population (45••). Importantly, an MPER− subset of this peripheral anergic population undergoes additional targeted SHM at 2F5 VH CDR residues (69••, Verkoczy L, Diaz M, Haynes BF, et al. unpublished data) presumably in an attempt to remove BnAb epitope-associated self-reactivity. A similar “affinity reversion/de-maturation” process has also been documented in the HEL KI model (Sabouri Z, Goodnow CC, personal communication).
Figure 2. Hypotheses for why BnAb+ B-cells acquire unusually high levels of VDJ mutations during chronic HIV-1 infection.
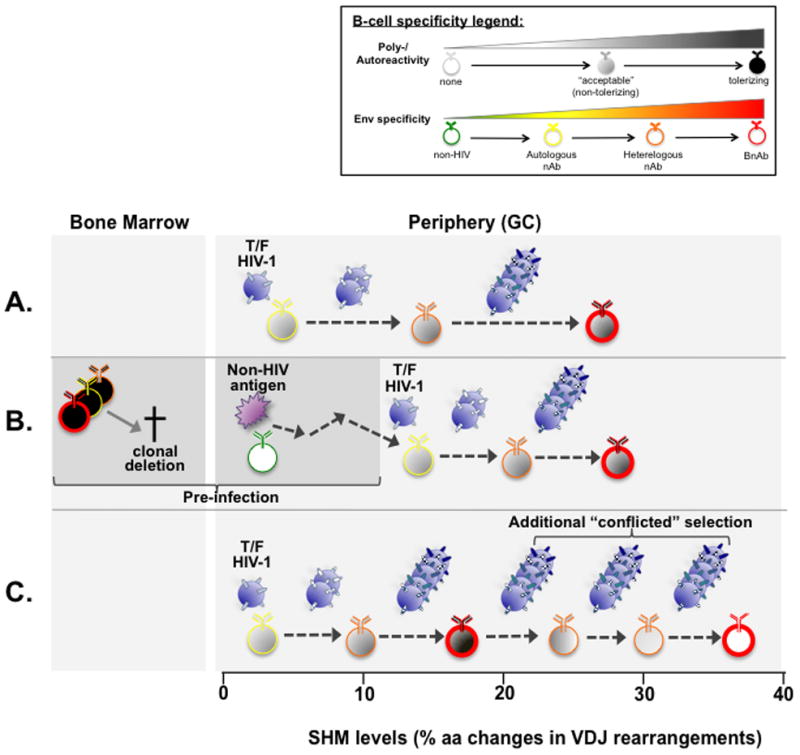
In a standard (non-tolerance) model (A), GC B cells with no/minimal reactivity to mature Envs (57••,65,66) interact with founder/transmitted Envs and undergo extensive AM, driven by viral escape/Env diversification pressure (21••), which demands difficult structural modifications to achieve both FRW flexibility and Env affinity (67•). Note that as shown, B-cells with “acceptable” (non-tolerizing) polyeractivity prior to entering the GC reaction may be selected for their ability to heteroligate Env (68), but poly-/self-reactivity may also be acquired during AM, as in the CH103-CH106 BnAb lineage (21••). In a model where self-tolerance indirectly impacts excess SHM (52••,57••) (B), B-cells with BnAb reactivity that mimic host antigens are clonally deleted in BM development, creating “holes in the repertoire”. In the absence of competition, weakly cross-reactive clones that already underwent non-Env driven AM can then participate in additional, Env-driven AM. Finally, in a model where self-tolerance directly drives excess SHM (69••)(C), B-cells that acquire both self and BnAb specificity via AM use additional SHM/selection to retain BnAb specificity, while removing tolerizing self-reactivity. Such a process would assume close, but not complete BnAb mimicry of self-epitopes. Thus, the degree of additional selection/SHM would be proportional to how closely a particular BnAb epitope mimics self-antigen(s). Note that these hypotheses are not mutually exclusive: for example, the “tolerance-based” models shown in (B–C) are shown in combination with the “standard” model, depicted in (A).
Examining the extent to which BnAb lineages are under host tolerance controls
Given that all BnAbs identified thus far have at least one of the three above-mentioned traits associated with negative selection (Fig. 1), another emerging consideration for vaccine strategies is understanding how often host tolerance limits BnAb production: namely, do they limit all MPER+ BnAb lineages, to what extent are BnAb lineages targeting other Env regions predisposed to such mechanisms, and what percentage of those target Env epitopes mimicking host structures, and finally, can such host controls be safely overcome by immunization? Thus, understanding how reliably the two BnAb traits most-associated with tolerance controls, in vitro autoreactivity and polyreactivity, i.e. as arbitrarily measured by common clinical assays (35,58) can predict in vivo tolerizing effects, is thus critical. Reassuringly, such in vitro poly-/autoreactivities exhibit reasonable concordance with immune tolerance, as suggested by the fact that in vitro poly-/autoreactivity of the normal human repertoire progressively wanes at developmental stages coinciding with previously-defined tolerance checkpoints in mice (58).
However, it is important to be mindful that in vitro poly-/autoreactivity assays can yield both “false positives” and “false negatives”. The former can occur because BCR/Ab affinities to host antigens in vivo are too low to effect tolerance (reviewed in 53). Given that ~15–20% of the mature T-dependent B2 population exhibit such “acceptable” in vitro poly/autoreactivity in healthy individuals’ repertoires, it may be fairly common (58). In addition, a large proportion of other B-cell subsets (i.e. MZ, B-1) (71,72), recently shown to harbor specificities resembling those of some BnAbs (73) also exhibit in vitro poly-/autoreactivity. “False negatives” can result from technical inter or intra-assay variation between laboratories. For example, the BnAb 10E8, initially reported to lack poly/autoreactivity measured by common assays (59) has strong affinity for a ubiquitous human protein, when detected in protein microarrays (Liu Y, Yang G, Kelsoe G, et al. unpublished data), and has also been reported to exhibit functionally-relevant lipid reactivity (74•). Furthermore, some relevant autoantigens could escape detection by existing technologies if they are tissue/cell-restricted, like insulin B-islets in the pancreas. Finally, “false negatives” could also result from the atypical biology of BnAbs with exceptionally high SHM, which, as products of BnAb lineages that otherwise exhibit prevalent self-reactivity, could represent atypical “snapshots”.
Ultimately, the “gold standard” for confirming the physiological relevance, i.e in vivo tolerizing effects of a given BnAb (or its lineage members’) in vitro poly-/autoreactivity measurements, is gene targeting of its/their V(D)J specificities. The power of the KI approach is evident in the vivo evaluation of 4E10, 2F5, and b12’s negative selection-associated traits (45••,46••,75•). While all these BnAbs have elongated HCDR3 regions and exhibit poly-/autoreactivity in vitro, b12 exhibits normal B-cell development when knocked into the mouse Ig locus (75•), unlike 4E10 and 2F5 (45••,46••). However, since b12 was generated via phage display, it remains to be seen if any BnAbs from naturally-derived H+L chain pairs represent “false positives” like notable exceptions in the general B-cell literature (76,77), and conversely, if candidate “false negative” BnAb lineages, like 10E8 or VRCO1, in fact do effect host tolerance controls in vivo. Higher-throughput KI approaches, based on RAG blastocyst-complementation (78) in which BnAb V(D)J rearrangements could be rapidly inserted, may facilitate this type of analysis. Finally, another independent, potentially fruitful avenue to exploring the prevalence of tolerance controls on BnAb production is examining frequencies and spectra of BnAbs produced by autoimmune patients. The first example of such an analysis is recent characterization of CH98, a CD4bs-specific BnAb isolated from an SLE patient (79).
Functional and practical aspects of BnAb traits associated with negative selection
Each of the above-mentioned traits of BnAbs are paradoxical because while they may predispose BnAbs to host tolerance controls, they may also be critical in conferring their function. Below, we discuss the potential practical relevance of each trait for vaccine development.
First, regarding BnAb polyeractivity, their degenerate recognition (and generally lower-affinity binding) to multiple distinct antigens, may have both general and specific relevance. General polyreactivity of Env-specific Abs may increase their overall affinity via heteroligation (68), a process proposed for dealing with the low density of Env trimer spikes found on HIV-1 (80,81). An elegant example of this is the structural analysis of bound 21c, a polyeractive Env Ab, which suggests interaction with not only its expected CD4i epitope, but also an adjacent CD4 receptor (82). Regarding BnAb function specifically, the lipid polyreactivities of 2F5, 4E10, 10E8, and m66 are believed to be an essential part of a general neutralization mechanism for all MPER-specific BnAbs, in which initial low-affinity contacts with the viral lipid membrane may extract lipid-immersed critical residues, thus allowing subsequent high-affinity interactions with exposed MPER epitopes (74•,83–86). There is growing consensus that for MPER-based vaccine strategies, appropriate presentation of 2F5/4E10 minimal peptide epitope in lipid membranes is required (87–90). Indeed, in 2F5 and 4E10 KI mice, only immunization regimens in which 4E10/2F5’s neutralization epitopes are presented in virosomes, and in the presence of the lipid component/TLR agonist MPLA, can elicit robust serum BnAb responses (91). With respect to BnAb autoreactivity, this trait merits practical considerations only if a BnAb’s relevant (tolerizing) self-antigen(s) exhibits considerable structural overlap with its neutralization epitope, as is likely for BnAbs targeting the 2F5 epitope (44,45••,52••). In such instances, a key question is whether vaccines can trigger either residual anergic B-cells (that retain both BnAb specificity and self-reactivity), or more ideally, rare peripheral “escape” variants (which have de-coupled self-reactivity and BnAb specificity). Although it remains to be determined if such latter clones exist, evidence for the former is suggested by the coincident generation of 2F5-like BnAbs and auto-Abs reported in sera from an HIV-1 infected subject (92). Importantly, immunization studies in 2F5 KI mice demonstrate that such residual anergic B-cells can be appropriately activated, thus providing proof-of-principle that their autoreactivity can be overcome by vaccination (69••). Furthermore, concerns about the pathogenicity of eliciting 2F5-like BnAbs are alleviated by passive infusion studies of 4E10, 2F5, or 2G12, which have not shown clinically adverse effects (93–95), except anticoagulant-activity for 4E10 (96), which is consistent with its higher lipid affinity, relative to 2F5 (35).
With respect to elongated and/or hydrophobic BnAb HCDR3s, hydrophobic residues appear critical for neutralization-conferring lipid reactivity of all MPER-specific BnAbs examined thus far, including 2F5/4E10 (83,84,97,98), 10E8 (74•) and M66 (86), but for BnAbs targeting other Env regions, it is unknown if their hydrophobicities (36) have functional relevance. However, for BnAbs with exceptionally elongated (>25aa) HCDR3s, such as the glycan/V1V2-specific BnAbs PG9/PG16, this unusual length is required to form the “hammerhead” structures thought critical for accessing occluded residues found in complex, glycan-masked epitopes (99). Encouragingly, while the frequencies of B-cells bearing exceptionally long CDRH3 regions are severely limited either by pre-antigenic constraints (HC/LC pairing and N-addition/VDJ recombination events) and/or various tolerance checkpoints (reviewed in 36), they nonetheless are present in repertoires of most healthy individuals (100).
High SHM levels in BnAbs, represent a potentially more formidable roadblock for vaccination because conventional vaccines cannot recapitulate AM pathways during infection that generated such mutational levels; existing HIV-1 immunization regimens produce SHM levels (4–5%) more in-line with autologous nAbs (56•). However, the degree to which observed mutations in BnAbs are required for neutralization breadth/potency is unknown, although several recent studies have begun assessing this (67•,101•,102). In one, selective reversion of V(D)J framework regions (FRW) from a panel of BnAbs impacted breadth/potency in most (67•) leading to the proposal that exceptionally high SHM levels in BnAb FRWs provide structural flexibility. However, two recent in vitro mutagenesis-scanning studies found that only ~10–25% of reverted mutations impacted neutralization function for 10E8 and VRCO1 (two of the most mutated BnAbs) (101•,102). One explanation for the disparate results is that highly-mutated BnAbs may result from the stochastic nature of SHM driving numerous rounds of AM to create the desired combination of mutations that enhance structural flexibility (FRW’s), and epitope binding affinity. Alternatively, SHM above the minimum required for neutralization, may be result of conflicted selection pressure in the GC: negative (for tolerance) and positive (for affinity) (Fig. 2C). It is also possible that a component of “excess” SHM found in BnAbs, may be related to chronic HIV-1 infection (34•) and its potential effects on immune function (see 103, 104), since high mutation levels (although not as dramatic as in BnAbs) is also produced in other chronic infection settings, like influenza (57••,105). Ultimately, proof-of-concept immunization studies, including those in KI mice expressing reverted ancestors/intermediates of individual BnAb clonal lineages, cross-bred to strains with altered SHM levels (106) may provide some insight on this issue.
Assuming a considerable degree of the SHM generated in BnAbs during infection is required during immunization, new vaccine strategies will have to re-create disfavored and/or convoluted AM pathways that generated such extensive SHM. B-cell lineage design has been recently proposed for this purpose (21••,57••), an approach aimed to use immunogens that optimally bind a given BnAb lineage’s ancestors and AM intermediates, administered either sequentially (to help “guide” AM pathways) or in combination (to mimic HIV-1 diversity). Perhaps the most straightforward and physiological use of this approach involves using the actual Env proteins identified from sampling serial isolates of BnAb lineages generated during infection, to re-create similar pathways by vaccination (21••). However, other variations have also been proposed, involving use of computational and in vitro selection methodologies to engineer “super-immunogens” that can bind BCRs of multiple clonal members from BnAb lineages with common epitopes (107•,108•).
Conclusions
Recent studies using KI models demonstrate immune tolerance can profoundly limit BnAb induction, and traits associated with negative selection in many recently discovered BnAbs suggest this has relevance for BnAbs at distinct Env targets. Some aspects of tolerance can be overcome, for example immunization of 2F5 KI mice demonstrates overcoming deletion/anergy is possible, provided a structurally-compatible immunogen is used, and passive infusion studies suggest pathogenicity would not result from their elicitation. Other aspects, like overlap of self-reactivity with BnAb specificity, which may result in inadvertent selection against the latter, may be more problematic. Further examination of the prevalence of this issue across BnAb lineages will benefit from autoantigen discovery/modeling and high-throughput KI approaches. Finally, immunoregulatory factors other than immune tolerance, that may contribute to subdominance of BnAb responses, including shaping of the initial B-cell repertoire by incidental exogenous antigen exposure (28), or host genetic determinants like allelic variation of VH segments utilized by BnAb lineages (109) and MHC class II-restriction of overlapping CD4+ TH/BnAb epitopes (110) may also have to be considered for vaccine strategies.
A key for vaccine development will be determining the feasibility of immunization to elicit AM pathways that can generate breath/neutralization in various BnAb lineages. The development of novel immunization strategies like B-cell lineage design, more powerful KI-based immunization models (expressing limited repertoires of unrearranged germline segments from representative BnAb lineages) to more efficiently probe AM pathways in vivo, and isolation of new clonal lineages from both infection and immunization settings will be key. Finally, it should be noted that even when only considering the minimally-required mutations for neutralization in highly-mutated BnAbs like VRC01, vaccination schemes would still have to generate much higher SHM levels than existing regimens can elicit. We therefore submit that BnAbs with relatively less SHM may be more desirable/tractable targets.
Key Points.
Numerous broadly neutralizing antibodies (BnAbs) have recently been identified, many of which have unusually long, hydrophobic HCDR3 regions and/or in vitro poly-/autoreactivity, traits normally associated with negative B-cell selection.
The exceptional degree of somatic mutation found in all BnAbs isolated thus far suggest their generation via convoluted affinity maturation pathways, which directly or indirectly, may result from mimicry of self-antigens by BnAb epitopes.
Knock-in mice of BnAbs targeting the gp41 MPER region have provided evidence that BnAb poly-/autoreactivity can invoke host tolerance controls that profoundly impacts their production.
The unusual HCDR3 regions, poly-/autoreactivity, and excess somatic mutation of BnAbs may all be functionally relevant, thus necessitating novel vaccine strategies capable of triggering and/or generating B-cells having such features.
Acknowledgments
The authors thank Barton Haynes, Garnett Kelsoe, Munir Alam, as well as other colleagues at DHVI and NIEHS for helpful discussions.
This work was supported by NIH, National Institute of Environmental Health Sciences, Division of Intramural Research project Z01 ES101603, the Center for HIV/AIDS Vaccine Immunogen-Discovery (CHAVI-ID) grant UM1 AI100645, NIH grant R01 AI087202 and a Collaboration for AIDS Discovery Vaccine Development Center grant from the Bill and Melinda Gates Foundation.
Footnotes
Conflicts of interest
There are no conflicts of interest to declare.
References and Recommended Reading
- 1.Plotkin S. Correlates of protection induced by vaccination. Clin Vaccine Immunol. 2010;17:1055–1065. doi: 10.1128/CVI.00131-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wei X, Decker JM, Wang S, et al. Antibody neutralization and escape by HIV. Nature. 2003;422:307–312. doi: 10.1038/nature01470. [DOI] [PubMed] [Google Scholar]
- 3.Hessell AJ, Poignard P, Hunter M, et al. Effective, low-titer antibody protection against low-dose repeated mucosal SHIV challenge in macaques. Nat Med. 2009;15:951–954. doi: 10.1038/nm.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hessell AJ, Rakasz EG, Poignard P, et al. Broadly neutralizing human anti-HIV antibody 2G12 is effective in protection against mucosal SHIV challenge even at low serum neutralizing titers. PLoS Path. 2009;5:e1000433. doi: 10.1371/journal.ppat.1000433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Balazs AB, Chen J, Hong CM, et al. Antibody-based protection against HIV infection by vectored immunoprophylaxis. Nature. 2012;481:81–84. doi: 10.1038/nature10660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Simek MD, Rida W, Priddy FH, et al. Human immunodeficiency virus type 1 elite neutralizers: individuals with broad and potent neutralizing activity identified by using a high-throughput neutralization assay together with an analytical selection algorithm. J Virol. 2009;83:7337–7348. doi: 10.1128/JVI.00110-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gray ES, Madiga MC, Hermanus T, et al. The neutralization breadth of HIV-1 develops incrementally ver four years and is associated with CD4+ T cell decline and high viral load during acute infection. J Virol. 2011;85:4828–4840. doi: 10.1128/JVI.00198-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tomaras GD, Binley JM, Gray ES, et al. Polyclonal B cell responses to conserved neutralization epitopes in a subset of HIV-1-infected individuals. J Virol. 2011;85:11502–11519. doi: 10.1128/JVI.05363-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mikell I, Sather DN, Kalams SA, et al. Characteristics of the Earliest Cross-Neutralizing Antibody Response to HIV-1. PLoS Pathog. 2011;7:e1001251. doi: 10.1371/journal.ppat.1001251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schied JF, Mouquet H, Feldhahn N, et al. A method for identification of HIV gp140 binding memory B cells in human blood. J Immunol Methods. 2009;343:65–67. doi: 10.1016/j.jim.2008.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Morris L, Chen X, Alam M, et al. Isolation of a human HIV gp41 membrane proximal region neutralizing antibody by antigen-specific B cell sorting. PLoS One. 2011;6:e23532. doi: 10.1371/journal.pone.0023532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Walker LM, Phogat SK, Chan-Hui PY, et al. Broad and potent neutralizing antibodies from an African donor reveal a new HIV-1 vaccine target. Science. 2009;326:285–289. doi: 10.1126/science.1178746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bonsignori M, Hwang KK, Chen X, et al. Analysis of a clonal lineage of HIV-1 envelope V2/V3 conformational epitope-specific broadly neutralizing antibodies and their inferred unmutated common ancestors. J Virol. 2011;85:9998–10009. doi: 10.1128/JVI.05045-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wardemann H, Yurasov S, Schaefer A, et al. Predominant autoantibody production by early human B cell precursors. Science. 2003;301:1374–1377. doi: 10.1126/science.1086907. [DOI] [PubMed] [Google Scholar]
- 15.Liao HX, Levesque MC, Nagel A, et al. High-throughput isolation of immunoglobulin genes from single human B cells and expression as monoclonal antibodies. J Virol Methods. 2009;158:171–179. doi: 10.1016/j.jviromet.2009.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16••.Mascola JR, Haynes BF. HIV-1 neutralizing antibodies: understanding nature’s pathways. Immunol Rev. 2013;254:225–244. doi: 10.1111/imr.12075. Comprehensive recent overview of progress made in characterizing novel BnAbs and their epitopes, as well as in understanding structural and immunological issues relevant for their elicitation by vaccines. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Scheid JF, Mouquet H, Ueberheide B, et al. Sequence and structural convergence of broad and potent HIV antibodies that mimic CD4 binding. Science. 2011;333:1633–1637. doi: 10.1126/science.1207227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Walker LM, Simek MD, Priddy F, et al. A limited number of antibody specificities mediate broad and potent serum neutralization in selected HIV-1 infected individuals. PLoS Path. 2010;6:e1001028. doi: 10.1371/journal.ppat.1001028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bonsignori M, Montefiori DC, Wu X, et al. Two distinct broadly neutralizing antibody specificities of different clonal lineages in a single HIV-1-infected donor: implications for vaccine design. J Virol. 2012;86:4688–4692. doi: 10.1128/JVI.07163-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Doria-Rose NA, Louder MK, Yang Z, et al. HIV-1 neutralization coverage is improved by combining monoclonal antibodies that target independent epitopes. J Virol. 2012;86:3393–3397. doi: 10.1128/JVI.06745-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21••.Liao H-X, Lynch R, Zhou Z, et al. Co-evolution of a broadly neutralizing HIV-1 antibody and founder virus. Nature. 2013;496:469–76. doi: 10.1038/nature12053. Landmark article that comprehensively documents the extensive co-evolution that HIV-1 and clonal members of a B-cell lineage underwent in a subject which eventually generated (after ~2.5 years) a BnAb targeting the CD4 binding site through its CDRH3 loop. By molecularly elucidating the naturally derived Env proteins from successive viral escape mutants of the transmitted/founder virus, and proposing their use in “guiding” comparable (i.e. “CDRH3 loop class”) CD4bs-specific BnAb lineages by immunization, these data provide a physiologically-relevant framework for testing the B-cell lineage design vaccine approach. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wyatt R, Kwong PD, Desjardins E, et al. The antigenic structure of the HIV gp120 envelope glycoprotein. Nature. 1998;393:705–711. doi: 10.1038/31514. [DOI] [PubMed] [Google Scholar]
- 23.Kwong PD, Doyle ML, Casper DJ, et al. HIV-1 evades antibody-mediated neutralization through conformational masking of receptor-binding sites. Nature. 2002;420:678–682. doi: 10.1038/nature01188. [DOI] [PubMed] [Google Scholar]
- 24.Wei X, Decker JM, Wang S, et al. Antibody neutralization and escape by HIV-1. Nature. 2003;422:307–312. doi: 10.1038/nature01470. [DOI] [PubMed] [Google Scholar]
- 25.Schief WR, Ban YE, Stamatatos L. Challenges for structure-based HIV vaccine design. Curr Opin HIV AIDS. 2009;4:431–440. doi: 10.1097/COH.0b013e32832e6184. [DOI] [PubMed] [Google Scholar]
- 26.Stamatatos L, Morris L, Burton DR, et al. Neutralizing antibodies generated during natural HIV-1 infection: good news for an HIV-1 vaccine? Nature Med. 2009;15:866–870. doi: 10.1038/nm.1949. [DOI] [PubMed] [Google Scholar]
- 27.Tomaras GD, Yates NL, Liu P, et al. Initial B-Cell Responses to Transmitted Human Immunodeficiency Virus Type 1: Virion-Binding Immunoglobulin M (IgM) and IgG Antibodies Followed by Plasma Anti-gp41 Antibodies with Ineffective Control of Initial Viremia. J Virol. 2008;82:12449–12463. doi: 10.1128/JVI.01708-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liao HX, Chen X, Munshaw S, et al. Initial antibodies binding to HIV-1 gp41 in acutely infected subjects are polyreactive and highly mutated. J Exp Med. 2011;208:2237–2249. doi: 10.1084/jem.20110363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Burton DR, Poignard P, Stanfield RL, et al. Broadly neutralizing antibodies present new prospects to counter highly antigenically diverse viruses. Science. 2012;337:183–186. doi: 10.1126/science.1225416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kwong PD, Mascola JR. Human antibodies that neutralize HIV-1: identification, structures, and B cell ontogenies. Immunity. 2012;37:412–425. doi: 10.1016/j.immuni.2012.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McCoy LE, Weiss RA. Neutralizing antibodies to HIV-1 induced by immunization. J Exp Med. 2013;210:209–223. doi: 10.1084/jem.20121827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schiffner T, Sattentau QJ, Dorrell L. Development of prophylactic vaccines against HIV-1. Retrovirology. 2013;10:72–88. doi: 10.1186/1742-4690-10-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kong L, Sattentau QJ. Antigenicity and Immunogenicity in HIV-1 Antibody-Based Vaccine Design. J AIDS Clin Res. 2012;S8:3. doi: 10.4172/2155-6113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34•.Moir S, Fauci AS. Insights into B cells and HIV-specific B-cell responses in HIV-infected individuals. Immunol Rev. 2013;254:207–24. doi: 10.1111/imr.12067. Elegant and comprehensive recent review of factors during chronic HIV-1 infection that contribute to, or are associated with, B-cell dysfunction/dysregulation. [DOI] [PubMed] [Google Scholar]
- 35.Haynes BF, Fleming J, St Clair EW, Katinger H, Stiegler G, et al. Cardiolipin polyspecific autoreactivity in two broadly neutralizing HIV-1 antibodies. Science. 2005;308:1906–1908. doi: 10.1126/science.1111781. [DOI] [PubMed] [Google Scholar]
- 36.Verkoczy L, Kelsoe G, Moody MA, et al. Role of immune mechanisms in induction of HIV-1 broadly neutralizing antibodies. Curr Opin Immunol. 2011;23:383–390. doi: 10.1016/j.coi.2011.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Haynes BF, Moody MA, Verkoczy L, et al. Antibody polyspecificity and neutralization of HIV-1: a hypothesis. Hum Antibodies. 2005;14:59–67. [PMC free article] [PubMed] [Google Scholar]
- 38.Kaye BR. Rheumatologic manifestations of infection with human immunodeficiency virus (HIV) Ann Intern Med. 1989;111:158–67. doi: 10.7326/0003-4819-111-2-158. [DOI] [PubMed] [Google Scholar]
- 39.Barthel HR, Wallace DJ. False-positive human immunodeficiency virus testing in patients with lupus erythematosus. Semin Arthritis Rheum. 1993;23:1–7. doi: 10.1016/s0049-0172(05)80021-6. [DOI] [PubMed] [Google Scholar]
- 40.Mylonakis E, Paliou M, Greenbough TC, et al. Report of a false-positive HIV test result and the potential use of additional tests in establishing HIV serostatus. Arch Intern Med. 2000;160:2386–2388. doi: 10.1001/archinte.160.15.2386. [DOI] [PubMed] [Google Scholar]
- 41.Palacios R, Santos J, Valdivielso P, et al. Human immunodeficiency virus infection and systemic lupus erythematosus. An unusual case and a review of the literature. Lupus. 2002;11:60–63. doi: 10.1191/0961203302lu141cr. [DOI] [PubMed] [Google Scholar]
- 42.Calza L, Manfredi R, Colangeli V, et al. Systemic and discoid lupus erythematosus in HIV-infected patients treated with highly active antiretroviral therapy. Int J STD AIDS. 2003;14:356–359. doi: 10.1258/095646203321605585. [DOI] [PubMed] [Google Scholar]
- 43.Verkoczy L, Diaz M, Holl TM, et al. Autoreactivity in an HIV-1 broadly reactive neutralizing antibody variable region heavy chain induces immunologic tolerance. Proc Natl Acad Sci USA. 2010;107:181–186. doi: 10.1073/pnas.0912914107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Verkoczy L, Chen Y, Bouton-Verville H, et al. Rescue of HIV-1 broad neutralizing antibody-expressing B cells in 2F5 VH x VL knockin mice reveals multiple tolerance controls. J Immunol. 2011;187:3785–3797. doi: 10.4049/jimmunol.1101633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45••.Chen Y, Zhang J, Hwang KK, et al. Common tolerance mechanisms, but distinct cross-reactivities associated with gp41 and lipids, limit production of HIV-1 broad neutralizing antibodies 2F5 and 4E10. J Immunol. 2013;191:1260–1275. doi: 10.4049/jimmunol.1300770. Thorough comparison of 2F5 and 4E10 KI models, describing profound HC-mediated clonal deletion and anergy in both, and identifying a small subset of “escape” clones in each, indicative of distinct positive selection pressures, that along with Ref.52, suggest 2F5’s neutralization epitope and tolerizing autoantigen specificities substantially overlap. This study also presents analysis of a KI model expressing the HC of 48d (a CD4i-specific Ab with a normal HCDR3 and lacking in vitro poly-/autoreactivity) is also presented as an important control to exclude human/mouse chimeric effects, and along with Refs. 45 and 75, validates the Ab KI approach for generally evaluation of if/how tolerance affects generation of BnAb lineages. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46••.Doyle-Cooper C, Hudson KE, Cooper AB, et al. Immune Tolerance Negatively Regulates B Cells in Knock-In Mice Expressing Broadly Neutralizing HIV Antibody 4E10. J Immunol. 2013;191:3186–3191. doi: 10.4049/jimmunol.1301285. Key analysis of an independently generated 4E10 KI model, that like Ref.45, reveals a similar profound developmental blockade at the pre-B to immature B-cell transition and BCR down-modulation in residual B-cell populations. Furthermore, negative selection is exacerbated when the ability to undergo secondary LC rearrangements is genetically eliminated (by cross-breeding to either JH or JCκ-deleted and RAG-deficient mice); thus, as found in the 2F5 KI model (Ref. 44), 4E10 KI B-cells are under multiple tolerance controls, including deletion, anergy and LC editing. Interestingly, 4E10 KI serum IgGs in this analysis, unlike Ref.45, had co-varying MPER epitope and lipid reactivity. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47•.Finton KA, Larimore K, Larman HB, et al. Autoreactivity and exceptional CDR plasticity (but not unusual polyspecificity) hinder elicitation of the anti-HIV antibody 4E10. PLoS Pathog. 2013;9:e1003639. doi: 10.1371/journal.ppat.1003639. Recent study reporting a third independently-generated KI model expressing the original 4E10 VDJ rearrangement, confirming the profound blockade and compromised BCR signaling of residual B-cells observed in Refs.45,46. This paper also identifies a candidate autoantigen target for 4E10, IP3R, and, in contrast to Refs. 35 and 52, reports no/minimal polyreactivity for 4E10 by either HEp-2 IFA or in protein arrays, and proposes low-affinity lipid interactions are mediated non-specifically, rather than via hydrophobic interactions. These disparities in how in vitro polyreactivity and lipid binding are measured and interpreted for similar assays performed in distinct laboratories highlight the difficulties in defining and standardizing stringency thresholds for performing such assays. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nemazee DA, Burki K. Clonal deletion of B lymphocytes in a transgenic mouse bearing anti-MHC class I antibody genes. Nature. 1989;337:562–566. doi: 10.1038/337562a0. [DOI] [PubMed] [Google Scholar]
- 49.Hartley SB, Crosbie J, Brink R, et al. Elimination from peripheral lymphoid tissues of self-reactive B lymphocytes recognizing membrane-bound antigens. Nature. 1991;353:765–769. doi: 10.1038/353765a0. [DOI] [PubMed] [Google Scholar]
- 50.Chen C, Nagy ZM, Radic MZ, et al. The site and stage of anti-DNA B-cell deletion. Nature. 1995;373:252–255. doi: 10.1038/373252a0. [DOI] [PubMed] [Google Scholar]
- 51.Goodnow CC. Transgenic mice and analysis of B-cell tolerance. Ann Rev Immunol. 1992;10:489–518. doi: 10.1146/annurev.iy.10.040192.002421. [DOI] [PubMed] [Google Scholar]
- 52••.Yang G, Holl TM, Liu Y, et al. Identification of autoantigens recognized by the 2F5 and 4E10 broadly neutralizing HIV-1 antibodies. J Exp Med. 2013;210:241–256. doi: 10.1084/jem.20121977. Key article reporting the biochemical identification, using immunoprecipitation and protein microarrays, of two highly-conserved, high-affinity host protein targets of 2F5 and 4E10, respectively, the former which interestingly contains a motif with perfect sequence identity to the 2F5 neutralization epitope. Using 2F5 and 4E10 as an example, this study also shows how protein arrays can be used to more comprehensively characterize the poly/autoreactivity profiles of BnAbs and outlines affinity threshold criteria as well as appropriate controls for performing this type of analysis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shlomchik MJ. Sites and stages of autoreactive B cell activation and regulation. Immunity. 2008;28:18–28. doi: 10.1016/j.immuni.2007.12.004. [DOI] [PubMed] [Google Scholar]
- 54.Lang J, Jackson M, Teyton L, et al. B-cells are exquisitely sensitive to central tolerance and receptor editing induced by ultralow affinity, membrane bound antigen. J Exp Med. 1996;184:1685–1697. doi: 10.1084/jem.184.5.1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ota T, Doyle-Cooper C, Cooper AB, et al. Anti-HIV B Cell Lines as Candidate Vaccine Biosensors. J Immunol. 2012;189:4816–4824. doi: 10.4049/jimmunol.1202165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56•.Bonsignori M, Alam SM, Liao HX, Verkoczy L, et al. HIV-1 antibodies from infection and vaccination: insights for guiding vaccine design. Trends Microbiol. 2013;20:532–9. doi: 10.1016/j.tim.2012.08.011. Thorough review of progress in technologies for identifying novel BnAbs, analyzing clonal BnAb lineages, and profiling Env Ab responses, both during infection and in human vaccine trials. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57••.Haynes BF, Kelsoe G, Harrison SC, et al. B-cell-lineage immunogen design in vaccine development with HIV-1 as a case study. Nat Biotech. 2012;30:423–433. doi: 10.1038/nbt.2197. Comprehensive review of BnAb affinity maturation pathways and B-cell lineage design-based vaccine approaches. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wardemann H, Yurasov S, Schaefer A, et al. Predominant autoantibody production by early human B cell precursors. Science. 2003;301:1374–7. doi: 10.1126/science.1086907. [DOI] [PubMed] [Google Scholar]
- 59.Huang J, Ofek G, Laub L, et al. Broad and potent neutralization of HIV-1 by a gp41-specific human antibody. Nature. 2012;491:406–412. doi: 10.1038/nature11544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kepler TB, Perelson AS. Somatic hypermutation in B cells: an optimal control treatment. J Theor Biol. 1993;164:37–64. doi: 10.1006/jtbi.1993.1139. [DOI] [PubMed] [Google Scholar]
- 61.Zhang J, Shakhnovich EI. Optimality of mutation and selection in germinal centers. PLoS Comput Biol. 2010;6:e1000800. doi: 10.1371/journal.pcbi.1000800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shlomchik M, Mascelli M, Shan H, et al. Anti-DNA antibodies from autoimmune mice arise by clonal expansion and somatic mutation. J Exp Med. 1990;171:265–92. doi: 10.1084/jem.171.1.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tiller T, Tsuiji M, Yurasov S, et al. Autoreactivity in human IgG+ memory B cells. Immunity. 2007;26:205–13. doi: 10.1016/j.immuni.2007.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mietzner B, Tsuiji M, Scheid J, et al. Autoreactive IgG memory antibodies in patients with systemic lupus erythematosus arise from nonreactive and polyreactive precursors. Proc Natl Acad Sci U S A. 2008;105:9727–32. doi: 10.1073/pnas.0803644105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Moore PL, Gray ES, Wibmer CK, et al. Evolution of an HIV glycan-dependent broadly neutralizing antibody epitope through immune escape. Nat Med. 2012;18:1688–92. doi: 10.1038/nm.2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hoot S, McGuire AT, Cohen KW, et al. Recombinant HIV envelope proteins fail to engage germline versions of anti-CD4bs bNAbs. PLoS Path. 2013;9:e1003106. doi: 10.1371/journal.ppat.1003106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67•.Klein F, Diskin R, Scheid JF, et al. Somatic mutations of the immunoglobulin framework are generally required for broad and potent neutralization. Cell. 2013;153:126–138. doi: 10.1016/j.cell.2013.03.018. First paper to examine the potential functional role that high SHM levels play in generating neutralizing function/breadth, demonstrating a key role for highly-mutated VDJ framework regions (FRW) in BnAbs for their breadth/potency, and leading to the proposal that such high FRW-specific SHM is generally required for structural flexibility. An intriguing aspect of this analysis is that FRW regions of the MPER+ BnAbs 2F5 and 4E10 (which have relatively less SHM than other BnAbs assessed) are also less critical for neutralization function. For 2F5 in particular, which already has lipid reactivity to initiate its neutralization mechanism [Ref.84] encoded in its inferred germline-reverted BnAb [Ref.108], this raises the possibility that it may employ a more targeted route of AM, acting selectively on HCDR residues to increase MPER epitope affinity. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mouquet H, Scheid JF, Zoller MJ, et al. Polyreactivity increases the apparent affinity of anti-HIV antibodies by heteroligation. Nature. 2010;467:591–595. doi: 10.1038/nature09385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69••.Verkoczy L, Chen Y, Zhang J, et al. Induction of HIV-1 broad neutralizing antibodies in 2F5 knockin mice: selection against MPER-associated autoreactivity limits T-dependent responses. J Immunol. 2013;191:2538–2550. doi: 10.4049/jimmunol.1300971. Key proof-of-concept study in which residual pre-existing B-cells in 2F5 KI mice which escape central clonal deletion (but functionally-silenced by tolerance) can be re-activated by immunization with a TLR-MPER peptide-liposome conjugate immunogen and produce robust serum IgG titers with the same specificity/neutralization profiles as original 2F5. Thus, in this model, host tolerance controls, when dissociated from other potential factors limiting BnAb induction (eg: pre-antigenic constraints, clonal competition by B-cells expressing dominant, nonneutralizing epitopes) can be overcome with an appropriate immunization regimen. However, the T-independent nature of this response also implies residual MPER epitope-associated self-reactivity may be avidly purged in T-dependent compartments by peripheral V-modifying mechanisms. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Diaz M, Klinman N. Relative roles of somatic and Darwinian evolution in shaping the antibody response. Immunol Res. 2000;21:89–102. doi: 10.1385/IR:21:2-3:89. [DOI] [PubMed] [Google Scholar]
- 71.Martin F, Kearney JF. Marginal-zone B cells. Nat Rev Immunol. 2002;2:323–35. doi: 10.1038/nri799. [DOI] [PubMed] [Google Scholar]
- 72.Hardy RR. B-1 B cell development. J Immunol. 2006;177:2749–54. doi: 10.4049/jimmunol.177.5.2749. [DOI] [PubMed] [Google Scholar]
- 73.Pujanauski LM, Janoff EN, McCarter MD, et al. Mouse marginal zone B cells harbor specificities similar to human broadly neutralizing HIV antibodies. Proc Natl Acad Sci USA. 2013;110:1422–7. doi: 10.1073/pnas.1213713110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74•.Chen J, Frey G, Peng H, et al. Mechanism of HIV-1 Neutralization by Antibodies Targeting a Membrane-Proximal Region of gp41. J Virol. 2014;88:1249–58. doi: 10.1128/JVI.02664-13. Recent study demonstrating, in conflict with original description of 10E8 [ref 59], 10E8’s functionally-relevant affinity for lipids, and providing data that is consistent with 10E8 being essentially a more potent version of 4E10, the latter whose neutralization potency can be enhanced by an alteration selectively made to its lipid-interacting CDHR3 that does not impact its MPER. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75•.Ota T, Doyle-Cooper C, Cooper AB, et al. B cells from knock-in mice expressing broadly neutralizing HIV antibody b12 carry an innocuous B cell receptor responsive to HIV vaccine candidates. J Immunol. 2013;191:3179–85. doi: 10.4049/jimmunol.1301283. Important companion article to Ref 45, demonstrating that a KI model of b12, one of the original BnAbs identified to have traits associated with negative selection (including in vitro poly-/autoreactivity), exhibits normal B-cell development and undergoes no/minimal diversification of its original specificity, and thus is not subjected to immune tolerance in vivo. Notwithstanding the caveat that b12 was generated by phage display, this study along with Refs 45,46, shows the general power of KI technology both for testing the physiological relevance of in vitro measurements of BnAb poly-/autoreactivity and as immunization platforms, for evaluating Env immunogens. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ota T, Aoki-Ota M, Tsunoda K, et al. Auto-reactive B cells against peripheral antigen, desmoglein 3, escape from tolerance mechanism. Int Immunol. 2004;16:1487–95. doi: 10.1093/intimm/dxh150. [DOI] [PubMed] [Google Scholar]
- 77.Aplin BD, Keech CL, de Kauwe AL, et al. Tolerance through indifference: autoreactive B cells to the nuclear antigen La show no evidence of tolerance in a transgenic model. J Immunol. 2003;171:5890–900. doi: 10.4049/jimmunol.171.11.5890. [DOI] [PubMed] [Google Scholar]
- 78.Chen J, Lansford R, Stewart V, et al. RAG-2-deficient blastocyst complementation: an assay of gene function in lymphocyte development. Proc Natl Acad Sci USA. 1993;90:4528–32. doi: 10.1073/pnas.90.10.4528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bonsignori MK, Wiehe R, Lynch M, et al. Isolation and Characterization of an Autoreactive CD4bs Broad Neutralizing Antibody from a Chronic HIV-1 Controller with Systemic Lupus Erythematosus. AIDS Res Human Retr. 2013;29:A45–A46. [Google Scholar]
- 80.Zhu P, Liu J, Bess J, Jr, et al. Distribution and three-dimensional structure of AIDS virus envelope spikes. Nature. 2006;441:847–52. doi: 10.1038/nature04817. [DOI] [PubMed] [Google Scholar]
- 81.Liu J, Bartesaghi A, Borgnia MJ, et al. Molecular architecture of native HIV-1 gp120 trimers. Nature. 2008;455:109–13. doi: 10.1038/nature07159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.West AP, Jr, Diskin R, Nussenzweig MC, et al. Structural basis for germ-line gene usage of a potent class of antibodies targeting the CD4-binding site of HIV-1 gp120. Proc Natl Acad Sci USA. 2012;109:E2083–2090. doi: 10.1073/pnas.1208984109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Alam SM, Morelli M, Dennison SM, et al. Role of HIV membrane in neutralization by two broadly neutralizing antibodies. Proc Natl Acad Sci USA. 2009;106:20234–9. doi: 10.1073/pnas.0908713106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sun ZY, Oh KJ, Kim M, et al. HIV-1 broadly neutralizing antibody extracts its epitope from a kinked gp41 ectodomain region on the viral membrane. Immunity. 2008;28:52–63. doi: 10.1016/j.immuni.2007.11.018. [DOI] [PubMed] [Google Scholar]
- 85.Shen X, Dennison SM, Liu P, Gao F, Jaeger F, et al. Prolonged exposure of the HIV-1 gp41 membrane proximal region with L669S substitution. Proc Natl Acad Sci USA. 2010;107:5972–5977. doi: 10.1073/pnas.0912381107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ofek G, Zirkle B, Yang Y, et al. Structural Basis for HIV-1 Neutralization by 2F5–like Antibodies m66 and m66.6. J Virol. 2014 doi: 10.1128/JVI.02837-13. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Alam SM, McAdams M, Boren D, et al. The role of antibody polyspecificity and lipid reactivity in binding of broadly neutralizing anti-HIV-1 envelope human monoclonal antibodies 2F5 and 4E10 to glycoprotein 41 membrane proximal envelope epitopes. J Immunol. 2007;178:4424–35. doi: 10.4049/jimmunol.178.7.4424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Dennison SM, Stewart SM, Stempel KC, et al. Stable docking of neutralizing human immunodeficiency virus type 1 gp41 membrane-proximal external region monoclonal antibodies 2F5 and 4E10 is dependent on the membrane immersion depth of their epitope regions. J Virol. 2009;83:10211–10223. doi: 10.1128/JVI.00571-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Dennison SM, Sutherland LL, Jaeger FH, et al. Induction of antibodies in rhesus macaques that recognize a fusion-intermediate conformation of HIV-1 gp41. PLoS One. 2011;6:e27824. doi: 10.1371/journal.pone.0027824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kim M, Song L, Moon J, et al. Immunogenicity of membrane-bound HIV-1 gp41 membrane-proximal external region (MPER) segments is dominated by residue accessibility and modulated by stereochemistry. J Biol Chem. 2014;288:31888–31901. doi: 10.1074/jbc.M113.494609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Chen Y, Zhang J, Bouton-Verville H, et al. Lipid Components in MPER-Based Immunization Regimens Are Critical for Inducing Broadly Neutralizing Antibody Responses in 2F5 and 4E10 Knockin Mice. AIDS Res Human Retr. 2013;29:A20–A20. [Google Scholar]
- 92.Shen X, Parks RJ, Montefiori DC, Kirchherr JL, Keele BF, et al. In vivo gp41 antibodies targeting the 2F5 monoclonal antibody epitope mediate human immunodeficiency virus type 1 neutralization breadth. J Virol. 2009;83:3617–3625. doi: 10.1128/JVI.02631-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Trkola A, Kuster H, Rusert P, et al. Delay of HIV-1 rebound after cessation of antiretroviral therapy through passive transfer of human neutralizing antibodies. Nat Med. 2005;11:615–622. doi: 10.1038/nm1244. [DOI] [PubMed] [Google Scholar]
- 94.Vcelar B, Stiegler G, Wolf HM, et al. Reassessment of autoreactivity of the broadly neutralizing HIV antibodies 4E10 and 2F5 and retrospective analysis of clinical safety data. AIDS. 2007;21:2161–2170. doi: 10.1097/QAD.0b013e328285da15. [DOI] [PubMed] [Google Scholar]
- 95.Mehandru S, Vcelar B, Wrin T, et al. Adjunctive passive immunotherapy in human immunodeficiency virus type 1-infected individuals treated with antiviral therapy during acute and early infection. J Virol. 2007;81:11016–11031. doi: 10.1128/JVI.01340-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Li Y, O’Dell S, Walker LM, et al. Mechanism of neutralization by the broadly neutralizing HIV-1 monoclonal antibody VRC01. J Virol. 2011;85:8954–67. doi: 10.1128/JVI.00754-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ofek G, McKee K, Yang Y, et al. Relationship between antibody 2F5 neutralization of HIV-1 and hydrophobicity of its heavy chain third complementarity-determining region. J Virol. 2010;84:2955–62. doi: 10.1128/JVI.02257-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Scherer EM, Leaman DP, Zwick MB, et al. Aromatic residues at the edge of the antibody combining site facilitate viral glycoprotein recognition through membrane interactions. Proc Natl Acad Sci USA. 2010;107:1529–34. doi: 10.1073/pnas.0909680107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Pejchal R, Walker LM, Stanfield RL, et al. Structure and function of broadly reactive antibody PG16 reveal an H3 subdomain that mediates potent neutralization of HIV-1. Proc Natl Acad Sci USA. 2010;107:11483–11488. doi: 10.1073/pnas.1004600107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100•.Briney BS, Willis JR, Crowe JE., Jr Human peripheral blood antibodies with long HCDR3s are established primarily at original recombination using a limited subset of germline genes. PLoS One. 2012;7:e36750. doi: 10.1371/journal.pone.0036750. Important study using pyrosequencing to determine the frequencies of PBL B-cells in healthy individuals that express elongated HCDR3 regions like those found in the PG9, PG16, 2F5 and 4E10 BnAbs. This analysis also provides important genetic insight regarding the impact of both N-region addition and choice of D and J segments used to generate such specificities. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101•.Georgiev IS, Rudicell RS, Saunders KO, et al. Antibodies VRC01 and 10E8 Neutralize HIV-1 with High Breadth and Potency Even with Ig-Framework Regions Substantially Reverted to Germline. J Immunol. 2014;192:1100–1106. doi: 10.4049/jimmunol.1302515. Analysis of the minimal residues in framework regions (FRW) of two highly mutated BnAbs, 10E8 and VRCO1, required for neutralization function. This study found ~80% of 10E8 and 90% of VRC01 FRW-specific mutations reverted to germline are dispensable for neutralization breadth and potency. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Jardine JG, Sok D, Kalyuzhniy O, et al. Identifying the Minimal Mutations in VRC01 Required for Neutralization Potency and Breadth to Inform Vaccine Design. AIDS Res Human Retr. 2013;29:A14–A14. [Google Scholar]
- 103.Jelicic K, Cimbro R, Nawaz F, et al. The HIV-1 envelope protein gp120 impairs B cell proliferation by inducing TGF-β1 production and FcRL4 expression. Nat Immunol. 2013;14:1256–65. doi: 10.1038/ni.2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Levesque MC, Moody MA, Hwang KK, et al. Polyclonal B cell differentiation and loss of gastrointestinal tract germinal centers in the earliest stages of HIV-1 infection. PLoS Med. 2009;6:e1000107. doi: 10.1371/journal.pmed.1000107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105•.Corti D, Lanzavecchia A. Broadly neutralizing antiviral antibodies. Annu Rev Immunol. 2013;31:705–742. doi: 10.1146/annurev-immunol-032712-095916. Key recent review comparing characteristics and epitopes of HIV-1 BnAbs with broadly neutralizing antibodies specific for other highly variable viral pathogens. [DOI] [PubMed] [Google Scholar]
- 106.Daly J, Bebenek K, Watt DL, et al. Altered Ig hypermutation pattern and frequency in complementary mouse models of DNA polymerase ζ activity. J Immunol. 2012;188:5528–37. doi: 10.4049/jimmunol.1102629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107•.Jardine J, Julien J-P, Menis S, et al. Rational HIV immunogen design to target specific germline B cell receptors. Science. 2013;340:711–6. doi: 10.1126/science.1234150. This study describes an alternative variation of the approach proposed in (21••), involving structural/computational inference and in vitro library screening to engineer a synthetic immunogen capable of binding and activating B-cells expressing either germline reverted (unmutated) or mature (mutated) BCRs from CD4bs-specific BnAbs, this time those in the “VRCO1-class”. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108•.McGuire AT, Hoot S, Dreyer AM, et al. Engineering HIV envelope protein to activate germline B cell receptors of broadly neutralizing anti-CD4 binding site antibodies. J Exp Med. 2013;210:655–63. doi: 10.1084/jem.20122824. Similar approach to the one described for Ref [105•], except targeted mutation, involving selected removal of an N-glycosylation site, was used instead, to select for immunogens that could bind/activate B-cells expressing germline reverted “VRCO1-class” CD4bs-specific BnAbs as BCRs. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Alam SM, Liao HX, Dennison SM, et al. Differential reactivity germ line allelic variants of a broadly neutralizing HIV-1 antibody to a gp41 fusion intermediate conformation. J Virol. 2011;85:11725–11731. doi: 10.1128/JVI.05680-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Zhang J, Alam SM, Bouton-Verville H, et al. Modulation of non-neutralizing gp41 responses by an MHC-restricted TH epitope overlapping those of MPER broad neutralizing antibodies. J Immunol. 2014 doi: 10.4049/jimmunol.1302511. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]