Abstract
Mutations in the neurofibromatosis type 1 (NF1 tumor suppressor gene are common in cancer, and can cause resistance to therapy. Using transcriptome analysis we identified MAF as an NF1 regulated transcription factor, and verified MAF regulation through RAS/MAPK/AP-1 signaling in malignant peripheral nerve sheath tumor (MPNST) cell lines. MAF was also downregulated in human MPNST. Acute re-expression of MAF promoted expression of glial differentiation markers in MPNST cells in vitro, decreased self-renewal of embryonic precursors and transiently affected tumor cell phenotypes in vitro by increasing MPNST cell death and reducing metabolic activity and anchorage independent growth. Paradoxically, chronic MAF overexpression enhanced MPNST cell tumor growth in vivo, correlating with elevated pS6 in vitro and in vivo. RAD001 blocked MAF-mediated tumor growth, and MAF regulated the mTOR pathway through DEPTOR. MAPK inhibition with NF1 loss of function is predicted to show limited efficacy due to reactivation of mTOR signaling via MAF.
Keywords: Neurofibromatosis, MPNST, Schwann, resistance, DEPTOR, differentiation
INTRODUCTION
NF1 mutation or loss in humans causes Neurofibromatosis type 1 (NF1). NF1 predisposes affected individuals to develop benign nerve tumors (neurofibromas) that can transform into life threatening malignant sarcoma (MPNST). NF1 is a tumor suppressor gene, and biallelic NF1 mutations are characteristic of MPNST (1). MPNST are a leading cause of death in adult NF1 patients; the lifetime risk of MPNST in NF1 patients is 8–13%, versus 0.001% in the general population (2). Therapies that are effective in NF1 patients may be relevant to treating other diseases, because NF1 mutations are common in sporadic human cancers including glioma, neuroblastoma, lung adenocarcinoma, and squamous cell carcinoma (3–6). Furthermore, NF1 mutations have recently been shown to mediate resistance to therapy, and understanding how NF1 mutations cause resistance is a goal of current studies (7, 8).
NF1 is a GTPase activating protein (GAP); GAPs serve as off signals for Ras proteins so that patient MPNST cells lacking NF1 have elevated levels of Ras-GTP (9). Loss of neurofibromin alters growth and differentiation of MPNST cells through increased levels of Ras-GTP (2, 10, 11). Current efforts to develop therapies for MPNST are focused on Ras pathways, although no MPNST therapy has advanced to clinical practice. Ras signaling in MPNST cells includes activation of pERK and pAKT and pS6K and p4EBP1, downstream effectors of the mTOR kinase (10–12). MPNST cells transiently slow growth in response to MEK inhibition (13), and in response to compounds which block mTOR signaling (12, 14). Efforts to identify effective drug combinations for MPNST cells are ongoing (15).
The idea that cancer cells arise from and/or adopt the self-renewal and properties of precursor and stem-like cells is increasingly accepted (16, 17). Tumor initiating cells with stem cell properties are common in MPNST (18) and may derive from peripheral nerve Schwann cell lineage cells or their multipotent neural crest cell precursors. Nf1 regulates Schwann cell precursor cell numbers in embryonic dorsal root ganglia (19). Use of Cre-drivers for cell type specific Nf1 deletion in Schwann cell precursors enabled formation of MPNST, consistent with Schwann cell precursors as one cell of origin for MPNST (20, 21). MPNST may derive from or assume characteristics of neural crest cells as neural crest gene expression marks MPNST (22, 23). Transcriptome analysis identified SOX9, a neural crest transcription factor required for stem cell survival, as critical for MPNST cell survival (24) supporting the idea that loss or suppression of Schwan cell differentiation is characteristic of MPNST. However, the molecular mechanisms that underlie the failure of MPNST cells to differentiate into Schwann cell precursors and then Schwann cells are not known.
MAF (c-MAF) is a member of the large MAF family, which includes MafA, MafB and c-Maf 26). MAF transcription factors drive cell specification and differentiation in T cells, the lens and retina, and sensory neurons (26, 27). MAF is a bZip transcription factor of the AP-1 family. MAF factors homo- or heterodimerize with other bZip factors or other transcription factors to regulate gene expression (26, 28). In cartilage MAF binds SOX9, regulating common transcriptional target genes and controlling differentiation (29). MAF is expressed in the developing nervous system of the chicken, in mature rat peripheral nerve (26), and in mouse embryonic neurons (27), but its expression in developing glia has not been characterized. MAF can act as an oncogene (26), but can also counteract Ras-induced transformation (30). One MAF target gene implicated in cancer is DEPTOR, an mTOR interacting protein that negatively regulates TORC1 in multiple myeloma cells (31, 32).
We found that MAF expression is low in NF1 tumors and mouse Schwann cell precursors and hypothesized that low MAF expression contributes to maintenance of a dedifferentiated state in MPNST tumor cells. We report that elevating MAF expression in MPNST cells promotes differentiation and increases tumor growth in xenografts, correlating with a decrease in DEPTOR and elevated mTOR signaling, and rendering cells sensitive to mTOR antagonists.
RESULTS
The NF1 GTPase activating protein (GAP)-related domain (GRD) normalizes MAF expression
The NF1-GRD accelerates conversion of active Ras-GTP to inactive Ras-GDP. To define transcriptional changes downstream of NF1-GRD expression we conducted gene expression Affymetrix microarray analysis on triplicate samples of NF1-deficient MPNST cells (ST88-14 cells) 32 h after infection with NF1-GRD adenovirus or GFP control adenovirus. At 32h GRD expression significantly reduces Ras-GTP and downstream P-MEK and P-ERK (33), while cell death induced by exogenous high-level expression of NF1-GRD is not yet present (not shown).
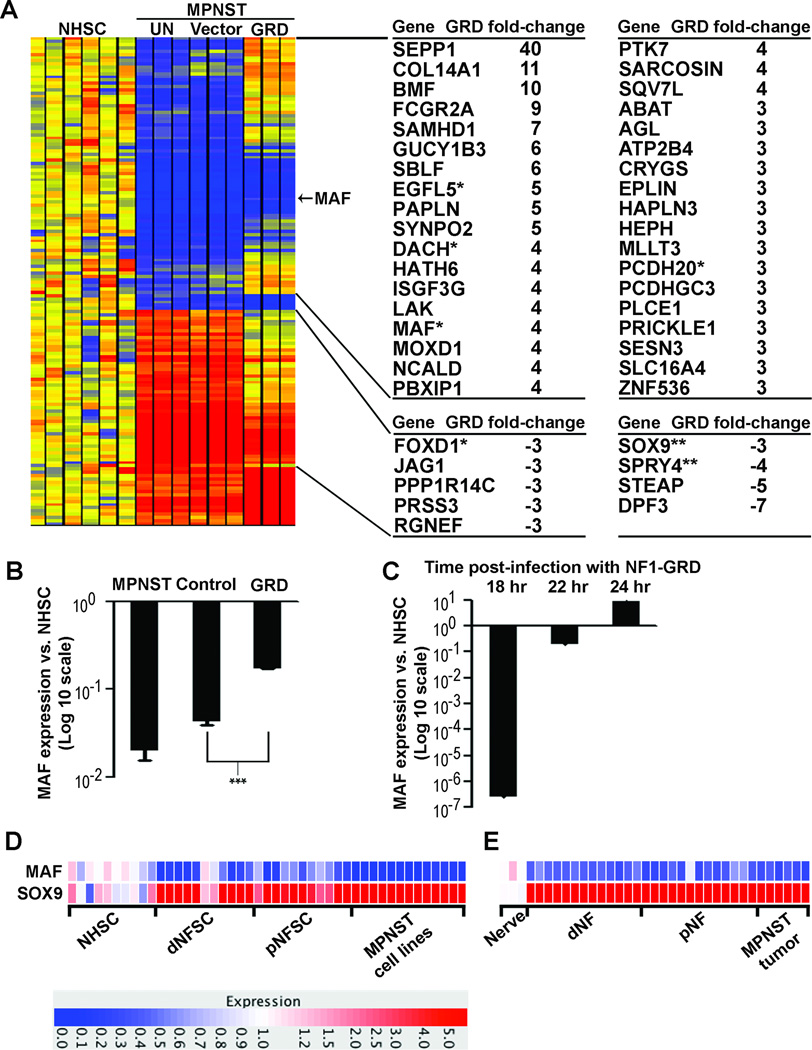
To identify genes with expression dysregulated in MPNST compared to human Schwann cells (NHSC) and normalized by the NF1-GRD, we first normalized gene expression in MPNST cells infected with NF1-GRD or control to gene expression in NHSC. Second, we identified 152 probe sets with differential expression ≥ 2.3-fold in MPNST cells expressing the NF1-GRD and not differentially expressed in GFP-expressing cells (Figure 1A). Third, we identified 128 of 152 probe sets with gene expression brought closer to levels in NHSC by NF1-GRD. Finally, we identified 45 known genes with expression restored to ≥ 3-fold levels of NHSC in MPNST cells infected with NF1-GRD relative to control. Microarray differential gene expression induced by GRD was confirmed by RT-PCR analysis for DACH1 (24), EGFL5, MAF, PCDH20, and FOXD1; SOX9 and SPRY4 were tested but failed to confirm, perhaps due to timing of transcriptional regulation.
Fig. 1. The NF1-GRD normalizes MAF expression in MPNST cells.
(A) Heatmap of gene expression microarray data comparing normal human Schwann cells (NHSC), ST8814 NF1 patient-derived malignant peripheral nerve sheath tumor (MPNST) cells (UN), ST8814 cells infected with an adenoviral vector expressing green fluorescent protein (Vector) or the NF1-GRD (GRD). Samples were analyzed in triplicate, and expression is shown relative to the mean gene expression in 6 NHSC samples. Yellow = normal; red = increased expression; blue = reduced expression. A list of 45 genes brought closer to levels in normal human Schwann cells (NHSC) by the NF1-GRD is displayed to the right of the heatmap. The top cluster includes 36 genes with expression downregulated at least 2.3-fold in MPNST relative to NHSC and increased at least 3-fold in MPNST cells expressing NF1-GRD relative to MPNST cells expressing GFP control, shown as positive GRD fold-change. The bottom cluster includes 9 genes with expression upregulated at least 2.3-fold in MPNST relative to NHSC and decreased at least 3-fold in MPNST cells expressing NF1-GRD relative to MPNST cells expressing GFP control, shown as negative GRD fold-change. * GRD regulation data confirmed by RT-PCR; ** GRD regulation not confirmed by RT-PCR. (B) Quantification of microarray expression data relative to NHSC, showing a trend toward normalization of MAF expression in MPNST cells expressing the NF1-GRD. Expression levels in MPNST cells infected with GFP control are not statistically different than uninfected MPNST cells; expression level differences in MPNST cells infected with NF1-GRD versus GFP control are statistically different (***; p = 0.001). (C) Time course RT-PCR analysis in MPNST cells at 18, 22, and 24 hours post-infection with NF1-GRD relative to NHSC, showing rapid induction of MAF expression by NF1-GRD. (D) Heatmap of gene expression microarray data comparing normal human Schwann cells (NHSCs), dermal (dNFSC) and plexiform (pNFSC) neurofibroma derived Schwann cells, and patient-derived malignant peripheral nerve sheath tumor cell lines (MPNST). Red = overexpression; blue = under-expression. (E) Heatmap of gene expression microarray data comparing normal human nerve, dermal (dNF) and plexiform (pNF) neurofibromas, and malignant peripheral nerve sheath tumors (MPNST). Red = overexpression; blue = under-expression.
In a previous study, MAF was in a cluster of genes significantly downregulated in primary tumors and cell lines derived from human MPNSTs and mouse MPNST models (13). Confirming these results, MAF expression was downregulated 24-fold in MPNST cells infected with control virus relative to NHSC, and increased 4-fold in MPNST cells expressing NF1-GRD (Figure 1B). We confirmed that MAF expression was induced rapidly in MPNST cells after infection with the NF1-GRD by RT-PCR (Fig 1C). Examination of dermal or plexiform neurofibroma- derived Schwann cells or MPNST cell lines, compared to normal human Schwann cells, or dermal and plexiform neurofibroma or MPNST solid tumors compared to nerve, underscore the consistently low or absent expression of MAF (Figure 1 D & E).
MAF is regulated by NF1 through a RAS/MAPK/AP-1 pathway
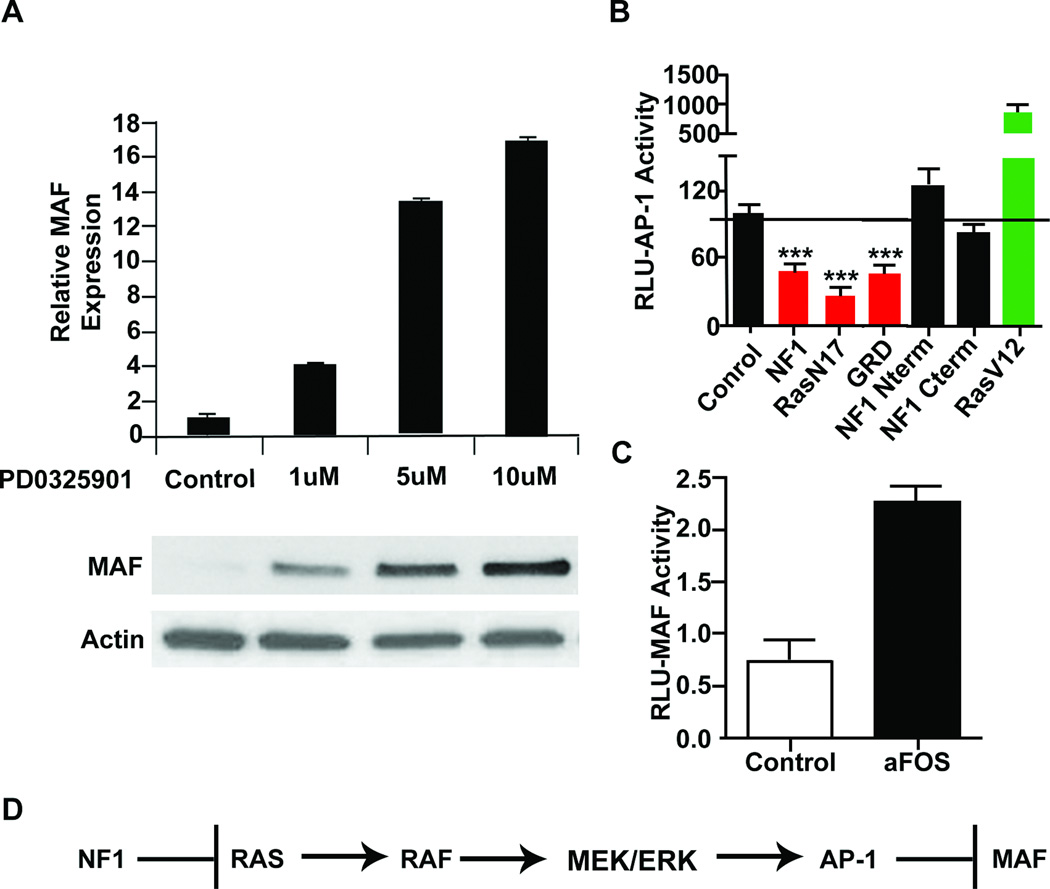
Increased MAPK signaling in MPNST cells is due to loss of NF1 RAS-Gap function (10, 11). Confirming that MAF is downstream of RAS-RAF-MAPK, MEK inhibitors restored MAF mRNA in MPNST cell lines and protein expression in S462TY cells (Figure 2A). AP-1 transcription factors are common effectors of the MAPK pathway (34) and MPNST cells have increased AP-1 activity (35). We tested whether NF1 regulates MAF via AP-1 activity. Transfecting MPNSTs with the GRD, dominant negative RasN17, or full length NF1 constructs decreased AP-1 luciferase reporter activity compared to the empty vector negative control, while constitutively active RasV12 activated AP-1 (Figure 2B). Mutant NF1 constructs lacking the GRD did not repress AP-1 activity, consistent with AP-1 regulation by RAS signaling (Figure 2B). Importantly, a dominant negative aFos (36) transfected into MPNST cell lines along with a MAF promoter luciferase reporter increased MAF activity, indicating that AP-1 represses MAF transcription in MPNST cells (Figure 2C) (37). Thus MAF is a novel NF1 target in MPNST, repressed by the RAS/MAPK/AP-1 pathway (Figure 2D).
Fig. 2. MAF is regulated by NF1 through a RAS/MAPK/AP-1 pathway.
(A) RT-PCR analysis showing mRNA expression of S462TY MPNST cells treated with the MEK inhibitor PD0325901 or vehicle control for 24 hours, relative to β-actin, with corresponding protein levels below. (B) Luciferase reporter assays for AP-1 response after transfecting full length human NF1 (NF1), dominant negative H-Ras (RasN17), the NF1 gap-related domain (GRD), an N-terminal NF1 fragment (NF1 N-term), a C-terminal NF1 fragment (NF1 c-term), or active RasG12V (RasV12) into MPNST cells. (C) Luciferase reporter assay for MAF response to dominant negative aFOS transfected into MPNST cell lines. (D) Model showing that NF1 loss through Ras activation promotes ERK activation and AP-1, thereby suppressing MAF expression.
MAF regulates glial differentiation markers and neural crest marker SOX9 in MPNST cells
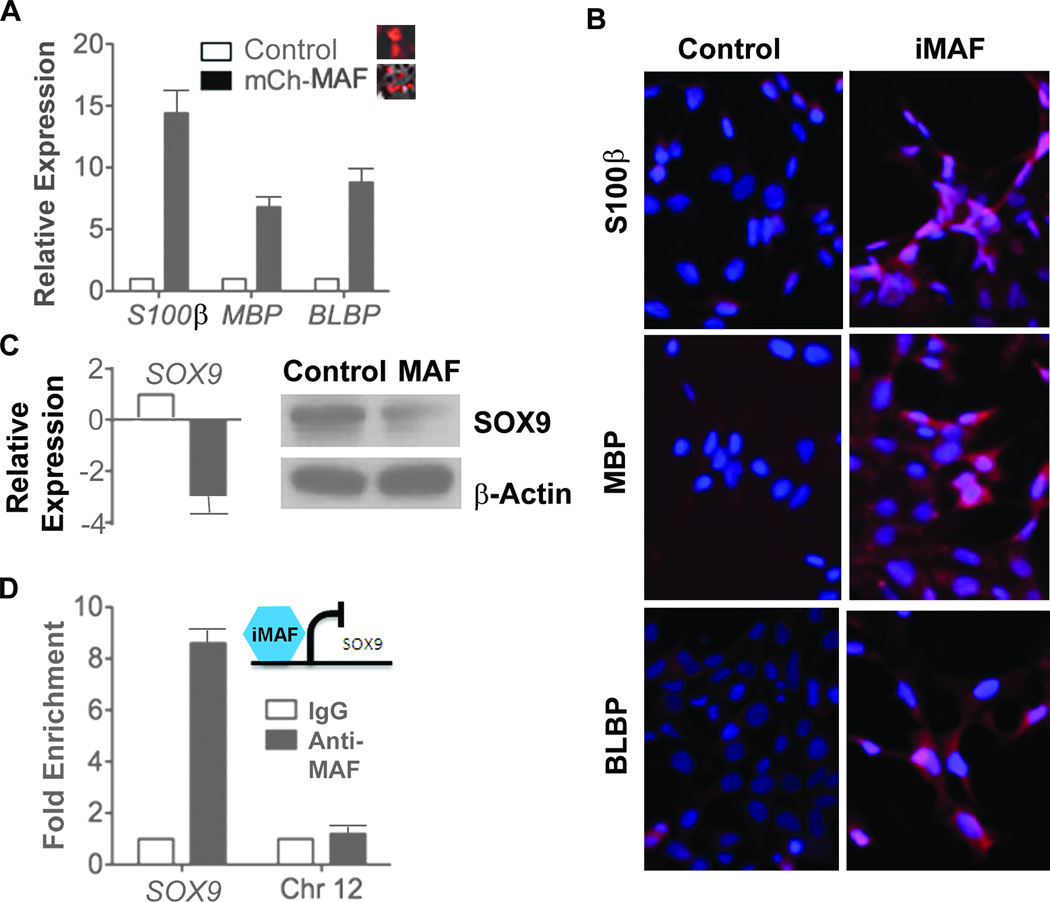
MAF can bind the MPNST biomarker SOX9 (29), and MAF is known to promote tissue specification and terminal differentiation in many cell types (26, 27, 29, 38). We explored whether low MAF expression was relevant to the failure of MPNST cells differentiation. RT-PCR revealed upregulation of glial lineage markers S100β, MBP, and BLBP in MPNST cells after transduction with MAF (Figure 3A). Immunohistochemical staining verified increased expression of these proteins in MAF transduced MPNST cells (Figure 3B). Because the neural crest marker gene SOX9 has MAF binding sites in its promoter (data not shown), we tested whether restoring MAF expression downregulates SOX9. RT-PCR and western bot analysis revealed decreased SOX9 mRNA and SOX9 protein when MAF was overexpressed (Figure 3C). Chromatin immunoprecipitation analysis showed MAF occupancy of the SOX9 promoter (Figure 3D), suggesting that MAF transcriptionally represses SOX9 in MPNST.
Fig. 3. MAF regulates glial differentiation markers and the neural crest marker SOX9.
(A) RT-PCR analysis of S462TY MPNST cells transduced with mCherry tagged MAF or mCherry control for 48 hours. mRNA levels are expressed relative to β-actin. (B) Immohistochemistry shows expression of differentiation markers (S100β, MBP, and BLBP) in MAF transduced MPNST cells. (C) RT-PCR and western blot analysis showing relative expression of SOX9 in MAF transduced MPNST cells before (white bar) and after (black bar) induction with doxycycline for 48 hours. (D) ChIP analysis of MAF occupancy of the SOX9 promoter.
In development, MAF expression correlates with Schwann cell differentiation status and reduces proliferation of Schwann cell precursors
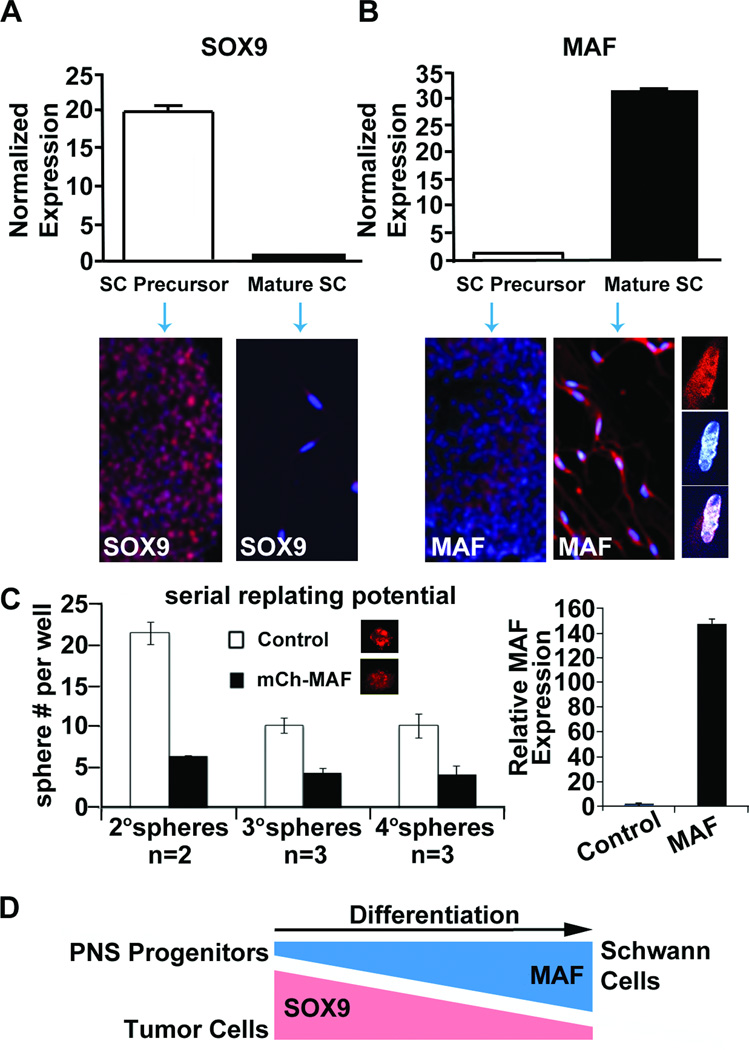
To determine if MAF expression is relevant to normal glial differentiation, we monitored expression levels in mouse Schwann cell precursors derived from mouse embryonic E12.5 dorsal root ganglia and in mature Schwann cells. RT-PCR and immunohistochemical analysis revealed high SOX9 expression in Schwann cell precursors normalized to Schwann cells (Figure 4A), and low MAF expression in Schwann cell precursors compared to Schwann cells (Figure 4B). Confocal imaging showed that MAF is present in Schwann cell cytoplasm and nuclei (Figure 4B). To determine if low MAF expression contributes to the precursor state, we transduced mouse Schwann cell precursors at clonal density with lenti-MAF. We observed a decrease in Schwann cell precursor self-renewal, which was maintained for up to three serial replatings (Figure 4C). The reciprocal expression pattern of SOX9 and MAF compared to mature Schwann cells, together with MAF’s ability to decrease precursor self-renewal, is consistent with the idea that low MAF expression in SC precursors sustains them in a progenitor state (Figure 4D).
Fig. 4. MAF expression increases with Schwann cell differentiation and MAF over-expression reduces formation of Schwann cell precursors.
(A & B) RT-PCR analysis of mouse Schwann cell precursors and mature Schwann cells showing mRNA expression relative to Gapdh and normalized to mature Schwann cells for Sox9 or Schwann cell precursors for Maf. Immunohistochemical staining of each cell type is displayed below the RT-PCR panels for Sox9 or Maf protein expression. Note partial nuclear localization (DAPI, blue) of MAF (red) in unmerged and merged higher confocal images at right. (C) mCherry tagged MAF infected Schwann cell precursors show reduced sphere forming potential in a self-renewal assay compared to mCherry infected control cells. Maf expression in 4° spheres was verified by RT-PCR (right). (D) Model showing that Maf expression increases as cells differentiate, while Sox9 expression decreases.
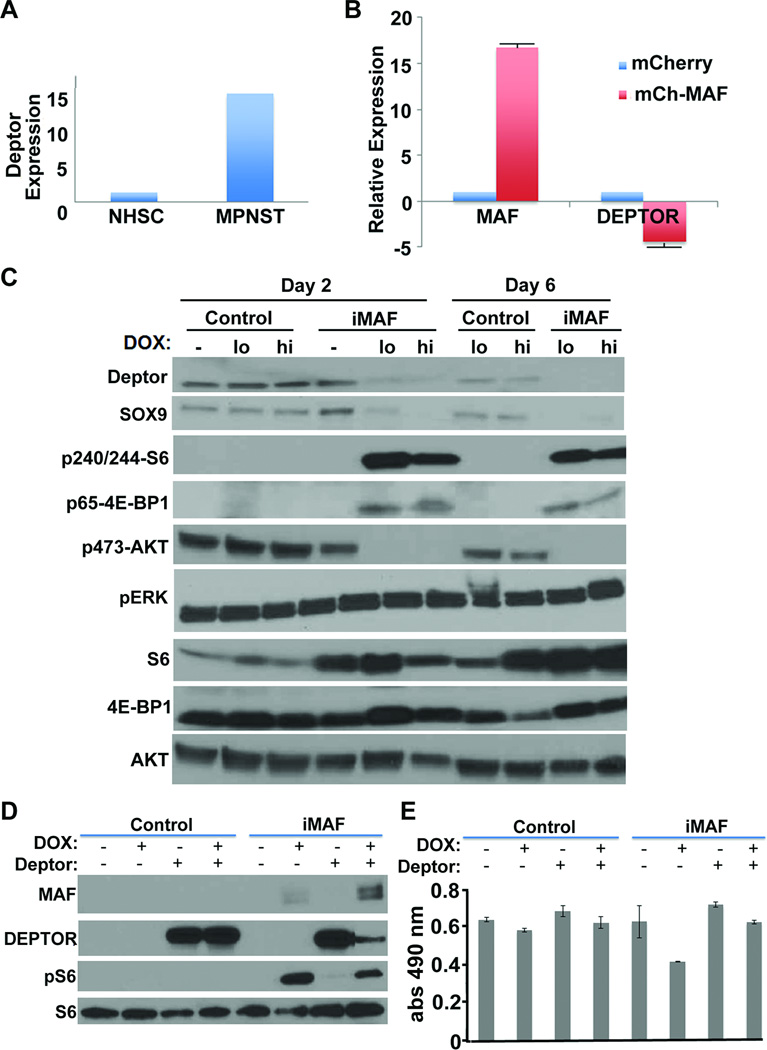
Acute over-expression of MAF in MPNST cells transiently increases cell death and reduces anchorage independent growth, while sustained lower MAF expressing cells proliferate and maintain increased TORC1 signaling through DEPTOR
If MAF promotes cell differentiation, then restoring MAF expression in MPNST cells might affect tumorigenic potential. S462-TY cells transduced with MAF for 48 hours significantly decreased metabolic activity as measured by MTS assay (Figure 5A). Flow cytometric analysis of PI and Annexin stained S462-TY cells transduced with MAF for 48 hours revealed increased apoptotic cell death (Figure 5B). Importantly, these cytostatic and cytotoxic effects were accompanied by decreased anchorage independent growth in soft agar in S462-TY cells overexpressing MAF constitutively or inducibly (Figure 5C). However, after these transient effects, MAF expressing cells were able to proliferate in culture, as confirmed by increased metabolic activity in MTS assays (Figure 5D) and BrdU incorporation (Figure 5E). Relative levels of MAF protein expression were higher 2 days after induction than at day 6, correlating with increased apoptosis evidenced by cleaved caspase 3 (Figure 5F). Low MAF levels by day 6 corresponded with increased proliferation. Thus, effects of MAF expression on MPNST cells are dose dependent.
Fig. 5. Acute over-expression of MAF in MPNST cells transiently increases cell death and reduces anchorage independent growth, while sustained lower MAF expressing cells proliferate and maintain increased TORC1 signaling through DEPTOR.
(A) Short term MTS assay on S462-TY cells transduced with mCherry tagged MAF for 48 hours. (B) Flow cytometry on PI and Annexin stained S462-TY cells transduced with inducible MAF 48 hours after doxycycline induction. (C) Anchorage independent growth of S462-TY cells assessed by colony formation in soft agar after overexpression of MAF using mCherry tagged MAF (mCh-MAF) or inducible MAF (iMAF). (D) Longer time course MTS assay on S462-TY cells transduced with inducible MAF showing recovery of cell growth by 6 days. (E) Flow analysis of BrdU incorporation in doxycycline induced pLVX (empty vector control) and iMAF transduced MPNST cells. (F) Western blot analysis of pLVX (empty vector control) or iMAF transduced S462-TY cells comparing uninduced, low (0.2 ug/ml) and high (2 ug/ml) doxycycline induced cells for 2 or 6 days, showing cell death (cleaved caspase 3) is transient and correlates with high levels of MAF expression.
As MAF can act as an oncogene in some cell types, we studied what survival mechanism(s) the MAF expressing population might exploit after initial growth suppression. We examined gene expression changes in NF1 tumors and cell lines compared to human Schwann cells from our Affymetrix array analysis (24). DEPTOR mRNA was 15-fold up-regulated in MPNST tumors and cell lines versus normal human Schwann cells (Figure 6A). DEPTOR is a MAF target and negative regulator of the AKT/mTOR pathway (31). Restoring MAF to S462-TY cells reduced DEPTOR mRNA expression (Figure 6B). Western blot analysis of S462-TY cells transduced with inducible MAF showed increased phosphorylation of S6 and 4E-BP1 and decreased phosphorylation of AKT (Figure 6C), readouts of TORC1 activity and its negative feedback onto AKT (39). Overexpression of MAF did not change levels of pERK, consistent with MAF acting downstream of MAPK signaling. Importantly, overexpression of MAF, plus simultaneous DEPTOR overexpression, attenuated DEPTOR levels and pS6 (Figure 6D), and DEPTOR blocked MAF-induced decreased cell survival in MTS assay (Figure 6E). These data suggest that MAF regulates AKT/mTOR signaling through DEPTOR.
Figure 6. Inducing MAF expression modulates DEPTOR.
(A) Relative expression of DEPTOR from gene expression array analysis comparing MPNST and NHSC. (B) RT-PCR analysis of DEPTOR expression in S462-TY cells after overexpression of MAF. (C) Western blot analysis (reprobes of blots from 5F) of pLVX empty vector (control) or iMAF transduced S462-TY cells comparing uninduced, low (0.2 ug/ml) and high (hi; 2 ug/ml) doxycycline induced cells for 2 or 6 days. Downstream of MAF expression, phosphorylation of the mTOR substrates S6 and 4EBP1 are elevated while P-AKT is reduced. (D) Western blot analysis of S462-TY cells showing induction of MAF with and without DEPTOR overexpression. Overexpression of MAF attenuated DEPTOR levels and DEPTOR reduced pS6. (E) MTS assay showing that the decreased cell numbers caused by MAF are rescued by co-expression of DEPTOR.
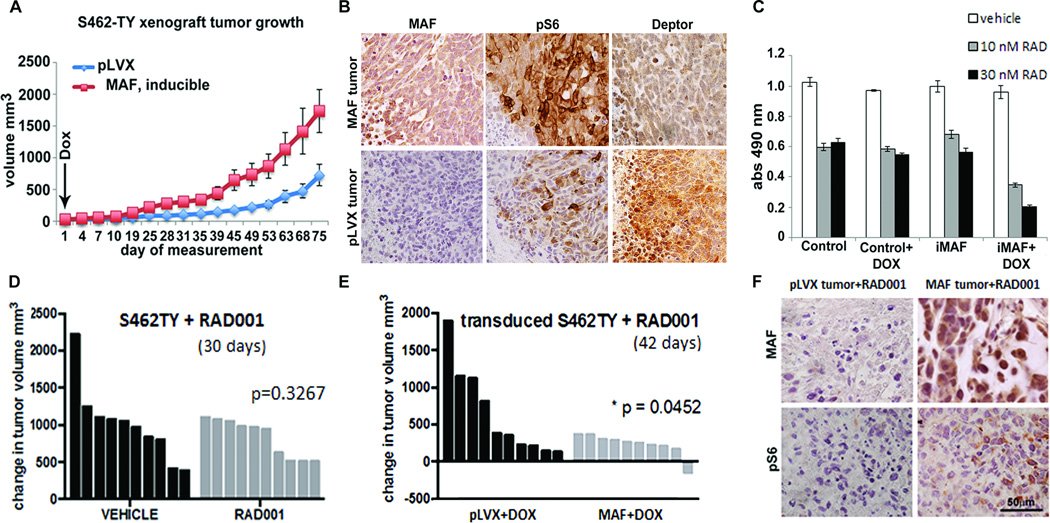
MAF enhances tumor growth in a xenograft model, and enhances response to RAD001
To test whether MAF-induced increased TORC1 activity affects tumorigenesis in vivo, we injected BALB/cnu/nu mice with S462-TY cells transduced with doxycycline-inducible MAF or vector-control lentivirus. Tumors overexpressing MAF were larger than vector control tumors (Figure 7A), consistent with MAF regulating an escape mechanism, conferring a survival advantage. IHC analysis of tumor xenografts confirmed crosstalk to the mTOR pathway seen in vitro. P-S6 increased and DEPTOR decreased in MAF expressing tumors (Figure 7B). Levels of pS6 and DEPTOR varied throughout the tumors and were most pronounced in regions of densely packed growing tumor cells.
Fig. 7. MAF enhances tumor growth in a xenograft model, and enhances response to RAD001.
(A) Growth curve shows average tumor volumes from S462-TY cells infected with vector (pLVX) or inducible MAF and injected into the right flanks of Balb/c nude mice (n=8 – 10 per group). Doxycycline (Dox) was administered after cell implantation. (B) Immunohistochemical staining of pLVX vector control and MAF expressing tumor paraffin sections for MAF, pS6 and DEPTOR. Brown indicates DAB reaction product.(C) Four day MTS assay using S462TY MPNST cells, comparing control and doxycycline inducible MAF (iMAF) expression with exposure to RAD001 at 10 or 30nM, showing enhanced drug effect with MAF expression. (D) Waterfall plot of change in tumor volume of S462-TY xenografts during treatment with either vehicle control or 10 mg/kg RAD001 showing absence of significant response of this MPNST cell line to RAD001 in vivo. (E) Waterfall plot of significant (p = 0.05) change in tumor volume of S462-TY xenografts transduced with inducible MAF (MAF) when 10 mg/kg RAD001 is also present. (F) Immunohistochemical staining of pLVX vector control and MAF expressing tumor paraffin sections with anti-MAF and anti-pS6K. Reaction product is brown.
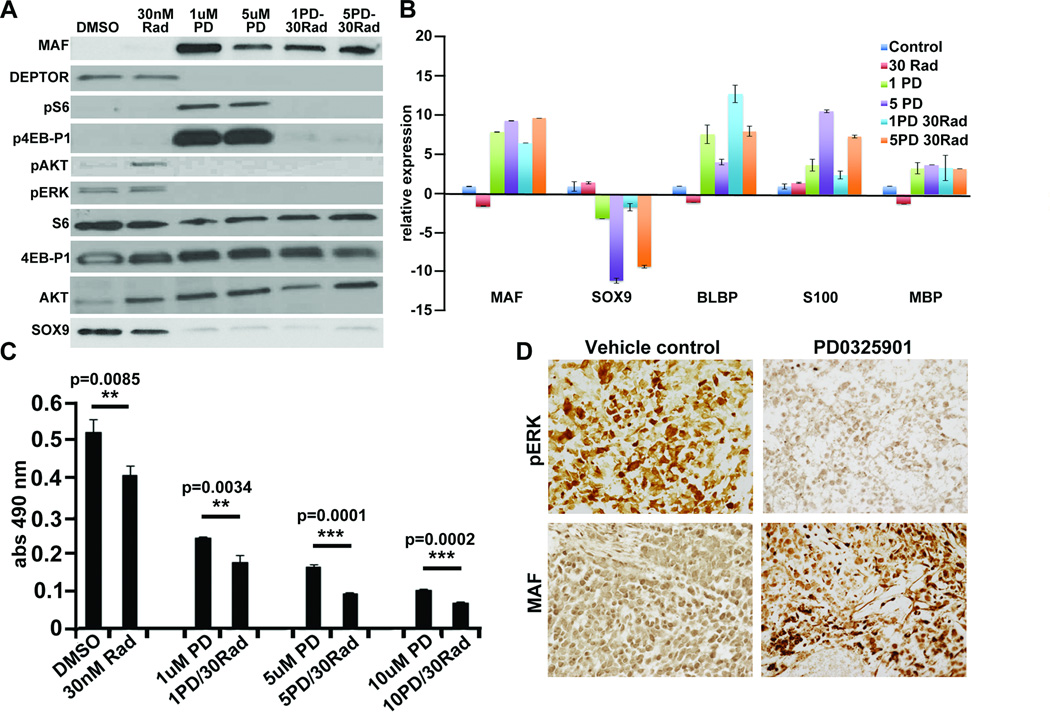
Since MAF regulated changes in DEPTOR affected TORC1 signaling, we reasoned that overexpressing MAF might enhance sensitivity to mTOR pathway inhibition. Normally, S462TY cells are poorly responsive to RAD001. However, the combination of treatment with 10 or 30 nM RAD001 in addition to MAF overexpression resulted in an enhanced response (Figure 7C). In xenograft models, the S462TY cells are also not significantly responsive to treatment with 10mg/kg RAD001 (Figure 7D), but a significant decrease in tumor volume compared to the vector control tumors was seen in combination with MAF overexpression (Figure 7E). IHC analysis confirmed a decrease in pS6 staining due to RAD001 inhibition of TORC1 activity in vector control and MAF expressing tumors (Figure 7F). Treatment with PD0325901 increased MAF expression (Figure 2A; 8A), decreased DEPTOR, increased pS6 and p4E-BP1 and decreased P-AKT (Figure 8A). The combination of PD0325901 with RAD001 abolished changes in pS6 and p4E-BP1 (Figure 8A). PD0325901 decreased mRNA and protein encoding the neural crest marker SOX9 (Figure 8A,B) and increased mRNA encoding Schwann cell differentiation markers BLBP/FABP7, S100β and MBP; RAD001 had no significant effects on cell differentiation. S462TY cells are resistant to RAD001 in MTS assays (Figure 8C), but responded to PD0325901 at 1uM (13). Combinations of these inhibitors had additive effects on cell viability (Figure 8C). S462-TY cells xenografted into immune-compromised mice delayed tumor growth when mice were exposed to 10mg/kg/day PD0325901 (13). We analyzed sections of tumor xenografts from mice treated with PD0325901 for 11 days. Tumors responding to drug (n=3) had little or no MAF expression, while faster growing tumors (n=2) showed MAF positivity (Figure 8D).
Figure 8. MEK inhibition recapitulates effects of MAF in MPNST cells.
(A) Western blot analysis of S462-TY cells treated with RAD001, PD0325901, or a combination showing downstream effects of restored MAF expression or drug treatment on DEPTOR and downstream signaling, and SOX9 expression. (B) RT-PCR analysis showing changes in neural crest marker SOX9 and differentiation factor mRNAs on exposure to MEK inhibition. (C) MTS assay of RAD001 or PD0325901 treated MPNST cells showing decreased proliferation with drug treatment, especially in response to the combination. (D) Immunohistochemical staining of vehicle or PD0325901 treated S462-TY xenograft tumors. Staining with anti-pERK and anti-MAF showing that PD0325901 resistant tumors show MAF expression. Reaction product is brown.
DISCUSSION
We identified 45 genes, including MAF, as aberrantly expressed in MPNST cells and normalized by expressing the NF1-GRD. We confirmed that MAF expression is low in MPNST cells and tumors, and that MAF expression is acutely regulated by NF1 through Ras/MAPK/AP1 signaling. High levels of MAF increased expression of Schwann cell differentiation markers in MPNST cells and killed MPNST cells in vitro, but lower levels of MAF expression were tolerated and correlated with enhanced growth in tumor xenografts. MAF re-expression in the setting of NF1 tumorigenesis resulted in elevation of mTOR signaling and concomitant decrease in DEPTOR, a negative regulator of the AKT/mTOR pathway (31). We conclude that MAF controls crosstalk between the MAPK and mTOR pathways in MPNST cells, helping to explain how NF1 loss confers resistance to therapies. In addition to these findings relevant to tumorigenesis our data support the idea that NF1 and MAF control self-renewal in Schwann cell precursors. The data suggest that MAF expression regulates cell phenotypes in a context dependent manner: MAF blocked Schwann cell precursor self-renewal, promoted MPNST cell differentiation, and paradoxically promoted tumorigenesis.
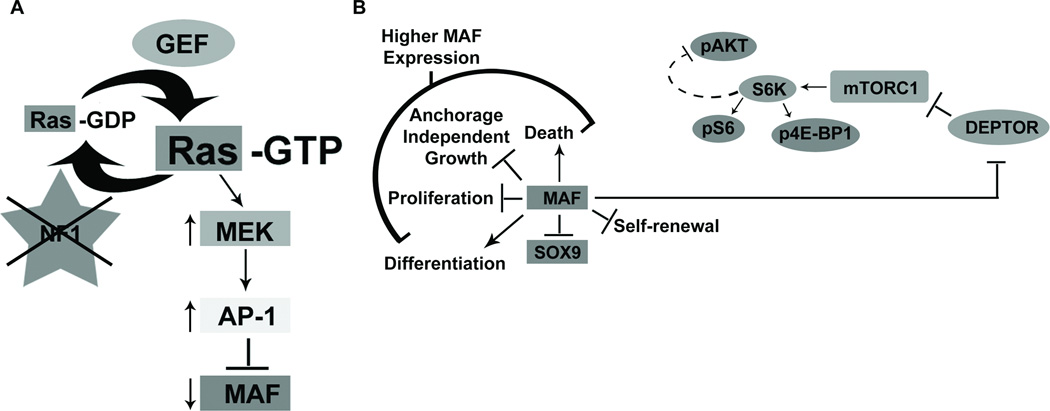
Our data identify MAF as an NF1-regulated target gene downstream of the RAS effector pathway RAS/MAPK/AP-1 (Figure 9A). To our knowledge, MAF is the first cancer relevant NF1 target identified. There is previous evidence supporting the regulation of AP-1 activity by NF1 through a MAPK pathway in the ST88-14 MPNST cell line (35) and MAF was previously reported to be regulated by a MEK-ERK-FOS pathway in myeloma cell lines (40). This suggests that NF1 regulation of MAF may be relevant to other cell types and other cancers.
Fig. 9. NF1 mediated control of Ras-MAPK signaling: cross-talk with mTOR signaling through DEPTOR.
(A) In the absence of NF1, MAF expression is suppressed through a RAS/MAPK/AP-1 pathway. The transient effects of high MAF re-expression include an increase in cell death and decreases in SOX9, proliferation, and anchorage independent growth. (B) MAF also regulates DEPTOR, which negatively regulates TORC1 activity. TORC1 activity can be read-out by the activity of its substrates, such as the phosphorylation of S6 by S6K. In turn, S6K activity causes negative feedback resulting in the decreased phosphorylation of AKT. Our studies suggest that sustained lower levels of MAF expression in vivo increase TORC1 activity and enhance tumor growth. This enhanced TORC1 signaling renders tumors vulnerable to TORC1 inhibition with RAD001.
A recent in vitro study confirmed our original finding that MAF expression is low in ST88-14 NF1 mutant MPNST cells relative to normal human Schwann cells (24), and that SOX9 is elevated (41). MAF expression is also reduced in prostate cancer (42). We found that levels of MAF are very low in all tested MPNST cell lines, including sporadic MPNST cells (24), possibly accounting for the fact that siRNA targeting NF1 in a sporadic (non-NF1 mutant) MPNST cell line did not alter MAF expression (41). Our finding that the NF1-GRD regulates MAF expression implies that activation of one or more Ras proteins suppresses MAF. In NF1 MPNST cells, MAF was not normalized by NRAS siRNA (41); other Ras proteins (e.g. K- or H-Ras) may regulate MAF expression.
MAF expression suppressed expression of the neural crest marker SOX9 (Figure 3 A, B &C, Figure 7A). Consistent with overexpression of MAF transiently lowering SOX9 expression and resulting in cell death, knockdown of SOX9 killed MPNST cells (24). As MAF binds the SOX9 promoter, the effect on SOX9 is most likely direct. It is possible that MAF competes with SOX9 for binding sites and/or that MAF partners with SOX9 to regulate transcription, as in cartilage (29). MAF also upregulated expression of glial differentiation markers BLBP, MBP, and S100β; it remains to be determined whether these effects are direct or indirect. Downregulation of MAF and Schwann cell differentiation markers is characteristic of, and thus likely relevant to, both sporadic and NF1 MPNST; notably, both cell types exhibit activation of the MAPK pathway (43).
Our studies support a role for MAF in suppressing Schwann cell precursor self-renewal. It will be essential to confirm this MAF role by loss of function in vivo. Roles for MAF in tissue specification and cell differentiation are also supported by studies in T cells, lens, retina, and endochondral bone. For example, MAF maintains cell quiescence in the lens (26). MAF showed a pro-differentiation role in MPNST cells and reduced anchorage independent growth in MPNST cell lines expressing MAF (Figure 7A), suggesting that MAF can act as a tumor suppressor in MPNST. A tumor suppressive role for MAF is supported findings that MafA counteracts the transforming properties of oncogenic RasV12 and B-Raf /MEK in neuroretina cells (30). MAF may have tumor suppressive roles through regulating p53-dependent cell death, inhibition of MYB, and/or induction of the cell cycle inhibitor p27 (26).
However, low levels of MAF expression correlated with proliferation in vitro and increased tumor growth in vivo, implying that MAF can act as an oncogene in the context of NF1 loss. MAF can transform fibroblasts, is activated by translocation in multiple myeloma, and is overexpressed in angioimmunoblastic T-cell lymphomas (26). The difference between acute MAF expression in vitro and our in vivo studies may be explained by levels of MAF expression: high MAF expressing cells die, and are survived by cells expressing lower levels of MAF with sustained expression of pS6. Possibly MAF interacts with different partner transcription factors depending on expression level, altering target gene expression. For example, increased expression of the MAF target gene integrinβ7 allowed myeloma cancer cells to adhere more efficiently to bone marrow stroma cells, mediating adaptation to the tumor microenvironment (26).
MAF regulates DEPTOR and represses mTOR signaling in multiple myeloma (31). We demonstrated that MAF also regulates DEPTOR, but in MPNST cells MAF expression increases TORC1 activity (Figure 7B). The AKT/mTOR pathway is activated in some MPNSTs, and enhanced activation of the pathway correlated with malignant progression (43). In MPNST cells the pro-survival effects of TORC1 (39) are likely to explain the tumor promoting effect caused by overexpressing MAF in MPNST xenografts, as the ineffectiveness of RAD001 treatment alone in S462TY MPNST cells was reversed when combined with MAF expression (Figure 6F).
Due to NF1 loss in MPNST, Ras-GTP is activated and inhibition of MAPK and mTOR pathways have been evaluated in MPNST preclinical studies (2, 12–14, 43–45); as single agents, no tested inhibitor caused tumor regression, revealing the need for combinatorial approaches in MPNST. Our finding that MAF expression and downstream effects are regulated by MAPK signaling in MPNST (Figures 2A; 8A,B) and in multiple myeloma (40) and that MAF expression enhances responses to Rapalogs (Figures 7C,E,F; 8C) provides further rationale for dual inhibition of the MAPK and AKT/mTOR pathways (7). Our results are consistent with a study in which DEPTOR accumulated in breast cancer cells on knockdown of its ubiquitin ligase, resulting in resistance to rapamycin and high pAKT in vitro (46). Examining PD0325901 treated xenografts revealed that less responsive tumors showed MAF positivity, suggesting that resistance may be in part due to the effects of MAF related crosstalk (Figure 8D).
Crosstalk between the MAPK and AKT/mTOR pathways is increasingly well documented. MEK inhibitors induce epidermal growth factor-induced AKT activation, ERK phosphorylates TSC2 and RAPTOR to promote TORC1 activity, and AKT can phosphorylate Raf at inhibitory sites that negatively regulate MEK activity (39). Further, drug studies in breast, melanoma, and colon cancer patient samples and a mouse model of prostate cancer showed MAPK activation after RAD001 treatment through a PI3K-dependent feedback loop (47). The reliance on both pathways in many cancer types, including NF1-deficient AML cell lines which became cytarabine sensitive when the GRD was restored or MEK or mTOR inhibitors were used (48), underscores the need for combinatorial therapeutic approaches.
In summary, we identified crosstalk between the MAPK and AKT/mTOR pathways through MAF and DEPTOR in NF1 mutant cells (Figure 9B). Strategies to upregulate MAF and/or downregulate DEPTOR in NF1 mutant cells, including MEK inhibition, may enhance response to well-tolerated rapalogs.
Materials & Methods
Microarray analyses
Collection of samples, RNA isolation, probe generation, Affymetrix HU133 Plus 2.0 microarray hybridization and data normalization were described (24). To compare MPNST cells infected with adenovirus, we normalized gene expression to average expression in three independent NHSC samples. Statistical analyses (analysis of variance; FDR=0.001) identified gene expression changes between uninfected MPNST cells, MPNST cells infected with control (GFP) virus and MPNST cells infected with NF1-GRD virus as described (33).
Cell culture
Normal human Schwann cells (NHSC) from trauma victims and MPNST cell lines were cultured as described (24). For cells transduced with doxycycline inducible lentivirus, we used certified Tet-free FBS (Hyclone).
Viral Infection
For microarray analysis, MPNST cells were infected with purified Ad-NF1-GRD or control Ad-GFP, in triplicate, for 2 h in serum-free medium at 200pfu/cell. Viruses were removed and cells cultured in normal medium for 30h, then RNA isolated for analysis of gene expression by RT–PCR analysis as described (24).
MPNST cells were transduced with lentiviral particles at 70 – 90% confluence. Constitutive expression used mCherry tagged MAF (EX-Y2046-Lv111, GeneCopoeia) and mCherry control lentivirus. Inducible expression used lentiviral vectors containing the MAF ORF or empty pLVX-Tight-Puro vector (GeneCopoeia) in a modified pTight backbone (Clonetech) with pLVX-Tet-On doxycycline inducible vector (Clonetech). CCHMC Viral Vector Core produced virus with 4-plasmid packaging http://www.cincinnatichildrens.org/research/div/exphematology/translational/vpf/vvc/default.htm. Lentiviral particles were incubated with MPNST cells (MOI of 10) in 8 µg/mL polybrene (Sigma) for 24h followed by selection in 2 µg/mL puromycin, which killed uninfected cells within three days.
Luciferase reporter assays
MPNST cells (5×104/ well) in 96-well plates were serum starved (0.1 % FBS) overnight. Cells were transfected using FuGENE HD (Roche) with 100ng per well of an AP-1 luciferase (Stratagene) reporter vector, 5ng of a renilla luciferase reference control, and 50ng of plasmid DNA expressing full length NF1, N-term and C-term constructs lacking the GRD (gifts of Frank McCormick), RasN17, the GRD, RasV12, aFos, and empty control (49). Cells were stimulated in 10% FBS for 24 h and lysed for luciferase assay (Promega). The MAF reporter was described (37).
Realtime-PCR
Total RNA was isolated from cells using the RNeasy kit (Qiagen) and used as a template for cDNA synthesis (ABI High capacity archive kit) and RT-PCR (ABI 7500 Sequence Detection System) as described (24). Data were normalized to β-Actin or Gapdh. Human primers included: MAF f-cgagtgggctcagttatgaa and r-cgagtgggctcagttatgaa, SOX9 f-gtaatccgggtggtccttct and r-gacgctgggcaagctct, S100β f-tccacaacctcctgctcttt and r-ggagacaagcacaagctgaa, MBP f-ggagccgtagtgagcagttc and r-gagccctctgccctctcat, BLBP f-aacagcaaccacatcaccaa and r-acagaaatgggatggcaaag, DEPTOR f-cgacaaaacagtttgggtagg and r-atggctgaggtcttggtcac, β-Actin f-gttgtcgacgacgagcg and r-gcacagagcctcgcctt. Mouse primers included: Maf f-tctcctgcttgaggtggtct and r-aaggaggaggtgatccgact, Sox9 f-tccacgaagggtctcttctc and r-aggaagctggcagaccagta, Gapdh f-ttgatggcaacaatctccac and r-cgtcccgtagacaaaatggt.
Immunocytochemistry
Cultured cells were fixed, permeabilized with 0.2% Triton-X 100 and blocked with 10% normal serum for 1h at room temperature (19). Primary antibodies used were S100β (Dako), MBP (Chemicon), and BLBP (Millipore). Secondary incubations used host-appropriate TRITC conjugated antibodies (Jackson Immunoresearch). Nuclei were labeled with DAPI (Sigma-Aldrich) and microscopic images acquired with ImageJ software (NIH) on a Zeiss Axiovert 200M. Tumor paraffin sections stained for MAF (Imgenex), pS6, pERK (Cell Signaling), and DEPTOR (Novus) were visualized with DAB.
Immunoblot
Cell lysates were created and Western blotting conducted as described (24), blocking without detergent. Membranes were probed with antibodies for MAF (Imgenex), SOX9 (Santa Cruz), S100β (Dako), MBP (Chemicon), BLBP (Millipore), DEPTOR (Novus), cleaved-caspase 3, pS6, S6, p4E-BP1, 4E-BP1, p473AKT, AKT, and β-ACTIN (Cell Signaling) as a loading control. Horseradish peroxidase-conjugated secondary antibodies (BioRad) were used with ECL Plus developing system (Amersham Biosciences).
Chromatin immunoprecipitation (ChIP)
MPNST 88-14 cells were cross-linked with 1% formaldehyde for 10 minutes on ice. Glycine was added to 0.125 M. Cells were pelleted by centrifugation and washed 3X with ice-cold PBS with 1× Complete inhibitor (Roche). Cells were lysed 1% SDS, 50 mm Tris-HCl, pH 8.1, 10 mm EDTA, 1× Complete inhibitor. A Bioruptor generated soluble chromatin (Cosmo Bio USA, Carlsbad, CA) with DNA fragment sizes of 200 to 800 bp. We precleared cell lysates with protein A/G-Sepharose beads (Santa Cruz). Anti-MAF (Imgenex) and control mouse IgG (Santa Cruz) immunoprecipitated chromatin-protein complexes, and cross-linking reversed with 0.3 M NaCl at 65°C for 12 hours. RNaseA and proteinase K were added for 1h at 37°C. We purified DNA fragments with the QIAquick PCR purification kit (Qiagen). Input chromatin was used as a positive control in RT-PCR with the primers: GPH1005991(-)01A for MAF binding sites on SOX9 promoter and GPH100001C(-)01A for a chromosome 12 promoter desert as a negative control (SABiosciences).
Sphere progenitor and Schwann cell culture
DRG from E12.5–13.5 embryos were dissociated with 0.25% Trypsin (Mediatech) for 20 min. at 37°C; single-cell suspensions were obtained with a narrow-bore pipette and 40 µm strainer (BD-Falcon). Live cells (500/well) in 24-well plates in sphere medium were counted after 7 days as described (19). To passage, spheres were pelleted, treated with 0.05% Trypsin-EDTA for 3 minutes, dissociated, and plated at clonal density. Schwann cells were derived from DRG single cell suspensions as described (19).
MTS assay
Transduced MPNST cells (500 cells/well) were seeded in triplicate and incubated overnight in 96-well plates in the presence or absence of puromycin (2µg/ml). Doxycycline, RAD001, PD0325901 or selection media was added the next day and subsequently daily. Absorbance was read day 4 post-infection or doxycycline induction using the CellTiter 96® Aqueous One Solution Cell Proliferation Assay (Promega).
Flow cytometry
Approximately 100,000 MPNST cells transduced with MAF or vector control were induced with doxycycline for 24h and fixed in 0.5% paraformaldehyde. Cells were stained with propidium iodide and annexin according to Apoptosis kit instructions (Invitrogen). Flow cytometry was performed on a FACSCanto (Becton Dickinson) and analyzed using FlowJo software.
Soft agar
Agar (6% in DMEM, 10% FBS) was plated in 35 mm dishes. 5x103 MPNST cells were mixed with 3.6% agar and overlaid on solidified agar. Cells were cultured at 37°C in 7.5% CO2 in puromycin or doxycycline. Colonies were counted after 10 days. Experiments were performed in triplicate.
Mouse xenograft
Mouse xenografts of NF1 patient derived MPNST cells were conducted as described (13). 1.5 ×106 MPNST S462-TY cells suspended in Matrigel (BD) and expressing pLVX control or inducible-MAF were injected subcutaneously into the right flanks of 6 to 8-week-old female athymic nude (nu/nu) mice (Harlan). Mice were maintained on 2000 ppm doxycycline feed. Tumor volumes were measured twice weekly and mice were sacrificed before tumor volume reached 10% of total body weight.
Acknowledgements
We thank Frank McCormick (UCSF) for providing NF1 constructs and Kohsuke Kataoka for providing the MAF reporter. We thank the Viral Vector Core in the Translational Core Laboratories at Cincinnati Children’s Hospital Medical Center for viral vector production. We thank Monica DeLay for assistance with flow cytometry from the Research Flow Cytometry Core in the Division of Rheumatology at Cincinnati Children’s Hospital Medical Center, supported in part by NIH AR-47363, NIH DK78392 and NIH DK90971. This work was supported by R01 NS28840 (to NR) and NIH-P50-NS057531 (to NR and TPC). MB was partially supported by a training grant, T32HD07463.
Footnotes
Conflict of Interest
The authors declare no conflict of interest.
References
- 1.Legius E, Marchuk DA, Collins FS, Glover TW. Somatic deletion of the neurofibromatosis type 1 gene in a neurofibrosarcoma supports a tumour suppressor gene hypothesis. Nature genetics. 1993 Feb;3(2):122–126. doi: 10.1038/ng0293-122. PubMed PMID: 8499945. [DOI] [PubMed] [Google Scholar]
- 2.Brems H, Beert E, de Ravel T, Legius E. Mechanisms in the pathogenesis of malignant tumours in neurofibromatosis type 1. The lancet oncology. 2009 May;10(5):508–515. doi: 10.1016/S1470-2045(09)70033-6. PubMed PMID: 19410195. [DOI] [PubMed] [Google Scholar]
- 3.Cancer Genome Atlas Research N. Comprehensive genomic characterization of squamous cell lung cancers. Nature. 2012 Sep 27;489(7417):519–525. doi: 10.1038/nature11404. PubMed PMID: 22960745. Pubmed Central PMCID: 3466113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ding L, Getz G, Wheeler DA, Mardis ER, McLellan MD, Cibulskis K, et al. Somatic mutations affect key pathways in lung adenocarcinoma. Nature. 2008 Oct 23;455(7216):1069–1075. doi: 10.1038/nature07423. PubMed PMID: 18948947. Pubmed Central PMCID: 2694412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Holzel M, Huang S, Koster J, Ora I, Lakeman A, Caron H, et al. NF1 is a tumor suppressor in neuroblastoma that determines retinoic acid response and disease outcome. Cell. 2010 Jul 23;142(2):218–229. doi: 10.1016/j.cell.2010.06.004. PubMed PMID: 20655465. Pubmed Central PMCID: 2913027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Verhaak RG, Hoadley KA, Purdom E, Wang V, Qi Y, Wilkerson MD, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer cell. 2010 Jan 19;17(1):98–110. doi: 10.1016/j.ccr.2009.12.020. PubMed PMID: 20129251. Pubmed Central PMCID: 2818769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maertens O, Johnson B, Hollstein P, Frederick DT, Cooper ZA, Messiaen L, et al. Elucidating distinct roles for NF1 in melanomagenesis. Cancer discovery. 2013 Mar;3(3):338–349. doi: 10.1158/2159-8290.CD-12-0313. PubMed PMID: 23171796. Pubmed Central PMCID: 3595355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Whittaker SR, Theurillat JP, Van Allen E, Wagle N, Hsiao J, Cowley GS, et al. A genome-scale RNA interference screen implicates NF1 loss in resistance to RAF inhibition. Cancer discovery. 2013 Mar;3(3):350–362. doi: 10.1158/2159-8290.CD-12-0470. PubMed PMID: 23288408. Pubmed Central PMCID: 3606893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Donovan S, See W, Bonifas J, Stokoe D, Shannon KM. Hyperactivation of protein kinase B and ERK have discrete effects on survival, proliferation, and cytokine expression in Nf1-deficient myeloid cells. Cancer cell. 2002 Dec;2(6):507–514. doi: 10.1016/s1535-6108(02)00214-3. PubMed PMID: 12498719. [DOI] [PubMed] [Google Scholar]
- 10.Basu TN, Gutmann DH, Fletcher JA, Glover TW, Collins FS, Downward J. Aberrant regulation of ras proteins in malignant tumour cells from type 1 neurofibromatosis patients. Nature. 1992 Apr 23;356(6371):713–715. doi: 10.1038/356713a0. PubMed PMID: 1570015. [DOI] [PubMed] [Google Scholar]
- 11.DeClue JE, Papageorge AG, Fletcher JA, Diehl SR, Ratner N, Vass WC, et al. Abnormal regulation of mammalian p21ras contributes to malignant tumor growth in von Recklinghausen (type 1) neurofibromatosis. Cell. 1992 Apr 17;69(2):265–273. doi: 10.1016/0092-8674(92)90407-4. PubMed PMID: 1568246. [DOI] [PubMed] [Google Scholar]
- 12.Johannessen CM, Johnson BW, Williams SM, Chan AW, Reczek EE, Lynch RC, et al. TORC1 is essential for NF1-associated malignancies. Current biology : CB. 2008 Jan 8;18(1):56–62. doi: 10.1016/j.cub.2007.11.066. PubMed PMID: 18164202. [DOI] [PubMed] [Google Scholar]
- 13.Jessen WJ, Miller SJ, Jousma E, Wu J, Rizvi TA, Brundage ME, et al. MEK inhibition exhibits efficacy in human and mouse neurofibromatosis tumors. The Journal of clinical investigation. 2013 Jan 2;123(1):340–347. doi: 10.1172/JCI60578. PubMed PMID: 23221341. Pubmed Central PMCID: 3533264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johansson G, Mahller YY, Collins MH, Kim MO, Nobukuni T, Perentesis J, et al. Effective in vivo targeting of the mammalian target of rapamycin pathway in malignant peripheral nerve sheath tumors. Molecular cancer therapeutics. 2008 May;7(5):1237–45. doi: 10.1158/1535-7163.MCT-07-2335. PubMed PMID: 18483311. Pubmed Central PMCID: 2855168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.De Raedt T, Walton Z, Yecies JL, Li D, Chen Y, Malone CF, et al. Exploiting cancer cell vulnerabilities to develop a combination therapy for ras-driven tumors. Cancer cell. 2011 Sep 13;20(3):400–413. doi: 10.1016/j.ccr.2011.08.014. PubMed PMID: 21907929. Pubmed Central PMCID: 3233475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Frank NY, Schatton T, Frank MH. The therapeutic promise of the cancer stem cell concept. The Journal of clinical investigation. 2010 Jan;120(1):41–50. doi: 10.1172/JCI41004. PubMed PMID: 20051635. Pubmed Central PMCID: 2798700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pardal R, Clarke MF, Morrison SJ. Applying the principles of stem-cell biology to cancer. Nature reviews Cancer. 2003 Dec;3(12):895–902. doi: 10.1038/nrc1232. PubMed PMID: 14737120. [DOI] [PubMed] [Google Scholar]
- 18.Buchstaller J, McKeever PE, Morrison SJ. Tumorigenic cells are common in mouse MPNSTs but their frequency depends upon tumor genotype and assay conditions. Cancer cell. 2012 Feb 14;21(2):240–252. doi: 10.1016/j.ccr.2011.12.027. PubMed PMID: 22340596. Pubmed Central PMCID: 3285409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Williams JP, Wu J, Johansson G, Rizvi TA, Miller SC, Geiger H, et al. Nf1 mutation expands an EGFR-dependent peripheral nerve progenitor that confers neurofibroma tumorigenic potential. Cell stem cell. 2008 Dec 4;3(6):658–669. doi: 10.1016/j.stem.2008.10.003. PubMed PMID: 19041782. Pubmed Central PMCID: 3487385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Keng VW, Rahrmann EP, Watson AL, Tschida BR, Moertel CL, Jessen WJ, et al. PTEN and NF1 inactivation in Schwann cells produces a severe phenotype in the peripheral nervous system that promotes the development and malignant progression of peripheral nerve sheath tumors. Cancer research. 2012 Jul 1;72(13):3405–3413. doi: 10.1158/0008-5472.CAN-11-4092. PubMed PMID: 22700876. Pubmed Central PMCID: 3428071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu J, Patmore DM, Jousma E, Eaves DW, Breving K, Patel AV, et al. EGFR-STAT3 signaling promotes formation of malignant peripheral nerve sheath tumors. Oncogene. 2013 Jan 14; doi: 10.1038/onc.2012.579. PubMed PMID: 23318430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pytel P, Karrison T, Can G, Tonsgard JH, Krausz T, Montag AG. Neoplasms with schwannian differentiation express transcription factors known to regulate normal schwann cell development. International journal of surgical pathology. 2010 Dec;18(6):449–457. doi: 10.1177/1066896909351698. PubMed PMID: 20034979. [DOI] [PubMed] [Google Scholar]
- 23.Vogel KS, Klesse LJ, Velasco-Miguel S, Meyers K, Rushing EJ, Parada LF. Mouse tumor model for neurofibromatosis type 1. Science. 1999 Dec 10;286(5447):2176–2179. doi: 10.1126/science.286.5447.2176. PubMed PMID: 10591653. Pubmed Central PMCID: 3079436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miller SJ, Jessen WJ, Mehta T, Hardiman A, Sites E, Kaiser S, et al. Integrative genomic analyses of neurofibromatosis tumours identify SOX9 as a biomarker and survival gene. EMBO molecular medicine. 2009 Jul;1(4):236–248. doi: 10.1002/emmm.200900027. PubMed PMID: 20049725. Pubmed Central PMCID: 3378132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mo W, Chen J, Patel A, Zhang L, Chau V, Li Y, et al. CXCR4/CXCL12 mediate autocrine cell- cycle progression in NF1-associated malignant peripheral nerve sheath tumors. Cell. 2013 Feb 28;152(5):1077–1090. doi: 10.1016/j.cell.2013.01.053. PubMed PMID: 23434321. Pubmed Central PMCID: 3594500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Eychene A, Rocques N, Pouponnot C. A new MAFia in cancer. Nature reviews Cancer. 2008 Sep;8(9):683–693. doi: 10.1038/nrc2460. PubMed PMID: 19143053. [DOI] [PubMed] [Google Scholar]
- 27.Wende H, Lechner SG, Cheret C, Bourane S, Kolanczyk ME, Pattyn A, et al. The transcription factor c-Maf controls touch receptor development and function. Science. 2012 Mar 16;335(6074):1373–1376. doi: 10.1126/science.1214314. PubMed PMID: 22345400. [DOI] [PubMed] [Google Scholar]
- 28.Kerppola TK, Curran T. Maf and Nrl can bind to AP-1 sites and form heterodimers with Fos and Jun. Oncogene. 1994 Mar;9(3):675–684. PubMed PMID: 8108109. [PubMed] [Google Scholar]
- 29.Huang W, Lu N, Eberspaecher H, De Crombrugghe B. A new long form of c-Maf cooperates with Sox9 to activate the type II collagen gene. The Journal of biological chemistry. 2002 Dec 27;277(52):50668–50675. doi: 10.1074/jbc.M206544200. PubMed PMID: 12381733. [DOI] [PubMed] [Google Scholar]
- 30.Pouponnot C, Sii-Felice K, Hmitou I, Rocques N, Lecoin L, Druillennec S, et al. Cell context reveals a dual role for Maf in oncogenesis. Oncogene. 2006 Mar 2;25(9):1299–1310. doi: 10.1038/sj.onc.1209171. PubMed PMID: 16247450. [DOI] [PubMed] [Google Scholar]
- 31.Peterson TR, Laplante M, Thoreen CC, Sancak Y, Kang SA, Kuehl WM, et al. DEPTOR is an mTOR inhibitor frequently overexpressed in multiple myeloma cells and required for their survival. Cell. 2009 May 29;137(5):873–886. doi: 10.1016/j.cell.2009.03.046. PubMed PMID: 19446321. Pubmed Central PMCID: 2758791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang Z, Zhong J, Inuzuka H, Gao D, Shaik S, Sarkar FH, et al. An evolving role for DEPTOR in tumor development and progression. Neoplasia. 2012 May;14(5):368–375. doi: 10.1593/neo.12542. PubMed PMID: 22745583. Pubmed Central PMCID: 3384424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miller SJ, Lan ZD, Hardiman A, Wu J, Kordich JJ, Patmore DM, et al. Inhibition of Eyes Absent Homolog 4 expression induces malignant peripheral nerve sheath tumor necrosis. Oncogene. 2010 Jan 21;29(3):368–379. doi: 10.1038/onc.2009.360. PubMed PMID: 19901965. Pubmed Central PMCID: 2809821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Karin M. The regulation of AP-1 activity by mitogen-activated protein kinases. The Journal of biological chemistry. 1995 Jul 14;270(28):16483–16486. doi: 10.1074/jbc.270.28.16483. PubMed PMID: 7622446. [DOI] [PubMed] [Google Scholar]
- 35.Kraniak JM, Sun D, Mattingly RR, Reiners JJ, Jr, Tainsky MA. The role of neurofibromin in N-Ras mediated AP-1 regulation in malignant peripheral nerve sheath tumors. Molecular and cellular biochemistry. 2010 Nov;344(1–2):267–276. doi: 10.1007/s11010-010-0551-1. PubMed PMID: 20680410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Olive M, Krylov D, Echlin DR, Gardner K, Taparowsky E, Vinson C. A dominant negative to activation protein-1 (AP1) that abolishes DNA binding and inhibits oncogenesis. The Journal of biological chemistry. 1997 Jul 25;272(30):18586–18594. doi: 10.1074/jbc.272.30.18586. PubMed PMID: 9228025. [DOI] [PubMed] [Google Scholar]
- 37.Kataoka K, Handa H, Nishizawa M. Induction of cellular antioxidative stress genes through heterodimeric transcription factor Nrf2/small Maf by antirheumatic gold(I) compounds. The Journal of biological chemistry. 2001 Sep 7;276(36):34074–34081. doi: 10.1074/jbc.M105383200. PubMed PMID: 11429414. [DOI] [PubMed] [Google Scholar]
- 38.Ho IC, Hodge MR, Rooney JW, Glimcher LH. The proto-oncogene c-maf is responsible for tissue-specific expression of interleukin-4. Cell. 1996 Jun 28;85(7):973–983. doi: 10.1016/s0092-8674(00)81299-4. PubMed PMID: 8674125. [DOI] [PubMed] [Google Scholar]
- 39.Mendoza MC, Er EE, Blenis J. The Ras-ERK and PI3K-mTOR pathways: cross-talk and compensation. Trends in biochemical sciences. 2011 Jun;36(6):320–328. doi: 10.1016/j.tibs.2011.03.006. PubMed PMID: 21531565. Pubmed Central PMCID: 3112285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Annunziata CM, Hernandez L, Davis RE, Zingone A, Lamy L, Lam LT, et al. A mechanistic rationale for MEK inhibitor therapy in myeloma based on blockade of MAF oncogene expression. Blood. 2011 Feb 24;117(8):2396–2404. doi: 10.1182/blood-2010-04-278788. PubMed PMID: 21163924. Pubmed Central PMCID: 3062408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sun D, Haddad R, Kraniak JM, Horne SD, Tainsky MA. RAS/MEK-Independent Gene Expression Reveals BMP2-Related Malignant Phenotypes in the Nf1-Deficient MPNST. Molecular cancer research : MCR. 2013 Jun;11(6):616–627. doi: 10.1158/1541-7786.MCR-12-0593. PubMed PMID: 23423222. [DOI] [PubMed] [Google Scholar]
- 42.Watson JE, Doggett NA, Albertson DG, Andaya A, Chinnaiyan A, van Dekken H, et al. Integration of high-resolution array comparative genomic hybridization analysis of chromosome 16q with expression array data refines common regions of loss at 16q23-qter and identifies underlying candidate tumor suppressor genes in prostate cancer. Oncogene. 2004 Apr 22;23(19):3487–3494. doi: 10.1038/sj.onc.1207474. PubMed PMID: 15007382. [DOI] [PubMed] [Google Scholar]
- 43.Endo M, Yamamoto H, Setsu N, Kohashi K, Takahashi Y, Ishii T, et al. Prognostic significance of AKT/mTOR and MAPK pathways and antitumor effect of mTOR inhibitor in NF1-related and sporadic malignant peripheral nerve sheath tumors. Clinical cancer research : an official journal of the American Association for Cancer Research. 2013 Jan 15;19(2):450–461. doi: 10.1158/1078-0432.CCR-12-1067. PubMed PMID: 23209032. [DOI] [PubMed] [Google Scholar]
- 44.Lau N, Feldkamp MM, Roncari L, Loehr AH, Shannon P, Gutmann DH, et al. Loss of neurofibromin is associated with activation of RAS/MAPK and PI3-K/AKT signaling in a neurofibromatosis 1 astrocytoma. Journal of neuropathology and experimental neurology. 2000 Sep;59(9):759–767. doi: 10.1093/jnen/59.9.759. PubMed PMID: 11005256. [DOI] [PubMed] [Google Scholar]
- 45.Zou CY, Smith KD, Zhu QS, Liu J, McCutcheon IE, Slopis JM, et al. Dual targeting of AKT and mammalian target of rapamycin: a potential therapeutic approach for malignant peripheral nerve sheath tumor. Molecular cancer therapeutics. 2009 May;8(5):1157–1168. doi: 10.1158/1535-7163.MCT-08-1008. PubMed PMID: 19417153. [DOI] [PubMed] [Google Scholar]
- 46.Zhao Y, Xiong X, Sun Y. DEPTOR, an mTOR inhibitor, is a physiological substrate of SCF(betaTrCP) E3 ubiquitin ligase and regulates survival and autophagy. Molecular cell. 2011 Oct 21;44(2):304–316. doi: 10.1016/j.molcel.2011.08.029. PubMed PMID: 22017876. Pubmed Central PMCID: 3216641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Carracedo A, Ma L, Teruya-Feldstein J, Rojo F, Salmena L, Alimonti A, et al. Inhibition of mTORC1 leads to MAPK pathway activation through a PI3K-dependent feedback loop in human cancer. The Journal of clinical investigation. 2008 Sep;118(9):3065–3074. doi: 10.1172/JCI34739. PubMed PMID: 18725988. Pubmed Central PMCID: 2518073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yin B, Morgan K, Hasz DE, Mao Z, Largaespada DA. Nfl gene inactivation in acute myeloid leukemia cells confers cytarabine resistance through MAPK and mTOR pathways. Leukemia. 2006 Jan;20(1):151–154. doi: 10.1038/sj.leu.2404033. PubMed PMID: 16307021. [DOI] [PubMed] [Google Scholar]
- 49.Stowe IB, Mercado EL, Stowe TR, Bell EL, Oses-Prieto JA, Hernandez H, et al. A shared molecular mechanism underlies the human rasopathies Legius syndrome and Neurofibromatosis-1. Genes & development. 2012 Jul 1;26(13):1421–1426. doi: 10.1101/gad.190876.112. PubMed PMID: 22751498. Pubmed Central PMCID: 3403010. [DOI] [PMC free article] [PubMed] [Google Scholar]