Abstract
Müller glia are the resident radial glia in the vertebrate retina. The response of mammalian Müller glia to retinal damage often results in a glial scar and no functional replacement of lost neurons. Adult zebrafish Müller glia, in contrast, are considered tissue-specific stem cells that can self-renew and generate neurogenic progenitors to regenerate all retinal neurons after damage. Here, we demonstrate that regulation of TGFβ signaling by the corepressors Tgif1 and Six3b is critical for the proliferative response to photoreceptor destruction in the adult zebrafish retina. When function of these corepressors is disrupted, Müller glia and their progeny proliferate less, leading to a significant reduction in photoreceptor regeneration. Tgif1 expression and regulation of TGFβ signaling are implicated in the function of several types of stem cells, but this is the first demonstration that this regulatory network is necessary for regeneration of neurons.
Keywords: Müller glia, photoreceptor, tgif1, six3b, stem cell
Introduction
Müller glia are the radial glia of the adult vertebrate retina and provide physical and homeostatic support for proper neuronal function. Mammalian Müller glia respond to retinal injury with reactive gliosis, characterized by increased glial fibrillary acidic protein (GFAP), hypertrophy, and in some cases mitotic activity. These cellular changes are initially neuroprotective, but typically result in a glial scar and no neuron regeneration (Bringmann et al., 2009; Reichenbach and Bringmann, 2013). Despite this, mammalian Müller glia express low levels of two neural progenitor markers, Sox2 and Pax6 (Roesch et al., 2008), and, in response to neuronal damage, increase expression of brain lipid binding protein (BLBP), a marker of young Müller glia in zebrafish retina (Raymond et al., 2006), radial glia, and a subtype of neural precursors in the developing mammalian brain (Chang et al., 2007; Hartfuss et al., 2001). These studies suggest that mammalian Müller glia have a latent ability to regenerate neurons (Karl and Reh, 2010). In fact, cultured mammalian Müller glia proliferate in response to exogenous growth factors (Ikeda and Puro, 1995; Ueki et al., 2012), and rodent Müller glia have a limited capacity to regenerate retinal neurons after damage, but only in the perinatal retina (Karl and Reh, 2010).
In contrast to mammals, adult zebrafish regenerate all retinal neurons through the activation, dedifferentiation, and proliferation of Müller glia. Like vertebrate neural stem cells in the brain, zebrafish Müller glia divide asymmetrically to produce a proliferative neurogenic progenitor, which then forms a neurogenic cluster that ultimately regenerates lost neurons to restore retinal structure and function (Bernardos et al., 2007). Early stages of regeneration in these retinal stem cells are characterized by upregulation of GFAP, BLBP, and proliferating cell nuclear antigen (PCNA), and expression of numerous retinal progenitor genes including rx1, pax6, and acsl1a (Bernardos et al., 2007; Bringmann et al., 2009; Fausett et al., 2008; Hartfuss et al., 2001; Raymond et al., 2006; Thummel et al., 2010).
TGFβ signaling, mediated through Smad2 and Smad3, controls proliferation of mammalian Müller glia and retinal progenitors in vivo (Close et al., 2005; Satoh and Watanabe, 2008) and in vitro (Ichida et al., 2009; Ikeda and Puro, 1995), and neural stem cells in the brain (Aigner and Bogdahn, 2008). During mammalian retinal development, TGFβ signaling increases when retinal cells differentiate, and Müller glia proliferation continues when TGFβ signaling is inhibited (Close et al., 2005). Mammalian astrocytes respond to TGFβ signaling by becoming gliotic and secreting extracellular matrix proteins, resulting in glial scars (Bringmann et al., 2009; Reichenbach and Bringmann, 2013; Robel et al., 2011), and inhibiting TGFβ prevents scarring (Moon and Fawcett, 2001). Preventing glial scars is likely a critical step in promoting regeneration in the central nervous system (Robel et al., 2011).
Several TGFβ signaling pathway members are differentially regulated in zebrafish Müller glia during retina regeneration: Two corepressors, tgif1 and six3b, are rapidly upregulated prior to the initial mitotic division (Inbal et al., 2007; Kassen et al., 2007; Qin et al., 2009; Wotton et al., 1999) and thus may have overlapping roles to repress TGFβ signaling in Müller glia. Tgif1 has been implicated in maintaining a stem cell identity, as tgif1 transcripts are enriched in murine organs containing stem cell populations, such as the ovary and testes, and embryonic stem cells (ESC) (Thorrez et al., 2008). Six3b binds to the DNA replication inhibitor Geminin during eye development and promotes proliferation in neural development in fish (Singh and Tsonis, 2010).
In this study, we tested the hypothesis that downregulation of Smad2/3-mediated TGFβ signaling by the transcriptional corepressors Tgif1 and Six3b is necessary for the injury-induced proliferative response of Müller glia and Müller-glial derived neurogenic progenitors in zebrafish retinas following photoreceptor destruction. We used tgif1 and six3b genetic mutants and Six3a/b translational knock-down to show that these corepressors function after acute photoreceptor damage to promote the proliferative response of Müller glia stem cells that is required for photoreceptor regeneration.
Materials and Methods
Zebrafish Lines and Light Lesions
We maintained fish under standard conditions (Westerfield, 2000). We used transgenic fish lines in which Müller glia are the only retinal cells that express green fluorescent protein (GFP) or enhanced green fluorescent protein (EGFP) (Bernardos et al., 2007): in Tg(gfa-p:EGFP)mi2002, differentiated Müller glia express GFP, and in Tg(gfap:nGFP)mi2004, immature Müller glia and Müller glia that are activated in response to retinal damage express nuclear GFP. We identified mutant fish lines with the tgif1fh258 allele (Y143→Stop) and the six3bvu87 allele (E109→Stop; Inbal et al., 2007) in a cryo-preserved TILLING library (Draper et al., 2004). For experiments with tgif1fh258 and six3bvu87 lines, we destroyed photoreceptors with an acute light exposure (Bernardos et al., 2007). Mutant allele annotations have been abbreviated as tgif1−/− and six3b−/−. For Six3 morpholino experiments, we destroyed photoreceptors in albino zebrafish using constant light treatment (Thummel et al., 2010). All procedures were approved by the Committees on Use and Care of Animals at the University of Michigan and Wayne State University.
Morpholinos
We injected 3′ lissamine-tagged morpholinos intravitreously and electroporated immediately prior to light lesion (Thummel et al., 2010). We used the Gene Tools standard control morpholino (5′-CCTCCTACCTCAGTTACAATTTATA-3′), and a morpholino that recognizes both six3a and six3b in the experimental treatment (5′-GCTCTAAAGGAGACCTGAAAACCAT-3′; Ando et al., 2005).
Immunohistochemistry and Cell Counting
Tissue was collected and processed as previously described (Bernardos et al., 2007). Antibody dilutions and antigen retrieval are provided in Supporting Information Table S1. We sectioned retinas through the dorsoventral axis and counted PCNA-positive nuclei in 100 µm linear length of the lesion using the cell counter in ImageJ (National Institutes of Health, Bethesda, MD). We counted cells in the lesioned area of the central retina in an area within 400 µm of the optic nerve head along the nasotemporal axis. We analyzed 8 to 16 sections per fish in sections at least 24 µm apart. PCNA-positive nuclei spanning the outer plexiform layer were included in the outer nuclear layer (ONL). We determined the lesioned area by increased expression of BLBP or GFP in transgenic gfap:EGFP fish, in combination with loss of cone nuclei. In morpholino experiments, we counted PCNA-positive nuclei in a 300 µm linear length of 2–4 sections per fish in peripheral regions of the dorsal retina marked with the morpholino lissamine tag, avoiding areas that were damaged by electroporation. To determine the extent of cone regeneration, we counted cone nuclei whose profiles extended more than 50% outside of the outer limiting membrane (OLM), which was visualized by GFP expression in gfap:EGFP fish or by ZO-1 immunoreactivity.
Western Blots
We immediately froze isolated retinas from dark-adapted fish on dry ice and then homogenized them in cold lysis buffer (50 µL/retina; 50 mM Tris-HCl pH 7.5, 150 mM NaCl, 1 mM EDTA pH 7.0, 0.1% Triton X-100) with complete mini protease inhibitor cocktail (Roche). We triturated retinas with a 200 µL micropipette tip, incubated them on ice for 30 min, and triturated again (~20 times). We centrifuged samples at 13,000 rpm for 12 min at 4°C, ran protein gels under reducing and denaturing conditions, and performed Western blots using standard protocols. For densitometric analysis, we digitized the film (AlphaEase FC 6.0, Alpha Innotech), subtracted an image of the light box alone from each blot image, and used the gel analysis tool in ImageJ. For presentation of blot images (Fig. 2, S2), the color levels were changed uniformly to the image in Photoshop. Antibodies are provided in Supporting Information Table S2.
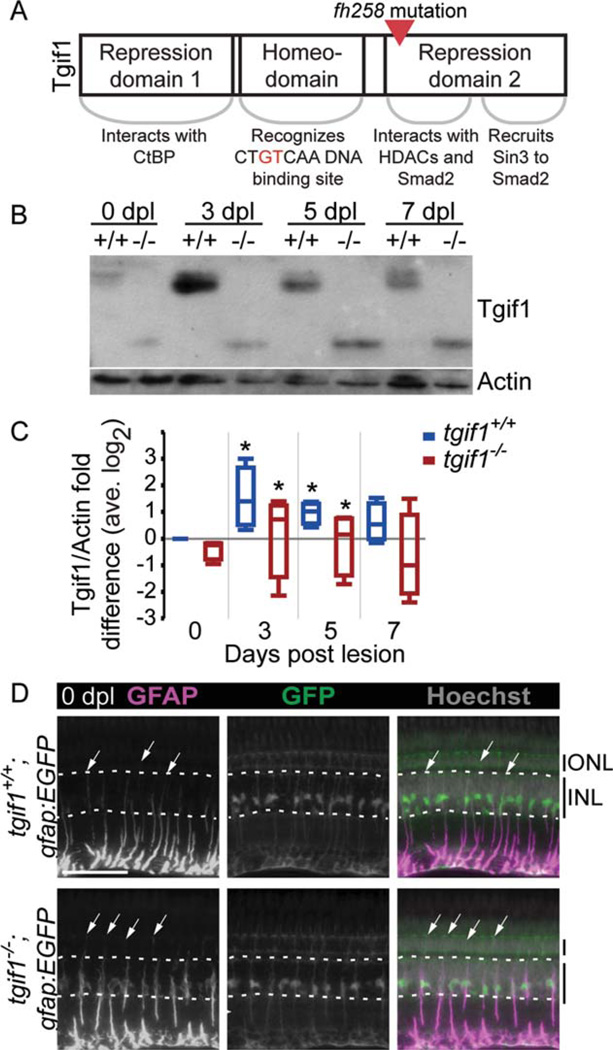
FIGURE 2.
Fish homozygous for the fh258 allele of tgif1 express a truncated Tgif1 and GFAP localization in Müller glia is altered. (A) Schematic of the Tgif1 protein and the point mutation in the fh258 allele. (B) Representative Tgif1 Western blot of whole retina extract from tgif1+/+ and tgif1−/− fish. (C) Densitometric analysis of Tgif1 protein in control and light lesioned retinas from tgif1+/+ and tgif1−/− fish normalized to actin and relative to 0 dpl tgif1+/+ (retinas pooled from two fish, n = 4 replicates). * P<0.05 relative to 0 dpl of the same genotype, Student t-test. (D) GFAP immunolocalization (first column) in the unlesioned retina of tgif1+/+;gfap:EGFP (top) and tgif1−/−;gfap:EGFP (bottom) fish expressing GFP only in the Müller glia. We used the same exposure time and did not alter the GFAP images postcapture. Arrows = top of GFAP distribution in individual Müller glia; dotted white lines delineate the base and top of the INL. Abbreviations as in Fig. 1. Scale bar = 50µm.
Real-Time Reverse Transcription PCR (qRT-PCR)
For transcriptional analysis of Müller glia, we isolated the Müller glia from gfap:EGFP fish using fluorescence-activated cell sorting (Qin et al., 2009). For whole retina analysis, we pooled retinas from individual fish in TRIzol (75 µL/retina) immediately after dissection. We isolated total RNA according to the manufacturer’s instructions and resuspended RNA in 20 µL of RNAse-free water. We treated DNase and extracted the RNA with phenol-chloroform. We generated cDNA with the Superscript III First-Strand Synthesis SuperMix for qRT-PCR kit (Invitrogen). For qRT-PCR, we supplemented iQ Sybr Green Supermix (BioRad) with 10 nM ROX and ran samples on a 7500 Fast Real-Time PCR System using Sequence Detection Software v.1.3.1 (Applied Biosystems). Primers are provided in Supporting Information Table S3.
Imaging and Statistical Analysis
All images were taken on an AxioImager epifluorescent compound microscope with Apotome using Zeiss AxioVision V4.8.0.0. We used Adobe PhotoShop to adjust the brightness and contrast of images equally to the entire image unless otherwise noted. We examined a minimum of three fish for each histological analysis. We generated box plots (indicating 75 and 25% quantiles, median, and outliers outside of whiskers) and performed all statistical analyses using JMP 9.0 (SAS Institute, Inc.).
Results
Tgif1 is Specifically Expressed in Activated Müller Glia After Photoreceptor Destruction
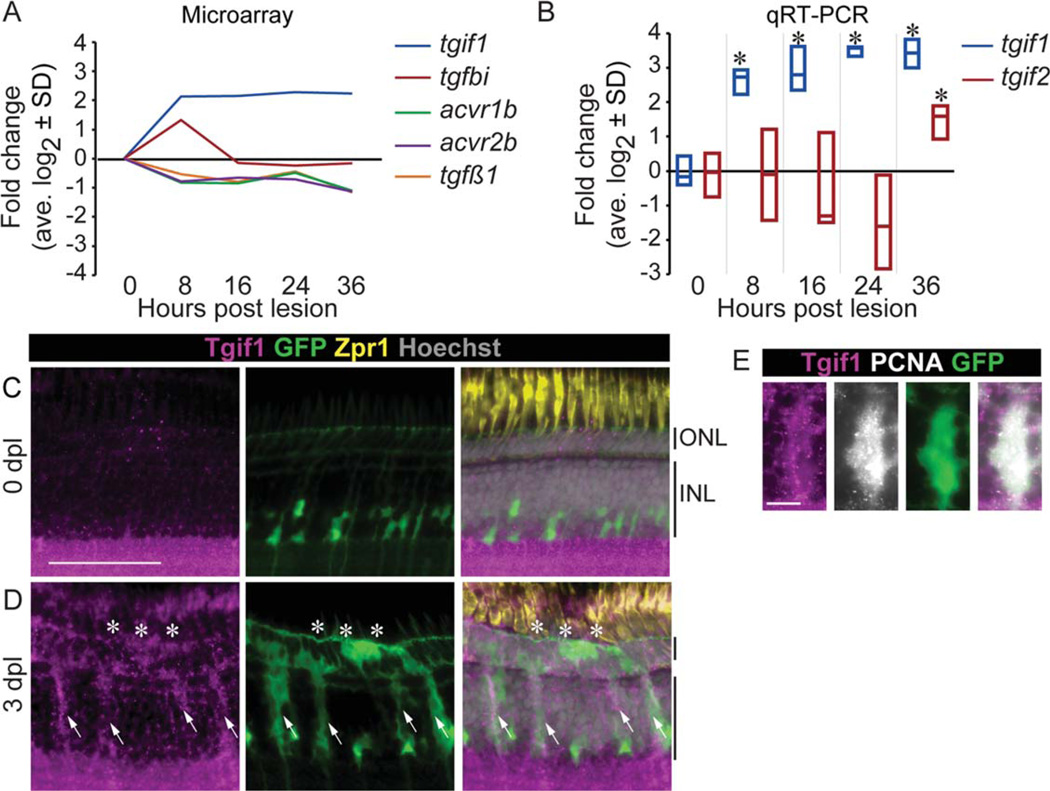
In the adult zebrafish retina, exposure to intense light destroys photoreceptors and leads to rapid upregulation of tgif1 transcription in Müller glia from 8 through 36-h post lesion (hpl) (Qin et al., 2009). Transcription levels of several other members of the Smad2/3 signaling pathway are also differentially regulated within 8 hpl (Fig. 1A): activin receptors, acvr1b and acvr2b, and the tgfβ1 ligand are downregulated, and conversely, the TGFβ target gene tgfβ- induced (tgfbi) is upregulated at 8 hpl, then downregulated by 16 hpl. We used the cDNA generated from isolated Müller glia in our earlier transcriptional analysis (Qin et al., 2009) to confirm the increased expression of tgif1 with qRT-PCR (analysis of variance (ANOVA) F-ratio = 9.72, P < 0.05; Fig. 1B). Because the mammalian Tgif2 is believed to compensate for Tgif1 in Tgif1 mouse knock-outs (Shen and Walsh, 2005), we analyzed expression of the putative zebrafish tgif2 gene (Ensembl CABZ01084922.1–201, Zv9). tgif2 expression increased in Müller glia but not until 36 hpl (ANOVA F-ratio = 3.53, P= 0.054; Fig. 1B). The overall pattern of transcriptional changes in members of the Smad2/3 signaling pathway suggests that TGFβ signaling initially is upregulated in the Müller glia after light lesion, but then is quickly suppressed.
FIGURE 1.
Smad2/3 signaling is downregulated and the corepressor Tgif1 upregulated after photoreceptors destruction. (A) Changes in expression of Smad2/3 signaling pathway in isolated Müller glia after acute light lesion, compared with uninjured retina, by microarray analysis (GEO GSE14495; Qin et al., 2009). (B) Changes in expression of tgif1 and tgif2 in isolated Müller glia measured by qRT-PCR. * P<0.05 relative to 0 hpl, Student t-test. Tgif1 immunoreactivity in an undamaged retina (C) and at 3 dpl (D) in gfap:EGFP fish that have GFP in the Müller glia and progenitors (arrows = Müller glia; *= lesion). (E) Colocalization of Tgif1 and PCNA in GFP-positive Müller glia in gfap:EGFP fish at 3 dpl. Scale bars = 50 µm in (C,D); 10 µm in (E). ONL = outer nuclear layer; INL = inner nuclear layer; hpl = hours post lesion; dpl 5 days post lesion.
In undamaged retinas, Tgif1 protein (Fig. 1C) and transcripts (Supp. Info. Fig. S1A) are expressed in the ganglion cell layer, more weakly in the inner nuclear layer (INL) and ONL, and in the ciliary marginal zone (Supp. Info. Fig. S1B), which contains retinal stem cells and progenitors in the adult zebrafish retina. Immunostaining showed a subset of amacrine cells expressing higher levels of Tgif1 (Fig. 1C, Supp. Info. S1C–E). At 3 dpl, when Müller glia-derived progenitor cells are migrating from the INL into the ONL to regenerate photoreceptors (Bernardos et al., 2007), cells expressing tgif1 spanned the INL and ONL in the lesion area (Fig. 1D, Supp. Info. S1A). By 5 dpl, tgif1 expression decreased, and by 14 dpl, expression further reduced to near control levels in the INL where the Müller glia reside (Supp. Info. Fig. S1A). The cells expressing higher levels of tgif1 at 3 dpl are proliferating Müller glial-derived progenitors, as Tgif1 colocalized with PCNA and the Müller glia GFP reporter in gfap:EGFP fish (arrows Fig. 1D,E).
tgif1−/− Fish Upregulate TGIF1 Expression in Response to Photoreceptor Damage
The N-terminus of Tgif1 contains domains that repress reti-noic acid signaling (Bartholin et al., 2006), and the C-terminus binds to pSmad2/3 and recruits histone deacetylases (HDACs) to repress TGFβ signaling (Fig. 2A; Powers et al., 2010; Wotton et al., 1999). Because TGFβ signaling regulates Müller glia and retinal progenitor proliferation, we focused on the potential importance of Tgif1 repression of TGFβ signaling during photoreceptor regeneration with a genetic functional analysis. We used TILLING (Draper et al., 2004) to generate a mutation in the tgif1 gene which introduces a stop codon (Y143X) and produces a truncated protein (~18 kDa, Figs. 2A,B). tgif1+/+ fish expressed only the full-length Tgif1 protein (~35 kDa, Fig. 2B).
We performed co-immunoprecipitation experiments in vitro and determined that the truncated protein can bind to pSmad2 (Supp. Info. Fig. S2A). Due to the loss of the HDAC binding domain in the truncated protein (Wotton et al., 2001), we predict that the Y143X mutation cannot recruit HDACs to repress TGFβ target genes. In Western blot analysis, both tgif1+/+ and tgif1−/− retinas significantly increased Tgif1 expression at 3 and 5 dpl relative to unlesioned retinas and decreased expression by 7 dpl (Fig. 2B,C). We did not make direct comparisons between the levels of the full-length and truncated proteins due to potential differences in how they present in Western blot. Immunocytochemistry showed that at 3 dpl in tgif1−/− fish, Tgif1 protein localized to the Müller glia-derived progeny (supp. Info. Fig. S2B).
tgif1−/− Fish Have a Normal Number of Müller Glia that Exhibit Features of Gliosis
Because TGFβ signaling limits Müller glia proliferation during development (Close et al., 2005), we asked if tgif1−/− fish had fewer Müller glia than tgif1+/+ fish. We found no difference between the number of Müller glia in adult whole-mount retinas of tgif1−/− gfap:EGFP and tgif1+/+;gfap:EGFP fish (Supp. Info. Fig. S2C). GFAP is an intermediate filament that is upregulated in both mammalian and fish Müller glia in reaction to retinal injury and is a component of retinal glial scars (Bernardos et al., 2007; Bringmann et al., 2009; Robel et al., 2011). Normally, GFAP localizes more strongly to Müller glia basal processes (Fig. 2D, top). We examined GFAP distribution in Müller glia of tgif1−/−;gfap:EGFP fish to determine if they exhibited characteristics of gliosis. GFAP protein is distributed more apically along the radial process in tgif1−/− Müller glia relative to wild-type siblings (arrows, Fig. 2D, n = 3), which suggests that Müller glia in tgif1−/− fish are in a more reactive state. GFAP expression also increases in response to TGFβ signaling (Robel et al., 2011), so we predicted it may be more highly expressed in tgif1−/− fish due to derepression of TGFβ signaling. Unlesioned retinas from Lenkowski et al.: Tgif1, Six3b, and Retinal Neuron Regeneration tgif1−/− fish tend to have higher levels of GFAP by Western blot analysis, but the difference is nonsignificant (Supp. Info. Fig. S2D, One-way Kruskal-Wallis Rank Sum test P = 0.15, n = 7).
tgif1−/− Fish Have Decreased Proliferation After Photoreceptor Destruction
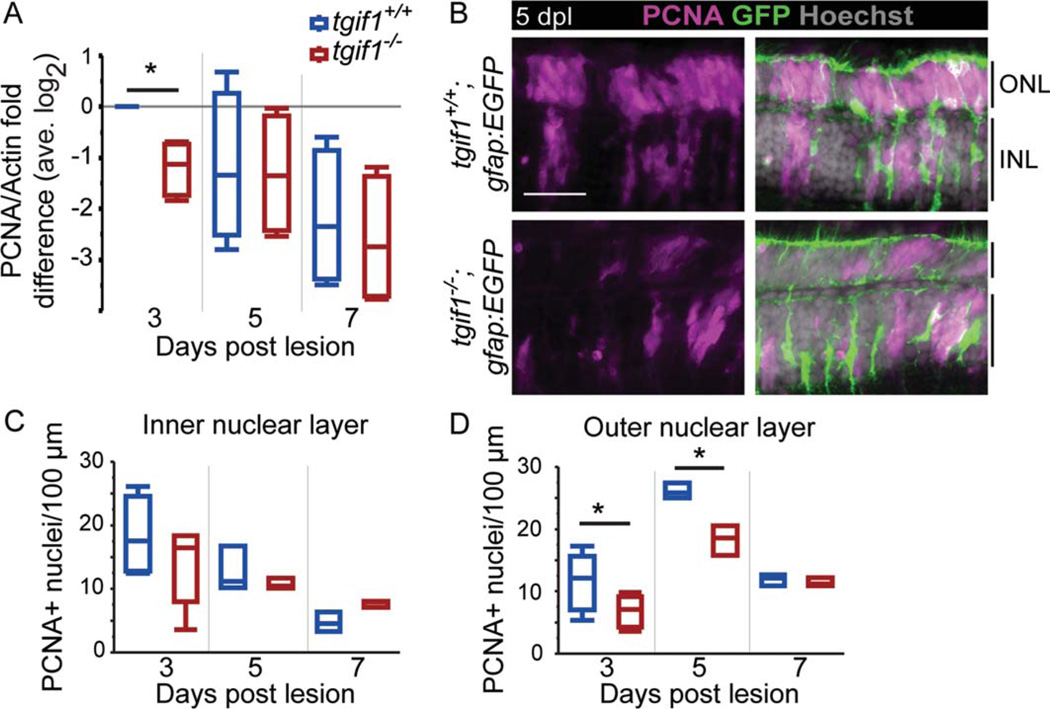
To compare the amount of proliferation in tgif1+/+ and tgif1−/− fish, we first analyzed the level of PCNA in whole retinas by Western blot (two-way ANOVA F-ratio = 5.13, P = 0.0011; Fig. 3A). At 3 dpl, tgif1−/− retinas have significantly less PCNA than tgif1+/+ retinas (Fig. 3A). We then counted PCNA immunolabeled nuclei in the lesioned area in cryosections (Fig. 3B – D). Although there was a significant difference among the genotypes across time points (two-way ANOVA F-ratio = 4.29, P < 0.05 and F-ratio = 20.09, P < 0.0001 in the INL and ONL, respectively), only the number of PCNA-positive nuclei in the ONL was significantly reduced in tgif1−/− fish compared with tgif1+/+ fish at 3 and 5 dpl (Fig. 3C,D). The proliferating cells in the ONL at 3 and 5 dpl are likely cone progenitors (Bernardos et al., 2007). Our analysis of proliferation indicates that fewer neurogenic progenitors are generated by injury-induced Müller glia in the retinas of tgif1−/− fish compared with tgif1+/+ fish.
FIGURE 3.
Proliferation is reduced in tgif1−/− fish compared with tgif1+/+ fish. (A) Densitometric analysis of PCNA in Western blots normalized to actin and relative to 3 dpl tgif1+/+ (retinas pooled from two fish, n = 4 replicates). (B) Representative images of PCNA immu-noreactivity at 5 dpl in tgif1+/+ and tgif1−/− fish with the gfap:EGFP transgene. The average number of PCNA-positive nuclei in the INL (C) and ONL (D) in tgif1+/+ and tgif1−/− fish; 8–17 sections per fish; n = 5 at 3 dpl; n = 3 at 5 and 7 dpl. Scale bars=25 µm. * P<0.05, Student t-test comparing genotypes at the same lesion interval. Abbreviations as in Fig. 1.
Müller Glia are Activated After Acute Light Lesion, but Smad2/3 Targets are Misregulated in tgif1−/− Retinas
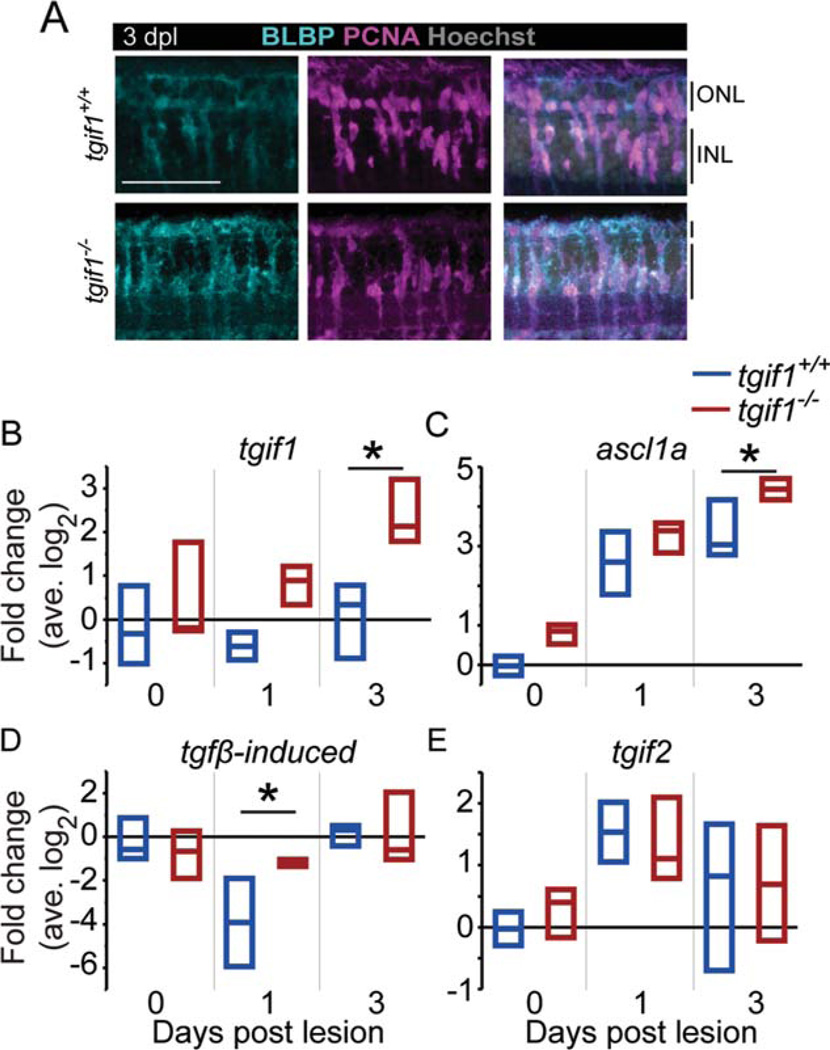
At 1 dpl, the same number of Müller glia were activated in tgif1+/+;gfap:nGFP and tgif1−/−;gfap:nGFP retinas (Supp. Info. Fig. S2E). BLBP is a marker of neural stem cells and radial glia in the brain (Götz and Barde, 2005) and increases in adult zebrafish Müller glia by 1 hpl (Nagashima, Barthel, Raymond, submitted). In both tgif1+/+ and tgif1−/− retinas, BLBP expression increased in the lesioned area at 3 dpl where there is proliferation (Fig. 4A). Therefore, Müller glia in tgif1−/− fish are activated and re-express the retinal progenitor markers BLBP and Pax6 (data not shown) in response to acute light lesion.
FIGURE 4.
After acute light lesion, Müller glia in tgif1−/− fish express a dedifferentiation marker similar to tgif1+/+ but misre-gulate Smad2/3 target genes. (A) BLBP and PCNA immunoreac-tivity in tgif1+/+ and tgif1−/− fish at 3 dpl in the lesioned area, as determined by the absence of cone nuclei. (B–E) qRT-PCR of whole retina extracts (retinas pooled from individual fish, n=3 replicates). We normalized gene expression to gpia and calculated fold change relative to the average tgif1+/+ 0 dpl level. * P<0.05, Student t-test comparing genotypes at the same lesion interval. Abbreviations as in Fig. 1.
Because Müller glia in tgif1−/−;gfap:EGFP fish upregulated retinal progenitor markers, indicative of dedifferentiation, we used qRT-PCR to examine the expression of genes that are regulated by TGFβ signaling and important in neural stem cells or Müller glia-mediated retina regeneration: tgif1 (Liu et al., 2011; Qin et al., 2009), tgfbi (Thapa et al., 2007), and ascl1a (Fausett et al., 2008; Liu et al., 2011). As controls, we examined the expression of several genes that are not regulated by Smad2/3 but are critical for retinal neuron regeneration: dkk1b (Ramachandran et al., 2011); hb-egfa (Wan et al., 2012); pax6a and pax6b (Thummel et al., 2010); and six3b (Fig. 5). Compared with tgif1+/+ siblings, tgif1−/− fish have significantly higher levels of Smad2/3 targets after lesion (Fig. 4B – D): tgif1 (two-way ANOVA F-ratio = 4.48, P< 0.05), ascl1a (two-way ANOVA F-ratio = 26.28, P< 0.001), and tgfbi (two-way ANOVA F-ratio = 4.61, P< 0.05). These data confirm that Smad2/3-mediated TGFβ signaling is increased in tgif1−/− fish during photoreceptor regeneration.
FIGURE 5.
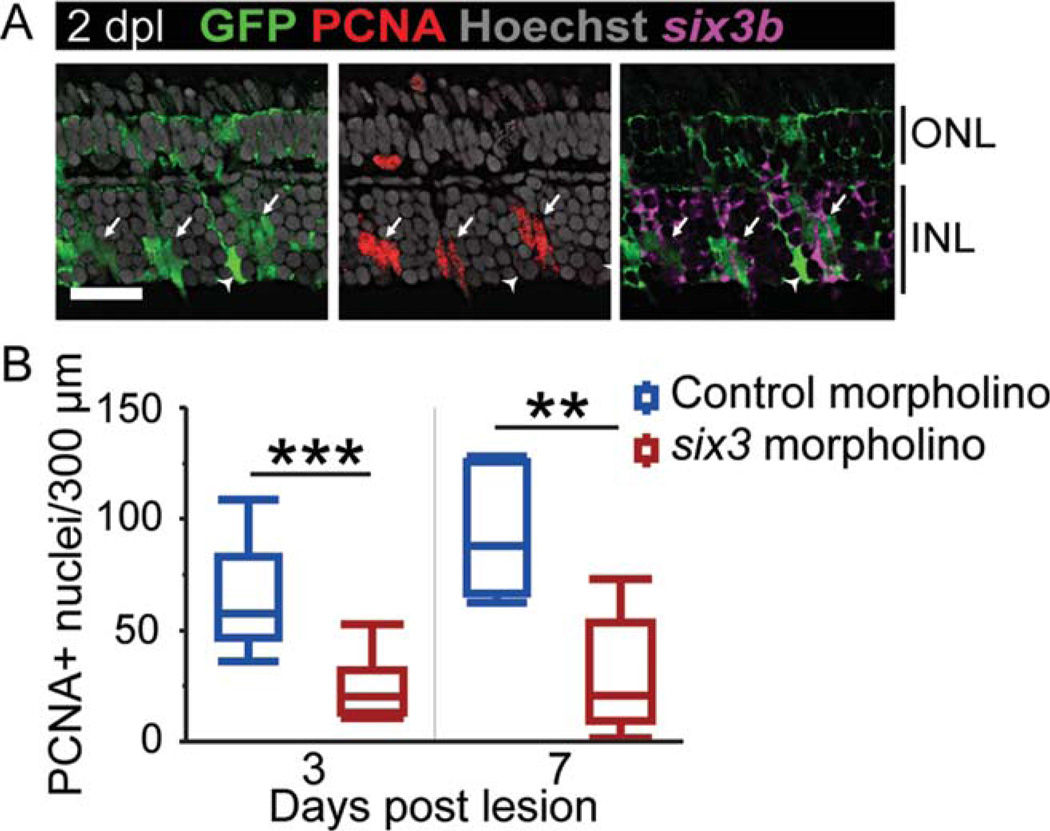
Repression of the Smad2/3 corepressor six3 inhibits proliferation after photoreceptor lesion. (A) six3b in situ hybridization and PCNA immunoreactivity in GFP-positive Müller glia and their progeny (arrows) at 2 dpl in gfap:EGFP fish. The arrowhead indicates a PCNA-negative Müller glial cell that is not expressing six3b. (B) The number of PCNA-positive nuclei at 3 and 7 dpl after electroporation of morpholinos targeting six3a/b. n=11 for control morpholino, n=10 for six3 morpholino at 3 dpl, and n=5 at 7 dpl. Scale bar=20 µm. ** P<0.01; *** P<0.0001, Student t-test comparing genotypes at the same lesion interval. Abbreviations as in Fig. 1.
In contrast, we detected no significant difference between tgif1+/+ and tgif1−/− retinas in the levels of several non-Smad2/3-regulated genes (Fig. 4E, Supp. Info. Fig. S3): hb-egfa (two-way ANOVA F-ratio = 1.44, P=0.28), pax6a (two-way ANOVA F-ratio = 0.94, P= 0.49), pax6b (two-way ANOVA F-ratio = 0.73, P= 0.62), six3b (two-way ANOVA F-ratio = 1.04, P=0.44), and tgif2 (two-way ANOVA F-ratio =0.61, P = 0.70). These data suggest that tgif2 is not upregulated to compensate for the defective Tgif1 protein in tgif1−/− fish. Previous studies showed that whole retina qRT-PCR analysis can detect changes in expression in Müller glia of ascl1a, dkk1b, hb-egfa, pax6a, and pax6b (Ramachandran et al., 2010; Ramachandran et al., 2011; Wan et al., 2012). We also saw no difference in levels of six3b between tgif1−/− and wild-type siblings, however, it is possible that changes in six3b transcript levels in the Müller glia could not be detected in whole retina extracts because they are masked by high levels of expression in retinal neurons (Supp. Info. Fig. S3E). Interestingly, undamaged retinas of tgif1−/− fish express significantly lower levels of the Wnt/βcatenin inhibitor dkk1b than wild-type siblings (two-way ANOVA F-ratio = 9.82, P< 0.001; Supp. Info. Fig. S3A), indicating that other signaling pathways in addition to TGFβ are altered in the retinas of tgif1−/− fish.
Concurrent Inhibition of Two six3-Related Genes Reduces Proliferation After Light Lesion
The TGFβ corepressor six3b is also rapidly upregulated in the Müller glia of light-lesioned adult zebrafish (Qin et al., 2009). Zebrafish have three six3-related genes that are differentially expressed in the unlesioned adult retina (Supp. Info. Fig. S4): six3a, six3b, and six7 (Inbal et al., 2007). We confirmed that only six3b, not six3a or six7, is upregulated in the lesion area (Supp. Info. Fig. S4) and that six3b colocalizes with GFP-positive, PCNA-positive Müller glia, and their neurogenic progenitors (Fig. 5A). We did not detect a regeneration defect in preliminary experiments using zebrafish homozygous for a six3b null allele, perhaps due to compensation by six3a as in early development (Inbal et al., 2007). Therefore, we used morpholinos that inhibit translation of both six3a and six3b paralogs (Ando et al., 2005) to ask whether Six3 is important for proliferation during retinal regeneration (Fig. 5B). We found a significant difference in PCNA-positive nuclei among the groups (ANOVA F-ratio = 14.77, P< 0.0001) and between control and six3 morpholino at 3 dpl (Student t-test P < 0.0001) and 7 dpl (Student t-test P < 0.01). As with our tgif1 results, perturbation of six3 function significantly reduced proliferation of neurogenic progenitors after light lesion.
Regeneration of Photoreceptors is Impaired in tgif1−/− and tgif1−/−; six3b−/− Fish
At 14 dpl, cone regeneration in wild-type fish is largely complete, but rod regeneration is continuing (Raymond et al., 2006). We asked whether the reduced proliferation in fish with impaired Tgif1 or Six3b function has a consequence for cone regeneration by exposing fish to BrdU from 1 through 7 dpl and examining BrdU-labeled nuclei at 14 dpl (Supp. Info. Fig. S5A). tgif1+/+;gfap:EGFP fish had BrdU-labeled photoreceptors in the ONL (arrowheads, Supp. Info. Fig. S5A) and BrdU-labeled Pax6-positive and Crx1-positive neurons in the INL (Supp. Info. Fig. S5B,C). tgif1−/−;gfa-p:EGFP fish had BrdU-labeled regenerated neurons in the ONL and the INL (arrows and arrowheads, respectively, supp. Info. Fig. S5), but there were fewer BrdU-positive nuclei in the INL, reinforcing our observations of reduced proliferation in tgif1−/− fish (Fig. 3). Müller glia in tgif+/+ fish had resumed a normal morphology and were labeled with BrdU at 14 dpl (arrows, Supp. Info. Fig. S5A). Interest-ingly, in tgif1−/− ;gfap:EGFP fish occasional Müller glia were still highly GFP-positive with dense processes at the OLM (double arrow, Supp. Info. Fig. S5A). This morphology was never observed in tgif1+/+ ;gfap:EGFP retinas and is reminiscent of gliotic hypertrophy of mammalian Müller glia, further supporting our hypothesis that normal levels of tgif1 are critical for regulating neurogenic proliferation of Müller-glia after light lesion.
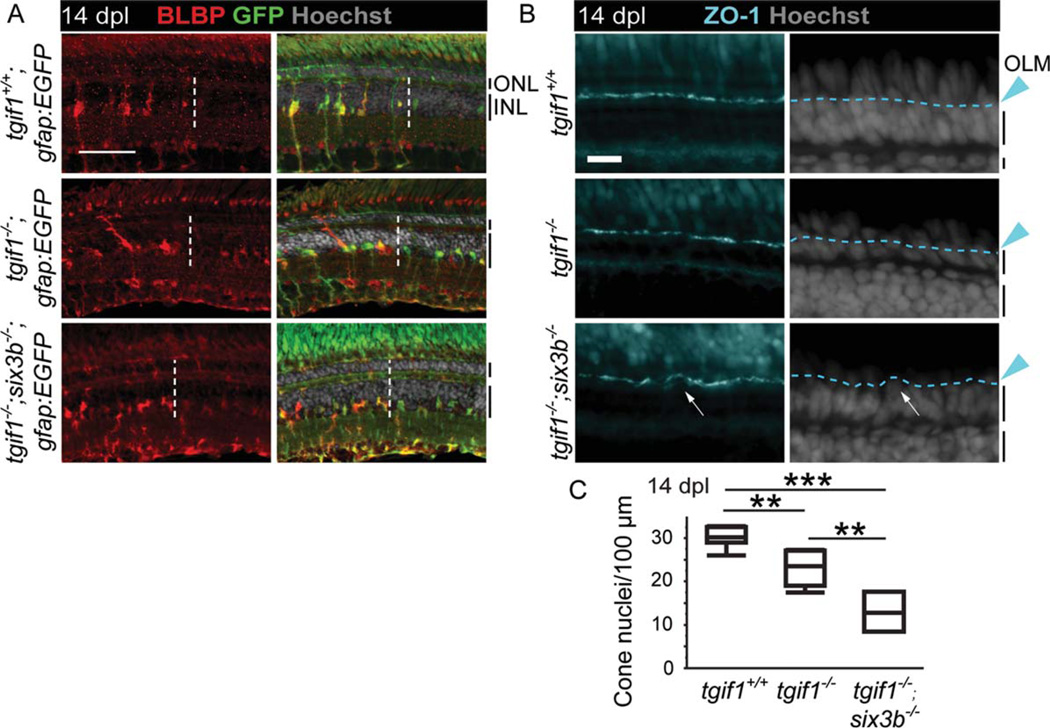
We predicted that the proliferation defect in tgif1−/− fish should result in reduced regeneration of cone photoreceptors. We quantified the number of cone nuclei in the lesioned area, as demarcated by increased BLBP expression in Müller glia (Fig. 6A). Cone nuclei were distinguished from rods by the position of their nuclei apical to the OLM, which we labeled with zonula occludens-1 (ZO-1) (Fig. 6B). There were significantly fewer regenerated cone photoreceptors in tgif1−/− fish compared with tgif1+/+ fish (Student t-test P< 0.005; Fig. 6B,C).
FIGURE 6.
Regeneration is significantly impaired in fish homozygous for tgif1−/− or both tgif1−/− and six3b−/− relative to tgif1+/+ fish. (A) BLBP immunoreactivity at 14 dpl in tgif1+/+;gfap:EGFPtgif1−/−;gfap:EGFP, and tgif1−/−;six3b−/−;gfap:EGFP fish expressing GFP in the Müller glia (lesioned area is left of dotted line). (B) ZO-1 immunolocalization at 14 dpl (panels at left) to visualize the OLM (dashed line in panels at right) of tgif1+/+tgif1−/−, and tgif1−/−;six3b−/− fish. (C) The number of photoreceptor nuclei in the regenerated area at 14 dpl. Scale bars=50 mm in (A), 10 µm in (B). ** P<0.005, *** P<0.0001, Student t-test pairwise comparisons of genotypes. OLM=outer limiting membrane. Other abbreviations as in Fig. 1.
Finally, we crossed tgtf1fh258 and six3bvu87 alleles into the gfap:EGFP Müller glia reporter line to test if tgif1 and six3b interact genetically. The cytoarchitecture of unlesioned retinas in adult tgif1−/− ;six3b−/− ;gfap:EGFP is normal (Supp. Info. Fig. S6). However, at 14 dpl, tgif1−/− ;six3b−/− ;gfa-p:EGFP fish exhibit an irregular OLM in the lesioned area (arrow, Fig. 6B) and significantly fewer regenerated cones compared with tgif1+/+ (Student t-test p< 0.0001) and tgif1−/− fish (Student t-test p< 0.005) (Fig. 6C. Therefore, loss of Six3b function enhances the regeneration defect we observed in tgif1−/− fish.
Discussion
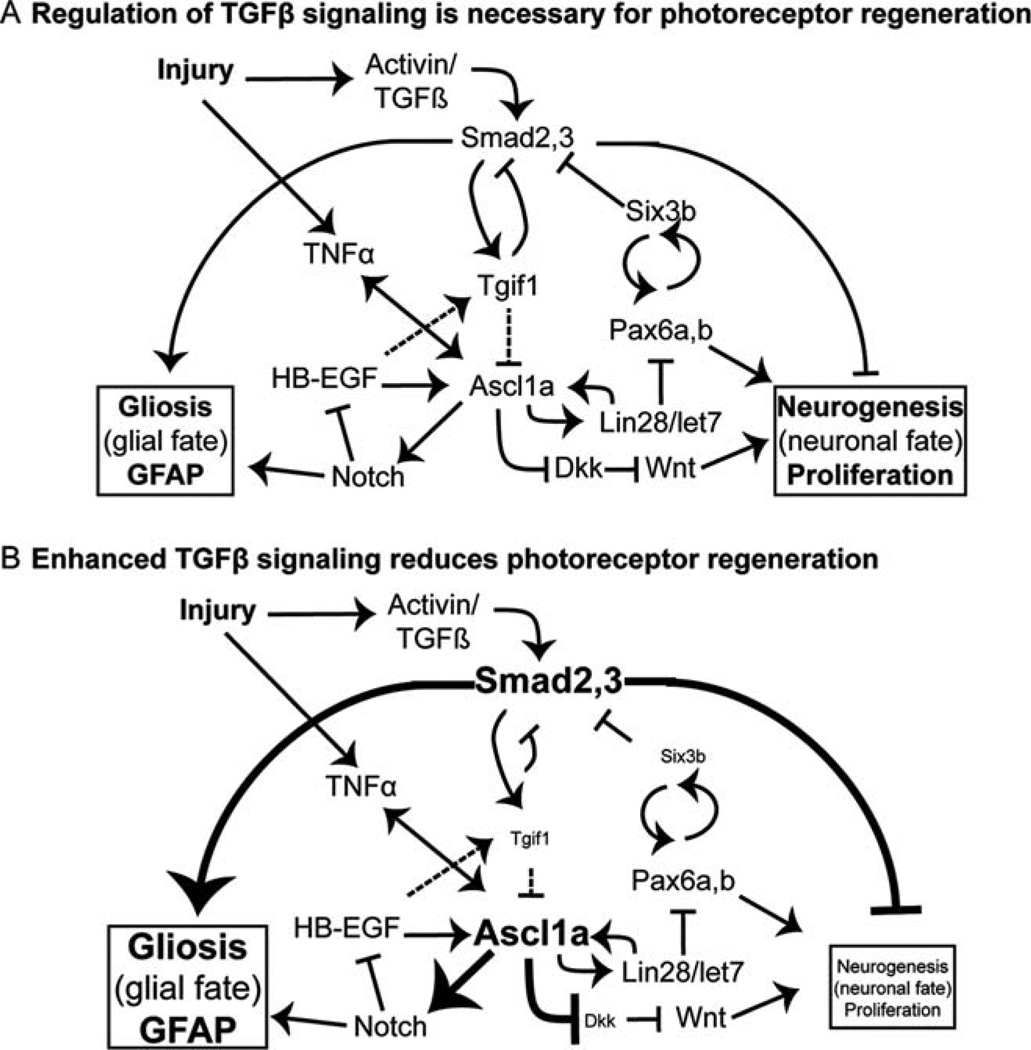
Adult zebrafish Müller glia have the capacity to self-renew and generate multipotent neuronal progenitors, characteristics typical of neural stem cells, and as such are a potential target for tissue-specific, regenerative stem cell therapies in the eye (Karl and Reh, 2010). To identify therapeutic strategies to promote stem cell properties in human Müller glia, it is necessary to identify both inductive and inhibitory cues (Locker et al., 2009). Our results indicate that Smad2/3-mediated TGFβ signaling acts to inhibit proliferation of neuronal progenitors following photoreceptor destruction in the adult zebrafish retina and may bias the damage response toward gliosis (Fig. 7). The mechanisms by which TGFβ signaling regulates proliferation are likely complex. TGFβ signaling-induced scarring is necessary for heart regeneration in adult zebrafish (Chablais and Jaźwińska, 2012), and similarly, TGFβ-induced deposition of extracellular matrix is critical for fin regeneration (Jaźwińska et al., 2007). However, studies of the vertebrate central nervous system suggest that the TGFβ signaling pathway normally promotes glial scarring in damaged neural tissues and inhibits cell proliferation (Bringmann et al., 2009; Robel et al., 2011).
FIGURE 7.
Signaling pathways that regulate Müller glial-derived regeneration of retinal neurons. Signaling pathways implicated in Müller glial response to retinal injury and retinal regeneration based on the present work and published studies (Bernardos et al., 2005; Ikeda and Puro, 1995; Inbal et al., 2007; Liu et al., 2006; Liu et al., 2011; Meyers et al., 2012; Nelson et al., 2012, 2013; Ramachandran et al., 2010; Robel et al., 2011; Thummel et al., 2010; Wan et al., 2012). Regeneration requires a dynamically balanced regulation of signals and transcriptional regulators promoting proliferation and differentiation, as well as cell fate choices between a glial fate (gliosis) and a neuronal fate (neurogenesis). See text for details.
TGFβ signaling influences retinal development and disease, but a role in the regulation of stem cells during retinal regeneration has not been explored. Small changes in the levels of Smad2/3 signaling in ESCs changes their transcriptional profile, leading to cell fate changes (Lee et al., 2011). Our results are consistent with this finding in that perturbations in the function of two TGFβ corepressors, tgif1 and six3b, inhibited the proliferative response of Müller glial-derived neuronal progenitors and, more importantly, diminished photoreceptor regeneration following acute light lesion (Fig. 7). The best understood mechanism by which Tgif1 inhibits TGFβ signaling is through binding to pSmad2/3 and then recruiting HDACs (Wotton et al., 1999), and Six3 is a Smad2/3 transcriptional corepressor (Inbal et al., 2007; Powers et al., 2010). Photoreceptor regeneration is not completely inhibited in tgif1−/− fish, perhaps because other functions of Tgif1 were not perturbed or because of functional redundancy with other genes that repress TGFβ signaling. Consistent with the later interpretation, we found that disruption of either six3 or tgif1 caused reduced proliferation after acute light lesion, and loss-of-function in both had a synergistic, negative effect on cone photoreceptor regeneration. This suggests that they cooperate in the same pathway to block TGFβ signaling in Müller glia and their neurogenic progeny.
Several Smad2/3 targets are rapidly upregulated in Müller glia in response to light damage, including ascl1a, tgfβ-induced, and tgif1 (Qin et al., 2009). Expression of the TGFβ ligand activin-βb increases dramatically in the ONL within 12 h after the initiation of a chronic light lesion in adult zebrafish (Craig et al., 2008). This ligand could be secreted by injured photoreceptors or activated microglia in the lesion and could induce the initial increase in Smad2/3 signaling that we observed. Interestingly, both of the corepressors we studied, tgif1 and six3b, maintain high levels of expression through the first few days after light lesion, whereas tgfβi, a secreted protein component of the extracellular matrix, is only transiently increased. Therefore, different mechanisms must regulate the transcription of cofactors and downstream targets after the initial increase in Smad2/3 signaling.
A growing body of research suggests that the growth factor EGF plays a critical role in the ability of Müller glia to become mitotically active. Murine Müller glia in retinal explants become mitotically active in response to EGF via signal transduction requiring bone morphogenetic protein/Smad signaling (Ueki and Reh, 2013). In the adult zebrafish retina, heparin-binding EGF (HB-EGF) is necessary in the repair of stab wounds and acts by mediating expression of critical genes such as pax6b and ascl1a (Wan et al., 2012), although a recent study of photoreceptor regeneration in adult zebrafish indicates that hb-egfa is not necessary for Müller glia-derived photoreceptor regeneration (Nelson et al., 2013). We found that retinas of tgif1−/− fish had similar levels of hb-egfa expression after acute light lesion relative to wild-type fish. This suggests that HB-EGF and canonical TGFβ signaling act in parallel pathways in retinal regeneration. It is also known that the mitotic effect of growth factors on Müller glia and neural stem cells is inhibited by TGFβ signaling both in vitro (Aigner and Bogdahn, 2008; Ikeda and Puro, 1995) and in vivo during rat retina development (Close et al., 2005). Additionally, EGF signaling increases phosphorylation and stabilization of Tgif1 in vitro (Lo et al., 2001), which would further suppress TGFβ signaling and allow for proliferation.
Consistent with these antiproliferative effects of TGFβ signaling, we propose that upregulation of the TGFβ corepressors Tgif1 and Six3b promotes the proliferation of Müller glia and Müller gliaderived neurogenic progenitors. The time course of Tgif1 and Six3b expression in the initial response in Müller glia suggests their importance: They reach a maximum level of expression before the initial mitotic division of Müller glia (Fig. 1) and earlier than other genes necessary for retina regeneration reach their maximum expression, such as ascl1a (Nelson et al., 2012; Qin et al., 2009; Ramachandran et al., 2010). Carefully controlled regulation of transcription factors and signaling pathways is very important in determining stem cell behavior and progenitor fate (Hsieh, 2012), and TGFβ signaling interacts with transcription factors that have critical roles in retinal development and regeneration: Pax6a and Ascl1a both have Smad2 binding sites (Liu et al., 2011); Six3b can inhibit Smad2/3-mediated signaling (Inbal et al., 2007); and Six3 activates Pax6, which is later necessary for maintenance of Six3 expression in lens development (Liu et al., 2006). Although tgif1−/− fish have significantly higher levels of ascl1a after light damage, they also have significantly less proliferation and fewer regenerated cone photoreceptors, indicating that ascl1a expression alone is not sufficient for retinal regeneration. The expression of ascl1a also indirectly inhibits expression of the Wnt signaling pathway inhibitor dkk1b (Ramachandran et al., 2011), and our study indicates that ascl1a and dkk1b expression are both downstream of Tgif1 transcriptional regulation. Consistent with increased ascl1a after lesion in tgif1−/− mutants, expression of dkk1b was reduced (Fig. 7). Wnt signaling is known to regulate aspects of Müller glia dedifferentiation, proliferation, differentiation, and cell survival in the retina of mice (Liu et al., 2013). In acute light lesions of larval zebrafish retina, hyper-activation of Wnt signaling does not increase proliferation of Müller glia (Meyers et al., 2012), but following stab wounds of the adult retina, inhibition of Wnt signaling reduces proliferation (Ramachandran et al., 2011). Further studies are necessary to clarify the role of Wnt signaling in retinal regeneration and the potential differences in lesion paradigms.
The proposition that dynamic and carefully controlled regulation of TGFβ signaling is particularly important for stem cell behavior and proliferation is consistent with methods developed for production of neural retinal precursors from ESCs: ESCs are first treated with LeftyA, a TGFβ signaling inhibitor, and Dkk1, then activin to upregulate TGFβ signaling (Ikeda et al., 2005). A small molecule inhibitor of TGFβ signaling can even replace Sox2 or cMyc to reprogram fibroblasts to become induced pluripotent stem cells (iPSC) (Ichida et al., 2009). Zebrafish Müller glia-derived progenitors express the pluripotent factors oct4 and lin28 following retina injury, suggesting they are similar to iPSCs (Ramachandran et al., 2010). Increased Tgif1 expression is associated with higher levels of proliferation and migration in several cancers as well (Castro et al., 2010; Yeh et al., 2012), and this may indicate the importance of functional separation of TGFβ inhibition of proliferation and induction of epithelial–mesenchymal transition (EMT) (Barrios-Rodiles et al., 2005; Hamid and Brandt, 2009). During Müller glia-based photoreceptor regeneration, dynamic regulation of TGFβ is likely critical in this environment: An initial upregulation of TGFβ signaling would allow for EMT and migration of Müller glia-derived neuronal progenitors, while subsequent inhibition of Smad2/3-mediated signaling by corepressors Tgif1 and Six3b would allow for proliferation of progenitors committed to a neuronal, not glial, fate.
Unlike neurogenesis in the localized and specialized stem cell niches of the adult vertebrate brain, Müller glia are mature differentiated cells embedded within the complex neuronal environment of the retina. After damage, Müller glia must actively repress glial differentiation signals and upregulate a transcriptome that will allow for controlled proliferation and generation of neuronal progenitors. Like development of the retina (Swaroop et al., 2010) and control of stem cells (Hsieh, 2012), this transcriptional program likely consists of a complex gene regulatory network in which the precise levels of many signals are critical for a successful regenerative outcome. An intriguing hypothesis is that an initial increase in TGFβ signaling is important for the damage response (gliosis) in Müller glia, and that subsequent inhibition of TGFβ signaling in adult zebrafish Müller glia distinguishes them from mammalian Müller glia and allows for a neurogenic response to damage (Fig. 7). Both fish and mammalian Müller glia react to retinal injury by upregulating GFAP, Pax6, and BLBP (Bernardos et al., 2007; Joly et al., 2011; Thummel et al., 2010). These are classic signs of gliosis (Joly et al., 2011), and yet adult zebrafish can regenerate lost neurons, whereas adult mammals generate glial scars. As more research is directed at the transcriptional and regulatory processes that allow for retinal regeneration in adult vertebrates, differences in the gene regulatory networks activated in damage responses in mammalian and nonmammalian Müller glia will be revealed. Our studies suggest that down regulation of Smad2/3 signaling in the Müller glia is particularly important for the proliferative, neurogenic, response of Müller glia to light-induced destruction of photoreceptors in the adult zebrafish.
Supplementary Material
Acknowledgment
Grant sponsor: National Institute of Health; Grant number: R01EY004318 (P.A.R.); T32EY013934-08 (University of Michigan); F32EY021659-01 (J.R.L.); R21EY019401 (R.T.); R01HG002995 (TILLING of Tgif1: C.B.M.).
The authors thank A. Fjose (six3a, six3b), A. Waskiewicz tgif1), and L. Solnica-Krezel (six3bvu87 fish line) for sharing reagents, D. Pawar for fish care, and L. Barthel for histology.
Footnotes
Additional Supporting Information may be found in the online version of this article.
References
- Aigner L, Bogdahn U. TGF-beta in neural stem cells and in tumors of the central nervous system. Cell Tissue Res. 2008;331:225–241. doi: 10.1007/s00441-007-0466-7. [DOI] [PubMed] [Google Scholar]
- Ando H, Kobayashi M, Tsubokawa T, Uyemura K, Furuta T, Okamoto H. Lhx2 mediates the activity of Six3 in zebrafish forebrain growth. Dev Biol. 2005;287:456–468. doi: 10.1016/j.ydbio.2005.09.023. [DOI] [PubMed] [Google Scholar]
- Barrios-Rodiles M, Brown KR, Ozdamar B, Bose R, Liu Z, Donovan RS, Shinjo F, Liu Y, Dembowy J, Taylor IW, Luga V, Przulj N, Robinson M, Suzuki H, Hayashizaki Y, Jurisica I, Wrana JL. High-throughput mapping of a dynamic signaling network in mammalian cells. Science. 2005;307:1621–1625. doi: 10.1126/science.1105776. [DOI] [PubMed] [Google Scholar]
- Bartholin L, Powers SE, Melhuish TA, Lasse S, Weinstein M, Wotton D. TGIF inhibits retinoid signaling. Mol Cell Biol. 2006;26:990–1001. doi: 10.1128/MCB.26.3.990-1001.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernardos RL, Barthel LK, Meyers JR, Raymond PA. Late-stage neuronal progenitors in the retina are radial Müller glia that function as retina stem cells. J Neurosci. 2007;27:7028–7040. doi: 10.1523/JNEUROSCI.1624-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernardos RL, Lentz SI, Wolfe MS, Raymond PA. Notch-delta signaling is required for spatial patterning and Müller glia differentiation in the zebra-fish retina. Dev Biol. 2005;278:381–395. doi: 10.1016/j.ydbio.2004.11.018. [DOI] [PubMed] [Google Scholar]
- Bringmann A, Iandiev I, Pannicke T, Wurm A, Hollborn M, Wiedemann P, Osborne NN, Reichenbach A. Cellular signaling and factors involved in Müller cell gliosis: Neuroprotective and detrimental effects. Prog Retin Eye Res. 2009;28:423–451. doi: 10.1016/j.preteyeres.2009.07.001. [DOI] [PubMed] [Google Scholar]
- Castro M, Grau L, Puerta P, Gimenez L, Venditti J, Quadrelli S, Sanchez-Carbayo M. Multiplexed methylation profiles of tumor suppressor genes and clinical outcome in lung cancer. J Transl Med. 2010;8:86. doi: 10.1186/1479-5876-8-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chablais F, Jaźwińska A. The regenerative capacity of the zebrafish heart is dependent on TGFβ signaling. Development. 2012;139:1921–1930. doi: 10.1242/dev.078543. [DOI] [PubMed] [Google Scholar]
- Chang M-L, Wu C-H, Jiang-Shieh Y-F, Shieh J-Y, Wen C-Y. Reactive changes of retinal astrocytes and Müller glial cells in kainate-induced neuro-excitotoxicity. J Anat. 2007;210:54–65. doi: 10.1111/j.1469-7580.2006.00671.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Close JL, Gumuscu B, Reh TA. Retinal neurons regulate proliferation of postnatal progenitors and Müller glia in the rat retina via TGF beta signaling. Development. 2005;132:3015–3026. doi: 10.1242/dev.01882. [DOI] [PubMed] [Google Scholar]
- Craig SE, Calinescu AA, Hitchcock PF. Identification of the molecular signatures integral to regenerating photoreceptors in the retina of the zebrafish. J Ocul Biol Dis Infor. 2008;1:73–84. doi: 10.1007/s12177-008-9011-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draper BW, McCallum CM, Stout JL, Slade AJ, Moens CB. A high-throughput method for identifying N-ethyl-N-nitrosourea (ENU)-induced point mutations in zebrafish. Methods Cell Biol. 2004;77:91–112. doi: 10.1016/s0091-679x(04)77005-3. [DOI] [PubMed] [Google Scholar]
- Fausett BV, Gumerson JD, Goldman D. The proneural basic helix-loop-helix gene ascl1a is required for retina regeneration. J Neurosci. 2008;28:1109–1117. doi: 10.1523/JNEUROSCI.4853-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Götz M, Barde YA. Radial glial cells defined and major intermediates between embryonic stem cells and CNS neurons. Neuron. 2005;46:369–372. doi: 10.1016/j.neuron.2005.04.012. [DOI] [PubMed] [Google Scholar]
- Hamid R, Brandt SJ. Transforming growth-interacting factor (TGIF) regulates proliferation and differentiation of human myeloid leukemia cells. Mo Oncol. 2009;3:451–463. doi: 10.1016/j.molonc.2009.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartfuss E, Galli R, Heins N, Gotz M. Characterization of CNS precursor subtypes and radial glia. Dev Biol. 2001;229:15–30. doi: 10.1006/dbio.2000.9962. [DOI] [PubMed] [Google Scholar]
- Hsieh J. Orchestrating transcriptional control of adult neurogenesis. Genes Dev. 2012;26:1010–1021. doi: 10.1101/gad.187336.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichida JK, Blanchard J, Lam K, Son EY, Chung JE, Egli D, Loh KM, Carter AC, Di Giorgio FP, Koszka K, Huangfu D, Akutsu H, Liu DR, Rubin LL, Eggan K. A small-molecule inhibitor of Tgf-β signaling replaces Sox2 in reprogramming by inducing Nanog . Cell Stem Cell. 2009;5:491–503. doi: 10.1016/j.stem.2009.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda H, Osakada F, Watanabe K, Mizuseki K, Haraguchi T, Miyoshi H, Kamiya D, Honda Y, Sasai N, Yoshimura N, et al. Generation of Rx+/Pax6+ neural retinal precursors from embryonic stem cells. Proc Natl Acad Sci USA. 2005;102:11331–11336. doi: 10.1073/pnas.0500010102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda T, Puro DG. Regulation of retinal glial cell proliferation by antiproliferative molecules. Exp Eye Res. 1995;60:435–443. doi: 10.1016/s0014-4835(05)80100-9. [DOI] [PubMed] [Google Scholar]
- Inbal A, Kim SH, Shin J, Solnica-Krezel L. Six3 represses nodal activity to establish early brain asymmetry in zebrafish. Neuron. 2007;55:407–415. doi: 10.1016/j.neuron.2007.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jazwinska A, Badakov R, Keating MT. Activin-βA signaling is required for zebrafish fin regeneration. Curr Biol. 2007;17:1390–1395. doi: 10.1016/j.cub.2007.07.019. [DOI] [PubMed] [Google Scholar]
- Joly S, Pernet V, Samardzija M, Grimm C. Pax6-positive Müller glia cells express cell cycle markers but do not proliferate after photoreceptor injury in the mouse retina. Glia. 2011;59:1033–1046. doi: 10.1002/glia.21174. [DOI] [PubMed] [Google Scholar]
- Karl MO, Reh TA. Regenerative medicine for retinal diseases: Activating endogenous repair mechanisms. Trends Mol Med. 2010;16:193–202. doi: 10.1016/j.molmed.2010.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassen SC, Ramanan V, Montgomery JE, C TB, Liu CG, Vihtelic TS, Hyde DR. Time course analysis of gene expression during light-induced photoreceptor cell death and regeneration in albino zebrafish. Dev Neurobiol. 2007;67:1009–1031. doi: 10.1002/dneu.20362. [DOI] [PubMed] [Google Scholar]
- Lee KL, Lim SK, Orlov YL, Yit le Y, Yang H, Ang LT, Poellinger L, Lim B. Graded Nodal/Activin signaling titrates conversion of quantitative phospho-Smad2 levels into qualitative embryonic stem cell fate decisions. PLoS Genet. 2011;7:e1002130. doi: 10.1371/journal.pgen.1002130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Hunter DJ, Rooker S, Chan A, Paulus YM, Leucht P, Nusse Y, Nomoto H, Helms JA. Wnt signaling promotes Muller cell proliferation and survival after injury. Invest Ophthalmol Vis Sci. 2013;54:444–453. doi: 10.1167/iovs.12-10774. [DOI] [PubMed] [Google Scholar]
- Liu W, Lagutin OV, Mende M, Streit A, Oliver G. Six3 activation of Pax6 expression is essential for mammalian lens induction and specification. EMBO J. 2006;25:5383–5395. doi: 10.1038/sj.emboj.7601398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Lin X, Cai Z, Zhang Z, Han C, Jia S, Meng A, Wang Q. Global identification of SMAD2 target genes reveals a role for multiple co-regulatory factors in zebrafish early gastrulas. J Biol Chem. 2011;286:28520–28532. doi: 10.1074/jbc.M111.236307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo RS, Wotton D, Massague J. Epidermal growth factor signaling via Ras controls the Smad transcriptional co-repressor TGIF. EMBO J. 2001;20:128–136. doi: 10.1093/emboj/20.1.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locker M, Borday C, Perron M. Stemness or not stemness? Current status and perspectives of adult retinal stem cells. Curr Stem Cell Res Ther. 2009;4:118–130. doi: 10.2174/157488809788167382. [DOI] [PubMed] [Google Scholar]
- Meyers JR, Hu L, Moses A, Kaboli K, Papandrea A, Raymond PA. β-cat-enin/Wnt signaling controls progenitor fate in the developing and regenerating zebrafish retina. Neural Dev. 2012;7:30. doi: 10.1186/1749-8104-7-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon LD, Fawcett JW. Reduction in CNS scar formation without concomitant increase in axon regeneration following treatment of adult rat brain with a combination of antibodies to TGFβ1 and β2. Eur J Neurosci. 2001;14:1667–1677. doi: 10.1046/j.0953-816x.2001.01795.x. [DOI] [PubMed] [Google Scholar]
- Nelson CM, Ackerman KM, O’Hayer P, Bailey TJ, Gorsuch RA, Hyde DR. Tumor necrosis factor-alpha is produced by dying retinal neurons and is required for Muller glia proliferation during zebrafish retinal regeneration. J Neurosci. 2013;33:6524–6539. doi: 10.1523/JNEUROSCI.3838-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson CM, Gorsuch RA, Bailey TJ, Ackerman KM, Kassen SC, Hyde DR. Stat3 defines three populations of Muller glia and is required for initiating maximal Müller glia proliferation in the regenerating zebrafish retina. J Comp Neurol. 2012;520:4294–4311. doi: 10.1002/cne.23213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers SE, Taniguchi K, Yen W, Melhuish TA, Shen J, Walsh CA, Sutherland AE, Wotton D. Tgif1 and Tgif2 regulate Nodal signaling and are required for gastrulation. Development. 2010;137:249–259. doi: 10.1242/dev.040782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin Z, Barthel LK, Raymond PA. Genetic evidence for shared mechanisms of epimorphic regeneration in zebrafish. Proc Natl Acad Sci USA. 2009;106:9310–9315. doi: 10.1073/pnas.0811186106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran R, Fausett BV, Goldman D. Ascl1a regulates Muller glia dedifferentiation and retinal regeneration through a Lin-28-dependent, let-7 microRNA signalling pathway. Nat Cell Biol. 2010;12:1101–1107. doi: 10.1038/ncb2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran R, Zhao X-F, Goldman D. Ascl1a/Dkk/β-catenin signaling pathway is necessary and glycogen synthase kinase-3β inhibition is sufficient for zebrafish retina regeneration. Proc Natl Acad Sci USA. 2011;108:15858–15863. doi: 10.1073/pnas.1107220108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond PA, Barthel LK, Bernardos RL, Perkowski JJ. Molecular characterization of retinal stem cells and their niches in adult zebrafish. BMC Dev Biol. 2006;6:36. doi: 10.1186/1471-213X-6-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichenbach A, Bringmann A. New functions of Müller cells. Glia. 2013;61:651–678. doi: 10.1002/glia.22477. [DOI] [PubMed] [Google Scholar]
- Robel S, Berninger B, Götz M. The stem cell potential of glia: lessons from reactive gliosis. Nat Rev Neurosci. 2011;12:88–104. doi: 10.1038/nrn2978. [DOI] [PubMed] [Google Scholar]
- Roesch K, Jadhav AP, Trimarchi JM, Stadler MB, Roska B, Sun BB, Cepko CL. The transcriptome of retinal Müller glial cells. J Comp Neurol. 2008;509:225–238. doi: 10.1002/cne.21730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh S, Watanabe S. TGIF, a homeodomain transcription factor, reguates retinal progenitor cell differentiation. Exp Eye Res. 2008;87:571–579. doi: 10.1016/j.exer.2008.09.007. [DOI] [PubMed] [Google Scholar]
- Shen J, Walsh CA. Targeted disruption of Tgif, the mouse ortholog of a human holoprosencephaly gene, does not result in holoprosencephaly in mice. Mol Cell Biol. 2005;25:3639–3647. doi: 10.1128/MCB.25.9.3639-3647.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A, Tsonis PA. Focus on molecules: Six3-master or apprentice? Exp Eye Res. 2010;90:535–536. doi: 10.1016/j.exer.2010.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swaroop A, Kim D, Forrest D. Transcriptional regulation of photoreceptor development and homeostasis in the mammalian retina. Nat Rev Neurosci. 2010;11:563–576. doi: 10.1038/nrn2880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thapa N, Lee BH, Kim IS. TGFBIp/βig-h3 protein: A versatile matrix molecule induced by TGF-β. Int J Biochem Cell Biol. 2007;39:2183–2194. doi: 10.1016/j.biocel.2007.06.004. [DOI] [PubMed] [Google Scholar]
- Thorrez L, Van Deun K, Tranchevent L-C, Van Lommel L, Engelen K, Marcha K, Moreau Y, Van Mechelen I, Schuit F. Using ribosomal protein genes as reference: A tale of caution. PLoS One. 2008;3:e1854. doi: 10.1371/journal.pone.0001854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thummel R, Enright JM, Kassen SC, Montgomery JE, Bailey TJ, Hyde DR. Pax6a and Pax6b are required at different points in neuronal progenitor cell proliferation during zebrafish photoreceptor regeneration. Exp Eye Res. 2010;90:572–582. doi: 10.1016/j.exer.2010.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueki Y, Karl MO, Sudar S, Pollak J, Taylor RJ, Loeffler K, Wilken MS, Reardon S, Reh TA. P53 is required for the developmental restriction in Muller glial proliferation in mouse retina. Glia. 2012;60:1579–1589. doi: 10.1002/glia.22377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueki Y, Reh TA. EGF stimulates Muller glial proliferation via a BMP-dependent mechanism. Glia. 2013;61:778–789. doi: 10.1002/glia.22472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan J, Ramachandran R, Goldman D. HB-EGF is necessary and sufficient for Müller glia dedifferentiation and retina regeneration. Dev Cell. 2012;22:334–347. doi: 10.1016/j.devcel.2011.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westerfield M. The zebrafish book: A guide for the laboratory use of zebrafish Danio (Brachydanio) rerio. Eugene, OR: M. Westerfield; 2000. [Google Scholar]
- Wotton D, Knoepfler PS, Laherty CD, Eisenman RN, Massagué J. The Smad transcriptional corepressor TGIF recruits mSin3. Cell Growth Differ. 2001;12:457–463. [PubMed] [Google Scholar]
- Wotton D, Lo RS, Swaby LA, Massague J. Multiple modes of repression by the Smad transcriptional corepressor TGIF. J Biol Chem. 1999;274:37105–37110. doi: 10.1074/jbc.274.52.37105. [DOI] [PubMed] [Google Scholar]
- Yeh BW, Wu WJ, Li WM, Li CC, Huang CN, Kang WY, Liu ZM, Huang HS. Overexpression of TG-interacting factor is associated with worse prognosis in upper urinary tract urothelial carcinoma. Am J Pathol. 2012;181:1044–1055. doi: 10.1016/j.ajpath.2012.05.024. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.