Abstract
The process of spermatogenesis in Drosophila melanogaster provides a powerful model system to probe a variety of developmental and cell biological questions, such as the characterization of mechanisms that regulate stem cell behavior, cytokinesis, meiosis, and mitochondrial dynamics. Classical genetic approaches, together with binary expression systems, FRT-mediated recombination, and novel imaging systems to capture single cell behavior, are rapidly expanding our knowledge of the molecular mechanisms regulating all aspects of spermatogenesis. This methods chapter provides a detailed description of the system, a review of key questions chapter that have been addressed or remain unanswered thus far, and an introduction to tools and techniques available to probe each stage of spermatogenesis.
1. Introduction
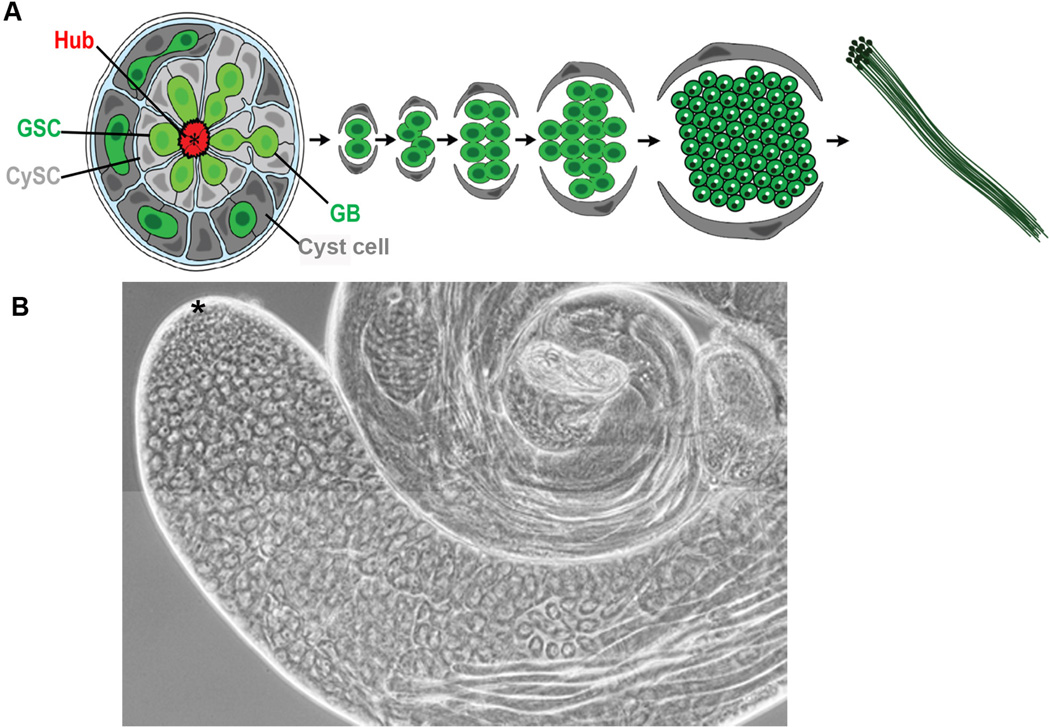
The Drosophila testis is a blunt-ended, coiled tube that supports the production of sperm throughout the life of the fly (Figure 1). Sustained spermatogenesis is accomplished by maintenance of a small number of stem cells that divide approximately once every 24 hours [1]. Both germline and somatic stem cells are located at the apical tip of the testis, and under homeostatic conditions, stem cells divide asymmetrically to produce two cells: one cell maintains stem cell characteristics, while the other daughter cell initiates differentiation. In the germ line, the differentiating daughter cell is called a gonialblast (GB), and the GB undergoes mitotic amplification (transit amplifying; TA) divisions to generate a cyst of spermatogonia, which will differentiate into spermatocytes and, ultimately, mature sperm. In D. melanogaster, four rounds of mitotic divisions and two meiotic divisions yield a total of 64 haploid spermatids [2] (Figure 1A). Elongation and maturation of the spermatids, spermiogenesis, is the final stage of spermatogenesis, after which the sperm is transported to the seminal vesicle [2, 3].
Figure 1. Spermatogenesis in Drosophila melanogaster.
A) Schematic of spermatogenesis. Germline stem cells (GSC, light green) and somatic cyst stem cells (CySC, light grey) contact hub cells (red). GSCs divide to self-renew and generate a daughter gonialblast (GB). GBs undergo 4 rounds of mitosis with incomplete cytokinesis to generate a cyst of 16 spermatogonia. Spermatogonial cysts (dark green) are surrounded by early cyst cells (dark grey). Spermatogonia initiate terminal differentiation as spermatocytes, which undergo 2 meiotic divisions with incomplete cytokinesis to generate 64 haploid spermatids. B) Phase contrast micrograph of a wild type testis. Asterisk denotes apical tip.
Thanks to efforts of the Drosophila community, reagents are readily available, including antibodies, reporter lines, and transgenic GAL4 ‘driver’ lines, which can be used to study the different stages of spermatogenesis (Tables 1 and 2). The use of specific drivers, in combination with immunofluorescence microscopy, permits characterization of specific cell types within the testis, even in a spatio-temporal manner. This chapter will provide an overview of spermatogenesis and discuss the tools and techniques available that facilitate a detailed analysis of the various cell biological processes in the male germ line.
Table 1.
Antibodies and transgenic lines useful for studying spermatogenesis
| Cell type /structure |
Marker | Type | Expression/Localization | Source/References |
|---|---|---|---|---|
| Soma | Fasciclin 3 (Fas3) | Antibody | Hub cells | DSHB [130] |
| Armadillo | Antibody | Hub cells | DSHB,[131] | |
| upd-lacZ | P-element insertion | Hub cells | [121, 132–134] | |
| esg-lacZ (M5–4) | P-element insertion | Hub cells | [57] | |
| esg-lacZ (esgG66B) | P-element insertion | Hub cells, cyst cells | [57, 135] | |
| Traffic Jam (Tj) | Antibody | CySCs and early cyst cells | [136] | |
| Zfh-1 | Antibody | CySCs and early cyst cells | [19, 137] | |
| Eyes Absent (Eya) | Antibody | Late cyst cells | DSHB [138, 139] | |
| esg-GFP | P-element insertion | Soma and germline | [140] | |
| Germline | UASt-Stat-GFP | GAL4/UAS | GSCs | [141, 142] |
| STAT92E | Antibody | GSCs | [143] | |
| GFP-Nanos | Transgene | GSCs and early germline cysts | [144] | |
| bam-GFP | Transgene | Mitotic germline cysts | [144] | |
| UASp-Bam-GFP | GAL4/UAS | Mitotic germline cysts | [145] | |
| Bam | Antibody | Mitotic germline cysts | DSHB, [77] | |
| Vasa-GFP | Transgene | Germline | [146] | |
| Vasa | Antibody | Germline | Santa Cruz Biotechnology #sc-26877, [147] | |
| Cytoskeleton | UASp-E-Cadherin-GFP | GAL4/UAS | Cell adhesion | [148] |
| UASp-Lifeact-GFP | GAL4/UAS | Actin, RCs | BDSC #35544, [149] | |
| UASp-Actin-GFP | GAL4/UAS | Actin, RCs | [150] | |
| GFP-Moesin | Transgene | Binds actin | [151] | |
| UASp-GFP-Tubulin | GAL4/UAS | Microtubules | [152] | |
| EB1-GFP | Transgene | Microtubule plus-ends | [153] | |
| UAS-IVS-myrtdTomato | GAL4/UAS | Plasma membranes | BDSC #32223, [154] | |
| α-Spectrin | Antibody | Fusome | DSHB, [155] | |
| Cytokinesis | GFP-Anillin | Transgene | Contractile rings, RCs, nuclei | [101] |
| mRFP-Anillin | Transgene | Contractile rings, RCs, nuclei | [101] | |
| Anillin | Antibody | Contractile rings, RCs, nuclei | [102, 156] | |
| GFP-Pavarotti | Transgene | Spindle midzone, RCs, nuclei | [102, 157] | |
| Pavarottii | Antibody | Spindle midzone, RCs | [157] | |
| Cindr | Antibody | Contractile rings and RCs | [102, 156] | |
| pTyr | Antibody | RCs | SIGMA #T1325, Santa Cruz Biotechnology #sc-508, [68] | |
| Zipper-GFP | P-element insertion | Contractile rings and RCs | BDSC #51564, [102, 146] | |
| Sqh-RFP | Transgene | Contractile rings and RCs | [158] | |
| Sqh-GFP | Transgene | Contractile rings and RCs | [159] | |
| Nucleus | His2Av-mRFP | Transgene | Histones | BDSC #23650 & #23651[160] |
| His2Av-EGFP | Transgene | Histones | BDSC #24163, [161] | |
| Sa-GFP | Transgene | Nucleolus in spermatocytes | [162] | |
| Polycomb-GFP | Transgene | Chromatin | [162] |
Table 2.
Commonly used Gal4 lines for studying spermatogenesis
| Cell type | Name | Insertion | Chr | BDSC # | Kyoto DGRC # |
Expression | References |
|---|---|---|---|---|---|---|---|
| Hub | Upd-Gal4 | P{upd-Gal4.U} | 1 | Hub | [132] | ||
| Hh-Gal4 | P{GAL4}hhGal4 | 3 | Hub | [163] | |||
| Fas3-Gal4 | P{GawB}NP1233 | 1 | 103948 | Hub | [164] | ||
| Soma | Tj-Gal4 | P{GawB}NP1624 | 2 | 104055 | CySCs and early cyst cells | [165] | |
| c587-Gal4 | P{GAL4}C587 | 1 | CySCs and early cyst cells | [166, 167] | |||
| Dpp-Gal4 | P{dpp-GAL4.PS} | 3 | 7007 | Hub and CySCs | [168] | ||
| Eya-Gal4 | P{eya-GAL4.A3} | 2 | Late cyst cells | [19] | |||
| Ptc-Gal4 | P{GawB}ptc559.1 | 2 | 2017 | 106629 | CySCs and cyst cells | [169] | |
| Germ line | Nanos-Gal4 | P{GalAL4-nos.NGT}40 | 2 | 4442 | 107748 | GSCs and early germline cysts | [170] |
| Nanos-Gal4-VP16 | P{GAL4::VP16-nos.UTR} CG6325[MVD1] | 3 | 4937 | GSCs and early germline cysts | [171] | ||
| Nanos-Gal4-VP16 | {GAL4::VP16-Nos.UTR}MVD2 | 1 | 7303 | GSCs and early | [171] | ||
| germline cysts | |||||||
| Bam-Gal4-VP16 | P{bam-GAL4:VP16} | 2 | Mitotic germline cysts | [144] | |||
| Vasa-Gal4 | P{vas-Gal4.2.6} | 2 | Germline | [172] | |||
| Germline and soma | Gal4 | P{tubP-Gal4}LL7 | 3 | 5138 | 108069 | Germline and soma | [173] |
1.1. The testis stem cell niche
Stem cells reside in specialized microenvironments called ‘niches’. These microenvironments are quite dynamic and provide structural and chemical support to the residing stem cells [4]. Examples of well-characterized stem cell niches can be found in many different animals. For example, in the gonad of the nematode Caenorhabditis elegans, the distal tip cell (DTC) participates in a niche, providing support to the germline stem cells via Notch signaling [5]. In both D. melanogaster and Mus musculus, more complex niches have been identified for germline, blood and intestinal stem cells, among others [6–17]. In most cases, one or more cell types and/or a basement membrane are integral components of the niche, providing structure and polarity to the system, and acting as a source of growth factors for stem cell maintenance and survival [4].
The Drosophila testis niche is composed of three highly interdependent cell types: the hub, the somatic cyst stem cells (CySCs) and the germline stem cells (GSCs) [18] (Figures 1A and 2A). The hub is comprised of a cluster of ten to twelve somatic cells that act as a signaling center for the GSCs and the CySCs. For example, hub cells secrete the ligand Unpaired (Upd), which activates the Janus kinase-Signal Transducer and Activator of Transcription (JAK-STAT) pathway in adjacent stem cells to regulate their behavior [8, 9, 19]. CySCs also strongly influence GSC behavior via the JAK-STAT pathway (discussed below in section 1.3) [20]. Additional factors, such as the bone morphogenetic protein (BMP) and hedgehog (Hh) pathways, also regulate stem cell behavior within the testis niche [21–26]. Indeed, the role of hub cells as an integral signaling center can be compared directly to the role of cap cells in the Drosophila ovary and the DTC in the C. elegans gonad [27]. However, in addition to acting as a signaling center, the hub is an important structural component that supports asymmetric GSC divisions by facilitating orientation of the mitotic spindle [28, 29].
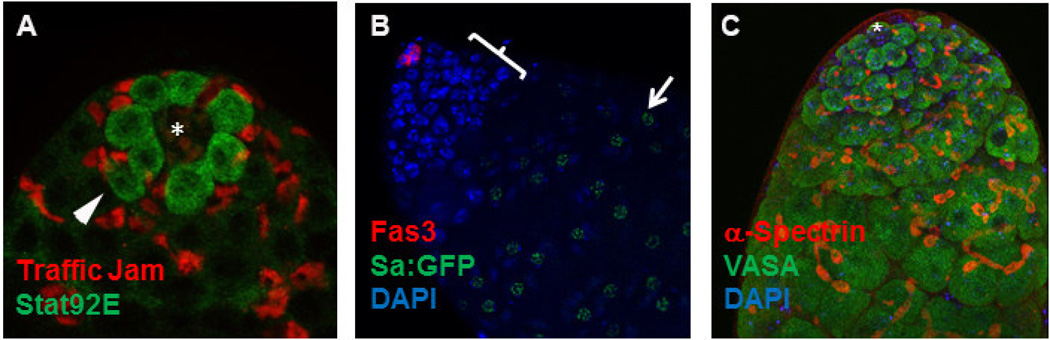
Figure 2. Immunofluorescence images of testis tips highlighting different tools used study early steps in spermatogenesis.
A) Immunofluorescence (IF) micrograph depicting GSCs and early cyst cells surrounding the apical hub (asterisk). Stat92E protein (green) is stabilized in GSCs, and early cyst cells (including CySCs) express the transcription factor Traffic Jam (red). B) IF image showing the hub marked by antibody staining for the cell surface marker Fasciclin3 (Fas3; red) and spermatocytes expressing Sa:GFP (arrow). Brackets denote the transit-amplification zone recognizable by dense DAPI staining of DNA in spermatogonia. C) IF image showing the progression of germline development as marked by the extension of a-spectrin+ fusomes in interconnected spermatogonia and spermatocytes. Germ cells are stained with an antibody to the RNA helicase Vasa (green). Asterisk denotes the hub.
Together with the Drosophila ovary, the Drosophila testis is one of the most readily available models for studying a niche in vivo [30]. Although there are tremendous similarities between mechanisms regulating stem cell behavior in the Drosophila ovary and testis [31, 32], in the testis, both germline and somatic stem cells are maintained within the same niche at the apical tip. Therefore, studies in the male germ line have facilitated our understanding of how stem cell populations cooperate and co-regulate each other. The interdependence of the three cell types – hub cells, CySCs and GSCs – makes this an important model system for stem cell researchers who are interested in how stem cells compete for niche occupancy in vivo [33–35]. Factors have been identified that are involved in regulating maintenance of hub cell fate and function[36, 37]. Such studies have permitted an analysis of how niche size influences stem cell number and how loss of niche function, over time, can contribute to loss of tissue homeostasis [38–40]. Recent work has also shed light into the plasticity of the stem cell niche, showing that quiescent hub cells can become ‘activated’ and contribute to the CySC pool upon loss of somatic stem cells or forced activation of the cell cycle in the hub [37, 41].
Hub cells can be recognized easily by staining with antibodies such as Fas3 or with reporters of unpaired expression (ie., upd-lacZ) (Table 1). Antibody staining for either N or E-cadherin will also exhibit enriched staining in hub cells, although reduced levels of staining are also observed in somatic cyst cells (Table 1). Importantly, either knock-down or overexpression of genes can be manipulated exclusively in hub cells by using drivers such as FasIII-Gal4 and upd-Gal4 (Table 2). Available tools for GSC and CySC identification and genetic manipulation are discussed below in sections 1.2 and 1.3, respectively.
1.2. Male germline stem cells
Although the average number varies by genotype, approximately 8 GSCs lie adjacent to and in direct contact with the hub cells, supporting continuous production of sperm throughout the lifetime of the fly. Each GSC can divide asymmetrically to give rise to one daughter cell that remains in contact with hub cells and maintains stem cell identity, while the GB is displaced away from the hub and begins differentiation (Figures 1A and 2A). However, recent advances in live-imaging have revealed that GSCs are also capable of dividing symmetrically, indicating that the dynamics of GSC divisions are more complicated that previously appreciated[1, 42] To ensure fidelity in stem cell maintenance and genome protection, multiple strategies (discussed below) control GSC self-renewal and gene expression [43].
GSCs initiate differentiation directly when signals to self-renew are abrogated, suggesting a mechanism whereby stem cell fate is actively maintained [44]. Indeed, an asymmetric outcome of male GSC divisions is guided largely by external cues [45–47]. Attachment of GSCs to hub cells via adherens junctions provides polarity cues that direct centrosomal anchoring and mitotic spindle orientation perpendicular to the GSC-hub interface during cell division [28, 29]. This ensures faithful asymmetric GSC division to maintain the GSC pool, as well as of differentiating progenitor cells [28]. Interestingly, proteins, chromosomes, and chromatin-associated factors have been shown to segregate preferentially into either the GSC or the GB, indicating that intrinsic cellular components may also contribute to self-renewal and maintenance of GSC fate [42, 48–52]. In addition, this may allow segregation of damaged components into daughter cells, such that the stem cell is protected. Furthermore, because each GSC has the potential to contribute to the next generation of flies, it is critical that the genome of the germ cells be kept intact and protected from damage. Indeed, transposon activity is considered a major deleterious event and is actively silenced in germ cells by members of the PIWI subfamily of RNA-binding proteins [53].
Lastly, during cytokinesis, the GSC-GB pair remains interconnected by a midbody ring until G2 of the following cell cycle, when finally abscission occurs [1]. The reason behind this delay in progression of differentiation of the GB is unclear, and unknown signals mediate closure and final abscission of the two cells [54]. Recent studies have shown that GSC-GB cytokinesis is asymmetric, such that the midbody ring is preferentially inherited by the GB and may eventually be released into somatic cells, where it is degraded [51].
GSCs can be distinguished using a combination of tools, coupled with the physical placement adjacent to hub cells (Figures 1A, 2A, 2C). For instance, antibodies to the conserved RNA helicase, Vasa, stain all germ cells; however, GSCs contain a spherical spectrosome, which is detected using antibodies to α-spectrin (Table 1; Figures 1A, 2C). Therefore, Vasa+ cells that are in physical contact with hub cells (Fas3+) are considered GSCs. In addition, GSCs often appear to be enriched for STAT92E (Figure 2A). Driver lines, such as nanos-Gal4 (Table 2), allow for specific gene manipulation in early germ cells, including GSCs.
Recent technological developments allowing genome-wide mutational or RNAi screening, as well as live imaging, have great potential to contribute to our knowledge about the complex regulation of GSC division and gene regulatory processes. A large RNAi screen identified more than 300 relevant factors that contribute to GSC maintenance in the Drosophila ovary, with many overlapping genes also required in neural stem cells[55]. A thorough comparison and validation of these genes in male GSC maintenance would enhance the current knowledge in the field, and likely increase the number of reagents available for the study of early germ cells. In sum, Drosophila GSCs serve as a useful model for studies of stem cell homeostasis and self-renewal, asymmetric cell division, cytokinesis and genome protection in vivo.
1.3 The somatic cyst cell lineage
Like the GSCs, somatic CySCs are also in physical contact with the hub and divide to both self-renew and give rise to cyst cells. In addition, recent work has shown that both the JAK-STAT and Hh pathways regulate CySC maintenance in a cell autonomous fashion [19, 22–24, 56]. Two cyst cells will encapsulate each GB and differentiate in concert with the germline cysts throughout spermatogenesis (Figures 1A and 2A) [18, 57, 58], functioning in an analogous way to the mammalian Sertoli cells. Indeed, communication between the germ line and soma ensures proper germline differentiation and, ultimately, adequate production of mature, functional gametes. Consequently, the study of germline-soma interactions in the Drosophila testis has provided valuable insight into cell-cell communication during development and maintenance of the tissue.
Encapsulation of the germ cells is required for efficient signaling between the germ line and soma, via epidermal growth factor (EGF) receptor signaling, to ensure differentiation of the developing germ cell cyst [59–62]. When EGFR signaling is blocked, early germ cells harboring characteristics of GSCs and GBs accumulate at the apical tip of the testis, at the expense of differentiating spermatogonia and spermatocytes. Aside from serving an instructive role during spermatogonial divisions, CySCs also regulate GSC proliferation and maintenance in a non-autonomous manner via the JAK-STAT and BMP pathways [19, 56, 63]. Precisely how the JAK-STAT, Hh, BMP, and EGFR signaling pathways are integrated to regulate the balance between self-renewal and differentiation remains an exciting question for scientists in the field.
As described above, GSCs require signals from both the hub and somatic cyst cells for proper homeostasis; however, upon loss of the germ line, such as in agametic mutants (ie., oskar mutants), the number of somatic cyst cells expands, due to increased proliferation, and somatic cyst cells acquire hub cell characteristics [57]. The pathways by which the germline restricts cyst cell proliferation and influences cyst cell versus hub cell fate have not been fully characterized and will also likely be a focus of future studies in the system.
CySCs and the cyst cells that encapsulate spermatogonial cysts (commonly referred to as ‘early’ cyst cells) can be visualized by staining with antibodies to proteins such as Traffic Jam (early cysts cells) (Table 1; Figure 2A). Staining for the transcription factor, Zfh-1 stains a more restricted subset of early cyst cells, presumably the CySCs and their immediate daughters [19]. By contrast, Eya is a transcription factor expressed in late cysts cells-those surrounding 16-cell spermatogonial cysts, as well as differentiating spermatocytes (Table 1). Unfortunately, no specific markers exist for CySCs. Therefore, a combination of Zfh-1 staining, placement adjacent to the hub, and markers of cell division, such as Bromodeoxyuridin (BrdU) incorporation or staining for phosphorylated histone H3, can be used to obtain information about the number and activity of CySCs.
Manipulation of gene expression in early cyst cells can be achieved with drivers such as c587-Gal4 and Tj-Gal4 (Table 2). However, each of these driver lines exhibits low-level, sporadic activation in hub cells (S. Landais, S. Sandall, and L. Jones, unpublished data). As signaling events between CySCs and GSCs appear critical for correct germline development, technological advances to allow expression of distinct transgenes in cyst cells (or hub cells) and germ cells, simultaneously, would benefit the field greatly. For example, transgenic flies could be generated that utilize both the GAL4/UAS and LexA/Op systems [64, 65]. In this way, pairs of interacting ligands and receptors may be identified and characterized, and a more detailed understanding of the interplay between the two different cell types in vivo can be gained.
1.4. Spermatogonia and mitotic amplification divisions
Following completion of GSC-GB cytokinesis, the GB enters the mitotic amplification (TA) division program. In D. melanogaster, the GB will undergo four rounds of mitotic divisions with incomplete cytokinesis, to generate a cyst of 16 spermatogonia that remain interconnected via stable intercellular bridges called ring canals (RCs) [2, 66]. Maintenance of these stable RCs are critical for germ cell development and are required for fertility in insects and mammals [54, 67, 68].|
For the precise development of cysts containing 16 spermatogonia, cell division must be controlled via networks of molecular interactions and signaling pathways [69, 70]. The fusome is a sub-cellular organelle that is a membrane and cytoskeletal-rich structure that traverses the RCs and progressively grows during the mitotic germ cell divisions to connect cells in the developing cyst [68] [71–74] (Figure 2C). Multiple cytoskeletal and signaling components display cell cycle-dependent localization to the fusome, suggesting that it may play a role in cell synchrony and coordination [75–77]. Incomplete cytokinesis during the germ cell divisions is also thought to contribute to synchronizing the mitotic cell divisions [54, 67]. However, the molecular mechanisms by which incomplete cytokinesis is controlled to prevent physical separation of the dividing cells remain largely unknown [54].
In addition, accumulation of the differentiation factor bag of marbles (bam) is required for germ cell differentiation [77, 78]. Through tight regulation of mRNA translation, a threshold level of Bam protein is reached to complete the TA divisions and initiate spermatocyte growth. Partial loss of bam function or modulation of Bam protein levels by expression of a non-degradable bamΔPEST transgene alters the dynamics of protein accumulation, and germ cell differentiation initiates either delayed or premature. Recently, additional layers of mRNA regulation have increased the network of players involved in initiation, maintenance and precise regulation of this gradient switch [69, 70, 79, 80].
Taken together, the TA zone of the testis provides a great model for understanding the cell and molecular machineries underlying lineage amplification. Spermatogonia within 4–16 cell cysts can be identified by staining with antibodies to Bam [77, 78]. In addition antibodies to α-spectrin are used to highlight fusome elongation as the degree of branching is often indicative of the number of successful TA divisions (Table 1). Furthermore, since this is a region of active cell division, BrdU incorporation and staining for phosphorylated histone H3 antibodies can also be utilized to recognize cells in S phase and mitosis, respectively, and to indicate that all cells within the cyst and progressing through the cell cycle in synchrony. Unfortunately, there is a dearth of tools to manipulate gene expression specifically in spermatogonia (Table 2). However, the bam-Gal4:VP16 driver permits strong, consistent staining in 4–16 cell spermatogonial cysts.
1.5. Spermatogonial de-differentiation
The plasticity of mitotic germ cells in the Drosophila testis is an exciting feature of this model system. Studies have shown that developing spermatogonia can dedifferentiate and re-acquire stem cell properties in order to contribute to the stem cell pool [34, 44]. Differentiation and de-differentiation of spermatogonia can be accurately timed and controlled when using genetic manipulations to reversibly force GSCs into differentiation or block self-renewal signaling. These powerful genetic manipulations rely on modulating the activity of either Stat92E or Bam [44, 81].
Fragmentation of spermatogonial cysts, coupled with re-attachment to the hub, replenishes the GSC pool in both these paradigms, although the detailed mechanisms remain unclear. To this end, several interesting questions regarding reversal of signaling gradients and incomplete cytokinesis remain unanswered. For example, what signals instruct the initial reversal of mitotic amplification divisions, and where does the signal originate? Secondly, what control mechanisms ensure fidelity and quality control when resetting self-renewal factors and stem cell identity genes? Approaching this plasticity with novel techniques and concepts will greatly add to the understanding of how tissues develop and how stem cells are coordinated in vivo.
The combination of a temperature-sensitive allele of Stat92E (StatFrankenstein, called StatF) and null alleles of Stat92E (ie., StatF /Stat06346, hereafter referred to as StatTS) allows for a temporary downregulation of STAT activity in all cells in the animal. JAK-STAT signaling in StatTS flies is permissive at 18oC, but when males are shifted to 29oC, Stat92E activity is lost and GSCs initiate differentiation [44]. Upon return to permissive temperatures, the stem cell pool is repopulated by de-differentiation of spermatogonial cysts [44, 82]. The second genetic tool available is a transgene carrying a heat-shock inducible bam (hs-bam), which permits transient overexpression [77]. In a series of heat shocks, bam is overexpressed and GSCs enter TA divisions while leaving the niche [81]. After a recovery period, the stem cell pool is again repopulated by dedifferentiation of spermatogonial cysts [34]. Whereas Bam is presumed to have an exclusive role in the germline, Stat92E activity is required in both the germline and somatic lineages for maintenance. Therefore, loss of Stat92E activity in the StatTS background will also affect somatic cells of the niche, which will have additional effects on the how germ cell behavior is controlled in this background [34].
1.6. Spermatocytes, meiosis and spermiogenesis
Spermatogonial cysts of 16 cells complete a pre-meiotic S-phase prior to enlargement and growth as primary spermatocytes. These cells grow in volume approximately 20–25 times, and multiple changes in transcription and chromatin organization can be detected, such as the separation of the chromosomes and the nucleolus into distinct compartments within the nucleus (Figure 2B) [83–85].
Because spermatocytes are large and progression through meiosis can easily be visualized, live imaging of dividing spermatocytes by phase contrast microscopy has resulted in several interesting discoveries [86–88]. Coupling phase contrast and fluorescence microscopy has also allowed for tracking of tagged molecules during meiosis, for example to address the separation of sister chromatids in vivo[89]. After two meiotic divisions, again with incomplete cytokinesis, cysts of 64 haploid spermatids are formed, sperm tails are elongated through extensive dynamics of actin and tubulin fibers, and these cells ultimately mature into sperm (Figure 1) [3]. Haploid spermatids display a morphology easily detected by phase contrast microscopy [83]. Small phase-light nuclei are paired with individual hyperfused mitochondria, forming a dense phase-dark structure called the nebenkern. This late stage of spermatogenesis, also referred as the ‘onion stage’, can be easily observed and has served as the basis for screens to identify factors required for regulation of chromosomal separation and functional cytokinesis [90–92]. If the chromosomes are not divided equally between the daughter cells, the size of the nuclei will differ [83]. Likewise, defects in contractile ring assembly or function will result in cleavage furrow regression and merging of the two nebenkerns into a single, larger black structure with two or four nuclei attached, depending on whether one or both of the meiotic divisions failed [92]. As spermatids undergo their dramatic morphological changes to acquire tails and begin to move, regulation of mitochondrial morphology, nuclear packaging and spermatid individualization is a highly specialized process [3]. Emergence of polarity within a developing cyst, assembly and growth of the flagellar axoneme and membrane dynamics during this multistep process must be coordinated and efficiently regulated to produce healthy spermatids in a fertile male [93, 94].
A number of beautiful experiments have revealed the complex waves of tissue specific transcription that occurs in spermatocytes, during terminal differentiation [43]. Furthermore, studies of spermatocytes and spermatids in Drosophila have contributed to the characterization of many conserved factors and processes that regulate the production of functional gametes . For instance, in the mitosis-to-meiosis transition, a class of mRNA-binding proteins called DAZ (deleted in azoospermia) was discovered to be the relevant gene in Y-chromosome azoospermia factor (AZF) thanks to the study of the fly homologue boule. Mutations in boule led to spermatocytes that fail to enter meiosis due to an impairment of meiotic-specific gene expression [95]. In addition, genetic screens in this system have led to the discovery of several regulators of cytokinesis, including the implication of central spindle microtubules in cleavage furrow formation [91, 96]. Because of the weak spindle assembly checkpoint present in spermatocytes, the functional uncoupling of genes required both in spindle assembly and later in cell division (such as the Polo kinase [97]) has been successfully investigated. Another example of a major discovery based on this system was of the gene fuzzy onions (fzo) which, when disrupted, resulted in the improper fusion of mitochondria into the nebenkern [98]. Fzo was the first characterized Mitofusin – a class of dynamin-like GTPases that promotes mitochondrial fusion, with many homologs throughout species [99].
Despite the importance of studies in spermatocytes and spermatids, few effective GAL4 driver lines exist for manipulation of gene expression after the TA mitotic divisions [100]. Sustained effects of transgene expression from the bam promoter (ie., using the bamGal4:VP16 driver line) into spermatocytes (and possibly further into meiotic divisions) may result in modest phenotypes, but the lack of a strong, specific and reliable binary system, such as the GAL4/UAS system, for probing meiosis and spermatid elongation has limited the approaches available [101, 102]. The development of efficient ways for genetic manipulation during these processes would be beneficial for further investigations into sperm differentiation.
2. Genetic manipulation and analysis of spermatogenesis
Thanks to the immense work done in the fly community over the years, a plethora of tools is available for the manipulation of genes in many Drosophila tissues, including the testis. Reagents and resources can be obtained from several stock centers, including the Developmental Studies Hybridoma Bank (DSHB) [103], FlyTED [104], the Exelixis Collection [105] and TRiP Collection [106] at Harvard Medical School, the Bloomington Drosophila Stock Center [107], the Vienna Drosophila RNAi Center [108], the Drosophila Genetic Resource Center (DGRC) at Kyoto [109], and FlyTrap [110], among others. In addition to these collections, several groups have performed extensive screens leading to the isolation of mutant alleles and P-element insertions that carry reporters and/or disrupt gene function [81, 111–113]. In this section, we will highlight and discuss some of the most used tools in the study of spermatogenesis. A previous review by Helen White-Cooper [100] offers a detailed analysis of the targeted ectopic gene manipulation methods available for the Drosophila testis.
2.1. Transgene expression
Most genetic manipulations done in Drosophila tissues and cells use the bipartite GAL4/UAS system from yeast [114]. This tool has been invaluable in the characterization of molecules and pathways important for development and tissue maintenance in the adult fly. GAL4 driver lines for transgene expression in the Drosophila male germ line are listed in Table 2. The first generation of UAS vectors used, named pUAST, was poorly expressed in the female germline. Developed by Brand and Perrimon in 1993, pUAST contains five GAL4-binding sites upstream of a basal hsp70 promoter, followed by a multiple-cloning site, and 3’ sequences from the SV40 virus [114]. Subsequently, Pernille Rørth developed a second generation of UAS vectors, named pUASP, in which expression in the female germ line was optimized [115]. Following a similar design as pUAST, pUASP contained fourteen GAL4-binding sites, followed by the P transposase promoter (including the first intron), restriction enzyme recognition sites, and stabilizing sequences for germline expression: the 3’ UTR and terminator of the K10 gene [115]. In contrast to the female germ line, male germ cells do not appear to have the same barrier for effective transgene expression from pUAST vectors. Nevertheless, spermatogenesis studies often make use of pUASP-based vectors for comparison to the female germ line and/or availability of constructs.
While in the past, transgenes were made primarily via random P-element insertion, new transgenic lines take advantage of phiC31-mediated insertion [116]. In this case, pUAS vectors contain an attB site that can be precisely recombined with an attP site previously inserted in the fly genome, allowing for site-directed insertion of trangenes into the genome. Collections of attP insertions have been generated, made by the Perrimon, Rubin and Bellen groups, among others, allowing for the possibility to choose where new transgenes will be integrated. Targeted integration of transgenes eliminates effects resulting from nearby enhancer, promoter or insulator elements. Therefore, expression derived from transgenes that are inserted into identical sites can be compared more directly than transgenes inserted in different, relatively uncharacterized, sites in the genome.
As noted above, the ability to manipulate independent signaling events in the germline and soma simultaneously would increase the types of questions that could be asked in this system tremendously. Therefore, adapting relatively new tools, such as the bipartite LexA/LexAop system (analogous to the GAL4/UAS system), to the germ line and somatic lineages of the testis should be a priority [64, 65]. In addition to existing driver lines (Table 2), which could be converted to other binary expression systems, the Rubin group at Janelia farm has a large collection of LexA enhancers used for neuronal studies that could potentially be screened for cell type-specific drivers in the testis [117].
2.2 RNA interference
All of the advances in Drosophila transgenesis can be appreciated when looking at the evolution of the RNAi libraries available. Two major stock centers maintaining transgenic flies harboring RNAi constructs, the Vienna Drosophila RNAi Center (VDRC [108]) and the Transgenic RNAi Project (TRiP [106]) at Harvard Medical School, carry an extensive number of RNAi-based transgenic lines. The VDRC has generated the P-Element RNAi Library (GD) and the phiC31 RNAi Library (KK); while the TRiP has generated the first and second generations of VALIUM (Vermilion-AttB-Loxp-Intron-UAS-MCS) vectors - both based on site-directed integration of transgenes, with the difference that the second generation evokes the microRNA pathway to deliver siRNAs. As exemplified in several instances [100, 101], the effects of RNAi-mediated knockdown vary based on the cell type in which the transgene is being expressed. Therefore, one should take advantage of transgenes that have been optimized for expression in the germline, when available.
2.3. FRT-mediated recombination, lineage-tracing, and clonal analysis
Although studies using conventional gain and loss-of-function alleles have been valuable, many strong hypomorphic or null alleles result in lethality during development. Using FRT-mediated recombination to generate cells that are homozygous mutant for a specific gene, in otherwise heterozygous animals, has been a powerful approach for studying cell-type specific roles for essential genes. In addition to analyzing the function of a particular gene within a cell, FRT-mediated recombination can be used to generate ‘marked’ cells that express a visible marker (ie., GFP) in order to follow the progeny of these cells, which is known as “clonal analysis” or “lineage-tracing” [118].
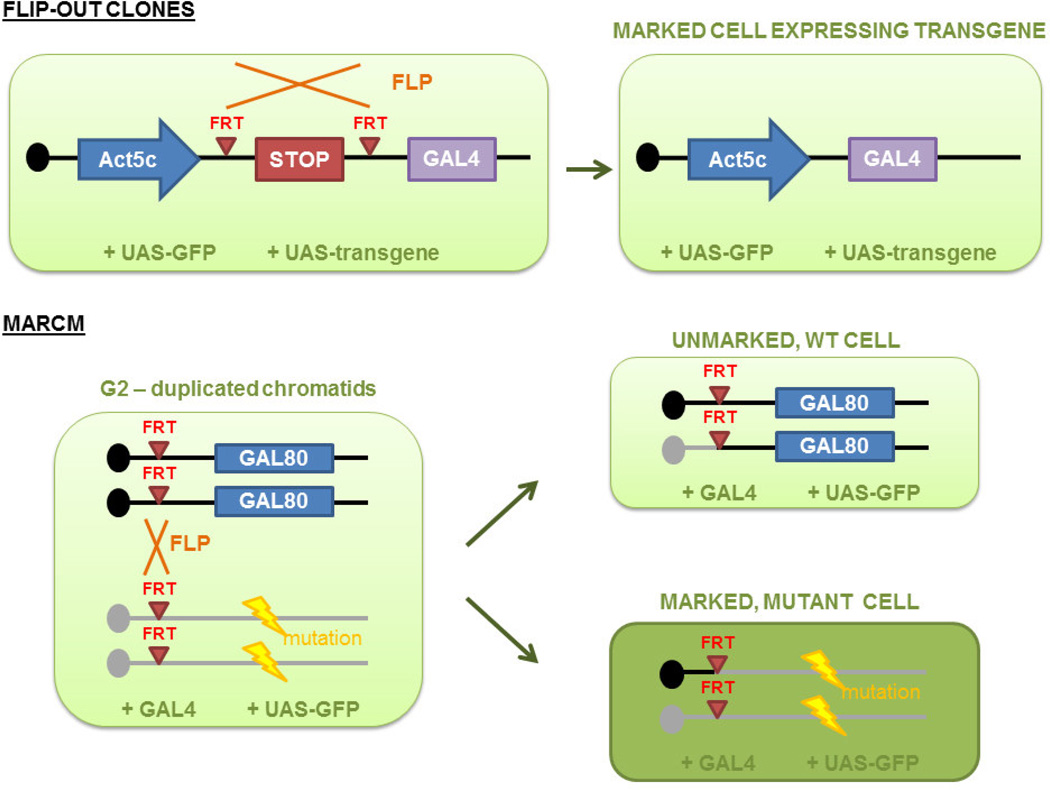
Instead of the traditional approach, in which clones are identified by the absence of a marker, systems like MARCM (mosaic analysis with a repressible cell marker) are based on marking the cell with a traceable molecule [119]. Specifically, MARCM is based on expression of a marker (i.e., UAS-GFP) resulting from the loss of the GAL80 repressor upon FLP/FRT-based recombination (Figure 3). More recently, derivations of mosaic analysis systems, such as G-TRACE [120], have been developed to optimize lineage analysis.
Figure 3. Use of FRT-mediated recombination strategies in Drosophila melanogaster.
A) FRT (flippase recognition target) sites located on the same chromosome permits generation of ‘flip-out’ clones. Upon expression of Flippase (FLP), recombination occurs, removing intervening sequences and permitting expression of ^Gal4. Resultant “clones” will be marked by the expression of the GAL4-responding transgene (i.e., UAS-GFP). If recombination occurs in a stem cell, progeny of that stem cell will be similarly marked. This strategy does not require mitosis. B) Use of the MARCM system permits the generation of clones and lineage-tracing studies. As shown above, upon expression of FLP in mitotic cells, FRT sites from homologous chromosomes, one of which contains a specific mutation, will recombine in a sitespecific manner. Wild type chromosomes also carry the GAL80 repressor, which represses GAL4 activity such that expression of the GAL4-responding transgene (UAS-GFP) is suppressed. Upon loss of GAL80, cells containing chromosomes carrying mutations will be marked by the expression of GFP.
When working with alleles on autosomes, clonal analysis in dividing germline and somatic stem cells is no different than in any other tissue. Mutations on the X chromosome, however, add an extra challenge to using FRT-mediated recombination in males, as recombination occurs between homologous chromosomes, and there is only one X in males. To overcome this hurdle, one can make use of genomic rescue constructs recombined onto an FRT-containing autosome. In this scenario, all cells would carry the loss-of-function allele on the X, and the rescue construct could be ‘flipped-out’ using the traditional FLP-FRT system.
One variation of the clonal analysis system allows for transgene overexpression and/or RNAi-mediated knockdown in a mosaic fashion [121]. In this system, any transgene that is under control of a UAS promoter can be used. The cassette consists of a strong promoter (i.e., Act5p) upstream of a pair of FRT sites flanking a spacer gene, often a visible marker, with a stop codon, followed by Gal4 (Act5p–FRT-CD2-FRT-Gal4). In this way, Gal4 is not expressed in the fly. Flies carrying this cassette in combination with a marker transgene, such as UAS-GFP, can be crossed to flies carrying a heat-shock inducible flippase (hs-flp) and the test transgene of choice (i.e., UAS-geneXRNAi). Upon a series of heat shocks, the resulting genotype (hs-flp; UAS-GFP; UAS-geneXRNAi/ Act5p–FRT-CD2-FRT-Gal4) will produce clones of cells expressing the marker and transgenes stochastically (Figure 3).
3. Imaging
Dissection of the Drosophila testis is relatively simple, especially from young, wild type flies. As the fly ages, testes decrease in size while the accessory glands grow, occasionally making it difficult to distinguish between the two. In addition to the characteristic coil of the testis that allows it to be detected, training and experience allows rapid and efficient dissections to be performed. The white gene produces bright yellow pigment cells that ensheath the testis. The use of genotypes that are wild-type for the white locus (w+), such as Oregon R can thus facilitate recognition of the testis‥ The tissue can be dissected in regular phosphate buffered saline (PBS) or modified buffers, such as TBS (testis buffer saline) or serum-enriched culture medium [86]. From here, the tissue can be prepared for fixation of whole mount or squashed preparatins, for observation in a phase contrast microscope, or for live imaging [122–124].
3.1 Fixed tissue
One protocol for fixation of testis tissue was developed in which the testis is gently squashed between a glass slide and a cover slip, and then quickly frozen in liquid nitrogen before proceeding with fixation and immunostaining (BOX 1) [83]. This method compresses (‘squashes’) the tissue to two to three cell layers at the mitotic zone, while many of the spermatocyte cysts are released from the tissue and can be studied without the confinements of the surrounding muscle layers [68, 101]. Fixation in methanol and acetone preserves microtubules well, while ethanol and formaldehyde fixation preserves F-actin (BOX 1) [83, 123–126].
BOX 1 – Protocols for tissue fixation.
The protocol described below may be adapted to optimize antibody detection and preservation of fluorescent tags. Steps are performed at room temperature (RT) unless otherwise noted. Hoechst or DAPI may be used to visualize nuclei before mounting in appropriate medium to protect the fluorophores from photo-bleaching.
A. Standard formaldehyde/paraformaldehyde fixation protocol
Reagents
Formaldehyde, 4%, methanol free, diluted in PBS
PBT (0.3%BSA/0.3%Triton X-100/PBS)
Dissect tissue in PBS and transfer to fixation solution.
Fix in 4% FA, 20–30 min.
Wash 3×10 min in PBT.
Block 30–45 min in PBT.
Incubate in primary antibody in PBT at 4˚C overnight.
Wash 3×10 min in PBT.
Secondary antibody in PBT 2 hours at RT.
Wash 3×10 min in PBT.
B. Testis squash protocol, from [83, 125]
Reagents
Methanol, water free
Acetone
Ethanol, 96 %
Formaldehyde, 4%, methanol free, diluted in PBS
PBST (0.1% Triton X-100/PBS)
Permeabilization buffer (0.3% Triton X-100/PBS)
PBT (5%BSA/0.3% Triton X-100/PBS).
Coverslips and glass slides
Dissect testes in PBS, transfer to a drop (~25 µL) of PBS on the glass slide. Puncture testes at the base, or cut 1/3 distance from apical tip, to allow cysts of cells to be released. Apply coverslip, and gently aspirate PBS from beneath the coverslip using a small piece of filter paper, or Kimwipe™, to squash the preparations. Watch under phase contrast microscope to control amount of ‘squashing’ achieved by surface tension between the coverslip and the slide. Do not over-squash the tissue.
Quick-freeze the slide in liquid nitrogen. Quickly remove coverslip using a scalpel or razorblade, and immediately place slide in appropriate fixation solution:
- Standard protocol to allow effective visualization of microtubules:
- Incubate in ice-cold methanol for 5 min at −20˚C.
- Transfer to ice-cold acetone for 30 sec-1 min at −20˚C.
- Incubate in PBS for 5 min.
- Modified protocol to allow visualization of actin filaments:
- Incubate in ice-cold ethanol for 10 min at −20˚C.
- Transfer to freshly prepared 3.7% formaldehyde for 7 min at RT.
- Incubate twice in PBS for 5 min.
Incubate in permeabilization buffer for 30 min.
Wash once in PBST, 5 min.
Incubate with primary antibody in PBT for 1 hour.
Wash 2×5 min in PBST.
Incubate with secondary antibodies in PBT for 1 hour.
Wash once in PBST.
As described for preparation of ovaries, most of the current work on the testis is performed using formaldehyde fixation of whole tissue at room temperature, and many of the same conditions and concerns apply to both systems [127]. Modifications to this standard protocol include the source of formaldehyde (with or without methanol), duration of fixation (5–30 minutes), fixation at room temperature or on ice, antigen blocking and the duration of the antibody labeling [128]. These adjustments influence antigen stabilization, penetrance into the tissue and background detection, and should be determined experimentally for antibodies that display suboptimal labeling.
3.2 Imaging spermatogenesis in real time
By gentle squashing or simply cutting the tissue open with a tungsten needle or tweezers, spermatocyte cysts can be released from the tissue and imaged under a phase contrast microscope, coupled to fluorescent lamps and detectors [87, 88, 96].
High resolution live imaging, using confocal or spinning disk systems (BOX 2), has also been adopted for visualizing events in the testis, with a focus on the stem cell niche at the apical tip [1, 34]. Avoiding damage caused by strong lasers is critical and must be monitored. Also, since the tissue is still intact, peristaltic muscle contractions and healthy movement can make long term imaging difficult, and thus, requires some form of stabilization or attachment of the tissue to the surface. The ability to provide necessary nutrients from a tissue culture medium, gas exchange with the surroundings and minimizing tissue movement can be accomplished using halocarbon oil as a spacer between the glass slide and the cover slip [1]. The protocol published for live imaging of processed during Drosophila oogenesis [129] provides additional insight to the precision required for maintaining a healthy tissue at physiological conditions for longer periods of time.
BOX 2 – Protocol for Live Imaging, adapted from[1].
Live Imaging of testesex vivo
Reagents
Halocarbon oil 27
Schneider’s insect medium
Fetal Bovine Serum (FBS)
Penicilin/Streptomycin (P/S)
Coverslips and glass slides
Dissect testes in Drosophila Culture Medium (15% FBS, 0.5× P/S, Schneider’s insect medium, DCM), rinse it twice and transfer to a glass slide containing a drop of DCM. Surround the drop with halocarbon oil and gently apply the coverslip on top. Experiments with extended imaging periods done by the Matunis lab have used 1cm2 Teflon sheets (YSI Life Sciences 5793) instead of glass slides [1]. Image using appropriate laser scanning or spinning disk microscopes.
Acknowledgements
We would like to thank the Drosophila community for its generosity with tools and reagents, Monica Boyle for contributing images used in Figure 2, and an anonymous reviewer for critical reading of the manuscript. In addition, we apologize to those colleagues whose work could not be referenced directly due to space constraints. Å.E. is funded by The Norwegian Cancer Society, and K.H is funded by a career grant from the South-Eastern Norway Regional Health Authority and acknowledges an EMBO Short Term Fellowship. D.L.J. is funded by the Eli and Edythe Broad Center of Regenerative Medicine and Stem Cell Research at the University of California-Los Angeles, CIRM, and the NIH.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Sheng XR, Matunis E. Live imaging of the Drosophila spermatogonial stem cell niche reveals novel mechanisms regulating germline stem cell output. Development. 2011;138:3367–3376. doi: 10.1242/dev.065797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fuller MT. Spermatogenesis. In: Bate MaAMA., editor. The Development of Drosophila. Cold Spring Harbor, NY: Cold Spring Harbor Press; 1993. pp. 71–147. [Google Scholar]
- 3.Fabian L, Brill JA. Drosophila spermiogenesis: Big things come from little packages. Spermatogenesis. 2012;2:197–212. doi: 10.4161/spmg.21798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Voog J, Jones DL. Stem cells and the niche: a dynamic duo. Cell Stem Cell. 2010;6:103–115. doi: 10.1016/j.stem.2010.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kimble J. Alterations in cell lineage following laser ablation of cells in the somatic gonad of Caenorhabditis elegans. Dev Biol. 1981;87:286–300. doi: 10.1016/0012-1606(81)90152-4. [DOI] [PubMed] [Google Scholar]
- 6.Margolis J, Spradling A. Identification and behavior of epithelial stem cells in the Drosophila ovary. Development. 1995;121:3797–3807. doi: 10.1242/dev.121.11.3797. [DOI] [PubMed] [Google Scholar]
- 7.Xie T, Spradling AC. decapentaplegic is essential for the maintenance and division of germline stem cells in the Drosophila ovary. Cell. 1998;94:251–260. doi: 10.1016/s0092-8674(00)81424-5. [DOI] [PubMed] [Google Scholar]
- 8.Kiger AA, Jones DL, Schulz C, Rogers MB, Fuller MT. Stem cell self-renewal specified by JAK-STAT activation in response to a support cell cue. Science. 2001;294:2542–2545. doi: 10.1126/science.1066707. [DOI] [PubMed] [Google Scholar]
- 9.Tulina N, Matunis E. Control of stem cell self-renewal in Drosophila spermatogenesis by JAK-STAT signaling. Science. 2001;294:2546–2549. doi: 10.1126/science.1066700. [DOI] [PubMed] [Google Scholar]
- 10.Rizki TM. Blood cells of Drosophila as related to metamorphosis. In: Campbell FL, editor. Physiology of Insect Development. Chicago, IL: Chicago University Press; 1956. pp. 91–94. [Google Scholar]
- 11.Evans CJ, Liu T, Banerjee U. Methods. 2014. Drosophila hematopoiesis: Markers and methods for molecular genetic analysis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weissman IL, Anderson DJ, Gage F. Stem and progenitor cells: origins, phenotypes, lineage commitments, and transdifferentiations. Annual review of cell and developmental biology. 2001;17:387–403. doi: 10.1146/annurev.cellbio.17.1.387. [DOI] [PubMed] [Google Scholar]
- 13.Wagers AJ. Stem cell grand SLAM. Cell. 2005;121:967–970. doi: 10.1016/j.cell.2005.06.017. [DOI] [PubMed] [Google Scholar]
- 14.Garrett RW, Emerson SG. Bone and blood vessels: the hard and the soft of hematopoietic stem cell niches. Cell Stem Cell. 2009;4:503–506. doi: 10.1016/j.stem.2009.05.011. [DOI] [PubMed] [Google Scholar]
- 15.Micchelli CA, Perrimon N. Evidence that stem cells reside in the adult Drosophila midgut epithelium. Nature. 2006;439:475–479. doi: 10.1038/nature04371. [DOI] [PubMed] [Google Scholar]
- 16.Ohlstein B, Spradling A. The adult Drosophila posterior midgut is maintainedlby pluripotent stem cells. Nature. 2006;439:470–474. doi: 10.1038/nature04333. [DOI] [PubMed] [Google Scholar]
- 17.Barker N, van Es JH, Kuipers J, Kujala P, van den Born M, Cozijnsen M, Haegebarth A, Korving J, Begthel H, Peters PJ, et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007;449:1003–1007. doi: 10.1038/nature06196. [DOI] [PubMed] [Google Scholar]
- 18.Hardy RW, Tokuyasu KT, Lindsley DL, Garavito M. The germinal proliferation center in the testis of Drosophila melanogaster. Journal of ultrastructure research. 1979;69:180–190. doi: 10.1016/s0022-5320(79)90108-4. [DOI] [PubMed] [Google Scholar]
- 19.Leatherman JL, Dinardo S. Zfh-1 controls somatic stem cell self-renewal in ‘the Drosophila testis and nonautonomously influences germline stem cell self-renewal. Cell Stem Cell. 2008;3:44–54. doi: 10.1016/j.stem.2008.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leatherman JL, Dinardo S. Germline self-renewal requires cyst stem cells and stat regulates niche adhesion in Drosophila testes. Nature cell biology. 2010;12:806–811. doi: 10.1038/ncb2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shivdasani AA, Ingham PW. Regulation of stem cell maintenance and transit amplifying cell proliferation by tgf-beta signaling in Drosophila spermatogenesis. Curr Biol. 2003;13:2065–2072. doi: 10.1016/j.cub.2003.10.063. [DOI] [PubMed] [Google Scholar]
- 22.Amoyel M, Sanny J, Burel M, Bach EA. Hedgehog is required for CySC self-renewal but does not contribute to the GSC niche in the Drosophila testis. Development. 2013;140:56–65. doi: 10.1242/dev.086413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Michel M, Kupinski AP, Raabe I, Bokel C. Hh signalling is essential for somatic stem cell maintenance in the Drosophila testis niche. Development. 2012;139:2663–2669. doi: 10.1242/dev.075242. [DOI] [PubMed] [Google Scholar]
- 24.Zhang Z, Lv X, Jiang J, Zhang L, Zhao Y. Dual roles of Hh signaling in the regulation of somatic stem cell self-renewal and germline stem cell maintenance in Drosophila testis. Cell Res. 2013;23:573–576. doi: 10.1038/cr.2013.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Michel M, Raabe I, Kupinski AP, Perez-Palencia R, Bokel C. Local BMP receptor activation at adherens junctions in the Drosophila germline stem cell niche. Nat Commun. 2011;2:415. doi: 10.1038/ncomms1426. [DOI] [PubMed] [Google Scholar]
- 26.Kawase E, Wong MD, Ding BC, Xie T. Gbb/Bmp signaling is essential for maintaining germline stem cells and for repressing bam transcription in the Drosophila testis. Development. 2004;131:1365–1375. doi: 10.1242/dev.01025. [DOI] [PubMed] [Google Scholar]
- 27.Kimble J, Crittenden SL. Controls of germline stem cells, entry into meiosis, and the sperm/oocyte decision in Caenorhabditis elegans. Annual review of cell and developmental biology. 2007;23:405–433. doi: 10.1146/annurev.cellbio.23.090506.123326. [DOI] [PubMed] [Google Scholar]
- 28.Yamashita YM, Jones DL, Fuller MT. Orientation of asymmetric stem cell division by the APC tumor suppressor and centrosome. Science. 2003;301:1547–1550. doi: 10.1126/science.1087795. [DOI] [PubMed] [Google Scholar]
- 29.Inaba M, Yuan H, Salzmann V, Fuller MT, Yamashita YM. E-cadherin is required for centrosome and spindle orientation in Drosophila male germline stem cells. PLoS One. 2010;5:e12473. doi: 10.1371/journal.pone.0012473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fuller MT, Spradling AC. Male and female Drosophila germline stem cells: two versions of immortality. Science. 2007;316:402–404. doi: 10.1126/science.1140861. [DOI] [PubMed] [Google Scholar]
- 31.de Cuevas M, Matunis EL. The stem cell niche: lessons from the Drosophila testis. Development. 2011;138:2861–2869. doi: 10.1242/dev.056242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xie T. Control of germline stem cell self-renewal and differentiation in the Drosophila ovary: concerted actions of niche signals and intrinsic factors. Wiley interdisciplinary reviews. Developmental biology. 2013;2:261–273. doi: 10.1002/wdev.60. [DOI] [PubMed] [Google Scholar]
- 33.Issigonis M, Tulina N, de Cuevas M, Brawley C, Sandler L, Matunis E. JAK-STAT signal inhibition regulates competition in the Drosophila testis stem cell niche. Science. 2009;326:153–156. doi: 10.1126/science.1176817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sheng XR, Brawley CM, Matunis EL. Dedifferentiating spermatogonia outcompete somatic stem cells for niche occupancy in the Drosophila testis. Cell Stem Cell. 2009;5:191–203. doi: 10.1016/j.stem.2009.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stine RR, Matunis EL. Stem cell competition: finding balance in the niche. Trends in cell biology. 2013;23:357–364. doi: 10.1016/j.tcb.2013.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Resende LP, Boyle M, Tran D, Fellner T, Jones DL. Headcase promotes cell survival and niche maintenance in the Drosophila testis. PLoS One. 2013;8:e68026. doi: 10.1371/journal.pone.0068026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Voog JSS, Hime G, Resende LPF, Aslanan A, Loza-Coll M, Hunter T, Fuller MT, Jones DL. Escargot restricts niche cell-stem cell conversion in the Drosophila testis. Cell reports. 2014 doi: 10.1016/j.celrep.2014.04.025. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Toledano H, D’Alterio C, Czech B, Levine E, Jones DL. The let-7-Imp axis regulates ageing of the Drosophila testis stem-cell niche. Nature. 2012;485:605–610. doi: 10.1038/nature11061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wallenfang MR, Nayak R, DiNardo S. Dynamics of the male germline stem cell population during aging of Drosophila melanogaster. Aging Cell. 2006;5:297–304. doi: 10.1111/j.1474-9726.2006.00221.x. [DOI] [PubMed] [Google Scholar]
- 40.Boyle M, Wong C, Rocha M, Jones DL. Decline in self-renewal factors contributes to aging of the stem cell niche in the Drosophila testis. Cell Stem Cell. 2007;1:470–478. doi: 10.1016/j.stem.2007.08.002. [DOI] [PubMed] [Google Scholar]
- 41.Hetie P, de Cuevas M, Matunis E. Conversion of Quiescent Niche Cells to Somatic Stem Cells Causes Ectopic Niche Formation in the Drosophila Testis. Cell reports. 2014 doi: 10.1016/j.celrep.2014.03.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Salzmann V, Inaba M, Cheng J, Yamashita YM. Lineage tracing quantification reveals symmetric stem cell division in Drosophila male germline stem cells. Cell Mol Bioeng. 2013;6:441–448. doi: 10.1007/s12195-013-0295-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.White-Cooper H, Caporilli S. Transcriptional and post-transcriptional regulation of Drosophila germline stem cells and their differentiating progeny. Adv Exp Med Biol. 2013;786:47–61. doi: 10.1007/978-94-007-6621-1_4. [DOI] [PubMed] [Google Scholar]
- 44.Brawley C, Matunis E. Regeneration of male germline stem cells by spermatogonial dedifferentiation in vivo. Science. 2004;304:1331–1334. doi: 10.1126/science.1097676. [DOI] [PubMed] [Google Scholar]
- 45.Yamashita YM, Yuan H, Cheng J, Hunt AJ. Polarity in stem cell division: asymmetric stem cell division in tissue homeostasis. Cold Spring Harbor perspectives in biology. 2010;2:a001313. doi: 10.1101/cshperspect.a001313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kohlmaier A, Edgar BA. Proliferative control in Drosophila stem cells. Curr Opin Cell Biol. 2008;20:699–706. doi: 10.1016/j.ceb.2008.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gonzalez C. Spindle orientation, asymmetric division and tumour suppression in Drosophila stem cells. Nat Rev Genet. 2007;8:462–472. doi: 10.1038/nrg2103. [DOI] [PubMed] [Google Scholar]
- 48.Tran V, Lim C, Xie J, Chen X. Asymmetric division of Drosophila male germline stem cell shows asymmetric histone distribution. Science. 2012;338:679–682. doi: 10.1126/science.1226028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yadlapalli S, Yamashita YM. Chromosome-specific nonrandom sister chromatid segregation during stem-cell division. Nature. 2013;498:251–254. doi: 10.1038/nature12106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bufalino MR, DeVeale B, van der Kooy D. The asymmetric segregation of damaged proteins is stem cell-type dependent. J Cell Biol. 2013;201:523–530. doi: 10.1083/jcb.201207052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Salzmann V, Chen C, Chiang CY, Tiyaboonchai A, Mayer M, Yamashita YM. Centrosome-dependent asymmetric inheritance of the midbody ring in Drosophila germline stem cell division. Mol Biol Cell. 2014;25:267–275. doi: 10.1091/mbc.E13-09-0541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Matunis E, Tran J, Gonczy P, Caldwell K, DiNardo S. punt and schnurri regulate a somatically derived signal that restricts proliferation of committed progenitors in the germline. Development. 1997;124:4383–4391. doi: 10.1242/dev.124.21.4383. [DOI] [PubMed] [Google Scholar]
- 53.Siomi MC, Miyoshi T, Siomi H. piRNA-mediated silencing in Drosophila germlines. Semin Cell Dev Biol. 2010;21:754–759. doi: 10.1016/j.semcdb.2010.01.011. [DOI] [PubMed] [Google Scholar]
- 54.Haglund K, Nezis IP, Stenmark H. Structure and functions of stable intercellular bridges formed by incomplete cytokinesis during development. Communicative & integrative biology. 2011;4:1–9. doi: 10.4161/cib.4.1.13550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yan D, Neumuller RA, Buckner M, Ayers K, Li H, Hu Y, Yang-Zhou D, Pan L, Wang X, Kelley C, et al. A regulatory network of Drosophila germline stem cell self-renewal. Dev Cell. 2014;28:459–473. doi: 10.1016/j.devcel.2014.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Flaherty MS, Salis P, Evans CJ, Ekas LA, Marouf A, Zavadil J, Banerjee U, Bach EA. chinmo is a functional effector of the JAK/STAT pathway that regulates eye development, tumor formation, and stem cell self-renewal in Drosophila. Dev Cell. 2010;18:556–568. doi: 10.1016/j.devcel.2010.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gonczy P, DiNardo S. The germ line regulates somatic cyst cell proliferation and fate during Drosophila spermatogenesis. Development. 1996;122:2437–2447. doi: 10.1242/dev.122.8.2437. [DOI] [PubMed] [Google Scholar]
- 58.Cheng J, Tiyaboonchai A, Yamashita YM, Hunt AJ. Asymmetric division of cyst stem cells in Drosophila testis is ensured by anaphase spindle repositioning. Development. 2011;138:831–837. doi: 10.1242/dev.057901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kiger AA, White-Cooper H, Fuller MT. Somatic support cells restrict germline stem cell self-renewal and promote differentiation. Nature. 2000;407:750–754. doi: 10.1038/35037606. [DOI] [PubMed] [Google Scholar]
- 60.Tran J, Brenner TJ, DiNardo S. Somatic control over the germline stem cell lineage during Drosophila spermatogenesis. Nature. 2000;407:754–757. doi: 10.1038/35037613. [DOI] [PubMed] [Google Scholar]
- 61.Schulz C, Wood CG, Jones DL, Tazuke SI, Fuller MT. Signaling from germ cells mediated by the rhomboid homolog stet organizes encapsulation by somatic support cells. Development. 2002;129:4523–4534. doi: 10.1242/dev.129.19.4523. [DOI] [PubMed] [Google Scholar]
- 62.Hudson AG, Parrott BB, Qian Y, Schulz C. A temporal signature of epidermal growth factor signaling regulates the differentiation of germline cells in testes of Drosophila melanogaster. PLoS One. 2013;8:e70678. doi: 10.1371/journal.pone.0070678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Leatherman JL, Dinardo S. Germline self-renewal requires cyst stem cells and stat regulates niche adhesion in Drosophila testes. Nat Cell Biol. 2010;12:806–811. doi: 10.1038/ncb2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lai SL, Lee T. Genetic mosaic with dual binary transcriptional systems in Drosophila. Nat Neurosci. 2006;9:703–709. doi: 10.1038/nn1681. [DOI] [PubMed] [Google Scholar]
- 65.del Valle Rodriguez A, Didiano D, Desplan C. Power tools for gene expression and clonal analysis in Drosophila. Nat Methods. 2012;9:47–55. doi: 10.1038/nmeth.1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Spradling A, Fuller MT, Braun RE, Yoshida S. Germline stem cells. Cold Spring Harbor perspectives in biology. 2011;3:a002642. doi: 10.1101/cshperspect.a002642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Robinson DN, Cooley L. Stable intercellular bridges in development: the cytoskeleton lining the tunnel. Trends in cell biology. 1996;6:474–479. doi: 10.1016/0962-8924(96)84945-2. [DOI] [PubMed] [Google Scholar]
- 68.Hime GR, Brill JA, Fuller MT. Assembly of ring canals in the male germ line from structural components of the contractile ring. J Cell Sci109. 1996;(Pt 12):2779–2788. doi: 10.1242/jcs.109.12.2779. [DOI] [PubMed] [Google Scholar]
- 69.Insco ML, Leon A, Tam CH, McKearin DM, Fuller MT. Accumulation of a differentiation regulator specifies transit amplifying division number in an adult stem cell lineage. Proc Natl Acad Sci U S A. 2009;106:22311–22316. doi: 10.1073/pnas.0912454106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Insco ML, Bailey AS, Kim J, Olivares GH, Wapinski OL, Tam CH, Fuller MT. A self-limiting switch based on translational control regulates the transition from proliferation to differentiation in an adult stem cell lineage. Cell Stem Cell. 2012;11:689–700. doi: 10.1016/j.stem.2012.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lin H, Yue L, Spradling AC. The Drosophila fusome, a germline-specific organelle, contains membrane skeletal proteins and functions in cyst formation. Development. 1994;120:947–956. doi: 10.1242/dev.120.4.947. [DOI] [PubMed] [Google Scholar]
- 72.de Cuevas M, Spradling AC. Morphogenesis of the Drosophila fusome and its implications for oocyte specification. Development. 1998;125:2781–2789. doi: 10.1242/dev.125.15.2781. [DOI] [PubMed] [Google Scholar]
- 73.Lighthouse DV, Buszczak M, Spradling AC. New components of the Drosophila fusome suggest it plays novel roles in signaling and transport. Dev Biol. 2008;317:59–71. doi: 10.1016/j.ydbio.2008.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Miyauchi C, Kitazawa D, Ando I, Hayashi D, Inoue YH. Orbit/CLASP is required for germline cyst formation through its developmental control of fusomes and ring canals in Drosophila males. PLoS One. 2013;8:e58220. doi: 10.1371/journal.pone.0058220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lilly MA, de Cuevas M, Spradling AC. Cyclin A associates with the fusome during germline cyst formation in the Drosophila ovary. Dev Biol. 2000;218:53–63. doi: 10.1006/dbio.1999.9570. [DOI] [PubMed] [Google Scholar]
- 76.Mathe E, Inoue YH, Palframan W, Brown G, Glover DM. Orbit/Mast, the CLASP orthologue of Drosophila, is required for asymmetric stem cell and cystocyte divisions and development of the polarised microtubule network that interconnects oocyte and nurse cells during oogenesis. Development. 2003;130:901–915. doi: 10.1242/dev.00315. [DOI] [PubMed] [Google Scholar]
- 77.McKearin D, Ohlstein B. A role for the Drosophila bag-of-marbles protein in the differentiation of cystoblasts from germline stem cells. Development. 1995;121:2937–2947. doi: 10.1242/dev.121.9.2937. [DOI] [PubMed] [Google Scholar]
- 78.McKearin DM, Spradling AC. bag-of-marbles: a Drosophila gene required to initiate both male and female gametogenesis. Genes Dev. 1990;4:2242–2251. doi: 10.1101/gad.4.12b.2242. [DOI] [PubMed] [Google Scholar]
- 79.Monk AC, Siddall NA, Volk T, Fraser B, Quinn LM, McLaughlin EA, Hime GR. HOW is required for stem cell maintenance in the Drosophila testis and for the onset of transit-amplifying divisions. Cell Stem Cell. 2010;6:348–360. doi: 10.1016/j.stem.2010.02.016. [DOI] [PubMed] [Google Scholar]
- 80.Li Y, Zhang Q, Carreira-Rosario A, Maines JZ, McKearin DM, Buszczak M. Mei-p26 cooperates with Bam, Bgcn and Sxl to promote early germline development in the Drosophila ovary. PLoS One. 2013;8:e58301. doi: 10.1371/journal.pone.0058301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Schulz C, Kiger AA, Tazuke SI, Yamashita YM, Pantalena-Filho LC, Jones DL, Wood CG, Fuller MT. A misexpression screen reveals effects of bag-of-marbles and TGF beta class signaling on the Drosophila male germ-line stem cell lineage. Genetics. 2004;167:707–723. doi: 10.1534/genetics.103.023184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wong C, Jones DL. Efficiency of spermatogonial dedifferentiation during aging. PLoS One. 2012;7:e33635. doi: 10.1371/journal.pone.0033635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Cenci G, Bonaccorsi S, Pisano C, Verni F, Gatti M. Chromatin and microtubule organization during premeiotic, meiotic and early postmeiotic stages of Drosophila melanogaster spermatogenesis. J Cell Sci. 1994;107(Pt 12):3521–3534. doi: 10.1242/jcs.107.12.3521. [DOI] [PubMed] [Google Scholar]
- 84.Hiller MA, Lin TY, Wood C, Fuller MT. Developmental regulation of transcription by a tissue-specific TAF homolog. Genes Dev. 2001;15:1021–1030. doi: 10.1101/gad.869101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hiller M, Chen X, Pringle MJ, Suchorolski M, Sancak Y, Viswanathan S, Bolival B, Lin TY, Marino S, Fuller MT. Testis-specific TAF homologs collaborate to control a tissue-specific transcription program. Development. 2004;131:5297–5308. doi: 10.1242/dev.01314. [DOI] [PubMed] [Google Scholar]
- 86.Bonaccorsi S, Giansanti MG, Cenci G, Gatti M. Preparation of live testis squashes in Drosophila. Cold Spring Harbor protocols. 2011;2011:pprot5577. doi: 10.1101/pdb.prot5577. [DOI] [PubMed] [Google Scholar]
- 87.Rebollo E, Gonzalez C. Visualizing the spindle checkpoint in Drosophila spermatocytes. EMBO reports. 2000;1:65–70. doi: 10.1093/embo-reports/kvd011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Rebollo E, Gonzalez C. Time-lapse imaging of male meiosis by phase-contrast and fluorescence microscopy. Methods Mol Biol. 2004;247:77–87. doi: 10.1385/1-59259-665-7:77. [DOI] [PubMed] [Google Scholar]
- 89.Barbosa V, Gatt M, Rebollo E, Gonzalez C, Glover DM. Drosophila dd4 mutants reveal that gammaTuRC is required to maintain juxtaposed half spindles in spermatocytes. J Cell Sci. 2003;116:929–941. doi: 10.1242/jcs.00295. [DOI] [PubMed] [Google Scholar]
- 90.Brill JA, Hime GR, Scharer-Schuksz M, Fuller MT. A phospholipid kinase regulates actin organization and intercellular bridge formation during germline cytokinesis. Development. 2000;127:3855–3864. doi: 10.1242/dev.127.17.3855. [DOI] [PubMed] [Google Scholar]
- 91.Giansanti MG, Farkas RM, Bonaccorsi S, Lindsley DL, Wakimoto BT, Fuller MT, Gatti M. Genetic dissection of meiotic cytokinesis in Drosophila males. Mol Biol Cell. 2004;15:2509–2522. doi: 10.1091/mbc.E03-08-0603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Giansanti MG, Fuller MT. What Drosophila spermatocytes tell us about the mechanisms underlying cytokinesis. Cytoskeleton (Hoboken) 2012;69:869–881. doi: 10.1002/cm.21063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Fabian L, Wei HC, Rollins J, Noguchi T, Blankenship JT, Bellamkonda K, Polevoy G, Gervais L, Guichet A, Fuller MT, et al. Phosphatidylinositol 4,5-bisphosphate directs spermatid cell polarity and exocyst localization in Drosophila. Mol Biol Cell. 2010;21:1546–1555. doi: 10.1091/mbc.E09-07-0582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.White-Cooper H, Bausek N. Evolution and spermatogenesis. Philos Trans R Soc Lond B Biol Sci. 2010;365:1465–1480. doi: 10.1098/rstb.2009.0323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Eberhart CG, Maines JZ, Wasserman SA. Meiotic cell cycle requirement for a fly homologue of human Deleted in Azoospermia. Nature. 1996;381:783–785. doi: 10.1038/381783a0. [DOI] [PubMed] [Google Scholar]
- 96.Giansanti MG, Bonaccorsi S, Gatti M. The role of anillin in meiotic cytokinesis of Drosophila males. J Cell Sci. y1999;112(Pt 14):2323–2334. doi: 10.1242/jcs.112.14.2323. [DOI] [PubMed] [Google Scholar]
- 97.Carmena M, Riparbelli MG, Minestrini G, Tavares AM, Adams R, Callaini G, Glover DM. Drosophila polo kinase is required for cytokinesis. J Cell Biol. 1998;143:659–671. doi: 10.1083/jcb.143.3.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hales KG, Fuller MT. Developmentally regulated mitochondrial fusion mediated by a conserved, novel, predicted GTPase. Cell. 1997;90:121–129. doi: 10.1016/s0092-8674(00)80319-0. [DOI] [PubMed] [Google Scholar]
- 99.Westermann B. Mitochondrial dynamics in model organisms: what yeasts, worms and flies have taught us about fusion and fission of mitochondria. Semin Cell Dev Biol. 2010;21:542–549. doi: 10.1016/j.semcdb.2009.12.003. [DOI] [PubMed] [Google Scholar]
- 100.White-Cooper H. Tissue, cell type and stage-specific ectopic gene expression and RNAi induction in the Drosophila testis. Spermatogenesis. 2012;2:11–22. doi: 10.4161/spmg.19088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Goldbach P, Wong R, Beise N, Sarpal R, Trimble WS, Brill JA. Stabilization of the actomyosin ring enables spermatocyte cytokinesis in Drosophila. Mol Biol Cell. 2010;21:1482–1493. doi: 10.1091/mbc.E09-08-0714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Eikenes AH, Brech A, Stenmark H, Haglund K. Spatiotemporal control of Cindr at ring canals during incomplete cytokinesis in the Drosophila male germline. Dev Biol. 2013;377:9–20. doi: 10.1016/j.ydbio.2013.02.021. [DOI] [PubMed] [Google Scholar]
- 103.Developmental Studies Hybridoma Bank. http://dshb.biology.uiowa.edu. [Google Scholar]
- 104.Zhao J, Klyne G, Benson E, Gudmannsdottir E, White-Cooper H, Shotton D. FlyTED: the Drosophila Testis Gene Expression Database. Nucleic Acids Res. 2010;38:D710–715. doi: 10.1093/nar/gkp1006. http://www.fly-ted.org/. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.The Exelixis Collection at the Harvard Medical School. http://drosophila.med.harvard.edu.
- 106.TRiP. http://www.flyrnai.org/TRiP-HOME.html.
- 107.Bloomington Stock Center. http://flystocks.bio.indiana.edu.
- 108.VDRC Stock Center. http://stockcenter.vdrc.at.
- 109.Kyoto DGRC. http://kyotofly.kit.jp/stocks/
- 110.FlyTrap. http://flytrap.med.yale.edu/
- 111.Wakimoto BT, Lindsley DL, Herrera C. Toward a comprehensive genetic analysis of male fertility in Drosophila melanogaster. Genetics. 2004;167:207–216. doi: 10.1534/genetics.167.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Castrillon DH, Gonczy P, Alexander S, Rawson R, Eberhart CG, Viswanathan S, DiNardo S, Wasserman SA. Toward a molecular genetic analysis of spermatogenesis in Drosophila melanogaster: characterization of male-sterile mutants generated by single P element mutagenesis. Genetics. 1993;135:489–505. doi: 10.1093/genetics/135.2.489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Gonczy P, Viswanathan S, DiNardo S. Probing spermatogenesis in Drosophila with P-element enhancer detectors. Development. 1992;114:89–98. doi: 10.1242/dev.114.1.89. [DOI] [PubMed] [Google Scholar]
- 114.Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- 115.Rorth P. Gal4 in the Drosophila female germline. Mech Dev. 1998;78:113–118. doi: 10.1016/s0925-4773(98)00157-9. [DOI] [PubMed] [Google Scholar]
- 116.Bateman JR, Lee AM, Wu CT. Site-specific transformation of Drosophila via phiC31 integrase-mediated cassette exchange. Genetics. 2006;173:769–777. doi: 10.1534/genetics.106.056945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Pfeiffer BD, Ngo TT, Hibbard KL, Murphy C, Jenett A, Truman JW, Rubin GM. Refinement of tools for targeted gene expression in Drosophila. Genetics. 2010;186:735–755. doi: 10.1534/genetics.110.119917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Golic KG, Lindquist S. The FLP recombinase of yeast catalyzes site-specific recombination in the Drosophila genome. Cell. 1989;59:499–509. doi: 10.1016/0092-8674(89)90033-0. [DOI] [PubMed] [Google Scholar]
- 119.Lee T, Luo L. Mosaic analysis with a repressible cell marker (MARCM) for Drosophila neural development. Trends in neurosciences. 2001;24:251–254. doi: 10.1016/s0166-2236(00)01791-4. [DOI] [PubMed] [Google Scholar]
- 120.Evans CJ, Olson JM, Ngo KT, Kim E, Lee NE, Kuoy E, Patananan AN, Sitz D, Tran P, Do MT, et al. G-TRACE: rapid Gal4-based cell lineage analysis in Drosophila. Nat Methods. 2009;6:603–605. doi: 10.1038/nmeth.1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Pignoni F, Zipursky SL. Induction of Drosophila eye development by decapentaplegic. Development. 1997;124:271–278. doi: 10.1242/dev.124.2.271. [DOI] [PubMed] [Google Scholar]
- 122.Bonaccorsi S, Giansanti MG, Cenci G, Gatti M. Paraformaldehyde fixation of Drosophila testes. Cold Spring Harbor protocols. 2012;2012:102–104. doi: 10.1101/pdb.prot067330. [DOI] [PubMed] [Google Scholar]
- 123.Bonaccorsi S, Giansanti MG, Cenci G, Gatti M. F-actin staining of Drosophila testes. Cold Spring Harbor protocols. 2012;2012:105–106. doi: 10.1101/pdb.prot067348. [DOI] [PubMed] [Google Scholar]
- 124.Bonaccorsi S, Giansanti MG, Cenci G, Gatti M. Formaldehyde fixation of Drosophila testes. Cold Spring Harbor protocols. 2012 doi: 10.1101/pdb.prot067355. 2012. [DOI] [PubMed] [Google Scholar]
- 125.Gunsalus KC, Bonaccorsi S, Williams E, Verni F, Gatti M, Goldberg ML. Mutations in twinstar, a Drosophila gene encoding a cofilin/ADF homologue, result in defects in centrosome migration and cytokinesis. J Cell Biol. 1995;131:1243–1259. doi: 10.1083/jcb.131.5.1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Bonaccorsi S, Giansanti MG, Cenci G, Gatti M. Methanol-acetone fixation of Drosophila testes. Cold Spring Harbor protocols. 2011;2011:1270–1272. doi: 10.1101/pdb.prot065763. [DOI] [PubMed] [Google Scholar]
- 127.Hudson AM, Cooley L. Methods for studying oogenesis. Methods. 2014 doi: 10.1016/j.ymeth.2014.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.White-Cooper H. Spermatogenesis: analysis of meiosis and morphogenesis. Methods Mol Biol. 2004;247:45–75. doi: 10.1385/1-59259-665-7:45. [DOI] [PubMed] [Google Scholar]
- 129.Prasad M, Jang AC, Starz-Gaiano M, Melani M, Montell DJ. A protocol for culturing Drosophila melanogaster stage 9 egg chambers for live imaging. Nat Protoc. 2007;2:2467–2473. doi: 10.1038/nprot.2007.363. [DOI] [PubMed] [Google Scholar]
- 130.Patel NH, Snow PM, Goodman CS. Characterization and cloning of fasciclin III: a glycoprotein expressed on a subset of neurons and axon pathways in Drosophila. Cell. 1987;48:975–988. doi: 10.1016/0092-8674(87)90706-9. [DOI] [PubMed] [Google Scholar]
- 131.Riggleman B, Schedl P, Wieschaus E. Spatial expression of the Drosophila segment polarity gene armadillo is posttranscriptionally regulated by wingless. Cell. 1990;63:549–560. doi: 10.1016/0092-8674(90)90451-j. [DOI] [PubMed] [Google Scholar]
- 132.Halder G, Callaerts P, Gehring WJ. Induction of ectopic eyes by targeted expression of the eyeless gene in Drosophila. Science. 1995;267:1788–1792. doi: 10.1126/science.7892602. [DOI] [PubMed] [Google Scholar]
- 133.Sun YH, Tsai CJ, Green MM, Chao JL, Yu CT, Jaw TJ, Yeh JY, Bolshakov VN. White as a reporter gene to detect transcriptional silencers specifying position-specific gene expression during Drosophila melanogaster eye development. Genetics. 1995;141:1075–1086. doi: 10.1093/genetics/141.3.1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Chao JL, Tsai YC, Chiu SJ, Sun YH. Localized Notch signal acts through eyg and upd to promote global growth in Drosophila eye. Development. 2004;131:3839–3847. doi: 10.1242/dev.01258. [DOI] [PubMed] [Google Scholar]
- 135.Whiteley M, Noguchi PD, Sensabaugh SM, Odenwald WF, Kassis JA. The Drosophila gene escargot encodes a zinc finger motif found in snail-related genes. Mech Dev. 1992;36:117–127. doi: 10.1016/0925-4773(92)90063-p. [DOI] [PubMed] [Google Scholar]
- 136.Li MA, Alls JD, Avancini RM, Koo K, Godt D. The large Maf factor Traffic Jam controls gonad morphogenesis in Drosophila. Nat Cell Biol. 2003;5:994–1000. doi: 10.1038/ncb1058. [DOI] [PubMed] [Google Scholar]
- 137.Lai ZC, Fortini ME, Rubin GM. The embryonic expression patterns of zfh-1 and zfh-2, two Drosophila genes encoding novel zinc-finger homeodomain proteins. Mech Dev. 1991;34:123–134. doi: 10.1016/0925-4773(91)90049-c. [DOI] [PubMed] [Google Scholar]
- 138.Fabrizio JJ, Boyle M, DiNardo S. A somatic role for eyes absent (eya) and sine oculis (so) in Drosophila spermatocyte development. Dev Biol. 2003;258:117–128. doi: 10.1016/s0012-1606(03)00127-1. [DOI] [PubMed] [Google Scholar]
- 139.Bonini NM, Leiserson WM, Benzer S. The eyes absent gene: genetic control of cell survival and differentiation in the developing Drosophila eye. Cell. 1993;72:379–395. doi: 10.1016/0092-8674(93)90115-7. [DOI] [PubMed] [Google Scholar]
- 140.Brown S, Zeidler MP, Hombria JE. JAK/STAT signalling in Drosophila controls cell motility during germ cell migration. Dev Dyn. 2006;235:958–966. doi: 10.1002/dvdy.20709. [DOI] [PubMed] [Google Scholar]
- 141.Yan R, Small S, Desplan C, Dearolf CR, Darnell JE., Jr Identification of a Stat gene that functions in Drosophila development. Cell. 1996;84:421–430. doi: 10.1016/s0092-8674(00)81287-8. [DOI] [PubMed] [Google Scholar]
- 142.Hou XS, Melnick MB, Perrimon N. Marelle acts downstream of the Drosophila HOP/JAK kinase and encodes a protein similar to the mammalian STATs. Cell. 1996;84:411–419. doi: 10.1016/s0092-8674(00)81286-6. [DOI] [PubMed] [Google Scholar]
- 143.Rangan P, DeGennaro M, Lehmann R. Regulating gene expression in the Drosophila germ line. Cold Spring Harb Symp Quant Biol. 2008;73:1–8. doi: 10.1101/sqb.2008.73.057. [DOI] [PubMed] [Google Scholar]
- 144.Chen D, McKearin DM. A discrete transcriptional silencer in the bam gene determines asymmetric division of the Drosophila germline stem cell. Development. 2003;130:1159–1170. doi: 10.1242/dev.00325. [DOI] [PubMed] [Google Scholar]
- 145.Nakamura A, Amikura R, Hanyu K, Kobayashi S. Me31B silences translation of oocyte-localizing RNAs through the formation of cytoplasmic RNP complex during Drosophila oogenesis. Development. 2001;128:3233–3242. doi: 10.1242/dev.128.17.3233. [DOI] [PubMed] [Google Scholar]
- 146.Kelso RJ, Buszczak M, Quinones AT, Castiblanco C, Mazzalupo S, Cooley L. Flytrap, a database documenting a GFP protein-trap insertion screen in Drosophila melanogaster. Nucleic Acids Res. 2004;32:D418–D420. doi: 10.1093/nar/gkh014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Hay B, Ackerman L, Barbel S, Jan LY, Jan YN. Identification of a component of Drosophila polar granules. Development. 1988;103:625–640. doi: 10.1242/dev.103.4.625. [DOI] [PubMed] [Google Scholar]
- 148.Oda H, Tsukita S. Nonchordate classic cadherins have a structurally and functionally unique domain that is absent from chordate classic cadherins. Dev Biol. 1999;216:406–422. doi: 10.1006/dbio.1999.9494. [DOI] [PubMed] [Google Scholar]
- 149.Huelsmann S, Ylanne J, Brown NH. Filopodia-like actin cables position nuclei in association with perinuclear actin in Drosophila nurse cells. Dev Cell. 2013;26:604–615. doi: 10.1016/j.devcel.2013.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Kelso RJ, Hudson AM, Cooley L. Drosophila Kelch regulates actin organization via Src64-dependent tyrosine phosphorylation. J Cell Biol. 2002;156:703–713. doi: 10.1083/jcb.200110063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Sano H, Renault AD, Lehmann R. Control of lateral migration and germ cell elimination by the Drosophila melanogaster lipid phosphate phosphatases Wunen and Wunen 2. J Cell Biol. 2005;171:675–683. doi: 10.1083/jcb.200506038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Grieder NC, de Cuevas M, Spradling AC. The fusome organizes the microtubule network during oocyte differentiation in Drosophila. Development. 2000;127:4253–4264. doi: 10.1242/dev.127.19.4253. [DOI] [PubMed] [Google Scholar]
- 153.Shimada Y, Yonemura S, Ohkura H, Strutt D, Uemura T. Polarized transport of Frizzled along the planar microtubule arrays in Drosophila wing epithelium. Dev Cell. 2006;10:209–222. doi: 10.1016/j.devcel.2005.11.016. [DOI] [PubMed] [Google Scholar]
- 154.Morais-de-Sa E, Sunkel C. Adherens junctions determine the apical position of the midbody during follicular epithelial cell division. EMBO reports. 2013;14:696–703. doi: 10.1038/embor.2013.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Pesacreta TC, Byers TJ, Dubreuil R, Kiehart DP, Branton D. Drosophila spectrin: the membrane skeleton during embryogenesis. J Cell Biol. 1989;108:1697–1709. doi: 10.1083/jcb.108.5.1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Haglund K, Nezis IP, Lemus D, Grabbe C, Wesche J, Liestol K, Dikic I, Palmer R, Stenmark H. Cindr interacts with anillin to control cytokinesis in Drosophila melanogaster. Curr Biol. 2010;20:944–950. doi: 10.1016/j.cub.2010.03.068. [DOI] [PubMed] [Google Scholar]
- 157.Adams RR, Tavares AA, Salzberg A, Bellen HJ, Glover DM. pavarotti encodes a kinesin-like protein required to organize the central spindle and contractile ring for cytokinesis. Genes Dev. 1998;12:1483–1494. doi: 10.1101/gad.12.10.1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Martin AC, Kaschube M, Wieschaus EF. Pulsed contractions of an actin-myosin network drive apical constriction. Nature. 2009;457:495–499. doi: 10.1038/nature07522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Royou A, Sullivan W, Karess R. Cortical recruitment of nonmuscle myosin II in early syncytial Drosophila embryos: its role in nuclear axial expansion and its regulation by Cdc2 activity. J Cell Biol. 2002;158:127–137. doi: 10.1083/jcb.200203148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Schuh M, Lehner CF, Heidmann S. Incorporation of Drosophila CID/CENP-A and CENP-C into centromeres during early embryonic anaphase. Curr Biol. 2007;17:237–243. doi: 10.1016/j.cub.2006.11.051. [DOI] [PubMed] [Google Scholar]
- 161.Ji JY, Squirrell JM, Schubiger G. Both cyclin B levels and DNA-replication checkpoint control the early embryonic mitoses in Drosophila. Development. 2004;131:401–411. doi: 10.1242/dev.00944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Chen X, Hiller M, Sancak Y, Fuller MT. Tissue-specific TAFs counteract Polycomb to turn on terminal differentiation. Science. 2005;310:869–872. doi: 10.1126/science.1118101. [DOI] [PubMed] [Google Scholar]
- 163.Tanimoto H, Itoh S, ten Dijke P, Tabata T. Hedgehog creates a gradient of DPP activity in Drosophila wing imaginal discs. Mol Cell. 2000;5:59–71. doi: 10.1016/s1097-2765(00)80403-7. [DOI] [PubMed] [Google Scholar]
- 164.Wolfstetter G, Holz A. The role of LamininB2 (LanB2) during mesoderm differentiation in Drosophila. Cellular and molecular life sciences : CMLS. 2012;69:267–282. doi: 10.1007/s00018-011-0652-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Hayashi S, Ito K, Sado Y, Taniguchi M, Akimoto A, Takeuchi H, Aigaki T, Matsuzaki F, Nakagoshi H, Tanimura T, et al. GETDB, a database compiling expression patterns and molecular locations of a collection of Gal4 enhancer traps. Genesis. 2002;34:58–61. doi: 10.1002/gene.10137. [DOI] [PubMed] [Google Scholar]
- 166.Zhu CH, Xie T. Clonal expansion of ovarian germline stem cells during niche formation in Drosophila. Development. 2003;130:2579–2588. doi: 10.1242/dev.00499. [DOI] [PubMed] [Google Scholar]
- 167.Kai T, Spradling A. An empty Drosophila stem cell niche reactivates the proliferation of ectopic cells. Proc Natl Acad Sci U S A. 2003;100:4633–4638. doi: 10.1073/pnas.0830856100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Takaesu NT, Johnson AN, Newfeld SJ. Posterior spiracle specific GAL4 lines: new reagents for developmental biology and respiratory physiology. Genesis. 2002;34:16–18. doi: 10.1002/gene.10109. [DOI] [PubMed] [Google Scholar]
- 169.Joti P, Ghosh-Roy A, Ray K. Dynein light chain 1 functions in somatic cyst cells regulate spermatogonial divisions in Drosophila. Sci Rep. 2011;1:173. doi: 10.1038/srep00173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Tracey WD, Jr, Ning X, Klingler M, Kramer SG, Gergen JP. Quantitative analysis of gene function in the Drosophila embryo. Genetics. 2000;154:273–284. doi: 10.1093/genetics/154.1.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Van Doren M, Williamson AL, Lehmann R. Regulation of zygotic gene expression in Drosophila primordial germ cells. Curr Biol. 1998;8:243–246. doi: 10.1016/s0960-9822(98)70091-0. [DOI] [PubMed] [Google Scholar]
- 172.Zhao S, Chen D, Geng Q, Wang Z. The highly conserved LAMMER/CLK2 protein kinases prevent germ cell overproliferation in Drosophila. Dev Biol. 2013;376:163–170. doi: 10.1016/j.ydbio.2013.01.023. [DOI] [PubMed] [Google Scholar]
- 173.Lee T, Luo L. Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron. 1999;22:451–461. doi: 10.1016/s0896-6273(00)80701-1. [DOI] [PubMed] [Google Scholar]