Abstract
We seek fluorogenic small molecules that generate a fluorescent conjugate signal if and only if they react with a given protein-of-interest (i.e., small molecules for which non-covalent binding to the protein-of-interest is insufficient to generate fluorescence). Consequently, it is the new chemical entity afforded by the generally irreversible reaction between the small molecule and the protein-of-interest that enables the energy of an electron occupying the lowest unoccupied molecular orbital (LUMO) of the chromophore to be given off as a photon instead of being dissipated by non-radiative mechanisms in complex biological environments. This category of fluorogenic small molecules is created by starting with environmentally sensitive fluorophores that are modified by an essential functional group that efficiently quenches the fluorescence until a chemoselective reaction between that functional group and the protein-of-interest occurs, yielding the fluorescent conjugate. Fluorogenic small molecules are envisioned to be useful for a wide variety of applications, including live cell imaging without the requirement for washing steps and pulse-chase kinetic analyses of protein synthesis, trafficking, degradation, etc.
Introduction
The environmental sensitivity of fluorescence, combined with the ability to monitor changes in fluorescence with both spatial and temporal resolution, has led to its widespread use in the imaging of biological systems1. Fluorescence research initially focused on organic small molecule fluorophores in an attempt to understand the physical-chemical underpinnings of the phenomenon.2–5 Subsequently, numerous small molecule fluorophore applications were developed based on this mechanistic information.2,6–10 A significant advance in the area of biological imaging came with the discovery of green fluorescent protein (GFP) and its analogs, which allowed one to genetically encode fluorescent tags as fusion proteins with a protein-of-interest.11–13 The ease of employing GFPs and analogous proteins for biological imaging brought fluorescence imaging to the scientific masses. Efforts to develop novel small molecule fluorescent chromophores have continued in parallel.4,6,7,14–16 The development of protein tags that react with these small molecule fluorophores has diversified the fluorescence spectrum available for biological imaging and expanded the possible applications, including super-resolution microscopy imaging.17–25 These small molecule fluorophores are generally fluorescent before and after reaction with the protein tag,17,18,26 which is problematic for use in some applications—exceptions will be discussed later.27–30
Our objective is to create a range of fluorogenic small molecules that only display fluorescence upon reaction with the protein or protein tag of interest.21,22 One key advantage of this approach is that the chromophore can be tuned through chemical modification to create a molecule with appropriate photophysical properties, e.g., desirable excitation/emission spectra, resistance to photobleaching, blinking properties, etc.21,22 A chemoselective reaction between the fluorogenic small molecule and the protein-of-interest allows for the post-translational regulation of fluorescence, which is useful for pulse-chase kinetic analysis and the like where probe washout steps using conventional non-fluorogenic probes are too slow. By mixing and matching genetically encoded protein binding site functionality and geometry with small molecule regulated fluorescence, a high level of control can be achieved for a plethora of applications.
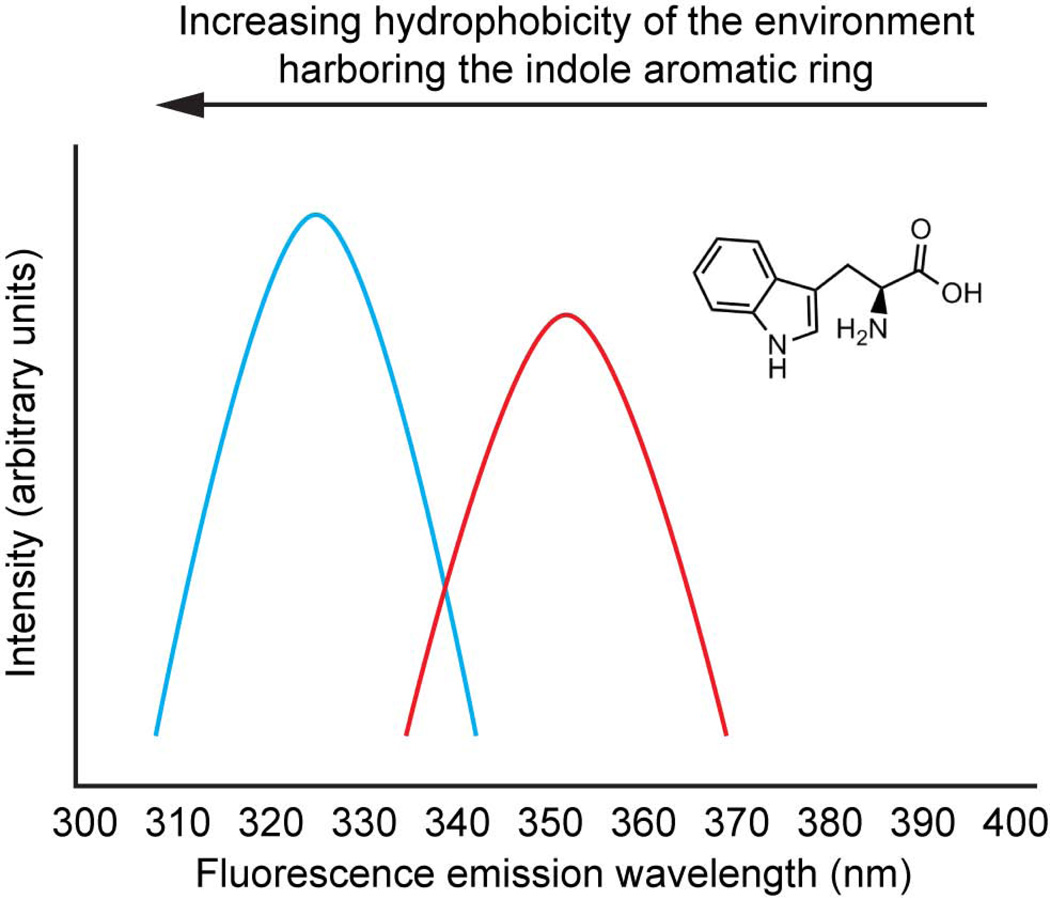
Environmentally-sensitive fluorophores are a category of molecules whose properties (excitation and emission wavelengths, fluorescence lifetimes, quantum yields, changes in dipole moments upon excitation etc.) are highly dependent on their microenvironment.14,31–37 The natural amino acid tryptophan is a classic example of an environmentally-sensitive fluorophore.38,39 The fluorescence associated with the indole side chain is red shifted in the denatured state of a protein, owing to water binding to and stabilization of the excited state, and blue shifted when it is positioned in the hydrophobic core of the protein in the folded state, owing to excited state destabilization (Figure 1).40,41 The fluorescence intensity of Trp, while sensitive to its environment, is not easily predicted. Chemists have made numerous environmentally-sensitive fluorophores over the past 50 years with more predictable intensity changes.42 Examples include the fluorophore 1,1'-bis(4-anilino)naphthalene-5,5'-disulfonate (ANS) that exhibits a very low quantum yield in aqueous solutions and a high quantum yield and a blue-shifted fluorescence when the chromophore is bound in the hydrophobic core of a partially folded protein.43 The advantage of using environmentally-sensitive fluorophores as a starting point for the creation of fluorogenic small molecules is that both the small molecule (as described two paragraphs below) as well as the protein binding pocket can be modified to ensure that binding alone does not result in fluorescence.
Figure 1.
The indole aromatic side chain of tryptophan is a good example of an environmentally-sensitive fluorophore, one that exhibits a difference between the dipole moment of the ground and electronic excited states. Thus, its emission maximum wavelength is environment sensitive. When placed in the hydrophobic environment of a folded hypothetical protein, the indole ring often emits around 330 nm (although the emission can be blue shifted to as far as 308 nm) owing to a relatively destabilized excited state free energy. Upon chaotrope-mediated hypothetical protein denaturation, a red-shifted indole ring fluorescence emission spectrum is generally observed, owing to the ability of water to lower the excited state free energy, relative to the higher electronic excited state free energy of the indole placed in the hydrophobic core of a protein.
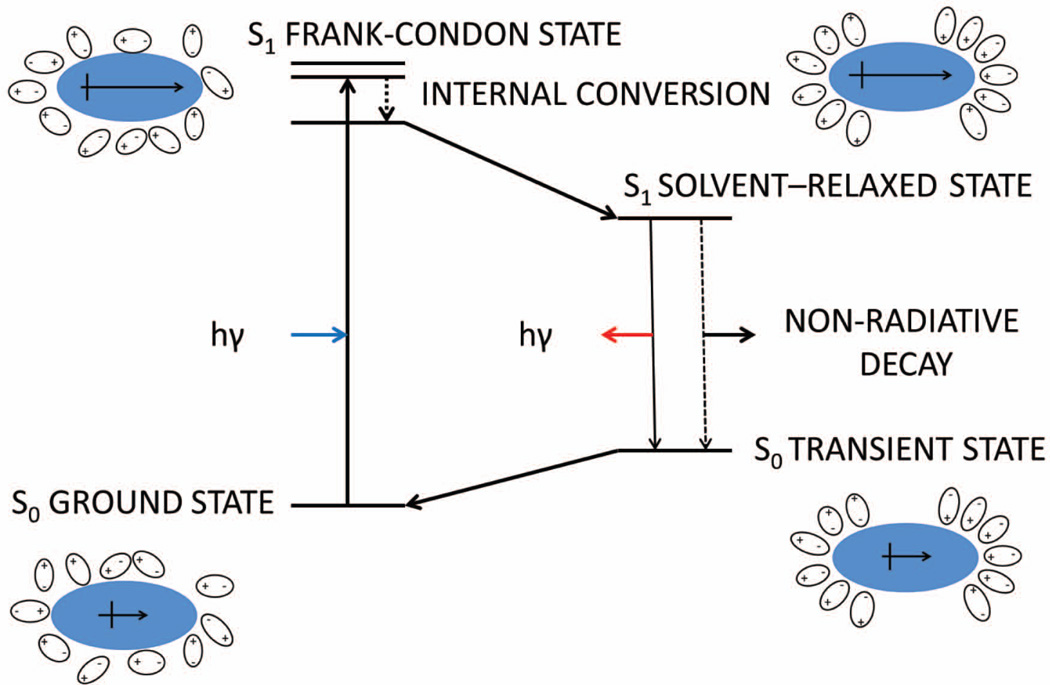
So-called push-pull fluorophores are a sub-category of environmentally-sensitive fluorophores.36,37,44–52 They feature an electron donating substituent (typically a hydroxyl, methoxy, or amino group) and an electron accepting substituent (often an ester, nitrile, carbonyl group, sulfonamide etc.) in remote positions on the chromophore. As a consequence of intramolecular charge transfer in the excited state upon absorption of a photon (S0→S1), there is a large increase in the dipole moment of these molecules (Figure 2). Following relaxation to S1solv resulting from polarization of solvent molecules, there is a decrease in the energy of the excited state and a red shift in the emission spectrum. The red shift tends to be most striking in protic solvents, where additional hydrogen bond formation further decreases the energy of S1solv. Most solvatochromic or environmentally-sensitive fluorophores exhibit very poor fluorescence properties in water (high polarity, strong hydrogen bonding capacity) and therefore are dark until they encounter a protein with a complementary binding site. Thus, these push-pull fluorophores can be used as the basis of fluorogenic small molecules, and of course the binding site can be tuned to maximize the red shift and potentially other photophysical properties, which is useful for biological imaging.
Figure 2.
Origin of solvatochromism or environment-sensitive fluorescence resulting from the interaction of S1 with its surrounding environment, which controls the energy of the solvent-relaxed electronic excited state or LUMO. Figure adapted from36,37.
The key to rendering environmentally-sensitive fluorophores fluorogenic is to modify the chromophore with a substituent or functional group that very efficiently quenches their fluorescence (even when bound to the protein-of-interest) until that functional group reacts with a protein-of-interest. The functional group employed is envisioned to be converted from an efficient fluorescence quencher (efficiently dissipates the energy of the absorbed photon through internal conversion, or an analogous process) into a functional group that no longer quenches fluorescence as efficiently, as a consequence of a chemoselective chemical reaction with the protein-of-interest. Our laboratory has now created fluorogenic small molecules that bind to and chemoselectively react with specific proteins-of-interest using this strategy.21,22 We need to discover more functional groups that are quenchers of fluorescence and that are switchable into non-quenchers after chemoselective protein reactions, and better understand their mechanism(s) of action. The dissociation constant of the fluorogenic small molecule (KD) and the conjugation rate constant (kconjugation) together dictate how selectively a fluorescent conjugate can be created in the context of a complex biological environment. The binding selectivity of fluorogenic molecules need not be exceptional, as long as the chemoselectivity of conjugate formation is very high.
The selectivity of fluorescent conjugate formation can easily be assessed by treating a cell lysate or a complex biological fluid lacking the protein-of-interest with the fluorogenic small molecule. Fluorogenic small molecules exhibiting high binding selectivity to the protein-of-interest and a highly chemoselective reaction with the protein-of-interest will exhibit very low background fluorescence when analyzed by microscopic imaging and/or after gel electrophoresis of the complex sample visualized employing a fluorescence gel scanner. Then the analogous experiment is done as a function of adding a known concentration of the protein-of-interest and the fluorogenic small molecule to discern the fluorescence signal-to-background ratio in that particular biological sample.
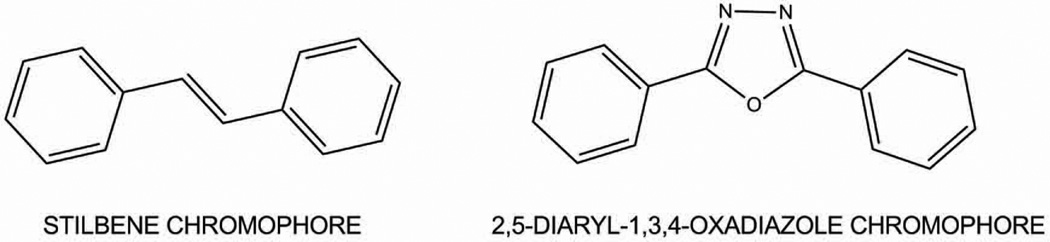
Herein using both the stilbene and 2,5-diaryl-1,3,4-oxadiazole chromophores (Figure 3), we will explain what we know and what we would like to learn about two fluorogenic small molecules that covalently modify the protein transthyretin.
Figure 3.
Backbone structure of chromophores that can be further modified to selectively bind to and then react with transthyretin, while exhibiting the photophysical properties of interest.
Transthyretin (TTR) is a 55-kDa homotetrameric protein composed of four 127-amino-acid β-sheet rich subunits (Figure 4).53 TTR is secreted from the liver and the choroid plexus into the blood and the cerebrospinal fluid (CSF) respectively.54 TTR transports both the thyroid hormone thyroxine (T4) and holo-retinol binding protein.55 In the blood, more than 99% of the T4 binding sites are unoccupied because of the presence of two major T4 carrier proteins, thyroid binding globulin and albumin.56
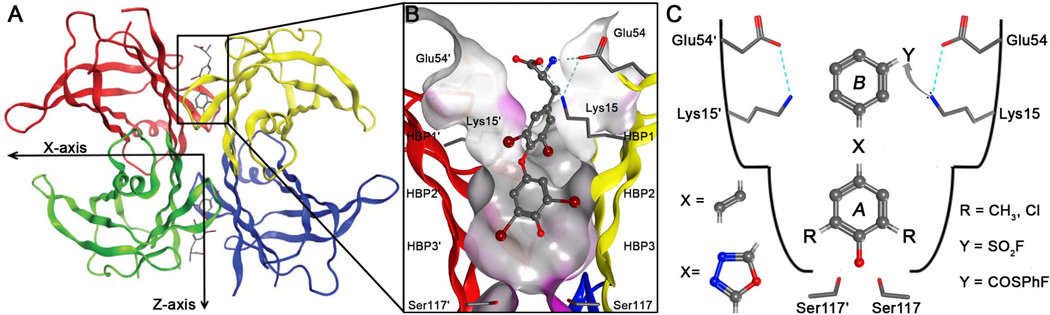
Figure 4.
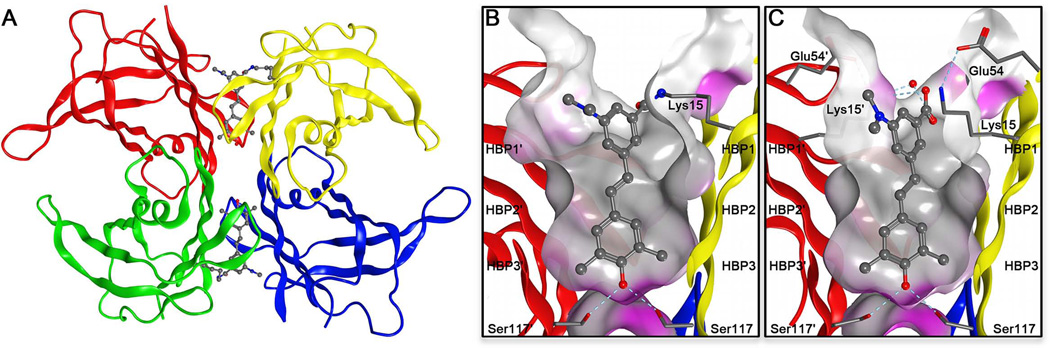
Structure of homotetrameric WT-TTR with a focus on the T4 binding pocket and pKa-perturbed Lys15 and 15′ residues. (A) Crystal structure of WT-TTR in complex with T4 (PDB accession code 2ROX53)(B) Close-up view of one of the two identical T4 binding sites showing a ribbon depicted tetramer (colored by chain) with a “Connelly” molecular surface applied to residues within 8 Å of T4 (hydrophobic = grey, polar = purple). The innermost halogen binding pockets (HBPs) 3 and 3′ are composed of the methyl and methylene groups of Ser117/117′, Thr119/119′, and Leu110/110′. HBPs 2 and 2′ are made up by the side chains of Leu110/110′, Ala109/109′, Lys15/15′, and Leu17/17′. The outermost HBPs 1 and 1′ are lined by the methyl and methylene groups of Lys15/15′, Ala108/108′, and Thr106/106′. These figures were generated using the program MOE (2011.10), Chemical Computing Group, Montreal, Canada. (C) Schematic representation of the T4 binding pocket and the amino acids that are targeted by the substituted (R = CH3 or Cl) aryl ring A connected by linker X (stilbene or 1,3,4-oxadiazole ring linker) to aryl ring B that contains an electrophile Y (sulfonyl fluoride or fluorophenol thioester) for attack by the pKa-perturbed nucleophilic ε-amino group of Lys15.
The energetically weaker dimer-dimer interface of TTR, bisected by the crystallographic C2 or Z axis, creates two T4 binding pockets (Figure 4A). Each T4 binding site is characterized by a series of symmetric hydrophobic depressions referred to as halogen binding pockets (HBPs), wherein the iodine atoms of T4 reside (Figure 4B).53 While the T4 binding site is predominantly hydrophobic, there are threonine and serine residues at the base of the pocket available for hydrogen bonding. At the periphery of the T4 binding site, there is a pair of lysine and glutamic acid residues that participate in salt bridges and H-bonding with water (Figure 4C).53 Recently, our laboratory conducted studies to systematically optimize the structures of ligands that bind to TTR.57–61 Using this experience in TTR ligand design, both of the aforementioned chromophores which complement the T4 binding site structure were modified using a variety of substituents to permit selective binding and chemoselective reactivity towards the pKa-perturbed Lys15 to render these molecules fluorogenic (Figure 4C).21,22,62
Stilbene chromophores
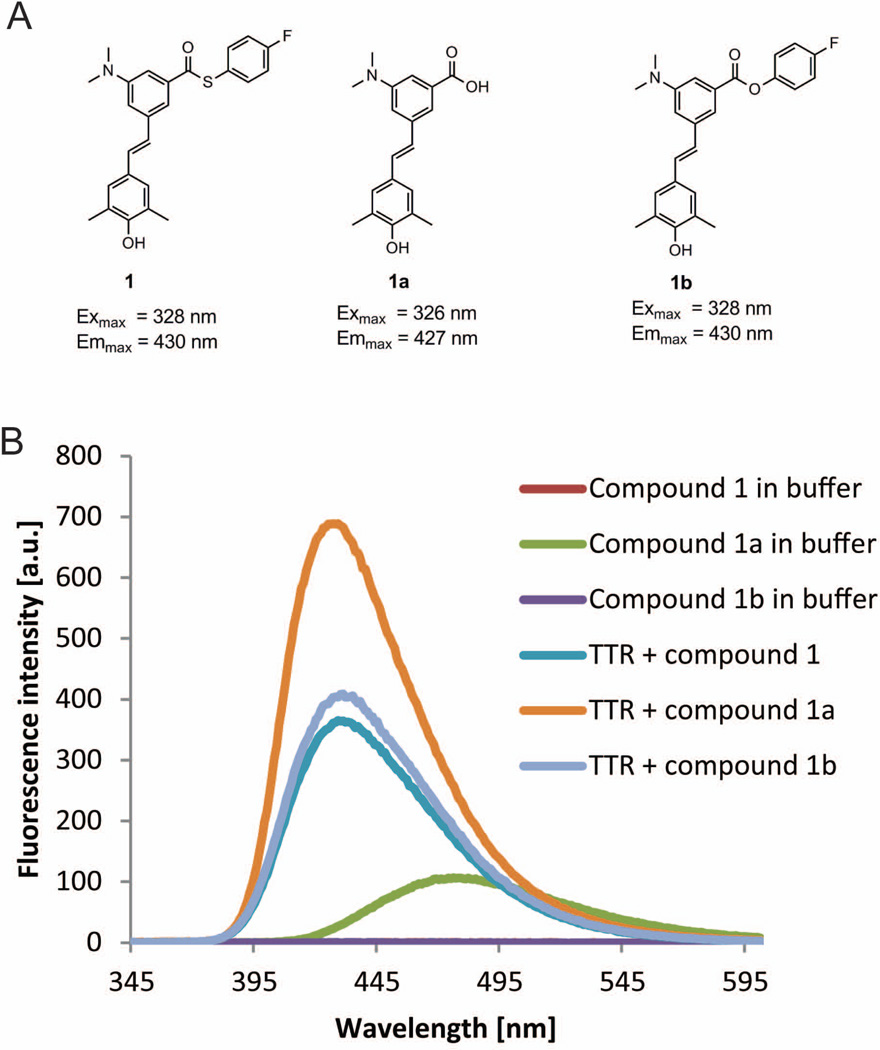
We first describe stilbene 1 (Figure 5A) that binds selectively to TTR in complex biological environments and remains dark until it undergoes a chemoselective reaction with the pKa-perturbed Lys15 ε-amino group of TTR to create a bright blue fluorescent conjugate, as described previously.21 Stilbene 1 is nonfluorescent in aqueous buffers (Figure 5B) and exhibits low background fluorescence in cell lysates lacking TTR, as previously described, in spite of 1 almost certainly binding to at least some proteins in the proteome.21 This is a highly desirable property for the type of fluorogenic molecules we seek. Moreover, when 1 is incubated with a Lys15Ala mutant of TTR, i.e., a variant lacking the required nucleophile for conjugation, the complex between 1 and TTR remains non-fluorescent, as previously reported.21
Figure 5.
(A) Structures of stilbene chromophores 1, 1a, 1b that bind to or bind to and then react with WT TTR to form fluorescent conjugates, together with their excitation and emission maxima when bound to or after reaction with TTR. (B) Fluorescence emission spectra of stilbenes 1, 1a and1b (7.2 µM) in phosphate buffer and when bound to or after reaction with WT TTR (3.6 µM).
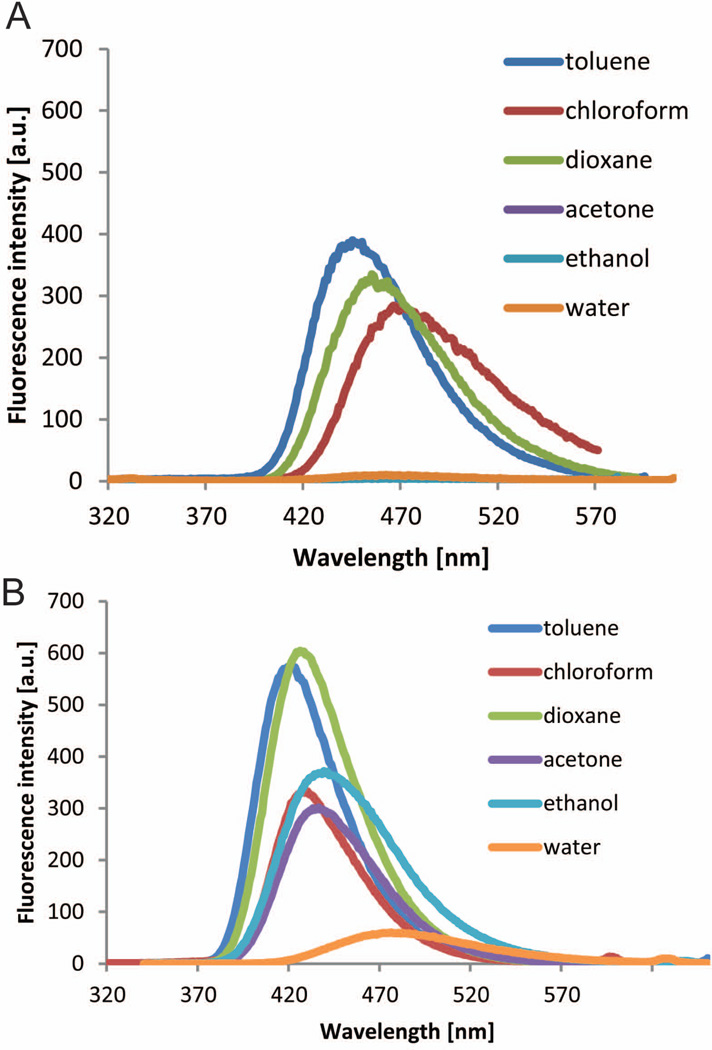
Stilbene 1 is clearly a solvatochromic or environmentally-sensitive fluorophore in that it fluoresces in organic solvents of low polarity (e.g., toluene, chloroform, dioxane), but becomes “dark” as the polarity and hydrogen-bond-donor strength of the solvent increases (e.g., when 1 is dissolved in ethanol, acetone, water) (Figure 6A). The TTR binding pocket (Figure 4) is not sufficiently hydrophobic on its own to cause 1 to become fluorescent, that is, binding of 1 to TTR does not create a fluorescent complex. A chemoselective reaction with TTR is required to afford a fluorescent conjugate, as previously reported.21 It appears that the thioester functional group is able to efficiently quench fluorescence before it is converted to an amide linkage. The chemoselective transformation of the thioester to an amide by Lys15 attack seems to be critical for rendering the conjugate fluorescent.21
Figure 6.
Fluorescence emission spectra of compound 1 (A) and compound 1b (B) (7.2 µM) in solvents of variable polarity. See also Table S1.
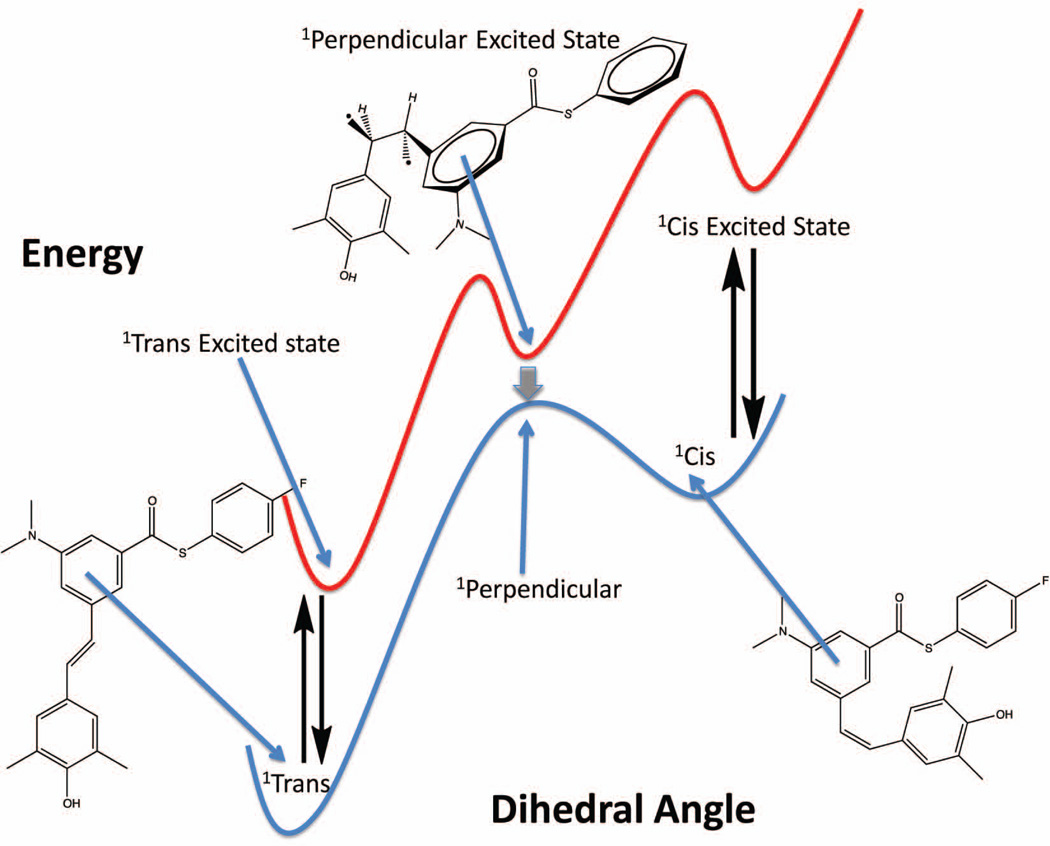
The chromophore in 1 is a stilbene (Figures 3 and 5). Upon promotion of an electron from the highest occupied molecular orbital (HOMO) to the lowest unoccupied molecular orbital (LUMO) of most stilbenes, a singlet excited state is formed that repopulates the ground state by isomerization to afford cis and trans isomers, with some fluorescence emission depending upon the exact structure (Figure 7). Stilbene 1 does not appear to photoisomerize in aqueous buffer after 10 sec of irradiation, as published previously, for reasons we do not fully understand. It appears that the presence of the thioester allows the stilbene to dissipate energy by a non-radiative, non-isomerization mechanism.21
Figure 7.
Structures and energetics associated with stilbene photochemistry and photophysics. Access to the perpendicular singlet excited state conformation is required for the trans to cis photoisomerization of stilbenes. When stilbenes are bound to or tethered to TTR (red curve), the perpendicular singlet excited state and the cis singlet excited state are strongly destabilized by the constraints imposed by the TTR binding site. Figure adapted from21.
The lack of fluorescence when 1 is non-covalently bound to Lys15Ala TTR suggests that its thioester functionality along with the surrounding microenvironment of the Lys15Ala TTR binding site (Figure 4) very efficiently quenches its fluorescence.21 However upon binding of 1 to wild type (WT) TTR, the pKa-perturbed Lys15 ε-amino group attacks the carbonyl group of the thioester to form an amide bond. The resulting conjugate exhibits bright blue fluorescence, largely because the thioester functional group is converted into an amide bond, covalently linking the stilbene chromophore to TTR.21 Converting the thioester in 1 to a propyl amide renders the molecule weakly fluorescent in aqueous buffer. Upon binding to WT and Lys15Ala TTR the non-covalent complex between the propyl amide derived from 1 exhibits a blue shift and a large increase in fluorescence quantum yield, as previously published,21 supporting the hypothesis that the conjugation chemistry is the fluorogenic switch.
The stilbene tethered to TTR by an amide bond makes non-covalent interactions (both hydrophobic and electrostatic) with the T4 binding pocket of WT TTR (Figure 8, to be described in more detail below) in such a fashion that the perpendicular singlet excited state (Figure 7) cannot form to any appreciable extent; thus precluding cis-trans double bond isomerization as a means of energy dissipation from the excited state. While TTR can bind two aromatic rings connected by a linker up to 40° out of coplanarity in the ground state, there is no evidence in the over 100 crystal structures reported to date that aromatic rings can be bound in a perpendicular orientation.57,61 It would likely be even more difficult to bind the aromatic rings substantially out of plane after amide bond tethering of the stilbene chromophore derived from 1 to TTR. That the roughly trans singlet excited state conformation of the stilbene is the only energetically accessible conformation of the chromophore in the covalent conjugate (Figure 7) likely explains why the fluorescence quantum yield goes from ~0 to 0.27 upon conjugate formation.21
Figure 8.
Crystal structures of the TTR ligand complex with 1a and the TTR conjugate resulting from the reaction between TTR and 1. (A) Quaternary structure of the fluorescent conjugate resulting from the reaction between TTR and 1, shown as a ribbon representation with monomers colored individually. (PDB accession code 4L1S) (B) Close up view of the T4 pocket depicting the substructure of 1 covalently attached to the Lys15 residue via an amide bond. (C) Close up view of the T4 pocket showing 1a in complex with TTR. (PDB accession code 4L1T). A Connolly molecular surface was applied to residues within 10 Å of ligand in the T4 binding pocket depicting a hydrophobic surface as gray and a polar surface as purple. HBPs 3 and 3′ are composed of the methyl and methylene groups of S117/117′, T119/119′, and L110/110′. HBPs 2 and 2′ are made up by the side chains of L110/110′, A109/109′, K15/15′, and L17/17′. The outermost HBPs 1 and 1′ are lined by the methyl and methylene groups of K15/15′, A108/108′, and T106/106′.
The sulfur atom in the leaving group of 1 appears to be the dominant quencher of fluorescence (cf. compounds 1 and 1b in Figure 5B). A comparison between compound 1 and its ester analog 1b (Figure 5A), also an environmentally-sensitive fluorophore (Figure 6B), indicates the much stronger quenching capacity of the sulfur atom vs. an oxygen atom, even in hydrophobic solvents. This observation is further supported by the fact that compound 1b exhibits a fluorescence signal when bound to Lys15Ala TTR as reported previously.21
Stilbene fluorescence can be quenched in trans by the addition of mM concentrations of thiophenol, as reported previously,21 or by S-ethyl benzothioate (Figure S1), supporting the hypothesis that the sulfur atom in the thioester of 1 plays a key role in quenching, as is also the case in thioamides.63 However, the precise explanation for how the thioester functional group quenches stilbene fluorophore fluorescence, and how general this effect is, remains unclear and merits further investigation. It seems likely that the application of theoretical/computational chemistry will be required in order to better understand the mechanism of fluorescence quenching and inhibition of photoisomerization.
Herein we report the crystal structure of the conjugate resulting from the reaction between 1 and WT TTR at 1.50 Å resolution, as well as the non-covalent complex between 1a (Figure 5, results from hydrolysis of 1) and WT TTR at 1.16 Å resolution (Figure 8 and Table S2). The conjugate crystal structure not only further substantiates the chemoselective chemical modification of TTR by 1, but also provides additional insight into the mechanism of the fluorogenicity by comparing the conjugate to the complex between WT TTR and 1a.
The 3,5-dimethyl-4-hydroxyphenyl substructures in the conjugate between 1 and TTR and in the TTR•1a complex occupy the inner T4 binding cavity of TTR (Figures 8B and C, respectively), consistent with the preferred binding orientation of noncovalent stilbenes harboring this substructure.57,61 The methyl substituents reside in the two symmetry-related halogen binding pockets (HPBs) 3 and 3′. The 4-OH substituent on the aryl ring in the inner T4 binding cavity makes bridging hydrogen bonds with the Ser117 and Ser117′ side chains of adjacent TTR subunits (Figures 8B and C). The trans –CH=CH– linker is placed into the hydrophobic environment of HBP 2 and 2′ which orients the other aromatic ring occupying the outer T4 binding cavity into HBP 1 and 1′.57
The placement of the thioester functional group at the 3-position on the aromatic ring occupying the outer T4 binding site enables the pKa-perturbed ε-amino group of Lys15 (the nucleophile) to approach the carbonyl at the Bürgi–Dunitz angle64 facilitating amide bond conjugation, as discussed previously in the case of phenyl esters.62 Amide bond conjugation orders the ordinarily flexible Lys15 side chain and the stilbene substructure of 1 such that their B-values (34.0 Å2) approach those of the protein (24.2 Å2). This data is consistent with the stilbene chromophore being rigidly held in the inner and outer T4 binding pockets, which could influence the fluorescent quantum yield. The carboxylate group of 1a makes a non-covalent interaction with the ε-amino group of Lys15 through bridging water molecules, as has been observed with other carboxylate containing compounds, including the regulatory agency approved TTR kinetic stabilizer drug tafamidis,65,66 which prevents TTR aggregation by preventing tetramer dissociation.
Interestingly, a comparison of the stilbene chromophore orientation in the conjugate and the TTR•1a complex shows the chromophore to be oriented in the T4 binding pocket almost identically (Figure 8 and Figure S2). This provides further evidence that the fluorogenic properties of 1 are likely due to the loss of the quenching thiophenol ester group and not a consequence of conformational alterations, although this assumes the noncovalent complex between 1 and TTR positions the chromophore identically as well. Differential solvation could also contribute, which cannot be discerned from the structure, thus the importance of quantum mechanical calculations to understand this result. Further analysis of the electron density maps and consideration of the mass spectrometry data indicate that none of the other seven lysine residues in TTR were modified by 1, consistent with the high chemoselectivity of such covalent modifiers.21
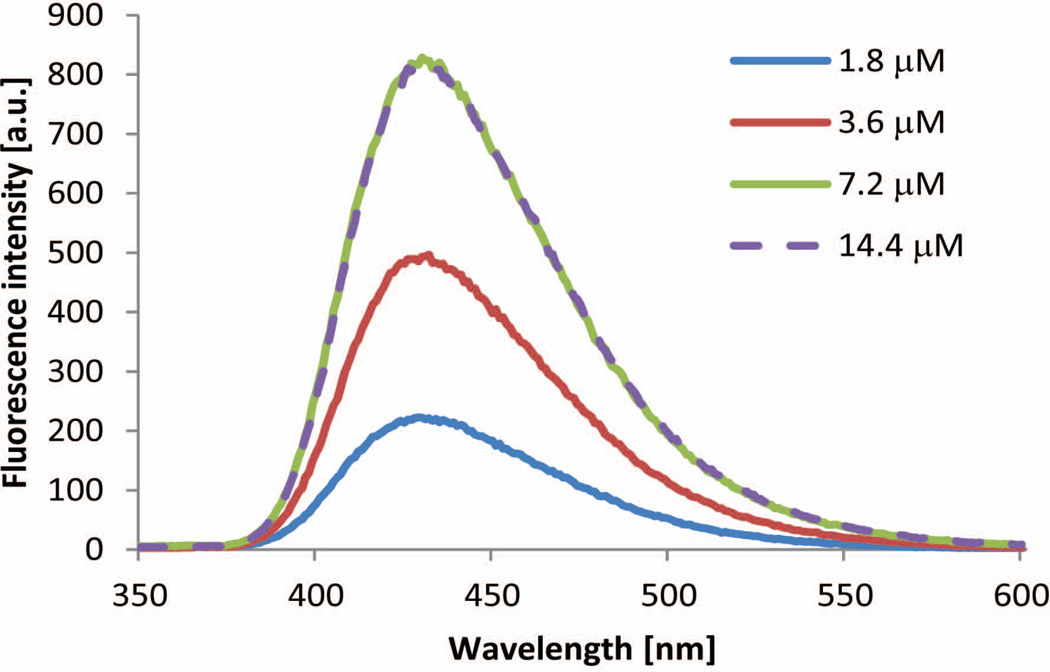
Since there are two T4 binding sites in each TTR tetramer interconverted by a C2 axis perpendicular to the crystallographic two-fold axis (x-axis, Figure 4A), it is important to understand what happens to the fluorescence of the conjugate afforded after TTR reacts with 0.5, 1, or 2 equivalents of 1.67 Here we report that the intensity of the fluorescence conjugate signal scales linearly with the chemical yield of the conjugate as discerned by HPLC. The fluorescence intensity doubles as the concentration of the conjugate doubles, resulting from the incubation of 1 with WT TTR for 24 h (Figure 9). Addition of more than 2 equivalents of 1 does not further increase the fluorescence of the conjugate, as would be expected based on the chemoselectivity of 1, i.e., that 1 does not react with the other Lys residues in TTR.
Figure 9.
Fluorescence emission spectra resulting from increasing concentrations of compound 1 reacting with WT TTR (3.6 µM) in 10mM phosphate buffer pH 7.0.
2,5-diaryl-1,3,4-oxadiazole chromophores
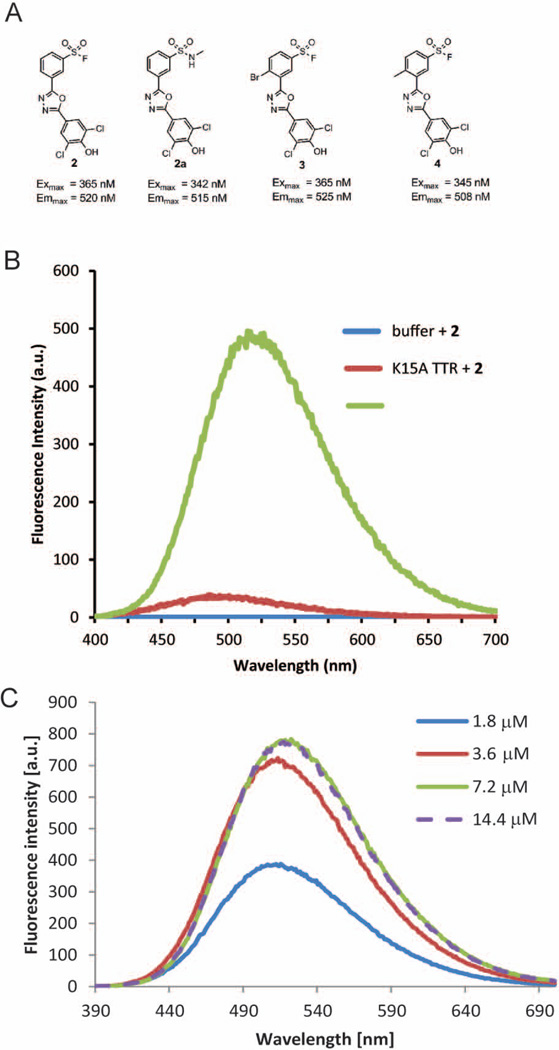
We next assessed whether we could create another fluorogenic small molecule TTR covalent modifier using a different chromophore and employing a distinct functional group, albeit one comprising a sulfur atom, to achieve fluorescence quenching until the functional group undergoes reaction with Lys15 of TTR. The 1,3,4-oxadiazole chromophores substituted with aryl rings at the 2 and 5 positions were conceived of by structure-based design to complement the shape of the two thyroid hormone binding sites in TTR, Figures 3 and 4.22 The aryl ring substituted at the 2 position of the 1,3,4-oxadiazole harbors a meta-sulfonyl fluoride substituent that was hoped to efficiently quench the fluorescence of the non-covalent complex between 2 and TTR, Figure 10A. The aryl ring attached at the 5 position of the 1,3,4-oxadiazole is substituted at the meta or meta and para positions with substituents that bind strongly to the inner binding site of TTR and thus enhance binding selectivity by lowering the KD.22 Attack of the pKa-perturbed Lys15 ε-amino group on the sulfur atom to form a sulfonamide covalent link to TTR by the displacement of fluoride apparently relieves efficient fluorescence quenching, thereby affording a fluorescent conjugate, as previously reported.22
Figure 10.
(A) Structures of 2,5-bisaryl-1,3,4-oxadiazole chromophores that bind to and then react with TTR to form fluorescent conjugates, together with the excitation and emission maxima of the conjugates. (B) Fluorescence emission spectra resulting from compound 2 alone or after reacting with WT TTR (3.6 µM) or K15A TTR in 10 mM phosphate buffer pH 7.0. (C) Fluorescence emission spectra resulting from increasing concentrations of compound 2 reacting with WT TTR (3.6 µM) in 10 mM phosphate buffer pH 7.0.
Homologs 2, 3 and 4 (Figure 10A) functionalized with a para-hydroxyl group in the aryl ring attached at the 5 position of the 1,3,4-oxadiazole reacted with TTR via their sulfonyl fluoride functional group to form covalent fluorescent conjugates exhibiting an average Stokes shift of 160 nm, as previously reported.22 The auxochromic OH substituent on the ring occupying the inner binding pocket of TTR and the electrophilic sulfonyl substituent on the ring occupying the outer TTR binding pocket exemplify a classic push-pull fluorophore, molecules generally exhibiting large Stokes shifts.6
It was previously reported that the rate of emergence of fluorescence upon incubation of 2 with TTR directly correlates with the rate of conjugate formation, as determined by HPLC analysis, suggesting that a chemoselective reaction between 2 and TTR is required for fluorescence.22 The sulfonyl fluoride reacts with TTR ≈ 20× faster than the thioester described above, and modifies TTR ≈ 1400× faster than the hydrolysis of 2, as previously reported.22 Thus the chemoselective reaction with TTR seems to be key to the fluorogenic properties of 2.
Sulfonyl fluorides, unlike sulfonyl chlorides, are relatively inert towards hydrolysis or other reactions in aqueous buffers until they are protein bound.22,68,69 Protein binding activates the sulfonyl fluoride functional group through H-bonding to the fluoride component of the sulfonyl fluoride and possibly by protein H-bond donor interactions with the sulfonyl oxygen substructures.22
While compound 2 exhibited no fluorescence in buffer (Figure 10B), it exhibited weak blue-shifted fluorescence when bound to Lys15Ala TTR, unlike the situation with 1, which remained dark under these conditions. While 2 could exhibit weak environmentally-sensitive fluorescence when bound to WT TTR, this binding-associated fluorescence is not observed in the previously published time courses of 2 reacting with WT TTR;22 if it were, fluorescence would be observed in the period required for binding, which is very fast. Collectively, these observations suggest that covalent conjugation through sulfonamide tethering is required to achieve the 12.5 fold more intense red-shifted fluorescence with WT TTR, but not with Lys15Ala TTR where binding seems to be sufficient to achieve weak fluorescence probably because the Lys15Ala binding site is not hydrophilic enough to achieve complete fluorescence quenching. These previously published observations support the hypothesis that both the quenching functional group and the exact protein micro-environment determine whether fluorophore 2 remains dark after binding and prior to its chemoselective reaction with WT TTR.22 This hypothesis is consistent with the fact that synthesis of an N-methyl sulfonamide derivative of 2, compound 2a (Figure 10A), is sufficient to afford the intense red-shifted fluorescence simply by binding WT or Lys15Ala TTR as previously reported.22 Exactly how covalent reaction with sulfonyl fluoride renders the conjugate fluorescent should be revealed by future theoretical/computational studies.
It is important to understand what happens to the fluorescence of the conjugate afforded after WT TTR reacts with 0.5, 1, or 2 equivalents of 2. Here we report that like the situation with fluorogenic small molecule 1, the fluorescence intensity of the conjugate signal scales linearly with the chemical yield of the conjugate formed from the reaction of 2 with WT TTR up to occupancy of one of the two thyroid hormone binding sites, as discerned by HPLC (Figure 10C). In contrast to the situation with fluorogenic small molecule 1, the reaction of fluorogenic compound 2 with the second thyroid hormone binding site of WT TTR does not increase the fluorescence intensity of the bis-conjugate further (Figure 10C). The apparent lack of fluorescence from occupancy of the second binding site of WT TTR by the chromophore derived from 2 may be caused by the established difference in structure between the allosterically connected sites in TTR.70,71 In other words, when 2 reacts with the low affinity TTR binding site after reacting with the high affinity TTR binding site, the chromophore derived from 2 in the low affinity site cannot adopt the necessary conformation to exhibit fluorescence (almost all TTR ligands bind negatively cooperatively to TTR).61,72 Another possible, but less likely, explanation is that occupancy of both sites fortuitously affords half the fluorescence intensity per bound ligand of the occupancy of one site.
Other strategies to create fluorogenic molecules by way of chemoselective reactions with proteins based on intramolecular quenching of fluorophores
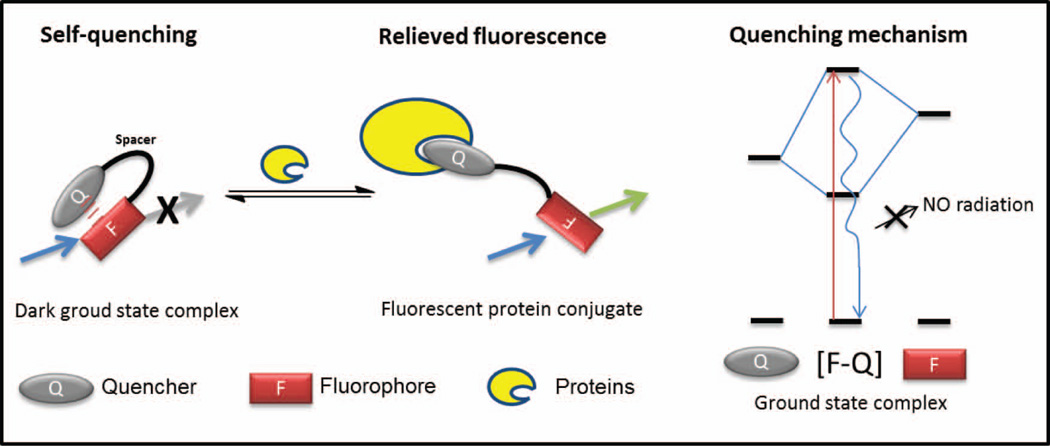
In addition to the concepts that we have discussed above to create fluorogenic small molecules for TTR, we would like to review other general strategies for creating fluorogenic small molecules that undergo a chemoselective reaction with a protein-of-interest to create a fluorescent conjugate. This category of small molecule fluorophores is kept in the “off” state via intramolecular self-quenching of the fluorophore through aromatic–aromatic interactions or Förster resonance energy transfer mechanism (FRET) (Figure 11). The fluorogenic molecule is switched “on” by protein binding to one of the aromatic components or via protein binding followed by a chemoselective reaction with the protein-of-interest (Figure 11).
Figure 11.
Small molecule fluorophores are kept in their “off” state via intramolecular quenching of the fluorophore through intramolecular aromatic–aromatic interactions or FRET and switched “on” by protein binding to one of the aromatic components or via protein binding followed by a chemoselective reaction with one of the aromatic components. Figure adapted from85.
In one scenario, the aromatic quencher (Q)–aromatic fluorophore (F) intramolecular static fluorescence quenching mechanism mediated by the formation of a dark ground-state aromatic Q–aromatic F interaction is taken advantage of (Figure 11).73–75 Protein binding to one of the aromatic components of this dark complex between fluorophore and intramolecular quencher relieves quenching and allows the fluorophore to fluoresce.76,77 In this scenario, the spacer between Q and F has to be optimized to achieve a predominant intramolecular aromatic Q–aromatic F closed conformation with an appropriate open conformation population to allow protein binding. A strategy to render such a bis-aromatic small molecule fluorogenic is to modify the aromatic F with a substituent or functional group that very efficiently quenches its fluorescence until that functional group reacts with a protein-of-interest.
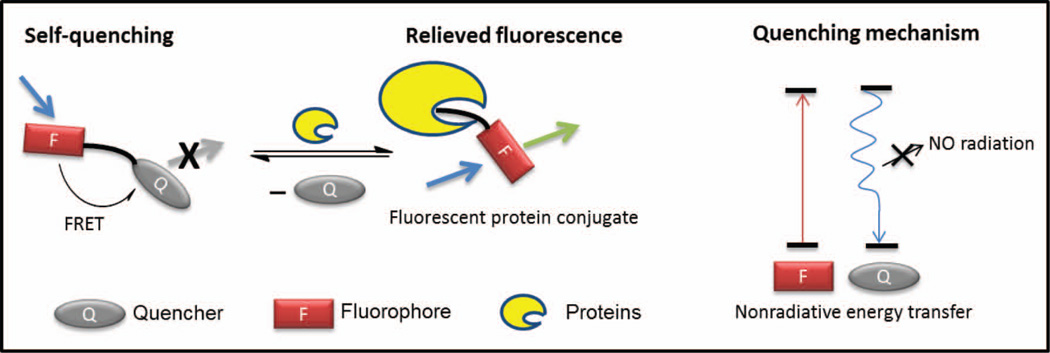
In the second scenario, a dynamic quenching mechanism, usually FRET, keeps the bis-aromatic molecule dark in the absence of binding to and reacting with a protein-of-interest (Figure 12).27–29 The designed fluorogenic small molecule consists of three components: a fluorophore, a spacer and a quencher. The absorption spectrum of the quencher substantially overlaps with the emission spectrum of the fluorophore and therefore quenches its fluorescence by intramolecular FRET, leading to a nonradiative energy transfer. Upon binding to and chemoselectively reacting with a protein-of-interest, the quencher is displaced as a leaving group by a chemical reaction, generating a fluorescent conjugate covalently modified by the fluorophore. This strategy has been successfully employed in the design of the fluorogenic versions of protein-based chemical tags for biological imaging applications.27–30
Figure 12.
FRET self-quenching and induction of fluorogenicity by a chemoselective reaction with a protein-of-interest. Upon binding to and chemoselectively reacting with a protein-of-interest, the quencher is displaced by a chemical reaction as a leaving group, generating a covalent fluorescent conjugate. Figure adapted from85.
Conclusion
Creating fluorogenic small molecules that chemoselectively react with a given protein and only then become fluorescent is highly desirable. Much research needs to be done in the future if we are to fully realize the promise of such molecules as fluorescence probes for live organism biological imaging. We need to discover more functional groups that are quenchers of fluorescence before chemoselective protein reactions, but not afterwards, and better understand their mechanism of action by recruiting theoretical chemists. We also need to work on chromophore development so as to get the longest excitation and emission wavelengths from small drug-like molecules that selectively bind a given protein and exhibit high quantum yields after the quenching functional group is modified or displaced by a chemoselective protein conjugation reaction.
Materials and Methods
Fluorimetric Assay with Recombinant Wild-Type Transthyretin
The covalent TTR modifiers (5 µL, 0.36 mM, 0.72 mM, 1.44 mM, 2.88 mM solution in DMSO, final concentration 1.8 µM, 3.6 µM, 7.2 µM, 14.4 µM respectively) were added to WT-TTR homotetramer (1 mL, 0.2 mg/mL, 3.6 µM solution in 10 mM phosphate, 100 mM KCl, and 1 mM EDTA, pH 7.0) in a microfuge tube. The samples were vortexed and incubated at 25 °C. After 24 h, the fluorescence changes were monitored using a Varian Cary 50 spectrofluorometer at 25 °C in a 1-cm path length quartz cell. The excitation slit was set at 5 nm, and the emission slit was set at 5 nm. Emission spectra were collected from 340 nm to 650 nm for compound 1 and from 390 to 700 nm for compound 2, using the excitation wavelengths: 328 nm for compound 1 and 365 nm for compound 2.
Fluorimetric Assay of Compounds in Solvents with Variable Polarity
Compounds 1 and 1b (5 µL, 1.44 mM solution in DMSO, final concentration 7.2 µM) were dissolved in solvents (1 mL) with variable polarity (toluene, chloroform, dioxane, acetone, ethanol, water) in a microfuge tube. The samples were vortexed. The fluorescence changes were monitored using a Varian Cary 50 spectrofluorometer at 25 °C in a 1-cm path length quartz cell. The excitation slit was set at 5 nm, and the emission slit was set at 5 nm. Emission spectra were collected from 320 nm to 650 nm, using the appropriate excitation wavelengths.
Crystallization and Structure Solution of Transthyretin Ligand Complexes
WT TTR protein was concentrated to 6 mg/mL in 10 mM sodium phosphate buffer and 100 mM KCl (pH 7.6) and co-crystallized at room temperature with a 5 molar excess of 1 or the hydrolyzed acid analog using the vapor-diffusion sitting drop method. The crystals were grown from 1.395 M sodium citrate, 3.5% v/v glycerol at pH 5.5 and cryo-protected with 10% v/v glycerol. Data were collected at beam-lines 12-2 at the Stanford Synchrotron Radiation Lightsource (SSRL) at a wavelength of 0.9795 Å. All diffraction data were indexed, integrated, and scaled using XDS in space group P21212 with two subunits observed per asymmetric unit. The ligand and modified lysine-ligand coordinates and restraints files were generated using JLigand.78 The covalently modified lysine residue was assigned an occupancy of 0.5 to account for the position on the incident 2-fold symmetry axis, leaving an unmodified lysine as an alternate conformation (z- or C2). Further model building and refinement was completed using Coot79 and Refmac.80 Hydrogens were added during refinement, and anisotropic B-values were calculated. Final models were validated using the JCSG quality control server incorporating Molprobity,81 ADIT (http://rcsb-deposit.rutgers.edu/validate) WHATIF,82 Resolve,83 and Procheck.84 Data collection and refinement statistics are displayed in Table S2.
Protein Data Bank Accession Codes
Atomic coordinates and structure factors for TTR in complex with 1a ((E)-3-(dimethylamino)-5-(4-hydroxy-3,5-dimethylstyryl)benzoic acid) and for the conjugate resulting from the reaction between TTR and two equivalents of 1 have been deposited in the Protein Data Bank (www.pdb.org) and are available under the accession codes 4L1T and 4L1S, respectively.
Supplementary Material
Acknowledgements
This work was supported by the Skaggs Institute for Chemical Biology, the Lita Annenberg Hazen Foundation, and by the National Institutes of Health grant DK046335 (JWK). Portions of this research were carried out at the Stanford Synchrotron Radiation Lightsource, a Directorate of SLAC National Accelerator Laboratory and an Office of Science User Facility operated for the U.S. Department of Energy Office of Science by Stanford University. The SSRL Structural Molecular Biology Program is supported by the DOE Office of Biological and Environmental Research, and by the National Institutes of Health, National Institute of General Medical Sciences (including P41GM103393) and the National Center for Research Resources (P41RR001209). The authors also wish to thank Jason Gilberg for assistance in preparing the protein ligand complexes, for their crystallization and with the data collection and initial model building.
References
- 1.Tsien RY. Integr Biol. 2010;2:77–93. doi: 10.1039/b926500g. [DOI] [PubMed] [Google Scholar]
- 2.Marmor MF, Ravin JG. Arch Ophthalmol. 2011;129:943–948. doi: 10.1001/archophthalmol.2011.160. [DOI] [PubMed] [Google Scholar]
- 3.Jablonski A. Z Phys. 1935;94:38–46. [Google Scholar]
- 4.Fernandez-Suarez M, Ting AY. Nat Rev Mol Cell Biol. 2008;9:929–943. doi: 10.1038/nrm2531. [DOI] [PubMed] [Google Scholar]
- 5.Lavis LD, Raines RT. ACS Chem Biol. 2008;3:142–155. doi: 10.1021/cb700248m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lord SJ, Lee H-lD, Samuel R, Weber R, Liu N, Conley NR, Thompson MA, Twieg RJ, Moerner WE. J Phys Chem B. 2010;114:14157–14167. doi: 10.1021/jp907080r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Conley NR, Biteen JS, Moerner WE. J Phys Chem B. 2008;112:11878–11880. doi: 10.1021/jp806698p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee H-lD, Lord SJ, Iwanaga S, Zhan K, Xie H-X, Williams JC, Wang H, Bowman GR, Goley ED, Shapiro L, Twieg RJ, Rao J-H, Moerner WE. J Am Chem Soc. 2010;132:15099–15101. doi: 10.1021/ja1044192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Peterman EJG, Sosa H, Moerner WE. Annu Rev Phys Chem. 2004;55:79–96. doi: 10.1146/annurev.physchem.55.091602.094340. [DOI] [PubMed] [Google Scholar]
- 10.Chan J, Dodani SC, Chang CJ. Nat Chem. 2012;4:973–984. doi: 10.1038/nchem.1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shimomura O, Johnson FH, Saiga Y. J Cell Comp Physiol. 1962;59:223–239. doi: 10.1002/jcp.1030590302. [DOI] [PubMed] [Google Scholar]
- 12.Chalfie M, Tu Y, Euskirchen G, Ward WW, Prasher DC. Science. 1994;263:802–805. doi: 10.1126/science.8303295. [DOI] [PubMed] [Google Scholar]
- 13.Tsien RY. Annu Rev Biochem. 1998;67:509–544. doi: 10.1146/annurev.biochem.67.1.509. [DOI] [PubMed] [Google Scholar]
- 14.Loving G, Imperiali B. J Am Chem Soc. 2008;130:13630–13638. doi: 10.1021/ja804754y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goncalves MS. Chem Rev. 2009;109:190–212. doi: 10.1021/cr0783840. [DOI] [PubMed] [Google Scholar]
- 16.Grimm JB, Heckman LM, Lavis LD. Prog Mol Biol Transl Sci. 2013;113:1–34. doi: 10.1016/B978-0-12-386932-6.00001-6. [DOI] [PubMed] [Google Scholar]
- 17.Brecht A, Gibbs T. BIOforum Eur. 2005;9:50–51. [Google Scholar]
- 18.Regoes A, Hehl AB. BioTechniques. 2005;39:809–810. 812. doi: 10.2144/000112054. [DOI] [PubMed] [Google Scholar]
- 19.Chen Z, Jing C, Gallagher SS, Sheetz MP, Cornish VW. J Am Chem Soc. 2012;134:13692–13699. doi: 10.1021/ja303374p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yao JZ, Uttamapinant C, Poloukhtine A, Baskin JM, Codelli JA, Sletten EM, Bertozzi CR, Popik VV, Ting AY. J Am Chem Soc. 2012;134:3720–3728. doi: 10.1021/ja208090p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Choi S, Ong DS, Kelly JW. J Am Chem Soc. 2010;132:16043–16051. doi: 10.1021/ja104999v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grimster NP, Connelly S, Baranczak A, Dong J, Krasnova LB, Sharpless KB, Powers ET, Wilson IA, Kelly JW. J Am Chem Soc. 2013;135:5656–5668. doi: 10.1021/ja311729d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jing C, Cornish VW. Acc Chem Res. 2011;44:784–792. doi: 10.1021/ar200099f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wombacher R, Cornish VW. J Biophotonics. 2011;4:391–402. doi: 10.1002/jbio.201100018. [DOI] [PubMed] [Google Scholar]
- 25.Lee MK, Williams J, Twieg RJ, Rao J, Moerner WE. Chem Sci. 2013;42:220–225. doi: 10.1039/C2SC21074F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Weinstain R, Kanter J, Friedman B, Ellies LG, Baker ME, Tsien RY. Bioconjugate Chem. 2013;24:766–771. doi: 10.1021/bc3006418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mizukami S, Watanabe S, Akimoto Y, Kikuchi K. J Am Chem Soc. 2012;134:1623–1629. doi: 10.1021/ja208290f. [DOI] [PubMed] [Google Scholar]
- 28.Mizukami S, Watanabe S, Hori Y, Kikuchi K. J Am Chem Soc. 2009;131:5016-+. doi: 10.1021/ja8082285. [DOI] [PubMed] [Google Scholar]
- 29.Sun X, Zhang A, Baker B, Sun L, Howard A, Buswell J, Maurel D, Masharina A, Johnsson K, Noren CJ, Xu MQ, Correa IR., Jr Chembiochem. 2011;12:2217–2226. doi: 10.1002/cbic.201100173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Watanabe S, Mizukami S, Hori Y, Kikuchi K. Bioconjug Chem. 2010;21:2320–2326. doi: 10.1021/bc100333k. [DOI] [PubMed] [Google Scholar]
- 31.Slavik J. Proc SPIE-Int Soc Opt Eng. 1996;2926:143–150. [Google Scholar]
- 32.Wang K, Ma H. Huaxue Jinzhan. 2010;22:1633–1640. [Google Scholar]
- 33.Demchenko AP, Mely Y, Duportail G, Klymchenko AS. Biophys J. 2009;96:3461–3470. doi: 10.1016/j.bpj.2009.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Saroja G, Soujanya T, Ramachandram B, Samanta A. J Fluoresc. 1998;8:405–410. [Google Scholar]
- 35.Kirtley ME, Koshland DE., Jr 1972:578–601. doi: 10.1016/s0076-6879(72)26027-x. [DOI] [PubMed] [Google Scholar]
- 36.Klymchenko AS, Mely Y. Prog Mol Biol Transl Sci. 2013;113:35–58. doi: 10.1016/B978-0-12-386932-6.00002-8. [DOI] [PubMed] [Google Scholar]
- 37.Loving GS, Sainlos M, Imperiali B. Trends Biotechnol. 2010;28:73–83. doi: 10.1016/j.tibtech.2009.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Feitelson J. Isr J Chem. 1970;8:241–252. [Google Scholar]
- 39.Giancotti V, Quadrifoglio F, Cowgill RW, Crane-Robinson C. Biochim Biophys Acta, Protein Struct. 1980;624:60–65. doi: 10.1016/0005-2795(80)90225-1. [DOI] [PubMed] [Google Scholar]
- 40.Leaback DH. J Fluoresc. 1997;7:3S–5S. [Google Scholar]
- 41.Sato S, Religa TL, Fersht AR. J Mol Biol. 2006;360:850–864. doi: 10.1016/j.jmb.2006.05.051. [DOI] [PubMed] [Google Scholar]
- 42.Reichardt C. Chem Rev. 1994;94:2319–2358. [Google Scholar]
- 43.Rosen CG, Weber G. Biochemistry. 1969;8:3915–3920. doi: 10.1021/bi00838a006. [DOI] [PubMed] [Google Scholar]
- 44.Cornec A-S, Baudequin C, Fiol-Petit C, Ple N, Dupas G, Ramondenc Y. Eur J Org Chem. 2013;2013:1908–1915. [Google Scholar]
- 45.Didier P, Ulrich G, Mely Y, Ziessel R. Org Biomol Chem. 2009;7:3639–3642. doi: 10.1039/b911587k. [DOI] [PubMed] [Google Scholar]
- 46.Dong Y, Bolduc A, McGregor N, Skene WG. Org Lett. 2011;13:1844–1847. doi: 10.1021/ol200353k. [DOI] [PubMed] [Google Scholar]
- 47.Jacquart A, Williams RM, Brouwer AM, Ishow E. Chemistry. 2012;18:3706–3720. doi: 10.1002/chem.201103411. [DOI] [PubMed] [Google Scholar]
- 48.Lord SJ, Conley NR, Lee H-lD, Nishimura SY, Pomerantz AK, Willets KA, Lu Z, Wang H, Liu N, Samuel R, Weber R, Semyonov A, He M, Twieg RJ, Moerner WE. ChemPhysChem. 2009;10:55–65. doi: 10.1002/cphc.200800581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lord SJ, Conley NR, Lee H-lD, Samuel R, Liu N, Twieg RJ, Moerner WE. J Am Chem Soc. 2008;130:9204–9205. doi: 10.1021/ja802883k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mata G, Luedtke NW. Org Lett. 2013;15:2462–2465. doi: 10.1021/ol400930s. [DOI] [PubMed] [Google Scholar]
- 51.Qian X, Xiao Y. Tetrahedron Lett. 2002;43:2991–2994. [Google Scholar]
- 52.Ventelon L, Blanchard-Desce M, Moreaux L, Mertz J. Chem Commun. 1999:2055–2056. [Google Scholar]
- 53.Wojtczak A, Cody V, Luft JR, Pangborn W. Acta Crystallogr D Biol Crystallogr. 1996;52:758–765. doi: 10.1107/S0907444996003046. [DOI] [PubMed] [Google Scholar]
- 54.Johnson SM, Wiseman RL, Sekijima Y, Green NS, Adamski-Werner SL, Kelly JW. Acc Chem Res. 2005;38:911–921. doi: 10.1021/ar020073i. [DOI] [PubMed] [Google Scholar]
- 55.Monaco HL, Rizzi M, Coda A. Science. 1995;268:1039–1041. doi: 10.1126/science.7754382. [DOI] [PubMed] [Google Scholar]
- 56.Purkey HE, Dorrell MI, Kelly JW. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:5566–5571. doi: 10.1073/pnas.091431798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Johnson SM, Connelly S, Wilson IA, Kelly JW. J Med Chem. 2008;51:6348–6358. doi: 10.1021/jm800435s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Johnson SM, Connelly S, Wilson IA, Kelly JW. J Med Chem. 2008;51:260–270. doi: 10.1021/jm0708735. [DOI] [PubMed] [Google Scholar]
- 59.Johnson SM, Connelly S, Wilson IA, Kelly JW. J Med Chem. 2009;52:1115–1125. doi: 10.1021/jm801347s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Choi S, Reixach N, Connelly S, Johnson SM, Wilson IA, Kelly JW. J Am Chem Soc. 2010;132:1359–1370. doi: 10.1021/ja908562q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Connelly S, Choi S, Johnson SM, Kelly JW, Wilson IA. Curr Opin Struct Biol. 2010;20:54–62. doi: 10.1016/j.sbi.2009.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Choi S, Connelly S, Reixach N, Wilson IA, Kelly JW. Nat Chem Biol. 2010;6:133–139. doi: 10.1038/nchembio.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Goldberg JM, Speight LC, Fegley MW, Petersson EJ. J Am Chem Soc. 2012;134:6088–6091. doi: 10.1021/ja3005094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Burgi HB, Dunitz JD, Lehn JM, Wipff G. Tetrahedron. 1974;30:1563–1572. [Google Scholar]
- 65.Bulawa CE, Connelly S, DeVit M, Wang L, Weigel C, Fleming JA, Packman J, Powers ET, Wiseman RL, Foss TR, Wilson IA, Kelly JW, Labaudiniere R. Proc Natl Acad Sci U S A. 2012;109:9629–9634. S9629/9621–S9629/9629. doi: 10.1073/pnas.1121005109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Razavi H, Palaninathan SK, Powers ET, Wiseman RL, Purkey HE, Mohamedmohaideen NN, Deechongkit S, Chiang KP, Dendle MT, Sacchettini JC, Kelly JW. Angewandte Chemie. 2003;42:2758–2761. doi: 10.1002/anie.200351179. [DOI] [PubMed] [Google Scholar]
- 67.Choi S, Kelly JW. Bioorg Med Chem. 2011;19:1505–1514. doi: 10.1016/j.bmc.2010.12.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Baker BR. Acc Chem Res. 1969;2:129–136. [Google Scholar]
- 69.Baker BR. Annu Rev Pharmacol. 1970;10:35–50. doi: 10.1146/annurev.pa.10.040170.000343. [DOI] [PubMed] [Google Scholar]
- 70.Trivella DBB, Bleicher L, Palmieri LdC, Wiggers HJ, Montanari CA, Kelly JW, Lima LMTR, Foguel D, Polikarpov I. J Struct Biol. 2010;170:522–531. doi: 10.1016/j.jsb.2010.03.002. [DOI] [PubMed] [Google Scholar]
- 71.Trivella DBB, Sairre MI, Foguel D, Lima LMTR, Polikarpov I. J Struct Biol. 2011;173:323–332. doi: 10.1016/j.jsb.2010.10.003. [DOI] [PubMed] [Google Scholar]
- 72.Klabunde T, Petrassi HM, Oza VB, Raman P, Kelly JW, Sacchettini JC. Nat Struct Biol. 2000;7:312–321. doi: 10.1038/74082. [DOI] [PubMed] [Google Scholar]
- 73.Hirano T, Akiyama J, Mori S, Kagechika H. Org Biomol Chem. 2010;8:5568–5575. doi: 10.1039/c0ob00207k. [DOI] [PubMed] [Google Scholar]
- 74.Kim TW, Park JH, Hong JI. B Kor Chem Soc. 2007;28:1221–1223. [Google Scholar]
- 75.Valdesaguilera O, Neckers DC. Acc Chem Res. 1989;22:171–177. [Google Scholar]
- 76.Hori Y, Nakaki K, Sato M, Mizukami S, Kikuchi K. Angew Chem Int Edit. 2012;51:5611–5614. doi: 10.1002/anie.201200867. [DOI] [PubMed] [Google Scholar]
- 77.Hori Y, Ueno H, Mizukami S, Kikuchi K. J Am Chem Soc. 2009;131:16610–16611. doi: 10.1021/ja904800k. [DOI] [PubMed] [Google Scholar]
- 78.Lebedev AA, Young P, Isupov MN, Moroz OV, Vagin AA, Murshudov GN. Acta Crystallogr D Biol Crystallogr. 2012;68:431–440. doi: 10.1107/S090744491200251X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Emsley P, Cowtan K. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 80.Storoni LC, McCoy AJ, Read RJ. Acta Crystallogr D Biol Crystallogr. 2004;60:432–438. doi: 10.1107/S0907444903028956. [DOI] [PubMed] [Google Scholar]
- 81.Lovell SC, Davis IW, Arendall WB, 3rd, de Bakker PI, Word JM, Prisant MG, Richardson JS, Richardson DC. Proteins. 2003;50:437–450. doi: 10.1002/prot.10286. [DOI] [PubMed] [Google Scholar]
- 82.Vriend G. J Mol Graph. 1990;8:52–56. 29. doi: 10.1016/0263-7855(90)80070-v. [DOI] [PubMed] [Google Scholar]
- 83.Terwilliger TC. Acta Crystallogr D Biol Crystallogr. 2003;59:38–44. doi: 10.1107/S0907444902018036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Laskowski RA, Macarthur MW, Moss DS, Thornton JM. J Appl Crystallogr. 1993;26:283–291. [Google Scholar]
- 85.Johansson MK. Methods Mol Biol. 2006;335:17–29. doi: 10.1385/1-59745-069-3:17. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.