ABSTRACT
Uncultured and therefore uncharacterized Bacteroidetes lineages are ubiquitous in many natural ecosystems which specialize in lignocellulose degradation. However, their metabolic contribution remains mysterious, as well-studied cultured Bacteroidetes have been shown to degrade only soluble polysaccharides within the human distal gut and herbivore rumen. We have interrogated a reconstructed genome from an uncultured Bacteroidetes phylotype that dominates a switchgrass-associated community within the cow rumen. Importantly, this characterization effort has revealed the first preliminary evidence for polysaccharide utilization locus (PUL)-catalyzed conversion of cellulose. Based on these findings, we propose a further expansion of the PUL paradigm and the saccharolytic capacity of rumen Bacteroidetes species to include cellulose, the most abundant terrestrial polysaccharide on Earth. Moreover, the perspective of a cellulolytic PUL lays the foundation for PULs to be considered an alternative mechanism for cellulose degradation, next to cellulosomes and free-enzyme systems.
Opinion/Hypothesis
Uncultured Bacteroidetes lineages dominate many lignocellulose-degrading communities. A comprehensive understanding of how plant biomass deconstruction occurs in nature has far-reaching implications related to mammalian health and nutrition, as well as development of sustainable bio-based economies. Our current understanding is severely impeded by the inability to cultivate and thus examine the majority of microbes that perform the key metabolic processes of interest. For example, the rumen of herbivores represents one of nature’s most proficient plant biomass-degrading ecosystems; however, it is controlled largely by uncharacterized microbes that belong to a limited number of frequently observed bacterial phyla (1). Degradation of the most abundant plant polysaccharide (cellulose) within ruminal ecosystems has for the most part been attributed to the metabolic capabilities of species affiliated with the bacterial phyla Firmicutes and Fibrobacteres. These species produce one or more well-known cellulases that are structurally assembled on the cell surface as a cellulosome or secreted as free enzymes (2). The ruminal Bacteroidetes represent another numerically dominating phylum; it is not associated with cellulose degradation, but its saccharolytic reputation is based on limited case studies of noncellulolytic Prevotella rumen isolates (3) and renowned culturable human gut representatives, such as Bacteroides thetaiotaomicron and Bacteroides ovatus (4). The saccharolytic machineries of gastrointestinal Bacteroidetes species have thus far been attributed to polysaccharide utilization loci (PULs), gene clusters that encode cell envelope-associated enzyme systems that enable the bacterium to respond to, bind, and degrade specific glycans and import released oligosaccharides (5).
The numerical predominance of uncultured Bacteroidetes species in lignocellulose-degrading ecosystems (1, 6) and the observed abundance and diversity of PUL-encoded carbohydrate-active enzymes within Bacteroidetes genomes suggest that there is much to learn about the contribution of these enzymatic complexes to polysaccharide metabolism. Here we propose an alternative hypothesis regarding cellulose degradation, which was generated by the biochemical characterization of a simplistic cellulase-encoding PUL previously annotated in a high-coverage uncultured Bacteroidetes phylotype (here referred to as AC2a) inherent to the cow rumen microbiome (6, 7). The gene organization of the AC2a PUL indicates a direct targeting of cellulose, which is unique for PULs that have been described and characterized to date (Fig. 1, and see Fig. S1 in the supplemental material).
FIG 1 .
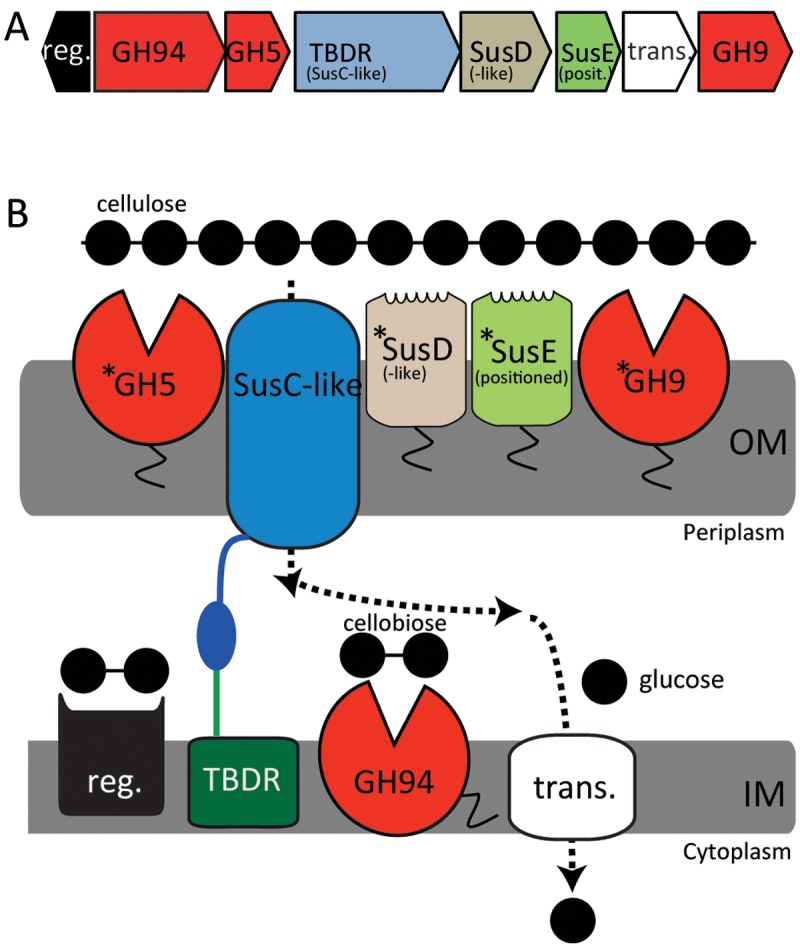
Putatively cellulolytic PUL recovered from the AC2a genome inherent to the cow rumen. (A) Gene organization of a cellulase (GH5 and GH9)-containing PUL identified in AC2a, which was selected for in-depth biochemical characterization. Gene identification numbers can be found in Table S2. (B) A hypothetical model, based on predicted protein locations and analogies to the model starch utilization system (Sus) of B. thetaiotaomicron (5), depicts a process in which glucans are bound and hydrolyzed via outer membrane lipoproteins and enzymes, whereas the generated cellobiose is transported to the periplasm, converted to glucose, and imported to the cytoplasm for cellular metabolism (see the text for more details). Proteins marked with an asterisk were subjected to biochemical characterization. Proteins marked with a “tail” are predicted to be membrane associated. The annotations of SusC (TonB-dependent receptor) and SusD are based on significant hits using Pfam, and these two proteins are therefore referred to as “SusC-like” and “SusD-like” in the main text. SusE is not recognized by Pfam, and its annotation is thus based on position only; this protein is referred to as “SusE positioned” (17). reg., regulator; trans., transporter; OM, outer membrane; IM, inner membrane; TBDR, TonB-dependent receptor.
EXPERIMENTAL RATIONALE
Biochemical characterization of a putative cellulolytic PUL within the uncultured AC2a Bacteroidetes phylotype.
The AC2a draft genome sequence (~76% complete) was one of 16 genomes previously binned using tetranucleotide signatures from a switchgrass-degrading metagenome recovered from a cow’s rumen (7). The high assembly coverage of the genome (284-fold coverage; third highest) indicated that AC2a is likely a numerically abundant organism in the rumen microbiome. Our own de novo predictions using Support Vector Machine classifiers (8) identified AC2a as a potential cellulolytic Bacteroidetes species, which challenges the current idea that Bacteroidetes drive only noncellulosic metabolism in the rumen. AC2a’s cellulolytic capabilities were predicted to be dependent on a relatively simple eight-gene PUL encoding two putative cellulases (GH5 and GH9 of the glycoside hydrolase [GH] family) and a cellobiose phosphorylase (GH94) (Fig. 1A) (6). Sequence analysis and comparisons of gene organization with the model starch utilization system (Sus) of B. thetaiotaomicron led to identification of a SusC-like (TonB-dependent) outer membrane transporter, SusD-like and SusE-positioned lipoproteins that putatively bind to the substrate (9, 10), an inner membrane sugar transporter, and an inner membrane sensor (4) (Fig. 1). We predicted that GH5 and GH9 could degrade cellulose to cellobiose, which would be transported to the periplasm via the SusC-positioned transporter, where the well-known GH94 activity would generate monomeric sugars for transport into the cytoplasm (Fig. 1B). To test this prediction, we have biochemically characterized the GH5 and GH9 enzymes and determined the functionality of the putative SusD-like and SusE-positioned glycan-binding lipoproteins.
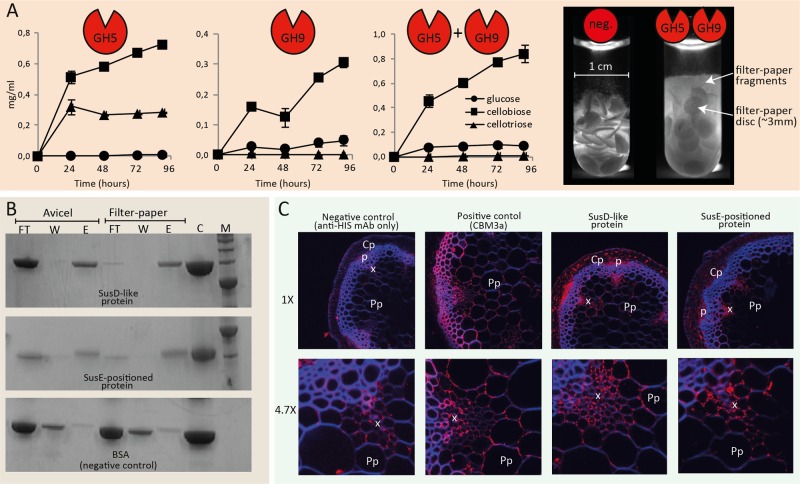
Initial screens with chromogenic substrates showed that the GH5 and GH9 glycoside hydrolases in the AC2a PUL are active on β-(1,4)-linked glucose units in amorphous cellulose and β-glucan (barley) (Fig. S2). The enzymes showed weak side activities on xyloglucan and xylans (Fig. S2) and were not active on β-(1,3) glucan (pachyman). These observations are consistent with the subfamily 4 classification of the GH5 enzyme (11), which typically encompasses extracellular bacterial enzymes that exhibit one or more activities categorized as endoglucanase, xyloglucan-specific endoglucanase, xylanase, and licheninase. Typically for endoglucanases, both GH5 and GH9 demonstrated higher activity on soluble cellulose and more highly accessible natural β-glucan substrates than recalcitrant crystalline cellulose (Table S1). The AC2a PUL is distinct from barley β-glucan PULs characterized from Bacteroides cellulosilyticus (12) and B. ovatus (4) in that it does not contain GH3 [β-(1-3)-glucosidase] or GH16 [β-(1,3)-glucanase] (Fig. S1C). High-pH anion-exchange chromatography–pulsed amperometric detection (HPAEC-PAD) analysis demonstrated that GH5 hydrolysis of filter paper produced dimer and trimer cellodextrins, whereas GH9 hydrolysis produced dimers and monomers (Fig. 2A). Interestingly, upon combination of the two enzymes, the filter paper was converted to dimers and monomers only (Fig. 2A), indicating synergism to produce cellobiose, which ultimately would be degraded by the periplasmic GH94 cellobiose phosphorylase. Further analysis of cellodextrin (DP2-6) hydrolysis revealed that GH5 cannot degrade cellotriose or cellobiose and produces only cellobiose from cellotetraose (Fig. S3). GH5 hydrolysis of cellopentaose and cellohexaose produced dimers and trimers. In contrast, GH9 degraded cellotriose and produced cellobiose and a small amount of glucose from DP4-6. This indicates that the two cellulases have different, complementary roles. The degradative effect of the AC2a PUL enzymes could easily be observed by monitoring the partial solubilization of filter paper discs (Fig. 2A).
FIG 2 .
Biochemical characterization of cellulases and binding proteins encoded within the AC2a PUL. (A) Enzymatic activities of the GH5 and GH9 proteins determined by HPAEC-PAD analysis of products generated from filter paper (5%, wt/vol, 3 µM total enzyme concentration, pH 6.6). Error bars represent standard deviations between results of three replicates. The image to the right visualizes partial solubilization of filter paper discs after a 6-day incubation (discs were diluted 3:1 prior to image capture). neg., negative. (B) SDS-PAGE analysis of fractions from pulldown assays using cellulosic substrates. Lanes: FT, unbound protein from supernatant fractions collected after a 1-h incubation and centrifugation; W, the wash fraction, containing protein washed off the substrate; E, eluted protein fractions where protein was released from the polysaccharides by boiling them in urea; C, control, where only the protein was loaded on the gel; M, a molecular weight standard. (C) Immunofluorescence labeling of A. thaliana cross sections using crystalline cellulose-binding CBM3a (positive control) as well as the SusD-like and SusE-positioned proteins from the AC2a cellulose-active PUL. Fluorescence from anti-His (in red) indicates bound protein, while autofluorescence, mainly in the interfascicular tissue, is blue. The SusD-like protein bound to cortical parenchyma (Cp)-, phloem (p)-, xylem (x)-, and pith parenchyma (Pp)-adjacent cell walls. The SusE-positioned protein showed weaker binding than the SusD-like protein and particularly targeted the intercellular junctions in the xylem tissue. The positive-control images were taken at lower gain due to high signals, while negative-control images were taken at higher gain. mAb, monoclonal antibody.
Pulldown binding assays showed that SusD-like and SusE-positioned proteins from the AC2a PUL bind to crystalline cellulose (Fig. 2B) and also bind weakly to β-glucan (Fig. S4). To further visualize the ability of the SusD-like and SusE-positioned proteins to interact with plant cell walls, both proteins were used for indirect immunofluorescence labeling of Arabidopsis thaliana cross sections. The two proteins demonstrated clear binding to various sections of the plant cell walls, including xylem, phloem, and cortical parenchyma, with the SusE-positioned protein giving weaker signals than the SusD-like protein (Fig. 2C). CBM3a from Clostridium thermocellum (13) was included as a positive control and showed binding to cellulose-rich secondary cell walls (SCW) in the xylem and adhered to faces of adjacent pith parenchyma (PP) cells. The SusD-like protein bound to similar regions, whereas binding of the SusE-positioned protein appeared limited to the intercellular junctions of adjacent PP cells (Fig. 2C).
HYPOTHESIS
PULs represent an alternative mechanism for cellulose degradation, next to cellulosomes and free-enzyme systems.
The insatiable interest in the human gastrointestinal microbiome has provided detailed accounts of the diversity and mechanisms of PULs that are central to plant polysaccharide degradation. However, this understanding has been limited by a reliance on well-known cultivated Bacteroidetes species, which represent a significant minority in many saccharolytic ecosystems. By specifically targeting uncultured microbiota resident in the cow rumen with approaches that go beyond predictive annotation, we reveal a possible alternative mechanism for microbial cellulose degradation, which implies that rumen Bacteroidetes utilize PUL-based machinery, rather than (or in addition to) well-known mechanisms such as cellulosomes and free-enzyme systems. Broader genomic comparisons of the AC2a PUL with publicly available metagenomes and Bacteroidetes genomes identified sequence homology and synteny with a partial metagenomic fragment derived from the tammar wallaby foregut, a marsupial herbivore whose diet is rich in lignocellulose (14) (Fig. S1A). Partial synteny was observed with the “core” components of a well-characterized xyloglucan PUL that encodes both GH5 and GH9 representatives but which targets only xyloglucans, while lacking activity against any other hemicellulose or cellulose substrate (Fig. S1A) (15). Closer inspection of the proteins occurring in both these PULs revealed low sequence similarity (Fig. S1A) as well as different Pfam-predicted domain organizations for GH5 (AC2a lacking the BACON [Bacteroidetes-associated carbohydrate-binding often N-terminal] domain at the N terminus) and the SusD-like lipoprotein (AC2a with PF12771, BACOVA_02651; PF14322/PF07980). While degrading activity for soluble cellulose-analogues has been described for several endoglucanases encoded within large hemicellulosic PULs, these enzymes are devoid of activity on recalcitrant cellulose and the PULs in question bear no resemblance to the AC2a PUL (Fig. S1B) (6, 14). Interestingly the SusD-like lipoprotein from the AC2a PUL exhibited very low sequence identity to two cellulose-binding SusD-like representatives that we previously characterized from a hemicellulose-degrading PUL reconstructed from an uncultured phylotype (both exhibited less than 23% alignment coverage and 31% sequence identity) (10). This suggests that functional differences cannot necessarily be detected by the binding assays done in the present and in past studies.
Collectively, these findings expand current perceptions regarding the overall saccharolytic capacity of rumen Bacteroidetes affiliates, which so far have been coupled only to the degradation of noncellulosic polysaccharides. Furthermore, it adds the Earth’s most abundant organic polymer to an already impressive catalogue of PUL target substrates, including starch, alginate, various hemicelluloses, and host mucin glycans (3, 4, 15–17). To conclusively determine that AC2a is indeed capable of sustaining cell metabolism and growth on recalcitrant cellulosic substrates, knockout mutagenesis studies of pure culture representatives are required. While isolating deeply branched novel affiliates of the Bacteroidetes has proved extremely difficult in herbivore microbiomes, the ability to mine the AC2a genome for growth requirements provides a unique opportunity to reconstruct a custom enrichment medium and isolation strategy. Similar metagenome-directed isolation approaches have ultimately proved successful for gut microbiomes in the past (18) and form the basis of our ongoing efforts.
PROCEDURES
Gene annotation of the AC2a genome.
The AC2a genome was previously reconstructed from metagenome sequencing data generated from the microbiota in a cow’s rumen (pH 7.0) (7). Assembled and unprocessed DNA reads previously assigned to AC2a based on tetranucleotide frequencies were retrieved from http://portal.nersc.gov/project/jgimg/CowRumenRawData/submission/ and annotated via the RAST server (19). The cellular localization of proteins was predicted using PSORTb 3.0 (20) and LipoP 1.0 (21).
Heterologous expression and purification of enzymes.
Genes encoding signal peptide-free versions of AC2a GH5, GH9, SusD-like, and SusE-positioned proteins were synthesized and cloned into the pNIC-CH expression vector by ligation-independent cloning (LIC) using the primers listed in Table S2 in the supplemental material (22). Transformants were verified by sequencing. Escherichia coli BL21 harboring the plasmids was precultured for 8 h in Luria-Bertani broth and inoculated to 1% in an overnight culture at 18°C. Expression was induced by adding isopropyl-β-d-thiogalactopyranoside (IPTG) to a final concentration of 0.75 mM at an optical density at 600 nm (OD600) of 0.5 to 1.0, followed by incubation for 24 h at 18°C. Cells were harvested by centrifugation (5,000 rpm, 10 min) and resuspended in lysis buffer (100 mM Tris-HCl, pH 8.0, 500 mM NaCl, 5 mM imidazole, 0.1 mg/ml lysozyme) before a 30-min incubation on ice. Cells were disrupted by pulsed sonication, and debris was removed by centrifugation (8,000 × g, 10 min), with the supernatant filtered using 0.45- and 0.22-µm syringe filters. Proteins were loaded onto 5-ml HisTrap HP Ni Sepharose columns (GE Healthcare) and eluted with a linear gradient of 100 mM Tris-HCl, pH 8.0, 500 mM NaCl, 500 mM imidazole. The eluted fractions were concentrated, and the buffer was changed to 100 mM Tris-HCl, pH 8.0, using Sartorius Vivaspin concentrators with a 10-kDa cutoff. Further purification steps were performed using ion-exchange chromatography (GH5, GH9, and SusD-like proteins) with a 5-ml HiTrap diethylaminoethyl (DEAE) fast-flow (FF) column (GE Healthcare) and gel filtration (HiLoad Superdex 75; GE Healthcare) in 50 mM Tris-HCl with 200 mM NaCl (SusE). Proteins were concentrated, and the buffer was changed to 10 mM Tris-HCl, pH 7.5. Protein purity was analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and protein concentration was estimated by measuring the A280 and using the proteins’ molar extinction coefficients.
Chromogenic substrates.
Azurine cross-linked labeled (AZCL) substrates (Table S3) partly dissolved in isopropanol (10 mg/ml) were added to 135 µl buffer (50 mM potassium phosphate, pH 7.5, or the positive control’s preferred pH). Plates were sealed with adhesive PCR plate seals (Thermo Scientific; AB-0558) and incubated with overhead rotation (~20 rpm, room temperature) for 1 h. Plates were spun down (4,000 rpm, 10 min), and the absorption of the filtrate was measured against that of negative controls at 590 nm. Values reported are relative absorbance values calculated against the absorbance values of the positive controls listed in Table S3.
Enzymatic assays.
The optimum pH was determined to be approximately 6.6 for both enzymes, and 20 mM Bis-Tris, pH 6.6, was used for all enzyme assays. Enzyme activities were determined for carboxymethyl-cellulose (CMC) (Sigma-Aldrich), filter paper (Whatman no. 1), Avicel (Sigma-Aldrich), and barley β-glucan (Megazyme). CMC (1%, wt/vol) and β-glucan (0.5%, wt/vol) were incubated at 40°C (900-rpm horizontal shaking) with 25 nM and 10 nM GH5, respectively, for 10 min in a total volume of 500 µl. The reactions were stopped by adding an equal amount of 3,5-dinitrosalicylic acid (DNS) reagent, and the amounts of reducing sugars relative to a glucose standard curve were determined using the DNS assay (23). A unit of enzyme activity was defined as the amount of enzyme releasing 1 µmol of reducing sugars per minute. For GH9, the enzyme concentration was increased to 100 nM, and the incubation time was 15 min. For filter paper and Avicel, the conditions were 1% substrate (wt/vol) and 100 nM GH5 or GH9, with an incubation time of 30 min. The reactions were stopped by boiling the mixtures (5 min) before soluble cellodextrins were quantified by HPAEC-PAD as described below. A unit of enzyme activity was defined as the amount of enzyme releasing 1 µmol of soluble products per minute. The time course analysis of degradation of filter paper was performed using a 5% (wt/vol) concentration of the substrate and 3 µM enzyme (GH5 plus GH9, 1.5 plus 1.5 µM enzyme). Soluble cellodextrin products were quantified against a standard curve of cellodextrins (DP1-3) by HPAEC-PAD using a Dionex ICS-3000 system with a CarboPac PA1 column at 0.25 ml/min and 0.1 M NaOH. Oligosaccharides were eluted in a multistep linear gradient going from 0.1 M NaOH to 0.1 M NaOH-0.1 M sodium acetate (NaOAc) in 10 min, to 0.1 M NaOH-0.18 M NaOAc in 8 min, to 0.1 M NaOH-0.3 M NaOAc in 1 min, and to 0.1 M NaOH-1.0 M NaOAc in 1 min, before column reconditioning by 0.1 M NaOH for 14 min. Visual assessment of the degradation of filter paper discs was performed using the same conditions as described above in glass tubes in a total volume of 1 ml, with 0.8 U/ml β-glucosidase (Megazyme) added to avoid potential cellobiose inhibition.
Binding assays.
Filter paper (Whatman no. 1) milled to a 0.5-mm size, Avicel (Sigma-Aldrich), and the insoluble fraction (room temperature) of barley β-glucan (Megazyme) were washed twice in MES (morpholineethanesulfonic acid) buffer (20 mM, pH 6.0), suspended to 6% (wt/vol) in a total volume of 200 µl along with 0.1 mg/ml protein, and incubated at 40°C with horizontal shaking (900 rpm). The substrate and bound protein were pelleted by centrifugation, and the supernatant containing unbound protein (referred to as the flowthrough) was carefully removed. The pellet was washed with 200 µl buffer for 15 min, and the supernatant was again removed by centrifugation. To elute the proteins, the pellets were resuspended in 200 µl 8 M urea and boiled for 10 min (filter paper and Avicel) or incubated with 200 µl 2% SDS and incubated with shaking for 10 min (β-glucan). The flowthrough, wash, and elution fractions were analyzed by SDS-PAGE.
Binding to plant material was tested by probing transverse sections through Arabidopsis thaliana stems. Hand-cut sections through the stems of 4- to 5-week-old plants were labeled using a His6 tag-based three-stage procedure essentially as previously described (24), in which binding was detected using a fluorescein isothiocyanate-conjugated tertiary antibody. Cellulose-binding CBM3a from Clostridium thermocellum (13) was included as a positive control.
SUPPLEMENTAL MATERIAL
Identification of partially homologous/syntenic PULs encoding GH5 and/or GH9 representatives. Genomic comparisons of the AC2a PUL were made with publicly available metagenomes, Bacteroidetes genomes, and previously characterized PULs that encode either a GH5 or GH9 representative. Download
Substrate specificity screening of AC2a outer membrane enzymes. Substrate specificities of the GH5 and GH9 enzymes were determined by AZCL substrate screening. Values are reported as relative absorbances calculated against the activity of the respective positive-control enzymes at 1 U/ml (for the controls, the absorbance value was set to 1.0). An overview of which enzymes were used as positive controls for the various substrates in the substrate specificity screens is provided in Table S3. HE-cellulose, hydroxyethylcellulose. Download
Analysis of products generated by AC2a GH5 and GH9 from oligomeric substrates. Shown are HPAEC-PAD chromatograms of product mixtures obtained from DP3-DP6 cellodextrins digested with GH5 or GH9 at pH 6.6. Enzyme assays were performed for 30 min at 40°C, and enzymes were then inactivated by boiling them for 5 min. Negative controls (nC) without added enzymes and containing 0.1 mg/ml cellodextrins are included. Download
Binding of AC2a SusD-like and SusE-positioned proteins to barley β-glucan. SDS-PAGE analysis of fractions from pulldown assays. Download
Activities of the AC2a PUL endoglucanases on various glycans. Previously calculated specificities for example endoglucanases characterized from rumen bacteria (Cel9B from Fibrobacter succinogenes and CelA from Butyrivibrio fibrisolvens) are listed.
Primers used to clone AC2a proteins.
Commercial enzymes used as positive controls for AZCL substrate specificity screening. The preferred pH of the enzyme was used for the respective positive controls during the assay.
ACKNOWLEDGMENTS
We are grateful for support from the Research Council of Norway’s FRIPRO program (grant 214042) and the European Commission Marie Curie International Incoming Fellowship (awarded to P.B.P.; PIIF-GA-2010-274303). A.K.M. was supported by a grant from the Norwegian Research Council (190965/S60).
Footnotes
Citation Naas AE, Mackenzie AK, Mravec J, Schückel J, Willats WGT, Eijsink VGH, Pope PB. 2014. Do rumen Bacteroidetes utilize an alternative mechanism for cellulose degradation? mBio 5(4):e01401-14. doi:10.1128/mBio.01401-14.
REFERENCES
- 1. Edwards JE, Mcewan NR, Travis AJ, Wallace RJ. 2004. 16S rDNA library-based analysis of ruminal bacterial diversity. Antonie Van Leeuwenhoek 86:263–281. 10.1023/B:ANTO.0000047942.69033.24 [DOI] [PubMed] [Google Scholar]
- 2. Flint HJ, Bayer EA, Rincon MT, Lamed R, White BA. 2008. Polysaccharide utilization by gut bacteria: potential for new insights from genomic analysis. Nat. Rev. Microbiol. 6:121–131. 10.1038/nrmicro1817 [DOI] [PubMed] [Google Scholar]
- 3. Dodd D, Moon YH, Swaminathan K, Mackie RI, Cann IK. 2010. Transcriptomic analyses of xylan degradation by Prevotella bryantii and insights into energy acquisition by xylanolytic Bacteroidetes. J. Biol. Chem. 285:30261–30273. 10.1074/jbc.M110.141788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Martens EC, Lowe EC, Chiang H, Pudlo NA, Wu M, McNulty NP, Abbott DW, Henrissat B, Gilbert HJ, Bolam DN, Gordon JI. 2011. Recognition and degradation of plant cell wall polysaccharides by two human gut symbionts. PLoS Biol. 9:e1001221. 10.1371/journal.pbio.1001221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Martens EC, Koropatkin NM, Smith TJ, Gordon JI. 2009. Complex glycan catabolism by the human gut microbiota: the Bacteroidetes Sus-like paradigm. J. Biol. Chem. 284:24673–24677. 10.1074/jbc.R109.022848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Pope PB, Mackenzie AK, Gregor I, Smith W, Sundset MA, McHardy AC, Morrison M, Eijsink VG. 2012. Metagenomics of the Svalbard reindeer rumen microbiome reveals abundance of polysaccharide utilization loci. PLoS One 7:e38571. 10.1371/journal.pone.0038571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hess M, Sczyrba A, Egan R, Kim TW, Chokhawala H, Schroth G, Luo S, Clark DS, Chen F, Zhang T, Mackie RI, Pennacchio LA, Tringe SG, Visel A, Woyke T, Wang Z, Rubin EM. 2011. Metagenomic discovery of biomass-degrading genes and genomes from cow rumen. Science 331:463–467. 10.1126/science.1200387 [DOI] [PubMed] [Google Scholar]
- 8. Weimann A, Trukhina Y, Pope PB, Konietzny SG, McHardy AC. 2013. De novo prediction of the genomic components and capabilities for microbial plant biomass degradation from (meta-)genomes. Biotechnol. Biofuels 6:24. 10.1186/1754-6834-6-24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cameron EA, Maynard MA, Smith CJ, Smith TJ, Koropatkin NM, Martens EC. 2012. Multidomain carbohydrate-binding proteins involved in Bacteroides thetaiotaomicron starch metabolism. J. Biol. Chem. 287:34614–36425. 10.1074/jbc.M112.397380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Mackenzie AK, Pope PB, Pedersen HL, Gupta R, Morrison M, Willats WG, Eijsink VG. 2012. Two SusD-like proteins encoded within a polysaccharide utilization locus of an uncultured ruminant Bacteroidetes phylotype bind strongly to cellulose. Appl. Environ. Microbiol. 78:5935–5937. 10.1128/AEM.01164-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Aspeborg H, Coutinho PM, Wang Y, Brumer H, Henrissat B. 2012. Evolution, substrate specificity and subfamily classification of glycoside hydrolase family 5 (GH5). BMC Evol. Biol. 12:186. 10.1186/1471-2148-12-186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. McNulty NP, Wu M, Erickson AR, Pan C, Erickson BK, Martens EC, Pudlo NA, Muegge BD, Henrissat B, Hettich RL, Gordon JI. 2013. Effects of diet on resource utilization by a model human gut microbiota containing Bacteroides cellulosilyticus WH2, a symbiont with an extensive glycobiome. PLoS Biol. 11:e1001637. 10.1371/journal.pbio.1001637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Blake AW, McCartney L, Flint JE, Bolam DN, Boraston AB, Gilbert HJ, Knox JP. 2006. Understanding the biological rationale for the diversity of cellulose-directed carbohydrate-binding modules in prokaryotic enzymes. J. Biol. Chem. 281:29321–29329. 10.1074/jbc.M605903200 [DOI] [PubMed] [Google Scholar]
- 14. Pope PB, Denman SE, Jones M, Tringe SG, Barry K, Malfatti SA, McHardy AC, Cheng JF, Hugenholtz P, McSweeney CS, Morrison M. 2010. Adaptation to herbivory by the tammar wallaby includes bacterial and glycoside hydrolase profiles different to other herbivores. Proc. Natl. Acad. Sci. U. S. A. 107:14793–14798. 10.1073/pnas.1005297107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Larsbrink J, Rogers TE, Hemsworth GR, McKee LS, Tauzin AS, Spadiut O, Klinter S, Pudlo NA, Urs K, Koropatkin NM, Creagh AL, Haynes CA, Kelly AG, Cederholm SN, Davies GJ, Martens EC, Brumer H. 2014. A discrete genetic locus confers xyloglucan metabolism in select human gut Bacteroidetes. Nature 506:498–502. 10.1038/nature12907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hehemann JH, Kelly AG, Pudlo NA, Martens EC, Boraston AB. 2012. Bacteria of the human gut microbiome catabolize red seaweed glycans with carbohydrate-active enzyme updates from extrinsic microbes. Proc. Natl. Acad. Sci. U. S. A. 109:19786–19791. 10.1073/pnas.1211002109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sonnenburg ED, Zheng H, Joglekar P, Higginbottom SK, Firbank SJ, Bolam DN, Sonnenburg JL. 2010. Specificity of polysaccharide use in intestinal Bacteroides species determines diet-induced microbiota alterations. Cell 141:1241–1252. 10.1016/j.cell.2010.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Pope PB, Smith W, Denman SE, Tringe SG, Barry K, Hugenholtz P, McSweeney CS, McHardy AC, Morrison M. 2011. Isolation of Succinivibrionaceae implicated in low methane emissions from tammar wallabies. Science 333:646–648. 10.1126/science.1205760 [DOI] [PubMed] [Google Scholar]
- 19. Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M, Meyer F, Olsen GJ, Olson R, Osterman AL, Overbeek RA, McNeil LK, Paarmann D, Paczian T, Parrello B, Pusch GD, Reich C, Stevens R, Vassieva O, Vonstein V, Wilke A, Zagnitko O. 2008. The RAST server: rapid annotations using subsystems technology. BMC Genomics 9:75. 10.1186/1471-2164-9-75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Yu NY, Wagner JR, Laird MR, Melli G, Rey S, Lo R, Dao P, Sahinalp SC, Ester M, Foster LJ, Brinkman FS. 2010. PSORTb 3.0: improved protein subcellular localization prediction with refined localization subcategories and predictive capabilities for all prokaryotes. Bioinformatics 26:1608–1615. 10.1093/bioinformatics/btq249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Juncker AS, Willenbrock H, Von Heijne G, Brunak S, Nielsen H, Krogh A. 2003. Prediction of lipoprotein signal peptides in gram-negative bacteria. Protein Sci. 12:1652–1662. 10.1110/ps.0303703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Aslanidis C, de Jong PJ. 1990. Ligation-independent cloning of PCR products (LIC-PCR). Nucleic Acids Res. 18:6069–6074. 10.1093/nar/18.20.6069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Miller GL. 1959. Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal. Chem. 31:426–428. 10.1021/ac60147a030 [DOI] [Google Scholar]
- 24. McCartney L, Gilbert HJ, Bolam DN, Boraston AB, Knox JP. 2004. Glycoside hydrolase carbohydrate-binding modules as molecular probes for the analysis of plant cell wall polymers. Anal. Biochem. 326:49–54. 10.1016/j.ab.2003.11.011 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Identification of partially homologous/syntenic PULs encoding GH5 and/or GH9 representatives. Genomic comparisons of the AC2a PUL were made with publicly available metagenomes, Bacteroidetes genomes, and previously characterized PULs that encode either a GH5 or GH9 representative. Download
Substrate specificity screening of AC2a outer membrane enzymes. Substrate specificities of the GH5 and GH9 enzymes were determined by AZCL substrate screening. Values are reported as relative absorbances calculated against the activity of the respective positive-control enzymes at 1 U/ml (for the controls, the absorbance value was set to 1.0). An overview of which enzymes were used as positive controls for the various substrates in the substrate specificity screens is provided in Table S3. HE-cellulose, hydroxyethylcellulose. Download
Analysis of products generated by AC2a GH5 and GH9 from oligomeric substrates. Shown are HPAEC-PAD chromatograms of product mixtures obtained from DP3-DP6 cellodextrins digested with GH5 or GH9 at pH 6.6. Enzyme assays were performed for 30 min at 40°C, and enzymes were then inactivated by boiling them for 5 min. Negative controls (nC) without added enzymes and containing 0.1 mg/ml cellodextrins are included. Download
Binding of AC2a SusD-like and SusE-positioned proteins to barley β-glucan. SDS-PAGE analysis of fractions from pulldown assays. Download
Activities of the AC2a PUL endoglucanases on various glycans. Previously calculated specificities for example endoglucanases characterized from rumen bacteria (Cel9B from Fibrobacter succinogenes and CelA from Butyrivibrio fibrisolvens) are listed.
Primers used to clone AC2a proteins.
Commercial enzymes used as positive controls for AZCL substrate specificity screening. The preferred pH of the enzyme was used for the respective positive controls during the assay.