Abstract
Objective
Determine the molecular characteristics of human spermatogonia and optimize methods to enrich spermatogonial stem cells (SSCs).
Design
Laboratory study using human tissues
Setting
Research institute
Patient(s)/Animal(s)
Normal adult human testicular tissue.
Interventions
Human testicular tissue was fixed or digested with enzymes to produce a cell suspension. Human testis cells were fractionated by FACS and MACS.
Main Outcome Measure(s)
Immunostaining for selected markers, human-to-nude mouse xenotransplantation assay.
Results
Immunohistochemistry co-staining revealed the relative expression patterns of SALL4, UTF1, ZBTB16, UCHL1 and ENO2 in human undifferentiated spermatogonia as well as the extent of overlap with the differentiation marker, KIT. Whole mount analyses revealed that human undifferentiated spermatogonia (UCHL1+) were typically arranged in clones of 1–4 cells while differentiated spermatogonia (KIT+) were typically arranged in clones of 8 or more cells. The ratio of undifferentiated to differentiated spermatogonia is greater in humans than in rodents. SSC colonizing activity was enriched in the THY1dim and ITGA6+ fractions of human testes sorted by FACS. ITGA6 was effective for sorting human SSCs by MACS; THY1 and EPCAM were not.
Conclusions
Human spermatogonial differentiation correlates with increased clone size and onset of KIT expression, similar to rodents. The undifferentiated to differentiated developmental dynamics in human spermatogonia is different than rodents. THY1, ITGA6 and EPCAM can be used to enrich human SSC colonizing activity by FACS, but only ITGA6 is amenable to high throughput sorting by MACS.
Keywords: Testis, stem cell, FACS, MACS, spermatogonial stem cell, human spermatogonia
Introduction
Spermatogenesis is a process that produces millions of sperm a day in postpubertal mammals (1–3). At the foundation of spermatogenesis are spermatogonial stem cells (SSCs) that balance self-renewing divisions with differentiating divisions in order to maintain the stem cell pool and fuel spermatogenesis, respectively (4–6). Despite their importance to male fertility, there is limited knowledge about the molecular characteristics of the human SSCs, which are typically described as Adark and Apale spermatogonia based on nuclear staining intensity with hematoxylin (6–8).
The majority of information about spermatogonia has been generated using rodent models and although no SSC specific marker has been identified some markers that are expressed by stem and/or progenitor cells have been described (e.g. GFRα1, POU3F1, POU5F1 (OCT4), ZBTB16 (PLZF), NGN3, NANOS2, NANOS3, SOHLH1, SOHLH2, FOXO1, ITGA6 (α6-integrin, CD49f), LIN28, ID4, UTF1, CDH1, GPR125, ITGB1 (β1-integrin, CD29), EPCAM (CD326), CD9 and THY1 (CD90) (9–38)). Rodent SSCs are only definitively identified by their ability to produce spermatogenesis when transplanted into the testes of infertile recipient mice, an assay that was first described by Brinster and colleagues (39, 40). In the transplant bioassay, each colony of spermatogenesis produced in the recipient testis arises from a single SSC and therefore allows quantification of the starting population of stem cells (41–44). The combination of the transplant technique with fluorescence activated cell sorting (FACS) has provided insights about additional phenotypic features that can be used to isolate and enrich mouse spermatogonia. Mouse spermatogonia have the phenotype: ITGA6+, ITGB1+, THY1+, CD9+, GFRα1+, mitochondrial membrane potentialhigh, Rhodamine 123 (Rho123) low, ITGAV (αv-Integrin, CD51)−, KIT (cKIT, CD117)−, MHC-I−, ALDH (aldehyde dehydrogenase) activity− and CD45− (16, 25, 27, 45–50). There is a lack of consensus about whether SSC activity can also be recovered in the Hoechst side population fraction of mouse testes (15, 51–53).
Over the past few years, several laboratories have started to describe the molecular characteristics of human SSCs. Some studies show that a number of SSC markers are conserved from mice to nonhuman primates and humans (Supplementary Table 1). Based on immunofluorescence and colorimetric staining of adult human testicular sections, human spermatogonia on the basement membrane of the seminiferous tubules express UTF1, SALL4, ZBTB16, GFRα1, UCHL1, GPR125, LIN28, EXOSC10, FGFR3, DSG2, CBL, SSX2 and OCT2 (22, 54–64). Less is known about cell surface markers that could be used to isolate and enrich human SSCs. THY1, a glycophosphatidylinositol anchored cell surface protein, that belongs to the immunoglobulin-like superfamily of genes (65), has been shown to be expressed by neuronal cells, CD34 positive hematopoietic stem cells, fibroblasts and endothelial cells (66–72). THY1 is involved in diverse processes, including cell migration, cell-cell/cell-matrix interactions (73) and T-cell activation (74). In testis, THY1 has been shown through transplantation assay to be a conserved spermatogonial stem cell marker in rodents (15) and non-human primates (75). However, the expression of THY1 in human spermatogonia has been contradictory. He et al. (60) showed that THY1 expression is limited to a few rare cells on the basement membrane of seminiferous tubules, whereas Izadyar et al. (76) showed staining in the germ cells located toward the lumen of the tubule and also in peritubular and interstitial cells. Both of these reports are based on immunofluorescence staining and no transplants were performed. Human to human transplants are not possible as a routine bioassay, but xenotransplants into the testes of infertile nude mice has emerged as a quantitative assay for human and nonhuman primate spermatogonia (22, 62, 75–83).
A few studies have reported enrichment of putative human SSCs by sorting based on cell surface marker expression (GPR125, SSEA4, EPCAM, ITGA6 and CD9 (60, 62, 76, 81, 84)) but currently only three studies have confirmed their results by demonstrating SSC colonizing activity in the xenotransplant assay. Magnetic activated cell sorting (MACS) revealed enrichment of SSC colonizing activity in the SSEA4+ and CD9+ fractions of human testis cells (62, 76) and FACS sorting for EPCAM resulted in a 6-fold enrichment of colonizing activity in the EPCAMdim fraction (81). Currently, no human data are available regarding whether spermatogonial markers used in FACS are also appropriate for MACS and vice versa. The choice of whether to use FACS or MACS depends on the desired output. FACS has limited throughput (~30 × 106 cells per day); it is fairly time consuming and requires specialized equipment and a skilled operator, but it allows high resolution selection of sorting gates. MACS has a lower resolving power, but is generally a faster and is a higher throughput sorting strategy that can be performed on the laboratory bench and does not require specialized equipment. A single adult human testis that can be obtained for research through an organ donor program can contain over 1 billion cells, which is far beyond the typical sorting capacity of FACS. MACS can easily be scaled to accommodate this number of cells and maximize the use of this valuable human tissue resource for fundamental research. In addition, MACS is technically accessible and affordable, which will facilitate application for enriching SSCs in the clinical setting.
Therefore, in this study, we evaluated FACS and MACS to isolate and enrich human SSCs based on cell surface marker expression of THY1 (CD90), ITGA6 (CD49f) (FACS and MACS) and EPCAM (MACS only; we previously reported FACS for EPCAM (81)). ITGA6 is the integrin alpha chain 6. Integrins are cell surface proteins that are made up of an alpha chain and a beta chain and they provide a link between extracellular matrix proteins and the cytoskeleton (85). ITGA6 has been shown to regulate glioblastoma stem cells (86) and is expressed by mouse mammary stem cells (87) and is crucial for the survival of the MCF-7 cell line stem cells(88). EPCAM (epithelial cell adhesion molecule) is a transmembrane glycoprotein that mediates homophilic cell-cell adhesion (89). Modulation of Epcam activity is thought to affect cell migration, proliferation and invasion (89, 90) and overexpression of Epcam plays a role in cancer development (90–92).
FACS fractions were analyzed by immunocytochemistry for the human spermatogonial marker SALL4 (56, 81) and human-to-nude mouse xenotransplantation. SALL4 is a member of sal-gene family of transcription factors that is highly conserved between species (93–99). SALL4 is expressed by the cells in an early embryo and is important for maintaining pluripotency of ES cells (100, 101). In addition SALL4 is a conserved marker of spermatogonia (56, 102, 103) and has been implicated in the regulation of spermatogonial differentiation in mice (102).
MACS fractions were analyzed by human-to-nude mouse xenotransplantation. Analyses of FACS fractions indicated that, similar to the EPCAMdim fraction that we previously described, ITGA6+ and THY1dim can be used to effectively isolate and enrich human SSCs from a heterogeneous testis cell suspension. In contrast, only ITGA6 was suitable for sorting human SSCs by MACS, as THY1 and EPCAM provided no enrichment.
Materials and Methods
Animals
All experiments utilizing animals were approved by the Institutional Animal Care and Use Committees of the Magee-Womens Research Institute and the University of Pittsburgh and were performed in accordance with the National Institute of Health guidelines for the care and use of animals (assurance # A3654-01).
Preparation of Human Testicular Tissue
Normal adult human testicular tissue was obtained through the University of Pittsburgh Health Sciences Tissue Bank and Center for Organ Recovery and Education (CORE) under University of Pittsburgh IRB #0506140. Following the removal of tissue, it was transported to the laboratory on ice in Lactated Ringer’s solution. Cells were recovered from human testicular parenchyma using a two-step enzymatic digestion described previously (75, 81, 82). Briefly, testicular tissue was digested with collagenase type IV for 5 minutes at 37°C on the shaker (250 rpm), then shaken vigorously and incubated for another 3 minutes and if necessary 2 additional minutes at 37°C on the shaker. The tubules were then sedimented by centrifugation at 200xg for 5 minutes and washed with Hank’s Balanced Salt Solution (HBSS, Gibco). The tubules were then digested with 0.25% trypsin/EDTA and DNase I. The suspension was triturated vigorously 3–5 times and incubated at 37°C for 5 minutes. The process was repeated in 5 minute increments for up to 15 minutes total. The digestion was stopped by adding 10% fetal bovine serum (FBS) and the cells were strained through 70μm strainer (Becton Dickson). The cells were pelleted by centrifugation at 600xg for 15 minutes. Cells were then suspended in minimal essential medium α (MEM α) + 10% FBS at a concentration of 40 × 106 cells/mL and aliquoted in cryovials. An equal volume of cryopreservation medium consisting of MEMα + 20% FBS + 20% dimethylsulphoxide (DMSO) was added drop-wise, making the final concentration 20 × 106/mL in MEMα/15% FBS/10% DMSO). The vials were frozen at a controlled rate using Nalgene freezing containers (Nalgene-Nunc International) or a CryoMed controlled-rate freezer (Thermo Scientific) and then stored in liquid nitrogen. For experiments, the cells were thawed rapidly at 37°C, washed and suspended in MEMα medium containing 10% FBS.
Fluorescence-activated cell sorting (FACS) and magnetic-activated cell sorting (MACS)
For FACS, human testis cell suspension was stained on ice in Dulbecco’s phosphate-buffered saline (D-PBS) containing 10% FBS for 20 min with fluorescent-conjugated antibodies (THY1-APC, clone 5E10, 0.5 μg/106 cells and ITGA6 -PE clone GoH3, 20 μl/106 cells; Becton Dickinson). The unbound primary antibody was washed away twice with D-PBS, the cells were filtered through a 35μm strainer (Becton Dickinson) and 0.5μg/ml propidium iodide (BD Bioscience) was added to distinguish between live and dead cells. FACS analysis was done using FACSvantage SE (Beckton, Dickinson) and the positive staining was identified by comparison to appropriate isotype control in order to correct for non-specific binding. Sorting gates were established based on level of marker expression as well as exclusion of dead cells stained with propidium iodide and exclusion of cells exhibiting non-specific binding or autofluorescence. MACS protocol was similar to that of FACS, except after fluorescent-conjugated antibody staining (THY1-PE and ITGA6–PE; Becton Dickson; and EPCAM-PE, clone 9C4, 20 μl/106 cells; BioLegend) and washes, anti-PE Microbeads (2 μl/106 cells; Miltenyi Biotec) were used to detect the fluorophore on the primary antibody. The cells were then sorted on a MACS column (Miltenyi Biotec) into positive and negative fractions.
Immunocytochemistry
Cells from FACS and MACS were spotted on Superfrost slides and fixed with methanol. The cells were then rehydrated with D-PBS and blocked with a buffer containing 3% bovine serum albumin and 5% normal goat serum in order to eliminate nonspecific binding. Rabbit anti- SALL4 (1:500; ab29112, Abcam) antibody was added to the cells and incubated for 90 min at room temperature. Isotype matched normal IgG was used as negative control. Primary antibody was detected using goat anti-rabbit AlexaFluor-488 conjugated secondary antibody (1:200, Invitrogen). The slides were mounted with VectaShield (Vector Laboratories) mounting medium containing DAPI for detection of all nuclei and the staining was observed with a Nikon Eclipse E600 Fluorescence microscope and images captured with MetaView Digital Imaging software.
Immunofluorescence
Human testicular tissue fragments were fixed with 4% paraformaldehyde (PFA) overnight, paraffin-embedded and sectioned (5 μm). The tissue slides were de-paraffinized, rehydrated, incubated for 30 minutes in sodium citrate buffer (10 mM sodium citrate, pH 6.0, 0.05% Tween-20) for antigen retrieval. The tissue was then blocked with a buffer containing 3% bovine serum albumin and 5% normal serum from the host species of the secondary antibody. Subsequently, sections were stained for 90 minutes at room temperature with the following primary antibodies in antibody diluent: mouse anti-UTF1 (1:50, MAB4337, Millipore) goat anti-ZBTB16 (1:50; AF2944, R&D Systems), rabbit anti- KIT; goat anti-KIT (1:400; A4502, DakoCytomation; 1:50; AF332, R&D Systems), rabbit anti-SALL4 (1:500; ab29112, Abcam; 1:40; ab181087, Abcam), mouse anti-ENO2 (1:500, LS-B2890, LSBio), rabbit anti-UCHL1 (1:1000, 7863-0507, Biogenesis), rabbit anti-EPCAM (1:200; ab71919, Abcam), rabbit anti-ITGA6 (1:100; ab75737, Abcam). Isotype matched normal IgG was used as negative control. Primary antibody was detected using AlexaFluor-488 or AlexaFluor-568 conjugated secondary antibodies (1:200, Invitrogen). The slides were mounted with VectaShield mounting medium containing DAPI (Vector Laboratories) for detection of nuclei. Sections were observed with a Nikon Eclipse E600 fluorescence microscope and images captured with MetaView Digital Imaging software. For the quantification of marker overlap, single-positive cells for each marker and double-positive cells were counted in cross-sections of seminiferous tubules. Total stained cell numbers were divided by the number of cross-sections (at least 100 per sample × 3 replicate samples).
Colorimetric immunohistochemistry
Human testicular tissue fragments were fixed with 4% PFA overnight, paraffin-embedded and sectioned (5 μm). The tissue slides were de-paraffinized, rehydrated, incubated for 30 minutes in sodium citrate buffer (10 mM sodium citrate, pH 6.0, 0.05% Tween-20) for antigen retrieval. The tissue was then incubated in peroxidase block for 10 minutes and washed in PBS and blocked with a buffer containing 3% bovine serum albumin and 5% normal goat serum. Subsequently, sections were stained for 90 minutes at room temperature with rabbit anti-UCHL1 (1:1000, 7863-0507, Biogenesis). Isotype matched normal IgG was used as negative control. Primary antibody was detected using goat anti-rabbit HRP conjugated secondary antibody (1:200, sc-2054, Santa Cruz Biotechnology) for 30 minutes. Metal enhanced DAB substrate kit was used to detect staining (Thermo Scientific). The tissue was then counterstained with Periodic acid-Schiff and hematoxylin (Sigma-Aldrich).
Whole mount immunohistochemistry
Human testicular tissue was teased apart using Collagenase type IV (1mg/mL) and DNase I (1mg/mL) in D-PBS. The tissue was then fixed overnight with 4% PFA. The tubules were permeabilized using PBS and 0.1% Triton-X and blocked with a blotto milk solution in D-PBS (D-PBS +0.02 mg/mL blotto dry milk powder + 5% Triton-X) and stained with a rabbit anti-UCHL1 (1:500, 7863-0507, Biogenesis) and goat anti-KIT (1:50; AF332, R&D Systems) primary antibodies overnight at 4°C. The primary antibodies were detected with donkey anti-rabbit IgG AlexaFluor568 and donkey anti-goat IgG AlexaFluor488 (1:200, Invitrogen). Finally, the seminiferous tubules were mounted with VectaShield mounting media containing DAPI (Vector Laboratories) with raised cover slips and imaged with fluorescent microscopy.
Xenotransplantation and whole mount immunofluorescent quantification of human SSC colonizing activity in mouse seminiferous tubules
The human-to-nude mouse xenotransplantion was performed as a biological assay to investigate colonizing activity of putative human SSCs. Following FACS and MACS, unsorted and sorted testicular cell fractions were transplanted into the testes of busulfan-treated (40 mg/kg; Sigma, at 5–6 weeks of age), immune-deficient nude mice (NCr nu/nu; Taconic, Germantown, NY) as previously described (75, 81–83). Briefly, xenotransplantation was performed 5 weeks after busulfan treatment by injecting cell suspensions containing 10% trypan blue (Invitrogen) into the seminiferous tubules of recipient mouse testes via the efferent ducts. Approximately 7 μl of cell suspension was injected per testis. For quantitative analysis of colonization by human donor spermatogonia, the testes were recovered 8 weeks following transplantation, the tunica was removed, and the intact seminiferous tubules were dispersed gently with Collagenase IV (1mg/mL) and DNase I (1mg/mL) in D-PBS. The tubules were fixed for 4 hours in 4% PFA and the whole mount immunofluorescence was carried out by dehydrating samples in a graded series of methanol dilutions before incubating in MeOH:DMSO:H2O2 (4:1:1) solution for three hours. The tubules were then rehydrated, blocked with a blotto milk solution in D-PBS (D-PBS + 0.02 mg/mL blotto dry milk powder + 5%Triton-X) and stained with a rabbit anti-primate testis cell primary antibody (82) at a 1:800 dilution overnight at 4°C. The primary antibody was detected with goat anti-rabbit IgG AlexaFluor488 (1:200, Invitrogen). Finally, the seminiferous tubules were mounted with VectaShield mounting media containing DAPI (Vector Laboratories) with raised cover slips and imaged with fluorescent microscopy. Spermatogonial colonies were counted based on the following criteria: at least 4 cells exhibiting spermatogonial morphology (ovoid shape with high nuclear to cytoplasmic ratio) and located on the basement membrane in a continuous area of recipient seminiferous tubule (≤100 μm between cells).
Statistical Analysis
We analyzed the data using linear mixed effect models, and performed Tukey’s tests, as described in (104), to compare differences among the percent of SALL4+ cells in unsorted versus sorted cell fractions in the immunocytochemistry experiments and colonizing activity in the human-to-nude mouse xenotransplant bioassay.
Results
Acquisition of human testicular tissue
Testicular tissues used in this study were obtained from a total of 12 post-pubertal organ donors (Age 14–50). Testes weighed 11.3 to 26.0 grams and produced a theoretical yield (after correcting for tissue removed for pathology and immunofluorescence studies) of 1.4 × 109 ± 0.14 × 109 cells per donor. All human testis cell suspensions used in this study were cryopreserved as described above and thawed at a later date for experimentation. Human testicular cells used in this study were frozen for periods of time ranging from 1 month to 15 months.
Immunohistochemical staining of human testicular sections
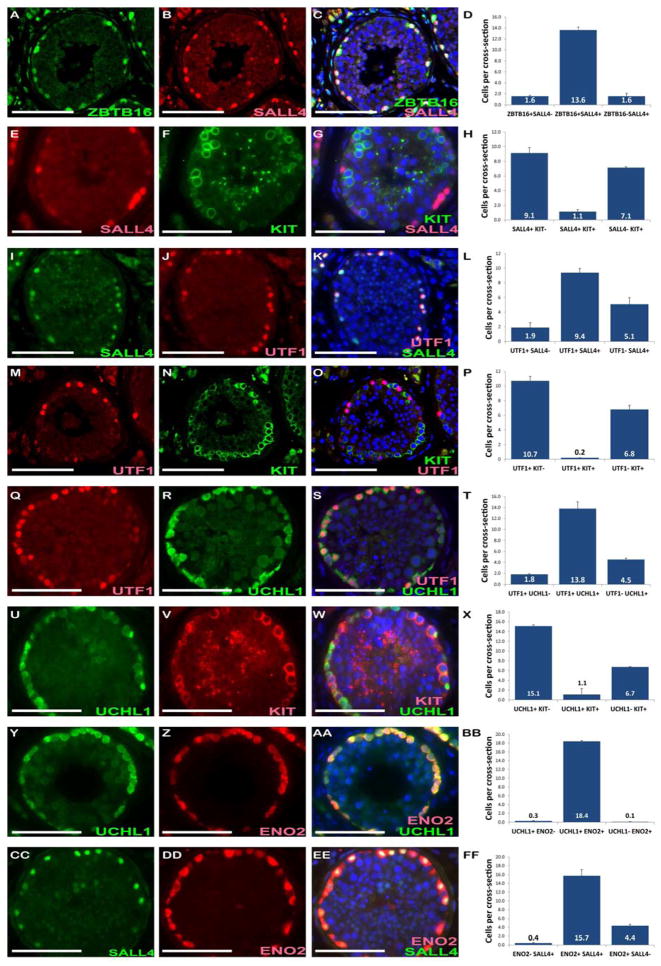
Immunohistochemical co-staining analysis was done to investigate the co-expression of known mouse and/or non-human primate spermatogonia markers in adult human testis. ZBTB16 and SALL4, which mark most stem and progenitor spermatogonia in rodents (103), were expressed in cells located on the basement membrane. Roughly 89% of ZBTB16 positive cells were also positive for SALL4 (Fig. 1A–D), but also a small population of ZBTB16 positive cells (11%) did not express SALL4. A sub-population of SALL4 positive cells also did not express ZBTB16 (11%) (Fig. 1D). Co-staining with KIT showed almost no overlap between the two markers (EH). UTF1 expression was also restricted to cells on the seminiferous tubule basement membrane (Fig. 1I–L). Co-staining with UTF1 and SALL4 indicated that 65% of the SALL4 positive cells express both markers, whereas 35% of expressed SALL4 only. Seventeen per cent of UTF1 positive cells express UTF1 only (Fig. 1L). To confirm that UTF1 is not expressed by differentiating spermatogonia, we co-stained UTF1 with a differentiation marker KIT (Fig. 1M–P) and found that there is almost no overlap between these two markers. Based on these results, we believe that UTF1 is a more restricted marker of stem and progenitor spermatogonia than SALL4. This interpretation is consistent with results of van Bragt and colleagues (24) who concluded that UTF1 is restricted to Asingle, Apaired and Aaligned4 spermatogonia in rats. Similar to SALL4, UCHL1 expression is less restricted than UTF1 (Fig. 1Q–T) with 75% of UCHL1 positive cells co-expressing both markers and 25% expressing UCHL1 only. UTF1 positive cells were UCHL1 positive 87% of the time and UTF1 only positive 13% (Fig. 1T). Co-staining with KIT, confirms that even though UCHL1 is less restricted than UTF1, it is not expressed by differentiating cells, demonstrated by a virtual absence of co-staining with KIT (Fig. 1U–X). We also analyzed the expression pattern of a novel marker, ENO2, which exhibited nearly complete overlap of expression with UCHL1 (Fig. 1Y-BB). By transitive logic, ENO2 is a marker of undifferentiated spermatogonia in humans because it exhibits nearly complete overlap with UCHL1, which has very little overlap with KIT. The overlap between ENO2 and SALL4 is less complete, with 78% of the ENO2 positive cells expressing SALL4 and 12% expressing ENO2 only (Fig. 1CC–FF). These results indicate that ENO2 expression is slightly broader than SALL4 expression in human undifferentiated spermatogonia. Supplementary Figure 2 summarizes our interpretation of these results in terms of the order and breadth of marker expression by human spermatogonia.
Figure 1. Expression of ZBTB16, UTF1, SALL4, UCHL1, ENO2 and KIT in human seminiferous epithelium.
Immunofluorescence co-staining for SALL4 and ZBTB16 (A–D), SALL4 and KIT (E–H), UTF1 and SALL4 (I–L), UTF1 and KIT (M–P), UTF1 and UCHL1 (Q–T), UCHL1 and KIT (U–X), UCHL1 and ENO2 (Y-BB) and SALL4 and ENO2 (CC–FF) in adult human testis. DAPI staining (blue) identifies all the nuclei. The bar graphs show quantification and relative proportion of each co-staining. The quantification is shown as the mean number of positive cells per cross-section of a seminiferous tubule. At least 100 seminiferous tubules were counted from 3 different organ donors. Bar graphs in D, H, L, P, T, X and BB indicate the mean percentage of marker positive cells. Error bars represent SEM. Scale bars = 100 μm.
Correlation of spermatogonial markers with dark and pale descriptions of nuclear morphology and clone size
To correlate molecular markers of human spermatogonia described in this study with classical descriptions of nuclear staining intensity (Adark and Apale), we performed colorimetric immunohistochemistry for UCHL1 followed by Periodic Acid-Schiff and hematoxylin counterstaining. The results in Figure 2A confirm that UCHL1 is expressed by Adark and Apale spermatogonia, which are considered the reserve and active stem cells of the human testis, respectively (105, 106). To correlate UCHL1 expression with clone size, we performed immunofluorescent analysis of UCHL1 expression in whole mount preparations of human seminiferous tubules. UCHL1 was expressed by cells located on the basement membrane of the seminiferous tubules and arranged as single cells and clones of 2, 4 and sometimes 8 interconnected cells. In contrast, KIT expressing cells were typically arranged in clones of 4, 8 and sometimes 16 interconnected cells (Figure 2B–F). The density of undifferentiated spermatogonia on the basement membrane of human seminiferous tubules appears greater than in rodents (Compare Fig. 2B to previous reports for mouse (17, 19, 30, 103)), whereas KIT+ differentiating spermatogonia are considerably less dense in human tubules than in mouse (Compare Fig. 2C to previous reports for mouse (17, 30, 103)).
Figure 2. UCHL1 expression in adult human testis.
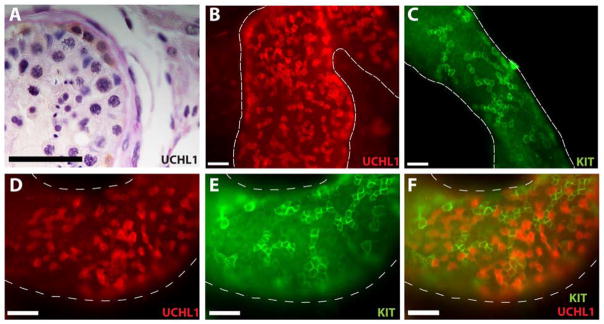
(A) UCHL1 staining in Periodic Acid-Schiff & Hematoxylin stained adult human testis section. UCHL1 is expressed by Adark and Apale spermatogonia. (B and D) UCHL1 and (C and E) KIT staining in whole mount staining of adult human testis. (F) UCHL1 clones are smaller (mostly 1–4 cells), whereas KIT clones tend to be bigger (more than 8). Scale bar = 50 μm.
Immunohistochemical evaluation of cell surface markers in adult human testes
THY1, ITGA6 and EPCAM are cell surface markers that have each been used to isolate and enrich spermatogonial stem cells in other species (15, 16, 25, 75). Previous studies indicated that these cell surface markers are conserved in human testes (60, 76, 81, 84) and we hypothesized that each could be used to isolate and enrich human SSCs by FACS and MACS. We were not able to confirm the expression of THY1 in adult human testes by immunohistochemistry in this study. However, others have reported that this marker is expressed in human testes (60, 76, 84). Immunohistochemical analysis of ITGA6 expression in normal adult human testis sections indicated that this antigen is expressed by many germ cells, including cells located on the basement membrane of seminiferous tubules (Supplementary Fig. 3A–C) and that EPCAM is expressed by cells on the basement membrane of the seminiferous tubules, as well as a few cells located more towards the lumen (Supplementary Fig. 3D–F).
Expression of THY1 in adult human testicular cell suspensions
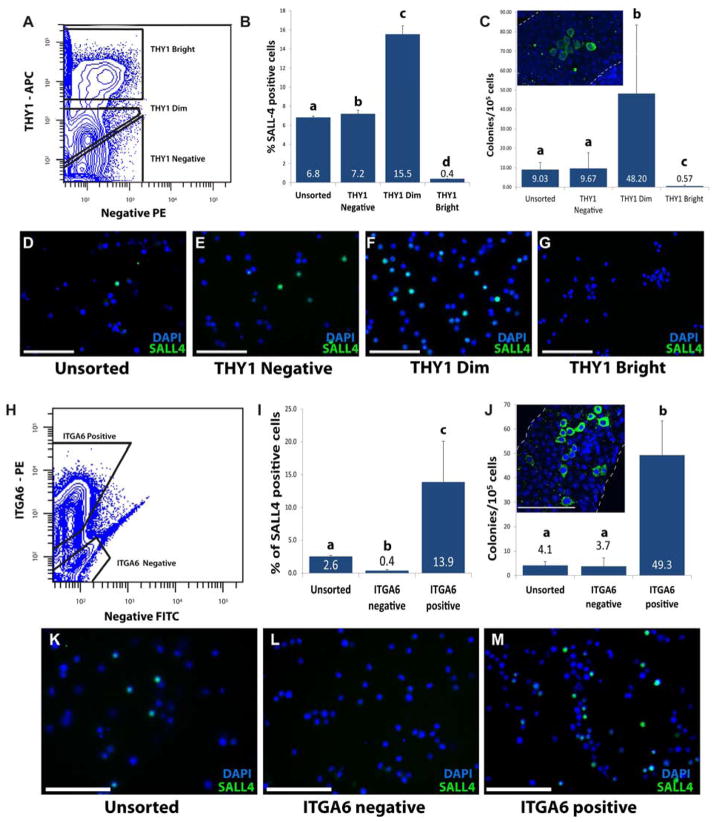
THY1 is a marker of mouse, rat and non-human primate SSCs (15, 25, 75) as well as a marker for mouse and human hematopoietic stem cells (107–109). Therefore, we hypothesized that THY1 is a marker for human SSCs and analyzed the expression on adult human testicular cells using FACS and MACS. Adult human testis cell suspensions stained with THY1 showed three populations of cells based on their level of fluorescence and on negative PE axis, which helps us plot the cells on a multi-color histogram and therefore eliminate autofluorescence. As indicated in Figure 3A, the three distinct populations were: THY1 bright, THY1 dim and THY1 negative. The THY1 bright, dim and negative fractions represented 12.2 ± 4.2, 19.0 ± 4.0 and 46.5 ± 7.0% of the live cells, respectively. Immunofluorescence staining revealed that 6.8 ± 0.1% of unsorted human testicular cells express human spermatogonia marker SALL4, compared to 7.2 ± 0.3% in the THY1 negative fraction (p<0.01), 15.5 ± 0.9% in the THY1 dim fraction (p<0.01) and only 0.4 ± 0% in the THY1 bright fraction (p<0.01) (Fig. 3B, 3D–G). To confirm the immunocytochemistry results and to functionally correlate THY1 expression in adult human testis to SSC colonizing activity, the human-to-nude mouse xenotransplantation assay was performed. The transplant results confirm that SSC colonizing activity was depleted from THY1 bright fraction (0.57 colonies/105 cells; p<0.01 compared to the unsorted controls). The majority of SSC colonizing activity was recovered in the THY1 dim fraction (48.2 colonies/105 cells; p<0.01 compared to the unsorted controls), compared to 9.03 and 9.67 colonies/105 cells in unsorted and THY1 negative fractions, respectively (Fig. 3C). Based on these results, there is roughly a 5-fold enrichment of SSC colonizing activity in the THY1 dim fraction of human testis cells. Immunohistochemical assessment of human colonizing events in recipient mouse testes indicate that colonizing cells are located on the basement membrane of seminiferous tubules and contain ENO2 positive undifferentiated human spermatogonia as well as ENO2 negative human cells that are presumably more differentiated germ cells (Supplementary Fig. 5). We previously reported that all cells in human colonizing events express the germ cell marker, VASA (81).
Figure 3. Expression of THY1 and ITGA6 in adult human testis.
(A) FACS was used to characterize and sort human testicular cells based on the level of THY1 expression. Based upon THY1-APC staining intensity and negative PE autofluorescence, three populations were identified – THY1bight, THY1dim and THY1neg. Negative gates were defined by analysis of human testis cells stained using APC-conjugated isotype control antibodies. (B) After the sort, all sorted fractions, as well as the unsorted cells, were fixed and immunocytochemistry for SALL4 was performed. SALL4 positive cells were enriched in the THY1 dim fraction compared to the unsorted cells. (C) To confirm the ICC results, human-to-nude mouse xenotransplants were also performed. Two months after transplant, colonies of human spermatogonia were identified in mouse recipient testes. (C and J insets) Examples of colonies of human spermatogonia in whole mount preparations of recipient mouse seminiferous tubules stained with the rabbit anti-primate antibody. Colonies in each recipient testis were counted and normalized to 105 viable cells transplanted per testis. (D–G) Representative images of SALL4 staining from each sorted fraction and unsorted cells. (H) FACS sorting for ITGA6 in human testis resulted in 2 different populations based upon ITGA6 -PE staining intensity and negative FITC autofluorescence – ITGA6 positive and ITGA6 negative. Negative gates were defined by analysis of human testis cells stained using PE-conjugated isotype control antibodies. (I) After the sort, all sorted fractions, as well as the unsorted cells, were fixed and immunocytochemistry for SALL4 was performed. SALL4 positive cells were enriched in the ITGA6 positive fraction compared to the unsorted cells. (J) To confirm the ICC results, human-to-nude mouse xenotransplants were also done. Two months after transplant, colonies of human spermatogonia were identified in mouse recipient testes. Colonies in each recipient testis were counted and normalized to 105 viable cells transplanted per testis. (K–M) Representative images of SALL4 staining from each sorted fraction and unsorted cells. At least 10 views were counted from each fraction based on DAPI staining and SALL-4 staining. Different letter indicate P < 0.01, same letters indicate P > 0.05. Bar graphs in B, C, I and J are presented as mean ± SEM. Scale bar = 100 μm.
Expression of ITGA6 in adult human testicular cell suspension
To determine whether ITGA6 is expressed on human spermatogonia and could be used as a positive selection marker to enrich human SSCs, adult human testicular cell suspensions were stained with a PE-conjugated antibody against ITGA6 and sorted by FACS. Two distinct populations of cells were gated; ITGA6 negative and ITGA6 positive (Fig. 3H), which represented 27.6 ± 7.6 and 11.6 ± 3.0 % of the live cells, respectively. Immunocytochemistry of the ITGA6 sorted fractions and unsorted cells revealed that 13.8 ± 6.2% of cells in the ITGA6 positive fraction were SALL4 positive (Fig. 3I and M), compared to 2.6 ± 0.2% in the unsorted cell population (p<0.01) (Fig. 3I and K). SALL4 positive cells were depleted from the ITGA6 negative fraction (0.38 ± 0.1%; p<0.01 compared to the unsorted controls; Fig. 3I and L). To confirm the immunocytochemistry results, colonizing activity in ITGA6 sorted and unsorted cells was assessed by xenotransplantation into nude mouse testes. On average, 49.3 colonies/105 cells were found in mice transplanted with cells from the ITGA6 positive fraction (p<0.01 compared to the unsorted controls), whereas only 4.1 colonies/105 cells and 3.7 colonies/105 cells were observed from mice transplanted with unsorted and ITGA6 negative cells, respectively (Fig. 3J). Thus, SSC colonizing activity resides predominantly in the ITGA6 positive fraction of human testis cells and is enriched approximately 12-fold compared to the unsorted population.
Enrichment of human spermatogonia using MACS
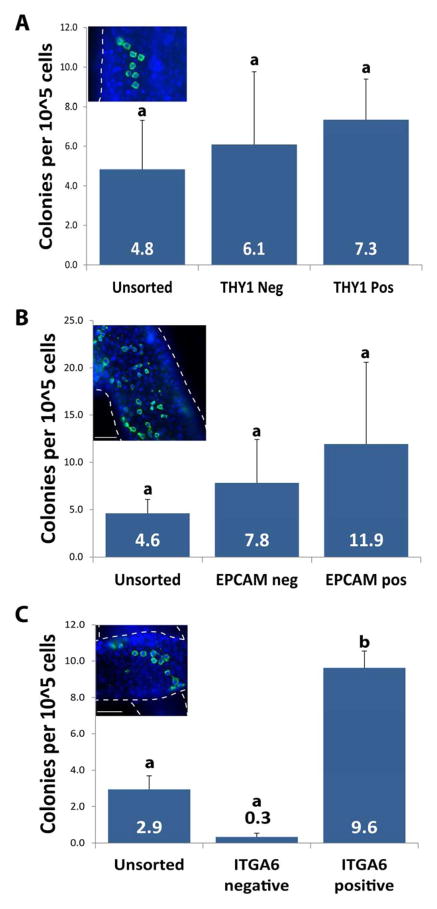
Analysis of FACS indicated that ITGA6 and THY1 can be used to effectively isolate and enrich human SSCs from a heterogeneous testis cell suspension. However, the FACS sorting approach has limited throughput (~30× 106 cells per day). Therefore, we decided to evaluate a higher throughput sorting approach (MACS) to maximize the use of human testicular cells and compare the results to FACS. We evaluated the fractionation of human testis cells by THY1 MACS where there is no option to distinguish between bright and dim expression of THY1. The cells were sorted into THY1 positive and negative fractions using MACS and then transplanted into nude mouse testes to analyze SSC colonizing activity relative to unsorted human testis cells. Unsorted cells produced 4.8 ± 2.5 colonies/105 cells, compared to 6.1± 2.0 and 7.3 ± 3.7 colonies/105 cells in THY1 negative and THY1 positive fractions, respectively (p> 0.05, compared to unsorted and each other), indicating that MACS did not effectively fractionate SSC colonizing activity based on THY1 expression (Fig. 4A). Similar to the THY1 FACS results in this study, we previously reported the SSC colonizing activity is enriched in the EPCAMdim fraction of human testis cells (81). Therefore, it is not surprising that MACS did not effectively fractionate SSC colonizing activity from human testis cells based on EPCAM expression (Fig. 4B).
Figure 4. MACS sorting of human testicular cells for THY1, EPCAM and ITGA6.
Human testicular cells were MACS sorted into 2 fractions – negative and positive. Both positive and negative fractions from MACS, as well as unsorted cells, were transplanted into nude mouse testis. (Inset A, B and C) Two months after transplant, colonies of human spermatogonia were identified in whole mount preparations of recipient mouse seminiferous tubules using the rabbit anti-primate antibody. Colonies in each recipient testis were counted and normalized to 105 viable cells transplanted per testis. (A and B) For THY1 and EPCAM, no significant difference was found between the unsorted cells and the sorted fractions (P>0.05). (C) ITGA6 positive fraction was enriched roughly 3 fold compared to unsorted cells (P<0.05). Bar graphs are presented as mean ± SEM. Scale bar = 100 μm.
In contrast, MACS was effective for isolation and enrichment of human SSC colonizing activity based on ITGA6 expression (Fig. 4C). SSC colonizing activity in the ITGA6 positive MACS fraction was enriched over 3-fold (9.6 ± 0.9 colonies/105 cells) compared relative to the unsorted fraction (2.9 colonies/105 cells ; p<0.05; Fig. 4C). SSC colonizing activity was nearly depleted in the ITGA6 negative fraction, which produced only 0.3 ± 0.2 colonies/105 cells, indicating that almost all SSCs were recovered in the ITGA6 positive fraction.
Discussion
In rodents, SSCs are defined by their ability to establish and maintain spermatogenesis when transplanted into infertile mouse testis (39, 40, 110, 111). Although there is no specific molecular marker of rodent SSCs (except possibly ID4 (18)), stem and progenitor spermatogonia can be described collectively by expression of some or all of the following markers GFRα1, POU3F1, POU5F1, ZBTB16, NGN3, NANOS2, NANOS3, SOHLH1, SOHLH2, FOXO1, ITGA6, LIN28, ID4, UTF1, CDH1, GPR125, ITGB1, EPCAM, CD9 and THY1(9–38, 112, 113)), and by their clonal arrangement on the basement membrane of seminiferous tubules (Asingle, Apaired, Aaligned; (114)). In humans, stem spermatogonia are described primarily as Adark and Apale based on the intensity of nuclear staining with hematoxylin (6–8). There is limited information about how dark and pale descriptions of nuclear morphology correlate with transplantation potential, molecular markers or clone size, although recent progress on the molecular details has been published by von Kopylow and colleagues (57, 58) and also Lim and colleagues (59)
Here we show that spermatogonia on the basement membrane of human seminiferous tubules have the phenotype of SALL4+, ZBTB16+, UTF+, UCHL1+ and ENO2+ (Figure 1). The expression of SALL4, ZBTB16, UTF1 and UCHL1 in human testes has been reported previously (22, 54–56, 60, 64). ENO2 is a gene that was identified by Oatley and co-workers because it is upregulated in ID4-GFP positive spermatogonia. This is the first study to demonstrate that ENO2 is expressed by human spermatogonia and co-expressed with established markers of human stem and progenitor spermatogonia (i.e., UCHL1 and SALL4) (56, 60) This is also the first study to quantify the expression of these markers at the cellular level and describe their expression relative to other stem and progenitor markers by co-staining. We believe this systematic molecular profiling will identify subpopulations of cells (e.g., putative stem, progenitor and differentiating cells) that will become the subject of future investigations.
The majority of cells that express SALL4, ZBTB16, UTF1, UCHL1 and ENO2, do not express the differentiation marker KIT, as demonstrated by direct co-staining (i.e., UCHL1/KIT, SALL4/KIT and UTF1/KIT) or transitive logic (UCHL1/ENO2; Figure 1). These results suggest that SALL4, ZBTB16, UTF1, UCHL1 and ENO2 mark human undifferentiated spermatogonia and immunohistochemical analysis confirms that UCHL1 is expressed by Adark and Apale spermatogonia, the putative SSCs in human testes (Figure 2). Examination of these markers in whole mount preparations of seminiferous tubules provides novel insights about human spermatogenic lineage development. Our results indicate that UCHL1 tended to be expressed by smaller clones (1–4 cells) while KIT is expressed in larger clones (usually 8 or more cells). Collectively, these results indicate that several markers of rodent stem and progenitor spermatogonia are conserved in humans and that spermatogonial differentiation in humans is correlated with increased clone size and initiation of KIT expression, similar to rodents (17, 103).
Spermatogenesis is an extremely productive system that produces millions of sperm per gram of testicular tissue each day in rodents and humans (1–3). However, our results suggest that the dynamics of spermatogenic lineage development in humans may be different than rodents. In rodents, rare undifferentiated spermatogonia are heavily outnumbered by transit-amplifying differentiated spermatogonia (115). In contrast, we found that number of undifferentiated spermatogonia in human testes was greater than the number of KIT+ differentiated spermatogonia (Fig. 2 and Supplementary Fig. 2). Thus, it appears that the highly productive spermatogenic system in rodents depends on a small pool of stem and progenitor spermatogonia and a large pool of transit-amplifying cells while the human spermatogenic lineage is characterized by a relatively larger pool of stem and progenitor cells and a smaller pool of transit amplifying cells.
FACS is suitable for characterizing relatively small cell populations (≤30 × 106) and can be used to achieve significant enrichment of spermatogonial stem cells (15, 25, 50, 75, 81, 112, 116–119). When coupled with molecular marker screening (using markers that are restricted to stem and progenitor spermatogonia) and the stem cell transplant assay to validate sorted fractions, FACS can be a powerful tool for dissecting the molecular phenotype of SSCs. In the current study, we used SALL4 immunocytochemistry (ICC) to screen sorted cell populations. We considered SALL4 an excellent marker for screening human stem and progenitor spermatogonia because it is conserved in mice (56, 102, 103), rats (Gassei and Orwig, unpublished), monkeys (56) and humans (56), including expression by human Adark and Apale spermatogonia (56). SALL4 ICC provided a rapid assessment of sorted fractions and was an excellent predictor of the results from human-to-nude mouse SSC xenotransplantation, which has an inherent two month delay to analysis. Based on the data presented here, we believe that UTF1, ZBTB16, UCHL1 and ENO2 would also be good markers to rapidly screen for human stem and progenitor spermatogonia.
SSC transplantation is the experimental “gold standard” for assaying spermatogonial stem cells (120, 121). SSC transplantation in humans may someday be feasible in the clinical setting (122), but cannot be used as a routine bioassay. However, Nagano and coworkers demonstrated that human SSCs can engraft the testes of infertile, immune compromised mice (123). Human SSCs do not produce complete spermatogenesis in mouse seminiferous tubules, but they do execute several functions that are consistent with the activity of SSCs: 1) they migrate to the basement membrane of seminiferous tubules without being phagocytosed by mouse Sertoli cells; 2) they proliferate to produce characteristic chains and networks of spermatogonia and 3) they persist for several months. Human-to-nude mouse xenotransplantation is becoming a routine bioassay for human SSCs (22, 62, 76, 79–81, 123).
Studies employing FACS followed by transplantation of sorted fractions have established that ITGA6, THY1 and EPCAM are markers of SSCs in rodents (15, 16, 25). Similar methodology with FACS or MACS sorting followed by human-to-mouse xenotransplantation has been used to demonstrate that EPCAM, CD9 and SSEA4 are markers of human SSCs (62, 76, 81). Human testis cells have also been fractionated by MACS based on expression of GPR125, THY1 and ITGA6 (60, 84, 124), but stem cell activity in sorted fractions was not tested by transplantation. Flow cytometry analyses in the current study identified two distinct THY1 positive populations in the human testis that we designated dim and bright. SALL4 staining as well as xenotransplant results suggested that the majority of the SSCs were in the THY1 dim fraction and SSC colonizing activity in that fraction was enriched approximately 5-fold compared to unsorted human testis cells (Fig. 3C). Almost no SSCs are found in the THY1bright fraction. Similar to the THY1 results reported here, we previously reported that SSC colonizing activity is recovered in the EPCAMdim fraction of human testis cells and depleted in the EPCAMbright and EPCAM− fractions (81). Interestingly, neither of these markers could be used to effectively fractionate and enrich SSC colonizing activity from the human testis using MACS. SSC colonizing activity was recovered in both the bound and flow through fractions and colonizing activity in each fraction was similar to unsorted controls (Fig. 4A and B). Perhaps this result can be attributed to the low expression level of these two antigens in human SSCs. Considering our MACS results, it is noteworthy that THY1 MACS is routinely used to sort SSCs from mouse testes (118, 125–129). These results may indicate that there are species-specific differences in the level of THY1 expression. Alternatively, these results may indicate technical differences between direct labeling with bead-conjugated THY1 primary antibodies (mouse) and indirect labeling using bead conjugated secondary antibodies (current study). The bead conjugated anti-mouse THY1 antibodies did not cross-react with the human THY1 antigen (data not shown). Flow cytometric analysis of ITGA6 in human testis cells revealed only two distinct populations, positive and negative, and the majority of SSC colonizing activity was recovered in the ITGA6+ fraction, which was enriched 12-fold compared to unsorted controls (Fig. 4J). In contrast to THY1 and EPCAM, cells with SSC colonizing activity could be effectively isolated and enriched from heterogeneous human testis cell suspensions using ITGA6 MACS. However, the level of enrichment achieved by ITGA6 MACS (3.3-fold) was less than ITGA6 FACS (12-fold). Sorting resolution by FACS is typically greater than MACS because FACS allows for gating of cell populations based on simultaneous evaluation several parameters, including viability (PI−), cell size (forward scatter of incident light), cell complexity (side scatter of incident light) and specific immunoreactivity (autofluorescent−, nonspecific binding−).
We identified several proteins with expression limited primarily to undifferentiated spermatogonia (KIT− cells) located on the basement membrane of seminiferous tubules in human testes. These markers may provide insights into the molecular mechanisms that regulate the function of human SSCs and can be used to screen human cell populations or tissues for putative SSCs. In addition they can be used to validate newly discovered markers of human stem and progenitor spermatogonia using co-staining approaches similar to those employed in the current study to validate the expression of ENO2 in human undifferentiated spermatogonia. In this study and a previous study (81), we demonstrated that human SSCs have the cell surface phenotype THY1dim, EPCAMdim, ITGA6+. SSEA4 and CD9 are also cell surface markers of human SSCs that have been validated by human-to-mouse xenotransplantation (62, 76). These markers can now be used alone or in combination to achieve significant enrichment of human SSCs for downstream studies. MACS can also be used for isolation and enrichment of SSCs prior to initiation of SSC cultures, as previously described for mice (118, 130). ITGA6 (current study), CD9 (62) and SSEA4 (76) are also amenable to immunomagnetic sorting, which has virtually unlimited cell sorting capacity and will facilitate isolation of SSCs from human testes that can contain over one billion cells.
Supplementary Material
Acknowledgments
Additional thanks to McGowan Institute for Regenerative Medicine FACS facility and Lynda Guzik for help with cell sorting, and to the Center for Organ Recovery and Education (CORE) and the Tissue and Research Pathology Services at the University of Pittsburgh Cancer Institute for human tissues.
This work was supported by NIH grants HD055475 and HD061289, Magee-Womens Research Institute, Foundation and the Richard King Mellon Foundation and the United States-Israel Binational Science Foundation
Footnotes
H.V. has nothing to disclose; M.S. has nothing to disclose; S.L.D. has nothing to disclose; K.A.P. has nothing to disclose; J.D. has nothing to disclose; C.A.C has nothing to disclose; T.C. has nothing to disclose; G.R.M. has nothing to disclose; K.E.O. has nothing to disclose.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Sharpe R. Regulation of spermatogenesis. In: Knobil ENJ, editor. The Physiology of Reproduction. New York: Raven Press, Ltd; 1994. pp. 1363–434. [Google Scholar]
- 2.Gupta G, Maikhuri JP, Setty BS, Dhar JD. Seasonal variations in daily sperm production rate of rhesus and bonnet monkeys. Journal of Medical Primatology. 2000;29:411–4. doi: 10.1111/j.1600-0684.2000.290605.x. [DOI] [PubMed] [Google Scholar]
- 3.Thayer KA, Ruhlen RL, Howdeshell KL, Buchanan DL, Cooke PS, Preziosi D, et al. Altered prostate growth and daily sperm production in male mice exposed prenatally to subclinical doses of 17α-ethinyl oestradiol. Human Reproduction. 2001;16:988–96. doi: 10.1093/humrep/16.5.988. [DOI] [PubMed] [Google Scholar]
- 4.de Rooij DG, Grootegoed JA. Spermatogonial stem cells. Current opinion in cell biology. 1998;10:694–701. doi: 10.1016/s0955-0674(98)80109-9. [DOI] [PubMed] [Google Scholar]
- 5.Tegelenbosch RA, de Rooij DG. A quantitative study of spermatogonial multiplication and stem cell renewal in the C3H/101 F1 hybrid mouse. Mutation research. 1993;290:193–200. doi: 10.1016/0027-5107(93)90159-d. [DOI] [PubMed] [Google Scholar]
- 6.Clermont Y. Kinetics of spermatogenesis in mammals: seminiferous epithelium cycle and spermatogonial renewal. Physiological reviews. 1972;52:198–236. doi: 10.1152/physrev.1972.52.1.198. [DOI] [PubMed] [Google Scholar]
- 7.Clermont Y. Renewal of spermatogonia in man. American Journal of Anatomy. 1966;118:509–24. doi: 10.1002/aja.1001180211. [DOI] [PubMed] [Google Scholar]
- 8.Clermont Y. The cycle of the seminiferous epithelium in man. American Journal of Anatomy. 1963;112:35–51. doi: 10.1002/aja.1001120103. [DOI] [PubMed] [Google Scholar]
- 9.Naughton CK, Jain S, Strickland AM, Gupta A, Milbrandt J. Glial Cell-Line Derived Neurotrophic Factor-Mediated RET Signaling Regulates Spermatogonial Stem Cell Fate. Biology of Reproduction. 2006;74:314–21. doi: 10.1095/biolreprod.105.047365. [DOI] [PubMed] [Google Scholar]
- 10.Hofmann M-C, Braydich-Stolle L, Dym M. Isolation of male germ-line stem cells; influence of GDNF. Developmental Biology. 2005;279:114–24. doi: 10.1016/j.ydbio.2004.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.He Z, Jiang J, Hofmann M-C, Dym M. Gfra1 Silencing in Mouse Spermatogonial Stem Cells Results in Their Differentiation Via the Inactivation of RET Tyrosine Kinase. Biology of Reproduction. 2007;77:723–33. doi: 10.1095/biolreprod.107.062513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ohmura M, Yoshida S, Ide Y, Nagamatsu G, Suda T, Ohbo K. Spatial analysis of germ stem cell development in Oct-4/EGFP transgenic mice. Archives of Histology and Cytology. 2004;67:285–96. doi: 10.1679/aohc.67.285. [DOI] [PubMed] [Google Scholar]
- 13.Costoya JA, Hobbs RM, Barna M, Cattoretti G, Manova K, Sukhwani M, et al. Essential role of Plzf in maintenance of spermatogonial stem cells. Nat Genet. 2004;36:653–9. doi: 10.1038/ng1367. [DOI] [PubMed] [Google Scholar]
- 14.Buaas FW, Kirsh AL, Sharma M, McLean DJ, Morris JL, Griswold MD, et al. Plzf is required in adult male germ cells for stem cell self-renewal. Nat Genet. 2004;36:647–52. doi: 10.1038/ng1366. [DOI] [PubMed] [Google Scholar]
- 15.Kubota H, Avarbock MR, Brinster RL. Spermatogonial stem cells share some, but not all, phenotypic and functional characteristics with other stem cells. Proceedings of the National Academy of Sciences. 2003;100:6487–92. doi: 10.1073/pnas.0631767100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shinohara T, Avarbock MR, Brinster RL. β1- and α6-integrin are surface markers on mouse spermatogonial stem cells. Proceedings of the National Academy of Sciences. 1999;96:5504–9. doi: 10.1073/pnas.96.10.5504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tokuda M, Kadokawa Y, Kurahashi H, Marunouchi T. CDH1 Is a Specific Marker for Undifferentiated Spermatogonia in Mouse Testes. Biology of Reproduction. 2007;76:130–41. doi: 10.1095/biolreprod.106.053181. [DOI] [PubMed] [Google Scholar]
- 18.Oatley MJ, Kaucher AV, Racicot KE, Oatley JM. Inhibitor of DNA Binding 4 Is Expressed Selectively by Single Spermatogonia in the Male Germline and Regulates the Self-Renewal of Spermatogonial Stem Cells in Mice. Biology of Reproduction. 2011;85:347–56. doi: 10.1095/biolreprod.111.091330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zheng K, Wu X, Kaestner K, Wang P. The pluripotency factor LIN28 marks undifferentiated spermatogonia in mouse. BMC Developmental Biology. 2009;9:38. doi: 10.1186/1471-213X-9-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Seandel M, James D, Shmelkov SV, Falciatori I, Kim J, Chavala S, et al. Generation of functional multipotent adult stem cells from GPR125+ germline progenitors. Nature. 2007;449:346–50. doi: 10.1038/nature06129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Seandel M, Falciatori I, Shmelkov SV, Kim J, James D, Rafii S. Niche players: Spermatogonial progenitors marked by GPR125. Cell Cycle. 2008;7:135–40. doi: 10.4161/cc.7.2.5248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu X, Schmidt JA, Avarbock MR, Tobias JW, Carlson CA, Kolon TF, et al. Prepubertal human spermatogonia and mouse gonocytes share conserved gene expression of germline stem cell regulatory molecules. Proceedings of the National Academy of Sciences. 2009;106:21672–7. doi: 10.1073/pnas.0912432106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kanatsu-Shinohara M, Takehashi M, Takashima S, Lee J, Morimoto H, Chuma S, et al. Homing of Mouse Spermatogonial Stem Cells to Germline Niche Depends on β1-Integrin. Cell Stem Cell. 2008;3:533–42. doi: 10.1016/j.stem.2008.08.002. [DOI] [PubMed] [Google Scholar]
- 24.van Bragt MPA, Roepers-Gajadien HL, Korver CM, Bogerd J, Okuda A, Eggen BJL, et al. Expression of the pluripotency marker UTF1 is restricted to a subpopulation of early A spermatogonia in rat testis. Reproduction. 2008;136:33–40. doi: 10.1530/REP-07-0536. [DOI] [PubMed] [Google Scholar]
- 25.Ryu B-Y, Orwig KE, Kubota H, Avarbock MR, Brinster RL. Phenotypic and functional characteristics of spermatogonial stem cells in rats. Developmental Biology. 2004;274:158–70. doi: 10.1016/j.ydbio.2004.07.004. [DOI] [PubMed] [Google Scholar]
- 26.Kanatsu-Shinohara M, Takashima S, Ishii K, Shinohara T. Dynamic changes in EPCAM expression during spermatogonial stem cell differentiation in the mouse testis. PLoS ONE. 2011;6:15. doi: 10.1371/journal.pone.0023663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kanatsu-Shinohara M, Toyokuni S, Shinohara T. CD9 is a surface marker on mouse and rat male germline stem cells. Biology of Reproduction. 2004;70:70–5. doi: 10.1095/biolreprod.103.020867. [DOI] [PubMed] [Google Scholar]
- 28.Yoshida S, Takakura A, Ohbo K, Abe K, Wakabayashi J, Yamamoto M, et al. Neurogenin3 delineates the earliest stages of spermatogenesis in the mouse testis. Developmental Biology. 2004;269:447–58. doi: 10.1016/j.ydbio.2004.01.036. [DOI] [PubMed] [Google Scholar]
- 29.Yoshida S, Sukeno M, Nakagawa T, Ohbo K, Nagamatsu G, Suda T, et al. The first round of mouse spermatogenesis is a distinctive program that lacks the self-renewing spermatogonia stage. Development. 2006;133:1495–505. doi: 10.1242/dev.02316. [DOI] [PubMed] [Google Scholar]
- 30.Suzuki H, Sada A, Yoshida S, Saga Y. The heterogeneity of spermatogonia is revealed by their topology and expression of marker proteins including the germ cell-specific proteins Nanos2 and Nanos3. Dev Biol. 2009;336:222–31. doi: 10.1016/j.ydbio.2009.10.002. [DOI] [PubMed] [Google Scholar]
- 31.Tsuda M, Sasaoka Y, Kiso M, Abe K, Haraguchi S, Kobayashi S, et al. Conserved Role of nanos Proteins in Germ Cell Development. Science. 2003;301:1239–41. doi: 10.1126/science.1085222. [DOI] [PubMed] [Google Scholar]
- 32.Suzuki A, Tsuda M, Saga Y. Functional redundancy among Nanos proteins and a distinct role of Nanos2 during male germ cell development. Development. 2007;134:77–83. doi: 10.1242/dev.02697. [DOI] [PubMed] [Google Scholar]
- 33.Lolicato F, Marino R, Paronetto MP, Pellegrini M, Dolci S, Geremia R, et al. Potential role of Nanos3 in maintaining the undifferentiated spermatogonia population. Developmental Biology. 2008;313:725–38. doi: 10.1016/j.ydbio.2007.11.011. [DOI] [PubMed] [Google Scholar]
- 34.Barrios F, Filipponi D, Campolo F, Gori M, Bramucci F, Pellegrini M, et al. SOHLH1 and SOHLH2 control Kit expression during postnatal male germ cell development. Journal of Cell Science. 2012;125:1455–64. doi: 10.1242/jcs.092593. [DOI] [PubMed] [Google Scholar]
- 35.Ballow DJ, Xin Y, Choi Y, Pangas SA, Rajkovic A. Sohlh2 is a germ cell-specific bHLH transcription factor. Gene Expression Patterns. 2006;6:1014–8. doi: 10.1016/j.modgep.2006.04.007. [DOI] [PubMed] [Google Scholar]
- 36.Toyoda S, Miyazaki T, Miyazaki S, Yoshimura T, Yamamoto M, Tashiro F, et al. Sohlh2 affects differentiation of KIT positive oocytes and spermatogonia. Developmental Biology. 2009;325:238–48. doi: 10.1016/j.ydbio.2008.10.019. [DOI] [PubMed] [Google Scholar]
- 37.Wu X, Oatley JM, Oatley MJ, Kaucher AV, Avarbock MR, Brinster RL. The POU Domain Transcription Factor POU3F1 Is an Important Intrinsic Regulator of GDNF-Induced Survival and Self-Renewal of Mouse Spermatogonial Stem Cells. Biology of Reproduction. 2010;82:1103–11. doi: 10.1095/biolreprod.109.083097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Goertz MJ, Wu Z, Gallardo TD, Hamra FK, Castrillon DH. Foxo1 is required in mouse spermatogonial stem cells for their maintenance and the initiation of spermatogenesis. The Journal of Clinical Investigation. 2011;121:3456–66. doi: 10.1172/JCI57984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brinster RL, Avarbock MR. Germline transmission of donor haplotype following spermatogonial transplantation. Proceedings of the National Academy of Sciences. 1994;91:11303–7. doi: 10.1073/pnas.91.24.11303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brinster RL, Zimmermann JW. Spermatogenesis following male germ-cell transplantation. Proceedings of the National Academy of Sciences. 1994;91:11298–302. doi: 10.1073/pnas.91.24.11298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dobrinski I, Ogawa T, Avarbock MR, Brinster RL. Computer assisted image analysis to assess colonization of recipient seminiferous tubules by spermatogonial stem cells from transgenic donor mice. Mol Reprod Dev. 1999;53:142–8. doi: 10.1002/(SICI)1098-2795(199906)53:2<142::AID-MRD3>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 42.Zhang X, Ebata KT, Nagano MC. Genetic analysis of the clonal origin of regenerating mouse spermatogenesis following transplantation. Biol Reprod. 2003;69:1872–8. doi: 10.1095/biolreprod.103.019273. [DOI] [PubMed] [Google Scholar]
- 43.Kanatsu-Shinohara M, Inoue K, Miki H, Ogonuki N, Takehashi M, Morimoto T, et al. Clonal origin of germ cell colonies after spermatogonial transplantation in mice. Biol Reprod. 2006;75:68–74. doi: 10.1095/biolreprod.106.051193. [DOI] [PubMed] [Google Scholar]
- 44.Nagano M, Avarbock MR, Brinster RL. Pattern and Kinetics of Mouse Donor Spermatogonial Stem Cell Colonization in Recipient Testes. Biology of Reproduction. 1999;60:1429–36. doi: 10.1095/biolreprod60.6.1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shinohara T, Orwig KE, Avarbock MR, Brinster RL. Spermatogonial stem cell enrichment by multiparameter selection of mouse testis cells. Proc Natl Acad Sci U S A. 2000;97:8346–51. doi: 10.1073/pnas.97.15.8346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lassalle B, Bastos H, Louis JP, Riou L, Testart J, Dutrillaux B, et al. ‘Side Population’ cells in adult mouse testis express Bcrp1 gene and are enriched in spermatogonia and germinal stem cells. Development. 2004;131:479–87. doi: 10.1242/dev.00918. [DOI] [PubMed] [Google Scholar]
- 47.Fujita K, Ohta H, Tsujimura A, Takao T, Miyagawa Y, Takada S, et al. Transplantation of spermatogonial stem cells isolated from leukemic mice restores fertility without inducing leukemia. Journal of Clinical Investigation. 2005;115:1855–61. doi: 10.1172/JCI24189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lo KC, Brugh VM, Parker M, Lamb DJ. Isolation and Enrichment of Murine Spermatogonial Stem Cells Using Rhodamine 123 Mitochondrial Dye. Biology of Reproduction. 2005;72:767–71. doi: 10.1095/biolreprod.104.033464. [DOI] [PubMed] [Google Scholar]
- 49.Buageaw A, Sukhwani M, Ben-Yehudah A, Ehmcke J, Rawe VY, Pholpramool C, et al. GDNF Family Receptor alpha1 Phenotype of Spermatogonial Stem Cells in Immature Mouse Testes. Biology of Reproduction. 2005;73:1011–6. doi: 10.1095/biolreprod.105.043810. [DOI] [PubMed] [Google Scholar]
- 50.Kanatsu-Shinohara M, Mori Y, Shinohara T. Enrichment of Mouse Spermatogonial Stem Cells Based on Aldehyde Dehydrogenase Activity. Biology of Reproduction. 2013;89:140, 1–10. doi: 10.1095/biolreprod.113.114629. [DOI] [PubMed] [Google Scholar]
- 51.Shinohara T, Ishii K, Kanatsu-Shinohara M. Unstable Side Population Phenotype of Mouse Spermatogonial Stem Cells In Vitro. Journal of Reproduction and Development. 2011;57:288–95. doi: 10.1262/jrd.10-168n. [DOI] [PubMed] [Google Scholar]
- 52.Falciatori I, Borsellino G, Haliassos N, Boitani C, Corallini S, Battistini L, et al. Identification and enrichment of spermatogonial stem cells displaying side-population phenotype in immature mouse testis. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2004;18:376–8. doi: 10.1096/fj.03-0744fje. [DOI] [PubMed] [Google Scholar]
- 53.Lassalle B, Bastos H, Louis JP, Riou L, Testart J, Dutrillaux B, et al. ‘Side Population’ cells in adult mouse testis express Bcrp1 gene and are enriched in spermatogonia and germinal stem cells. Development. 2004;131:479–87. doi: 10.1242/dev.00918. [DOI] [PubMed] [Google Scholar]
- 54.von Kopylow K, Kirchhoff C, Jezek D, Schulze W, Feig C, Primig M, et al. Screening for biomarkers of spermatogonia within the human testis: a whole genome approach. Hum Reprod. 2010;25:1104–12. doi: 10.1093/humrep/deq053. [DOI] [PubMed] [Google Scholar]
- 55.Kristensen DM, Nielsen JE, Skakkebaek NE, Graem N, Jacobsen GK, Meyts ER-D, et al. Presumed pluripotency markers UTF-1 and REX-1 are expressed in human adult testes and germ cell neoplasms. Human Reproduction. 2008;23:775–82. doi: 10.1093/humrep/den010. [DOI] [PubMed] [Google Scholar]
- 56.Eildermann K, Aeckerle N, Debowski K, Godmann M, Christiansen H, Heistermann M, et al. Developmental expression of the pluripotency factor sal-like protein 4 in the monkey, human and mouse testis: restriction to premeiotic germ cells. Cells Tissues Organs. 2012;196:206–20. doi: 10.1159/000335031. [DOI] [PubMed] [Google Scholar]
- 57.von Kopylow K, Staege H, Spiess A-N, Schulze W, Will H, Primig M, et al. Differential marker protein expression specifies rarefaction zone-containing human Adark spermatogonia. Reproduction. 2012;143:45–57. doi: 10.1530/REP-11-0290. [DOI] [PubMed] [Google Scholar]
- 58.von Kopylow K, Staege H, Schulze W, Will H, Kirchhoff C. Fibroblast growth factor receptor 3 is highly expressed in rarely dividing human type A spermatogonia. Histochem Cell Biol. 2012;138:759–72. doi: 10.1007/s00418-012-0991-7. [DOI] [PubMed] [Google Scholar]
- 59.Lim J, Goriely A, Turner GDH, Ewen KA, Jacobsen GK, Graem N, et al. OCT2, SSX and SAGE1 reveal the phenotypic heterogeneity of spermatocytic seminoma reflecting distinct subpopulations of spermatogonia. The Journal of Pathology. 2011;224:473–83. doi: 10.1002/path.2919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.He Z, Kokkinaki M, Jiang J, Dobrinski I, Dym M. Isolation, Characterization, and Culture of Human Spermatogonia. Biology of Reproduction. 2010;82:363–72. doi: 10.1095/biolreprod.109.078550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Grisanti L, Falciatori I, Grasso M, Dovere L, Fera S, Muciaccia B, et al. Identification of Spermatogonial Stem Cell Subsets by Morphological Analysis and Prospective Isolation. STEM CELLS. 2009;27:3043–52. doi: 10.1002/stem.206. [DOI] [PubMed] [Google Scholar]
- 62.Zohni K, Zhang X, Tan SL, Chan P, Nagano M. CD9 is expressed on human male germ cells that have a long-term repopulation potential after transplantation into mouse testes. Biol Reprod. 2012;87:27. doi: 10.1095/biolreprod.112.098913. [DOI] [PubMed] [Google Scholar]
- 63.Aeckerle N, Eildermann K, Drummer C, Ehmcke J, Schweyer S, Lerchl A, et al. The pluripotency factor LIN28 in monkey and human testes: a marker for spermatogonial stem cells? Molecular Human Reproduction. 2012;18:477–88. doi: 10.1093/molehr/gas025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kossack N, Terwort N, Wistuba J, Ehmcke J, Schlatt S, Schöler H, et al. A combined approach facilitates the reliable detection of human spermatogonia in vitro. Human Reproduction. 2013;28:3012–25. doi: 10.1093/humrep/det336. [DOI] [PubMed] [Google Scholar]
- 65.Williams AF. Immunology: Immunoglobulin-related domains for cell surface recognition. Nature. 1985;314:579–80. doi: 10.1038/314579a0. [DOI] [PubMed] [Google Scholar]
- 66.Seeger RC, Danon YL, Rayner SA, Hoover F. Definition of a Thy-1 determinant on human neuroblastoma, glioma, sarcoma, and teratoma cells with a monoclonal antibody. The Journal of Immunology. 1982;128:983–9. [PubMed] [Google Scholar]
- 67.Kemshead JT, Ritter MA, Cotmore SF, Greaves MF. Human Thy-1: Expression on the cell surface of neuronal and and glial cells. Brain research. 1982;236:451–61. doi: 10.1016/0006-8993(82)90727-2. [DOI] [PubMed] [Google Scholar]
- 68.Majeti R, Park CY, Weissman IL. Identification of a Hierarchy of Multipotent Hematopoietic Progenitors in Human Cord Blood. Cell Stem Cell. 2007;1:635–45. doi: 10.1016/j.stem.2007.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Craig W, Kay R, Cutler RL, Lansdorp PM. Expression of Thy-1 on human hematopoietic progenitor cells. The Journal of Experimental Medicine. 1993;177:1331–42. doi: 10.1084/jem.177.5.1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Haeryfar SM, Hoskin DW. Thy-1: more than a mouse pan-T cell marker. J Immunol. 2004;173:3581–8. doi: 10.4049/jimmunol.173.6.3581. [DOI] [PubMed] [Google Scholar]
- 71.Saalbach A, Kraft R, Herrmann K, Haustein UF, Anderegg U. The monoclonal antibody AS02 recognizes a protein on human fibroblasts being highly homologous to Thy-1. Arch Dermatol Res. 1998;290:360–6. doi: 10.1007/s004030050318. [DOI] [PubMed] [Google Scholar]
- 72.Saalbach A, Wetzig T, Haustein UF, Anderegg U. Detection of human soluble Thy-1 in serum by ELISA. Fibroblasts and activated endothelial cells are a possible source of soluble Thy-1 in serum. Cell and tissue research. 1999;298:307–15. doi: 10.1007/s004419900079. [DOI] [PubMed] [Google Scholar]
- 73.Rege TA, Hagood JS. Thy-1 as a regulator of cell-cell and cell-matrix interactions in axon regeneration, apoptosis, adhesion, migration, cancer, and fibrosis. The FASEB Journal. 2006;20:1045–54. doi: 10.1096/fj.05-5460rev. [DOI] [PubMed] [Google Scholar]
- 74.Barboni E, Gormley AM, Pliego Rivero FB, Vidal M, Morris RJ. Activation of T lymphocytes by cross-linking of glycophospholipid-anchored Thy-1 mobilizes separate pools of intracellular second messengers to those induced by the antigen-receptor/CD3 complex. Immunology. 1991;72:457–63. [PMC free article] [PubMed] [Google Scholar]
- 75.Hermann BP, Sukhwani M, Simorangkir DR, Chu T, Plant TM, Orwig KE. Molecular dissection of the male germ cell lineage identifies putative spermatogonial stem cells in rhesus macaques. Human Reproduction. 2009;24:1704–16. doi: 10.1093/humrep/dep073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Izadyar F, Wong J, Maki C, Pacchiarotti J, Ramos T, Howerton K, et al. Identification and characterization of repopulating spermatogonial stem cells from the adult human testis. Hum Reprod. 2011;26:1296–306. doi: 10.1093/humrep/der026. [DOI] [PubMed] [Google Scholar]
- 77.Nagano M, McCarrey JR, Brinster RL. Primate spermatogonial stem cells colonize mouse testes. Biol Reprod. 2001;64:1409–16. doi: 10.1095/biolreprod64.5.1409. [DOI] [PubMed] [Google Scholar]
- 78.Nagano M, Patrizio P, Brinster RL. Long-term survival of human spermatogonial stem cells in mouse testes. Fertil Steril. 2002;78:1225–33. doi: 10.1016/s0015-0282(02)04345-5. [DOI] [PubMed] [Google Scholar]
- 79.Sadri-Ardekani H, Mizrak SC, van Daalen SK, Korver CM, Roepers-Gajadien HL, Koruji M, et al. Propagation of human spermatogonial stem cells in vitro. JAMA : the journal of the American Medical Association. 2009;302:2127–34. doi: 10.1001/jama.2009.1689. [DOI] [PubMed] [Google Scholar]
- 80.Sadri-Ardekani H, Akhondi MA, van der Veen F, Repping S, van Pelt AM. In vitro propagation of human prepubertal spermatogonial stem cells. JAMA : the journal of the American Medical Association. 2011;305:2416–8. doi: 10.1001/jama.2011.791. [DOI] [PubMed] [Google Scholar]
- 81.Dovey SL, Valli H, Hermann BP, Sukhwani M, Donohue J, Castro CA, et al. Eliminating malignant contamination from therapeutic human spermatogonial stem cells. J Clin Invest. 2013;123:1833–43. doi: 10.1172/JCI65822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hermann BP, Sukhwani M, Lin C-C, Sheng Y, Tomko J, Rodriguez M, et al. Characterization, Cryopreservation, and Ablation of Spermatogonial Stem Cells in Adult Rhesus Macaques. STEM CELLS. 2007;25:2330–8. doi: 10.1634/stemcells.2007-0143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hermann BP, Sukhwani M, Salati J, Sheng Y, Chu T, Orwig KE. Separating spermatogonia from cancer cells in contaminated prepubertal primate testis cell suspensions. Human Reproduction. 2011;26:3222–31. doi: 10.1093/humrep/der343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Conrad S, Renninger M, Hennenlotter J, Wiesner T, Just L, Bonin M, et al. Generation of pluripotent stem cells from adult human testis. Nature. 2008;456:344–9. doi: 10.1038/nature07404. [DOI] [PubMed] [Google Scholar]
- 85.Wixler V, Laplantine E, Geerts D, Sonnenberg A, Petersohn D, Eckes B, et al. Identification of novel interaction partners for the conserved membrane proximal region of α-integrin cytoplasmic domains. FEBS letters. 1999;445:351–5. doi: 10.1016/s0014-5793(99)00151-9. [DOI] [PubMed] [Google Scholar]
- 86.Lathia JD, Gallagher J, Heddleston JM, Wang J, Eyler CE, Macswords J, et al. Integrin alpha 6 regulates glioblastoma stem cells. Cell Stem Cell. 2010;6:421–32. doi: 10.1016/j.stem.2010.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Stingl J, Eirew P, Ricketson I, Shackleton M, Vaillant F, Choi D, et al. Purification and unique properties of mammary epithelial stem cells. Nature. 2006;439:993–7. doi: 10.1038/nature04496. [DOI] [PubMed] [Google Scholar]
- 88.Cariati M, Naderi A, Brown JP, Smalley MJ, Pinder SE, Caldas C, et al. Alpha-6 integrin is necessary for the tumourigenicity of a stem cell-like subpopulation within the MCF7 breast cancer cell line. International Journal of Cancer. 2008;122:298–304. doi: 10.1002/ijc.23103. [DOI] [PubMed] [Google Scholar]
- 89.Litvinov SV, Velders MP, Bakker HA, Fleuren GJ, Warnaar SO. Ep-CAM: a human epithelial antigen is a homophilic cell-cell adhesion molecule. The Journal of Cell Biology. 1994;125:437–46. doi: 10.1083/jcb.125.2.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Osta WA, Chen Y, Mikhitarian K, Mitas M, Salem M, Hannun YA, et al. EpCAM Is Overexpressed in Breast Cancer and Is a Potential Target for Breast Cancer Gene Therapy. Cancer Research. 2004;64:5818–24. doi: 10.1158/0008-5472.CAN-04-0754. [DOI] [PubMed] [Google Scholar]
- 91.Maetzel D, Denzel S, Mack B, Canis M, Went P, Benk M, et al. Nuclear signalling by tumour-associated antigen EpCAM. Nat Cell Biol. 2009;11:162–71. doi: 10.1038/ncb1824. [DOI] [PubMed] [Google Scholar]
- 92.Litvinov SV, van Driel W, van Rhijn CM, Bakker HA, van Krieken H, Fleuren GJ, et al. Expression of Ep-CAM in cervical squamous epithelia correlates with an increased proliferation and the disappearance of markers for terminal differentiation. Am J Pathol. 1996;148:865–75. [PMC free article] [PubMed] [Google Scholar]
- 93.Jurgens G. Head and tail development of the Drosophila embryo involves spalt, a novel homeotic gene. Embo J. 1988;7:189–96. doi: 10.1002/j.1460-2075.1988.tb02799.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kuhnlein RP, Frommer G, Friedrich M, Gonzalez-Gaitan M, Weber A, Wagner-Bernholz JF, et al. spalt encodes an evolutionarily conserved zinc finger protein of novel structure which provides homeotic gene function in the head and tail region of the Drosophila embryo. Embo J. 1994;13:168–79. doi: 10.1002/j.1460-2075.1994.tb06246.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hollemann T, Schuh R, Pieler T, Stick R. Xenopus Xsal-1, a vertebrate homolog of the region specific homeotic gene spalt of Drosophila. Mechanisms of development. 1996;55:19–32. doi: 10.1016/0925-4773(95)00485-8. [DOI] [PubMed] [Google Scholar]
- 96.Camp E, Hope R, Kortschak RD, Cox TC, Lardelli M. Expression of three spalt (sal) gene homologues in zebrafish embryos. Development genes and evolution. 2003;213:35–43. doi: 10.1007/s00427-002-0284-6. [DOI] [PubMed] [Google Scholar]
- 97.Sweetman D, Smith T, Farrell ER, Chantry A, Munsterberg A. The conserved glutamine-rich region of chick csal1 and csal3 mediates protein interactions with other spalt family members. Implications for Townes-Brocks syndrome. The Journal of biological chemistry. 2003;278:6560–6. doi: 10.1074/jbc.M209066200. [DOI] [PubMed] [Google Scholar]
- 98.Ott T, Kaestner KH, Monaghan AP, Schutz G. The mouse homolog of the region specific homeotic gene spalt of Drosophila is expressed in the developing nervous system and in mesoderm-derived structures. Mechanisms of development. 1996;56:117–28. doi: 10.1016/0925-4773(96)00516-3. [DOI] [PubMed] [Google Scholar]
- 99.Kohlhase J, Schuh R, Dowe G, Kuhnlein RP, Jackle H, Schroeder B, et al. Isolation, characterization, and organ-specific expression of two novel human zinc finger genes related to the Drosophila gene spalt. Genomics. 1996;38:291–8. doi: 10.1006/geno.1996.0631. [DOI] [PubMed] [Google Scholar]
- 100.Elling U, Klasen C, Eisenberger T, Anlag K, Treier M. Murine inner cell mass-derived lineages depend on Sall4 function. Proceedings of the National Academy of Sciences. 2006;103:16319–24. doi: 10.1073/pnas.0607884103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zhang J, Tam WL, Tong GQ, Wu Q, Chan HY, Soh BS, et al. Sall4 modulates embryonic stem cell pluripotency and early embryonic development by the transcriptional regulation of Pou5f1. Nat Cell Biol. 2006;8:1114–23. doi: 10.1038/ncb1481. [DOI] [PubMed] [Google Scholar]
- 102.Hobbs Robin M, Fagoonee S, Papa A, Webster K, Altruda F, Nishinakamura R, et al. Functional Antagonism between Sall4 and Plzf Defines Germline Progenitors. Cell Stem Cell. 2012;10:284–98. doi: 10.1016/j.stem.2012.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Gassei K, Orwig KE. SALL4 Expression in Gonocytes and Spermatogonial Clones of Postnatal Mouse Testes. PLoS ONE. 2013;8:e53976. doi: 10.1371/journal.pone.0053976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Hothorn T, Bretz F, Westfall P. Simultaneous inference in general parametric models. Biometrical journal Biometrische Zeitschrift. 2008;50:346–63. doi: 10.1002/bimj.200810425. [DOI] [PubMed] [Google Scholar]
- 105.Amann RP. The Cycle of the Seminiferous Epithelium in Humans: A Need to Revisit? Journal of Andrology. 2008;29:469–87. doi: 10.2164/jandrol.107.004655. [DOI] [PubMed] [Google Scholar]
- 106.Dym M, Kokkinaki M, He Z. Spermatogonial stem cells: Mouse and human comparisons. Birth Defects Research Part C: Embryo Today: Reviews. 2009;87:27–34. doi: 10.1002/bdrc.20141. [DOI] [PubMed] [Google Scholar]
- 107.Goldschneider I, Gordon LK, Morris RJ. Demonstration of Thy-1 antigen on pluripotent hemopoietic stem cells in the rat. J Exp Med. 1978;148:1351–66. doi: 10.1084/jem.148.5.1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Spangrude GJ, Heimfeld S, Weissman IL. Purification and characterization of mouse hematopoietic stem cells. Science. 1988;241:58–62. doi: 10.1126/science.2898810. [DOI] [PubMed] [Google Scholar]
- 109.Baume CM, Weissman IL, Tsukamoto AS, Buckle A, Peault B. Isolation of a candidate human hematopoietic stem-cell population. ProcNatlAcadSciUSA. 1992;89:2804–8. doi: 10.1073/pnas.89.7.2804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Ogawa T, Arechaga JM, Avarbock MR, Brinster RL. Transplantation of testis germinal cells into mouse seminiferous tubules. The International journal of developmental biology. 1997;41:111–22. [PubMed] [Google Scholar]
- 111.Nagano M, Brinster RL. Spermatogonial transplantation and reconstitution of donor cell spermatogenesis in recipient mice. APMIS. 1998;106:47–55. doi: 10.1111/j.1699-0463.1998.tb01318.x. discussion 6–7. [DOI] [PubMed] [Google Scholar]
- 112.Shinohara T, Orwig KE, Avarbock MR, Brinster RL. Spermatogonial stem cell enrichment by multiparameter selection of mouse testis cells. Proceedings of the National Academy of Sciences. 2000;97:8346–51. doi: 10.1073/pnas.97.15.8346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Meng X, Lindahl M, Hyvonen ME, Parvinen M, de Rooij DG, Hess MW, et al. Regulation of cell fate decision of undifferentiated spermatogonia by GDNF. Science. 2000;287:1489–93. doi: 10.1126/science.287.5457.1489. [DOI] [PubMed] [Google Scholar]
- 114.de Rooij DG, Russell LD. All you wanted to know about spermatogonia but were afraid to ask. JAndrol. 2000;21:776–98. [PubMed] [Google Scholar]
- 115.de Rooij DG. Spermatogonial stem cell renewal in the mouse. I. Normal situation. Cell Tissue Kinet. 1973;6:281–7. doi: 10.1111/j.1365-2184.1973.tb01617.x. [DOI] [PubMed] [Google Scholar]
- 116.Maki CB, Pacchiarotti J, Ramos T, Pascual M, Pham J, Kinjo J, et al. Phenotypic and molecular characterization of spermatogonial stem cells in adult primate testes. Human Reproduction. 2009;24:1480–91. doi: 10.1093/humrep/dep033. [DOI] [PubMed] [Google Scholar]
- 117.Kanatsu-Shinohara M, Morimoto H, Shinohara T. Enrichment of Mouse Spermatogonial Stem Cells by Melanoma Cell Adhesion Molecule Expression. Biology of Reproduction. 2012;87:139, 1–10. doi: 10.1095/biolreprod.112.103861. [DOI] [PubMed] [Google Scholar]
- 118.Kubota H, Avarbock MR, Brinster RL. Culture Conditions and Single Growth Factors Affect Fate Determination of Mouse Spermatogonial Stem Cells. Biology of Reproduction. 2004;71:722–31. doi: 10.1095/biolreprod.104.029207. [DOI] [PubMed] [Google Scholar]
- 119.Paul C, Nagano M, Robaire B. Aging results in molecular changes in an enriched population of undifferentiated rat spermatogonia. Biol Reprod. 2013;89:147. doi: 10.1095/biolreprod.113.112995. [DOI] [PubMed] [Google Scholar]
- 120.Phillips BT, Gassei K, Orwig KE. Spermatogonial stem cell regulation and spermatogenesis. Philosophical transactions of the Royal Society of London Series B, Biological sciences. 2010;365:1663–78. doi: 10.1098/rstb.2010.0026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Brinster RL. Germline Stem Cell Transplantation and Transgenesis. Science. 2002;296:2174–6. doi: 10.1126/science.1071607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Valli H, Phillips BT, Shetty G, Byrne JA, Clark AT, Meistrich ML, et al. Germline stem cells: toward the regeneration of spermatogenesis. Fertility and sterility. 2014;101:3–13. doi: 10.1016/j.fertnstert.2013.10.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Nagano M, Patrizio P, Brinster RL. Long-term survival of human spermatogonial stem cells in mouse testes. Fertility and sterility. 2002;78:1225–33. doi: 10.1016/s0015-0282(02)04345-5. [DOI] [PubMed] [Google Scholar]
- 124.Chen B, Wang YB, Zhang ZL, Xia WL, Wang HX, Xiang ZQ, et al. Xeno-free culture of human spermatogonial stem cells supported by human embryonic stem cell-derived fibroblast-like cells. Asian J Androl. 2009;11:557–65. doi: 10.1038/aja.2009.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Yang Q-E, Racicot KE, Kaucher AV, Oatley MJ, Oatley JM. MicroRNAs 221 and 222 regulate the undifferentiated state in mammalian male germ cells. Development. 2013;140:280–90. doi: 10.1242/dev.087403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Yang Q-E, Kim D, Kaucher A, Oatley MJ, Oatley JM. CXCL12–CXCR4 signaling is required for the maintenance of mouse spermatogonial stem cells. Journal of Cell Science. 2013;126:1009–20. doi: 10.1242/jcs.119826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Oatley JM, Oatley MJ, Avarbock MR, Tobias JW, Brinster RL. Colony stimulating factor 1 is an extrinsic stimulator of mouse spermatogonial stem cell self-renewal. Development. 2009;136:1191–9. doi: 10.1242/dev.032243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Oatley JM, Avarbock MR, Brinster RL. Glial Cell Line-derived Neurotrophic Factor Regulation of Genes Essential for Self-renewal of Mouse Spermatogonial Stem Cells Is Dependent on Src Family Kinase Signaling. Journal of Biological Chemistry. 2007;282:25842–51. doi: 10.1074/jbc.M703474200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Yeh JR, Zhang X, Nagano MC. Wnt5a is a cell-extrinsic factor that supports self-renewal of mouse spermatogonial stem cells. Journal of Cell Science. 2011;124:2357–66. doi: 10.1242/jcs.080903. [DOI] [PubMed] [Google Scholar]
- 130.Kubota H, Avarbock MR, Brinster RL. Growth factors essential for self-renewal and expansion of mouse spermatogonial stem cells. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:16489–94. doi: 10.1073/pnas.0407063101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Kent Hamra F, Schultz N, Chapman KM, Grellhesl DM, Cronkhite JT, Hammer RE, et al. Defining the spermatogonial stem cell. Developmental Biology. 2004;269:393–410. doi: 10.1016/j.ydbio.2004.01.027. [DOI] [PubMed] [Google Scholar]
- 132.Sada A, Suzuki A, Suzuki H, Saga Y. The RNA-Binding Protein NANOS2 Is Required to Maintain Murine Spermatogonial Stem Cells. Science. 2009;325:1394–8. doi: 10.1126/science.1172645. [DOI] [PubMed] [Google Scholar]
- 133.Kusz KM, Tomczyk L, Sajek M, Spik A, Latos-Bielenska A, Jedrzejczak P, et al. The highly conserved NANOS2 protein: testis-specific expression and significance for the human male reproduction. Molecular Human Reproduction. 2009;15:165–71. doi: 10.1093/molehr/gap003. [DOI] [PubMed] [Google Scholar]
- 134.Dettin L, Ravindranath N, Hofmann M-C, Dym M. Morphological Characterization of the Spermatogonial Subtypes in the Neonatal Mouse Testis. Biology of Reproduction. 2003;69:1565–71. doi: 10.1095/biolreprod.103.016394. [DOI] [PubMed] [Google Scholar]
- 135.Eildermann K, Gromoll J, Behr R. Misleading and reliable markers to differentiate between primate testis-derived multipotent stromal cells and spermatogonia in culture. Human Reproduction. 2012;27:1754–67. doi: 10.1093/humrep/des091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Mitchell RT, Cowan G, Morris KD, Anderson RA, Fraser HM, Mckenzie KJ, et al. Germ cell differentiation in the marmoset (Callithrix jacchus) during fetal and neonatal life closely parallels that in the human. Human Reproduction. 2008;23:2755–65. doi: 10.1093/humrep/den295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Pesce M, Wang X, Wolgemuth DJ, Schöler HR. Differential expression of the Oct-4 transcription factor during mouse germ cell differentiation. Mechanisms of development. 1998;71:89–98. doi: 10.1016/s0925-4773(98)00002-1. [DOI] [PubMed] [Google Scholar]
- 138.Ohbo K, Yoshida S, Ohmura M, Ohneda O, Ogawa T, Tsuchiya H, et al. Identification and characterization of stem cells in prepubertal spermatogenesis in mice. Developmental Biology. 2003;258:209–25. doi: 10.1016/s0012-1606(03)00111-8. [DOI] [PubMed] [Google Scholar]
- 139.Oatley JM, Avarbock MR, Telaranta AI, Fearon DT, Brinster RL. Identifying genes important for spermatogonial stem cell self-renewal and survival. Proc Natl Acad Sci U S A. 2006;103:9524–9. doi: 10.1073/pnas.0603332103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Raverot G, Weiss J, Park SY, Hurley L, Jameson JL. Sox3 expression in undifferentiated spermatogonia is required for the progression of spermatogenesis. Developmental Biology. 2005;283:215–25. doi: 10.1016/j.ydbio.2005.04.013. [DOI] [PubMed] [Google Scholar]
- 141.Wang Y-L, Liu W, Sun Y-J, Kwon J, Setsuie R, Osaka H, et al. Overexpression of ubiquitin carboxyl-terminal hydrolase L1 arrests spermatogenesis in transgenic mice. Molecular Reproduction and Development. 2006;73:40–9. doi: 10.1002/mrd.20364. [DOI] [PubMed] [Google Scholar]
- 142.Suzuki H, Ahn HW, Chu T, Bowden W, Gassei K, Orwig K, et al. SOHLH1 and SOHLH2 coordinate spermatogonial differentiation. Dev Biol. 2012;361:301–12. doi: 10.1016/j.ydbio.2011.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Müller T, Eildermann K, Dhir R, Schlatt S, Behr R. Glycan stem-cell markers are specifically expressed by spermatogonia in the adult non-human primate testis. Human Reproduction. 2008;23:2292–8. doi: 10.1093/humrep/den253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Wyns C, Van Langendonckt A, Wese F-X, Donnez J, Curaba M. Long-term spermatogonial survival in cryopreserved and xenografted immature human testicular tissue. Human Reproduction. 2008;23:2402–14. doi: 10.1093/humrep/den272. [DOI] [PubMed] [Google Scholar]
- 145.Jarvis S, Elliott DJ, Morgan D, Winston R, Readhead C. Molecular markers for the assessment of postnatal male germ cell development in the mouse. Human Reproduction. 2005;20:108–16. doi: 10.1093/humrep/deh565. [DOI] [PubMed] [Google Scholar]
- 146.Maymon BB-S, Elliott DJ, Kleiman SE, Yogev L, Hauser R, Botchan A, et al. The contribution of RNA-binding motif (RBM) antibody to the histopathologic evaluation of testicular biopsies from infertile men. Human Pathology. 2001;32:36–41. doi: 10.1053/hupa.2001.20887. [DOI] [PubMed] [Google Scholar]
- 147.Elliott DJ, Ma K, Kerr SM, Thakrar R, Speed R, Chandley AC, et al. An RBM Homologue Maps to the Mouse Y Chromosome and is Expressed in Germ Cells. Human Molecular Genetics. 1996;5:869–74. doi: 10.1093/hmg/5.7.869. [DOI] [PubMed] [Google Scholar]
- 148.Österlund C, Ståbi B, Bhasin S, Kvist U, Pousette Å, Arver S. Specific localization of RBM1a in the nuclei of all cell types except elongated spermatids within seminiferous tubules of the human. International Journal of Andrology. 2001;24:272–7. doi: 10.1046/j.1365-2605.2001.00299.x. [DOI] [PubMed] [Google Scholar]
- 149.Elliott DJ, Millar MR, Oghene K, Ross A, Kiesewetter F, Pryor J, et al. Expression of RBM in the nuclei of human germ cells is dependent on a critical region of the Y chromosome long3arm. Proceedings of the National Academy of Sciences. 1997;94:3848–53. doi: 10.1073/pnas.94.8.3848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Gely-Pernot A, Raverdeau M, Celebi C, Dennefeld C, Feret B, Klopfenstein M, et al. Spermatogonia differentiation requires retinoic acid receptor gamma. Endocrinology. 2012;153:438–49. doi: 10.1210/en.2011-1102. [DOI] [PubMed] [Google Scholar]
- 151.Sa R, Miranda C, Carvalho F, Barros A, Sousa M. Expression of stem cell markers: OCT4, KIT, ITGA6, and ITGB1 in the male germinal epithelium. Syst Biol Reprod Med. 2013;59:233–43. doi: 10.3109/19396368.2013.804964. [DOI] [PubMed] [Google Scholar]
- 152.Manova K, Nocka K, Besmer P, Bachvarova RF. Gonadal expression of c-kit encoded at the W locus of the mouse. Development. 1990;110:1057–69. doi: 10.1242/dev.110.4.1057. [DOI] [PubMed] [Google Scholar]
- 153.Yoshinaga K, Nishikawa S, Ogawa M, Hayashi S, Kunisada T, Fujimoto T, et al. Role of c-kit in mouse spermatogenesis: identification of spermatogonia as a specific site of c-kit expression and function. Development. 1991;113:689–99. doi: 10.1242/dev.113.2.689. [DOI] [PubMed] [Google Scholar]
- 154.Schrans-Stassen BHGJ, van de Kant HJG, de Rooij DG, van Pelt AMM. Differential Expression of c-kit in Mouse Undifferentiated and Differentiating Type A Spermatogonia. Endocrinology. 1999;140:5894–900. doi: 10.1210/endo.140.12.7172. [DOI] [PubMed] [Google Scholar]
- 155.Unni SK, Modi DN, Pathak SG, Dhabalia JV, Bhartiya D. Stage-specific Localization and Expression of c-kit in the Adult Human Testis. Journal of Histochemistry & Cytochemistry. 2009;57:861–9. doi: 10.1369/jhc.2009.953737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Oulad-Abdelghani M, Bouillet P, Décimo D, Gansmuller A, Heyberger S, Dollé P, et al. Characterization of a premeiotic germ cell-specific cytoplasmic protein encoded by Stra8, a novel retinoic acid-responsive gene. The Journal of Cell Biology. 1996;135:469–77. doi: 10.1083/jcb.135.2.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Giuili G, Tomljenovic A, Labrecque N, Oulad-Abdelghani M, Rassoulzadegan M, Cuzin F. Murine spermatogonial stem cells: targeted transgene expression and purification in an active state. EMBO reports. 2002;3:753–9. doi: 10.1093/embo-reports/kvf149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Antonangeli F, Giampietri C, Petrungaro S, Filippini A, Ziparo E. Expression profile of a 400-bp Stra8 promoter region during spermatogenesis. Microscopy Research and Technique. 2009;72:816–22. doi: 10.1002/jemt.20724. [DOI] [PubMed] [Google Scholar]
- 159.Falender AE, Freiman RN, Geles KG, Lo KC, Hwang K, Lamb DJ, et al. Maintenance of spermatogenesis requires TAF4b, a gonad-specific subunit of TFIID. Genes Dev. 2005;19:794–803. doi: 10.1101/gad.1290105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Ruggiu M, Speed R, Taggart M, McKay SJ, Kilanowski F, Saunders P, et al. The mouse Dazla gene encodes a cytoplasmic protein essential for gametogenesis. Nature. 1997;389:73–7. doi: 10.1038/37987. [DOI] [PubMed] [Google Scholar]
- 161.Cooke HJ, Lee M, Kerr S, Ruggiu M. A Murine Homologue of the Human Daz Gene Is Autosomal and Expressed only in Male and Female Gonads. Human Molecular Genetics. 1996;5:513–6. doi: 10.1093/hmg/5.4.513. [DOI] [PubMed] [Google Scholar]
- 162.Tanaka SS, Toyooka Y, Akasu R, Katoh-Fukui Y, Nakahara Y, Suzuki R, et al. The mouse homolog of Drosophila Vasa is required for the development of male germ cells. Gene Dev. 2000;14:841–53. [PMC free article] [PubMed] [Google Scholar]
- 163.Castrillon DH, Quade BJ, Wang TY, Quigley C, Crum CP. The human VASA gene is specifically expressed in the germ cell lineage. Proc Natl Acad Sci U S A. 2000;97:9585–90. doi: 10.1073/pnas.160274797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Huszar JM, Payne CJ. MicroRNA 146 (Mir146) Modulates Spermatogonial Differentiation by Retinoic Acid in Mice. Biology of Reproduction. 2013;88:15, 1–10. doi: 10.1095/biolreprod.112.103747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.He Z, Jiang J, Kokkinaki M, Tang L, Zeng W, Gallicano I, et al. MiRNA-20 and mirna-106a regulate spermatogonial stem cell renewal at the post-transcriptional level via targeting STAT3 and Ccnd1. STEM CELLS. 2013;31:2205–17. doi: 10.1002/stem.1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Tong M-H, Mitchell D, Evanoff R, Griswold MD. Expression of Mirlet7 Family MicroRNAs in Response to Retinoic Acid-Induced Spermatogonial Differentiation in Mice. Biology of Reproduction. 2011;85:189–97. doi: 10.1095/biolreprod.110.089458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Zhang Y, Su H, Luo F, Wu S, Liu L, Liu T, et al. E-cadherin can be expressed by a small population of rat undifferentiated spermatogonia in vivo and in vitro. In Vitro Cellular & Developmental Biology - Animal. 2011;47:593–600. doi: 10.1007/s11626-011-9446-z. [DOI] [PubMed] [Google Scholar]
- 168.Anderson R, Schaible K, Heasman J, Wylie C. Expression of the homophilic adhesion molecule, Ep-CAM, in the mammalian germ line. Journal of Reproduction and Fertility. 1999;116:379–84. doi: 10.1530/jrf.0.1160379. [DOI] [PubMed] [Google Scholar]
- 169.Kokkinaki M, Djourabtchi A, Golestaneh N. Long-term Culture of Human SSEA-4 Positive Spermatogonial Stem Cells (SSCs) J Stem Cell Res Ther. 2011;2 doi: 10.4172/2157-7633.S2-003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.