Abstract
Chalcone synthase, a key regulatory enzyme in the flavonoid pathway, constitutes an eight-member gene family in Glycine max (soybean). Three of the chalcone synthase (CHS) gene family members are arranged as inverted repeats in a 10-kb region, corresponding to the I locus (inhibitor). Spontaneous mutations of a dominant allele (I or ii) to a recessive allele (i) have been shown to delete promoter sequences, paradoxically increasing total CHS transcript levels and resulting in black seed coats. However, it is not known which of the gene family members contribute toward pigmentation and how this locus affects CHS expression in other tissues. We investigated the unusual nature of the I locus using four pairs of isogenic lines differing with respect to alleles of the I locus. RNA gel blots using a generic open reading frame CHS probe detected similar CHS transcript levels in stems, roots, leaves, young pods, and cotyledons of the yellow and black isolines but not in the seed coats, which is consistent with the dominant I and ii alleles mediating CHS gene silencing in a tissue-specific manner. Using real-time RT-PCR, a variable pattern of expression of CHS genes in different tissues was demonstrated. However, increase in pigmentation in the black seed coats was associated with release of the silencing effect specifically on CHS7/CHS8, which occurred at all stages of seed coat development. These expression changes were linked to structural changes taking place at the I locus, shown to encompass a much wider region of at least 27 kb, comprising two identical 10.91-kb stretches of CHS gene duplications. The suppressive effect of this 27-kb I locus in a specific tissue of the G. max plant represents a unique endogenous gene silencing mechanism.
INTRODUCTION
Homology-dependent gene silencing (HDGS) is a comprehensive term that refers to epigenetic silencing effects based on recognition of nucleic acid sequence homology at either the DNA (transcriptional) or RNA (posttranscriptional) level (Meyer and Saedler, 1996). First discovered in transgenic plants of Petunia, where it was termed cosuppression (Jorgensen, 1990), it is now recognized that HDGS is a commonly observed outcome of transgenesis in plants. Originally proposed and documented as the immune system of the genome (reviewed in Plasterk, 2002; Wassenegger, 2002), several emerging lines of evidence are implicating the various components of the HDGS machinery to be a part of a sophisticated network of normal regulation of endogenous genes (Jacobsen et al., 1999; Reinhart et al., 2002; Carrington and Ambros, 2003).
Posttranscriptional gene silencing (PTGS) is one form of HDGS that is characterized by sequence-specific degradation of cytoplasmic RNA (Meins, 2000). A key conserved feature of PTGS is that it is triggered by double stranded RNA (dsRNA). The origin of dsRNA is diverse (Wassenegger, 2000). Some of the plant transgenic system studies have attributed synthesis of dsRNA to the RNA polymerase II transcription through inverted DNA repeats (IRs), which are multiple tandemly linked gene copies. However, endogenous plant genes forming the repeat structure also may be subjected to gene silencing (summarized in Muskens et al., 2000). The inability to synthesize anthocyanins in the seed coat of particular Glycine max (soybean) cultivars is one such example of a naturally occurring inverted repeat–associated gene silencing system (Todd and Vodkin, 1996).
Extensively studied in several plant species, anthocyanins form a group of secondary metabolites, serving a variety of functions in flowers, fruits, and seeds. In G. max, three genetic loci (I, R, and T) are known to control pigmentation of the seed coat (Bernard and Weiss, 1973; Palmer and Kilen, 1987). The classically defined I locus (inhibitor), characterized by its four alleles, inhibits the production and accumulation of anthocyanins and proanthocyanidins in the epidermal layer of the seed coat in a spatial manner. The absence of pigmentation, which is the dominant phenotype and results in a yellow seed coat at maturity, is brought about by the dominant I allele, whereas the homozygous recessive i allele gives rise to a fully pigmented seed coat. Pigmentation is confined to the hilum and saddle shaped regions by two other alleles, namely ii and ik, respectively.
Though all wild Glycine accessions have pigmented seed coats (recessive i allele), most cultivated G. max varieties have been selected for a yellow seed coat (homozygous I or ii). Several spontaneous independent mutations have frequently occurred in cultivars having either the I or ii alleles (yellow seed coats or yellow seed coats with pigmented hila) to give rise to the recessive i alleles (pigmented seed coats). Some of these mutations have been preserved as near isogenic lines differing only in respect to the alleles at the I locus (Wilcox, 1988).
Analysis of these isogenic pairs has shown that the I locus corresponds to a region of chalcone synthase (CHS) gene duplications (Todd and Vodkin, 1996; Senda et al., 2002a, 2002b). In 11 of the 15 such isogenic pairs studied by Todd and Vodkin (1996), mutations of a dominant allele (I or ii) to the recessive allele (i) delete promoter sequences, paradoxically resulting in increased total CHS transcript levels and thereby pigmented (black) seed coats. This case of suppression of pigmentation because of additional copies of CHS and its restoration upon deletion was suggestive of HDGS (Todd and Vodkin, 1996), established recently by documenting the presence of the diagnostic 21- to 26-nucleotide short-interfering RNAs (siRNAs) (Senda et al., 2004). We decided to pursue this intriguing endogenous gene silencing mechanism by using 4 of the 15 isogenic pairs (Table 1), representing the two classes of spontaneous mutations of the I locus studied before (Todd and Vodkin, 1996).
Table 1.
Genotype and Phenotype of the Four Isogenic or Near Isogenic Pairs of the I Locus Alleles Used for This Study
| Cultivar | Genotype | Phenotype | Source/Origin |
|---|---|---|---|
| Williams 43 | ii, R, T | Yellow seed coat pigmented hilum, tawny pubescence | Parent line, released 1971 |
| Williams 44 | i, R, T | Black seed coat, tawny pubescence | Mutation in Williams, 1980 |
| Williams 54 | ii, R, T | Yellow seed coat pigmented hilum, tawny pubescence | Parent line, released 1971 |
| Williams 55 | i, R, T | Pigmented seed coat, tawny pubescence | Mutation in Williams, 1973 |
| Richland | I, R, t | Yellow seed coat, gray pubescence | Parent line, released 1926 |
| T157 | i, R, t | Imperfect black seed coat, gray pubescence | Mutation in Richland, 1938 |
| UC31 | I, R, t | Yellow seed coat, gray pubescence | Backcrossed line |
| UC33 | i, R, t | Black seed coat, gray pubescence | Backcrossed line |
All cultivars are homozygous for the alleles indicated. The Williams and UC numbers are internal laboratory numbers, whereas the T number refers to the official line designation used in the USDA Germplasm Collection. UC31 and UC33 are independent lines created from repeated backcrossing of the I and i alleles from the lines T201 and UC9 into Clark ii, respectively. Both UC31 and UC33 maintain the CHS HindIII patterns of their nonrecurrent parents in showing the presence of the 12.1-kb fragment and the absence of the 2.3-kb fragment, respectively.
In this article, we demonstrate that silencing of chalcone synthase expression in the dominant I allele takes place in a tissue-specific manner. Secondly, we also determine the relative expression profile of the CHS gene family members in seed coats and cotyledons of the isogenic pairs and attribute the increase in seed coat pigmentation in the recessive i allele to increased expression of CHS7/CHS8. Finally, we corroborate these changes in CHS expression profile and consequential seed coat pigmentation to structural changes taking place at the chalcone synthase inverted repeat of not three but six CHS genes comprising the I locus.
RESULTS
The Dominant I Allele Inhibits CHS Expression Only in the Seed Coats
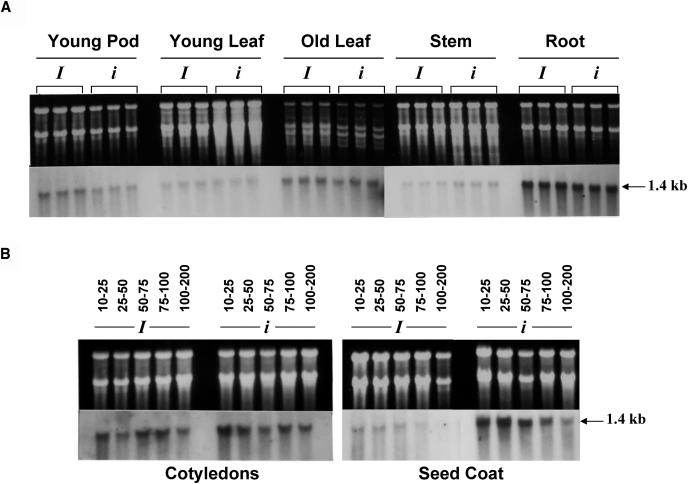
Previous work had shown that the I locus, corresponding to a region of chalcone synthase gene duplications, in its dominant form prevents the accumulation of anthocyanins in the G. max seed coat (Wang et al., 1994; Todd and Vodkin, 1996). Evidently, chalcone synthase, a key regulatory enzyme in the flavonoid pathway, is not only involved in the production of pigment in the seed coat, but plays a significant role in the synthesis of fundamental secondary metabolites functioning as UV protectants, phytoalexins, insect protectants, and symbiosis initiators in various plant tissues. Therefore, it was pertinent to analyze chalcone synthase expression levels in other plant tissues. We examined the steady state CHS mRNA levels in stem, root, young leaf, old leaf, and young pod of the isogenic pair Richland (IRt, yellow seed coat) and T157 (iRt, imperfect black seed coat) by probing RNA gels with an open reading frame CHS probe. The results shown in Figure 1A revealed that in both isolines, roots displayed the highest levels of CHS expression, and reasonable amounts also were detected in young pods and old leaves. Much weaker hybridization to the 1.4-kb RNA was detected in the young leaf and stem. Significantly, no obvious differences of expression were found in CHS transcript levels between the different tissues of the two isolines, thereby suggesting that the suppressive effect of the dominant I allele on CHS expression is absent at least in the vegetative plant parts and the young pod.
Figure 1.
RNA Gel Blot Analysis of CHS in Seven Different Tissues of the Isogenic Lines Richland (I) and T157 (i).
Ten micrograms of total RNA was electrophoresed through 1.2% agarose/3% formaldehyde gel, blotted to nitrocellulose, and probed with a generic CHS probe. The top panel is the ethidium bromide staining of the gel before blotting, whereas the bottom panel reflects the amount of the 1.4-kb CHS mRNA.
(A) CHS mRNA levels in stem, roots, young leaf, and old leaf tissue from 4-week-old plants and young pods from mature plants.
(B) CHS mRNA levels at various stages of seed development in the cotyledons and seed coat. Lanes are marked according to the milligram fresh weight of the seed.
To further elucidate the tissue-specific nature of the silencing effect of the I locus, we decided to analyze CHS expression levels in the developing cotyledons. It has been shown that steady state CHS mRNA levels are reduced in yellow seed coats relative to the pigmented seed coats for all developmental stages (Wang et al., 1994). We therefore compared levels of CHS expression in the seed coats and cotyledons at various stages of seed development for the two isolines, Richland (I) and T157 (i). Hybridization of RNA gel blots to the generic CHS probe revealed that the levels of CHS transcripts did not vary between the cotyledons of the two isolines at all stages of seed development (Figure 1B, panel 1). In both Richland and T157, CHS was expressed at the highest levels very early during seed development (10 to 75 mg) and gradually decreased as the seed matured (100 to 200 mg). However, in striking contrast with the cotyledons, there was a drastic reduction of the 1.4-kb CHS mRNA in seed coats of Richland (I) relative to T157 (i) throughout all developmental stages (Figure 1B, panel 2). CHS transcript levels were highest during very early stages of seed coat development (10 to 75 mg) and decreased as the seed started to reach maturity (75 to 200 mg) in the pigmented isoline T157 (Figure 1B, panel 2). These results suggest that the gene silencing effect of the dominant I allele is seed coat specific.
High Nucleotide Similarity in the CHS Open Reading Frames Necessitates Design of Gene-Specific Primers and Probes in the 5′ and 3′ Untranslated Regions of the Multigene Family
Chalcone synthase constitutes an eight-member gene family in G. max, and all its members have been cloned and sequenced (Akada et al., 1990a, 1990b, 1991, 1993a, 1993b, 1993c; Akada and Dube, 1995). However, the increase in CHS expression levels in pigmented seed coats, as shown in Figure 1, could not be attributed to one or more members of the multigene family because the probe used on RNA gel blots does not distinguish among the different CHS mRNAs. Consequently, it was imperative to obtain quantitative data on the expression of this multigene family using a much more sensitive and powerful technique, TaqMan RT-PCR (Holland et al., 1991). To design gene-specific primers and probes for TaqMan RT-PCR, a detailed investigation of the CHS nucleotide sequences was performed.
Nucleotide sequences of the eight CHS genes, entered into GenBank (Akada et al., 1990a, 1990b, 1991, 1993a, 1993b, 1993c; Akada and Dube, 1995), range primarily from 2 to 2.8 kb in length, with the only exception of CHS3 (4.04 kb). All genes are characterized by the presence of two exons separated by a small intron (122 bp in most of them). Using the sequence analysis program Sequencher, eight members of this multigene family were divided into two subfamilies based on the degree of nucleotide identity in the open reading frame (see supplemental data online). The two subfamilies were 80% identical, with CHS1 to CHS6 genes grouped together and CHS7 and CHS8 forming the second subfamily. As much as 93% nucleotide sequence identity was observed among CHS1 to CHS6 genes, with CHS6 being the more diverged member of this subfamily. The two members of the second subfamily, CHS7 and CHS8, were 97% identical. Using the protein sequence prediction option of another sequence analysis package, Gene Jockey (Biosoft, Firguson, MO), it was estimated that CHS1 to CHS6 genes result in a 388–amino acid polypeptide, whereas CHS7 and CHS8 result in a 389–amino acid polypeptide because of an extra amino acid encoded by the second exon. The 5′ upstream and the 3′ untranslated regions were not conserved in most of the members and showed very little similarity (see supplemental data online). Exceptions to this rule were the 3′ UTR sequences of CHS4 and CHS5, which were base-by-base identical.
The extreme homology among the gene family members necessitated the use of a sensitive and reproducible method, TaqMan RT-PCR. Gene-specific primers and probes were designed for CHS1 to CHS6 either in the 5′ upstream region or the 3′ untranslated region (Table 2). Considering nearly 97% homology between the other two members, CHS7 and CHS8, a common primer-probe set (CHS7/CHS8) spanning a portion of the second exon was used to quantify both CHS7 and CHS8. Additionally, primers and probes also were designed for phosphoenolpyruvate carboxylase (PEPC16) to be used as the endogenous control. Phosphoenolpyruvate carboxylase (PEPC), a ubiquitous cytosolic enzyme in higher plants, catalyzes the irreversible carboxylation of phosphoenolpyruvate to yield oxaloacetate and Pi. It forms a highly similar four-member gene family in G. max (Hata et al., 1998). Using a probe in the 3′ noncoding region, it was shown on RNA gel blots that the levels of one particular member, PEPC16, were similar in the different parts of the G. max plant (Sugimoto et al., 1992). We analyzed expression levels of PEPC16 in the seed coats, cotyledons, roots, and leaves of our standard cultivar, Williams 43, using TaqMan RT-PCR and consistent with the previous study, observed similar levels of PEPC16 in different tissues.
Table 2.
Sequences of Forward and Reverse Primers and Oligo Probes Used in TaqMan RT-PCR for Expression Analysis of CHS Gene Family Members
| Gene | GenBanka | Oligonucleotide | Sequence (5′ to 3′) |
|---|---|---|---|
| CHS1 | AI855764 | CHS1 forward | GCAAGAGAACAAATCTTTCTTTTTTCATAT |
| CHS1 reverse | CAGAAGCATTTGCAGGGCA | ||
| CHS1 probe | ATTCTTGGCTGGCCGGTTTGAAAAAA | ||
| CHS2 | X65636 | CHS2 forward | ACAACAAATCTTTCTTTTTTCATATGTATTG |
| CHS2 reverse | GAAGGCAGGGCAGGGAA | ||
| CHS2 probe | TGGCTGATCAAGGCTTATTCTGTCTTTTGATTT | ||
| CHS3 | X53958 | CHS3 forward | CCAAGTCCTTTTCTTTCTTATTCATTC |
| CHS3 reverse | AAGAAGCATGTGAGGGAAGCAG | ||
| CHS3 probe | TTCATGTTGAGTTTGAAAAATGTATTCTTTCTCTTCCTTT | ||
| CHS4 | X52097 | CHS4 forward | CCTTCCAAGCCACTTTGCA |
| CHS4 reverse | CTGGAGCAAAGGATGAAAGTGA | ||
| CHS4 probe | CATCCATCCAAGCCTTTTCTTTCGTAGATAGC | ||
| CHS5 | L07647 | CHS5 forward | CACTTTGCCACATTCATTCC |
| CHS5 reverse | TGTGAATGAACTAATGAAGCTATAGC | ||
| CHS5 probe | CCTCATACCCTTTTCTTTCGTGCCTAGCTA | ||
| CHS6 | L03352 | CHS6 forward | CGATCCCCATCATTCATATC |
| CHS6 reverse | CCTAATTTTCAATCTCTACCAACAA | ||
| CHS6 probe | TGAGTTTCATTAATTCTTGGGTTCAAAGAAGC | ||
| CHS7 | M98871 | CHS7/CHS8 forward | TAGGCAAGACATGGTGGTGG |
| CHS8 | AY237728 | CHS7/CHS8 reverse | CTTTGACTTTGGCTGGCCC |
| CHS7/CHS8 probe | CTAGGGAAAGAGGCTGCAGTAAAGGCCATAA | ||
| PEPC16 | D10717 | PEPC16 forward | TTCCTTTATCAGAAATAACGAGTTTAGCT |
| PEPC16 reverse | TGTCTCATTTTGCGGCAGC | ||
| PEPC16 probe | CCCTCCCCTGTACCCATGTTTCCATTATAA |
The GenBank accession numbers as shown for the CHS genes represent the genomic sequences submitted by Akada and Dube (1995) except for CHS1, which represents an EST from the Soybean EST collection. These sequences were used to design gene-specific primer-probe combinations for TaqMan RT-PCR.
CHS Gene Family Members Are Expressed in all Tissues but at Varying Levels
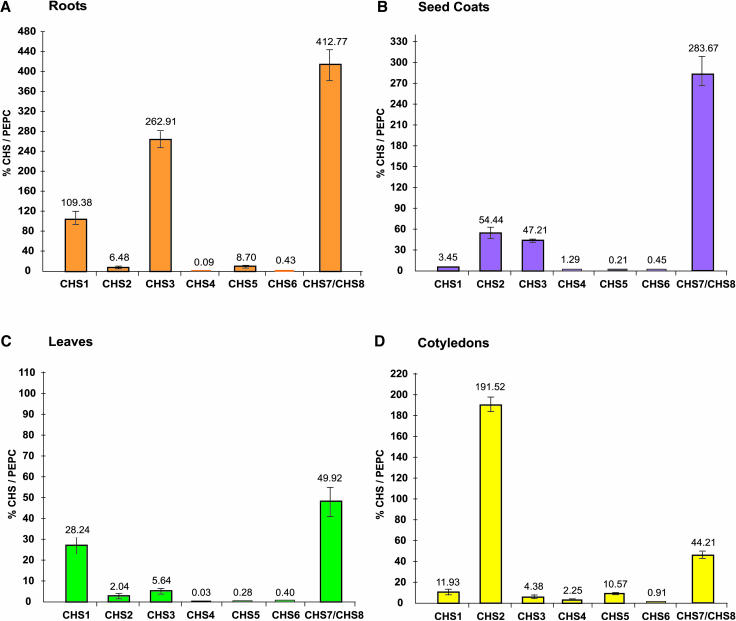
Though multiple CHS genes have been shown to be transcribed in various tissues in some studies, it has been reported for other plants that only one or two members of the gene family account for most of the mRNA produced (Koes et al., 1986; Fukada-Tanaka et al., 1997). This is in agreement with the presence of single CHS genes in Arabidopsis (Arabidopsis thaliana) and Petroselinum crispum (parsley). To ascertain the tissue-specific expression profile, we investigated the relative expression levels of the CHS gene family members in different tissues of our standard G. max cultivar, Williams 43 (ii, yellow seed coats with pigmented hilum) using the gene-specific primers and probes designed for the TaqMan assay. Figure 2 shows the expression levels of all CHS genes in four tissues, namely roots, young leaves, cotyledons, and seed coats. Our results indicate that all members of the CHS family are transcribed, though their relative mRNA amounts might vary considerably from tissue to tissue. On the whole, significantly higher total CHS transcript levels were detected in roots than in any other tissue (Figure 2A). These results substantiate those from RNA gel blot analysis on another yellow seed coat line (Richland, I) presented above (Figure 1). CHS transcripts also have been found in roots of Pisum sativum (pea) and parsley (Koes et al., 1989; Wingender et al., 1989; Harker et al., 1990; Estabrook and Sengupta-Gopalan, 1991). The higher CHS transcript levels in roots reflects the obvious function of synthesis of flavonoid derivatives in the legume roots, which are secreted into the rhizosphere and induce the nod genes of Rhizobium. Moderate accumulation of CHS RNA was observed in seed coats and cotyledons, and very little was found in leaves (Figures 2B to 2D).
Figure 2.
Tissue-Specific Expression Profile of the CHS Gene Family Members in G. max cv Williams 43 as Determined by TaqMan RT-PCR.
Total RNA was isolated from roots and leaves (from 4-week-old plants), seed coats, and cotyledons (50 to 75 mg seed fresh weight), reverse transcribed, and subjected to real-time PCR. Relative amounts were calculated and normalized with respect to PEPC transcript levels (=100%). Data shown represent mean values obtained from three independent amplification reactions, and the error bars indicate the se of the mean. Note that different scales are used in the graphs.
At the transcript level, maximal amount of CHS7/CHS8 expression was detected in roots, followed by seed coats (Figures 2A and 2B). In the cotyledons, on the other hand, CHS2 was nearly fourfold more abundant than CHS7/CHS8 (Figure 2D). The other green tissue, leaves, predominantly expressed CHS1 and CHS7/CHS8 (Figure 2C). Of the three CHS genes constituting the 10-kb inverted duplicated repeat (CHS1, CHS3, and CHS4) (Akada and Dube, 1995), CHS1 and CHS3 transcripts were low to moderate abundance in the four tissues. However, CHS4 transcripts were primarily low abundance in all the tissues.
Pigmentation in the Seed Coats Results from Release of the Silencing Effect Primarily on CHS7/CHS8 Expression
Having shown that we could conclusively differentiate the gene family members at the transcript level, we wanted to answer the key question: Which of the family members is affected by tissue-specific silencing in the seed coats? We performed this investigation on young seed coats (50 to 75 mg seed fresh weight) of the four isogenic pairs described in Table 1.
ii → i Mutations
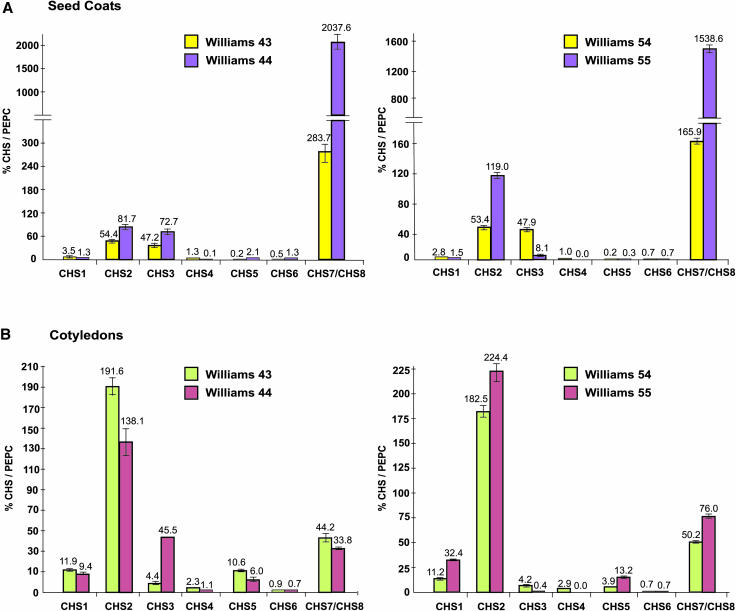
In the first class of mutations, resulting in a completely pigmented seed coat (i) from a pigmented hilum and yellow seed coat (ii), two pairs Williams 43-Williams 44 and Williams 54-Williams 55 were examined. In both of the isogenic pairs, significantly higher total CHS transcripts were detected in the seed coats of the pigmented (Williams 44 and Williams 55) than the hilum pigmented isolines (Williams 43 and Williams 54) (Figure 3A). Strikingly, CHS7/CHS8 transcripts were the primary contributors toward total CHS expression and were nearly sevenfold to ninefold more abundant in the seed coats of the i (Williams 44 and Williams 55) than the ii genotypes (Williams 43 and Williams 54) (Figure 3A). Though moderately expressed in the pigmented and nonpigmented seed coats, CHS2 and CHS3 transcript levels were significantly different only in one isogenic pair (Williams 54-Williams 55). CHS1, CHS4, CHS5, and CHS6 transcripts were the least abundant and did not show any significant differences in their levels of expression in the silenced and the nonsilenced seed coats (Figure 3A). Futhermore, absence of CHS4 transcripts in the isoline Williams 55 could be correlated to a deletion of the CHS4 promoter observed by Todd and Vodkin (1996).
Figure 3.
Expression Profile of the CHS Gene Family Members in Seed Coats and Cotyledons of Isogenic Lines of G. max with the Genotypes ii (Hilum Pigmented Seed Coat) and i (Completely Pigmented Seed Coat).
Total RNA was isolated from seed coats (A) (50 to 75 mg seed fresh weight) and cotyledons (B) (50 to 75 mg seed fresh weight), reverse transcribed, and subjected to real-time PCR. Relative amounts were calculated and normalized with respect to PEPC transcript levels (=100%). Data shown represent mean values obtained from three independent amplification reactions, and the error bars indicate the se of the mean. Note that different scales are used in the graphs. A break in the scale (=) has been incorporated to show the higher amounts of CHS7/CHS8 in the pigmented seed coats.
I → i Mutations
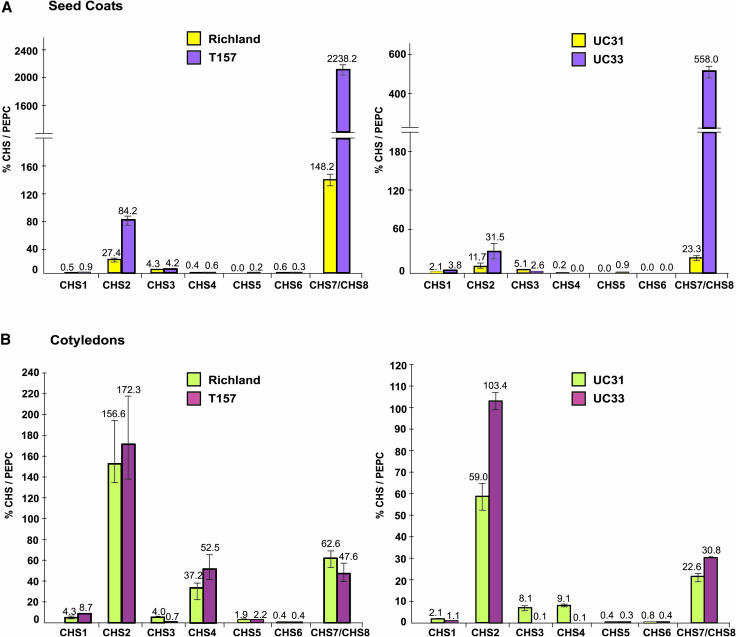
The second set of spontaneous mutations, whereby mutation of the dominant I allele (yellow seed coats) to the recessive i allele (pigmented seed coats) has been correlated with loss of the duplicated CHS1 promoter (Todd and Vodkin, 1996), were analyzed using the two pairs Richland-T157 and UC31-UC33 (Figure 4A). As observed in the first set of spontaneous mutations, there was a significant increase in total CHS transcripts in the pigmented seed coats upon release of silencing (Figure 4A). The increased seed coat pigmentation in the i isolines T157 and UC33 relative to their nonpigmented counterparts Richland and UC31, respectively, was attributed mainly to the higher expression of CHS7/CHS8 and CHS2, the former contributing to as much as a 15-fold increase in T157 and 25-fold increase in UC33 (Figure 4A). The other CHS genes were expressed at fairly low levels, with the exception of CHS6, which was not detected both in UC31 and UC33 (Figure 4A). Also, CHS5 transcripts, absent in the yellow seed coats, were detected at a very low level in the black seed coats. Of the two pairs, CHS4 transcripts were not detected in cultivar UC33, which has been shown to maintain the restiction fragment length polymorphism (RFLP) pattern of its nonrecurrent parent, UC9, exhibiting the loss of the 2.3-kb CHS4 fragment (Todd and Vodkin, 1996).
Figure 4.
Expression Profile of the CHS Gene Family Members in Seed Coats and Cotyledons of Isogenic Lines of G. max with the Genotypes I (Yellow Seed Coat) and i (Completely Pigmented Seed Coat).
Total RNA was isolated from seed coats (A) (50 to 75 mg seed fresh weight) and cotyledons (B) (50 to 75 mg seed fresh weight), reverse transcribed, and subjected to real-time PCR. Relative amounts were calculated and normalized with respect to PEPC transcript levels (=100%). Data shown represent mean values obtained from three independent amplification reactions, and the error bars indicate the se of the mean. Note that different scales are used in the graphs. A break in the scale (=) has been incorporated to show the higher amounts of CHS7/CHS8 in the pigmented seed coats.
TaqMan Expression Profiling of the CHS Gene Family Members in Cotyledons Confirms the Seed Coat–Specific Suppressive Effect of the I Locus
Having attributed pigmentation to increased expression of CHS7/CHS8 in the seed coats, we investigated relative transcript levels of members of this multigene family in a nonsilencing representative tissue, cotyledons (50 to 75 mg seed fresh weight), using TaqMan RT-PCR (Figures 3B and 4B). In cotyledons of both sets of spontaneous mutations, though no significant differences in the total CHS transcript levels were observed among the pigmented and nonpigmented isolines using RNA gel blots (Figure 1B), differences at the individual transcript level were detected in all four pairs using TaqMan RT-PCR (Figures 3B and 4B). Thus, a relative increase in a particular transcript level in the pigmented genotype was being compensated for by a decrease in expression of another member. Unlike the seed coats, CHS2 expression levels were predominantly higher than any of the other genes in all the genotypes, whereas CHS7/CHS8 and CHS4 were moderately expressed (Figures 3B and 4B). The relative levels of CHS2 decreased in the pigmented isoline of one pair (Williams 44) but increased in the other pigmented isoline (Williams 55), though not significantly. A similar kind of variation in expression was observed for many other genes (CHS1, CHS5, and CHS7/CHS8). However, no specific pattern of increase or decrease of transcripts could be identified among the four pairs. This analysis further substantiated the results from RNA gel blots presented above (Figure 1), thereby reinforcing that the suppressive effect of the I locus is seed coat specific.
Silencing Effect in the Seed Coats Is Mediated at All Stages of Seed Development
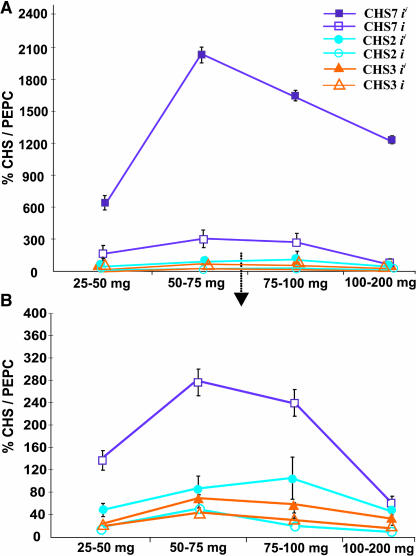
Once it was established that pigmentation in the young seed coats (50 to 75 mg fresh weight range) results from increased expression of CHS7/CHS8, a comparison of the ii (Williams 43) and i (Williams 44) genotypes at different stages of seed development was undertaken using TaqMan RT-PCR to determine if CHS7/CHS8 governs increased expression at all stages of development in the pigmented phenotype. Figure 5 presents the developmental profile of CHS gene family members in seed coats of the two genotypes using TaqMan RT-PCR. As seen with the RNA gel blots (Figure 1B), total CHS expression levels were relatively higher in the completely pigmented seed coats than the hilum pigmented seed coats at all the stages of development. In both of the isolines, CHS7/CHS8 transcripts accounted for most of the expression and CHS2 and CHS3 were moderately expressed, whereas the other genes were very low abundance (Figure 5B). Furthermore, after an initial increase in the developing seed coat (25 to 50 mg stage to the 50 to 75 mg stage), expression levels of all CHS genes tapered off at latter stages of development (Figures 5A and 5B).
Figure 5.
Developmental Profile of Members of the CHS Gene Family in Seed Coats of Williams 43 (ii) and Williams 44 (i).
Total RNA was isolated from seed coats (25 to 50 mg, 50 to 75mg, 75 to 100 mg, and 100 to 200 mg fresh seed weight), reverse transcribed, and subjected to real-time PCR. Relative amounts were calculated and normalized with respect to PEPC transcript levels (=100%). Data shown represent mean values obtained from three independent amplification reactions, and the error bars indicate the se of the mean.
(A) Relative levels of CHS7/CHS8 and other CHS transcripts in the ii and i genotypes over the course of seed development. CHS1, CHS4, CHS5, and CHS6 were expressed at minimal levels (from 0 to 2% CHS/PEPC) in both of the isolines and have not been included in the graph.
(B) An enlarged view of a portion of graph in (A) comparing the relative abundance levels of the CHS2 and CHS3 transcripts in the pigmented and hilum pigmented isolines.
The striking feature, however, was a significant difference in CHS7/CHS8 transcript levels between the pigmented and the hilum pigmented seed coats at all stages of development (Figure 5A). This difference in expression rose from as much as fourfold in the earliest stage of seed coat development (25 to 50 mg) to almost 24-fold in the mature seed. Contrary to significantly higher (twofold to threefold) transcript levels of CHS2 in the pigmented seed coats than the hilum pigmented ones, CHS3 expression differences were not considerable (Figure 5B). These results clearly illustrate that a release of the silencing effect of the ii allele is manifested via an increased expression of most CHS genes at all stages of seed coat development, with CHS7/CHS8 being the key player.
Structural Changes Associated with the 27-kb Duplication Region in Different Alleles of the I Locus
Of the three CHS genes, all arranged as inverted repeats in a 10-kb cluster, CHS1 and CHS3 are in a head-to-head inverted repeat whereas CHS4 and CHS3 are in a tail-to-tail orientation. To obtain an insight into the flanking regions of this 10-kb inverted repeat and be able to elucidate the structural changes associated with this locus, parallel efforts were undertaken to completely sequence a 103-kb BAC harboring an I allele (S.J. Clough, J. Tuteja, M. Li, L. Marek, R. Shoemaker, and L. Vodkin, unpublished data). Interestingly, this sequencing project revealed the existence of this 10-kb CHS cluster in a very gene-rich region. Furthermore, an exact base-by-base duplication of this 10.91-kb CHS cluster was found on the BAC clone. The presence of this duplicated inverted repeat was verified via PCR amplifications of 5′ and 3′ ends of the two clusters along with the spacer region in the genome of our standard G. max variety, Williams (ii allele), from which the BAC library was made (S.J. Clough, J. Tuteja, M. Li, L. Marek, R. Shoemaker, and L. Vodkin, unpublished data).
The presence of an exact duplication of the 10.91-kb CHS inverted repeat in the G. max line carrying the ii allele raised important questions about the presence of this structure in the other I locus alleles and how it might affect seed coat pigmentation. To look for the presence/absence of this 27-kb inverted repeat structure, PCR amplifications of genomic DNA from the four pairs of isogenic lines were performed.
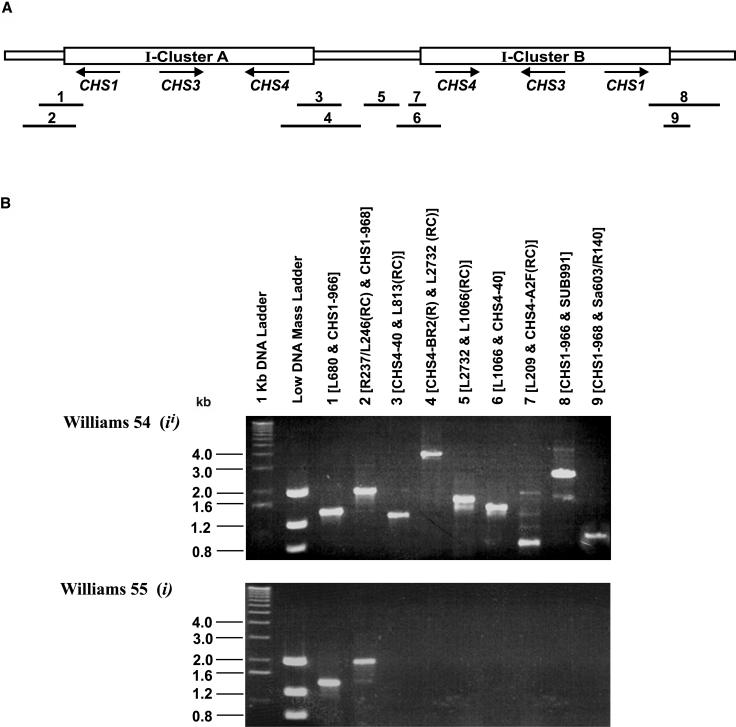
Of the two isogenic pairs analyzed for the first set of spontaneous mutations (ii to i mutation), Williams 55 (i) in comparison to Williams 54 (ii, which was found to contain an intact duplicated repeat) showed complete absence of the second cluster along with the spacer region (Figures 6 and 7). With the tested primers, the deletion extended into the first cluster to as close as 36 bases of the start codon for CHS4, thereby abolishing most of the promoter, including the HindIII site on its way. This is in agreement with the absence of the 2.3-kb HindIII fragment in the CHS RFLP patterns observed by Todd and Vodkin (1996).
Figure 6.
PCR Analysis of the 27-kb CHS Duplicated Inverted Repeat Structure in the Williams 54-Williams 55 Isogenic Pair.
(A) Diagram of the 27-kb duplicated inverted repeat of CHS as revealed by sequencing of the 103-kb BAC. The two 10.91-kb CHS regions are referred to as clusters A and B. The arrows indicate the direction of transcription. Fragments 1 to 9 indicate the relative location of the nine amplicons used to verify the existence of this inverted repeat structure in genomic DNA.
(B) PCR analysis of the 27-kb CHS duplicated inverted repeat in the isogenic pair Williams 54 (ii) and Williams 55 (i). PCR was used to amplify all four ends (5′ and 3′) of the two clusters (A and B) along with the spacer region from G. max genomic DNA using the nine primer combinations. The amplified reactions were separated on a 1% agarose gel, and bands were visualized by ethidium bromide staining. The amplicon number and forward and reverse primers used for each amplicon are indicated above the corresponding lane. Table 3 provides the sequences of the primers used to amplify the nine fragments.
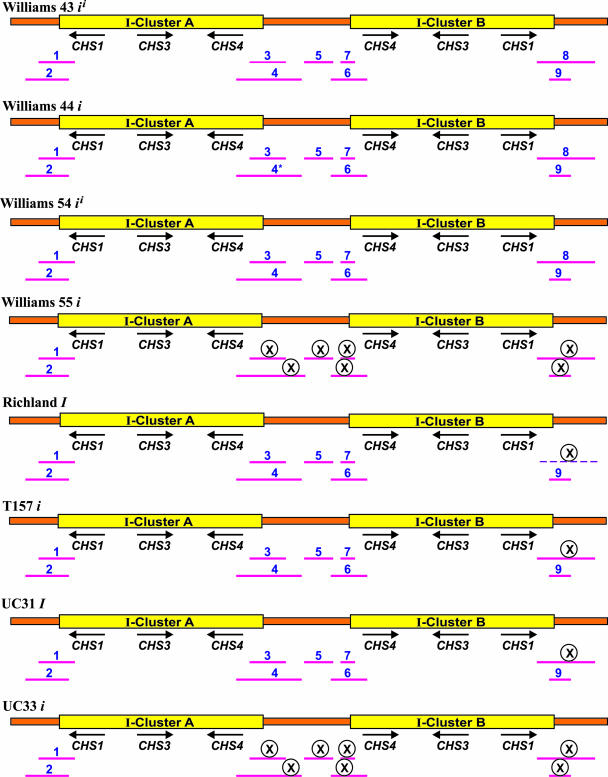
Figure 7.
Structural Analysis of Mutations from the Dominant I or ii Alleles to the Recessive i Allele.
PCR was used to verify the existence and extent of an inverted repeat duplication at the I locus alleles. Nine sets of forward and reverse primers (as given in Figure 6) spanning the 5′ and 3′ ends of the two clusters (A and B) and the spacer region were used to amplify genomic DNA from the eight isolines. The amplified reactions were separated on a 1% agarose gel, and bands were visualized by ethidium bromide staining. The figure is a depiction of the relative presence/absence of the nine fragments in the eight cultivars. The encircles Xs indicate the absence of the corresponding PCR fragment. The asterisk refers to the amplification of this large 3.8-kb region as contiguous subfragments. The dashed line denotes amplification of some but not all parts of this 2.59-kb region as smaller subfragments.
On the other hand, in the other isogenic pair, Williams 43-Williams 44, both the genotypes seemed to show the presence of the two clusters along with the spacer region. Though we could not amplify a 3.8-kb region extending from the promoter of CHS4 in the first cluster into the spacer region (fragment 4) as one intact band in Williams 44, we could successfully amplify this large stretch of DNA as three contiguous subfragments in both Williams 43 and Williams 44 (Figure 7).
PCR amplification studies of the second class of I locus mutations, for example, from a yellow seed coat (I) to a fully pigmented seed coat (i), were performed using the isogenic pair Richland-T157 and UC31-33. In the Richland-T157 pair, presence of the duplicated CHS clusters was verified by the amplification of all fragments except for fragment 8. The 2.59-kb fragment 8, extending further into the 3′ end of the second cluster, could not be amplified in both Richland and T157 (Figure 7). However, some subfragments within this 2.59-kb region amplified in Richland. The absence of the 2.59-kb fragment could potentially indicate some structural divergence downstream of the 3′ end of the second cluster, which may or may not play a role in relieving gene silencing. This hypothesis is supported from RFLP data on the dominant I allele (in Richland), demonstrating the presence of an extra 12.1-kb HindIII fragment that hybridizes to both the CHS1 promoter and the coding region probe, thereby indicating the existence of an extra CHS1 very close to the I locus. A deletion in the promoter of this duplicated CHS1 (referred to as dCHS1 or ICHS1) leading to a shift from 12.1 kb to either a 10.6- or 9.4-kb HindIII fragment, restored a higher level of CHS mRNA in the seed coats (Todd and Vodkin, 1996). Furthermore, the isolation of ICHS1 from another G. max cultivar with the I/I genotype (Miyagi shirome) substantiates this hypothesis (Senda et al., 2002a). Therefore, in light of the RFLP and the presented PCR amplification data, it appears that the site of action in the I allele is adjacent to either sides of the 27-kb inverted repeat present in the ii allele of Williams.
DNA from the second pair in this set of spontaneous mutations, UC31-UC33, also was amplified using the nine primer sets to verify the existence of the 27-kb inverted repeat structure (Figure 7). As mentioned in Table 1, UC31 and UC33 are near isogenic lines. Both these lines have been created from repeated backcrossing of the I and i alleles from the lines T201 (I) and UC9 (i) into Clark ii as the recurrent parent. UC31 and UC33 therefore showed the pattern of their respective nonrecurrent parents on PCR amplification of genomic DNA with the nine primer pairs. UC31 showed the absence of fragment 8 as seen in Richland, whereas UC33, harboring the recessive i allele, showed the complete absence of the 10.91-kb CHS cluster B and the spacer region, as seen in Williams 55.
DISCUSSION
Expression of Closely Related CHS Gene Family Members in Different G. max Tissues as Determined by TaqMan RT-PCR
Of considerable study in plants, chalcone synthase, a member of the plant polyketide superfamily, catalyzes the first committed step of flavonoid biosynthesis. In most dicots, particularly the legumes, chalcone synthase forms a 6- to 12-member multigene family, though it appears to consist of only two copies in the very limited number of grass species studied (Wienand et al., 1986; Rohde et al., 1991; Christensen et al., 1998).
Sequence analysis of CHS genes cloned from several plant species has shown them to share a high degree of nucleotide sequence homology. As part of this study, at least 80% nucleotide identity was observed among all the eight members of the G. max chalcone synthase gene family. Furthermore, sequence analysis classified the gene family into two highly homologous subgroups based on nucleotide similarity in the open reading frame. Previous phylogenetic analysis of ∼90 cDNA sequences encoding CHS from various plant species also classified the G. max genes CHS1, CHS2, CHS3, CHS4, CHS5, and CHS6 as a closely related cluster from the more distantly related CHS7 (Fukada-Tanaka et al., 1997). Interestingly, many other G. max multigene families contain two distinct subgroups of closely related genes (Lee et al., 1983; Hightower and Meagher, 1985; Nielsen et al., 1989), thereby supporting the hypothesis that G. max, subgenus soja, is an ancient tetraploid whose genome has become diploidized over time (Hadley and Hymowitz, 1973).
Although total CHS expression has been widely documented to be developmentally and environmentally regulated in many plant species (van der Meer et al., 1993; Dixon and Paiva, 1995), there are very few studies that have embarked upon the gene-specific expression profiling of this multigene family primarily because of extensive homology between the gene family members and, essentially, lack of a suitable technique. Tissue- and stimulus-specific regulation of the CHS multigene family in G. max has been investigated by different groups (Grab et al., 1985; Wingender et al., 1989; Todd and Vodkin, 1996; Shimizu et al., 1999). The earliest report suggesting the existence of multiple translatable CHS mRNAs came from elicitor-treated G. max cell suspension culture studies containing six different CHS isomers (Grab et al., 1985). Existence of at least three different CHS transcripts in seed coats of G. max was shown via RT-PCR and cDNA cloning (Todd and Vodkin, 1996). However, specific probing of seedling RNA for three different CHS transcripts (CHS1, CHS2, and CHS3) showed the expression of only CHS1 in response to UV light irradiation and elicitor treatment (Wingender et al., 1989). All these approaches, though successful in showing that quite a few CHS genes are transcribed, do not quantify the relative amounts of the gene family members. We have demonstrated effectively the use of a sensitive and highly specific technique, TaqMan RT-PCR, toward studying the relative quantitative expression profile of the closely related CHS gene family members.
Evolution of multigene families has been explained as a gene duplication process that involves the acquisition of new enzymatic activities. Our results with tissue-specific expression profiling show that all CHS genes are transcribed and have diversified expression profiles, although the predicted polypeptides encoded by this gene family are fairly constant. Presumably, this diversification is more closely associated with specialization of tissue-specific expression than to variation in enzymatic functions. Additionally, CHS7/CHS8 are the most abundant transcripts in roots. Confirmatory evidence for this is provided by the analysis of root EST libraries from the G. max EST project (Shoemaker et al., 2002). In libraries Gm-c1028 and Gm-c1004, ∼0.29 and 0.19% of the total ESTs, respectively, are represented by CHS7 and CHS8 (http://www.tigr.org/tdb/tgi/gmgi/). This study clearly illustrates the potential of complementary data from TaqMan expression studies and EST analysis toward aiding attempts to identify possible tissue- and stimulus-specific promoter functions.
Existence of Tandem CHS Clusters in the Different I Locus Isolines
Clustering of some members of the CHS multigene family is a unique feature of leguminous plants, such as P. sativum (An et al., 1993), Phaseolus vulgaris (bean) (Ryder et al., 1987), and Trifolium subterraneum (subterranean clover) (Arioli et al., 1994). In subterranean clover, four CHS genes are tightly clustered in a 15-kb region, whereas in P. sativum, a two gene cluster has been reported. In G. max, analysis of genomic clones from cultivar Williams (ii genotype) has shown the existence of a 10-kb cluster comprising three tandemly arranged chalcone synthase genes (Akada and Dube, 1995) corresponding to the I locus (Todd and Vodkin, 1996). Of the three CHS genes, all arranged as inverted repeats, CHS1 and CHS3 are in a head-to-head inverted repeat, whereas CHS4 and CHS3 are in a tail-to-tail orientation. Close association of the duplicated CHS1 (ICHS1) to CHS3 also has been observed in analysis of genomic clones from a G. max cultivar with I/I genotype, cv Miyagi shirome (Senda et al., 2002a).
Considering the dynamics of this chalcone synthase gene–rich region and its effect on G. max seed coat pigmentation, we sequenced and assembled a 103-kb BAC clone, BAC104J7, harboring the ii allele from cultivar Williams. This BAC clone was found to represent a very gene-rich region of the G. max genome, with ORFs separated on average by ∼3 kb (S.J. Clough, J. Tuteja, M. Li, L. Marek, R. Shoemaker, and L. Vodkin, unpublished data). In addition to showing the characteristic HindIII RFLP pattern for the I locus, complete sequencing of this BAC strikingly revealed the existence of two base-by-base identical, 10.91-kb CHS inverted repeat clusters separated by a 5.8-kb spacer region. PCR amplifications of genomic DNA from different G. max cultivars in our study clearly verified the existence of this 27-kb duplicated structure in the genome.
A recent study has presented evidence that recruitment of new duplicate CHS genes is occurring at a remarkably rapid rate in grass genomes, and these novel genes have been indicated to be functional (Oberholzer et al., 2000). It has been proposed that the different types of CHS gene clusters in the present-day G. max may have arisen through a combination of different processes, including gene duplication, tetraploidization of the genome, and nonallelic gene conversion (Akada and Dube, 1995). Large-scale genome segmental duplications have been demonstrated in both Arabidopsis (Vision et al., 2000) and maize (Zea mays) (Gaut, 2001) and are clearly a common feature of eukaryotic genome evolution. From a functional point of view, large-scale genome duplications create the potential for new diversity and possibility of subfunctionalization (Force et al., 1999).
Furthermore, though the two maize CHS genes, c2 and whp are believed to be the result of an allotetraploid event that occurred some 15 million years ago (Gaut and Doebley, 1997), the 10.91-kb CHS duplication in G. max most likely occurred and was selected for in the recent past, probably during early domestication of G. max, some 10,000 years ago. Also, the dominant yellow alleles (I) are not found in the nondomesticated G. soja, which crosses readily with G. max.
The Highly Structured 27-kb I Locus Directs Silencing of CHS7/CHS8 Specifically in the Seed Coats
Several findings suggest that inverted repeats, both transgenic and endogenous, can be particularly potent silencers of gene expression (Bender and Fink, 1995; Muskens et al., 2000). Many of the IRs are dominant silencing loci, and palindromic sequences up to 15 kb in mice and Petunia hybrida (petunia) appear to be relatively stable (Ehrlich, 1989; Stam et al., 1997). Silencing of CHS expression and consequential abrogation of seed coat pigmentation in G. max by the dominant I and ii loci is an example of an endogenous inverted repeat locus associated with gene silencing (Todd and Vodkin, 1996).
Previously, Todd and Vodkin had associated spontaneous mutations of the dominant I and ii to the nonsilencing i locus to deletions in the promoters of CHS4 and ICHS1, respectively (Todd and Vodkin, 1996). This is consistent with studies indicating that inverted repeats that are either perfect palindromes or quasipalindromes (containing a short spacer between the repeats) frequently undergo deletions and truncations to a much more stable form (Collins, 1980; Akgun et al., 1997). These spontaneous deletions restored a higher level of total CHS mRNA levels, thereby erasing the gene silencing phenomena and yielding pigmented seed coats. How these deletions in the CHS genes manifest themselves via restoration of seed coat pigmentation was intriguing.
Toward determining the precise nature of the I locus–mediated silencing mechanism operating in the G. max seed coat, in this article, our investigations on the relative expression profile of CHS gene family members show that the three CHS genes comprising the I locus are transcribed at comparable although low levels in both the pigmented and the nonpigmented isolines. It has been shown in IR-induced PTGS in transgenic P. hybrida that a high level of transcription of CHS genes is not essential (Stam et al., 1997, 2000). However, the increase in total CHS mRNA levels in the seed coats of both the ii → i and I → i mutations is primarily because of a 7- to 25-fold increase in CHS7/CHS8 transcript levels. Furthermore, this increase in expression of CHS7/CHS8 is seen at all stages of seed coat development in the pigmented isolines. Thus, both of the dominant I and ii alleles inhibit seed coat pigmentation in a trans-dominant manner. There is accumulating evidence that the trans effect might be mediated by very potent molecules, dsRNA, transcribed from the IR genes, and its derivative siRNAs, which induces sequence-specific RNA degradation and establishes PTGS.
Accordingly, it could be speculated that the transcriptional orientation of the two genes comprising the inverted repeat, CHS3 and CHS4, converges, which makes it possible to synthesize dsRNA by read-through transcription. Furthermore, it is not hard to envision the origin of dsRNA from this 27-kb stretch of DNA, comprising six genes in multiple tandem or inverted orientations. Consistent with this hypothesis of dsRNA generation from the inverted CHS repeats, we observed deletion of the second 10.91-kb CHS cluster, extending into the spacer region and further into as close as 36 bp of the start codon of CHS4 in the first cluster, thereby eliminating most of the promoter. This deletion event from the ii → i can then abolish the generation of dsRNA, consequently the silencing mechanism.
In the case of I → i mutations, Todd and Vodkin (1996) using RFLPs have associated the mutation to loss of the duplicated CHS1 (ICHS1) promoter. This CHS1 gene is yet another duplication of CHS1, outside of the 27-kb region containing clusters A and B, as shown in Figure 6. The existence of this duplicated CHS1 (ICHS1) has been confirmed by its isolation from a λ library screen of the G. max cultivar Miyagi shirome (I) (Senda et al., 2002a). Thus, we hypothesize that the dominant I allele encompasses even more than six CHS genes. Additionally, both studies have correlated the I → i mutation to a deletion event, likely to be adjacent to the 27-kb cluster, thereby pointing again to the importance of transcription from these inverted repeats and potential generation of dsRNA in silencing.
A widely accepted feature of dsRNA-induced gene silencing is that the dsRNA is cleaved into small RNAs of 21 to 25 nucleotides, known as siRNAs (Hamilton and Baulcombe, 1999). Consistent with this feature, in our system, putative siRNAs could induce a sequence-specific degradation process for CHS7/CHS8 transcripts, as can be inferred from nearly 7- to 25-fold decrease in their expression levels in the nonpigmented seed coats as determined by RT-PCR. Support for this hypothesis comes from the detection of siRNAs in another yellow seed coat cultivar of G. max, Toyohomare (I) (Senda et al., 2004).
Multiple alignments of the CHS gene family members reveal quite a few stretches of 21 to 25 bp with 100% identity. Furthermore, CHS7 and CHS8 are nearly 80% identical in the transcribed region to the other gene family members. Thus, it can be speculated that the siRNAs triggering sequence-specific degradation of CHS7/CHS8 transcripts originate from the CHS dsRNA from the CHS1/CHS3/CHS4 cluster regions. However, we do not observe complete absence of the CHS7/CHS8 or CHS2 transcripts in the yellow seed coats. This could possibly be explained by the ability of the cells to sense and maintain a critical threshold of transcription of CHS because it is a key metabolic enzyme in the phenylpropanoid pathway leading to the production of other functional proteins involved in plant defense.
Significantly, as part of this study, using two different techniques for measuring CHS transcript levels in the various plant tissues of both the black and yellow isolines, we have provided compelling evidence for the I locus–mediated silencing effect to be seed coat specific, showing IR-mediated tissue-specific silencing in an endogenous plant system. In many transgenic plants, variable and spatial patterns of PTGS have been observed, for example, the simple and complex vein patterns of flower pigmentation of CHS PTGS in P. hybrida (Que et al., 1997). Grafting experiments in transgenic Nicotiana tabacum (tobacco) have provided direct evidence for the transmission of PTGS over long distances, a process called systemic acquired silencing (Palauqui et al., 1997; Voinnet and Baulcombe, 1997). However, in our system, silencing is restricted to one tissue type.
Many studies have documented the ability of certain viral proteins in overcoming the plant antiviral defense mechanism and thereby suppressing RNA silencing (Anandalakshmi et al., 1998; Kasschau and Carrington, 1998; Guo and Ding, 2002). Release of CHS PTGS in the G. max seed coats by a Cucumber mosaic virus suppressor protein has been shown by Senda et al., 2004. If our system is similar to viral-induced gene silencing, any mechanism would have to explain localization of silencing to one tissue type. In our system, it is possible that specialized proteins that bind and traffic RNA (Xoconostle-Cazares et al., 1999) are lacking, or else there exists an endogenous barrier that impedes the systemic transmission of this signal via the plant vasculature to other tissues. Alternatively, even though there is a diffusible signal being transmitted from the silenced seed coats, it is not being accepted in other tissues. Competence to respond to silencing signals has been shown to be developmentally regulated (Palauqui et al., 1996). Despite some parallels to viral-induced gene silencing, our tissue-specific silencing does not completely mimic it.
To summarize, we have described an endogenous duplicated IR repeat system in G. max that drives silencing of CHS genes in a novel tissue-specific manner, thereby inhibiting pigmentation of the seed coats. Further study of this system should provide insight into the mechanism of tissue-specific gene silencing, which could potentially be of practical use to target silencing to a restricted tissue/cell type.
METHODS
Plant Materials and Genetic Nomenclature
All isolines of G. max used for this study are described in Table 1. They were obtained from the USDA Soybean Germplasm Collections (Department of Crop Sciences, USDA Agricultural Research Service, University of Illinois), which totals >18,000 plant introductions. All lines are homozygous for the loci indicated, and for brevity, only one allele is indicated in the text and figures. Plants were grown in the greenhouse, and tissue was harvested from at least four plants of each isoline. Shoot tips, leaves, and roots were harvested from 4-week-old plants and frozen in liquid nitrogen. For seed coats and cotyledons, pods of mature plants were first harvested and seeds extracted from the pods and divided into the following categories by fresh weight of the entire seed: 10 to 25 mg, 25 to 50 mg, 50 to 75 mg, 75 to 100 mg, and 100 to 200 mg. Seed coats and cotyledons were then dissected from the seeds and frozen in liquid nitrogen. The frozen tissues were then freeze dried (Multi-Dry Lyophilizer; FTS Systems, Stone Ridge, NY) and stored at −20°C.
RNA Extraction and Gel Blot Analysis
Total RNA was extracted from freeze-dried tissue using the SDS/phenol chloroform method and lithium chloride precipitation (McCarty, 1986; Wang et al., 1994). RNA samples were quantitated by spectrophotometry, and the integrity confirmed using agarose gel electrophoresis (Sambrook et al., 1989). RNA was stored at −70°C until further use.
For RNA gel blot analysis, 10 μg of total RNA containing 400 ng of ethidium bromide was electrophoresed through 1.2% agarose/3% formaldehyde gels (Sambrook et al., 1989). The RNA gels were then blotted onto nitrocellulose membranes (Schleicher and Schuell, Keene, NH) via capillary action with 10× SSC (1.5 M NaCl and 0.15 M sodium citrate, adjusted pH to 7.0) overnight. RNA was cross-linked to the nitrocellulose membranes with UV radiation by a UV cross-linker (Stratagene, La Jolla, CA). Nitrocellulose RNA gel blots were then prehybridized, hybridized, washed, and exposed to Hyperfilm (Amersham, Piscataway, NJ) as described by Todd and Vodkin (1996).
The probe for CHS was a 2.0-kb HindIII fragment prepared from a genomic CHS subclone, pC2H2.0, which was isolated with a P. vulgaris gene and identified by sequence data to have 90% sequence similarity to the P. vulgaris gene (Frank and Vodkin, 1988). The 2.0-kb HindIII fragment representing the CHS6 open reading frame hybridizes with all CHS genes. It contains an ∼600-bp intron. From 20 to 100 ng of purified DNA was labeled with [α-32P]dATP by random primer reaction method (Feinberg and Vogelstein, 1983). After labeling, unincorporated nucleotides were removed using a Bio-Spin chromatography column (Bio-Rad, Hercules, CA).
Real-Time Quantitative RT-PCR
RNA Purification and cDNA Synthesis
A 100-μg aliquot of extracted RNA was subjected to another round of acid phenol chloroform extraction, pH 4.7 (Ambion, Austin, TX), to eliminate residual genomic DNA present in the preparation. To further ensure the purity of the RNA samples, the acid phenol chloroform–extracted RNA was treated with DNase I using Ambion's DNA-free kit. Purified RNA was concentrated using Microcon YM-30 columns (Millipore, Bedford, MA). RNA extractions were stored at −70°C.
A 1-μg aliquot of purified RNA was then reverse transcribed in quadruplicate in a 20-μL reaction volume using the Superscript II first strand cDNA synthesis system for RT-PCR according to the manufacturer's instructions (Invitrogen, Carlsbad, CA). Parallel reactions for each RNA sample were run in the absence of Superscript II (no RT control) to assess any genomic DNA contamination. The reaction was terminated by heat inactivation at 70°C for 15 min. Subsequently, the cDNA product was treated with 2 units of RNase H for 20 min at 37°C. The cDNA samples for each tissue were pooled and stored at −20°C.
Design of Gene-Specific TaqMan Primers and Probes
The TaqMan 5′ nuclease assay enables the measurement of an accumulating PCR product by utilizing a dual-labeled TaqMan fluorogenic probe, designed specificly to the nucleotide sequence of the target RNA or transcript to be measured (Holland et al., 1991). TaqMan chemistry uses the endogenous 5′ to 3′ nuclease activity of Taq polymerase during strand elongation to sequentially cleave the probe, which is bound to the template. Upon probe cleavage, fluorescence from the reporter dye is emitted, and thus, a detectable signal is generated during amplification.
To design gene-specific primers and probes, a detailed analysis of the nucleotide sequences of the eight CHS genes was performed using Sequencher version 4 (Gene Codes, Ann Arbor, MI). Keeping in mind the extensive homology among the CHS gene family members, gene-specific regions (5′ upstream sequences or 3′ untranslated regions) were identified. Primer Express (Applied Biosystems, Foster City, CA) was then used to design gene-specific primer-probe sets for genes CHS1 to CHS6, whereas the seventh primer-probe set was used to quantify both CHS7 and CHS8 transcript levels. Additonally, primers and probes specific to the endogenous reference PEPC16 also were designed. The amplified products from these primer-probe sets ranged in size from 81 to 114 bp. The 5′ and 3′ ends of these probes were labeled with fluorescent dyes FAM (6-carboxyfluorescein) and TAMRA (6-carboxy-tetramethyl-rhodamine), respectively. Also, BLASTN searches against dbEST and nr (nonredundant set of GenBank, EMBL, and DDJB database sequences) were conducted to confirm the total gene specificity of the nucleotide sequences chosen for the primers and probes. The fluorescent probes were ordered from Applied Biosystems, and the forward and reverse primers were synthesized at the Keck Center (University of Illinois Biotechnology Center). Details of these are presented in Table 2.
PCR Amplification
TaqMan RT-PCR assays for each gene target per tissue were performed in triplicate on cDNA samples or no RT control samples on an ABI Prism 7700 sequence detection system (Applied Biosystems). Parallel amplifications using the same cDNA pools were performed using primers and probes to the endogenous reference and normalizer PEPC16. From the pooled cDNA, 2 μL of the RT reaction was used as a template in a 25-μL PCR reaction containing 1× TaqMan buffer, 0.4 μM forward and reverse primers, 0.2 μM probe, 0.2 mM dATP, dCTP, and dGTP, 0.4 mM dUTP, 3.5 mM MgCl2, and 0.025 units/μL of AmpliTaq Gold DNA polymerase. The PCR thermal cycling parameters were 50°C for 2 min, 95°C for 10 min followed by 40 cycles of 95°C for 15 s and 60°C for 1 min. This experiment was replicated thrice.
Data Analysis
Data were captured as amplification plots. Transcript levels of the CHS gene family members were measured relative to the endogenous reference PEPC. All calculations and statistical analysis were performed as described in the ABI 7700 sequence detection system User Bulletin 2 (Applied Biosystems). Amplification efficiency (75 to 95%) for the eight primer-probe sets was determined by amplification of cDNA reverse transcribed from a dilution series of a pool of RNA (Williams 43) using 2, 1, 0.5, and 0.25 μg per reaction (data not shown).
Confirmation of Primer/Probe Specificity
Specificity of the RT-PCR products was documented with high-resolution gel electrophoresis and resulted in a single product with desired length.
DNA Isolation and PCR Amplifications
G. max genomic DNA was isolated from lyophilized shoot tips of the eight cultivars using the method of Dellaporta et al. (1983) with minor modifications. The nuclease inhibitor O-phenanthroline (10 mM) was added to the extraction buffer, and the hexadecyltrimethylammonium bromide step was omitted. Sequencing of a G. max BAC, BACJ0147, harboring the ii allele, in our lab has shown the existence of a base-by-base duplication of the 10-kb inverted CHS repeat (S.J. Clough, J. Tuteja, M. Li, L. Marek, R. Shoemaker, and L. Vodkin, unpublished data). To look for the existence of this duplication in G. max lines containing different I locus alleles, nine sets of forward and reverse primers were designed to amplify the 5′ and 3′ ends of the two 10-kb repeats and the spacer region (as given in Table 3).
Table 3.
Sequences for the 15 Oligonucleotides Used as Forward or Reverse Primers for Verification of the Existence of the 27-kb CHS Duplicated Inverted Repeat Structure
| Primer | Sequence (5′ to 3′) |
|---|---|
| CHS1-966 | GGAGCACCATGTGACGGAGAAG |
| CHS1-968 | CTTACCCCCTCTACCAAACACACC |
| CHS4-40 | CTTATATCCCACAACTTCTTAAC |
| CHS4-A2 (F) | AAGCTTCATCACCCACTTAT |
| CHS4-BR2 (R) | TGAAGTAGTAGCTATCTACG |
| L1066 | CTTATTATCCACCCTCACTCC |
| L1066 (RC) | GGAGTGAGGGTGGATAATAAG |
| L209 | TTCCCGATCAGATTGTTGTTC |
| L2732 | AGCGAGAGGAGAGCGGAGTG |
| L2732 (RC) | CACTCCGCTCTCCTCTCGCT |
| L680 | GAAAATAGAAGAGTTGGTTGAGG |
| L813 (RC) | GGTGACGCTAGGGTTGAGGGAG |
| R237/L246(RC) | CACCATCAGTGACCATTAAAC |
| Sa603/R140 | GAATAAATGGAGCTTAGTTTG |
| SUB991 | CTGGAAGAAAGCTAGATTTGG |
PCR amplifications were performed in a 50-μL reaction volume using 500 ng of genomic DNA in the presence of 1× PCR buffer, 1.5 mM MgCl2, 0.2 mM deoxynucleotide triphosphate, 2.5 units of Taq polymerase (Invitrogen), and 0.4 μM forward and reverse primers. PCR reactions were conducted in a PTC-100 programmable thermocycler (MJ Research, Watertown, MA) via an initial denaturation step at 96°C for 2 min followed by 40 cycles of denaturing at 96°C for 20 s, annealing at 55°C for 1 min, and polymerization at 72°C for 2 min, to end with a 7-min extension at 72°C. The amplified reactions were separated on a 1% agarose gel and bands visualized via ethidium bromide staining.
Supplementary Material
Acknowledgments
We are grateful for the Physiological and Molecular Plant Biology Fellowship Award from the Interdisciplinary Physiological and Molecular Plant Biology Program, University of Illinois, to J.H.T. We thank Mark Band for advice on TaqMan RT-PCR. This work was supported by grants from the Illinois Soybean Program Operating Board, University of Illinois Research Board, and National Science Foundation (DBI9872565) to L.O.V.
Online version contains Web-only data.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Lila Vodkin (l-vodkin@uiuc.edu).
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.021352.
References
- Akada, S., and Dube, S.K. (1995). Organization of soybean chalcone synthase gene clusters and characterization of a new member of the family. Plant Mol. Biol. 29, 189–199. [DOI] [PubMed] [Google Scholar]
- Akada, S., Kung, S.D., and Dube, S.K. (1990. a). The nucleotide sequence of gene 3 of the soybean chalcone synthase multigene family. Nucleic Acids Res. 18, 5899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akada, S., Kung, S.D., and Dube, S.K. (1990. b). Nucleotide sequence of one member of soybean chalcone synthase multi-gene family. Nucleic Acids Res. 18, 3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akada, S., Kung, S.D., and Dube, S.K. (1991). The nucleotide sequence of gene 1 of the soybean chalcone synthase multigene family. Plant Mol. Biol. 16, 751–752. [DOI] [PubMed] [Google Scholar]
- Akada, S., Kung, S.D., and Dube, S.K. (1993. a). Nucleotide sequence of a soybean chalcone synthase gene with a possible role in ultraviolet-B sensitivity, Gmchs6. Plant Physiol. 102, 699–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akada, S., Kung, S.D., and Dube, S.K. (1993. b). Nucleotide sequence and putative regulatory elements of gene 2 of the soybean (Glycine max) chalcone synthase multigene family. Plant Physiol. 102, 317–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akada, S., Kung, S.D., and Dube, S.K. (1993. c). Nucleotide sequence and putative regulatory elements of a nodule-development-specific member of the soybean (Glycine max) chalcone synthase multigene family, Gmchs 7. Plant Physiol. 102, 321–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akgun, E., Zahn, J., Baumes, S., Brown, G., Liang, F., Romanienko, P.J., Lewis, S., and Jasin, M. (1997). Palindrome resolution and recombination in the mammalian germ line. Mol. Cell. Biol. 17, 5559–5570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An, C., Ichinose, Y., Yamada, T., Tanaka, Y., Shiraishi, T., and Oku, H. (1993). Organization of the genes encoding chalcone synthase in Pisum sativum. Plant Mol. Biol. 21, 789–803. [DOI] [PubMed] [Google Scholar]
- Anandalakshmi, R., Pruss, G.J., Ge, X., Marathe, R., Mallory, A.C., Smith, T.H., and Vance, V.B. (1998). A viral suppressor of gene silencing in plants. Proc. Natl. Acad. Sci. USA 95, 13079–13084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arioli, T., Howles, P.A., Weinman, J.J., and Rolfe, B.G. (1994). In Trifolium subterraneum, chalcone synthase is encoded by a multigene family. Gene 138, 79–86. [DOI] [PubMed] [Google Scholar]
- Bender, J., and Fink, G.R. (1995). Epigenetic control of an endogenous gene family is revealed by a novel blue fluorescent mutant of Arabidopsis. Cell 83, 725–734. [DOI] [PubMed] [Google Scholar]
- Bernard, R.L., and Weiss, M.G. (1973). Qualitative genetics. In Soybeans: Improvement, Production, and Uses, 1st ed., B.E. Caldwell, ed (Madison, WI: American Society of Agronomy), pp. 117–149.
- Carrington, J.C., and Ambros, V. (2003). Role of microRNAs in plant and animal development. Science 301, 336–338. [DOI] [PubMed] [Google Scholar]
- Christensen, A.B., Gregersen, P.L., Schroder, J., and Collinge, D.B. (1998). A chalcone synthase with an unusual substrate preference is expressed in barley leaves in response to UV light and pathogen attack. Plant Mol. Biol. 37, 849–857. [DOI] [PubMed] [Google Scholar]
- Collins, J. (1980). Instability of palindromic DNA in Escherichia coli. Cold Spring Harb. Symp. Quant. Biol. 45, 409–416. [DOI] [PubMed] [Google Scholar]
- Dellaporta, S.L., Wood, J., and Hicks, J.B. (1983). A rapid DNA minipreparation, version II. Plant Mol. Biol. Rep. 2, 21–42. [Google Scholar]
- Dixon, R.A., and Paiva, N.L. (1995). Stress-induced phenylpropanoid metabolism. Plant Cell 7, 1085–1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrlich, S.D. (1989). Illegitimate recombination in bacteria. In Mobile DNA, D.E. Berg and M.M Howe, eds (Washington, D.C.: American Society for Microbiology), pp. 799–832.
- Estabrook, E.M., and Sengupta-Gopalan, C. (1991). Differential expression of phenylalanine ammonia-lyase and chalcone synthase during soybean nodule development. Plant Cell 3, 299–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg, A.P., and Vogelstein, B. (1983). A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal. Biochem. 132, 6–13. [DOI] [PubMed] [Google Scholar]
- Force, A., Lynch, M., Pickett, F.B., Amores, A., Yan, Y.L., and Postlethwait, J. (1999). Preservation of duplicate genes by complementary, degenerative mutations. Genetics 151, 1531–1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank, R.L., and Vodkin, L. (1988). Identification of chalcone synthase and phenylalanine ammonia lyase genes in soybean. Soyb. Genet. Newsl. 15, 181. [Google Scholar]
- Fukada-Tanaka, S., Hoshino, A., Hisatomi, Y., Habu, Y., Hasebe, M., and Iida, S. (1997). Identification of new chalcone synthase genes for flower pigmentation in the Japanese and common morning glories. Plant Cell Physiol. 38, 754–758. [DOI] [PubMed] [Google Scholar]
- Gaut, B.S. (2001). Patterns of chromosomal duplication in maize and their implications for comparative maps of the grasses. Genome Res. 11, 55–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaut, B.S., and Doebley, J.F. (1997). DNA sequence evidence for the segmental allotetraploid origin of maize. Proc. Natl. Acad. Sci. USA 94, 6809–6814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grab, D., Loyal, R., and Ebel, J. (1985). Elicitor-induced phytoalexin synthesis in soybean cells: Changes in the activity of chalcone synthase mRNA and the total population of translatable mRNA. Arch. Biochem. Biophys. 243, 523–529. [DOI] [PubMed] [Google Scholar]
- Guo, H.S., and Ding, S.W. (2002). A viral protein inhibits the long range signaling activity of the gene silencing signal. EMBO J. 21, 398–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadley, H.H., and Hymowitz, T. (1973). Speciation and cytogenetics. In Soybeans: Improvement, Production, and Uses, 1st ed., B.E. Caldwell, ed (Madison, WI: American Society of Agronomy), pp. 97–116.
- Hamilton, A.J., and Baulcombe, D.C. (1999). A species of small antisense RNA in posttranscriptional gene silencing in plants. Science 286, 950–952. [DOI] [PubMed] [Google Scholar]
- Harker, C.L., Ellis, T.H., and Coen, E.S. (1990). Identification and genetic regulation of the chalcone synthase multigene family in pea. Plant Cell 2, 185–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hata, S., Izui, K., and Kouchi, H. (1998). Expression of a soybean nodule-enhanced phosphoenolpyruvate carboxylase gene that shows striking similarity to another gene for a house-keeping isoform. Plant J. 13, 267–273. [DOI] [PubMed] [Google Scholar]
- Hightower, R.C., and Meagher, R.B. (1985). Divergence and differential expression of soybean actin genes. EMBO J. 4, 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland, P.M., Abramson, R.D., Watson, R., and Gelfand, D.H. (1991). Detection of specific polymerase chain reaction product by utilizing the 5′----3′ exonuclease activity of Thermus aquaticus DNA polymerase. Proc. Natl. Acad. Sci. USA 88, 7276–7280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobsen, S.E., Running, M.P., and Meyerowitz, E.M. (1999). Disruption of an RNA helicase/RNAse III gene in Arabidopsis causes unregulated cell division in floral meristems. Development 126, 5231–5243. [DOI] [PubMed] [Google Scholar]
- Jorgensen, R. (1990). Altered gene expression in plants due to trans interactions between homologous genes. Trends Biotechnol. 8, 340–344. [DOI] [PubMed] [Google Scholar]
- Kasschau, K.D., and Carrington, J.C. (1998). A counter defensive strategy of plant viruses: Suppression of post-transcriptional gene silencing. Cell 95, 461–470. [DOI] [PubMed] [Google Scholar]
- Koes, R.E., Spelt, C.E., Reif, H.J., van den Elzen, P.J., Veltkamp, E., and Mol, J.N. (1986). Floral tissue of Petunia hybrida (V30) expresses only one member of the chalcone synthase multigene family. Nucleic Acids Res. 14, 5229–5239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koes, R.E., Spelt, C.E., van den Elzen, P.J., and Mol, J.N. (1989). Cloning and molecular characterization of the chalcone synthase multigene family of Petunia hybrida. Gene 81, 245–257. [DOI] [PubMed] [Google Scholar]
- Lee, J.S., Brown, G.G., and Verma, D.P. (1983). Chromosomal arrangement of leghemoglobin genes in soybean. Nucleic Acids Res. 11, 5541–5553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarty, D. (1986). A simple method for the extraction of RNA from maize tissue. Maize Genet. Coop. News Lett. 60, 61. [Google Scholar]
- Meins, F., Jr. (2000). RNA degradation and models for post-transcriptional gene-silencing. Plant Mol. Biol. 43, 261–273. [DOI] [PubMed] [Google Scholar]
- Meyer, P., and Saedler, H. (1996). Homology-dependent gene silencing in plants. Annu. Rev. Plant Physiol. 47, 23–48. [DOI] [PubMed] [Google Scholar]
- Muskens, M.W., Vissers, A.P., Mol, J.N., and Kooter, J.M. (2000). Role of inverted DNA repeats in transcriptional and post-transcriptional gene silencing. Plant Mol. Biol. 43, 243–260. [DOI] [PubMed] [Google Scholar]
- Nielsen, N.C., Dickinson, C.D., Cho, T.J., Thanh, V.H., Scallon, B.J., Fischer, R.L., Sims, T.L., Drews, G.N., and Goldberg, R.B. (1989). Characterization of the glycinin gene family in soybean. Plant Cell 1, 313–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberholzer, V., Durbin, M.L., and Clegg, M.T. (2000). Comparative genomics of chalcone synthase and Myb genes in the grass family. Genes Genet. Syst. 75, 1–16. [DOI] [PubMed] [Google Scholar]
- Palauqui, J.C., Elmayan, T., Deborne, F.D., Crete, P., Charles, C., and Vaucheret, H. (1996). Frequencies, timing, and spatial patterns of co-suppression of nitrate reductase and nitrite reductase in transgenic tobacco plants. Plant Physiol. 112, 1447–1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palauqui, J.C., Elmayan, T., Pollien, J.M., and Vaucheret, H. (1997). Systemic acquired silencing—Transgene-specific post-transcriptional silencing is transmitted by grafting from silenced stocks to non-silenced scions. EMBO J. 16, 4738–4745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer, R.G., and Kilen, T.C. (1987). Qualitative genetics. In Soybeans: Improvement, Production, and Uses, 2nd ed., J.R. Wilcox, ed (Madison, WI: American Society of Agronomy), pp. 135–209.
- Plasterk, R.H.A. (2002). RNA silencing: The genome's immune system. Science 296, 1263–1265. [DOI] [PubMed] [Google Scholar]
- Que, Q.D., Wang, H.Y., English, J.J., and Jorgensen, R.A. (1997). The frequency and degree of cosuppression by sense chalcone synthase transgenes are dependent on transgene promoter strength and are reduced by premature nonsense codons in the transgene coding sequence. Plant Cell 9, 1357–1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhart, B.J., Weinstein, E.G., Rhoades, M.W., Bartel, B., and Bartel, D.P. (2002). MicroRNAs in plants. Genes Dev. 16, 1616–1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohde, W., Dorr, S., Salamini, F., and Becker, D. (1991). Structure of a chalcone synthase gene from Hordeum vulgare. Plant Mol. Biol. 16, 1103–1106. [DOI] [PubMed] [Google Scholar]
- Ryder, T.B., Hedrick, S.A., Bell, J.N., Liang, X.W., Clouse, S.D., and Lamb, C.J. (1987). Organization and differential activation of a gene family encoding the plant defense enzyme chalcone synthase in Phaseolus vulgaris. Mol. Gen. Genet. 210, 219–233. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., Fritsch, E.F., and Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual, 2nd ed. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Senda, M., Jumonji, A., Yumoto, S., Ishikawa, R., Harada, T., Niizeki, M., and Akada, S. (2002. b). Analysis of the duplicated CHS1 gene related to the suppression of the seed coat pigmentation in yellow soybeans. Theor. Appl. Genet. 104, 1086–1091. [DOI] [PubMed] [Google Scholar]
- Senda, M., Kasai, A., Yumoto, S., Akada, S., Ishikawa, R., Harada, T., and Niizeki, M. (2002. a). Sequence divergence at chalcone synthase gene in pigmented seed coat soybean mutants of the Inhibitor locus. Genes Genet. Syst. 77, 341–350. [DOI] [PubMed] [Google Scholar]
- Senda, M., Masuta, C., Ohnishi, S., Goto, K., Kasai, A., Sano, T., Hong, J.-S., and MacFarlane, S. (2004). Patterning of virus-infected Glycine max seed coat is associated with suppression of endogenous silencing of chalcone synthase genes. Plant Cell 16, 807–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu, T., Akada, S., Senda, M., Ishikawa, R., Harada, T., Niizeki, M., and Dube, S.K. (1999). Enhanced expression and differential inducibility of soybean chalcone synthase genes by supplemental UV-B in dark-grown seedlings. Plant Mol. Biol. 39, 785–795. [DOI] [PubMed] [Google Scholar]
- Shoemaker, R., et al. (2002). A compilation of soybean ESTs: Generation and analysis. Genome 45, 329–338. [DOI] [PubMed] [Google Scholar]
- Stam, M., de Bruin, R., Kenter, S., van der Hoorn, R.A., van Blokland, R., Mol, J.N., and Kooter, J.M. (1997). Post-transcriptional silencing of chalcone synthase in Petunia by inverted transgene repeats. Plant J. 12, 63–82. [Google Scholar]
- Stam, M., de Bruin, R., van Blokland, R., van der Hoorn, R.A., Mol, J.N., and Kooter, J.M. (2000). Distinct features of post-transcriptional gene silencing by antisense transgenes in single copy and inverted T-DNA repeat loci. Plant J. 21, 27–42. [DOI] [PubMed] [Google Scholar]
- Sugimoto, T., Kawasaki, T., Kato, T., Whittier, R.F., Shibata, D., and Kawamura, Y. (1992). cDNA sequence and expression of a phosphoenolpyruvate carboxylase gene from soybean. Plant Mol. Biol. 20, 743–747. [DOI] [PubMed] [Google Scholar]
- Todd, J.J., and Vodkin, L.O. (1996). Duplications that suppress and deletions that restore expression from a chalcone synthase multigene family. Plant Cell 8, 687–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Meer, I.M., Mol, J.N., and Stuitje, A.R. (1993). Regulation of general phenylpropanoid and flavonoid gene expression. In Control of Plant Gene Expression, D.A.V Pal, ed (Boca Raton, FL: CRC Press), pp. 125–155.
- Vision, T.J., Brown, D.G., and Tanksley, S.D. (2000). The origins of genomic duplications in Arabidopsis. Science 290, 2114–2117. [DOI] [PubMed] [Google Scholar]
- Voinnet, O., and Baulcombe, D.C. (1997). Systemic signalling in gene silencing. Nature 389, 553. [DOI] [PubMed] [Google Scholar]
- Wang, C.S., Todd, J.J., and Vodkin, L.O. (1994). Chalcone synthase mRNA and activity are reduced in yellow soybean seed coats with dominant I alleles. Plant Physiol. 105, 739–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassenegger, M. (2000). RNA-directed DNA methylation. Plant Mol. Biol. 43, 203–220. [DOI] [PubMed] [Google Scholar]
- Wassenegger, M. (2002). Gene silencing-based disease resistance. Transgenic Res. 11, 639–653. [DOI] [PubMed] [Google Scholar]
- Wienand, U., Weydemann, U., Niesbach-Kloesgen, U., Peterson, P.A., and Saedler, H. (1986). Molecular cloning of the c2 locus of Zea mays, the gene coding for chalcone synthase. Mol. Gen. Genet. 203, 202–207. [Google Scholar]
- Wilcox, J.R. (1988). Performance and use of seed coat mutants in soybean. Crop Sci. 28, 30–32. [Google Scholar]
- Wingender, R., Rohrig, H., Horicke, C., Wing, D., and Schell, J. (1989). Differential regulation of soybean chalcone synthase genes in plant defence, symbiosis and upon environmental stimuli. Mol. Gen. Genet. 218, 315–322. [DOI] [PubMed] [Google Scholar]
- Xoconostle-Cazares, B., Yu, X., Ruiz-Medrano, R., Wang, H.L., Monzer, J., Yoo, B.C., McFarland, K.C., Franceschi, V.R., and Lucas, W.J. (1999). Plant paralog to viral movement protein that potentiates transport of mRNA into the phloem. Science 283, 94–98. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.