Abstract
Genetic and biochemical studies have revealed that the diversity of cell types and developmental patterns evident within the animal kingdom is generated by a handful of conserved, core modules. Core biological modules must be robust, able to maintain functionality despite perturbations, and yet sufficiently adaptable for random mutations to generate phenotypic variation during evolution. Understanding how robust, adaptable modules have influenced the evolution of eukaryotes will inform both evolutionary and synthetic biology. One such system is the MAP kinase module, which consists of a 3-tiered kinase circuit configuration that has been evolutionarily conserved from yeast to man. MAP kinase signal transduction pathways are used across eukaryotic phyla to drive biological functions that are crucial for life. Here we ask the fundamental question, why do MAPK modules follow a conserved 3-tiered topology rather than some other number? Using computational simulations, we identify a fundamental 2-tiered circuit topology that can be readily reconfigured by feedback loops and scaffolds to generate diverse signal outputs. When this 2-kinase circuit is connected to proximal input kinases, a 3-tiered modular configuration is created that is both robust and adaptable, providing a biological circuit that can regulate multiple phenotypes and maintain functionality in an uncertain world. We propose that the 3-tiered signal transduction module has been conserved through positive selection, because it facilitated the generation of phenotypic variation during eukaryotic evolution.
Keywords: MAP kinase, facilitated variation, robustness, signal transduction, evolution, systems biology
Introduction
The ability to generate useful phenotypic variation within organism populations is crucial for evolution. Useful variation often requires complex phenotypic change, yet if complex change depends on numerous small and random phenotypic variations, evolution is likely to be impeded because mutations are more likely to be deleterious than beneficial.1 Evolution is further constrained by the number of functions a component or pathway performs, as the more critical functions a component is involved in, the higher the probability that mutations will be deleterious and negatively impact on organism fitness.2 As the morphological diversity present within Eukaryotes, and in particular Metazoans, is regulated through the use of a few conserved core cellular pathways,3 how complex, yet viable, phenotypic variation is generated utilizing a handful of core processes remains an ongoing question in evolutionary biology.
A potential solution to this problem was proposed by Gerhart and Kirschner when they predicted that core processes driving Eukaryotic growth and development possess key properties such as weak linkage, compartmentalization, exploratory behavior, and robustness and adaptability; which together facilitate evolution by reducing constraints on change and allowing the accumulation of non-lethal variation.4 In this paradigm, called the theory of facilitated variation, much of the phenotypic variation acted upon by evolution results from mutations within the regulatory mechanisms that control robust and adaptable core processes. Thus the robust-yet-adaptable core modules are proposed to de-constrain evolution by enabling the generation of phenotypic variation through random changes in regulation, which can be acted upon by natural selection.2 The utility of this model is that although the fundamental structure and function of the core modules remains fixed, novel outputs and phenotypes may still be generated through random heritable change within regulatory networks. Under this paradigm, core modules can stochastically access a large number of phenotypes via non-lethal mutations within their regulatory networks, which increases the degree of phenotypic variation that is available for natural selection and evolution.
An important step in testing the theory of facilitated evolution is to first examine core processes for evidence of robustness and adaptability, and then characterize the network topologies that have been selected during evolution, which allow core modules to be both robust and adaptable. To that end we have focused on the signal transduction circuits known as mitogen activated protein kinase (MAPK) modules. MAPK modules are signaling pathways that are evolutionarily conserved across eukaryotic species.5 Each MAPK module is a 3-tiered kinase cascade containing a proximal MAP kinase kinase kinase (M3K), which phosphorylates and activate a MAP Kinase Kinase (M2K), which then phosphorylates and activates a terminal MAP kinase (MAPK/M1K).5 MAPK modules are embedded within signaling circuits that regulate many essential biological processes. In yeast the cellular functions regulated through MAPK modules includes mating, invasive growth, cell wall integrity and the osmolarity response.5 In mammals, the 4 characterized MAPK modules are involved in the regulation of proliferation, differentiation, motility, survival, apoptosis, inflammation and osmoregulation.6
The striking conservation of MAPK modules throughout the eukaryote kingdom, combined with their incorporation into regulatory circuits that control essential cellular functions, can be explained by 2 opposing hypotheses. The first is the null hypothesis, in which mutations within MAPK modules are highly maladaptive; once the system was discovered and incorporated into key signal transduction circuits it was conserved because structural variations came at a considerable fitness cost. In this scenario the 3-tiered module has been evolutionarily conserved through negative selection. The second hypothesis, derived from the theory of facilitated evolution, is that the topology of the core MAPK module de-constrains the evolution of signaling pathways in which they are embedded by facilitating the rapid acquisition of new phenotypes. In this scenario MAPK modules are actively maintained through positive selection due to their ability to enhance the evolutionary capacity of the organism.
Here we test the positive selection hypothesis in silico by exploring basic MAPK network topologies in an effort to identify configurations that facilitate the generation of phenotypic variation. Specifically, by building on established models of MAPK signal transduction, we asked the following question: what is the simplest circuit configuration that can produce a signaling module that is both robust and adaptable?
Results
Exploring Basic Model Parameters and validating model outputs
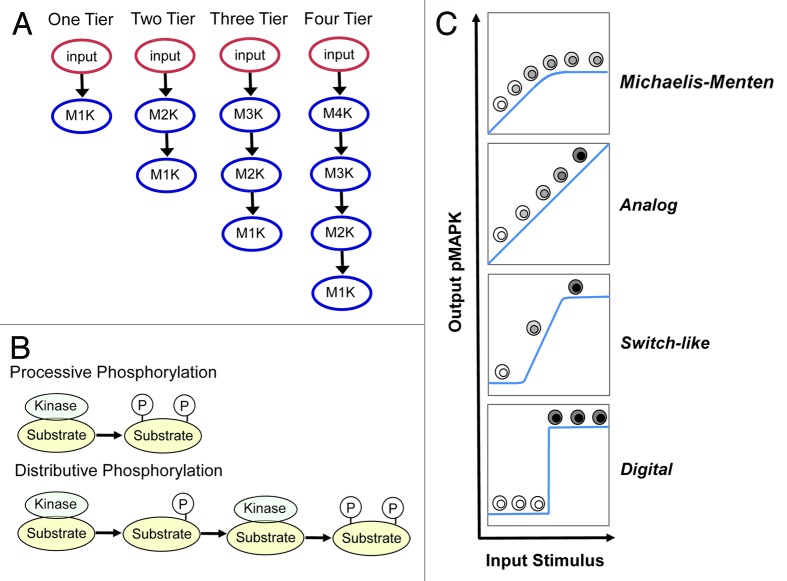
We began our in silico analyses by focusing on 3 primary variables: (1) the number of kinases in the module; (2) the role of processive phosphorylation; and (3) the role of distributive phosphorylation, and assessed how does the combination of these 3 variables determine the signal output of the MAPK module (summarized in Fig. 1). The starting point of our in silico analysis began with the simplest possible module, a single kinase, activated either through processive (P) or distributive (D) phosphorylation (Fig. 2A). In all simulations the activating input is graded/analog. As expected, a single kinase activated through processive phosphorylation displays Michaelis–Menten activation kinetics. In contrast, activation through distributive phosphorylation endowed the system with switch-like activation kinetics; the system is initially less responsive at low inputs and then displays a more rapid activation with a corresponding increase in the Hill co-efficient (Fig. 2B). An important difference between the outputs generated from the distributive systems compared with the processive system, is that distributive phosphorylation generates a large difference between the inactive and maximally active states compared with processive phosphorylation. Together, these results confirm that the processive Michaelis-Menten systems is far more sensitive and responsive toward low inputs, whereas a distributive switch-like system is much better at discriminating low vs. high input levels.7,8
Figure 1. Key variables and system outputs explored in mathematical models. (A) The number of kinases present within the module: one kinase = one-Tiered (1T), 2 kinases = 2-tiered (2T), 3 kinases = 3-tiered (3T) and 4 kinases = 4-tiered (4T). (B) Two modes of activating kinases in the MAPK module: processive phosphorylation [P] (upper panel), where phosphorylation sites within the activation loops are phosphorylated during a single binding step: vs. distributive phosphorylation [D] (lower panel), where only one phosphorylation occurs during binding, and the kinases must re-bind for the second phosphorylation to occur. (C) The 4 different outputs that can be generated from MAPK modules. Classic Michaelis–Menten (MM) kinetics72 where the system is initially linear but then becomes saturate; analog output, which transmit continuous information that is directly proportional to the input stimulus17; switch-like output, which follows a sigmoidal dose-response curve where initially the system is relatively unresponsive and then responds rapidly to input10,73; and digital output, where the system can only stably exists in one of 2 states, off or on15 with a threshold that defines the switch.
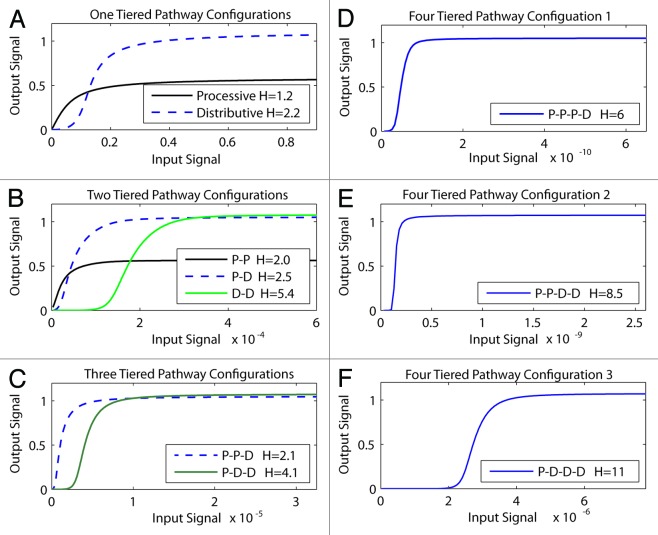
Figure 2. How increasing the number of kinases within the module and processive vs. distributive activating phosphorylation mechanisms change signal output. Different modules that contained increasing number of kinases (1T = one-tiered kinase module, 2T = 2-tiered kinase module etc), with each kinase activated through either processive [P], or distributive [D] phosphorylation, were constructed and their signal outputs tested in silico. The Hill co-efficient [H] for each system was calculated and is displayed in the box within each panel. (A) One-tiered processive (solid black lines) and one-tiered distributive (blue dashed line). (B) Tow tiered processive-procecessive (P-P solid black line), processive-distributive (P-D dashed blue line) and 2 tiered distributive-distributive (DD solid green line). (C) Three tiered processive-processive-distributive (P-P-D solid black line) and processive-distributive-distributive (P-D-D solid green line). (D) Four tiered processive-processive-processive-distributive. (E) Four tiered processive-processive-distributive-distributive. (F) Four tiered processive-distributive-distributive-distributive.
Next we looked at increasing the number of kinases within the module, as well as exploring the role of processive vs. distributive phosphorylation within these more complex module configurations. These simulations confirmed several important principles of module configuration. First, consistent with predictions from Kholodenko and Colleagues,9 we show that increasing the number of kinases within the module increases the sensitivity of the system (compare the input sensitivity displayed on the x-axis in Fig. 2A–E). Second, we confirm earlier work that the presence of distributive phosphorylation makes the module output more switch-like.10,11 Moreover this effect is additive; as more distributive activation steps are added to the module, the Hill co-efficient increases and the system approaches a true digital output. The combination of distributive phosphorylation plus increasing number of tiers within the module drives the module toward a digital output.9,12 In contrast to distributive phosphorylation, the presence of processive phosphorylation within the module makes the system less switch-like but more sensitive and responsive to low input levels.
Based on these results and historic studies cited above, we can formally propose the following general rules of MAPK module structure-function: (1) more tiers equals more sensitivity, fewer tiers equals less sensitivity; (2) processive phosphorylation increases sensitivity and responsiveness to low input but decreases the capacity of the module to discriminate between low and high input levels; (3) distributive phosphorylation makes the module more switch-like, increases the modules ability to discriminate between low vs. high input levels, but reduces the modules ability to accurately transmit low levels of activating input. These findings are in agreement with historical studies,8-11 and confirm that our model and model assumptions are both reasonable and able to generate signaling outputs that are consistent with the well characterized signaling properties of MAPK modules.
Defining robust module configurations
As functional robustness is a key property of any biological system and is strongly selected for during evolution,13 we interrogated our simulations to determine which of pathway configurations were functionally robust with respect to random perturbations within the module. The robustness property of a mathematical model with respect to a set of perturbations P is defined as the average of an evaluation function of the system over all perturbations.14 Here we use the Hill coefficient of the signal output as the evaluation function of the MAPK module, because the Hill coefficient can be used to define different signaling phenotypes, such as graded or switching-like outputs. To constrain our in silico robustness search to within achievable simulation times, we conducted the robustness analysis on the subset of modules that most closely represent the system found in nature, which are those modules that generated a switch-like output comparable to that established experimentally for the classical ERK module (Fig. 3).
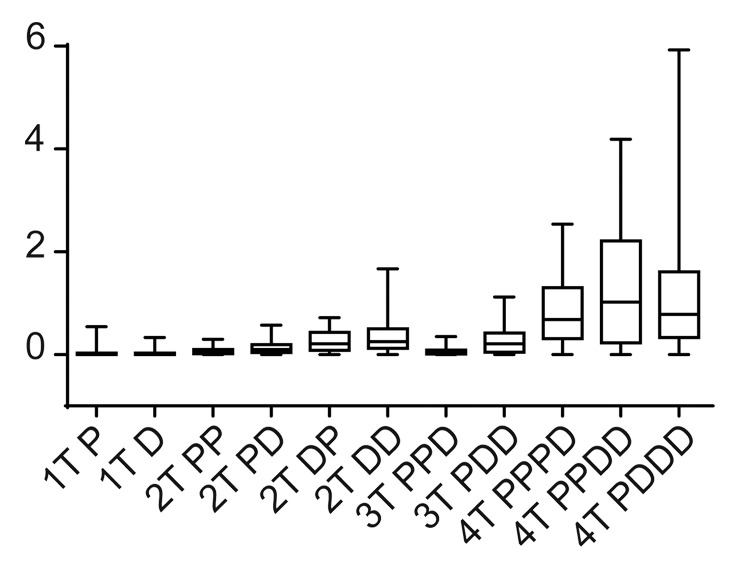
Figure 3. Testing module robustness in silico. We used the Hill coefficient to evaluate the robustness of the MAPK module, because the Hill coefficient can be used to define the system output. Robustness was assessed in silico by determining how the Hill co-efficient was maintained in the face of random perturbations to the system. Here we display the box plot of the system outputs (i.e., the value of the Hill co-efficient) in the face of random perturbations.
Our simulations revealed that modules with greater than 3-tiers were not robust, with random perturbations having a large effect on signal output (Fig. 3). This is because of signal amplification, which occurs within the 4-tiered systems as signal is transmitted down the cascade, causes small perturbations to have large effects on signal output. Modules with 3 kinases or below were significantly more robust, nevertheless some intriguing differences were revealed by our analyses. The single kinase "systems" were the most robust. This is to be expected, as by definition a single protein will be subject to less random variation (as there are less components to randomly perturb), however these data provide a suitable baseline robustness level, which we can use to compare more complex systems. More interesting were the results from 2 and 3 tiered model configurations. In these systems, the presence of processive phosphorylation tended to make the system more robust, whereas the presence of distributive phosphorylation tended to make the systems less robust. This is because the presence of distributive phosphorylation makes the systems more switch-like, and a switch-like system has the capacity to amplify random perturbations if they occur within the nonlinear range of the system. Processive phosphorylation generates a Michaelis–Menten output, which at low levels inputs provides a linear output, the system output is reduced to below linear is reduced to below linear. The processive systems can suppress the effects of random perturbations if those perturbations occur within the sub-linear range of the system, and it is this ability to dampen the effects to random perturbations that allows processive phosphorylation the increase the robustness of the overall module.
Based on these results, we conclude that the 2 most robust modules are the 2-tiered processive-processive, and the 3-tiered processive-processive-distributive module, with the 2-tiered processive-distributive and distributive-processive modules being slightly less robust. Intriguingly the robust, 3-tiered processive-processive-distributive module is the configuration found in nature (for example see refs. 15 and 16), a finding consistent with the hypothesis that evolution selects for robust systems.13
Defining robust modules that can generate digital outputs
Phenotypic variation can be generated from core MAPK modules through the judicious use of feedback control circuits (reviewed in refs. 17 and 18). We therefore mathematically define MAPK module adaptability as the capacity of feedback circuits to reconfigure the module to generate both graded and digital signal output with feedback regulation. We focused on the 4 most robust circuits identified above and asked the question, which of these 4 robust configurations could generate the most phenotypic variation (and is therefore adaptable) using feedback control?
First we asked whether these configurations could generate a digital output through the addition of a positive feedback loop, a mechanism known to drive digital MAPK activation within diverse biological settings.15,19,20 Digital signaling is formally defined as a system with a steep Hill co-efficient that can rest in only 2 stable states corresponding to off and on, and also displays hysteresis. Hysteresis occurs when, upon reducing the activating stimulus, the system remains activated until the input stimulus falls below the input level initially required to activate the circuit21 (and reviewed in refs. 8 and 22).
For all 3 robust 2T and 3T configurations, a true digital output (as defined by having a steep Hill co-efficient, possessing 2 stable steady-states, and displaying hysteresis) was obtained through the addition of a positive feedback loop (Fig. 4). However, the presence of distributive phosphorylation endowed the system with 2 additional properties. First, distributive phosphorylation amplified the final output approximately 2-fold relative to the processive only system, with the maximal “on” state of the 2T processive-processive system half that of the “on” state of the systems containing a distributive phosphorylation step (Fig. 5). Hence a bistable system containing a distributive phosphorylation step provides greater discrimination between the “off” and “on” stable states. Second, when the systems are switching off (red dashed curves in Fig. 5 indicated with the downward pointing arrow), the distributive systems rapidly transition between the “on” and “off” states, providing a sharp threshold that clearly discriminates between the 2 steady-states of the system. In contrast, when switching off the 2-tiered processive system generates a more graded output that does not provide an unambiguous threshold between the on and off steady-states. Instead, it gradually transitions from high to low activation, increasing the potential to generate a graded output and drive an erroneous and potentially deleterious phenotypic decision.
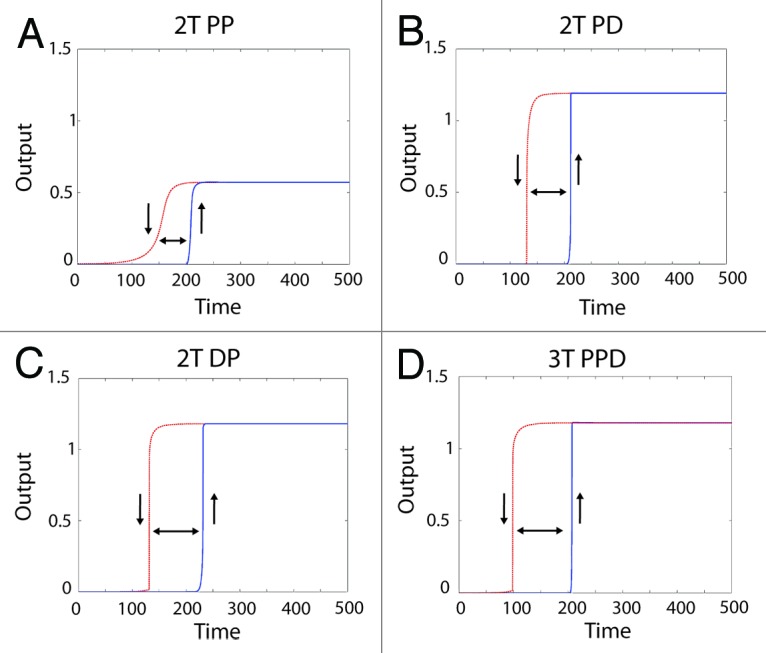
Figure 4. Generating a digital output from different MAPK module topologies using a positive feedback loop. Shown are the system outputs of the 4 most robust module configurations in the presence of a positive feedback loop. The blue solid curve shows the system being activated, with the upward pointing arrow indicating “switching on”. The red dashed line indicates the system becoming inactive after the input is removed, with a downward pointing arrow indicating 'switching off'. The double-headed arrow between the curves shows the presence of hysteresis, which combined with the sharp transition between “off” and “on” states indicates a bistable system output. The module configuration is indicated above each panel with a heading in bold.
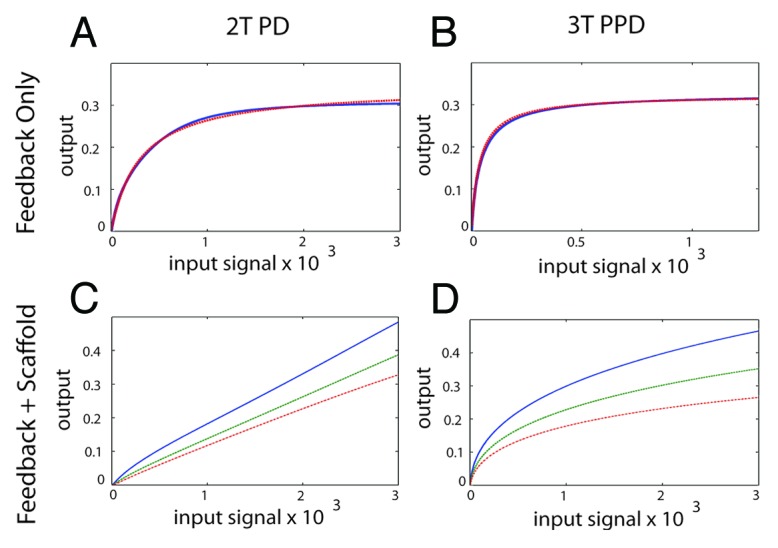
Figure 5. Generating an analog output from the MAPK module using feedback control and a scaffold protein. (A) The 2T P-D system (left panels) and the 3T P-P-D system (right panels) (B) in the presence of a negative feedback loop. The blue line shows the output generated in silico, the red dashed lines shows the output of an ideal Michaelis–Menten system, revealing that both the 2T P-D and 3T P-P-D system outputs closely approximate a Michaelis–Menten system in the presence of a negative feedback loop. (C) The signal output of the 2T processive-distributive module modified using a scaffold plus negative feedback loop (solid-blue-line; scaffold concentration = 1.0: dash-green-line; scaffold concentration = 1.2: dash-dot-red-line; scaffold concentration = 1.4). (D) The signal output of the 3T P-P-D module (solid-blue-line; scaffold concentration = 1.0: dash-green-line; scaffold concentration = 1.2: dash-dot-red-line; scaffold concentration = 1.4).
We conclude that, although all 3 systems can generate a digital output, only those systems that incorporate a distributive phosphorylation step provide a suitable digital configuration. This is because they combine high robustness (as assessed by maintaining constant output in the presence of random perturbation) with a superior ability to discriminate between the low “off” and high “on” stable steady-states that make up the biological switch.
Defining a robust core module that can generate an analog output
Next, we assessed which of the robust module configurations could be reconfigured to generate an analog output. Here we focused only on the robust 2T and 3T systems that contain a distributive phosphorylation step because they make the best switches (shown above). In addition, we focused on the systems with a distributive phosphorylation step on the terminal kinase because this is the system that is present in nature.15,16
First we attempted to generate an analog output using negative feedback control. Increasing the strength of negative feedback progressively reduced the Hill co-efficient of the 2T P-D system and converted a switch-like output into a Michalis-Menten output (Fig. 5A). Interesting, the inclusion of negative feedback control progressively increased the robustness of the system (Fig. S8). However, we could not realize an analog output solely using negative feedback.
It has been shown that processive phosphorylation contributes to the generation of analog signal output from MAPK modules in mammalian cells.23 To test this hypothesis in silico, we suppressed distributive phosphorylation in our models by invoking a scaffold function (Fig. S9). Similar to the established results reported by Levchenko et al.,24 at low concentrations the addition of a scaffold protein increased the values of the Hill coefficient. Consistent with previous work,24 when the amount of scaffold protein reached a concentration where the scaffold began to inhibit the productive interaction between the kinases in the module by forming dead-end complexes, the Hill coefficients decreased (Fig. S9). Further, the total signal output also decreased when the scaffold protein was present in excess. Thus our mathematical simulations of the MAPK scaffold protein captures the fundamental properties predicted for a scaffold protein.24 Although a passive scaffold protein was sufficient to suppress switch-like signal output and generate a Michaelis-Menten response, in and of itself it was insufficient to generate a true analog output (Fig. S9).
Next we tested the combination of negative feedback regulation plus a scaffold protein. Beginning with the 2T P-D core module, we found that when the concentration of the scaffold protein is large, we could realize a true analog signal output using the properly selected strength of negative feedback (Fig. 5C). Importantly, for each concentration of scaffold protein there was a wide range of negative feedback strengths that were capable of producing a graded output (not shown). Using the same assumptions with the 3T P-P-D core module produced a markedly different result using identical scaffold and feedback assumptions. Here, in contrast to the 2T system, the 3-tiered module failed to realize graded signal output, and the signal output always followed a Michaelis–Menten function under these simulation conditions even when the feedback regulation was very strong (Fig. 5D). This is because the increased signal amplification that occurs within a 3-tiered kinase cascade (due to amplification that occurs at each tier of the cascade9) was sufficient to overwhelm the effects of a scaffold protein combined with a negative feedback loop, preventing a true analog signal output from being realized in our simulations.
Under our simulation conditions, only the 2-tiered, processive distributive module could generate a true analog signal output. Therefore, we conclude that with respect to analog signaling, a 2-tiered scaffolded module is more adaptable than a 3-tiered scaffolded module, a result we did not expect.
Evolutionary conservation within the MAPK module
Using simulations we identified a series of MAPK module configurations that display robustness, a fundamental property of biological systems. Choosing the modules that most closely resembled those found in nature, we generated the unexpected result of a 2-tiered module being the most adaptable, robust module configuration of the systems tested.
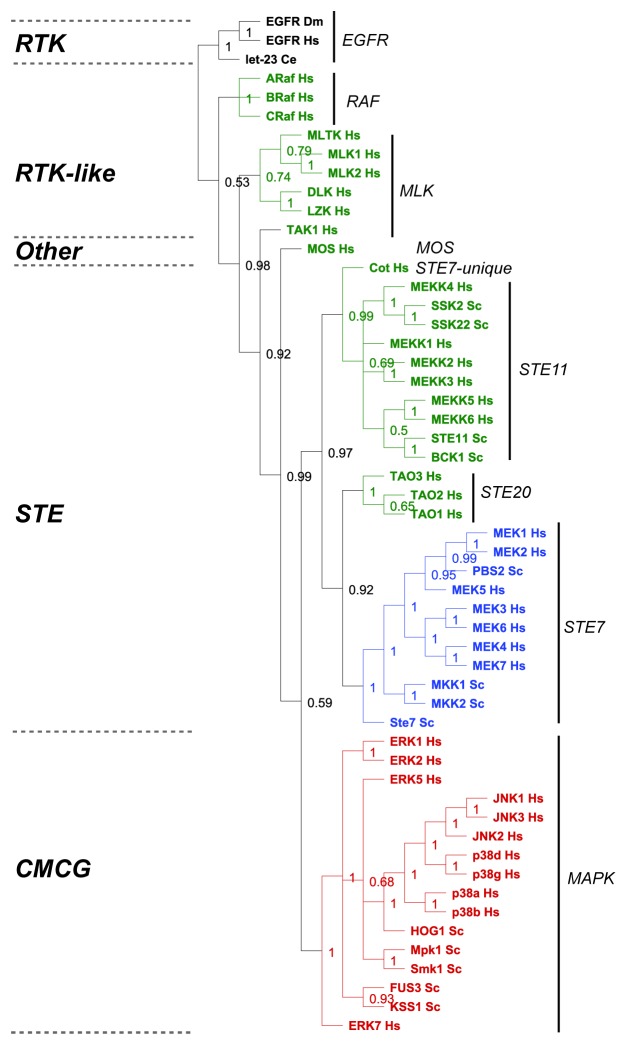
Adaptable, robust core processes that increase nonlethal variation and facilitate phenotypic variation are predicted to be continuously positively selected for during evolution.2 To validate or refute our surprising simulation results, we conducted a basic evolutionary analysis on the established MAPK modules present in humans and yeast and asked the question which components of the module display the lowest rate of evolutionary divergence? We restricted our analysis to MAPK modules present in baker’s yeast (Saccharomyces cerevisiae, Sc)5 and human (Homo sapiens, Hs),6 which have been experimentally validated to function as MAPK modules (Fig. 7). Despite the fact that fungi and mammals diverged over 900 million years ago,25 the M2K and M1Ks from both phyla all cluster within single, respective clades that correspond to specific kinase families (M2Ks fall within the Ste7 family, and M1Ks fall within the MAPK family) (Fig. 6). In contrast, the M3Ks are distributed across multiple clades (Fig. 6), consistent with established phylogenetic analyses showing that M3Ks are composed of diverse kinase families and groups.26-28 These data suggest the hypothesis that there has been strong continued selection (either positive or negative) upon the M2K and M3K tiers of MAPK modules, and this has maintained their exclusive distribution within the Ste7 and MAPK families, respectively. In contrast, the M3K tier is composed of a variety of different kinases groups, suggesting that these diverse kinases have been connected to the more highly conserved 2-tiered core modules at various times during eukaryotic evolution.
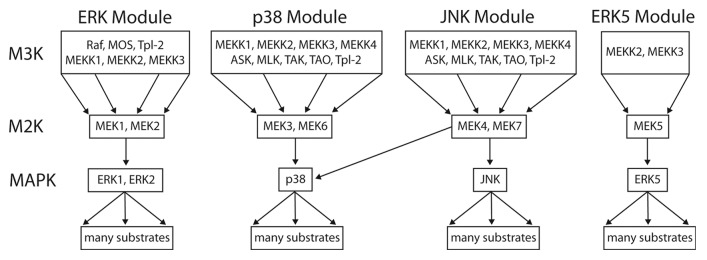
Figure 7. The 3-tiered MAPK module structure in mammals. Mammals have 4 established MAP kinase modules, the canonical ERK MAPK module, the p38, and JNK stress-activated modules, and the ERK5 module. All of the MAPK cascades follow a 3-tiered module configuration, with the proximal MAP Kinase Kinase Kinase (M3K), a central MAP Kinase (M2K) and a terminal MAP Kinase (MK). In general, each module regulates distinct cell phenotypes, with the ERK module controlling cellular proliferation and differentiation, the p38 and JNK modules controlling cellular response to various forms of stress, and the ERK5 module controlling both stress response and cell proliferation. However, MAPK module functions are often overlapping and opposing, and individual MAPK modules can also regulate non-canonical cell phenotypes under certain conditions. The proximal M3K consists of a group of evolutionarily diverse, promiscuous input kinases that link the evolutionarily conserved, non-promiscuous M2Ks to a variety of external stimuli. The M2Ks are tightly linked to their cognate, evolutionarily conserved MAPKs. Once phosphorylated the MAPKs dissociate from their M2K partners and function as the output node of the module, phosphorylating a diverse array of substrates that include transcription factors, phosphatases and other kinases. For detailed reviews of the form and function of mammalian MAPK modules the readers are referred to.6,29
Figure 6. Evolutionary analysis of MAP kinase modules from yeast and man. We restricted our evolutionary analyses to kinases that have been experimentally verified to belong to MAP Kinase modules in humans (Hs) and baker's yeast Saccharomyces cerevisiae (Sc) (reviewed in refs. 5, 6, 27, and 28). We conducted a Maximum Likelihood evolutionary analysis on the aligned sequences using Mr Bayes,70 using the standard options available in the Geneious software plugin.71 The Epithelial Growth Factor Receptor (EGFR) from human, fruit fly (Drosophila melanogaster DM) and worm (let-23 Caenorhabditis elegans Ce) were used as out-groups to root the tree (shown in black). The M3K tier of the module is shown in green, the M2K tier shown in blue, and the M1K tier shown in red. The kinase family groups are indicated by lines and named on the right, the kinase groups are shown on the left.
This result supports our simulation results, suggesting the hypothesis that the 2 highly conserved M2K and M1K kinases makes up the fundamental robust-yet-adaptable core of the MAPK module, which has been weakly linked to a variety of diverse kinases that function as input nodes, making up the third tier of MAPK module.
Discussion
Robustness and adaptability
The theory of facilitated evolution predicts that modules that are both robust and adaptable will be evolutionarily conserved throughout Eukaryotes, because they allow the generation of novel phenotypes using a few conserved pathways.4 Our aim was to explore this hypothesis by combining in silico simulations with evolutionary analyses in an effort to identify fundamental circuit configurations that are both robust and adaptable. By focusing on the evolutionarily conserved MAPK pathways, and building on established computational models, we have uncovered a simple 2-tiered circuit configuration within MAPK modules that is both robust, being able to maintain function despite perturbation, and adaptable, able to be readily reconfigured to generate either analog or digital outputs by using feedback loops and scaffold proteins. This finding is supported by evolutionary analyses which reveal that the 2 proximal M2K and M3K kinases are highly conserved across phyla.
Together, these data support the argument that the 2 terminal kinases within MAPK modules have been maintained during Eukaryotic evolution, at least in part, through positive selection of the key functional properties of robustness and adaptability that are inherent to a 2 kinase, processive-distributive circuit.
Weak regulatory linkage
If a simple 2 kinase module is both robust and flexible, why then do MAPK modules contain 3 kinases? We propose that the answer lies in the requirement for weak regulatory linkage. Weak regulatory linkage is a central hypothesis underpinning the theory of facilitated variation because ‘different core processes must become linked, by regulatory means, in different combinations, and operated in different amounts, states, times, and places for the generation of new anatomical and physiological traits’4. In MAPK modules the proximal MAP kinase kinase kinases function as input nodes, providing weak linkage between the core 2-tiered circuit and a variety of different external stimuli.26 Importantly, we have shown that the addition of a third input node activated by processive phosphorylation also increases the overall robustness of the module, further buffering the system against the effects of random perturbation. Based on our simulation and evolutionary analyses we propose the hypothesis that the fundamental 3-tiered MAPK module consists of a proximal input kinases weakly linked to a highly conserved 2 kinase core.
This hypothesis is entirely consistent with the known biology of MAPK modules in both yeast and man. Many of the proximal kinases of mammalian MAPK modules (i.e., M3Ks) activate multiple MAPK modules (reviewed in refs. 6, 26, and 29; summarized in Fig. 7). Some examples of promiscuous M3Ks include MEKK2 and MEKK3, which activate the JNK, p38 and ERK5 modules30-32; and MLTK and Tpl2, which can activate all 4 mammalian MAPK modules.33-35 The diversity of different kinases that make up the M3K tier, combined with their complex activation and regulation, allows the connection between MAPK modules and the multitude of inputs that direct of cell fate decisions (reviewed in ref. 29).
In contrast, the specificity of MAP kinase activation by their cognate M2Ks is high. The M2K and MK proteins are constitutively tightly linked, thanks to the presence of strong protein-protein interactions between the 2 kinases.36-39 This interaction is specific, with each M2K protein domain specific for its cognate MK.40 For the canonical ERK MAPK module, the activated MEKs display unique selectivity toward ERK.41 The same fidelity is present within the ERK5 MAPK module, with MEK5α and MEK5βb phosphorylating ERK5 but not ERK1/2 despite their similar activation loops (ref. 42 and reviewed in ref. 6). The primary M2K’s of the p38 MAPK module, MKK3 (MEK3) and MKK6 (MEK6), display high selectivity toward p38 and do not activate either JNK or ERK (refs. 43 and 44 and reviewed in ref. 29). MKK7 (MEK7) activates JNK and generally appears incapable of activating p38 (refs. 45 and 46 and reviewed in ref. 29). The exception appears to be MKK4 (MEK4), which is slightly promiscuous, being able to activate both JNK and p38.47,48 Nevertheless, the general rule appears to be that the M2K tier of MAPK modules is composed of kinases with high specificity for their MAPK substrates.
The terminal MAP kinases are highly promiscuous, capable of phosphorylating hundreds of different substrates including transcription factors, kinases and phosphatases, cytoskeletal proteins, regulators of apoptosis and inflammation, and other signaling proteins (reviewed in refs. 29 and 49). MAPK regulated transcriptional networks have evolved an exquisite sensitivity to changes in MAPK signal output.50,51 Thus, the 3 tiered MAPK module configuration is able to robustly drive divergent cell fate decisions utilizing the same core signaling components due to weak linkages between the input signal and internal outputs.
The role of scaffolds in compartmentation and the suppression of distributive phosphorylation
Our current simulations and seminal studies7,10 highlight the ability of distributive systems to amplify input signal. This amplification is particularly well suited to building digital systems, as it ensures that a large difference exists between the 2 system steady-states that represent the “on” and “off” of the biological switch. In addition, we show that the presence of distributive phosphorylation generates a sharp threshold between the “on” and “off” states occurs not only when the system is switching on, but also when the system is switching off in the absence of reinforcing feedback control. This ensures that the digital nature of the system is maintained throughout activation and inactivation cycle, thereby protecting the cell against the spurious generation of false analog outputs. The role of MAPK modules in generating critical all-or-nothing cell fate decisions is well established (for example see refs. 15, 19, 52, and 53), and our simulation results argue that a distributive phosphorylation step within the cascade has been positively selected and fixed during evolution because it enables reliable digital signal transduction to occur. However, this network topology provides a challenge for the generation of analog signaling when using the same core pathway components, as we found that even with strong negative feedback control, signal pathways that contain a distributive phosphorylation step cannot generate a true analog output.
We solved this problem in silico by utilizing the scaffold-dependent suppression of distributive phosphorylation, which successfully generated an analog output when combined with negative feedback. Scaffold proteins have been hypothesized to enable analog signaling in MAPK modules by holding the M2K and M3K proteins together to encourage processive phosphorylation,54,55 a prediction we realized in silico56 that was confirmed experimentally in yeast by Takahashi and Pryciak.57 Further, Matsuda and colleagues revealed experimentally that cells can also suppress distributive phosphorylation using molecular crowding, which contributes to analog signaling from the canonical MAPK pathway in mammalian cells.23 We show that scaffold-mediated suppression of distributive phosphorylation, in combination with negative feedback, allows the generation of an analog output from a pathway containing a distributive phosphorylation step.
Spatial constraint is another mechanism proposed to facilitate the generation of phenotypic variation from core components.4 Spatial compartmentalization has been proposed to facilitate the use of core processes in different biological contexts, while decreasing the chance of interference or leakage into other biological functions.4 An important additional role of scaffolds is to provide spatial localization signals to direct promiscuous MAK kinases toward specific substrates. For example, KSR functions as a scaffold protein for the canonical mammalian Raf-MEK-ERK cascade.58 Within the immune system, one function of the Raf-MEK-ERK pathway is to direct cytotoxic killing by natural killer cells and cytotoxic T-cells.58 Here, the scaffold protein KSR recruits activated ERK to the immunological synapse where it can phosphorylate key substrates that regulate cytotoxic immune cell function.58 Scaffolds can also directly regulate pathway output using complex allosteric mechanisms as has been discovered in yeast59 and mammalian scaffold MAPK scaffold proteins.60
Clearly, our simplified models do not realize the full depth and breadth of scaffold functionality. Nevertheless they highlight that the suppression of distributive phosphorylation by scaffolds, in combination with negative feedback, as one fundamental mechanism that can be used to generate MAPK module signaling flexibility in vivo. These findings may inform our understanding of scaffold evolutionary dynamics, as MAPK scaffolds are made up of diverse proteins and appear ubiquitous to MAPK signal transduction in vivo.61,62
Exploratory processes
Gerhart and Kirschner discussed how the exploratory behavior of robust-yet-adaptable modules has been crucial in the evolution of complex animal physiology.4 Here, core processes are proposed to “search and find targets in large spaces or molecular populations”.4 We propose that the modular, robust-yet-adaptable structure of MAPK modules enables exploratory behavior that enhances the generation of phenotypic variation. At the single cell level protein copy numbers are heterogeneous due to stochastic gene expression (reviewed in ref. 63). Heterogeneous protein expression can have a significant effect on cellular phenotype.63-65 Using a synthetic MAPK module, O’Shanaughnessy et al. demonstrated that changing concentration of components of a MAPK module can alter pathway output,16 providing proof-of-principle experimental data supporting the idea that changes in protein expression levels can be used to explore different phenotypes driven by MAPK pathway activation. In addition, heterogeneous expression of scaffolds such as KSR1, which alter pathway sensitivity, is also used to affect the distribution of cell phenotypes in populations of immune cells controlled by the MAPK module.52 Thus the flexibility of the core MAPK module is well positioned to allow cells to explore new phenotypes through the heterogeneous expression of kinases or key regulatory proteins within the module.
Conclusions
Based on our new simulation results combined with historic studies, we propose that MAP kinase modules are composed of promiscuous input kinases (M3Ks) converging on a conserved core (M2K → MAPK), which then fans out to connect to a highly diverse repertoire of outputs (Fig. 7). It has been argued that this type of “bow-tie” architecture is critical for the generation of robust yet evolvable networks,66,67 however the circuit logic selected for during evolution that enables robustness and evolvability inherent to bow-tie networks remain poorly characterized. Here we formally propose that the 3-tiered module is one such circuit topology, which is robust-yet-adaptable, allowing the generation of multiple output types from the same core components. We hypothesize that this robust-yet-adaptable circuit configuration, after its initial selection and fixation within the Eukaryotic genome, has served to facilitate the generation of phenotypic variation during the evolution of Eukaryotic signal transduction. This model goes some way toward explaining why the fundamental 3-tiered structure of MAPK modules has been conserved from yeast to man, and why it has been utilized to control many biological functions that are essential for life.
Materials and Methods
Mathematical modeling
Our proposed models of the MAP kinase pathway include Ras-GTP (denoted as UIS) as the signal input of the MAP kinase cascade. In the 3-tiered model, UIS activates MAP kinase kinase kinase (M3K) molecules in a single step. This activation is followed by sequential activation of the dual-specificity MAP kinase kinase (M2K) by M3Kp (i.e., the activated M3K) in either a single-step processive module or 2-step distributive module. The activated M2Kpp (i.e., phosphorylated M2K at 2 residue positions) in turn activates MAP kinase (M1K) in either a single-step processive module or 2-step distributive module. The activated M1Kpp (i.e., phosphorylated M1K at 2 residue positions) is the signal output of the MAK kinase module. In addition, phosphatases, termed as PM3K, PM2K, and PM1K, can deactivate the activated M3Kp, M2Kpp, and M1Kpp kinases, respectively. However, in the 2-tiered model, signal input UIS activates M2K directly; while in the one-tiered model, UIS activates M1K directly.
A set of chemical reactions was used to describe the detailed process of kinase activation. Briefly, the activated kinase (or phosphatase) K binds to its substrate S (or activated kinase Sp) to form a protein complex K-S (or K-Sp), which leads to the activated substrate Sp (or deactivated kinase S). Examples of these reactions are: the processive phosphorylation module of M2K kinase
and the distributive phosphorylation module of M1K kinase
Where ai, di, and ki are protein binding, dissociation and activation rate constants, respectively. All the chemical reactions are listed in the supplementary information.
A mathematical model was developed according to the chemical rate equations of these chemical reactions. For example, reaction (1) leads to the following differential equation for the dynamics of the M3Kp-M2K complex, given by:
Detailed information of the differential equations is given in the supplementary information.
Robustness analysis
We used the concept defined by Kitano14 to measure the robustness property of the proposed model. The robustness property of a mathematical model with respect to a set of perturbations P is defined as the average of an evaluation function
of the system over all perturbations p∈P, weighted by the perturbation probabilities prob(p), given by:
Here we proposed to use the following measure to evaluate the average behavior
that is the mean of kinase activities that should be close to the simulated kinase activity obtained from the unperturbed rate constants. In addition, the impact of perturbations on nominal behavior is defined by
Where xij(p) and xij are the simulated activities of kinase xi at time point tj with perturbed and unperturbed rate constants, respectively, and
is the mean of xij(p) over all the perturbated kinetic rates.
For each network model, we consider nine cases with the perturbation to different parameters. For each of the rate constants ai, di, and ki, we considered 3 cases for the phosphorylation rates, dephosphorylation rates, and both, respectively. In each case for the rate constant ki, the perturbation
is set to:
Evolutionary analyses
We restricted our evolutionary analyses to kinases that have been experimentally verified to belong to MAP Kinase modules in humans and baker’s yeast Saccharomyces cerevisiae (Table 10.1; reviewed in refs. 5, 6, 27, and 28). The amino acid sequences of the complete kinases and the kinase domains were downloaded from the kinase database (KinBase) at Sugen/Salk (http://kinase.com/kinbase/). Kinase Group and Family displayed in Table 10.1 were defined according to the KinBase classifications.27,28 We chose to conduct our evolutionary analyses on the kinase domains only because the accuracy of phylogenetic analyses can be critically dependent on the alignment quality.68 We aligned the kinase domains of the proteins by eye using the established kinase domain alignment, published by Hanks and Hunter, where the kinase domains are aligned according to residue function as well as sequence homology.69
We conducted a Maximum Likelihood phylogenetic analysis on the aligned sequences using the Mr Bayes Plugin in Geneious70 using the standard options available in the Geneious software plugin.71 The analysis was repeated 5 times, a representative tree is shown in Figure 4. The kinase domain of the EGFR from human, yeast and worm was included as an out-group to allow for out-group rooting. The reasonableness of the phylogenetic analyses were determined by comparing our results with evolutionary analyses published in the literature27,28 and more recent phylogenies published online at kinase.com.
Supplementary Material
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We are extremely grateful to our colleague Dr James Whitacre for his insightful comments during manuscript preparation. T.T. and A.H. are supported by an Australian Research Foundation (ARC) Project Grants (DP1094181 and DP120104460). T.T. is also a recipient of the Australian Research Council Future Fellowship (FT100100748). AH is also supported by The University of Queensland Research Foundation.
References
- 1.Kimura M, Ohta T. On some principles governing molecular evolution. Proc Natl Acad Sci U S A. 1974;71:2848–52. doi: 10.1073/pnas.71.7.2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kirschner M, Gerhart J. Evolvability. Proc Natl Acad Sci U S A. 1998;95:8420–7. doi: 10.1073/pnas.95.15.8420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gerhart J. 1998 Warkany lecture: signaling pathways in development. Teratology. 1999;60:226–39. doi: 10.1002/(SICI)1096-9926(199910)60:4<226::AID-TERA7>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 4.Gerhart J, Kirschner M. The theory of facilitated variation. Proc Natl Acad Sci U S A. 2007;104(Suppl 1):8582–9. doi: 10.1073/pnas.0701035104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Qi M, Elion EA. MAP kinase pathways. J Cell Sci. 2005;118:3569–72. doi: 10.1242/jcs.02470. [DOI] [PubMed] [Google Scholar]
- 6.Keshet Y, Seger R. The MAP kinase signaling cascades: a system of hundreds of components regulates a diverse array of physiological functions. Methods Mol Biol. 2010;661:3–38. doi: 10.1007/978-1-60761-795-2_1. [DOI] [PubMed] [Google Scholar]
- 7.Ferrell JE., Jr. Tripping the switch fantastic: how a protein kinase cascade can convert graded inputs into switch-like outputs. Trends Biochem Sci. 1996;21:460–6. doi: 10.1016/S0968-0004(96)20026-X. [DOI] [PubMed] [Google Scholar]
- 8.Kholodenko BN, Birtwistle MR. Four-dimensional dynamics of MAPK information processing systems. Wiley Interdiscip Rev Syst Biol Med. 2009;1:28–44. doi: 10.1002/wsbm.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brown GC, Hoek JB, Kholodenko BN. Why do protein kinase cascades have more than one level? Trends Biochem Sci. 1997;22:288. doi: 10.1016/S0968-0004(97)82216-5. [DOI] [PubMed] [Google Scholar]
- 10.Huang CY, Ferrell JE., Jr. Ultrasensitivity in the mitogen-activated protein kinase cascade. Proc Natl Acad Sci U S A. 1996;93:10078–83. doi: 10.1073/pnas.93.19.10078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Burack WR, Sturgill TW. The activating dual phosphorylation of MAPK by MEK is nonprocessive. Biochemistry. 1997;36:5929–33. doi: 10.1021/bi970535d. [DOI] [PubMed] [Google Scholar]
- 12.Ferrell JE., Jr. How responses get more switch-like as you move down a protein kinase cascade. Trends Biochem Sci. 1997;22:288–9. doi: 10.1016/S0968-0004(97)82217-7. [DOI] [PubMed] [Google Scholar]
- 13.Kitano H. Biological robustness. Nat Rev Genet. 2004;5:826–37. doi: 10.1038/nrg1471. [DOI] [PubMed] [Google Scholar]
- 14.Kitano H. Towards a theory of biological robustness. Mol Syst Biol. 2007;3:137. doi: 10.1038/msb4100179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ferrell JE, Jr., Machleder EM. The biochemical basis of an all-or-none cell fate switch in Xenopus oocytes. Science. 1998;280:895–8. doi: 10.1126/science.280.5365.895. [DOI] [PubMed] [Google Scholar]
- 16.O’Shaughnessy EC, Palani S, Collins JJ, Sarkar CA. Tunable signal processing in synthetic MAP kinase cascades. Cell. 2011;144:119–31. doi: 10.1016/j.cell.2010.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harding AS, Hancock JF. Using plasma membrane nanoclusters to build better signaling circuits. Trends Cell Biol. 2008;18:364–71. doi: 10.1016/j.tcb.2008.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shin SY, Rath O, Choo SM, Fee F, McFerran B, Kolch W, Cho KH. Positive- and negative-feedback regulations coordinate the dynamic behavior of the Ras-Raf-MEK-ERK signal transduction pathway. J Cell Sci. 2009;122:425–35. doi: 10.1242/jcs.036319. [DOI] [PubMed] [Google Scholar]
- 19.Bagowski CP, Ferrell JE., Jr. Bistability in the JNK cascade. Curr Biol. 2001;11:1176–82. doi: 10.1016/S0960-9822(01)00330-X. [DOI] [PubMed] [Google Scholar]
- 20.Santos SD, Verveer PJ, Bastiaens PI. Growth factor-induced MAPK network topology shapes Erk response determining PC-12 cell fate. Nat Cell Biol. 2007;9:324–30. doi: 10.1038/ncb1543. [DOI] [PubMed] [Google Scholar]
- 21.Xiong W, Ferrell JE., Jr. A positive-feedback-based bistable ‘memory module’ that governs a cell fate decision. Nature. 2003;426:460–5. doi: 10.1038/nature02089. [DOI] [PubMed] [Google Scholar]
- 22.Ferrell JE., Jr. Self-perpetuating states in signal transduction: positive feedback, double-negative feedback and bistability. Curr Opin Cell Biol. 2002;14:140–8. doi: 10.1016/S0955-0674(02)00314-9. [DOI] [PubMed] [Google Scholar]
- 23.Aoki K, Yamada M, Kunida K, Yasuda S, Matsuda M. Processive phosphorylation of ERK MAP kinase in mammalian cells. Proc Natl Acad Sci U S A. 2011;108:12675–80. doi: 10.1073/pnas.1104030108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Levchenko A, Bruck J, Sternberg PW. Scaffold proteins may biphasically affect the levels of mitogen-activated protein kinase signaling and reduce its threshold properties. Proc Natl Acad Sci U S A. 2000;97:5818–23. doi: 10.1073/pnas.97.11.5818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Douzery EJ, Snell EA, Bapteste E, Delsuc F, Philippe H. The timing of eukaryotic evolution: does a relaxed molecular clock reconcile proteins and fossils? Proc Natl Acad Sci U S A. 2004;101:15386–91. doi: 10.1073/pnas.0403984101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Widmann C, Gibson S, Jarpe MB, Johnson GL. Mitogen-activated protein kinase: conservation of a three-kinase module from yeast to human. Physiol Rev. 1999;79:143–80. doi: 10.1152/physrev.1999.79.1.143. [DOI] [PubMed] [Google Scholar]
- 27.Manning G, Plowman GD, Hunter T, Sudarsanam S. Evolution of protein kinase signaling from yeast to man. Trends Biochem Sci. 2002;27:514–20. doi: 10.1016/S0968-0004(02)02179-5. [DOI] [PubMed] [Google Scholar]
- 28.Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. The protein kinase complement of the human genome. Science. 2002;298:1912–34. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- 29.Kyriakis JM, Avruch J. Mammalian MAPK signal transduction pathways activated by stress and inflammation: a 10-year update. Physiol Rev. 2012;92:689–737. doi: 10.1152/physrev.00028.2011. [DOI] [PubMed] [Google Scholar]
- 30.Blank JL, Gerwins P, Elliott EM, Sather S, Johnson GL. Molecular cloning of mitogen-activated protein/ERK kinase kinases (MEKK) 2 and 3. Regulation of sequential phosphorylation pathways involving mitogen-activated protein kinase and c-Jun kinase. J Biol Chem. 1996;271:5361–8. doi: 10.1074/jbc.271.10.5361. [DOI] [PubMed] [Google Scholar]
- 31.Deacon K, Blank JL. MEK kinase 3 directly activates MKK6 and MKK7, specific activators of the p38 and c-Jun NH2-terminal kinases. J Biol Chem. 1999;274:16604–10. doi: 10.1074/jbc.274.23.16604. [DOI] [PubMed] [Google Scholar]
- 32.Nakamura K, Johnson GL. PB1 domains of MEKK2 and MEKK3 interact with the MEK5 PB1 domain for activation of the ERK5 pathway. J Biol Chem. 2003;278:36989–92. doi: 10.1074/jbc.C300313200. [DOI] [PubMed] [Google Scholar]
- 33.Salmeron A, Ahmad TB, Carlile GW, Pappin D, Narsimhan RP, Ley SC. Activation of MEK-1 and SEK-1 by Tpl-2 proto-oncoprotein, a novel MAP kinase kinase kinase. EMBO J. 1996;15:817–26. [PMC free article] [PubMed] [Google Scholar]
- 34.Gotoh I, Adachi M, Nishida E. Identification and characterization of a novel MAP kinase kinase kinase, MLTK. J Biol Chem. 2001;276:4276–86. doi: 10.1074/jbc.M008595200. [DOI] [PubMed] [Google Scholar]
- 35.Chiariello M, Marinissen MJ, Gutkind JS. Multiple mitogen-activated protein kinase signaling pathways connect the cot oncoprotein to the c-jun promoter and to cellular transformation. Mol Cell Biol. 2000;20:1747–58. doi: 10.1128/MCB.20.5.1747-1758.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bardwell L, Cook JG, Chang EC, Cairns BR, Thorner J. Signaling in the yeast pheromone response pathway: specific and high-affinity interaction of the mitogen-activated protein (MAP) kinases Kss1 and Fus3 with the upstream MAP kinase kinase Ste7. Mol Cell Biol. 1996;16:3637–50. doi: 10.1128/mcb.16.7.3637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bardwell L, Thorner J. A conserved motif at the amino termini of MEKs might mediate high-affinity interaction with the cognate MAPKs. Trends Biochem Sci. 1996;21:373–4. doi: 10.1016/0968-0004(96)30032-7. [DOI] [PubMed] [Google Scholar]
- 38.Fukuda M, Gotoh I, Gotoh Y, Nishida E. Cytoplasmic localization of mitogen-activated protein kinase kinase directed by its NH2-terminal, leucine-rich short amino acid sequence, which acts as a nuclear export signal. J Biol Chem. 1996;271:20024–8. doi: 10.1074/jbc.271.33.20024. [DOI] [PubMed] [Google Scholar]
- 39.Fukuda M, Gotoh Y, Nishida E. Interaction of MAP kinase with MAP kinase kinase: its possible role in the control of nucleocytoplasmic transport of MAP kinase. EMBO J. 1997;16:1901–8. doi: 10.1093/emboj/16.8.1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bardwell AJ, Frankson E, Bardwell L. Selectivity of docking sites in MAPK kinases. J Biol Chem. 2009;284:13165–73. doi: 10.1074/jbc.M900080200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Seger R, Seger D, Lozeman FJ, Ahn NG, Graves LM, Campbell JS, Ericsson L, Harrylock M, Jensen AM, Krebs EG. Human T-cell mitogen-activated protein kinase kinases are related to yeast signal transduction kinases. J Biol Chem. 1992;267:25628–31. [PubMed] [Google Scholar]
- 42.Kato Y, Kravchenko VV, Tapping RI, Han J, Ulevitch RJ, Lee JD. BMK1/ERK5 regulates serum-induced early gene expression through transcription factor MEF2C. EMBO J. 1997;16:7054–66. doi: 10.1093/emboj/16.23.7054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zheng CF, Guan KL. Properties of MEKs, the kinases that phosphorylate and activate the extracellular signal-regulated kinases. J Biol Chem. 1993;268:23933–9. [PubMed] [Google Scholar]
- 44.Raingeaud J, Whitmarsh AJ, Barrett T, Dérijard B, Davis RJ. MKK3- and MKK6-regulated gene expression is mediated by the p38 mitogen-activated protein kinase signal transduction pathway. Mol Cell Biol. 1996;16:1247–55. doi: 10.1128/mcb.16.3.1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lu X, Nemoto S, Lin A. Identification of c-Jun NH2-terminal protein kinase (JNK)-activating kinase 2 as an activator of JNK but not p38. J Biol Chem. 1997;272:24751–4. doi: 10.1074/jbc.272.40.24751. [DOI] [PubMed] [Google Scholar]
- 46.Tournier C, Whitmarsh AJ, Cavanagh J, Barrett T, Davis RJ. Mitogen-activated protein kinase kinase 7 is an activator of the c-Jun NH2-terminal kinase. Proc Natl Acad Sci U S A. 1997;94:7337–42. doi: 10.1073/pnas.94.14.7337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sánchez I, Hughes RT, Mayer BJ, Yee K, Woodgett JR, Avruch J, Kyriakis JM, Zon LI. Role of SAPK/ERK kinase-1 in the stress-activated pathway regulating transcription factor c-Jun. Nature. 1994;372:794–8. doi: 10.1038/372794a0. [DOI] [PubMed] [Google Scholar]
- 48.Dérijard B, Raingeaud J, Barrett T, Wu IH, Han J, Ulevitch RJ, Davis RJ. Independent human MAP-kinase signal transduction pathways defined by MEK and MKK isoforms. Science. 1995;267:682–5. doi: 10.1126/science.7839144. [DOI] [PubMed] [Google Scholar]
- 49.Yoon S, Seger R. The extracellular signal-regulated kinase: multiple substrates regulate diverse cellular functions. Growth Factors. 2006;24:21–44. doi: 10.1080/02699050500284218. [DOI] [PubMed] [Google Scholar]
- 50.Murphy LO, MacKeigan JP, Blenis J. A network of immediate early gene products propagates subtle differences in mitogen-activated protein kinase signal amplitude and duration. Mol Cell Biol. 2004;24:144–53. doi: 10.1128/MCB.24.1.144-153.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Murphy LO, Smith S, Chen RH, Fingar DC, Blenis J. Molecular interpretation of ERK signal duration by immediate early gene products. Nat Cell Biol. 2002;4:556–64. doi: 10.1038/ncb822. [DOI] [PubMed] [Google Scholar]
- 52.Lin J, Harding A, Giurisato E, Shaw AS. KSR1 modulates the sensitivity of mitogen-activated protein kinase pathway activation in T cells without altering fundamental system outputs. Mol Cell Biol. 2009;29:2082–91. doi: 10.1128/MCB.01634-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Malleshaiah MK, Shahrezaei V, Swain PS, Michnick SW. The scaffold protein Ste5 directly controls a switch-like mating decision in yeast. Nature. 2010;465:101–5. doi: 10.1038/nature08946. [DOI] [PubMed] [Google Scholar]
- 54.Burack WR, Shaw AS. Signal transduction: hanging on a scaffold. Curr Opin Cell Biol. 2000;12:211–6. doi: 10.1016/S0955-0674(99)00078-2. [DOI] [PubMed] [Google Scholar]
- 55.Ferrell JE., Jr. What do scaffold proteins really do? Sci STKE. 2000;2000:pe1. doi: 10.1126/stke.2000.52.pe1. [DOI] [PubMed] [Google Scholar]
- 56.Tian T, Harding A, Inder K, Plowman S, Parton RG, Hancock JF. Plasma membrane nanoswitches generate high-fidelity Ras signal transduction. Nat Cell Biol. 2007;9:905–14. doi: 10.1038/ncb1615. [DOI] [PubMed] [Google Scholar]
- 57.Takahashi S, Pryciak PM. Membrane localization of scaffold proteins promotes graded signaling in the yeast MAP kinase cascade. Curr Biol. 2008;18:1184–91. doi: 10.1016/j.cub.2008.07.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Giurisato E, Lin J, Harding A, Cerutti E, Cella M, Lewis RE, Colonna M, Shaw AS. The mitogen-activated protein kinase scaffold KSR1 is required for recruitment of extracellular signal-regulated kinase to the immunological synapse. Mol Cell Biol. 2009;29:1554–64. doi: 10.1128/MCB.01421-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bhattacharyya RP, Reményi A, Good MC, Bashor CJ, Falick AM, Lim WA. The Ste5 scaffold allosterically modulates signaling output of the yeast mating pathway. Science. 2006;311:822–6. doi: 10.1126/science.1120941. [DOI] [PubMed] [Google Scholar]
- 60.Brennan DF, Dar AC, Hertz NT, Chao WC, Burlingame AL, Shokat KM, Barford D. A Raf-induced allosteric transition of KSR stimulates phosphorylation of MEK. Nature. 2011;472:366–9. doi: 10.1038/nature09860. [DOI] [PubMed] [Google Scholar]
- 61.Morrison DK, Davis RJ. Regulation of MAP kinase signaling modules by scaffold proteins in mammals. Annu Rev Cell Dev Biol. 2003;19:91–118. doi: 10.1146/annurev.cellbio.19.111401.091942. [DOI] [PubMed] [Google Scholar]
- 62.Good MC, Zalatan JG, Lim WA. Scaffold proteins: hubs for controlling the flow of cellular information. Science. 2011;332:680–6. doi: 10.1126/science.1198701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li GW, Xie XS. Central dogma at the single-molecule level in living cells. Nature. 2011;475:308–15. doi: 10.1038/nature10315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Feinerman O, Veiga J, Dorfman JR, Germain RN, Altan-Bonnet G. Variability and robustness in T cell activation from regulated heterogeneity in protein levels. Science. 2008;321:1081–4. doi: 10.1126/science.1158013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cohen AA, Geva-Zatorsky N, Eden E, Frenkel-Morgenstern M, Issaeva I, Sigal A, Milo R, Cohen-Saidon C, Liron Y, Kam Z, et al. Dynamic proteomics of individual cancer cells in response to a drug. Science. 2008;322:1511–6. doi: 10.1126/science.1160165. [DOI] [PubMed] [Google Scholar]
- 66.Csete M, Doyle J. Bow ties, metabolism and disease. Trends Biotechnol. 2004;22:446–50. doi: 10.1016/j.tibtech.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 67.Oda K, Matsuoka Y, Funahashi A, Kitano H. A comprehensive pathway map of epidermal growth factor receptor signaling. Mol Syst Biol. 2005;1:0010. doi: 10.1038/msb4100014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang LS, Leebens-Mack J, Kerr Wall P, Beckmann K, dePamphilis CW, Warnow T. The impact of multiple protein sequence alignment on phylogenetic estimation. IEEE/ACM Trans Comput Biol Bioinform. 2011;8:1108–19. doi: 10.1109/TCBB.2009.68. [DOI] [PubMed] [Google Scholar]
- 69.Hanks SK, Hunter T. Protein kinases 6. The eukaryotic protein kinase superfamily: kinase (catalytic) domain structure and classification. FASEB J. 1995;9:576–96. [PubMed] [Google Scholar]
- 70.Huelsenbeck JP, Ronquist F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17:754–5. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- 71.Drummond AJAB, Buxton S, Cheung M, Cooper A, Duran C, Field M, Heled J, Kearse M, Markowitz S, Moir R, et al. Geneious v5.6. Available from http://wwwgeneiouscom 2012.
- 72.Michaelis L, Menten ML, Johnson KA, Goody RS. The original Michaelis constant: translation of the 1913 Michaelis-Menten paper. Biochemistry. 2011;50:8264–9. doi: 10.1021/bi201284u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Koshland DE, Jr., Goldbeter A, Stock JB. Amplification and adaptation in regulatory and sensory systems. Science. 1982;217:220–5. doi: 10.1126/science.7089556. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.