Abstract
Hematopoietic cell transplant (HCT) is a life-saving therapy for many malignant and non-malignant bone marrow diseases. Associated morbidities are often due to transplant-related toxicities and infections, exacerbated by regimen-induced immune suppression and systemic incursion of bacterial products. Patients undergoing myeloablative conditioning for HCT become endotoxemic and display blood/plasma changes consistent with lipopolysaccharide (LPS)-induced systemic innate immune activation. Herein, we addressed whether patients scheduled for HCT display differences in recognition/response to LPS ex vivo traceable to specific single nucleotide polymorphisms (SNPs). Two SNPs of LPS binding protein (LBP) were associated with changes in plasma LBP levels, with one LBP SNP also associating with differences in efficiency of extraction and transfer of endotoxin to myeloid differentiation factor-2 (MD-2), a step needed for activation of TLR4. None of the examined SNPs of CD14, bactericidal/permeability-increasing protein (BPI), TLR4 or MD-2 were associated with corresponding protein plasma levels or endotoxin delivery to MD-2, but CD14 and BPI SNPs significantly associated with differences in LPS-induced TNF-α release ex vivo and infection frequency, respectively. These findings suggest that specific LBP, CD14 and BPI SNPs might be contributory assessments in studies where clinical outcome may be affected by host response to endotoxin and bacterial infection.
Keywords: LPS-binding protein (LBP), bactericidal/permeability-increasing protein (BPI), bone marrow transplant, hematopoietic cell transplant (HCT), lipopolysaccharide (LPS), CD14, TNF
Introduction
Hematopoietic cell transplant (HCT) is a life-saving therapy for many malignant and non-malignant bone marrow diseases. HCT regimen-related toxicities and post-transplant infections contribute significantly to post-transplant morbidity and mortality.1,2 Post-transplant inflammation and infections are likely due to the combined effects of conditioning regimen-induced immune suppression and tissue injury, which permit translocation of bacteria and bacterial products to the systemic circulation.3,4 Lipopolysaccharide (LPS), often referred to as endotoxin, is an abundant and potently pro-inflammatory constituent of the outer membrane of Gram-negative bacteria, and humans undergoing myeloablative HCT are frequently endotoxemic.5 LPS leakage across damaged intestinal epithelium may be a significant contributor to the so-called ‘cytokine storm’ after myelo-ablative HCT.6 Systemic LPS induces production of a range of pro-inflammatory cytokines and chemokines, including TNF-α,7 CCL2, CXCL10 and CCL5,8 as well as activation of the inflammasome complex and IL-1 β secretion.9
The potential of pM concentrations of plasma endotoxin to contribute to immune pathology in HCT patients reflects the remarkable potency of the immune-stimulating properties of LPS in humans. This potency depends on the ordered actions of several host extracellular and cell surface proteins, including LPS-binding protein (LBP), membrane and soluble forms of CD14, secreted or receptor-associated myeloid differentiation factor-2 (MD-2) and TLR4, which together make possible host detection and response to pM concentrations of endotoxin. In brief, LBP catalyzes extraction and transfer of individual molecules of endotoxin from the outer membrane of Gram-negative bacteria (or from aggregates of purified endotoxin) to CD14, followed by transfer of activating endotoxin to MD-2 and engagement and activation of MD-2/ TLR4 complexes.10–12 This catalytic role of LBP is most pronounced at relatively low LBP concentrations. When present in higher concentrations, LBP can react stoichiometrically with endotoxin and endotoxin/CD14 complexes to promote clearance of endotoxin without engagement and activation of TLR4.13,14 While the ability of humans to respond sensitively and robustly to endotoxin is important for timely mobilization of host defense responses against invading Gram-negative bacteria, it also increases the risk of potentially harmful immune responses.
Given the potent bioactivity of LPS, humans express a variety of mechanisms to counteract the effects of endotoxin and endotoxemia; these serve to blunt TLR4 signaling and/or skew cellular responses toward production of anti-inflammatory rather than pro-inflammatory mediators. Such mechanisms include (a) modification of MD-2 and TLR4 surface expression;15 (b) alteration of relative molar ratios of LBP, CD14 and MD-2;11,16 (c) enzymatic detoxification of LPS;17 and (d) neutralization of endotoxin by cationic antimicrobial proteins and peptides,18 such as the bactericidal/permeability-increasing protein (BPI), which has high affinity for LPS12 and is found in high concentrations in human neutrophils. Other factors involved in the recognition of, and response to, bacterial compounds include TLR2 (recognizes bacterial lipoproteins), and TLR adaptor molecules myeloid differentiation primary response gene 88 (MyD88) and MyD88 adapter-like Mal/TIRAP, which enable downstream signaling following TLR engagement. The complexity of host mechanisms mediating and regulating host recognition and response to endotoxin is likely to result in variability between individuals in their response to plasma endotoxin.
One possible source of variability between individuals in levels and/or activity of pertinent LPS recognition and signaling proteins could originate from allelic differences deriving from single nucleotide polymorphisms (SNPs). Among these, certain SNPs within the BPI, CD14, LBP, Mal/TIRAP, MD-2, TLR2 and TLR4 genes are associated with genotype-dependent in vitro, ex vivo and/or in vivo (clinical) phenotypes (see Table 1). It is therefore conceivable that effects of endotoxemia in HCT (or other) patients could be influenced by differences between patients in their genotypes for specific LPS recognition or signaling proteins. To date, there have been no reported studies of the possible association of specific SNP genotypes of plasma-derived endotoxin recognition proteins with the efficiency of recognition and response to plasma LPS.
Table 1.
SNPs analyzed in Cohort II.
| Gene | SNP | Nucleotides | Phenotype | Findings/clinical correlations |
|---|---|---|---|---|
| BPI | rs1341023 | C/T | Ala16Val | – |
| rs4358188 | A/G | Glu216Lys | Rapid airflow decline after HSCT (Chien, Zhao et al. 2006) | |
| LessaGVHD after HCT (Wermke, Maiwald et al. 2010) | ||||
| rs1131847 | A/G | 3′ UTR | Plasma BPI protein levels (Gubern, Lopez-Bermejo et al. 2006) | |
| rs6099106 | C/T | intronic | COPD (Chen, Ou et al. 2012) LPS | |
| CD14 | rs2569190 | A/G | promoter | Airway inflammation with LPS exposure (Smit, Heederik et al. 2011) |
| sCD14 levels in blood (Levan, Michel et al. 2008; Munthe-Kaas, Torjussen et al. 2010) | ||||
| Increased promoter activity (LeVan, Bloom et al. 2001; Mertens, Bregadze et al. 2009) | ||||
| Survival in ICU patients (Fallavena, Borges et al. 2009) | ||||
| rs2569191 | C/T | – | sCD14 levels in blood(Munthe-Kaas, Torjussen et al. 2010) | |
| Airway inflammation (Smit, Heederik et al. 2011) | ||||
| LBP | rs2232571 | C/T | promoter | Severe sepsis (Flores, Perez-Mendez et al. 2009) |
| rs2232582 | C/T | promoter | Gram-negative bacteremia after HCT (Chien, Boeckh et al. 2008) | |
| rs2232613 | C/T | Pro333Leu | – | |
| rs2232618 | C/T | Phe436Leu | Susceptibility to sepsis and MOD (Zeng, Gu et al. 2012) | |
| Mal/TIRAP | rs8177374 | C/T | Ser180Leu | Severe infections after surgery (Kumpf, Giamarellos-Bourboulis et al. 2010) |
| MD-2 | rs6472812 | A/G | Gly56Arg | – |
| TLR2 | rs5743708 | A/G | Arg753Gln | Reduced responsiveness to TLR ligands (Lorenz, Mira et al. 2000) |
| rs1898830 | A/G | – | Bronchiolitis obliterans after lung transplantation (Kastelijn, van Moorsel et al. 2010) | |
| rs1816702 | C/T | – | Monocyte activation (Bielinski, Hall et al. 2011) | |
| TLR4 | rs4986790 | A/G | Asp299Gly | Severe infections after surgery (Kumpf, Giamarellos-Bourboulis et al. 2010) |
| Hypo-responsiveness to LPS (Arbour, Lorenz et al. 2000; Guo, Loke et al. 2009) | ||||
| Gram-negative bacterial infection (Agnese, Calvano et al. 2002) | ||||
| Allograft rejection (Palmer, Burch et al. 2003) | ||||
| rs4986791 | C/T | Thr399Ile | Severe infections after surgery (Kumpf, Giamarellos-Bourboulis et al. 2010) | |
| Hypo-responsiveness to LPS (Arbour, Lorenz et al. 2000; Guo, Loke et al. 2009) | ||||
| Gram-negative bacterial infection (Agnese, Calvano et al. 2002) | ||||
| Allograft rejection (Palmer, Burch et al. 2003) |
MOD: multiple organ dysfuction; COPD: chronic obstructive pulmonary disease; ICU: intensive care unit.
To address the hypothesis that individual HCT patients differ in their recognition of and response to plasma endotoxin due, at least in part, to SNP-derived genotypic differences in LPS recognition proteins, we designed a new observational cohort study and characterized potentially relevant SNPs of LBP, CD14, MD-2, TLR4, BPI and Mal/TIRAP, and the possible relation of specific SNP genotypes to (a) plasma protein levels; (b) plasma-dependent extraction and transfer of endotoxin to MD-2; and (c) responses to LPS in whole blood.
These studies have revealed specific LBP and CD14 SNPs that associate with alterations in plasma protein levels, plasma-dependent extraction and transfer of activating endotoxin to MD-2 (LBP), and activation of MD-2/TLR4 (CD14). In addition, our studies have revealed a potential association of a BPI SNP with post-transplant infection frequency, suggesting that these SNPs should be monitored in future clinical studies where clinical outcome may be affected by host response to plasma endotoxin.
Materials and methods
Patient characteristics
Cohort I
Patients (n = 48) undergoing myeloablative allogeneic HCT from 2005 to 2009 at Boston Children’s Hospital (BCH) or Brigham and Women’s Hospital (BWH) were recruited prospectively onto an Institutional Review Board (IRB) approved protocol. All participants and/or legal guardians gave consent or assent as appropriate. Patient and treatment characteristics have been published previously (Supplementary Table S1).19 Supportive care was per institutional routine.20,21 Prophylactic oral non-absorbable antibiotics were administered: bacitracin and polymyxin (BWH) or vancomycin (BCH). Blood counts and cultures were performed in clinical laboratories.
Cohort II
Patients (n = 201) being evaluated for myeloablative or non-myeloablative allogeneic, autologous or syngeneic HCT from 2009 to 2011 at the Dana-Farber Cancer Institute (DFCI), BWH, the University of Iowa and Veterans’ Administration Medical Center, Iowa City (UIVAMC) were recruited prospectively onto an IRB-approved study (Supplementary Table S2). Onetime peripheral blood samples were taken before transplant (B—baseline) for SNP genotyping and in vitro assays. DFCI/BWH care guidelines were identical to those of Cohort I. Supportive care was according to institutional routine20,21 at UIVAMC (prophylactic antibiotics used were levofloxacin 500 mg daily or ciprofloxacin 500 mg q12h). Infection data were derived from Center for International Blood & Marrow Transplant Research day 100 reports. We did not have the statistical power to analyze the effects of SNPs on veno-occlusive disease or diffuse alveolar hemorrhage/interstitial pneumonia owing to low frequencies of these clinical complications in our patient population (Supplementary Table S3).
Blood collection and plasma preparation
Peripheral blood samples were drawn into K2-EDTA or sodium heparin vacutainer tubes (Becton-Dickinson, San Jose, CA, USA). Cohort I samples were collected at baseline (B—before myelo-ablation), d 0 (d 0—day of transplant prior to cell infusion) and d 7, 14, 21 and 28, while Cohort II samples were drawn after enrollment and prior to onset of conditioning. Indicated volumes were centrifuged at 1200 g for 5 min at 4°C, and aliquots of the recovered plasma stored in pyrogen-free tubes (Denville Scientific, Metuchen, NJ, USA) at −20°C.
LPS-induced TNF-α release ex vivo
Samples of EDTA blood (Cohort II) were stimulated with 1 ng/ml LPS (List Laboratories, Campbell, CA, USA) in Modified Eagle Medium (Invitrogen, Grand Island, NY, USA) control or medium only, and incubated in pyrogen-free 1.5-ml tubes for 4 h at 37°C, rotating end-over-end, prior to collection of the extracellular medium for subsequent TNF-α ELISA (R&D Systems, Minneapolis, MN, USA or BD BioSciences, San Jose, CA, USA).
ELISAs and measurement of chemokines
ELISAs were used to measure TNF-α, sCD14, sTNF-RI, sTNF-RII (all R&D Systems), BPI and LBP (HyCult, Uden, the Netherlands), according to the manufacturers’ instructions. sMD-2 was measured by a customized ELISA as previously described.22 CCL2, CCL5 and CXCL10 were measured by flow cytometry (MoFlo; DakoCytomation, Glostrup, Denmark) using fluorescent beads covalently linked to corresponding mAbs (Cytometric Bead Array BD Flex Sets; BD BioSciences) and results analyzed with Summit v4.0 software (DakoCytomation).
Quantitative real-time PCR arrays
Peripheral blood (2.5 ml) was collected into PAXgene tubes (PreAnalytiX/Qiagen, Hilden, Germany). Total mRNA was isolated using the PAXgene blood RNA kit and the RNeasyMinElute cleanup kit, followed by cDNA synthesis using the RT2 First Strand Kit (all Qiagen) according to the manufacturer’s instructions. The equivalent of 1 ng RNA/well was assayed using a customized 84-gene signaling pathway PCR array (Qiagen), according to the manufacturer’s instructions. Total RNA was normalized using five housekeeping genes and data analyzed using the ΔΔCt method.23 Mean fold changes in gene expression in a given patient were calculated by comparing mRNA levels at baseline (B) with subsequent time-points (d 0 and d 14), with >fourfold increases and decreases classified as meaningful up- and down-regulation, respectively.
SNP genotyping
SNPs were genotyped by allelic discrimination using TaqMan assays following the manufacturer’s protocol (Applied Biosystems, Foster City, CA, USA). Two SNPs were genotyped by direct sequencing: rs6099106 as previously described.24 PCR reactions were conducted as previously described and sequencing was performed by Functional Biosciences (http://functionalbio.com/web/) Rationale for SNP selection included significant association in previous publications, known missense mutations or haplotype coverage in relevant pathway genes (Table 1). Observed SNP frequencies within Cohort II are shown in Table 2.
Table 2.
SNP frequencies in cohort II.
| Frequencies in current cohort |
||||||
|---|---|---|---|---|---|---|
| NCBI number | Gene | SNP | Phenotye | individuals (%) | individuals (n) | total (n) |
| rs1131847 | BPI | A/G | 3′ UTR | |||
| AA | 12.50% | 25 | 200 | |||
| AG | 48.50% | 97 | ||||
| GG | 39.00% | 78 | ||||
| rs1341023 | BPI | C/T | Ala16Val | |||
| CC | 22.00% | 44 | 198 | |||
| CT | 51.00% | 100 | ||||
| TT | 27.00% | 54 | ||||
| rs4358188 | BPI | A/G | Glu216Lys | |||
| AA | 26.00% | 51 | 198 | |||
| AG | 49.00% | 98 | ||||
| GG | 25.00% | 49 | ||||
| rs6099106 | BPI | C/T | intronic | |||
| CC | 13.00% | 26 | 200 | |||
| CT | 53.00% | 106 | ||||
| TT | 34.00% | 68 | ||||
| rs2569190 | CD14 | A/G | promoter region | |||
| AA | 28.00% | 54 | 192 | |||
| AG | 41.00% | 79 | ||||
| GG | 31.00% | 59 | ||||
| rs2569191 | CD14 | C/T | – | |||
| CC | 28.00% | 48 | 170 | |||
| CT | 40.00% | 68 | ||||
| TT | 32.00% | 54 | ||||
| rs2232571 | LBP | C/T | promoter region | |||
| CC | 3.00% | 6 | 194 | |||
| CT | 35.00% | 68 | ||||
| TT | 62.00% | 120 | ||||
| rs2232582 | LBP | C/T | promoter region | |||
| CC | 2.00% | 4 | 199 | |||
| CT | 30.00% | 59 | ||||
| TT | 68.00% | 136 | ||||
| rs2232613 | LBP | C/T | Pro333Leu | |||
| CC | 59.00% | 116 | 196 | |||
| CT | 37.00% | 72 | ||||
| TT | 4.00% | 8 | ||||
| rs2232618 | LBP | C/T | Phe436Leu | |||
| CC | 1.00% | 2 | 196 | |||
| CT | 15.00% | 29 | ||||
| TT | 84.00% | 165 | ||||
| rs8177374 | MAL/TIRAP | C/T | Ser180Leu | |||
| CC | 75.50% | 149 | 197 | |||
| CT | 23.00% | 45 | ||||
| TT | 1.50% | 3 | ||||
| rs6472812 | MD-2 | A/G | Gly56Arg | |||
| AA | 0.50% | 1 | 194 | |||
| AG | 8.00% | 16 | ||||
| GG | 91.50% | 177 | ||||
| rs5743708 | TLR2 | A/G | Arg753Gln | |||
| AA | 4.00% | 8 | 197 | |||
| AG | 6.00% | 11 | ||||
| GG | 90.00% | 178 | ||||
| rs1898830 | TLR2 | A/G | – | |||
| AA | 42.00% | 81 | 192 | |||
| AG | 46.00% | 89 | ||||
| GG | 12.00% | 22 | ||||
| rs1816702 | TLR2 | C/T | – | |||
| CC | 76.00% | 146 | 191 | |||
| CT | 23.50% | 44 | ||||
| TT | 0.50% | 1 | ||||
| rs4986790 | TLR4 | A/G | Asp299Gly | |||
| AA | 86.00% | 172 | 199 | |||
| AG | 13.00% | 26 | ||||
| GG | 1.00% | 1 | ||||
| rs4986791 | TLR4 | C/T | Thr399Ile | |||
| CC | 85.00% | 166 | 196 | |||
| CT | 14.00% | 29 | ||||
| TT | 1.00% | 1 | ||||
Assay of plasma-dependent extraction and transfer of endotoxin to MD-2
Typical incubation mixtures contained 85% (v/v) human plasma/EDTA spiked with 5% (v/v) purified aggregates of metabolically-labeled meningococcal lipooligosaccharide (LOS)26,27 {[3H]LOSagg (500 cpm; 100 pM (500 pg LOS/ml), final concentration} and 10% (v/v) insect cell (Hi5)-conditioned cell medium with or without His6-sMD-227–29 [~5 nM (100 ng bioactive sMD-2 monomer/ml), final concentration] in a total volume of 0.2 ml. Human plasma samples were either from individual donors/patients or from a pool of healthy donors used as a control for each assay. All plasma samples were stored in aliquots at −80°C and used after one freeze/thaw. Plasma + [3H]LOSagg±His6-sMD-2 were incubated for 15 min at 37°C followed by dilution 2 × with PBS, addition of 20 µl of a 1:1 slurry of Ni2+ FF-Sepharose and incubation on a rotating wheel at 4°C for 60 min. After sedimentation of the Ni2+ FF-Sepharose gel by centrifugation at 10,000 g for 30 s, the supernatant was removed. The gel was washed once with 0.4 ml ice-cold PBS. The recovered supernatants and resuspended gel (in PBS) were transferred to counting vials to measure the recovered [3H]LOS by liquid scintillation spectroscopy, using a Beckman LS6500 scintillation counter (Beckman Coulter, Indianapolis, IN, USA). Percent capture of [3H]LOS by the Ni2+FF-Sepharose gel was calculated as {cpm recovered in gel/[total cpm recovered (gel + combined supernatants)]} × 100. Percent capture of [3H]LOS in the absence of plasma (incubations contained PBS/1% human albumin instead of plasma) was < 1%. Percent capture of [3H]LOS in the absence of added His6-sMD-2 was 0.5±0.2 (range:<0.1–1.1) and was subtracted from percentage capture observed for the same plasma sample incubated with His6-sMD-2 to calculate His6-sMD-2-dependent co-capture of [3H]LOS by the Ni2+ FF-Sepharose gel (i.e. plasma-dependent extraction and transfer of [3H]LOS to His6-MD-2). Data for individual plasma samples are expressed as [% MD-2-dependent co-capture of [3H]LOS with the individual plasma/% MD-dependent co-capture of [3H]LOS with the healthy donor plasma pool] × 100 and, for each individual plasma sample, represent the mean of 2–4 independent determinations. MD-2-dependent co-capture of [3H]LOS with the healthy donor plasma pool was measured in each experiment (also testing several individual plasma samples) and was 15.2 ± 2.4 (SD) % (n = 25).
Statistical analyses
Samples with undetectable analytes were assigned a value at half the lower limit of detection.
Cohort I
For most assays, data were analyzed after logarithmic transformation, as this yielded distributions that were more approximately normal. For these data, values were then transformed back to original units and plotted on a logarithmic axis. The Wilcoxon signed rank test for matched pairs was used when comparing values for the same patients at different time points with values compared with baseline (B). The significance of fold-change relative to baseline in gene expression for each gene was evaluated with a paired t-test on the log-transformed fold-changes. All P-values are two-sided. mRNA levels were normalized to housekeeping genes and quantified using the ΔΔ comparative threshold (Ct) method,23 using an analysis tool from Qiagen (http://www.sabiosciences.com/pcr/arrayanalysis.php). Statistical analyses of mRNA expression employed SA Biosciences software (Qiagen) using a Student’s t-test of the replicate 2−ΔCt values for each gene compared with baseline (see Supplementary Table S4).
Cohort II
Owing to disproportionate distribution of certain SNP genotypes in our study population, only groups of n ≥ 8 were considered for statistical comparisons. For two-way comparisons, the two-tailed t-test or Mann–Whitney test was applied for parametric or non-parametric distributions, respectively. For three-group analyses, a Kruskal–Wallis test with Dunn’s Multiple Comparisons Test was applied (all non-parametric distributions). For both Cohorts I and II, results were considered significant at P < 0.05, and indicated as follows: *P < 0.05, **P< 0.01, ***P < 0.001. Statistical significance and graphic output were generated using Qiagen online analysis software (http://www.sabiosciences.com/pcr/arrayanalysis.php), Prism v. 4.0a (GraphPad Software, San Diego, CA, USA) and SAS v. 9.1 (SAS Institute, Cary, NC, USA).
Results
Plasma endotoxin during myelo-ablative HCT is followed by systemic inflammatory responses
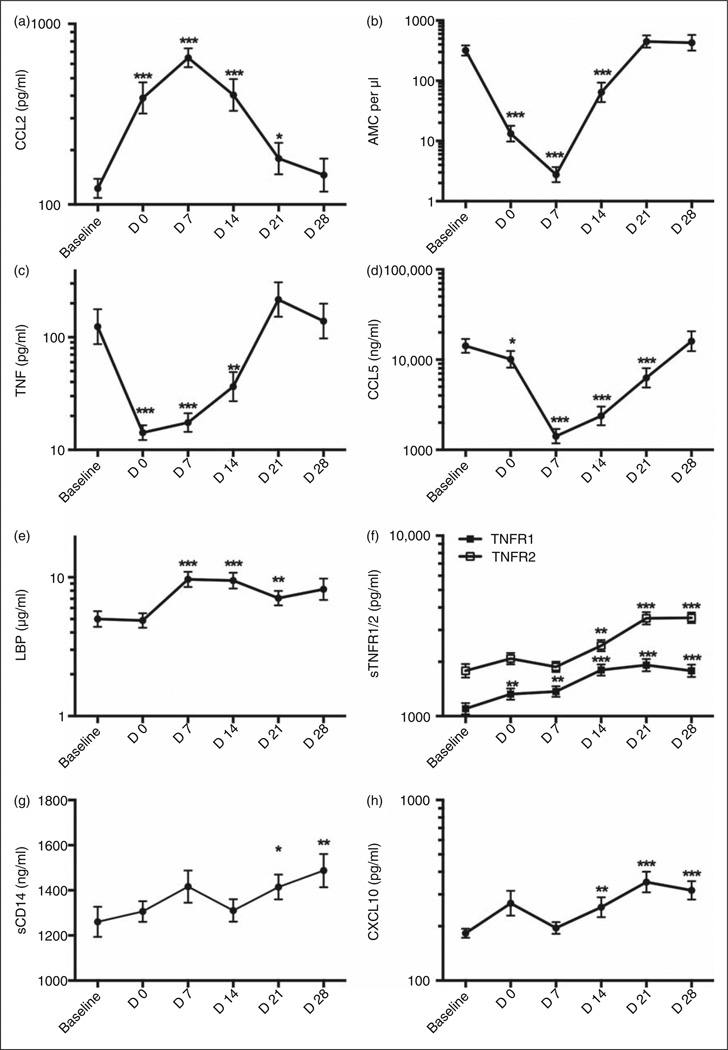
We recently reported a high incidence of endotoxemia in HCT patients enrolled in an observational cohort study19 (Cohort I) with elevated levels of plasma LPS manifest by the completion of myeloablative conditioning (d 0). Changes in monocyte cell surface levels of membrane (m)CD14 (diminished) and TLR4 (increased),19 and in plasma levels of monocyte chemotactic protein 2 (CCL2; Figure 1a) were also manifest on d 0, consistent with acute host responses to plasma endotoxin during myeloablative conditioning. The regimen-induced monocyte nadir at d 0 and d 7 corresponded with a significant decrease in levels of spontaneous TNF-α production in whole blood ex vivo on those days (Figure 1b, c), with both changes normalizing by d 21. Plasma levels of the chemokine CCL5 followed a similar pattern (Figure 1d), contemporaneous with nadirs and normalization in absolute neutrophil count, platelet counts and BPI, and a spike in IL-6 production.19 In contrast, plasma levels of LBP rose rapidly, peaking at d 7 and d 14, (Figure 1e), while levels of soluble TNF-α receptors 1 and 2 (sTNRF1/2), sCD14 and the IFN-responsive cytokine CXCL10 also increased. However, these changes were more gradual and sustained (Figure 1f – h). No changes in sMD-2 or TLR2 were observed (data not shown).
Figure 1.
In vivo and ex vivo cellular and plasma changes in inflammatory responses following onset of endotoxemia in myeloablative HCT. Cohort I: peripheral blood and plasma samples from patients undergoing HCT were collected at baseline (B—before myeloablative conditioning), d 0 (day of transplant) and subsequent d 7, 14, 21 and 28, and analyzed for (a) CCL2 protein (pg/ml), (b) absolute monocyte count (AMC) per µl, (c) spontaneously produced TNF-a protein (pg/ml), (d) CCL5 protein (ng/ml), (e) LBP protein (µg/ml), (f) TNF-α receptor 1 and 2 (TNFR1/2) protein (pg/ml), (g) soluble CD14 (sCD14) protein (ng/ml) and (h) CXCL10 protein (pg/ml) over time. Values are shown as mean ± SEM for n = 22–48 patients. Statistical significance compared to B was assessed using Wilcoxon signed rank test for matched pairs and is indicated as *P < 0.05 **P < 0.01 and ***P<0.001.
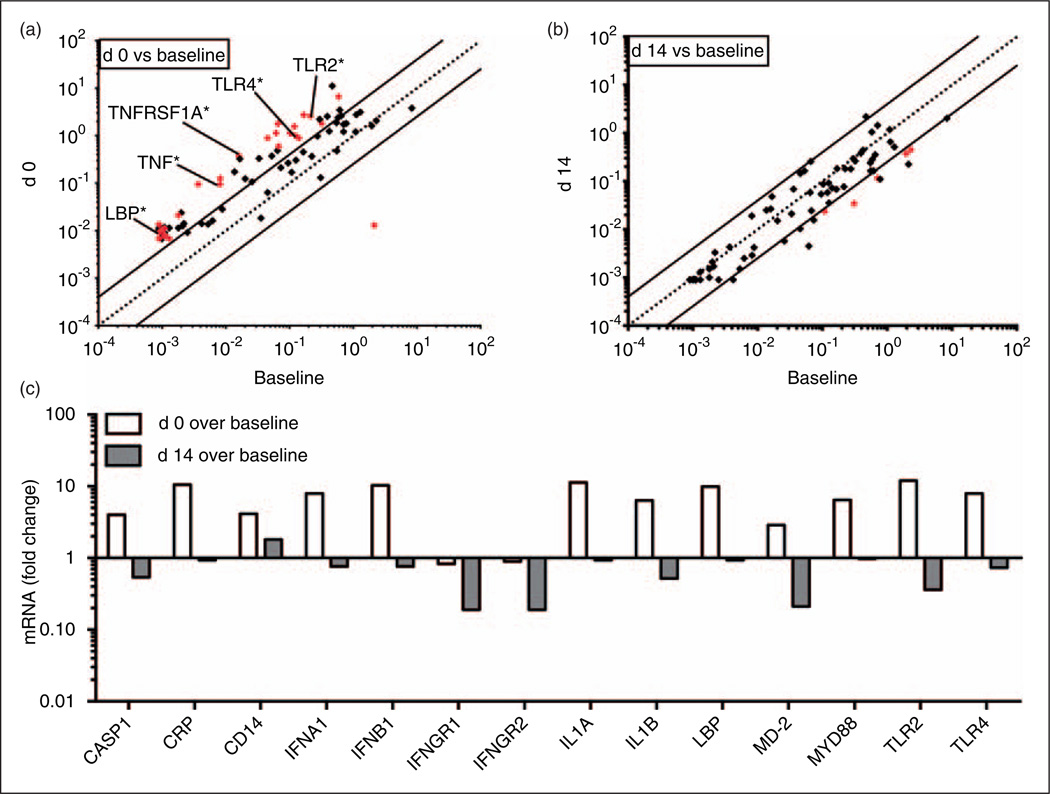
Measurement of peripheral blood levels of mRNA derived from a customized panel of 84 inflammatory genes provided further evidence suggesting in vivo activation of cellular inflammatory responses by d 0 (Figure 2a; Supplementary Table S4) that generally returned to baseline levels (i.e. mRNA levels prior to initiation of myeloablation) by d 14 (Figure 2b). Species of mRNA that were elevated at d 0 relative to cells collected at baseline and at d 14 included multiple genes involved in the LPS–TLR4 signaling axis (LBP, CD14, MD-2, TLR4 and MyD88), as well as inflamma-some signaling (CASP1, IL1B) and the acute phase response (CRP, LBP) (Figure 2c). The in vivo elevation of whole blood mRNA levels for these inflammatory genes is especially remarkable given the marked reduction in blood monocyte levels and ex vivo whole blood responses to systemic LPS (e.g. secretion of TNF-α) manifest by d 0 (Figure 1b, c). Taken together, these findings suggest acute in vivo endotoxin-induced innate immune activation in patients undergoing myelo-ablative conditioning prior to HCT.
Figure 2.
Changes in mRNA levels of selected inflammatory genes following onset of endotoxemia in myelo-ablative HCT. Cohort I: peripheral blood samples from patients undergoing HCT were collected at baseline (B—before myelo-ablative conditioning), d 0 (day of transplant) and d 14, and mRNA extracted and analyzed for selected inflammatory markers. Fold changes of mRNA levels over B (x-axis) on (a) d 0 (y-axis) and (b) d 14 (y-axis) are shown. (c) Differential regulation over B comparing d 0 and d 14 (from a and b) are shown as bar graphs for a selection of genes. Statistical significance comparing B to either d 0 or d 14 was assessed by Student’s t-test of the replicate 2−∆Ct values for each gene compared with baseline. Fold changes greater than fourfold were considered meaningful and statistically significant changes are shown in red for each individual gene (Supplementary Table S4).
Associations of specific genotypes of LBP with differences in LBP protein plasma levels
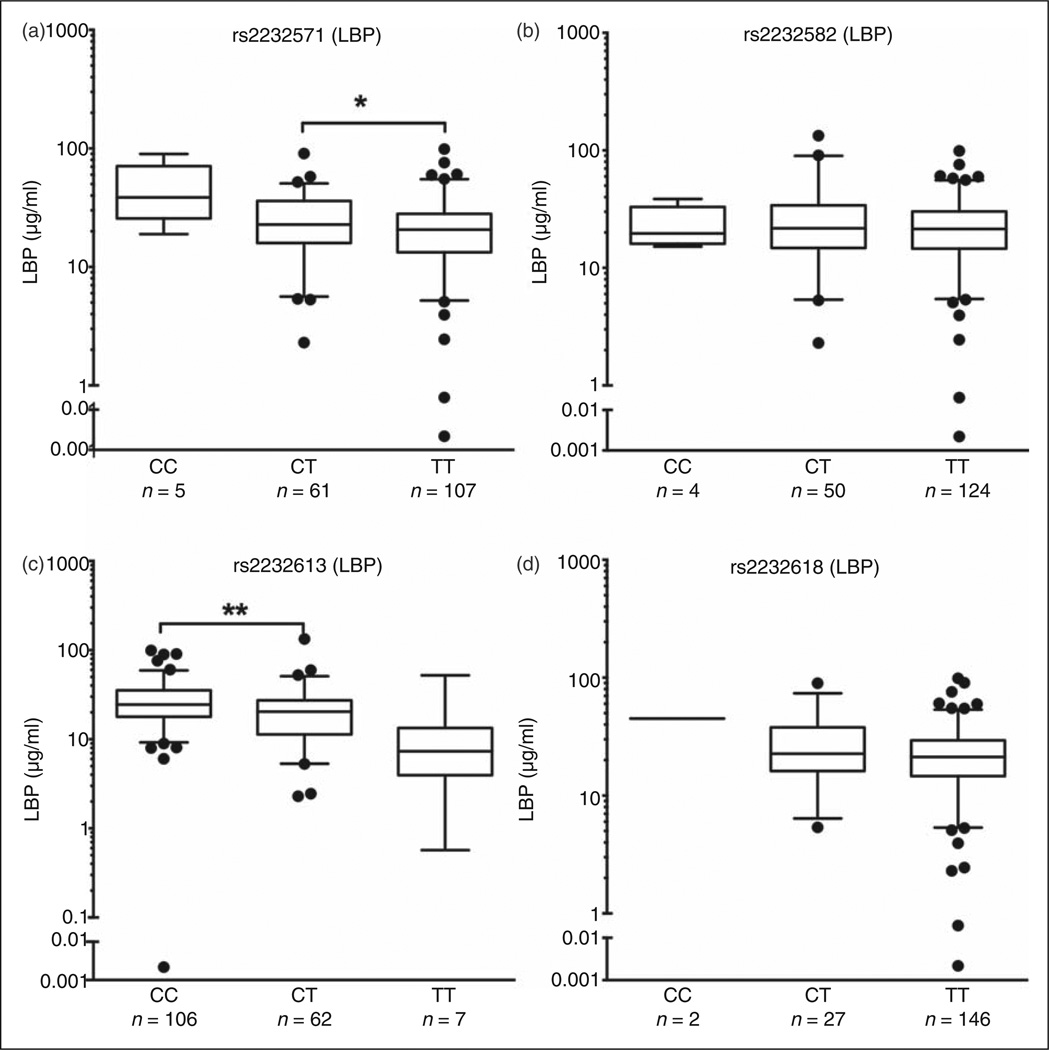
Four LBP SNPs were analyzed: two SNPs located within the structural gene of LBP, giving rise to structural alterations in LBP (rs2232613 Pro333Leu; rs2232618 Phe436Leu) and two SNPs located in the promoter region of LBP (rs2232571 and rs2232582; Table 1). Genotype-associated differences in plasma levels of LBP protein were observed for rs2232571 and rs2232613. These differences were statistically significant for the more frequently represented genotypes in the Cohort II patient population (rs2232571: CT>TT; rs2232613: CC>CT) (Figure 3a, c). In contrast, no statistically significant differences in plasma LBP protein levels were detected for the other two LBP SNPs examined (Figure 3b, d).
Figure 3.
LBP SNPs rs2232571 and rs2232613 are associated with plasma protein levels. Cohort II: plasma samples corresponding to LBP SNPs (a) rs2232571, (b) rs2232582, (c) rs2232613 and (d) rs2232618 are shown. LBP protein levels in plasma were assessed by ELISA. Values are represented as median and 5–95 percentiles, with individual outliers shown. Only groups of n ≥ 8 were considered for statistical analyses. Statistical significance was assessed by two-tailed t-test or Mann-Whitney test (parametric or non-parametric distributions, respectively) and indicated as *P<0.05 and **P<0.01.
Differences in plasma-dependent extraction and transfer of endotoxin monomer to MD-2 associate with specific LBP genotypes
Maximal potency of activation of TLR4 by endotoxin depends on sequential interactions of endotoxin with LBP, membrane (m) and/or soluble (s) CD14 and MD-2 (secreted or, more typically, associated with TLR4 on cell plasma membranes). LBP catalyzes extraction and transfer of individual molecules of endo-toxin from aggregated forms of endotoxin to sCD14 or mCD14, which, in turn, allows more efficient transfer of TLR4-activating endotoxin monomers to MD-2 and MD-2/TLR4 complexes.10–12
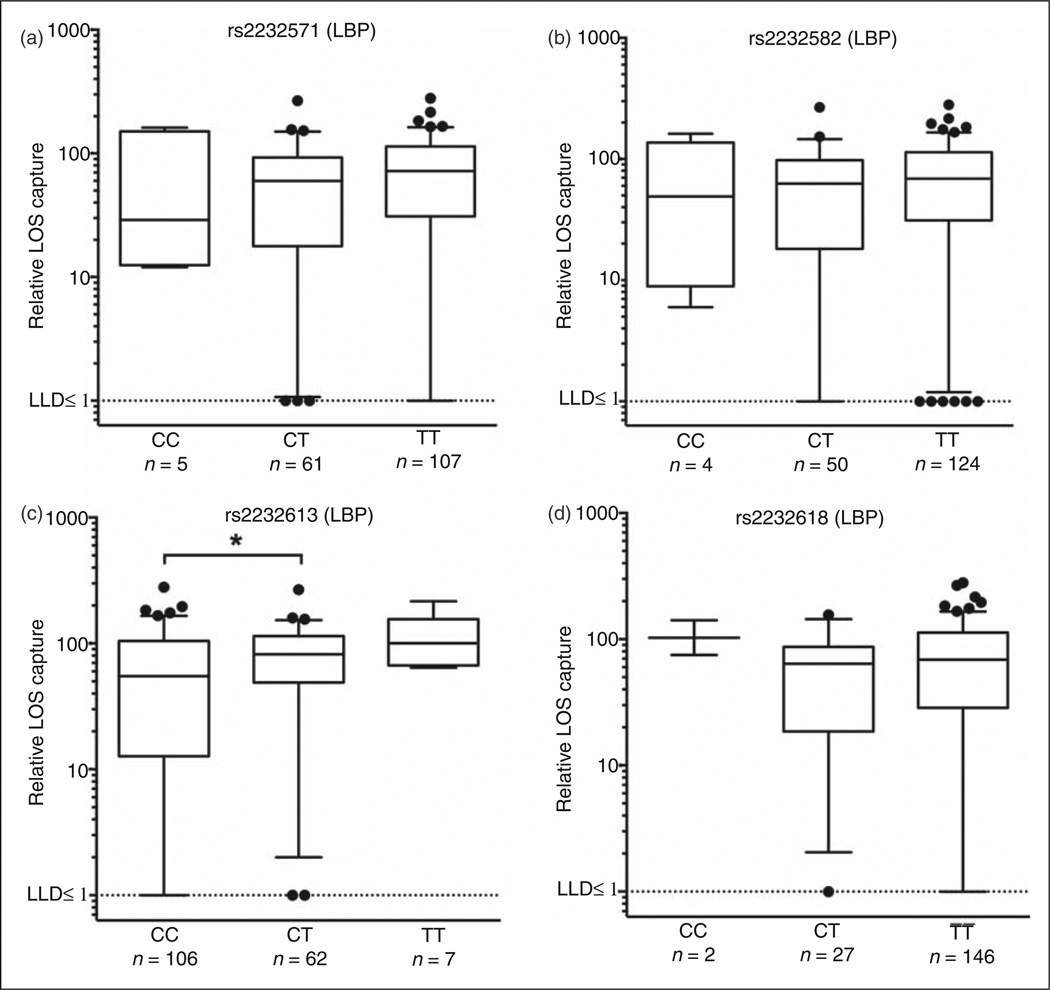
To measure the extraction and transfer of endotoxin to MD-2, we designed a novel assay in which individual plasma samples were spiked with recombinant His6-MD-2 and aggregates of metabolically labeled, purified [3H] endotoxin of very high and virtually uniform specific radioactivity (see ‘Materials and methods’).13,25,30 LBP SNP rs2232613 showed genotype-associated differences in endotoxin extraction and transfer to MD-2 that seemed inversely related to genotype-associated mean plasma LBP levels (compare Figures 3c and 4c). Only the differences in MD-2 transfer between the most frequently represented genotypes of LBP rs2232613 (CT > CC; Figure 4c) were statistically significant. The other three LBP SNPs were not associated with any significant differences in endotoxin extraction and transfer to MD-2 (Figure 4a, b, d).
Figure 4.
LBP SNP rs2232613 is associated with altered endotoxin extraction and delivery to MD-2. Cohort II: plasma samples corresponding to LBP SNPs (a) rs2232571, (b) rs2232582, (c) rs2232613 and (d) rs2232618 are shown. Plasma-dependent extraction and transfer of endotoxin monomer to MD-2 was measured and represented as relative LOS capture as described in the ‘Materials and methods’. Values are presented as median relative LOS capture (5–95 percentile with outliers shown). Assay detection limit is signified by dotted line (≤ 1%). Only groups of n≥ 8 were considered for statistical comparisons. Statistical significance was assessed by two-tailed t-test or Mann–Whitney test (parametric or non-parametric distributions, respectively) and indicated as *P < 0.05. LLD: lower limit of detection.
CD14 genotypes confer differences in cell activation by LPS in whole blood ex vivo
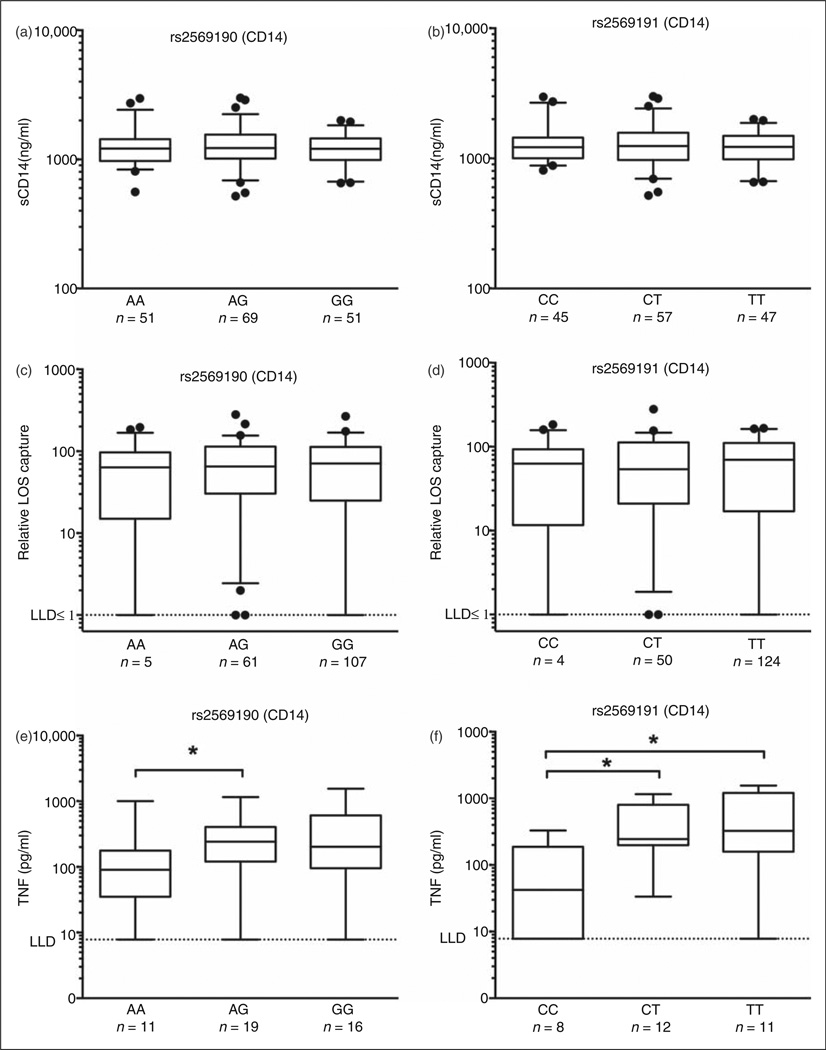
Two CD14 SNPs (rs2569190 and rs2569191) were analyzed, the former located within the promoter region of the CD14 gene (Table 1). Neither genotype-associated differences in CD14 protein levels in plasma nor in plasma-dependent extraction and transfer of [3H] endotoxin to His6-MD-2 were observed (Figure 5a – d). However, comparison of LPS-induced secreted TNF-α levels in vitro for CD14 SNPs showed a significant difference in TNF-α production between the AG and AA alleles for CD14 SNP rs2569190 (Figure 5e) and between the CC genotype of CD14 SNP rs2569191 compared with either the CT or the TT genotypes (Figure 5f). No effect on LPS-induced TNF-α production from peripheral blood was observed for any of the other SNPs investigated, including TLR4 SNPs rs4986790 and rs4986791 (data not shown).
Figure 5.
Association of CD14 SNPs rs2569190 and rs2569191 with altered LPS-induced TNF production ex vivo. Cohort II: plasma samples corresponding to CD14 SNPs rs2569190 (a, c, e) and rs2569191 (b, d, f) are shown. sCD14 protein levels were assessed by ELISA (a, b), ex vivo plasma-dependent extraction and transfer of endotoxin monomer to MD-2 was measured and represented as relative LOS capture as described in the ‘Materials and methods’ (assay detection limit signified by dotted line ≤ 1%) (c, d). Extracellular TNF-α accumulation measured by ELISA following ex vivo stimulation of whole blood with LPS (1 ng/ml) for 4h (assay detection limit signified by dotted line = 7.8125 pg/ml) (e, f). Values represent median and 5–95 percentile with outliers shown for raw data (a–d) and of log-transformed values labeled in original units (e, f). Only groups of n≥ 8 were considered for statistical comparisons. For two-way comparisons (c, d), two-tailed t-test or Mann–Whitney test was applied (parametric or non-parametric distributions, respectively). For three-group analyses (a, b, e, f), Kruskal-Wallis test with Dunn’s Multiple Comparisons Test was applied. Statistical significance is indicated as *P < 0.05. LLD: lower limit of detection.
BPI SNP rs4358188 genotype is associated with increased frequency of infection following autologous and syngeneic HCT
Infection outcome data for patients in Cohort II were available for all patients that underwent HCT (n= 188; Supplementary Table S2 and S3). In order to assess whether the SNPs studied were associated with differences in post-transplant infections, we restricted analysis to those patients receiving autologous and syngeneic transplants and, thus, in contrast to those receiving allogeneic cells, retained their SNP genotypes in the hematopoietic compartment (n=54). Of the 54 patients receiving autologous or syngeneic transplants, 21 patients had documented infections, all of whom experienced bacterial infection alone, or bacterial plus either viral or fungal infection (Table 3). Four BPI SNPs were analyzed: two located within the structural gene of BPI, giving rise to structural variants of BPI (rs1341023 Ala16Val; rs4358188 Glu216Lys), one located within the 3’-UTR (rs1131847) and one located within an intron (rs6099106) (Table 1). Consistent with prior studies linking BPI genotype with risks of infection,31,32 there was a decreasing association of infection rates with BPI SNP rs4358188 genotypes AA, AG and GG (n=54; Table 4). The limited numbers of patients with infections precluded a sub-group analysis by type of bacterial infection. No apparent differences in SNP genotype distributions and infection frequency in autologous and syngeneic HCT recipients were observed for the other three BPI SNPs or for CD14, LBP, Mal/TIRAP, MD-2, TLR2 or TLR4 SNPs (data not shown).
Table 3.
Cohort II infection data.
| All HCT patiens |
Allogeneic |
Autologous/Syngeneic |
||||||
|---|---|---|---|---|---|---|---|---|
| Patients with infections | (n/total) | (%) | (n/total) | (%) | (n/total) | (%) | ||
| Bacteria only | 45/188 | 24% | 25/134 | 19% | 20/54 | 37% | ||
| Bacteria total | 77/188 | 41% | 56/134 | 43% | 21/54 | 39% | ||
| Virus only | 9/188 | 5% | 9/134 | 7% | 0/54 | 0% | ||
| Virus total | 40/188 | 21% | 39/134 | 29% | 1/54 | 2% | ||
| Fungal only | 0/188 | 0% | 0/134 | 0% | 0/54 | 0% | ||
| Fungal total | 6/188 | 3% | 5/134 | 4% | 1/54 | 2% | ||
| Protozoan only | 0/188 | 0% | 0/134 | 0% | 0/54 | 0% | ||
| Protozoan total | 2/188 | 1% | 2/134 | 1% | 0/54 | 0% | ||
| Multiple types of infection | 32/188 | 17% | 31/134 | 23% | 1/54 | 2% | ||
| Total infections | 86/188 | 46% | 65/134 | 49% | 21/54 | 39% | ||
Table 4.
Cohort II BPI SNP rs4358188 infection data.
| BPI rs4358188 |
|||
|---|---|---|---|
| AA | AG | GG | |
| n/Total | 10/15 | 8/29 | 3/10 |
| (%) | (66.7%) | (27.6%) | (30%) |
Discussion
In this study we observed significant changes in multiple innate immune mediators, including TNF-α, CCL2 and CCL5, as well as components of the LPS-detection machinery at the mRNA (CD14, LBP, MD-2, MyD88, TLR4) and protein (LBP, sCD14) levels in patients undergoing myeloablative HCT. Based on these and our previous observations for BPI,19 we studied another prospective cohort of patients undergoing HCT to analyze the potential effects of different SNP genotypes in BPI, CD14, LBP, Mal/TIRAP, MD-2, TLR2 and TLR4 on protein levels, functionality and infection outcomes in this population.
We observed two distinct temporal patterns of change in Cohort I. There were components that increased or decreased at early-to-mid-range time points (d 0–d 14) with rapid recovery and those that increased at later time points (d 14–d 28). Components in the early d 0–d 14 group, which includes CCL2, CCL5 and spontaneously secreted TNF-α, corresponded closely to nadirs and recovery of platelets19 and monocytes, and signify shifts similar to those we have reported previously for neutrophils and neutrophil constituents.19 We hypothesize that changes following the second pattern, such as the d 14–d 28 increases in LPS receptor components CD14 and LBP, as well as sTNFR1/2 and CXCL10, which are up-regulated in response to Gram-negative bacteria,8 are suggestive of inflammatory responses that might be influenced by the pattern of hematopoietic ablation and recovery. These can be further exacerbated by the presence of systemic endotoxin,19 and potentially by clinically evident post-transplant infections.
It should be noted that levels of endotoxemia observed during this time in the HCT patients in Cohort I ranged from intermediate levels of endotoxin activity units (≥ 0.4 and < 0.6; 4/18, 22%) to high levels (≥ 0.6; n = 12/18, 67%),19 as classified for clinical relevance in the 2003 Multi-Center Endotoxin Detection in Critical illness (MEDIC) trial, where endotoxin units ≥ 0.6 conferred a significantly higher risk of developing severe sepsis within 24 h of testing in Intensive Care Unit patients.33 At the mid-to-low range of concentrations, the immune-stimulatory activity of plasma endotoxin is likely more dependent on the levels and activity of several host proteins that regulate delivery of LPS to MD-2/TLR4 and/or cell signaling following TLR4 activation. By monitoring several SNPs within genes encoding these host proteins in a second observational cohort (Cohort II), we have found certain SNP genotypes of LBP and CD14 that modify the plasma concentrationsof these proteins (LBP SNPs rs2232613 and rs2232571) and/or the handling (LBP rs2232613) and response to plasma endotoxin ex vivo (CD14 SNPs rs2569190 and rs2569191).
Our findings that LBP SNPs rs2232571 and rs2232613 were associated with changes in plasma LBP protein concentrations confirm two previous reports linking rs2232613 and rs2232571 to LBP serum34 and plasma35 levels, respectively. Detailed mapping and mutational studies of the LBP promoter region have suggested an effect of the C/T substitution in SNP rs2232571 on the function of the CAAT box at position —778 and thus on LBP promoter activity.36 By showing effects on mean plasma LBP concentrations, our findings (Figure 3a) and those of an earlier study35 support the hypothesis that LBP SNP rs2232571 affects the LBP promoter, with SNP genotype frequency ranking TT > CT > CC inversely relating to protein levels (CC > CT > TT). It is interesting to note that although a common four-SNP risk haplotype in the 5’-flanking region of the LBP gene (positions –1978 to –763, rs2232571 is at position –836) has been linked to an increased risk for severe sepsis,37 we did not find any association of this SNP with post-transplant infection frequency (albeit in a small cohort). How SNP rs2231613 confers alterations in circulating LBP protein levels (Figure 3c) is less clear, but it is conceivable that the Pro333Leu substitution leads to structural alterations affecting protein secretion, stability and/or plasma half-life. Whatever the basis of SNP rs2232613 effects on protein levels, this structural alteration appears to be associated with different handling of plasma endotoxin, as manifested ex vivo by genotype-associated differences in extraction and efficiency of TLR4-activating endotoxin monomer to MD-2 (Figure 4c). The apparently inverse relation within this LBP SNP of genotype-associated differences in mean plasma LBP levels and plasma-dependent endotoxin delivery to MD-2 is consistent with in vitro and in vivo data showing that elevated LBP concentrations blunt—rather than promote—delivery of activating endotoxin to MD-2/ TLR4 and endotoxin-induced inflammation.16
The elevated plasma LBP levels observed in all HCT patients by d 7–d 14 (Figure 1e) may also contribute to blunting of blood cell responses to endotoxemia that can persist for at least 28 d. It should be noted, however, that our data do not reveal an overall correlation between plasma LBP protein levels and plasma-dependent formation of endotoxin/MD-2 complexes (Spearman correlation, P = 0.5), suggesting a possibly added role of the structural alteration of LBP conferred by the rs2232613 SNP and/or other variables in plasma composition. As no genotype-associated differences in plasma-dependent endotoxin/MD-2 formation were observed within the examined SNPs of CD14, MD-2 and BPI, these three endotoxin-binding proteins—as plasma constituents—do not appear to contribute to differences at baseline in handling of plasma endotoxin by patients scheduled for HCT.
Others have shown both CD14 SNPs rs2569190 and rs2569191 to be associated with alterations in CD14 protein levels in blood38 and plasma39 without showing associations with altered LPS-induced TNF-α production ex vivo.40 Inversely, we did not observe any associations of CD14 SNPs rs2569190 and rs2569191 with sCD14 protein levels in plasma, but ex vivo stimulations of peripheral blood with LPS revealed significant differences in secreted TNF levels when comparing the AA and AG genotypes for rs2569190 and the CC genotype compared with either CT or TT for rs2569191. These apparent contradictions need to be considered in the context of differences in population demographics, age and disease context: Munthe-Kaas et al.39 showed that rs2569190 and rs2569191 were associated with serum sCD14 protein levels in Norwegian children up to 2 yrs of age, but not in those older than 2 yrs of age. Others have shown that increased CD14 promoter activity in vitro with rs2569190 and rs2569191 did not translate into increased mRNA levels in population of healthy Caucasian blood donors,41 and that rs2569190 association with increased sCD14 levels in blood was observed before, but not after, LPS exposure in healthy volunteers in Belgium.38
Nearly 15% of our study subjects showed heterozygous expression of TLR4 SNP variants Asp299Gly and Thr339Ile derived from the SNP variants A/G and C/T, respectively, but there were no significant differences in cellular response to LPS associated with expression of these TLR4 variants in our whole blood assays. This is in agreement with ex vivo human whole-blood stimulation assays42 and consistent with recent evidence indicating that the greatest functional effects of expression of these TLR4 variants are manifest in settings containing relatively low levels of plasma proteins and in cells expressing cell surface TLR4 without MD-2.27,43 Taken together, we speculate that the differences in cellular response to LPS in whole blood that we see associated with specific CD14 genotypes could reflect differences in mCD14 (rather than soluble CD14), thereby affecting the efficiency of delivery of activating endotoxin to MD-2/ TLR4 and/or cell signaling following TLR4 activation.
Among the BPI SNPs examined, we saw no genotype-associated differences in either plasma protein levels or ex vivo handling of plasma endotoxin. The examined SNPs include a SNP located within the 3′ UTR of the BPI gene previously reported to show genotype-associated differences in plasma BPI levels in a study of diabetics.44 Notably, the plasma BPI levels we measured were about five-fold lower than those reported in the aforementioned study, raising the possibility of additional ethnic and disease-specific or other pathophysiological variables that could affect plasma BPI levels. Given the very low plasma BPI concentrations we measured in HCT patients, it is not surprising that little or no effect of genetic variation in BPI was observed in either of the two ex vivo assays performed. In line with previous observations31,32 and within the limits of a relatively small number of autologous or syngeneic HCT patients (n=54) with documented infection (n = 21), we observed a potential association of one BPI variant (AA genotype within rs4358188) with overall infection frequency among HCT patients post-transplant that should be further validated independently in a larger cohort. If validated, such an effect could conceivably reflect extravascular roles of BPI following expression on mucosa/epithelia and/or secretion from recruited and activated neutrophils at sites of microbial infection.45,46
In summary, our findings suggest that recognition and response to plasma endotoxin may be modified by SNP-derived variables in the levels and/or activity of plasma LBP and cell surface CD14 manifest at baseline in patients scheduled for HCT. Additional and more detailed studies will be needed to test whether the associations we have observed reflect effects of the specific LBP and CD14 SNPs on protein expression, secretion, surface expression, half-life or on specific functional properties of these proteins. However, even in the absence of this more detailed mechanistic knowledge, we suggest that future clinical studies where outcome may be affected by host responses to endotoxin, including observational and treatment studies in myelo-ablative HCT,47 measure the specific LBP, CD14 and BPI SNPs functionally implicated in this study.
Supplementary Material
Acknowledgements
We would like to thank the patients and their families who consented to participate in this study. We are also grateful to Ms Betsy Golden for technical assistance and Ms Janice Russell for assistance with clinical data.
Funding
This study was funded by collaborative research in the Guinan and Levy laboratories supported by NIH 5R21 HL089659 and 5U19 AI067751, a Dana Foundation Human Immunology Award, a DartDose Center for Medical Countermeasures Against Radiation (1U19AI091173-01) Pilot Award and by NIH R01 AI059372 to JW. CDP received salary support from the American Heart Association (11POST7660006). LAK was partially supported by Harvard Catalyst/The Harvard Clinical and Translational Science Center (8UL1TR000170-05), and financial contributions from Harvard University and its affiliated academic health centers.
References
- 1.Bird GT, Farquhar-Smith P, Wigmore T, et al. Outcomes and prognostic factors in patients with haematological malignancy admitted to a specialist cancer intensive care unit: a 5 yr study. Br J Anaesth. 2012;108:452–459. doi: 10.1093/bja/aer449. [DOI] [PubMed] [Google Scholar]
- 2.Castagnola E, Faraci M. Management of bacteremia in patients undergoing hematopoietic stem cell transplantation. Exp Rev Anti Infect Ther. 2009;7:607–621. doi: 10.1586/eri.09.35. [DOI] [PubMed] [Google Scholar]
- 3.Packey CD, Ciorba MA. Microbial influences on the small intestinal response to radiation injury. Curr Opin Gastroenterol. 2010;26:88–94. doi: 10.1097/MOG.0b013e3283361927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brook I, Elliott TB, Ledney GD, et al. Management of postirra-diation infection: lessons learned from animal models. Mil Med. 2004;169:194–197. doi: 10.7205/milmed.169.3.194. [DOI] [PubMed] [Google Scholar]
- 5.Levy O, Teixeira-Pinto A, White ML, et al. Endotoxemia and elevation of lipopolysaccharide-binding protein after hematopoi-etic stem cell transplantation. Pediatr Infect Dis J. 2003;22:978–981. doi: 10.1097/01.inf.0000095196.19606.d2. [DOI] [PubMed] [Google Scholar]
- 6.Antin JH, Ferrara JL. Cytokine dysregulation and acute graft-versus-host disease. Blood. 1992;80:2964–2968. [PubMed] [Google Scholar]
- 7.Vogelsang GB, Lee L, Bensen-Kennedy DM. Pathogenesis and treatment of graft-versus-host disease after bone marrow transplant. Annu Rev Med. 2003;54:29–52. doi: 10.1146/annurev.med.54.101601.152339. [DOI] [PubMed] [Google Scholar]
- 8.Shibata T, Motoi Y, Tanimura N, et al. Intracellular TLR4/MD-2 in macrophages senses Gram-negative bacteria and induces a unique set of LPS-dependent genes. Int Immunol. 2011;23:503–510. doi: 10.1093/intimm/dxr044. [DOI] [PubMed] [Google Scholar]
- 9.Dowling DJ, Tan Z, Prokopowicz ZM, et al. The ultra-potent and selective TLR8 agonist VTX-294 activates human newborn and adult leukocytes. PloS One. 2013;8:e58164. doi: 10.1371/journal.pone.0058164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gioannini TL, Teghanemt A, Zhang D, et al. Isolation of an endotoxin-MD-2 complex that produces Toll-like receptor 4-dependent cell activation at picomolar concentrations. Proc Natl Acad Sci U S A. 2004;101:4186–4191. doi: 10.1073/pnas.0306906101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Viriyakosol S, Tobias PS, Kitchens RL, et al. MD-2 binds to bacterial lipopolysaccharide. J Biol Chem. 2001;276:38044–38051. doi: 10.1074/jbc.M105228200. [DOI] [PubMed] [Google Scholar]
- 12.Gioannini TL, Weiss JP. Regulation of interactions of Gram-negative bacterial endotoxins with mammalian cells. Immunol Res. 2007;39:249–260. doi: 10.1007/s12026-007-0069-0. [DOI] [PubMed] [Google Scholar]
- 13.Opal SM, Palardy JE, Marra MN, et al. Relative concentrations of endotoxin-binding proteins in body fluids during infection. Lancet. 1994;344:429–431. doi: 10.1016/s0140-6736(94)91767-1. [DOI] [PubMed] [Google Scholar]
- 14.Weiss J. Bactericidal/permeability-increasing protein (BPI) and lipopolysaccharide-binding protein (LBP): structure, function and regulation in host defence against Gram-negative bacteria. Biochem Soc Trans. 2003;31:785–790. doi: 10.1042/bst0310785. [DOI] [PubMed] [Google Scholar]
- 15.Fan H, Cook JA. Molecular mechanisms of endotoxin tolerance. J Endotoxin Res. 2004;10:71–84. doi: 10.1179/096805104225003997. [DOI] [PubMed] [Google Scholar]
- 16.Kitchens RL, Thompson PA. Modulatory effects of sCD14 and LBP on LPS-host cell interactions. J Endotoxin Res. 2005;11:225–229. doi: 10.1179/096805105X46565. [DOI] [PubMed] [Google Scholar]
- 17.Munford RS. Detoxifying endotoxin: time, place and person. J Endotoxin Res. 2005;11:69–84. doi: 10.1179/096805105X35161. [DOI] [PubMed] [Google Scholar]
- 18.Levy O. Antimicrobial proteins and peptides: Anti-infective molecules of mammalian leukocytes. J Leuk Biol. 2004;76:909–925. doi: 10.1189/jlb.0604320. [DOI] [PubMed] [Google Scholar]
- 19.Guinan EC, Barbon CM, Kalish LA, et al. Bactericidal/permeability-increasing protein (rBPI21) and fluoroquinolone mitigate radiation-induced bone marrow aplasia and death. Sci Transl Med. 2011;3:110ra118. doi: 10.1126/scitranslmed.3003126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thornley I, Lehmann LE, Sung L, et al. A multiagent strategy to decrease regimen-related toxicity in children undergoing allogen-eic hematopoietic stem cell transplantation. Biol Blood Marrow Transplant. 2004;10:635–644. doi: 10.1016/j.bbmt.2004.06.004. [DOI] [PubMed] [Google Scholar]
- 21.Armand P, Gannamaneni S, Kim HT, et al. Improved survival in lymphoma patients receiving sirolimus for graft-versus-host disease prophylaxis after allogeneic hematopoietic stem-cell transplantation with reduced-intensity conditioning. J Clin Oncol. 2008;26:5767–5774. doi: 10.1200/JCO.2008.17.7279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Viriyakosol S, McCray PB, Ashbaugh ME, et al. Characterization of monoclonal antibodies to human soluble MD-2 protein. Hybridoma (Larchmt) 2006;25:349–357. doi: 10.1089/hyb.2006.25.349. [DOI] [PubMed] [Google Scholar]
- 23.Bustin SA. Absolute quantification of mRNA using real-time reverse transcription polymerase chain reaction assays. J Mol Endocrinol. 2000;25:169–193. doi: 10.1677/jme.0.0250169. [DOI] [PubMed] [Google Scholar]
- 24.Chen CZ, Ou CY, Wang RH, et al. The role of bactericidal/ permeability-increasing protein in men with chronic obstructive pulmonary disease. COPD. 2012;9:197–202. doi: 10.3109/15412555.2011.654143. [DOI] [PubMed] [Google Scholar]
- 25.Mansilla MA, Cooper ME, Goldstein T, et al. Contributions of PTCH gene variants to isolated cleft lip and palate. Cleft Palate Craniofac J. 2006;43:21–29. doi: 10.1597/04-169R.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Giardina PC, Gioannini T, Buscher BA, et al. Construction of acetate auxotrophs of Neisseria meningitidis to study host-meningococ-cal endotoxin interactions. J Biol Chem. 2001;276:5883–5891. doi: 10.1074/jbc.M009273200. [DOI] [PubMed] [Google Scholar]
- 27.Prohinar P, Rallabhandi P, Weiss JP, et al. Expression of functional D299G.T399I polymorphic variant of TLR4 depends more on coexpression of MD-2 than does wild-type TLR4. J Immunol. 2010;184:4362–4367. doi: 10.4049/jimmunol.0903142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Teghanemt A, Widstrom RL, Gioannini TL, et al. Isolation of monomeric and dimeric secreted MD-2. Endotoxin.sCD14 and Toll-like receptor 4 ectodomain selectively react with the monomeric form of secreted MD-2. J Biol Chem. 2008;283:21881–21889. doi: 10.1074/jbc.M800672200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yu L, Phillips RL, Zhang D, et al. NMR studies of hexaacylated endotoxin bound to wild-type and F126A mutant MD-2 and MD-2TLR4 ectodomain complexes. J Biol Chem. 2012;287:16346–16355. doi: 10.1074/jbc.M112.343467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Prohinar P, Re F, Widstrom R, et al. Specific high affinity interactions of monomeric endotoxin protein complexes with Tolllike receptor 4 ectodomain. J Biol Chem. 2007;282:1010–1017. doi: 10.1074/jbc.M609400200. [DOI] [PubMed] [Google Scholar]
- 31.Liu L, Wang J, Zhao Q, et al. Genetic variation in exon 10 of the BPI gene is associated with Escherichia coli F18 susceptibility in Sutai piglets. Gene. 2013;523:70–75. doi: 10.1016/j.gene.2013.02.051. [DOI] [PubMed] [Google Scholar]
- 32.Wermke M, Maiwald S, Schmelz R, et al. Genetic variations of interleukin-23R (1143A>G) and BPI (A645G), but not of NOD2, are associated with acute graft-versus-host disease after allogeneic transplantation. Biol Blood Marrow Transplant. 2010;16:1718–1727. doi: 10.1016/j.bbmt.2010.06.001. [DOI] [PubMed] [Google Scholar]
- 33.Marshall JC, Foster D, Vincent JL, et al. Diagnostic and prognostic implications of endotoxemia in critical illness: results of the MEDIC study. J Infect Dis. 2004;190:527–534. doi: 10.1086/422254. [DOI] [PubMed] [Google Scholar]
- 34.LeVan TD, Slager RE, Romberger DJ, et al. Lipopolysaccharide-binding protein (LBP) polymorphisms are associated with serum levels of LBP in agricultural workers. Proc Am Thoracic Soc. 2010;7:151. [Google Scholar]
- 35.Chien JW, Boeckh MJ, Hansen JA, et al. Lipopolysaccharide binding protein promoter variants influence the risk for Gramnegative bacteremia and mortality after allogeneic hematopoietic cell transplantation. Blood. 2008;111:2462–2469. doi: 10.1182/blood-2007-09-101709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schumann RR, Kirschning CJ, Unbehaun A, et al. The lipopolysaccharide-binding protein is a secretory class 1 acute-phase protein whose gene is transcriptionally activated by APRF/ STAT/3 and other cytokine-inducible nuclear proteins. Mol Cell Biol. 1996;16:3490–3503. doi: 10.1128/mcb.16.7.3490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Flores C, Perez-Mendez L, Maca-Meyer N, et al. A common haplotype of the LBP gene predisposes to severe sepsis. Crit Care Med. 2009;37:2759–2766. doi: 10.1097/CCM.0b013e3181a57b90. [DOI] [PubMed] [Google Scholar]
- 38.Levan TD, Michel O, Dentener M, et al. Association between CD14 polymorphisms and serum soluble CD14 levels: effect of atopy and endotoxin inhalation. J Allergy Clin Immunol. 2008;121:434–440. doi: 10.1016/j.jaci.2007.08.050. [DOI] [PubMed] [Google Scholar]
- 39.Munthe-Kaas MC, Torjussen TM, Gervin K, et al. CD14 polymorphisms and serum CD14 levels through childhood: a role for gene methylation? J Allergy Clin Immunol. 2010;125:1361–1368. doi: 10.1016/j.jaci.2010.02.010. [DOI] [PubMed] [Google Scholar]
- 40.Smit LA, Heederik D, Doekes G, et al. Endotoxin exposure, CD14 and wheeze among farmers: a gene—environment interaction. Occup Environ Med. 2011;68:826–831. doi: 10.1136/oem.2010.060038. [DOI] [PubMed] [Google Scholar]
- 41.Mertens J, Bregadze R, Mansur A, et al. Functional impact of endotoxin receptor CD14 polymorphisms on transcriptional activity. J Mol Med. 2009;87:815–824. doi: 10.1007/s00109-009-0479-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.von Aulock S, Schroder NW, Gueinzius K, et al. Heterozygous toll-like receptor 4 polymorphism does not influence lipopolysac-charide-induced cytokine release in human whole blood. J Infect Dis. 2003;188:938–943. doi: 10.1086/378095. [DOI] [PubMed] [Google Scholar]
- 43.Guo J, Loke J, Zheng F, et al. Functional linkage of cirrhosis-predictive single nucleotide polymorphisms of Toll-like receptor 4 to hepatic stellate cell responses. Hepatology. 2009;49:960–968. doi: 10.1002/hep.22697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gubern C, Lopez-Bermejo A, Biarnes J, et al. Natural antibiotics and insulin sensitivity: the role of bactericidal/permeability-increasing protein. Diabetes. 2006;55:216–224. [PubMed] [Google Scholar]
- 45.Canny G, Levy O, Furuta GT, et al. Lipid mediator-induced expression of bactericidal/ permeability-increasing protein (BPI) in human mucosal epithelia. Proc Natl Acad Sci USA. 2002;99:3902–3907. doi: 10.1073/pnas.052533799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Levy O, Sisson RB, Fryer HE, et al. Neutrophil defense in patients undergoing bone marrow transplantation: bactericidal/ permeability-increasing protein (BPI) and defensins in graft-derived neutrophils. Transplantation. 2002;73:1522–1526. doi: 10.1097/00007890-200205150-00027. [DOI] [PubMed] [Google Scholar]
- 47.Palmer CD, Guinan EC, Levy O. Deficient expression of bactericidal/permeability-increasing protein in immunocompromised hosts: translational potential of replacement therapy. Biochem Soc Trans. 2011;39:994–999. doi: 10.1042/BST0390994. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.