Abstract
Background: MicroRNAs (miRNAs) are small, non-coding RNAs which can function as oncogenes or tumor suppressor genes in human cancers. Researchers have found that the expression level of miR-107 was decreased in human non-small cell lung cancer (NSCLC) tissues and cell lines, however, its clinicopathological and prognostic significance in NSCLC has not been investigated. Methods: Quantitative real-time PCR (qRT-PCR) was used to analyze the expression of miR-107 in 137 pairs of fresh NSCLC and matched adjacent normal tissue specimens. The chi-square test and Fishers exact tests were used to examine the associations between miR-107 expression and the clinicopathological characters. The overall survival (OS) and progression-free survival (PFS) were analyzed by log-rank test, and survival curves were plotted according to Kaplan-Meier. Results: The expression level of miR-107 was significantly lower in tumor tissues than that in corresponding noncancerous tissues (0.4676±0.2078 vs. 1.000±0.3953, P<0.001). Low expression of miR-107 was found to significantly correlate with TNM stage (p=0.001), regional lymph node involvement (p=0.04), and tumor differentiation (p=0.003). Kaplan-Meier analysis with the log-rank test indicated that low miR-107 expression had a significant impact on OS (35.2% vs. 69.3%; P=0.008) and PFS (30.0% vs. 56.2%; P=0.029). In a multivariate Cox model, we found that miR-107 expression was an independent poor prognostic factor for both 5-year OS (HR=2.57, 95% CI: 1.88-10.28; P=0.007) and 5-year PFS (HR=3.08, 95% CI: 2.01-8.92; P=0.003). Conclusion: The expression of miR-107 was decreased in NSCLC. Low expression of miR-107 was significantly associated with tumor progression and decreased survival in patients with NSCLC, indicating that miR-107 may serve as a novel prognostic marker in NSCLC.
Keywords: MicroRNA-107, non-small cell lung cancer, prognosis
Introduction
Lung cancer is the most common cause of cancer deaths worldwide [1]. Non-small cell lung cancer (NSCLC) is the most frequent type of lung cancer, accounting for over 80% of all lung cancer cases. It includes two predominant subtypes, adenocarcinoma and squamous cell carcinoma, which comprise 40% and 25%, respectively [2]. NSCLC has a low 5-year overall survival rate and a high recurrence rate [3]. Despite recent diagnostic and therapeutic advancements, poor prognosis is observed in a large portion of NSCLC patients, even when diagnosis is made early in the course of the disease [4].
MicroRNAs (miRNAs) are a large family of highly conserved short (~22 nucleotides in length) single stranded noncoding RNAs that regulate the translational inhibition of target messenger RNAs by binding to their 3’-untranslated region [5]. The dysregulation of miRNAs is common in various carcinomas and plays an important role in cancer progression by altering normal gene expression [6-9]. In a previous study, researchers found that miR-107 suppressed growth and induced a G1 cell cycle arrest in the human NSCLC cell lines [10]. However, the associations between the miR-107 expression, clinicopathological characteristics and the prognostic value of miR-107 in NSCLC have not yet been reported. In the present study, we investigated miR-107 expression in tumor tissues and matched adjacent normal tissues from 137 NSCLC patients to determine its clinicopathological and prognostic significance.
Materials and methods
Patients and controls’ samples
This study was approved by the Research Ethics Committee of Qilu Hospital, Shandong University. Written informed consent was obtained from all of the patients. All specimens were handled and made anonymous according to the ethical and legal standards. The selection criteria for patients with NSCLC were as follows: (1) pathologically confirmed patients with NSCLC; (2) the patients had no previous history of other cancers; (3) the patients haven’t received preoperative treatment such as radiation or chemotherapy. All patients were diagnosed and treated at the department of thoracic surgery, Qilu Hospital, Shandong University from May 2006 to March 2011. All subjects underwent clinical examination; plain chest radiograph; CT scan of the chest, upper abdomen, and brain; fiberoptic bronchoscopy; and bone scan. The pathologic diagnosis was conducted by two pathologists, and any different conclusions were resolved by careful study and discussion. Tumor stage was determined according to the 2009 TNM staging classification system. The duration of follow-up was calculated from the date of surgery to death or last follow-up, and patients were excluded if they had incomplete medical records or inadequate follow-up. Overall survival (OS) time was calculated from the date of the initial surgery to death. Progression-free survival (PFS) time was calculated from the date of the initial surgery until the first evidence of local, regional, or distant tumor progression of disease. For qRT-PCR, 137 pairs of fresh NSCLC and matched adjacent normal tissue specimens were collected from patients who underwent surgery in the department of thoracic surgery, Qilu Hospital, Shandong University. The fresh tissue specimens were collected and immediately placed in liquid nitrogen and then stored at -80°C until the isolation of RNA. Clinicopathological features of patients are summarized in Table 1.
Table 1.
Clinicopathological characteristics of 137 NSCLC patients
| Characteristics | No. of Patients | % |
|---|---|---|
| Age (years) | ||
| <65 | 65 | 47.45 |
| ≥65 | 72 | 52.55 |
| Sex distribution | ||
| Female | 59 | 43.07 |
| Male | 78 | 56.93 |
| Smoking history | ||
| Current | 87 | 63.50 |
| Former | 39 | 28.47 |
| Never | 11 | 8.03 |
| Weight loss | ||
| <10% | 108 | 78.83 |
| ≥10% | 29 | 21.17 |
| TNM stage | ||
| I | 56 | 40.88 |
| II | 55 | 40.15 |
| IIIa | 26 | 18.98 |
| Regional lymph node involvement | ||
| Absent | 89 | 64.96 |
| Present | 48 | 35.04 |
| Histology | ||
| Squamous cell carcinoma | 54 | 39.42 |
| Adenocarcinoma | 57 | 41.61 |
| Large cell carcinoma | 26 | 18.98 |
| Differentiation | ||
| Poor | 48 | 35.04 |
| Moderate | 59 | 43.07 |
| Well | 30 | 21.90 |
| Surgical margins | ||
| Free | 98 | 71.53 |
| Not free | 39 | 28.47 |
MicroRNA isolation and real-time quantitative RT-PCR assay
Total RNA was isolated from frozen specimens by homogenizing tissue in Trizol reagent (Invitrogen, Carlsbad, CA, USA), according to the manufacturer’s instructions. The purity and concentration of RNA were determined using NanoDrop 1000 spectrophotometer (Thermo Scientific, Wilmington, DE, USA). The differentially expressed amount of the miR-107 was validated in triplicate by quantitative reverse-transcription polymerase chain reaction (qRT-PCR). Briefly, 2 ug of RNA was added to RT reaction, and then, the cDNA served as the template for amplification of PCR with sequence-specific primers (Sangon Biotech, Shanghai, China) using SYBR PrimeScript miRNA RT-PCR kit (Takara Biotechnology Co. Ltd, Dalian, China) on the 7500 Real-Time PCR systems (Applied Biosystems, Carlsbad, CA, USA). The PCR cycling profile was denatured at 95°C for 30 s, followed by 40 cycles of annealing at 95°C for 5 s, and extension at 60°C for 34 s. Small nucleolar RNA U6 was used as an internal standard for normalization. The cycle threshold (CT) value was calculated. The 2-ΔCT (ΔCT=CTmiR107-CTU6 RNA) method was used to quantify relative amount of miR-107.
Statistical analysis
The comparison of the expression levels of miR-107 between NSCLC tissues and adjacent normal tissues were performed using the two-sample Student’s t test. The chi-square test and Fishers exact tests were used to examine the associations between miR-107 expression and the clinicopathological characters. The OS and PFS were analyzed by log-rank test, and survival curves were plotted according to Kaplan-Meier. Univariate Cox regression was performed on each clinical covariate to examine its influence on patient survival. Final multivariate models were based on step-wise addition. A Wald statistic of P<0.05 was used as the criterion for inclusion in final multivariate models. All tests were two tailed and results with P<0.05 were considered statistically significant. Statistical analyses were performed using SPSS 18.0 soft-ware (Chicago, Ill., USA) and GraphPad Prism 5 (GraphPad Software Inc., CA, USA).
Results
Expression levels of miR-107 in NSCLC
We examined miR-107 expression in 137 pairs of NSCLC tissues and the corresponding noncancerous tissues by qRT-PCR. As shown in Figure 1, the expression level of miR-107 was significantly lower in tumor tissues than that in corresponding noncancerous tissues (0.4676 ±0.2078 vs. 1.000±0.3953, P<0.001). Furthermore, the expression levels of miR-107 in patients with TNM stage II and IIIa were significantly lower than those in patients with TNM stage I (0.4119±0.1292 vs. 0.6023±0.2203, P<0.001; 0.2954±0.1171 vs. 0.6023±0.2203, P<0.001, respectively, shown in Figure 2).
Figure 1.
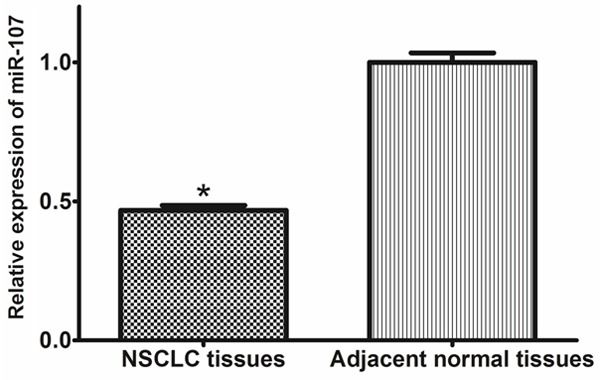
Comparison of miR-107 expression levels between NSCLC tissues and adjacent normal tissues.
Figure 2.
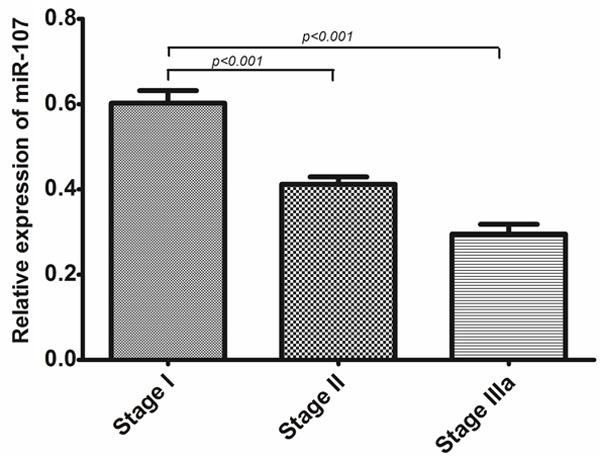
The expression level of miR-107 in NSCLC patients (n=137) at different clinical stages.
Expression levels of miR-107 and clinicopathological parameters in NSCLC
The miR-107 expression levels were classified as high or low in relation to the median value. Low expression of miR-107 was found to significantly correlate with TNM stage (p=0.001), regional lymph node involvement (p=0.04), and tumor differentiation (p=0.003). However, no significant difference in miR-107 expression was observed with age (p=0.89), sex distribution (p=0.74), smoking history (p=0.11), weight loss (p=0.06), histology (p=0.17), and surgical margins (p=0.07, shown in Table 2).
Table 2.
Correlation between the expression of miR-107 and clinicopathological parameters in NSCLC patients
| Variable | n | miR-132 expression | P value | |
|---|---|---|---|---|
|
| ||||
| High (n=69) | Low (n=68) | |||
| Age (years) | ||||
| <65 | 65 | 32 | 33 | 0.89 |
| ≥65 | 72 | 37 | 35 | |
| Sex distribution | ||||
| Female | 59 | 31 | 28 | 0.74 |
| Male | 78 | 38 | 40 | |
| Smoking history | ||||
| Current | 87 | 41 | 46 | 0.11 |
| Former | 39 | 23 | 16 | |
| Never | 11 | 5 | 6 | |
| Weight loss | ||||
| <10% | 108 | 59 | 49 | 0.06 |
| ≥10% | 29 | 10 | 19 | |
| TNM stage | ||||
| I | 56 | 36 | 20 | 0.001 |
| II | 55 | 24 | 31 | |
| IIIa | 26 | 9 | 17 | |
| Regional lymph node involvement | ||||
| Absent | 89 | 50 | 39 | 0.04 |
| Present | 48 | 19 | 29 | |
| Histology | ||||
| Squamous cell carcinoma | 54 | 29 | 25 | 0.17 |
| Adenocarcinoma | 57 | 26 | 31 | |
| Large cell carcinoma | 26 | 14 | 12 | |
| Differentiation | ||||
| Poor | 48 | 19 | 29 | 0.003 |
| Moderate | 59 | 24 | 35 | |
| Well | 30 | 26 | 4 | |
| Surgical margins | ||||
| Free | 98 | 52 | 46 | 0.07 |
| Not free | 39 | 17 | 22 | |
Low-expression level of miR-107 predicts poor prognosis in NSCLC patients
Kaplan-Meier analysis with the log-rank test indicated that low miR-107 expression had a significant impact on OS (35.2% vs. 69.3%; P=0.008; Figure 3) and PFS (30.0% vs. 56.2%; P=0.029; Figure 4). Univariate and multivariate analyses were utilized to evaluate whether the miR-107 expression level and various clinicopathological features were independent prognostic parameters. Multivariate analysis revealed that miR-107 expression (HR=2.57, 95% CI: 1.88-10.28; P=0.007), TMN stage (HR=3.37, 95% CI: 2.21-18.91; P=0.004), and tumor differentiation (HR=4.77, 95% CI: 3.62-16.46; P<0.001) were independently associated with the OS (shown in Table 3), and that miR-107 expression (HR=3.08, 95% CI: 2.01-8.92; P=0.003), TMN stage (HR=4.28, 95% CI: 3.11-16.21; P=0.001), regional lymph node involvement (HR=2.11, 95% CI: 1.39-5.31; P=0.01), and surgical margins (HR=2.09, 95% CI: 1.88-4.85; P=0.04) were independent prognostic factors for PFS (shown in Table 3).
Figure 3.
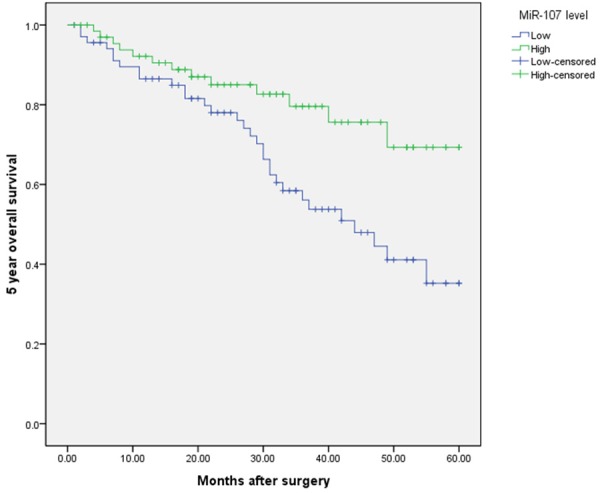
Kaplan-Meier curves of the overall survival of 137 NSCLC patients. Overall survival rate in patients with low miR-107 expression was significantly lower than that in patients with high miR-107 expression.
Figure 4.
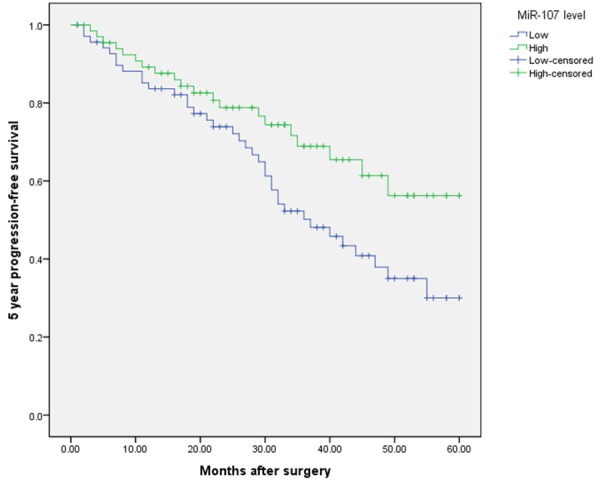
Kaplan-Meier curves of the progression-free survival of 137 NSCLC patients. Progression-free survival rate in patients with low miR-107 expression was significantly lower than that in patients with high miR-107 expression.
Table 3.
Multivariate analyses for progression-free survival and overall survival by Cox regression model
| Progression-free survival | Overall survival | |||||
|---|---|---|---|---|---|---|
|
|
|
|||||
| Variable | Hazard ratio | 95% CI | P-value | Hazard ratio | 95% CI | P-value |
| Age (years) | 0.91 | 0.45-1.89 | 0.56 | 1.22 | 0.23-2.81 | 0.61 |
| Gender | 0.69 | 0.25-3.27 | 0.69 | 0.97 | 0.48-3.11 | 0.34 |
| Smoking history | 1.47 | 0.89-7.28 | 0.09 | 1.83 | 0.72-8.77 | 0.07 |
| Weight loss | 1.38 | 0.77-4.81 | 0.12 | 1.69 | 0.79-4.99 | 0.08 |
| TNM stage | 4.28 | 3.11-16.2 | 0.001 | 3.37 | 2.21-18.91 | 0.004 |
| Regional lymph node involvement | 2.11 | 1.39-5.31 | 0.01 | 1.24 | 0.92-2.49 | 0.07 |
| Histology | 1.27 | 0.79-2.97 | 0.57 | 0.93 | 0.49-1.28 | 0.48 |
| Differentiation | 3.82 | 0.73-14.78 | 0.06 | 4.77 | 3.62-16.46 | <0.001 |
| Surgical margins | 2.09 | 1.88-4.85 | 0.04 | 1.63 | 0.73-6.63 | 0.06 |
| MiR-107 | 3.08 | 2.01-8.92 | 0.003 | 2.57 | 1.88-10.28 | 0.007 |
Discussion
The discovery of the first miRNA, lin-4 in Caenorhabditis elegans initiated a new era of miRNA biology. Since then, thousands of miRNAs have been identified and annotated. Furthermore, an increasing body of evidence indicates that miRNAs are differentially expressed between normal and tumor tissues, suggesting that dysregulation of miRNA expression is a key factor underlying tumorigenesis [11-13]. A large number of studies suggest that understanding of miRNA function will provide us broad prospects to understand and overcome tumor.
MiR-107, located on 10th chromosome, is differentially expressed in a number of metabolic pathways, including adipogenesis, hypoxia, cell cycle arrest, angiogenesis and neurodegenerative diseases. miR-103/miR-107 are highly conserved miRNAs that map to intron 5 of pantothenate kinase (Pank) genes. Pank2 and Pank3 host the pre-miRNA sequences of miR-103, whereas Pank1 encodes miR-107, which differs from miR-103 by a single nucleotide [14,15]. Pantothenate kinase is the rate limiting enzyme in the biosynthesis of coenzyme A, a cofactor that is involved in over 100 metabolic reactions. p53 can induce expression of PANK1 and miR-107, presumably, through a p53 element located ~1 kb upstream of the PANK1 transcriptional start site [16]. Wang et al demonstrated that expression of miR-107 was inversely associated with expression of hypoxia inducible factor-1β (HIF-1β) and could mediate p53 regulation of hypoxic signaling and tumor angiogenesis in colon cancer. Members of the miR-107 gene group have also been shown to repress granulin (GRN) protein, a potent mitogen and growth factor, in prostate cancer cells [17]. miR-107 induces cell cycle G1 arrest and inhibits invasion by targeting cyclin dependent kinase 6 (CDK6), thereby inhibiting tumor progression in gastric cancer, pancreatic cancer, and NSCLC cell lines [10,18,19]. Another study revealed that Toll-like Receptor-4 down-regulated miR-107, increasing macrophage adhesion via CDK6 [20]. On the contrary, miR-107 has also been implicated in tumor progression. miR-107 showed over-expression in pancreatic cancer and breast cancer suggesting some positive role in carcinogenesis and its expression was inversely correlated with let-7 expression in breast tumors and cancer cell lines [21,22].
Previously, Inoue et al found that the mean expression level of miR-107 was significantly higher in the gastric cancer tissues compared to that of normal tissues. In the comparison of clinicopathological factors,miR-107 expression showed significant association with depth of tumor invasion, lymph node metastasis and TNM stage. In Kaplan-Meier survival curve analysis, OS rates and PFS rates of patients with high miR-107 expressionwere significantly worse than those of patients with low miR-107 expression. In the Cox multivariate analysis, it was shown that miR-107 expression in gastric cancer tissues was an independent prognostic factor for OS and PFS [23]. However, the association between the miR-107 expression, clinicopathological characteristics and the prognostic value of miR-107 in NSCLC have not yet been reported.
In the present study, our results showed that miR-107 expression was significantly lower in NSCLC tissues compared with normal adjacent lung tissues. The relationship of the miR-107 with various clinical features of NSCLC was analyzed. The results revealed that a low level of miR-107 expression was significantly correlated with TNM stage (p=0.001), regional lymph node involvement (p=0.04), and tumor differentiation (p=0.003), suggesting that miR-107 might be involved in the carcinogenesis and metastasis of NSCLC. Further more, the 5-year OS of low miR-107 expression group was significantly shorter than that of high miR-107 expression group. Moreover, the 5-year PFS of low miR-107 expression group was also significantly shorter than that of high miR-107 expression group. In a multivariate Cox model, we found that miR-107 expression was an independent poor prognostic factor for both 5-year OS and 5-year PFS, indicating that low miR-107 level was a promising non-invasive biomarker for prognosis of patients with NSCLC.
In conclusion, the expression of miR-107 was decreased in NSCLC. Low expression of miR-107 was significantly associated with tumor progression and decreased survival in patients with NSCLC, indicating that miR-107 may serve asa novel prognostic marker in NSCLC.
Acknowledgements
This work was supported by Natural Science Foundation of Shandong Province (ZR2009CM044) and Foundation for Outstanding Young Scientist in Shandong Province (2006BS03066).
Disclosure of conflict of interest
None.
References
- 1.Siegel R, Ma J, Zou Z, Jemal A. Cancer statistics, 2014. CA Cancer J Clin. 2014;64:9–29. doi: 10.3322/caac.21208. [DOI] [PubMed] [Google Scholar]
- 2.Rosell R, Karachaliou N. Lung cancer: Maintenance therapy and precision medicine in NSCLC. Nat Rev Clin Oncol. 2013;10:549–550. doi: 10.1038/nrclinonc.2013.152. [DOI] [PubMed] [Google Scholar]
- 3.Miller YE. Pathogenesis of lung cancer: 100 year report. Am J Respir Cell Mol Biol. 2005;33:216–223. doi: 10.1165/rcmb.2005-0158OE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lin PY, Yu SL, Yang PC. MicroRNA in lung cancer. Br J Cancer. 2010;103:1144–1148. doi: 10.1038/sj.bjc.6605901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 6.Wu J, Wu G, Lv L, Ren YF, Zhang XJ, Xue YF, Li G, Lu X, Sun Z, Tang KF. MicroRNA-34a inhibits migration and invasion of colon cancer cells via targeting to Fra-1. Carcinogenesis. 2012;33:519–528. doi: 10.1093/carcin/bgr304. [DOI] [PubMed] [Google Scholar]
- 7.Zhang T, Liu M, Wang C, Lin C, Sun Y, Jin D. Down-regulation of MiR-206 promotes proliferation and invasion of laryngeal cancer by regulating VEGF expression. Anticancer Res. 2011;31:3859–3863. [PubMed] [Google Scholar]
- 8.Guz M, Rivero-Muller A, Okon E, Stenzel-Bembenek A, Polberg K, Slomka M, Stepulak A. MicroRNAs-Role in Lung Cancer. Dis Markers. 2014;2014:218169. doi: 10.1155/2014/218169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Daniels MG, Bowman RV, Yang IA, Govindan R, Fong KM. An emerging place for lung cancer genomics in 2013. J Thorac Dis. 2013;5:S491–S497. doi: 10.3978/j.issn.2072-1439.2013.10.06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Takahashi Y, Forrest AR, Maeno E, Hashimoto T, Daub CO, Yasuda J. MiR-107 and MiR-185 can induce cell cycle arrest in human non small cell lung cancer cell lines. PLoS One. 2009;4:e6677. doi: 10.1371/journal.pone.0006677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kong YW, Ferland-McCollough D, Jackson TJ, Bushell M. microRNAs in cancer management. Lancet Oncol. 2012;13:e249–258. doi: 10.1016/S1470-2045(12)70073-6. [DOI] [PubMed] [Google Scholar]
- 12.Esteller M. Non-coding RNAs in human disease. Nat Rev Genet. 2011;12:861–874. doi: 10.1038/nrg3074. [DOI] [PubMed] [Google Scholar]
- 13.Inui M, Martello G, Piccolo S. MicroRNA control of signal transduction. Nat Rev Mol Cell Biol. 2010;11:252–263. doi: 10.1038/nrm2868. [DOI] [PubMed] [Google Scholar]
- 14.Polster BJ, Westaway SK, Nguyen TM, Yoon MY, Hayflick SJ. Discordant expression of miR-103/7 and pantothenate kinase host genes in mouse. Mol Genet Metab. 2010;101:292–295. doi: 10.1016/j.ymgme.2010.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang WX, Rajeev BW, Stromberg AJ, Ren N, Tang G, Huang Q, Rigoutsos I, Nelson PT. The expression of microRNA miR-107 decreases early in Alzheimer’s disease and may accelerate disease progression through regulation of beta-site amyloid precursor protein-cleaving enzyme 1. J Neurosci. 2008;28:1213–1223. doi: 10.1523/JNEUROSCI.5065-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yamakuchi M, Lotterman CD, Bao C, Hruban RH, Karim B, Mendell JT, Huso D, Lowenstein CJ. P53-induced microRNA-107 inhibits HIF-1 and tumor angiogenesis. Proc Natl Acad Sci U S A. 2010;107:6334–6339. doi: 10.1073/pnas.0911082107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang WX, Wilfred BR, Madathil SK, Tang G, Hu Y, Dimayuga J, Stromberg AJ, Huang Q, Saatman KE, Nelson PT. miR-107 regulates granulin/progranulin with implications for traumatic brain injury and neurodegenerative disease. Am J Pathol. 2010;177:334–345. doi: 10.2353/ajpath.2010.091202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Feng L, Xie Y, Zhang H, Wu Y. miR-107 targets cyclin-dependent kinase 6 expression, induces cell cycle G1 arrest and inhibits invasion in gastric cancer cells. Med Oncol. 2012;29:856–863. doi: 10.1007/s12032-011-9823-1. [DOI] [PubMed] [Google Scholar]
- 19.Lee KH, Lotterman C, Karikari C, Omura N, Feldmann G, Habbe N, Goggins MG, Mendell JT, Maitra A. Epigenetic silencing of MicroRNA miR-107 regulates cyclin-dependent kinase 6 expression in pancreatic cancer. Pancreatology. 2009;9:293–301. doi: 10.1159/000186051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hennessy EJ, Sheedy FJ, Santamaria D, Barbacid M, O’Neill LA. Toll-like receptor-4 (TLR4) down-regulates microRNA-107, increasing macrophage adhesion via cyclin-dependent kinase 6. J Biol Chem. 2011;286:25531–25539. doi: 10.1074/jbc.M111.256206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Roldo C, Missiaglia E, Hagan JP, Falconi M, Capelli P, Bersani S, Calin GA, Volinia S, Liu CG, Scarpa A, Croce CM. MicroRNA expression abnormalities in pancreatic endocrine and acinar tumors are associated with distinctive pathologic features and clinical behavior. J Clin Oncol. 2006;24:4677–4684. doi: 10.1200/JCO.2005.05.5194. [DOI] [PubMed] [Google Scholar]
- 22.Li XY, Luo QF, Wei CK, Li DF, Li J, Fang L. MiRNA-107 inhibits proliferation and migration by targeting CDK8 in breast cancer. Int J Clin Exp Med. 2014;7:32–40. [PMC free article] [PubMed] [Google Scholar]
- 23.Inoue T, Iinuma H, Ogawa E, Inaba T, Fukushima R. Clinicopathological and prognostic significance of microRNA-107 and its relationship to DICER1 mRNA expression in gastric cancer. Oncol Rep. 2012;27:1759–1764. doi: 10.3892/or.2012.1709. [DOI] [PubMed] [Google Scholar]


