Abstract
Members of the Leprecan family of proteins include enzymes, prolyl 3-hydroxylase 1 (P3h1), P3h2 and P3h3, and non-enzymatic proteins, Crtap and Sc65. Mutations in CRTAP and LEPRE1 (encoding P3H1) have been associated with human disease such as recessive osteogenesis imperfecta, however, the function of Sc65 which is closely related and highly homologous to Crtap is unknown. Sc65 has been described as a synaptonemal complex protein, a nucleolar protein, and a cytoplasmic adapter protein. In light of its high sequence similarity with Crtap, an endoplasmic reticulum (ER)-associated protein, and the importance of post-translational modifications such as collagen prolyl 3-hydroxylation in bone metabolism, we hypothesized that Sc65 was an ER-resident protein that would have an important role in bone homeostasis. In this study, we demonstrate that Sc65 is a previously unrecognized ER protein, and that it does not localize in the nucleus of somatic cells. Moreover, Sc65 is expressed and functional during skeletal development since loss of Sc65 results in a progressive osteopenia that affects both trabecular and cortical bone. Bone loss is due to increased bone resorption mediated by a non-cell autonomous effect on osteoclasts. Therefore, Sc65, like its related family member Crtap, is an important modulator of bone homeostasis, acting as a negative regulator of osteoclastogenesis.
Keywords: Sc65, Crtap, bone, endoplasmic reticulum, osteogenesis imperfecta, osteoclasts
INTRODUCTION
Osteogenesis imperfecta (OI) is a congenital, generalized connective tissue disorder characterized by severe osteoporosis and bone fragility(1). Although OI is usually caused by dominant mutations in type I collagen genes, COL1A1 or COL1A2, the disorder is characterized by genetic heterogeneity and a broad spectrum of OI phenotypes.
We and others recently demonstrated that mutations in the CRTAP and LEPRE1 genes, encoding cartilage-associated protein (CRTAP) and prolyl 3-hydroxylase 1 (P3H1 or Leprecan) respectively, cause recessive forms of OI(2–5). The identification of two rough endoplasmic reticulum (rER) proteins associated with collagen post-translational modifications in the development of OI resulted in a transition away from the investigation of matrix components (i.e. type I collagens) and led to a focus on rER-resident proteins. Three additional rER genes, PPIB, FKBP65 and SERPINH1 encoding the proteins Cyclophilin B, FKBP65 and HSP47 respectively, were subsequently linked to recessive forms of OI(6–9). These novel findings significantly improved our understanding of the molecular basis of OI and have clearly demonstrated an essential role, in human skeletal dysplasias, for proteins resident in the rER.
At least where collagen synthesis is concerned, rER-associated proteins are involved in a host of functions including procollagen chain recognition and assembly, post-translational modification(s), folding of the triple helix and also inhibition of the inappropriate intracellular polymerization of procollagen monomers into fibrils(10). Members of three families of proteins have been identified as mediators to these critical collagen assembly functions. One family shares peptidyl-prolyl cis-trans isomerase activity, and includes the Cyclophilins and the FK-506 binding proteins (FKBPs)(11). Another family, the Serpins, is a heterogeneous group of proteins including HSP47, a specific ER collagen chaperone(12, 13). Finally, a third family of proteins called Leprecans (Leucine Proline-Enriched Proteoglycans)(14) that includes Crtap, P3h1, P3h2, P3h3 and Sc65, which is the focus of our investigation, has been identified. Interestingly, mutations in members of all three protein families have been associated with rare forms of OI(15).
Besides collagens, osteoblasts are active synthetic cells that produce a plethora of factors that are important for bone formation and resorption (e.g. osteonectin, osteocalcin, osteopontin, osteoprotegerin, RANKL, and others)(16). For the regulation of bone cell activities it is critical that these proteins are correctly modified, folded and chaperoned outside the cell to perform their function. Hence, the maintenance of proper homeostasis and function of the ER is very important, especially in highly secreting cells such as osteoblasts.
In the current study, in an effort to uncover the role of rER proteins in bone development and homeostasis, we discovered that Sc65 (Synaptonemal complex 65, Nucleolar autoantigen No55 or Leprecan-like protein 4) is a previously unrecognized rER-resident protein and not a nucleolar protein as described earlier(17). Importantly, Sc65 is the closest known family member to Crtap with which it shares ~55% amino acid identity. Similar to Crtap, Sc65 lacks the C-terminal 2-oxoglutarate and Fe(II)-dependent dioxygenase domain present in prolyl-3 hydroxylases but maintains a tetratricopeptide-like (TPR-like) repeat, which is the presumptive protein-protein interaction domain(18). Given the similarity between the Crtap and Sc65 proteins and the well-described role of CRTAP in the pathogenesis of recessive OI, we hypothesized that Sc65 would have an important role in skeletal formation and ultimately, in human skeletal dysplasias. Here we show that Sc65 is not only specifically localized in the ER but is also expressed in skeletal cells and that mice with insertional inactivation of the Sc65 gene and lack Sc65 expression have a low bone mass phenotype, due primarily to increased bone resorption.
SUBJECTS and METHODS
Cell Culture, Transfections, shRNA Transduction, Constructs
Human primary skin fibroblasts and HeLa cells were grown in DMEM; MC3T3-E1, OB6 and primary cells from murine calvariae, spleen or bone marrow were grown in α-MEM. All media contained 4500mg/L glucose and 110mg/ml sodium pyruvate (HyClone, Thermo Fisher Scientific, Pittsburgh, PA) and were supplemented with 10% FBS, L-Glutamine (2mM), 100 units/mL penicillin, and 100µg/mL streptomycin. Ascorbic acid (100µg/ml) and β-glycerophosphate (5mM) were added to the cell culture medium as indicated. Primary osteoblasts from calvaria were obtained using standard procedures as we have described(2). To induce ER stress, osteoblasts were treated with 2ug/ml tunicamycin for 0, 8, 12, 16, 20 h. Whole bone marrow cultures were established by harvesting femurs and tibiae, flushing out the marrow with a 25 gauge needle under sterile conditions, filtering the marrow through a 70µm mesh and removing red blood cell with a commercially available solution (Miltenyi Biotec, Auburn, CA). Bone marrow was plated at the density of 2×106 cells/well in a 24-well plate in presence of 1α,25 dihydroxyvitamin D3 (10−8M) (Sigma cat.# D1530). Similarly, splenocyte cultures were established by harvesting spleens, disaggregating the tissue and filtering the cells through a 70um mesh. Splenocytes (without red blood cells) were seeded on either plastic or Osteo Assay (Corning Inc, Corning, NY) 96-well plates at 1.75×105 cells/well in presence of M-CSF and RANKL (100ng/ml) and stained for tartrate-resistant acid phophatase (TRAP) activity after 5–7 days. Bone marrow macrophage cultures were prepared from cell culture plastic non-adherent bone marrow cells which were expanded for 24–48 hours with M-CSF and then harvested, counted and seeded on either plastic or dentine (ImmunoDiagnosticSystems – IDS, Scottsdale, AZ) in 96- or 48-well plates at 1×106 cells/well with M-CSF and RANKL (100ng/ml). Both M-CSF and RANKL used in all osteoclast assays were kindly provided by Dr. Bryant G. Darnay as conditioned medium from L929 cells (used at 5%) and as His-RANKL purified recombinant protein, respectively. Cells were removed from the dentine slices with fresh 20% bleach solution in PBS for 30 min. Bone resorption pit area was quantified using a Osteomeasure software (Osteometrics, Atlanta, GA) after the staining with 1:20 wheat germ agglutinin (WGA) - lectin conjugated to TRITC (Sigma Alrich, St.Louis, MO) for 1 h. Mouse Sc65 cDNA subcloned in pcDNA3.1 myc/His A (Invitrogen,Grand Island, NY), sequenced to confirm its identity and then transfected into HeLa cells using X-tremeGENE according to the manufacturer’s instructions (Roche Applied Science, Indianapolis, IN). Mission® Lentiviral Transduction particles encoding shRNA against Sc65 or TurboGFP were purchased from Sigma-Aldrich (St.Louis, MO) and used according to the recommended transduction protocol from the manufacturer. OB6 cells(19) were plated in a 12-well plate at 2 different densities (0.5 and 0.25×105 cells/well) and the next day transduced with lentiviral particles in the presence of hexadimethrine bromide (8µg/ml final concentration) at an MOI of 5. Selection with puromycin (2µg/ml) began on the second day following transduction and drug resistant OB6 cells were always maintained under puromycin selection.
Mice, Gene-trap Clone, Southern Blot, RT-PCR, Mouse PCR Genotyping and qPCR
The Sc65 mouse ES cell clone (129X1/SvJ × 129S1) with a proviral insertion (gene-trap) in the Sc65 gene was purchased from the Gene Trap Resource of the Phenogenomics Toronto Center (Toronto, Canada). The construct contains several elements, including an EGFP and a neomycin resistance gene and can be viewed at http://www.cmhd.ca/genetrap/how_to.html#vectors_UPA. The insertion into the Sc65 locus was verified by Southern analysis using an external flanking probe. C57BL6 blastocysts were injected with ES cells and chimeric mice were generated. Germ line transmission of the mutant allele was obtained by crossing to C57BL6. The use of laboratory mice was approved by the University of Arkansas for Medical Sciences (UAMS) IACUC committee. Total RNA was extracted from tissues or cells using Tripure reagent (Roche-Applied-Science, USA). cDNA synthesis was performed using the Transcriptor First Strand cDNA kit (Roche Applied Science, USA). PCR genotyping was done using the GoTaq PCR kit and reagents (Promega, Madison, WI) and a Master cycler PCR machine to run the samples (Eppendorf AG, Hamburg, Germany). Quantitative PCR was performed with the LightCycler® version 1.1 instrument using Roche Applied Science reagents according to manufacturer recommendations and as described earlier(2). Primer sequences are available upon request.
Immunofluorescence Staining
Cells were plated onto glass LAB-TEK 2-well chamber slides or onto glass disks placed inside wells of a 24-well plate and grown to 70–90% confluence. Paraffin embedded E17.5embryo tissue sections were pre-treated in antigen retrieval solution (10mM citrate buffer). Immunofluorescence staining was performed as described earlier(20). Primary antibodies were: anti-Sc65 (Proteintech Group, Chicago, IL), anti-protein Disulfide Isomerase (PDI - monoclonal RL90, Abcam, Cambridge, MA), anti-GM130 (a gift from Dr. Vladimir Lupashin), anti-myc (clone 9E10 from Developmental Studies Hybridoma Bank), anti-human α1(I) (LF-68, from Dr. Larry Fisher at NIDCR)(21). Secondary antibodies were goat anti-rabbit or anti-mouse secondary antibody conjugated to Alexa Fluor 488 or 594 (Invitrogen, Grand Island, NY), respectively. Slides or disks were mounted with Dapi Fluoromount-G (Southern Biotech Inc). Images were captured using a Zeiss Axio Imager M1 microscope (Carl Zeiss, Jena, Germany). Where indicated, optical sections along the z axis were acquired by structured epi-fluorescent illumination, using a Zeiss Apotome module and processed with Axiovision software (Carl Zeiss).
Western Blotting
Cells or tissues were lysed into RIPA buffer (50mM Tris-HCl pH 7.5, 150mM NaCl, 0.1% SDS, 1 mM EDTA, 0.5% Sodium Deoxycholate, and 1% Triton X-100) containing a cocktail of protease inhibitors including EDTA (Cat. # M250, AMRESCO, Solon, OH). Lysates were centrifuged (@18,000g) and supernatants collected and quantified using the Bio-Rad protein assay dye reagent (Bio-Rad, Hercules, CA). Primary antibodies: Sc65 (Proteintech Group, Chicago, IL), PDI, ERp57, ERp44 β, Bip (Cat.# 4759, Cell signaling, Danvers, MA), α-tubulin monoclonal antibody (Clone TBN06, Thermo Fisher Scientific, Pittsburgh, PA), β-actin (Cat.# A00702, GenScript, Piscataway, NJ). Secondary antibodies were goat anti-rabbit or anti-mouse IRDye 680LT (LI-COR Biosciences, Lincoln, NE). Membranes were scanned using a LICOR Odyssey instrument.
Tissue Sections, In Situ Hybridization, Bone Histomorphometry and Micro-CT
Embryonic tissues, hind limbs and spines at 10 weeks and 24 weeks of age were harvested, fixed in formalin, dehydrated and embedded in either paraffin after demineralizing in 5% formic acid or non-decalcified in methyl-methacrylate according to standard procedures(22). Following sectioning at 4–5µm, specimens were utilized for different staining procedures. In situ hybridization on paraffin embedded tissues was performed as described previously(23). The Sc65 RNA in situ probe corresponds to base pairs 963–1506 (accession number sequence BC031856). See supplementary method for bone histomorphometry and detailed micro-CT analysis.
Serum Quantification of CTX, RANKL and Cytokine Profile
The quantification of bone related degradation products from the C-terminal telopeptides of type I collagen was measured in serum using the RatLaps (CTX-I) EIA kit (IDS) according to the manufacturer’s recommendations. The quantification of RANKL in mouse serum was performed using the mouse TRANCE/RANKL/TNFSF11 Quantikine ELISA Kit (catalog #MTR00 - R&D Systems, Minneapolis, MN) according to the manufacturer’s instructions. A Proteome Profiler array (R&D Systems, Minneapolis, MN) was performed on sera from WT and Sc65Mut mice (n=3 each genotype). This kit consists of a Mouse Cytokine Array Panel of 40 selected capture antibodies spotted in duplicate on nitrocellulose membranes and includes 3 reference spots and a negative control. Spot intensity was normalized to reference spots using densitometry.
Statistical analyses
All parameters measured are presented as mean ± Standard Deviation and were analyzed with the Student’s t-test using a two-tailed distribution and two-sample equal variance as appropriate. P values <0.05 were considered statistically significant and reported as such.
RESULTS
Sc65 IS LOCALIZED IN THE ENDOPLASMIC RETICULUM
Sc65 was initially described as a rat synaptonemal complex protein of 65kDa (and hence its name) expressed in testis, brain, heart and, to a lesser extent, in the liver(24). Subsequently, it was also identified as a human tumor-associated autoantigen and as a 55 kDa nucleolar protein (also named NO55)(17, 25, 26). Sc65 was also described as a cytoplasmic ‘adapter protein’ capable of interacting directly with the cytoplasmic tail of myelin protein zero (P0) and the receptor for activated C kinase 1 (RACK1)(27). Based on this evidence, Sc65 would be the only Leprecan family member with a specific nuclear localization. Based on our analysis of the Leprecan family member Crtap(2–5), and our hypothesis that Sc65 was important in bone homeostasis, we first established the precise sub-cellular localization of Sc65.
Initially, a detailed bioinformatic analysis using algorithms for signal peptide (SignalP4.1, Phobius, Philius)(28–30) or protein sub-cellular localization prediction (Psort II, TargetP)(31, 32) was performed. The data suggested that Sc65 contained a signal peptide (aa 1–18) and the protein was predicted to be targeted to the secretory pathway (Supplementary Fig.1).
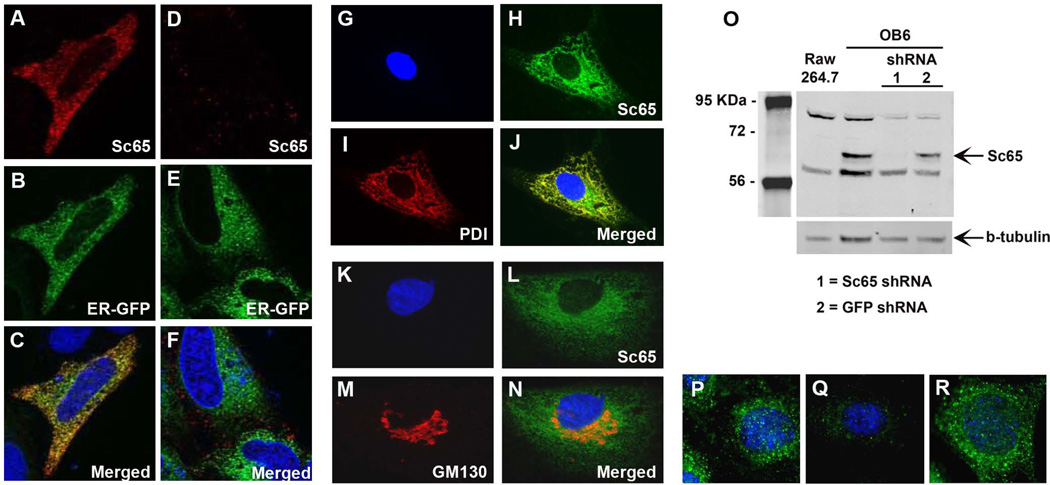
Next, Hela cells were co-transfected with an expression vector expressing Sc65 fused to an N-terminal myc-tag and with an ER-specific marker fused to green fluorescent protein (GFP). Immunostaining with a primary anti-myc antibody demonstrated a specific reticular distribution pattern that overlapped with the staining of the ER (Fig. 1A–F). Consistently, endogenous Sc65, immunolocalized with an affinity purified rabbit polyclonal antibody raised against the human full-length Sc65 recombinant protein (Proteintech Group, Inc.), was detected in the ER of primary human fibroblasts. In fact, Sc65 co-localized with protein disulfide isomerase (PDI), a known and specific ER resident protein (Fig. 1G–J). A similar immunolocalization in the ER was observed in osteoblastic MCT3T3-E1 cells. In addition, in these cells Sc65 was not immunolocalized to the Golgi compartment as shown by co-immunostaining with the Golgi marker protein GM130 (Fig. 1K–N).
Figure 1.
Sc65 is an ER-associated protein. A–F. Hela cells co-transfected with Sc65-myc (A–C) or empty vector (D–F) and an ER-GFP marker and then immunostained with anti-myc antibody (A, D). C and F show merged images of optical sections A–B and D–E, respectively. G–J and K–N. Representative images of human primary fibroblasts and MC3T3 cells stained with antibody against Sc65 and protein disulfide isomerase (a marker of the rER) or GM130 (a Golgi marker), respectively. O. Sc65 knock-down by lentiviral shRNA in OB6 osteoblastic cells demonstrating specificity of the Sc65 polyclonal antibody. Also note lack of Sc65 expression in Raw264.7 cells. P–R. Immunofluorescence on OB6 cells (P) and OB6 cells infected with Sc65 shRNA (Q) or GFP shRNA (R) showing specific down-regulation of Sc65 expression.
Although Sc65 and Crtap are >50% identical at the amino acid level, the lack of cross-reactivity of the Sc65 polyclonal antibody with Crtap was apparent (Supplementary Fig. 2A). We next confirmed the specificity of the Sc65 polyclonal antibody using the osteoblastic cell line, OB6(19) that were infected with a lentivirus expressing a Sc65 shRNA (short hairpin RNA) or a GFP shRNA as control (Fig. 1O). Specific downregulation of Sc65 expression by the shRNA was observed by both Western blot and IF analysis (Fig. 1O–R).
Collectively, these data demonstrated that Sc65, like the other leprecan family members is localized in the ER of somatic cells (including osteoblasts) and is not a nuclear protein.
Sc65 IS EXPRESSED IN THE SKELETON AND IN OTHER TISSUES
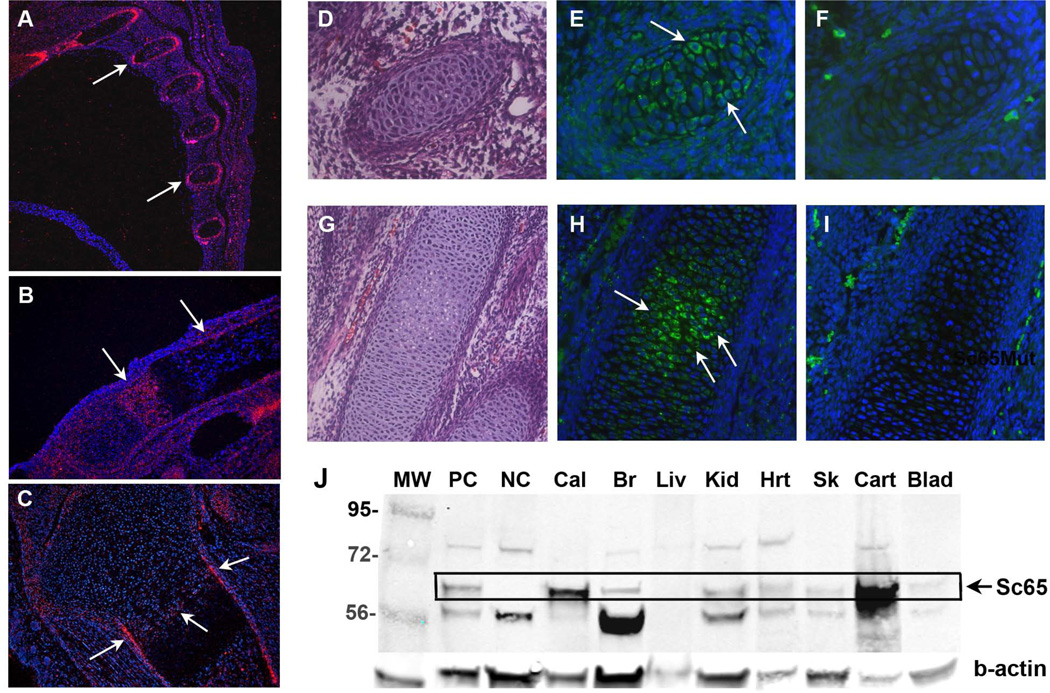
We next determined the temporal expression of Sc65 during mouse skeletal development. By mRNA in situ hybridization, Sc65 mRNA transcripts were detected in cross-section of fetal ribs at E14.5 and E16.5 (Fig. 2A). At postnatal day 2 (P2) Sc65 was expressed in the proliferating and pre-hypertrophic chondrocytes in the growth plate of long bones and in mineralizing chondro-osseus bone collar and cortical bone (Fig. 2B–C). Immunofluorescence confirmed the mRNA in situ hybridization localization and demonstrated specific expression in cross sections of embryonic ribs and long bone chondrocytes, with a clear intracellular protein localization (Fig. 2D–I). Moreover, multiple organ and tissues were harvested from 3 day-old mouse pups, homogenized and analyzed by Western blot. The specific expression of Sc65 protein was identified in calvaria, cartilage, skin, kidney, and to a lower extent in heart, and brain (Fig. 2J). Sc65 was also abundantly expressed in osteoblastic cell lines (OB6 and MCT3T3-E1) and in primary calvarial osteoblasts. These data suggest that Sc65 is expressed throughout skeletal development but similar to Crtap(33), Sc65 is expressed in non-bone tissues. Interestingly, the lack of Sc65 does not affect expression of Crtap and viceversa (Supplementary Fig. 2B), suggesting that there is no inter-dependence for protein expression between these two highly related ER proteins.
Figure 2.
Sc65 expression during mouse embryonic development. A–C In situ hybridization showing Sc65 mRNA expression (red signal) in mouse embryo sections of ribs at E16.5 (A) and in metatarsus (B) and tibia (C) at P2. D,G Hematoxilin and eosin stained sections of E13.5 rib and E15.5 metatarsus, respectively. E–F, H–I Serial sections of D and G stained with Sc65 antibody (E and H) or secondary antibody only (F and I). J. Qualitative Western Blot on several P3 tissue lysates. PC = positive control (osteoblast lysate), NC= negative control (Sc65Mut osteoblast lysate), Cal = calvaria, Br = brain, Liv = liver, Kid = kidney, Hrt = heart, Msc = muscle, Sk =skin, Cart = cartilage. The boxed region identifies Sc65.
Sc65 protein expression was not observed in the osteoclast precursor, monocyte/macrophage cell line RAW264.7(34) by immunoblot analysis (see Fig. 1O). In addition, Sc65 mRNA transcripts were not detected by RT-PCR in cDNA derived from CD11b+ murine bone marrow cells isolated by flow cytometry or in isolated splenocytes or bone marrow macrophages (BMM). Similarly, no Sc65 mRNA expression was observed by RT-PCR (35 cycles) performed on cDNAs derived from splenocytes or BMMs treated with M-CSF and RANKL for 1–2 days (osteoclast precursors) or 6 days (fully mature TRAP-positive multinucleated osteoclasts) (Supplementary Fig.3). These data demonstrate that Sc65 is not detectable in osteoclast precursors or mature osteoclasts.
GENERATION OF Sc65 MUTANT MICE
To determine the in vivo function of Sc65, a gene-trap ES cell clone that harbored a trap-vector insertion into the Sc65 gene was used. Southern blot of the ES cells DNA confirmed that the Sc65 locus was targeted (data not shown) and further sequencing showed that the trap vector had inserted at the 3’end of exon 5 (out of 8 total exons) and disrupted its integrity (Supplementary Fig. 3A). Chimeric mice were generated and germline transmission of the insertion was obtained. Homozygous mutant pups were obtained at the expected Mendelian ratio by mating mice that were heterozygous for the insertion (Supplementary Fig. 3B). The effect of the trap-vector insertion into the Sc65 locus was characterized both at the mRNA and protein level. RT-PCR from Sc65 homozygous mutant (Sc65Mut) calvarial osteoblast RNA detected the presence of two transcripts which were sequenced to determine the splicing consequence of the insertion at the single base pair level (Supplementary Fig. 3A): a bicistronic transcript coding for the 5’ portion of the Sc65 gene, including a truncated exon 5, and containing an open reading frame with a stop codon 5 amino acids into the LTR sequence followed by an IRES element and the EGFP gene; a second transcript coding for the Neo resistance gene followed by a truncated 3’ end of the gene. Although these transcripts were detected by RT-PCR, a Western blot using a specific Sc65 antibody generated against the entire recombinant protein demonstrated the absence of Sc65 protein or other Sc65-related protein products compared to WT calvarial osteoblast controls (Supplementary Fig. 3C). Furthermore, immunofluorescent staining of homozygous mutant embryonic bone sections confirmed the dramatic reduction of Sc65 expression compared to WT bone sections (Supplementary Fig. 3D). These data indicate that the insertion created a null allele.
PHENOTYPE OF THE Sc65 MUTANT MICE
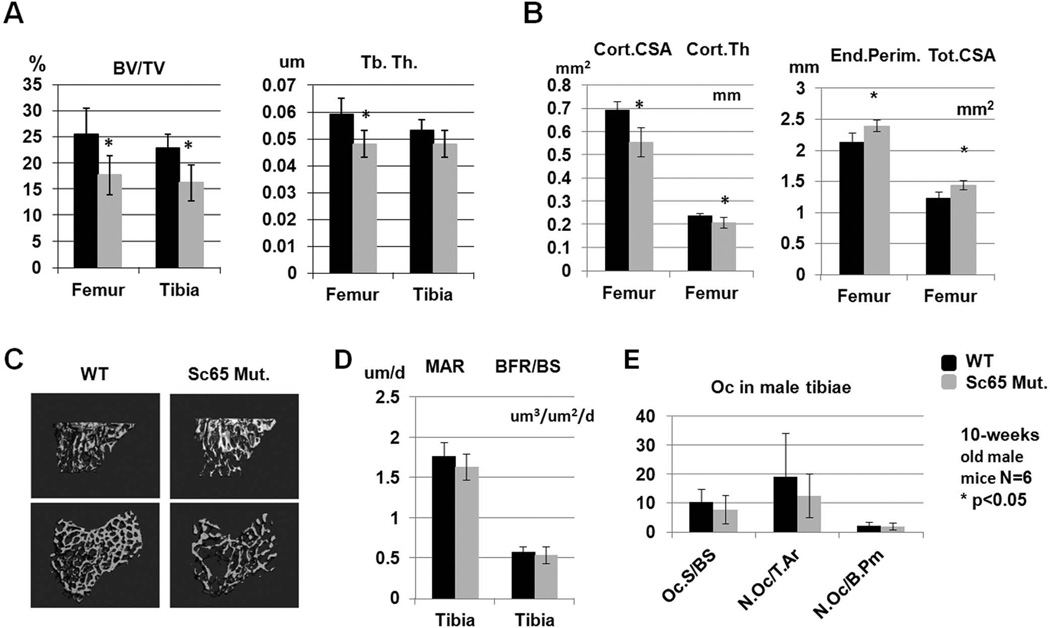
The skeletal phenotype of the homozygous Sc65Mut mice was characterized in detail as we have described(22, 35). Skeletal preparations of newborn pups did not show any macroscopic differences or patterning defects between Sc65Mut and WT genotypes (data not shown). Micro-CT analysis of tibiae and femurs at 10 weeks of age revealed a statistically significant decrease in bone volume/tissue volume (BV/TV) in male Sc65Mut mice (N=6, p=0.004 and p=0.013, respectively) compared to WT littermates (Fig. 3A, C), with reduced femoral trabecular thickness. Moreover, micro-CT analysis of femur cortical bone geometry (males, N=6) identified a significantly decreased cortical cross-sectional area and cortical thickness, with increased endosteal perimeter and total cross sectional area in the Sc65Mut mice, indicating a larger bone with thinner cortices (Fig. 3B). The micro-CT analysis was repeated in a second cohort of mice (both males and females, n=7 each group) at 6 months of age and confirmed the low bone mass phenotype in both male and female Sc65Mut compared to WT mice (Supplementary Fig. 5). Osteoblast numbers, surfaces and dynamic parameters of bone formation measured at 10 weeks of age were similar between the two genotypes indicating normal osteoblast counts and function (data not shown and Fig. 3D). Osteoclast parameters measured in tibiae at 10 weeks were not significantly different between the two genotypes (Fig. 3E).
Figure 3.
Static and dynamic bone histomorphometric measurements. A. 10-week old Sc65Mut male mice (n=6) showed significantly reduced BV/TV (as shown by micro-CT rendering of proximal tibia in C), femoral trabecular thickness and femoral cortical geometry with reduced cortical cross sectional area and thickness and increased endosteal perimeter and total cross sectional area (B). Both dynamic indices of bone formation and osteoclast parameters were not significantly different between the two genotypes (D–E). Asterisks indicate statistical significant differences (p<0.05).
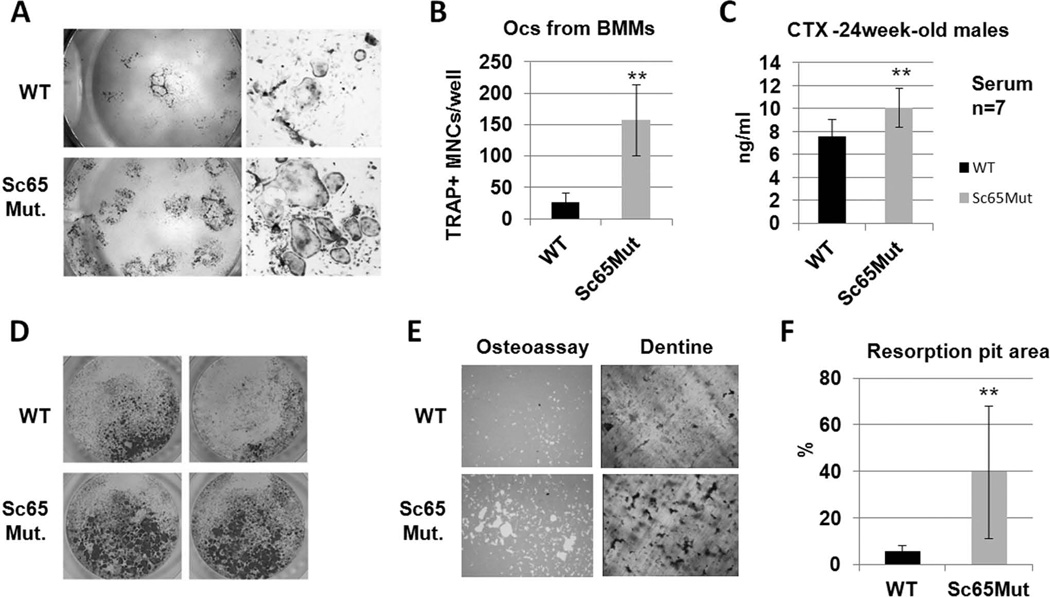
We next performed ex vivo whole bone marrow cultures derived from 1 month-old and adult mice (n=4 and n=5 mice per group, respectively). In all cases, a significant increase in osteoclast numbers was observed in cultures derived from Sc65Mut compared to WT marrow (Fig. 4A). Similarly, ex vivo bone marrow macrophage (BMM) cultures (n=5) and primary splenocytes (n=5) treated with M-CSF and RANKL gave rise to increased numbers of TRAP positive mature multinucleated osteoclasts when derived from adult or even 1 month-old (in the case of BMMs) Sc65Mut compared to WT mice (Fig. 4B,D).
Figure 4.
Increased osteoclastogenesis in Sc65 mutant mice. Ex-vivo whole bone marrow (A) and bone marrow macrophages (BMM) (B) cultures (n=3 each) from 5 month-old or older mice showed increased number of TRAP-positive multinucleated cells (TRAP+ MNC/well) in Sc65Mut compared to WT mice (C). Serum levels of CTX were significantly elevated in 6-month old Sc65Mut compared to WT mice (n = 7, p = 0.012). Increased numbers of TRAP-positive MNCs (D) and resorption areas (E) were observed when Sc65Mut splenocytes or BMMs were plated either on plastic (96-well plate), osteo-assay plates or dentine slices, compared to WT controls (F). Quantification of resorption pits area from Osteassay plates (p = 0.015).
In addition, when murine osteoclast differentiation was induced from splenocytes or BMMs cultured on OsteoAssay plates or on dentine slices for 6–8 days in the presence of RANKL and M-CSF, significantly increased resorbed area was observed in Sc65Mut cells compared to WT control mice (Fig. 4E–F). This is likely the result of the higher number of osteoclasts in Sc65Mut cultures and not of an increased resorption capacity of single cells. Supporting these in vitro observations, serum measurement of 6 month-old males demonstrated significantly higher levels of cross-linked C-telopeptide of type I collagen in Sc65Mut compared to WT control mice (Fig. 4C, n=7 males, p<0.05). These data support the conclusion that increased bone resorption underlies the observed low bone mass phenotype of Sc65Mut mice.
WHAT IS THE FUNCTION OF Sc65?
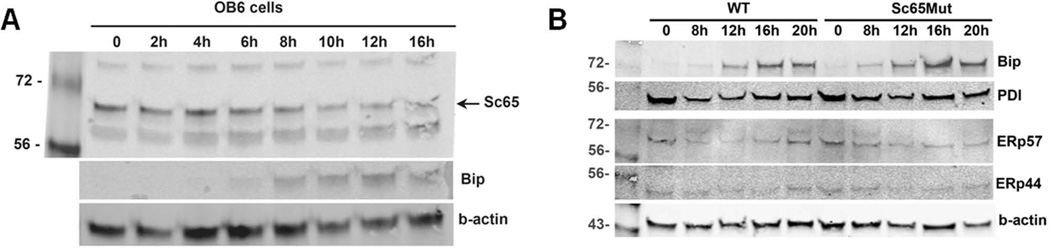
Since Sc65 is not expressed by osteoclasts, we surmised that the increased bone resorption was likely due to a non-cell autonomous osteoclastic defect. Sc65 is expressed in osteoblasts and these cells, among others, are known to secrete factors that affect the differentiation and function of osteoclasts. We hypothesized that, as Sc65 is localized in the ER, its absence might cause abnormal processing, accumulation and/or unfolding of polypeptides and thereby induce ER stress or an impaired response to ER stress affecting normal cross-talk between osteoblasts and osteoclasts. To test this hypothesis, cultures of primary murine fibroblasts or murine OB6 osteoblastic cells were treated with 2µg/ml tunicamycin (a stress inducer) and then harvested at different time points. As shown, levels of BiP expression increased with time, confirming increasing ER stress, but the expression of endogenous Sc65 was not significantly affected (Fig. 5A). These data demonstrated that Sc65 is not regulated during ER stress. Next, ER stress was induced in primary calvarial osteoblast cultures from Sc65Mut and WT mice (n=2) and the expression levels of Bip, PDI (PDa1), ERp57 (PDIa3), ERp44 analyzed after 0, 8, 12, 16 and 20 hours of tunicamycin treatment (2µg/ml). No significant differences between the two genotypes were observed (Fig. 5B). Moreover, qPCR analyses of ER-stress sensors and response molecules such as PDI, ERp57, Bip, Perk, Chop, Xbp1, ERp44, ERp72, ERp57 were not significantly different (data not shown).
Figure 5.
Sc65 and ER stress. A. Western blot of OB6 cells treated with tunicamycin for the indicated time. While levels of Bip expression rose indicating induction of ER stress, Sc65 expression level did not change significantly. B. Western blots of Sc65Mut and WT primary osteoblast treated with tunicamycin for the indicated time points and showing expression of several proteins involved in protein folding and ER stress. No significant differences were detected.
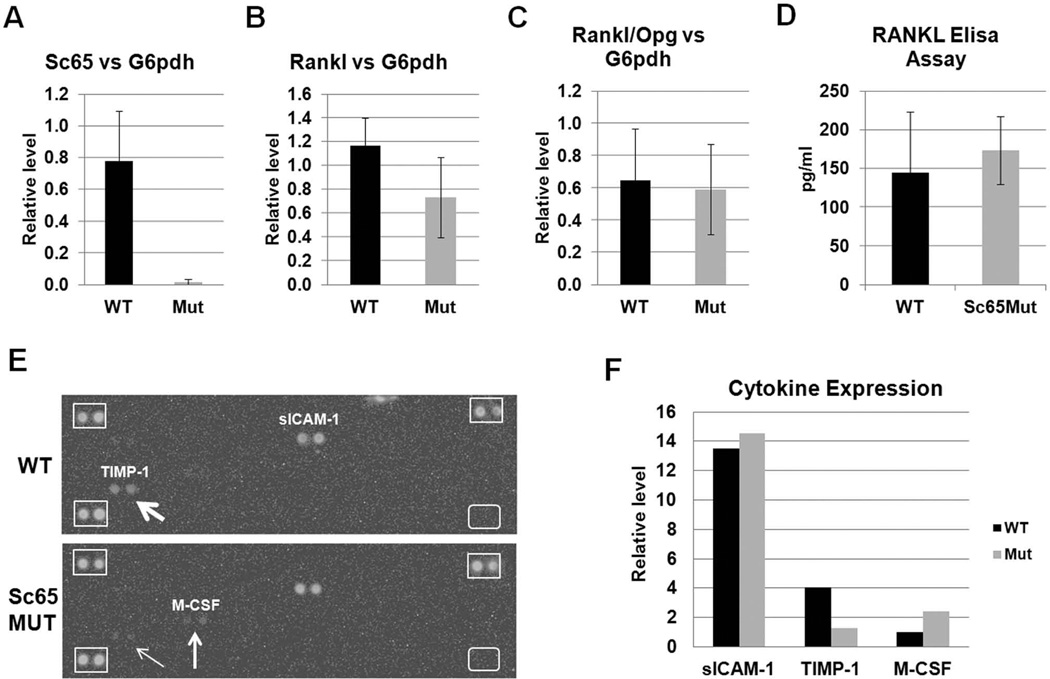
An alternative mechanism for the increased bone resorption observed is that lack of Sc65 could alter synthesis of the receptor activator for NF-kB ligand (RANKL)(36). While quantitative PCR analysis confirmed the dramatic decrease in Sc65 transcripts in Sc65Mut osteoblasts, no significant differences in RANKL, osteoprotegerin (Opg) expression or their ratio was observed in Sc65Mut compared to WT osteoblasts (Fig. 6A–C).
Figure 6.
Measurements of RANKL, Opg and other cytokines. qPCR analysis showed reduction of Sc65 (A) but no significant changes in either RANKL (B) or osteoprotegerin (Opg) transcripts or their ratio (C) in cDNA from Sc65Mut or WT primary osteoblasts. Similar results were obtained when normalized against multiple housekeeping genes, including Gapdh, b-actin and G6pdh. D. Elisa assay for serum RANKL did not reveal significant differences between WT and Sc65Mut adult (4–6 month-old) male mice (n = 7–8, p = 0.3). E. Image of the Cytokine Array panels hybridized with pooled sera from Sc65Mut or WT mice (n=3) showing differential expression of both TIMP-1 and M-CSF between the two genotypes. Differences of expression are quantified by densitometry and shown in F.
Next, an in vivo assay was used to measure circulating RANKL in serum from adult male mice. No significant difference between the two genotypes was observed (Fig. 6D). Since whole bone marrow cultures from Sc65Mut mice demonstrated increased osteoclastogenesis, we next measured the expression of circulating cytokines capable of stimulating osteoclastogenesis. A Mouse Cytokine Array Proteome Profiler array was probed with pooled aliquots of freshly prepared sera from one month-old WT and Sc65Mut mice (n=3). As shown (Fig. 6E–F) a 2.5 fold increase in circulating M-CSF and a 3.2 fold decrease in tissue inhibitor of metalloprotease-1 (TIMP-1) was observed in Sc65Mut compared to WT sera. These data suggest that the circulating serum cytokine profile is supportive of increased osteoclast precursor survival and potentially bone resorption.
Finally, to determine whether Sc65, like Crtap, has a role in the modification of collagen at the post-translational level, mass spectrometric analysis of types I and V collagens extracted from Sc65Mut mouse bone and type II collagen from Sc65Mut mouse cartilage was performed. A normal level of occupancy of 3-Hyp at α1(I) Pro986, α1(II) Pro986, α2(V) Pro986 and α2(I) Pro707 was observed (data not shown). Moreover, type I collagen accumulation in the ER was normal in permeabilized WT and Sc65Mut primary calvarial osteoblasts, suggesting no overt alterations in collagen synthesis/secretion (Supplementary Fig. 6). These data are entirely consistent with the complete lack of changes in BFR and MAR measured histomorphometrically.
DISCUSSION
The Leprecan family of proteins currently consists of five known members(14), Crtap, Sc65, P3h1, P3h2 and P3h3. Mutations in CRTAP and LEPRE1 (encoding P3H1) genes have been shown to cause recessive forms of OI(2, 4, 5) while mutations in LEPREL1 (LEPRECAN LIKE-1, encoding P3H2) have been associated with severe myopia(37). LEPREL2, encoding P3H3, also contains the conserved 2-oxoglutarate and Fe(II)-dependent dioxygenase domain(38) and is likely responsible for the 3-hydroxylation of proline residues with its own collagen substrate and tissue specificity. However, the Sc65 protein member of this family is the subject of somewhat conflicting data. In fact, Sc65 was initially described as a synaptonemal complex protein during meiosis in the rat(24) and subsequently proposed as a human nucleolar antigen(17). Interestingly, the corresponding cDNA was isolated by an immunoscreening approach, however, no validation of the specificity of the antisera was provided(17, 24). Another report recently suggested that Sc65 was an adapter protein, capable of direct interaction with myelin protein zero (P0) and the receptor for activated C kinase 1 (RACK1)(27). Our interest in the members of the Leprecan family as regulators of bone development led us to challenge these reports. We performed an in silico prediction analysis that suggested the presence of a signal peptide in Sc65. Furthermore, robust immunostaining of both cells lines and primary cells demonstrated that Sc65 is an ER protein, just like Crtap and the P3hs and is not nuclear as had been suggested. We have yet to immunostain for Sc65 in testis and thus cannot completely exclude a role for Sc65 during certain phases of meiosis(24). However, the clear negative staining in the nucleolus or the nucleus of somatic cells strongly suggests that Sc65 is indeed a new ER-resident protein.
The insertion of the gene-trap vector into the Sc65 locus resulted in a null allele with complete absence of Sc65 protein in tissues from homozygous mutant mice. Sc65Mut mice were significantly osteopenic and showed reduced BV/TV in the trabecular bone compartment, in addition to altered cortical geometry with decreased cortical thickness in the appendicular skeleton. Female mice appeared less affected and osteopenic compared to males, however the phenotype worsened with age in both genders. Dynamic indices of bone formation, as well as osteoblast numbers and surfaces measured in tibiae of 10 weeks old male mice, showed no significant differences between the two genotypes. Surprisingly, measurements of osteoclast parameters in vivo, such as osteoclast surface and numbers per bone surface were not significantly different in either the tibiae or spines of 10 week old mice. However, serum levels of type I collagen breakdown product (CTX) were significantly elevated in 6-month old Sc65Mut mice compared to WT controls, suggesting increased bone resorption with age. It is likely that significant differences in osteoclast parameters in vivo may not have reached significance at 10 weeks and older age mice may reveal the difference. Alternatively, it is possible that a small increase in osteoclastogenesis at 10 weeks of age would have required a larger cohort of mice (>6) to be detected. Despite the lack of histomorphometric data demonstrating altered osteoclast parameters at 10 weeks, significantly increased osteoclastogenesis and bone resorption was observed in whole bone marrow cultures, BMM and splenocytes derived from both 1 month-old and adult Sc65Mut mice treated with M-CSF and RANKL compared with WT control mice.
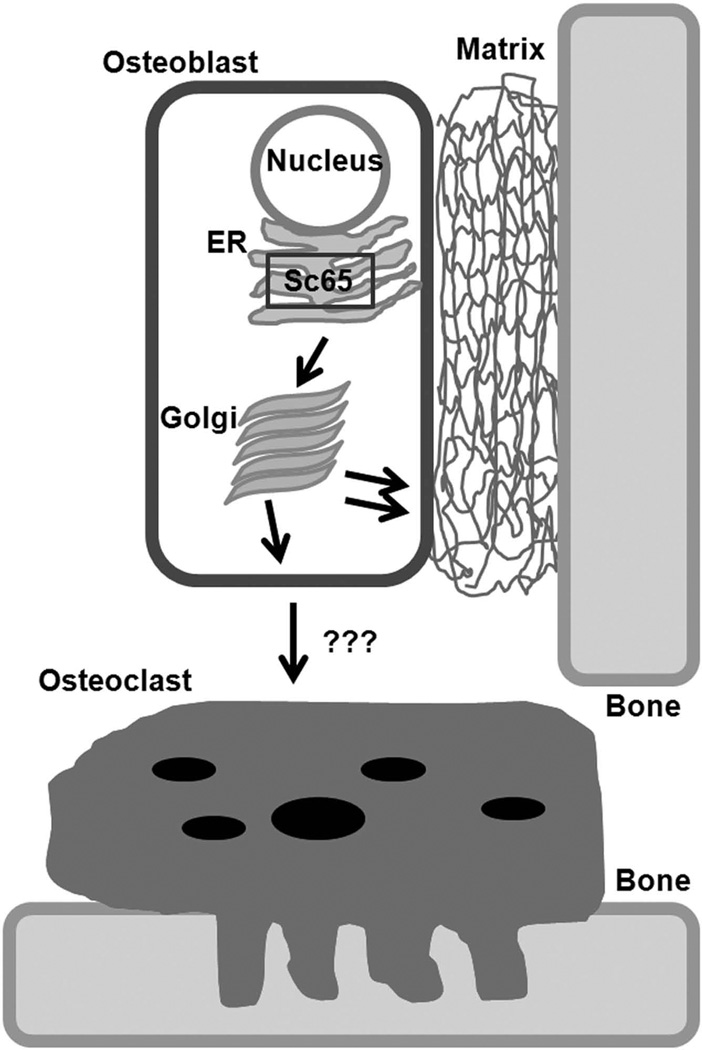
While Crtap is expressed in all bone cells, chondrocytes, osteoblasts and osteoclasts(2), Sc65 is found only in chondrocytes and osteoblasts. Using Western blot and RT-PCR, we could not detect expression of Sc65 in the monocyte/macrophage cell line RAW264.7 or in cDNA derived from bone marrow CD11b+ cells isolated by flow cytometry. Similarly, purified splenocytes or BMMs did not express Sc65 mRNA, even when cultured with M-CSF and RANKL for 24 hours or for 6 days. These data suggest that the osteopenic phenotype of Sc65Mut mice is mediated by a lack of Sc65 expression in other cell types, not osteoclasts. Our hypothesis is that the lack of Sc65 in osteoblasts causes an increased osteoclastogenesis (Fig.7A), though it is possible that the observed low bone mass and increased bone resorption are secondary to the lack of Sc65 in non-skeletal cells. However, several lines of evidence argue against such a scenario: 1) Sc65, like the highly related Crtap, is expressed in several tissues but the primary phenotype of the Crtap−/− mice is manifested in the skeleton(2, 33); 2) histological survey of 10 week old Sc65 mutant organs did not show any obvious macroscopic abnormalities; 3) isolated bone marrow cultures produce many more osteoclasts compared to WT control bone marrow cultures; 4) we have been unable to generate double homozygous Crtap−/−;Sc65Mut mice (total mice generated = 170; expected double mutants=22; obtained 0 – data not shown); 5) importantly, WT BMMs co-cultured with Sc65Mut osteoblasts showed a 5-fold increase in number of TRAP-positive osteoclasts compared to the same cells cultured with WT osteoblasts (p<0.05, Supplementary Fig.7). Collectively, the data suggest that Crtap and Sc65 may lie in the same genetic pathway or affect bone mass in a compounding way that is incompatible with postnatal life. Conclusive answers will come only from cell lineage-specific conditional inactivation of the Sc65 allele, an effort that is currently underway in our laboratory.
Figure 7.
Current working hypothesis. Current model postulates an osteoblast-mediated increased osteoclastogenesis with loss of Sc65.
The hypothesis that Sc65 may be involved in ER stress or that an unfolded protein response (UPR) could be impaired in its absence was reasonable, since Sc65 localizes to the ER and a bioinformatics prediction suggested putative folding similarity to ER stress related molecules (data not shown). However, induction of ER stress using tunicamycin demonstrated that Sc65 expression was not regulated and no significant differences in several ER stress response genes were detected between the two genotypes. This demonstrated no obvious role for Sc65 in the unfolded protein response (UPR).
Furthermore, our efforts to determine whether the lack of Sc65 altered secretion of RANKL were negative both in vitro and in vivo. It is still plausible that a small local rise in the concentration of RANKL in the bone marrow milieu or on osteoblast/stromal cell surface could enhance osteoclastogenesis.
To begin to understand if systemic changes in pro-osteoclastic cytokines may be responsible for the increased osteoclastogenesis and low bone mass, we profiled the levels of cytokines in the serum of 1 month old mice. The analysis revealed a significant down-regulation of TIMP-1 and up-regulation of M-CSF expression in Sc65Mut mice compared to WT. These observations are compelling and argue for a systemic change that produced an osteoclast-supportive environment in the absence of Sc65. We anticipate that the increased expression of M-CSF likely sustains an increased pool of monocyte/macrophage lineage cells in vivo that differentiates into more mature osteoclasts in vitro compared to controls. Similarly, the down-regulation of TIMP-1, an MMP-9 inhibitor, would also support a pro-osteolytic resorption environment in vivo. Indeed, TIMP-1 has been shown to inhibit bone resorption, decrease bone turnover and reduce bone loss in a mouse model when over-expressed by osteoblasts(39–41). Thus, the cytokine profile supports the idea that Sc65 is a negative regulator of osteoclastogenesis. These data also suggest that further studies of the role of Sc65 are warranted and are ongoing in additional cohorts of mice.
Finally, although we could not demonstrate an effect of the lack of Sc65 on fibril-forming collagens, known proline residues that become 3-hydroxylated in the ER(2, 42), including Pro986, appeared to be modified normally in type I collagen extracted from Sc65Mut bones. While this confirms a non-overlapping role for Sc65 with the function of Crtap, it does not exclude the possibility that Sc65 interacts with either P3h2 or P3h3 and may be involved in the hydroxylation of other prolyl residues, perhaps of type IV collagen(42). Indeed, Sc65 is expressed in the kidney and recent work has linked Sc65 auto-immune antibodies with idiopathic human membranous nephropathy(43).
In conclusion, Sc65 is a novel ER-resident protein and not a nuclear/nucleolar antigen in somatic cells. Inactivation of the Sc65 gene in the mouse causes a low bone mass phenotype affecting both trabecular and cortical compartments; this is mediated by increased bone resorption that progresses with age. Therefore Sc65, similar to other Leprecan family members, has a protective effect on the skeleton. Interestingly, while the lack of Crtap causes an osteoblast functional defect, the lack of Sc65 appears to act in a non cell-autonomous fashion on the osteoclast compartment. Current efforts are focused on the identification of Sc65 interacting proteins in an attempt to gain further insights into the mechanism of activity and function of Sc65 in skeletal disease and OI.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr. Charles O’Brien (UAMS, Little Rock, AR) for helpful discussions and for providing cDNA from CD11B+ bone marrow cells and Dr. Bryant G. Darnay (UT-MD Anderson Cancer Center, Houston, TX) for providing L929 cells and His-RANKL recombinant protein. The GMP130 antibody and the OB6 cell line were a kind gift of Dr. Vladimir Lupashin and Dr. Robert Jilka, respectively (UAMS, Little Rock, AR). The 9E10 anti-myc monoclonal antibody was obtained from the Developmental Studies Hybridoma Bank. This work was supported by UAMS Institutional Funds, Arkansas Biosciences Institute (Arkansas Tobacco Settlement Proceeds Act of 2000), ASBMR 2010 Junior Faculty Osteoporosis Research Award (RM) and in part supported by the National Institute of Arthritis and Musculoskeletal and Skin Diseases Award Number 1 R01 AR060823-01A1 (RM). Support also came from the NIH National Clinical and Translational Science Award (CTSA) grant UL1 TR000039 and grants P30 GM103450 and P20 GM103429. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Supplemental data are included with this submission.
DISCLOSURES
All authors state that they have no conflicts of interest to disclose.
Authors’ role: All authors contributed, read and approved this manuscript. KG, PC, RB and MD performed IF, Western Blots, mouse genotyping, cell culture in vitro experiments. YQ did all work on ES cells, microinjections and mouse chimera generation. NS, FS, RS and LS generated all bone histological preparations, special stainings, performed micro-CT and bone histomorphometric measurements, and helped in the interpretation. DE performed mass-spectrometry on collagens. PC, DE, DG, LJS, and RM contributed to the design and interpretation of the experiments. RM takes responsibility for the integrity of the data analysis.
REFERENCES
- 1.Rauch F, Glorieux FH. Osteogenesis imperfecta. Lancet. 2004 Apr 24;363(9418):1377–1385. doi: 10.1016/S0140-6736(04)16051-0. [DOI] [PubMed] [Google Scholar]
- 2.Morello R, Bertin TK, Chen Y, Hicks J, Tonachini L, Monticone M, Castagnola P, Rauch F, Glorieux FH, Vranka J, Bachinger HP, Pace JM, Schwarze U, Byers PH, Weis M, Fernandes RJ, Eyre DR, Yao Z, Boyce BF, Lee B. CRTAP is required for prolyl 3- hydroxylation and mutations cause recessive osteogenesis imperfecta. Cell. 2006 Oct 20;127(2):291–304. doi: 10.1016/j.cell.2006.08.039. [DOI] [PubMed] [Google Scholar]
- 3.Barnes AM, Chang W, Morello R, Cabral WA, Weis M, Eyre DR, Leikin S, Makareeva E, Kuznetsova N, Uveges TE, Ashok A, Flor AW, Mulvihill JJ, Wilson PL, Sundaram UT, Lee B, Marini JC. Deficiency of cartilage-associated protein in recessive lethal osteogenesis imperfecta. New Engl J Med. 2006 Dec 28;355(26):2757–2764. doi: 10.1056/NEJMoa063804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cabral WA, Chang W, Barnes AM, Weis M, Scott MA, Leikin S, Makareeva E, Kuznetsova NV, Rosenbaum KN, Tifft CJ, Bulas DI, Kozma C, Smith PA, Eyre DR, Marini JC. Prolyl 3-hydroxylase 1 deficiency causes a recessive metabolic bone disorder resembling lethal/severe osteogenesis imperfecta. Nat Genet. 2007 Mar;39(3):359–365. doi: 10.1038/ng1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baldridge D, Schwarze U, Morello R, Lennington J, Bertin TK, Pace JM, Pepin MG, Weis M, Eyre DR, Walsh J, Lambert D, Green A, Robinson H, Michelson M, Houge G, Lindman C, Martin J, Ward J, Lemyre E, Mitchell JJ, Krakow D, Rimoin DL, Cohn DH, Byers PH, Lee B. CRTAP and LEPRE1 mutations in recessive osteogenesis imperfecta. Hum Mutat. 2008 Dec;29(12):1435–1442. doi: 10.1002/humu.20799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Christiansen HE, Schwarze U, Pyott SM, AlSwaid A, Al Balwi M, Alrasheed S, Pepin MG, Weis MA, Eyre DR, Byers PH. Homozygosity for a missense mutation in SERPINH1, which encodes the collagen chaperone protein HSP47, results in severe recessive osteogenesis imperfecta. Am J Hum Genet. 2010 Mar 12;86(3):389–398. doi: 10.1016/j.ajhg.2010.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van Dijk FS, Nesbitt IM, Zwikstra EH, Nikkels PG, Piersma SR, Fratantoni SA, Jimenez CR, Huizer M, Morsman AC, Cobben JM, van Roij MH, Elting MW, Verbeke JI, Wijnaendts LC, Shaw NJ, Hogler W, McKeown C, Sistermans EA, Dalton A, Meijers-Heijboer H, Pals G. PPIB mutations cause severe osteogenesis imperfecta. Am J Hum Genet. 2009 Oct;85(4):521–527. doi: 10.1016/j.ajhg.2009.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Alanay Y, Avaygan H, Camacho N, Utine GE, Boduroglu K, Aktas D, Alikasifoglu M, Tuncbilek E, Orhan D, Bakar FT, Zabel B, Superti-Furga A, Bruckner-Tuderman L, Curry CJ, Pyott S, Byers PH, Eyre DR, Baldridge D, Lee B, Merrill AE, Davis EC, Cohn DH, Akarsu N, Krakow D. Mutations in the gene encoding the RER protein FKBP65 cause autosomal-recessive osteogenesis imperfecta. Am J Hum Genet. 2010 Apr 9;86(4):551–559. doi: 10.1016/j.ajhg.2010.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barnes AM, Carter EM, Cabral WA, Weis M, Chang W, Makareeva E, Leikin S, Rotimi CN, Eyre DR, Raggio CL, Marini JC. Lack of cyclophilin B in osteogenesis imperfecta with normal collagen folding. New Engl J Med. 2010 Feb 11;362(6):521–528. doi: 10.1056/NEJMoa0907705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Myllyharju J, Kivirikko KI. Collagens, modifying enzymes and their mutations in humans, flies and worms. Trends Genet. 2004 Jan;20(1):33–43. doi: 10.1016/j.tig.2003.11.004. [DOI] [PubMed] [Google Scholar]
- 11.Gothel SF, Marahiel MA. Peptidyl-prolyl cis-trans isomerases, a superfamily of ubiquitous folding catalysts. Cell Mol Life Sci. 1999 Mar;55(3):423–436. doi: 10.1007/s000180050299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Law RH, Zhang Q, McGowan S, Buckle AM, Silverman GA, Wong W, Rosado CJ, Langendorf CG, Pike RN, Bird PI, Whisstock JC. An overview of the serpin superfamily. Genome Biol. 2006;7(5):216. doi: 10.1186/gb-2006-7-5-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van Gent D, Sharp P, Morgan K, Kalsheker N. Serpins: structure, function and molecular evolution. Int J Biochem Cell Biol. 2003 Nov;35(11):1536–1547. doi: 10.1016/s1357-2725(03)00134-1. [DOI] [PubMed] [Google Scholar]
- 14.Vranka JA, Sakai LY, Bachinger HP. Prolyl 3-hydroxylase 1, enzyme characterization and identification of a novel family of enzymes. J Biol Chem. 2004 May 28;279(22):23615–23621. doi: 10.1074/jbc.M312807200. [DOI] [PubMed] [Google Scholar]
- 15.Forlino A, Cabral WA, Barnes AM, Marini JC. New perspectives on osteogenesis imperfecta. Nat Rev Endocrinol. 2011;7(9):540–557. doi: 10.1038/nrendo.2011.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mackie EJ. Osteoblasts: novel roles in orchestration of skeletal architecture. Int J Biochem Cell Biol. 2003 Sep;35(9):1301–1305. doi: 10.1016/s1357-2725(03)00107-9. [DOI] [PubMed] [Google Scholar]
- 17.Ochs RL, Stein TW, Jr, Chan EK, Ruutu M, Tan EM. cDNA cloning and characterization of a novel nucleolar protein. Mol Biol Cell. 1996 Jul;7(7):1015–1024. doi: 10.1091/mbc.7.7.1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Andrade MA, Perez-Iratxeta C, Ponting CP. Protein repeats: structures, functions, and evolution. J Struct Biol. 2001 May-Jun;134(2–3):117–131. doi: 10.1006/jsbi.2001.4392. [DOI] [PubMed] [Google Scholar]
- 19.Lecka-Czernik B, Gubrij I, Moerman EJ, Kajkenova O, Lipschitz DA, Manolagas SC, Jilka RL. Inhibition of Osf2/Cbfa1 expression and terminal osteoblast differentiation by PPARgamma2. J Cell Biochem. 1999 Sep 1;74(3):357–371. [PubMed] [Google Scholar]
- 20.Amor IM, Rauch F, Gruenwald K, Weis M, Eyre DR, Roughley P, Glorieux FH, Morello R. Severe osteogenesis imperfecta caused by a small in-frame deletion in CRTAP. Am J Med Genet. Part A. 2011 Sep 30; doi: 10.1002/ajmg.a.34269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fisher LW, Stubbs JT, 3rd, Young MF. Antisera and cDNA probes to human and certain animal model bone matrix noncollagenous proteins. Acta Orthop Scand Suppl. 1995 Oct;266:61–65. [PubMed] [Google Scholar]
- 22.Suva LJ, Hartman E, Dilley JD, Russell S, Akel NS, Skinner RA, Hogue WR, Budde U, Varughese KI, Kanaji T, Ware J. Platelet dysfunction and a high bone mass phenotype in a murine model of platelet-type von Willebrand disease. Am J Pathol. 2008 Feb;172(2):430–439. doi: 10.2353/ajpath.2008.070417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Morello R, Zhou G, Dreyer SD, Harvey SJ, Ninomiya Y, Thorner PS, Miner JH, Cole W, Winterpacht A, Zabel B, Oberg KC, Lee B. Regulation of glomerular basement membrane collagen expression by LMX1B contributes to renal disease in nail patella syndrome. Nat Genet. 2001 Feb;27(2):205–208. doi: 10.1038/84853. [DOI] [PubMed] [Google Scholar]
- 24.Chen Q, Pearlman RE, Moens PB. Isolation and characterization of a cDNA encoding a synaptonemal complex protein. Biochem Cell Biol. 1992 Oct-Nov;70(10–11):1030–1038. doi: 10.1139/o92-147. [DOI] [PubMed] [Google Scholar]
- 25.Fossa A, Siebert R, Aasheim HC, Maelandsmo GM, Berner A, Fossa SD, Paus E, Smeland EB, Gaudernack G. Identification of nucleolar protein No55 as a tumour-associated autoantigen in patients with prostate cancer. Br J Cancer. 2000 Sep;83(6):743–749. doi: 10.1054/bjoc.2000.1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Comtesse N, Zippel A, Walle S, Monz D, Backes C, Fischer U, Mayer J, Ludwig N, Hildebrandt A, Keller A, Steudel WI, Lenhof HP, Meese E. Complex humoral immune response against a benign tumor: frequent antibody response against specific antigens as diagnostic targets. Proc Natl Acad Sci. USA. 2005 Jul 5;102(27):9601–9606. doi: 10.1073/pnas.0500404102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gaboreanu AM, Hrstka R, Xu W, Shy M, Kamholz J, Lilien J, Balsamo J. Myelin protein zero/P0 phosphorylation and function require an adaptor protein linking it to RACK1 and PKC alpha. J Cell Biol. 2007 May 21;177(4):707–716. doi: 10.1083/jcb.200608060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Petersen TN, Brunak S, von Heijne G, Nielsen H. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods. 2011;8(10):785–786. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- 29.Kall L, Krogh A, Sonnhammer EL. A combined transmembrane topology and signal peptide prediction method. J Mol Biol. 2004 May 14;338(5):1027–1036. doi: 10.1016/j.jmb.2004.03.016. [DOI] [PubMed] [Google Scholar]
- 30.Reynolds SM, Kall L, Riffle ME, Bilmes JA, Noble WS. Transmembrane topology and signal peptide prediction using dynamic bayesian networks. PLoS Comput Biol. 2008 Nov;4(11):e1000213. doi: 10.1371/journal.pcbi.1000213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Emanuelsson O, Nielsen H, Brunak S, von Heijne G. Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J Mol Biol. 2000 Jul 21;300(4):1005–1016. doi: 10.1006/jmbi.2000.3903. [DOI] [PubMed] [Google Scholar]
- 32.Nakai K, Horton P. PSORT: a program for detecting sorting signals in proteins and predicting their subcellular localization. Trends Biochem Sci. 1999 Jan;24(1):34–36. doi: 10.1016/s0968-0004(98)01336-x. [DOI] [PubMed] [Google Scholar]
- 33.Baldridge D, Lennington J, Weis M, Homan EP, Jiang MM, Munivez E, Keene DR, Hogue WR, Pyott S, Byers PH, Krakow D, Cohn DH, Eyre DR, Lee B, Morello R. Generalized connective tissue disease in Crtap−/ − mouse. PloS one. 2010;5(5):e10560. doi: 10.1371/journal.pone.0010560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Raschke WC, Baird S, Ralph P, Nakoinz I. Functional macrophage cell lines transformed by Abelson leukemia virus. Cell. 1978 Sep;15(1):261–267. doi: 10.1016/0092-8674(78)90101-0. [DOI] [PubMed] [Google Scholar]
- 35.Fowler TW, McKelvey KD, Akel NS, Vander Schilden J, Bacon AW, Bracey JW, Sowder T, Skinner RA, Swain FL, Hogue WR, Leblanc DB, Gaddy D, Wenger GR, Suva LJ. Low bone turnover and low BMD in Down syndrome: effect of intermittent PTH treatment. PloS one. 2012;7(8):e42967. doi: 10.1371/journal.pone.0042967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Suda T, Takahashi N, Udagawa N, Jimi E, Gillespie MT, Martin TJ. Modulation of osteoclast differentiation and function by the new members of the tumor necrosis factor receptor and ligand families. Endocr Rev. 1999 Jun;20(3):345–357. doi: 10.1210/edrv.20.3.0367. [DOI] [PubMed] [Google Scholar]
- 37.Mordechai S, Gradstein L, Pasanen A, Ofir R, El Amour K, Levy J, Belfair N, Lifshitz T, Joshua S, Narkis G, Elbedour K, Myllyharju J, Birk OS. High myopia caused by a mutation in LEPREL1, encoding prolyl 3-hydroxylase 2. Am J Hum Genet. 2011 Sep 9;89(3):438–445. doi: 10.1016/j.ajhg.2011.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vranka JA, Sakai LY, Bachinger HP. Prolyl 3-hydroxylase 1: Enzyme characterization and identification of a novel family of enzymes. J Biol Chem. 2004 Mar 24; doi: 10.1074/jbc.M312807200. [DOI] [PubMed] [Google Scholar]
- 39.Hill PA, Reynolds JJ, Meikle MC. Inhibition of stimulated bone resorption in vitro by TIMP-1 and TIMP-2. Biochim Biophys Acta. 1993 May 8;1177(1):71–74. doi: 10.1016/0167-4889(93)90159-m. [DOI] [PubMed] [Google Scholar]
- 40.Merciris D, Schiltz C, Legoupil N, Marty-Morieux C, de Vernejoul MC, Geoffroy V. Over-expression of TIMP-1 in osteoblasts increases the anabolic response to PTH. Bone. 2007 Jan;40(1):75–83. doi: 10.1016/j.bone.2006.07.013. [DOI] [PubMed] [Google Scholar]
- 41.Schiltz C, Prouillet C, Marty C, Merciris D, Collet C, de Vernejoul MC, Geoffroy V. Bone loss induced by Runx2 over-expression in mice is blunted by osteoblastic over-expression of TIMP-1. J Cell Physiol. 2010 Jan;222(1):219–229. doi: 10.1002/jcp.21941. [DOI] [PubMed] [Google Scholar]
- 42.Weis MA, Hudson DM, Kim L, Scott M, Wu JJ, Eyre DR. Location of 3-hydroxyproline residues in collagen types I, II, III, and V/XI implies a role in fibril supramolecular assembly. J Biol Chem. 2010 Jan 22;285(4):2580–2590. doi: 10.1074/jbc.M109.068726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cavazzini F, Magistroni R, Furci L, Lupo V, Ligabue G, Granito M, Leonelli M, Albertazzi A, Cappelli G. Identification and characterization of a new autoimmune protein in membranous nephropathy by immunoscreening of a renal cDNA library. PloS one. 2012;7(11):e48845. doi: 10.1371/journal.pone.0048845. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.