Abstract
Covalent attachment of ubiquitin to target proteins is one of the most pervasive post-translational modifications in eukaryotes. Target proteins are often modified with polymeric ubiquitin chains of defined lengths and linkages that may further undergo dynamic changes in composition in response to cellular signals. Biochemical characterization of the enzymes responsible for building and destroying ubiquitin chains is often thwarted by the lack of methods for preparation of the appropriate substrates containing probes for biochemical or biophysical studies. We have discovered that a yeast ubiquitin C-terminal hydrolase (Yuh1) also catalyzes transamidation reactions that can be exploited to prepare site-specifically modified polyubiquitin chains produced by thiol-ene chemistry. We have used this chemoenzymatic approach to prepare dual-functionalized ubiquitin chains containing fluorophore and biotin modifications. These dual-functionalized ubiquitin chains enabled the first real-time assay of ubiquitin chain disassembly by a human deubiquitinase (DUB) enzyme by single molecule fluorescence microscopy. In sum, this work provides a powerful new toolkit for elucidating the mechanisms of DUBs and other ubiquitin processing enzymes.
Keywords: ubiquitin oligomers, single molecule fluorescence, bifunctional, deubiquitinases
Protein ubiquitination is a reversible posttranslational modification that marks proteins for degradation and provides a mechanism to modulate enzymatic activity and protein-protein interactions.[1] The reversibility of ubiquitination is mediated by a family of ~90 hydrolytic enzymes known as deubiquitinases (DUBs) that catalyze the removal of ubiquitin (Ub) from target proteins or the disassembly of polymeric Ub chains.[2] The mechanisms by which DUBs perform these activities largely remain unknown. The two primary challenges in studying mechanisms of DUBs are limited access to poly-Ub chain substrates and the regulation of DUB activity by a number of trans-acting factors.[3] These accessory proteins and the DUBs can reside in large, multicomponent complexes, which are rather dynamic and heterogeneous, with multiple post-translational modifications on different subunits. The result is a mixture of species each with potentially different kinetic properties that can obfuscate bulk biochemical assays.
Our labs are interested in meeting these challenges by using single molecule fluorescence (SMF) microscopy to deconvolute DUB kinetics and chemical approaches to prepare modified poly-Ub chain substrates for these experiments. SMF offers several advantages over traditional bulk biochemical assays.[4] For instance, kinetic heterogeneity in enzymatic mixtures can be detected and molecules grouped according to similar patterns of activity. SMF also facilitates the segregation of complexes with deubiquitination activity from enzymatically inactive species. While there are many different SMF techniques,[4b, 4c] immobilization of poly-Ub chains and following DUB activity by total internal reflection fluorescence (TIRF) is well-suited for studying the complete reaction trajectories of single DUB enzymes over the minutes required for Ub chain disassembly. Herein, we describe a chemoenzymatic approach for preparation of bifunctional Ub chains containing both fluorophores suitable for SMF imaging and functional groups for surface immobilization. These molecules can be used to image DUB activity in real time by SMF microscopy.
We envisioned that poly-Ub substrates for DUBs could be immobilized for SMF experiments by inclusion of biotin[5] on one Ub monomer and visualized using a fluorophore on a second Ub subunit. Previously, we have shown that Yuh1 is capable of modifying the C-terminus of Ub with allylamine. Using this modification, we prepared Ub chains of defined lengths and linkages using thiol-ene coupling (TEC).[6](Scheme 1A). The resulting polymers contain an N-terminal subunit with a free thiol and the C-terminal subunit decorated with allylamine. Since the free thiol is well suited for fluorophore labeling, but installation of an immobilization tag requires the replacement of the allylamine moiety, we hypothesized that a biotin tag could be installed on Ub polymers through the action of the yeast Ub C-terminal hydrolase Yuh1 (Scheme 1B).
Scheme 1.
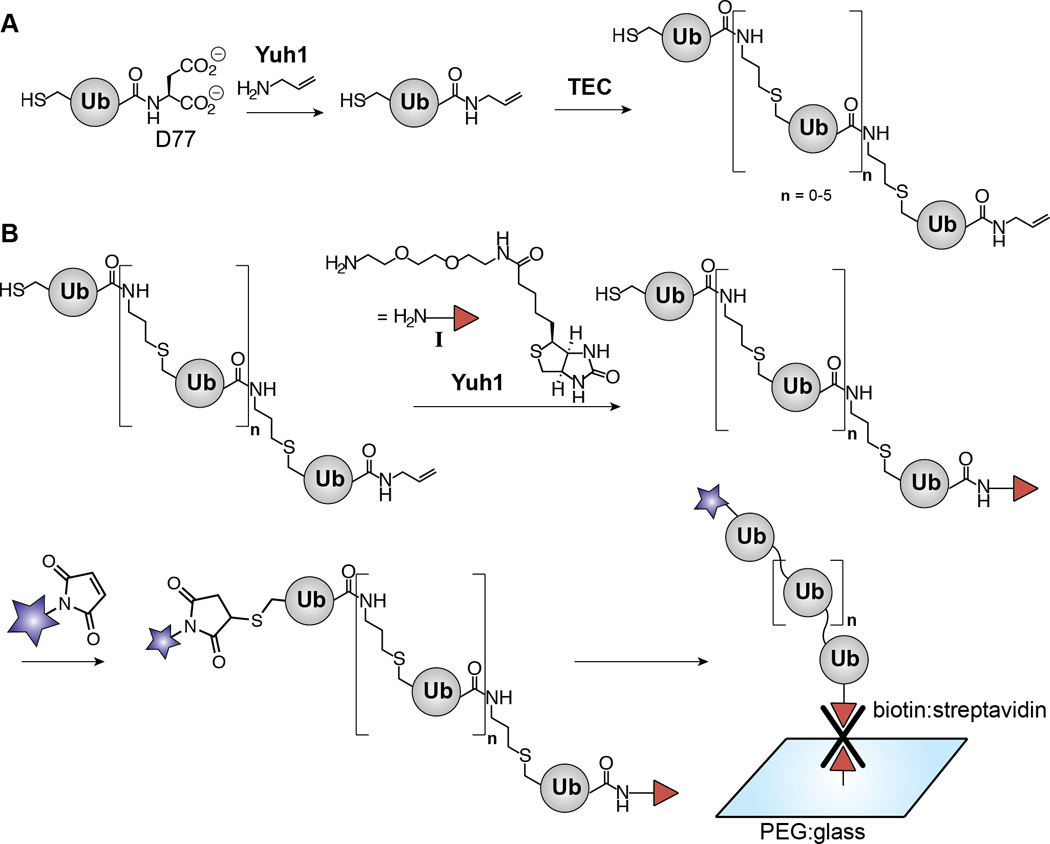
Synthesis of dually functionalized Ub polymers by exploiting the transamidase activity of Yuh1. (A) Scheme depicting our previous work using Yuh1 to replace D77 of Ub with allylamine and subsequent formation of Ub polymers via thiol-ene coupling (TEC). The linkage afforded by TEC is a functional mimic of the native isopeptide bond. For the purpose of differentiating between native isopeptide bonds and TEC-derived non-native linkages, “Kx-linked” represents the former and “x-linked” represents the latter where×is any of the seven native lysine positions. (B) Scheme showing the studies described here in which Yuh1 catalyzes the C-terminal installation of amine-biotin I. The polymers are subsequently tagged with a fluorophore (blue star) and immobilized on a polyethylene glycol (PEG)-modified glass surface for detection by SMF.
Normally a DUB itself, Yuh1 indiscriminately cleaves a variety of small C-terminal adducts from Ub through the intermediacy of a thioester acyl-enzyme.[7] In the presence of high concentrations of amine nucleophiles, acyl-enzyme intermediates undergo aminolysis instead of hydrolysis.[8] Given the lack of activity many Ub C-terminal hydrolases display towards the isopeptide bonds linking Ub subunits in polymers, [7c, 9] we reasoned that aminolysis by Yuh1 could be used to site-specifically modify the only subunit of a Ub chain containing a small molecule adduct, leaving the remaining chain intact. In the case of Ub chains prepared by TEC, this reaction (a net transamidation) would result in rapid exchange between a C-terminal allylamine moiety, a remnant of TEC that allows the chain to serve as substrate, and any primary amine nucleophile (Scheme 1B). The successful application of this synthetic sequence would address the need for additional methods capable of chemically modifying Ub chains, as most strategies currently employed require multi-step syntheses and are limited to 1,2-aminothiols as nucleophiles.[10]
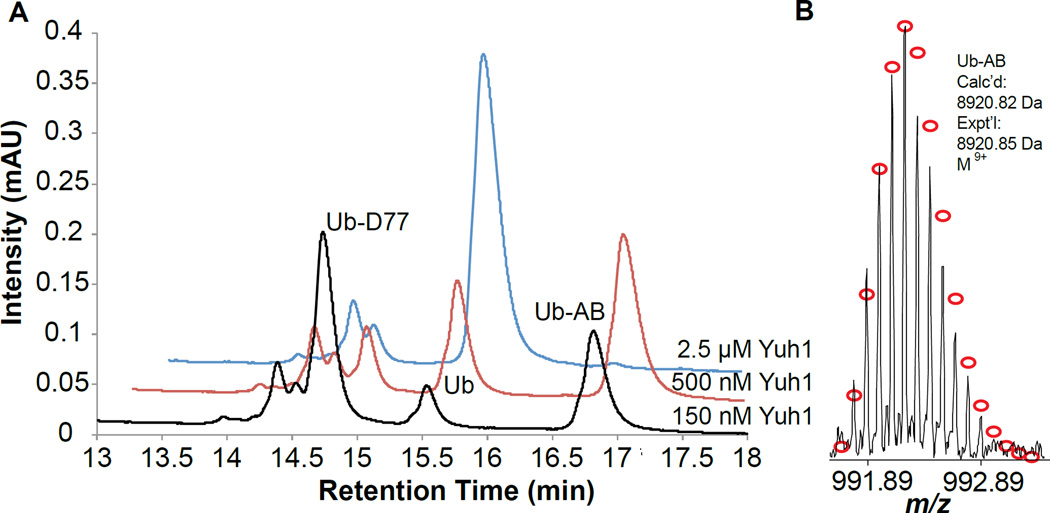
To understand whether aminolysis by Yuh1 could be exploited for the surface immobilization of Ub polymers, we first sought to test the ability of this DUB to modify the C-terminus of Ub with a biotin-derivatized amine. The amine-biotin derivative I provides an effective handle for monitoring the transamidase activity of Yuh1, and more importantly, is routinely used to immobilize biomolecules[5a] for SMF assays.[4b, 11] Ub containing an additional aspartate residue at the C-terminus (Ub-D77), which has been historically used as a UCH substrate,[7] was also employed as the substrate for Yuh1. Ub-D77 was incubated with I in the presence of different concentrations of Yuh1 overnight. The relative amounts of hydrolysis (Ub) and transamidation product (Ub-AB), which arises from aminolysis of the acyl-enzyme intermediate, were then measured using HPLC (Figure 1A). At lower concentrations of Yuh1 (150 nM), aminolysis prevailed over hydrolysis and a Ub-AB/Ub ratio of 3.6 was obtained, corresponding to a yield of 34%. When the concentration of Yuh1 was increased to 500 nM the yield of transamidation improved to 55 %, but this occurred at the expense of the Ub-AB/Ub ratio, which decreased to 1.8. Nevertheless, reactions employing 500 nM Yuh1 proved optimal considering higher concentrations of Yuh1 shifted the product distribution to entirely favor hydrolysis. A Fourier-transform ion cyclotron resonance mass spectrometer (FT-ICR) was then used to characterize the transamidation product Ub-AB. As shown in Figure 1B, the isotopic distribution of Ub-AB matches the theoretical distribution of isotope abundance for the same species in the M9+ charge state. These results demonstrate that while Yuh1 is normally a hydrolase, it can also be used as a transamidase to biotinylate the C-terminus of a Ub monomer.
Figure 1.
Yuh1 catalyzes the C-terminal exchange between D77 and amine-biotin I. (A) HPLC traces showing the formation of transamidation product (Ub-AB) and hydrolysis product (Ub) as a function of Yuh1 concentration. Reaction conditions: Ub-D77 (750 µM), amine-biotin I (250 mM), Yuh1 (indicated in plot), pH 10.4, RT, overnight. (B) The isotopic distribution of Ub-AB in a single charge state (z=9 or M9+). Red circles correspond to the theoretical distribution of isotopic abundance. Calc’d: calculated most abundance molecular weight. Expt’l: experimental most abundant molecular weight.
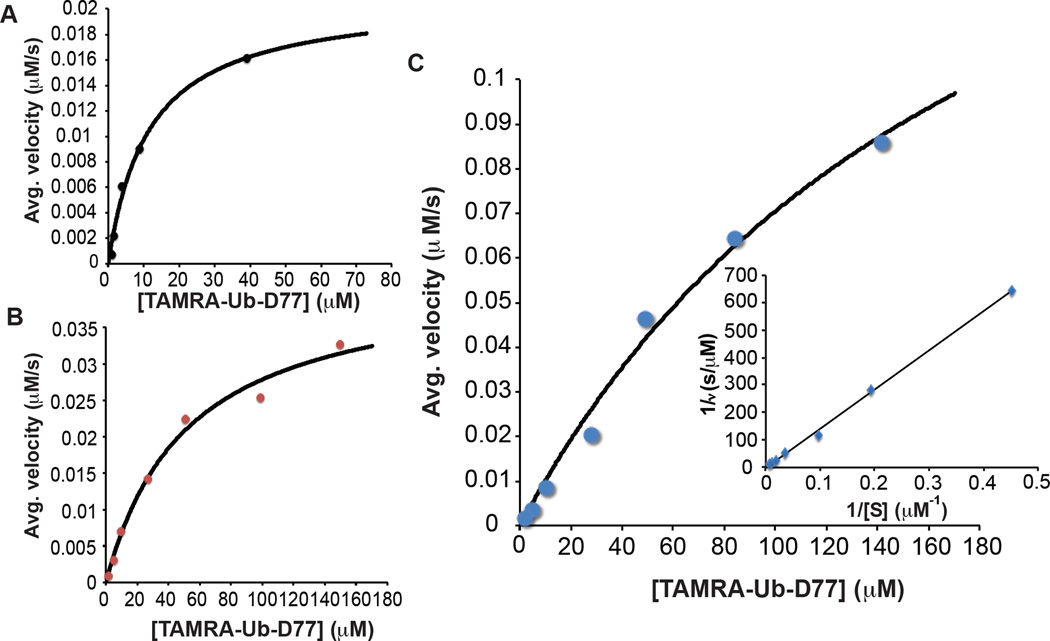
To further evaluate the efficiency with which Yuh1 replaces the C-terminal aspartate residue (D77) of Ub with I, the kinetics of transamidation were investigated. Initial rate measurements were acquired by HPLC using a fluorescent form of Ub-D77 in which K63 is substituted with cysteine (K63C) and subsequently labeled with TAMRA (TAMRA-Ub-D77) to enhance the UV-Vis absorbance. Depending on the reaction, initial rates of formation were measured for the production of TAMRA-Ub (hydrolysis) or TAMRA-Ub-AB (transamidation) at alkaline pH (this pH is required to increase the abundance of unprotonated amine). The kinetics of hydrolysis reveals that the shift to a higher pH causes a 5-fold increase in Km, but the kcat remains the same (Table 1 and Figures 2A and 2B), suggesting the loss of electrostatic interactions impedes the ability of Yuh1 to engage Ub. As for the kinetics of transamidation, the presence of high amine concentrations further suppresses the interaction between Yuh1 and Ub, as evidenced by the inability to completely saturate Yuh1 with Ub-D77 (Figure 2C). According to the measured bimolecular rate constant for the transamidation (~103 M−1s−1), however, Yuh1 does indeed serve as an efficient catalyst for the replacement of D77 with I (Table 1).
Table 1.
Kinetic parameters for Yuh1-catalyzed hydrolysis and transamidation reactions with amine-biotin I
| Reaction | Km (µM) | kcat (s−1) | kcat/Km (M−1 • s−1) |
|---|---|---|---|
| TAMRA-Ub-D77 hydrolysis (pH 7.5)a | 11.5 ± 2.3 | 4.2 ± 0.3 | (3.6 ± 0.7)×105 |
| TAMRA-Ub-D77 hydrolysis (pH 10.4)b | 52.0 ± 10.4 | 4.5 ± 0.4 | (8.6 ± 1.7)×104 |
| TAMRA-Ub-D77 transamidation with amine-biotin I (pH 10.4)c | NDd | NDd | (7.9 ± 2.7)×103 |
Reaction was run in HEPES pH 7.5 (50 mM) and EDTA (1 mM) with 5 nM Yuh1
Reaction was run in CAPS pH 10.4 (50 mM) with 9.5 nM Yuh1
Reaction was run in CAPS pH 10.4 (50 mM) with 135 nM Yuh1 and amine-biotin I (250 mM)
Values could not be determined due to the lack of data points at substrate concentrations close to the saturation of the enzyme
Figure 2.
Yuh1 catalyzes the installation of amine-biotin I at the C-terminus of TAMRA-Ub. (A) Plot of average velocity (µM•s−1) with which TAMRA-Ub is formed as a function of TAMRA-Ub-D77(µM) concentration at pH 7.5 by Yuh1 (5 nM). Each data point is an average of two independent measurements. The black line represents the fit to the Michaelis-Menten equation. (B) Plot of average velocity (µM•s−1) with which TAMRA-Ub is formed as a function of TAMRA-Ub-D77 (µM) concentration at pH 10.4 by Yuh1 (9.5 nM). Each data point is an average of two independent measurements. The black line represents the fit to the Michaelis-Menten equation. (C) Plot of average velocity (µM•s−1) with which TAMRA-Ub-AB is formed as a function of TAMRA-Ub-D77 concentration (µM) by Yuh1 (135 nM). Each data point is an average of two independent measurements. The black line represents the fit to the Michaelis-Menten equation. Inset shows Lineweaver-Burk plot used to confirm the kinetic parameters for transamidation in Table 1.
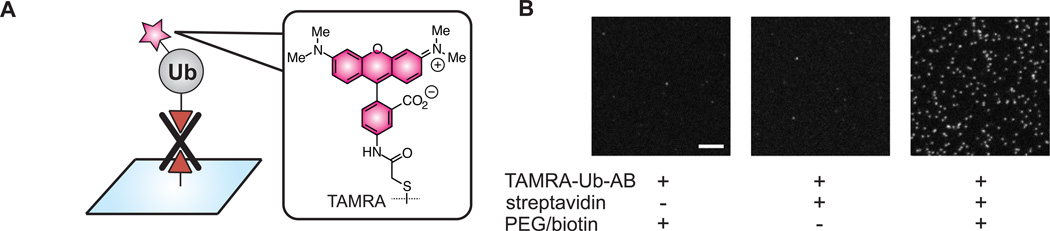
With biotin installed on the C-terminus of TAMRA-Ub, we explored whether this derivative could be immobilized on a glass surface and detected by SMF. Glass slides were derivatized with a mixture of polyethylene glycol (PEG) and PEG-biotin.[12] Streptavidin was then flowed over the surface and TAMRA-Ub-AB was loaded on the slide to allow visualization of single molecules by TIRF microscopy (Figure 3A). To increase photostability (i.e., fluorophore lifetime), an O2 scavenging system was employed.[13] Under these conditions, single molecules of Ub can be readily detected upon excitation at 532 nm (Figure 3B). In controls where either streptavidin or the initial PEG/biotin mixture was omitted these same isolated spots were not observed.
Figure 3.
Immobilization of TAMRA-Ub-AB and visualization by TIRF microscopy. (A) Experimental scheme. (B) TIRF microscope fields showing fluorescence of single TAMRA-Ub-AB molecules (white spots). All images are at the same magnification (scale bar = 10 µm) and adjusted to the same contrast level.
Building on our results with mono-Ub, we next sought to investigate the ability of Yuh1 to catalyze the modification of TEC-derived Ub polymers. Due to the nature of the polymerization, each TEC polymer contains a C-terminal allylamine moeity. We assessed the ability for Yuh1 to treat a C-terminal moiety other than aspartate. Mono Ub containing a C-terminal allylamine adduct (Ub-AA, 0.75 mM)) was incubated with I (250 mM) and Yuh1 (500 nM) at room temperature for 1.5 hours. Yuh1 was capable of modifying Ub-AA more effectively than Ub-D77, yielding the amine-biotin adduct Ub-AB in 69% compared to 17% with Ub-D77. (Figure S2)
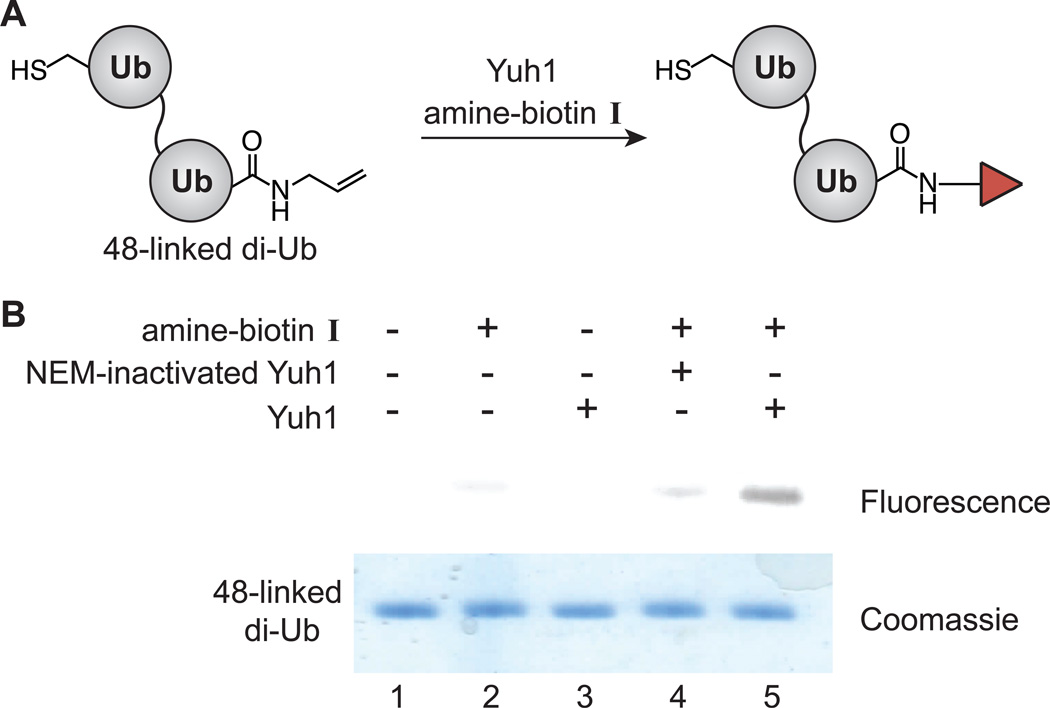
Our initial studies focused on dimers linked through position-48, as these linkages are the most abundant in eukaryotic cells[14] and act as the primary signals of proteasomal degradation. [15] Dimers were synthesized using a dually functionalized Ub-K48C and were synthesized using a dually functionalized Ub-K48C and separated from higher molecular weight oligomers by size exclusion chromatography. Substitution of the C-terminal allylamine moiety with amine-biotin I was performed in the presence or absence of active Yuh1 (Figure 4A). Reactions were analyzed by transferring the dimers to a PVDF membrane and detected with TAMRA-labeled streptavidin (Figure 4B). A fluorescent band, in which the dimer interacts with TAMRA-streptavidin, is only detected in the presence of both biotin and active Yuh1. These results not only confirm Yuh1 is unable to serve as an isopeptidase, but also show that this enzyme can site-specifically modify a Ub oligomer. These results also imply that Yuh1 chemistry could readily be applied to the remaining topoisomers, i.e., those linked through positions 6, 11, 27, 29, 33, and 63 of Ub.
Figure 4.
Yuh1 site-specifically modifies Ub dimers. 48-linked di-Ub (700 µM) was incubated with I (250 mM) and Yuh1 (150 nM) overnight.(A) Experimental scheme. (B) Biotinylated dimers are detected by their interaction with TAMRA-labeled streptavidin. Fluorescence was detected with a green laser (532 nm) and a 580±15 nm filter on a Typhoon imager FLA 9500 (GE Healthcare). Coomassie-stained SDS-PAGE is shown for the purposes of a loading control. NEM; N-ethylmaleimide.
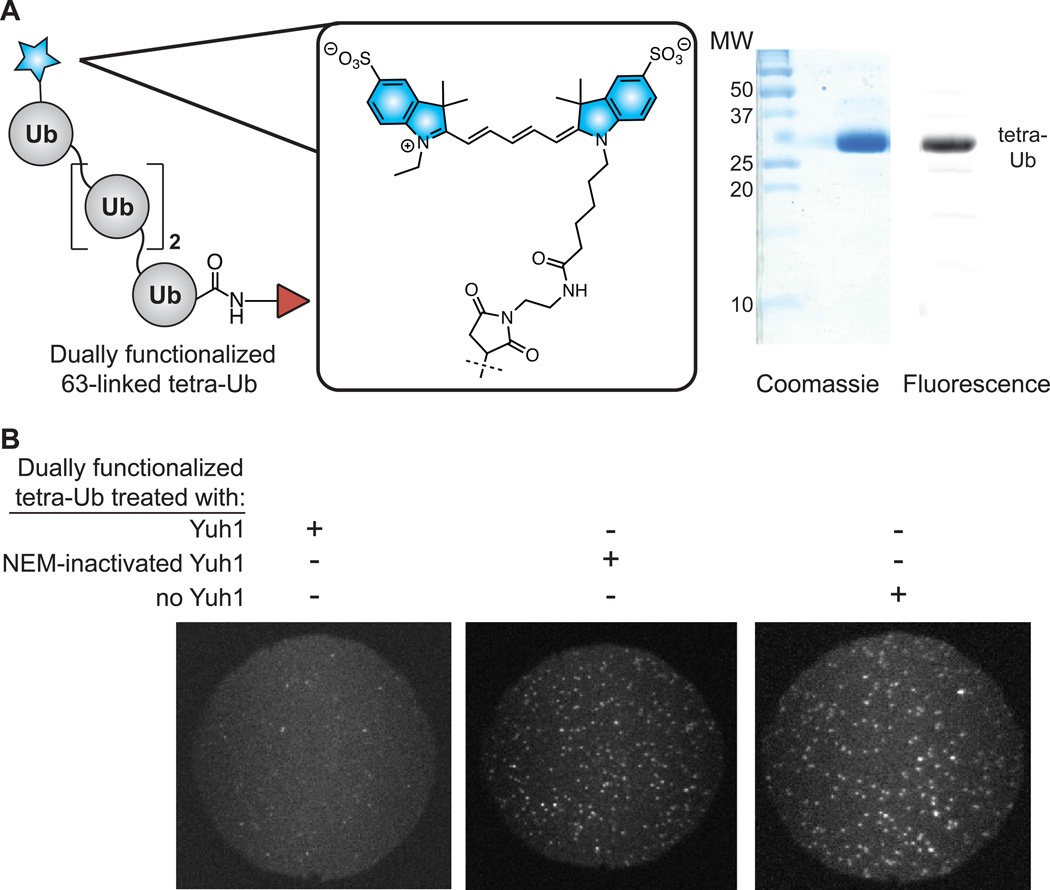
To further demonstrate the utility of Yuh1 transamidase activity in the context of different chain lengths and linkages, we developed a synthetic route towards bifunctional 63-linked Ub tetramers. K63-linked chains serve as the preferred substrates of a number of DUBs that reside in multicomponent complexes, most of which are members of the JAMM (Jab1/Mov34/Mpr1 Pad1 N-terminal+) metalloprotease family.[16] For example, the K63-specific DUBs Poh1,[17] BRCC36,[17–18] and AMSH[19] are members of the 26S proteasome, [20] BRCA1-A complex, [21] and the ESCRT machinery,[22] respectively. As in our synthesis of 48-linked dimers, TEC was again used to construct the 63-linked tetramers. Once the tetra-Ub chains were purified, the C-terminal allylamine appendage was exchanged with I using Yuh1 prior to installation of a Cy5 fluorophore on the N-terminal subunit. Excess dye and Yuh1 were subsequently removed by ion-exchange chromatography. The dually functionalized, fluorescent Ub tetramer (Figure 5A) was then flowed over a streptavidin/PEG-treated glass slide. Upon excitation at 633 nm, punctate spots of fluorescence were observed corresponding to single molecules of the tetramer (Figure 5B, image on right). As before, the spots of fluorescence were dependent on prior transamidation of the Ub chain with biotin by Yuh1. Additionally, the Cy5 tetra-Ub was reacted with Yuh1 at pH 7.5 to hydrolytically cleave the C-terminal biotin from the fluorescent protein and the resultant solution applied to the slide (Figure 5B, left image). Under these conditions, very few spots of fluorescence were observed. Collectively, these results confirm that the Cy5-labeled chains are tethered to the glass slide through a C-terminal biotin moiety. Leveraging TEC and the transamidase activity of Yuh1 therefore enables facile preparation of Ub chain molecules detectable by TIRF microscopy.
Figure 5.
Synthesis and SMF detection of Cy5-labeled, 63-linked tetra-Ub chains. (A) Coomassie and fluorescence gels showing purified, dually functionalized, 63-linked tetra-Ub chains. (B) TIRF microscope fields (each 50 µm in diameter) showing fluorescence of dually functionalized chains. The three images represent different solutions applied to the glass surface. The surface is densely decorated with spots representing single molecules of the Ub-chains only in the absence of treatment with active Yuh1 (center and right panels). If Yuh1 is added to hydrolytically remove the C-terminal biotin adduct, very few spots of fluorescence are observed (left panel).
With the tetramer Ub substrate in hand, we tested whether or not the cleavage of 63-linked chains by AMSH could be followed by SMF. We chose to start with AMSH because this DUB has previously been shown to be functional in vitro when purified. An important first step in our investigations was to establish that AMSH retains its bulk solution activity in the presence of oxygen-scavenging systems and triplet quenchers used to improve fluorophore photostability.[13,23] We observed that neither oxygen-scavengers nor triplet quenchers affect the extent of chain cleavage by AMSH in bulk enzymatic assays (Figure S11). Moreover, AMSH readily hydrolyzes Cy5-labeled 63-linked tetra-Ub chains, indicating the fluorophore also does not have adverse effects on DUB activity (Figure S12).
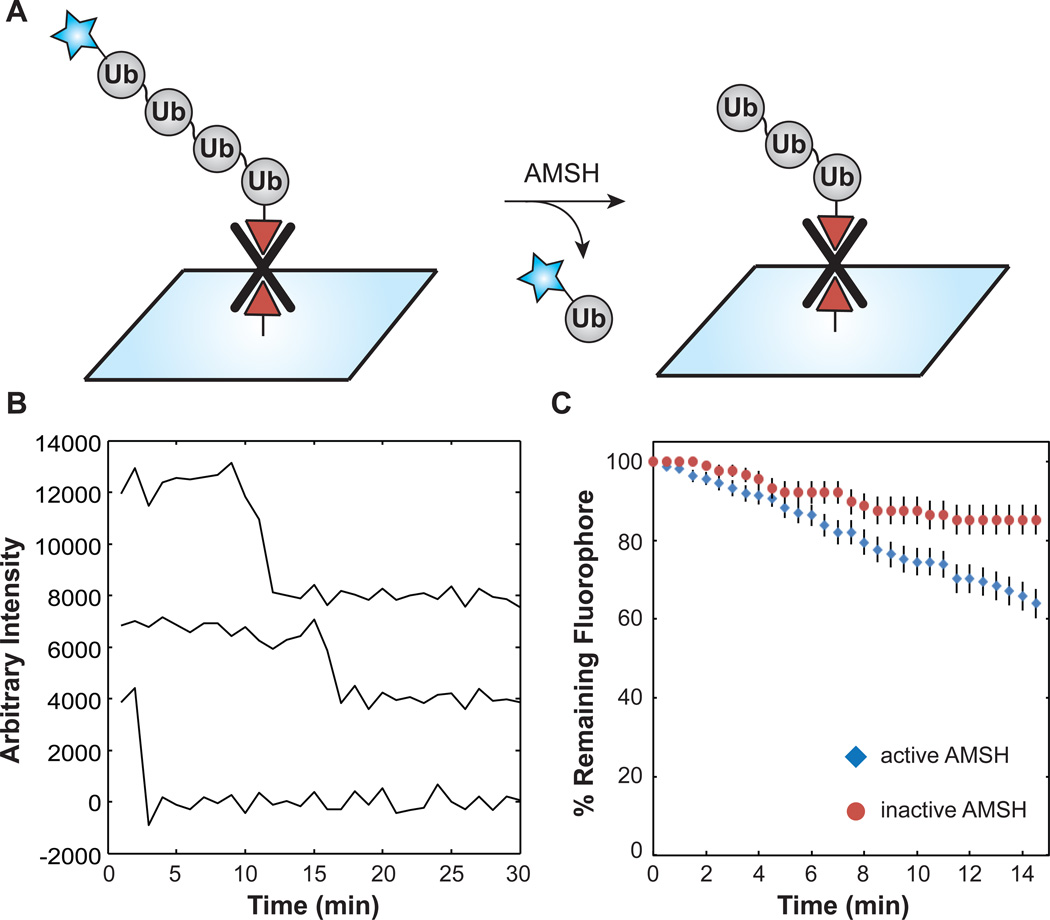
To then detect DUB isopeptidase activity by SMF, biotinylated 63-linked tetra-Ub chains were tethered to a streptavidin-derivatized glass surface, and then individual fields of fluorophores were monitored under cleavage conditions by TIRF microscopy (Figure 6A). Time-lapse recording was used to monitor Cy5 fluorescence at regular intervals throughout the experiment to reduce laser-induced photodamage during the reaction. Spots that appeared initially in the experiment disappeared over time via a single step loss in fluorescence consistent with single tetra-Ub chains containing a single Cy5 fluorophore (Figure 6B). The number of spots present at each time point was plotted as the surviving fraction of the spots originally present in the field of view. In the presence of active AMSH, ~40% of the spots vanished during the experiment over 15 min (Figure 6C). In contrast, only ~20% of spots vanished in a photobleaching control carried out under identical conditions but with chemically inactivated AMSH. These results are consistent with the DUB activity of AMSH cleaving a fraction of the tethered tetra-Ub chains present on the slide and releasing Cy5-Ub monomers into solution. From these experiments we conclude that AMSH displays DUB activity against immobilized substrates bearing cyanine fluorophores prepared by TEC chemistry and that the reaction can be followed by SMF. We predict that real-time assays of DUB activity such as these will prove useful for elucidating kinetic mechanisms of Ub chain remodeling.
Figure 6.
Detection of DUB activity at the single-molecule level. (A) Reaction scheme. (B) Representative single molecule intensity time courses from AMSH hydrolysis assays. (C) Remaining fluorescence versus time. Consistent with DUB activity, the greatest loss in surface fluorescence occurred in reactions containing both dually functionalized 63-linked tetra-Ub chains and active AMSH ( , N = 160 molecules). This rate was ~2-fold greater than the photobleaching rate measured in the presence of 1,10-phenanthroline-inactivated AMSH (
, N = 160 molecules). This rate was ~2-fold greater than the photobleaching rate measured in the presence of 1,10-phenanthroline-inactivated AMSH ( , N = 88 molecules). Error bars represent the sampling error calculated as a binomial proportion confidence interval.
, N = 88 molecules). Error bars represent the sampling error calculated as a binomial proportion confidence interval.
In summary, we demonstrate that the hydrolytic activity of DUBs can be interrogated at the single molecule level using surface-tethered Ub polymers as substrates. Central to these studies is the use of Yuh1, a DUB, in preparation of bifunctional Ub chains. This unique reactivity exploits two important features of Yuh1. First, Yuh1 can act as a transamidase in the presence of high concentrations of amine nucleophiles to facilitate the exchange of C-terminal amides with other functional groups such as biotin. Second, Yuh1 modifies only the subunit in a Ub chain that is not tethered to another Ub molecule through its C-terminus and leaves the remaining chain intact. We predict that the bifunctional Ub oligomers prepared using these methods will enable a number of biochemical and biophysical experiments for elucidating Ub biology. In combination with SMF, these Ub chains may prove particularly useful in answering questions regarding the mechanism of DUBs that reside in large, multicomponent enzyme complexes.
Supplementary Material
Acknowledgments
Financial support was provided by NSF (VHT, graduate fellowship), UW-Madison (AAH and ERS), the Greater Milwaukee Shaw Scientist Program (ERS), Susan G. Komen for the Cure (ERS, KG110081), the Wisconsin Alumni Research Foundation (AAH and ERS), NIH R00 GM086471 (AAH), the Molecular Biophysics Training Program (MLR, NIH T32-GM08293), the Arnold and Mabel Beckman Foundation (AAH), and the NIH (Department of Chemistry, NCRR 1S10RR024601-01).
Footnotes
Supporting information for this article is available on the WWW under http://www.chembiochem.org or from the author.
Contributor Information
Aaron A. Hoskins, Email: ahoskins@wisc.edu.
Eric R. Strieter, Email: strieter@chem.wisc.edu.
References
- 1.a Hershko A, Ciechanover A. Annu. Rev. Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]; b Grabbe C, Husnjak K, Dikic I. Nat. Rev. Mol. Cell Biol. 2011;12:295–307. doi: 10.1038/nrm3099. [DOI] [PMC free article] [PubMed] [Google Scholar]; c Weissman AM. Nat. Rev. Mol. Cell Biol. 2001;2:169–178. doi: 10.1038/35056563. [DOI] [PubMed] [Google Scholar]
- 2.a Amerik AY, Hochstrasser M. Biochim. Biophys. Acta. 2004;1695:189–207. doi: 10.1016/j.bbamcr.2004.10.003. [DOI] [PubMed] [Google Scholar]; b Komander D, Clague MJ, Urbe S. Nat. Rev. Mol. Cell Biol. 2009;10:550–563. doi: 10.1038/nrm2731. [DOI] [PubMed] [Google Scholar]; c Nijman SM, Luna-Vargas MP, Velds A, Brummelkamp TR, Dirac AM, Sixma TK, Bernards R. Cell. 2005;123:773–786. doi: 10.1016/j.cell.2005.11.007. [DOI] [PubMed] [Google Scholar]; d Reyes-Turcu FE, Ventii KH, Wilkinson KD. Annu. Rev. Biochem. 2009;78:363–397. doi: 10.1146/annurev.biochem.78.082307.091526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ventii KH, Wilkinson KD. Biochem. J. 2008;414:161–175. doi: 10.1042/BJ20080798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.a Kelley AM, Michalet X, Weiss S. Science. 2001;292:1671–1672. doi: 10.1126/science.1060096. [DOI] [PubMed] [Google Scholar]; b Joo C, Balci H, Ishitsuka Y, Buranachai C, Ha T. Annu. Rev. Biochem. 2008;77:51–76. doi: 10.1146/annurev.biochem.77.070606.101543. [DOI] [PubMed] [Google Scholar]; c Walter NG, Huang CY, Manzo AJ, Sobhy MA. Nat. Methods. 2008;5:475–489. doi: 10.1038/nmeth.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.a Rusmini F, Zhong ZY, Feijen J. Biomacromolecules. 2007;8:1775–1789. doi: 10.1021/bm061197b. [DOI] [PubMed] [Google Scholar]; b Chen YX, Triola G, Waldmann H. Acc. Chem. Res. 2011;44:762–773. doi: 10.1021/ar200046h. [DOI] [PubMed] [Google Scholar]
- 6.a Trang VH, Valkevich EM, Minami S, Chen YC, Ge Y, Strieter ER. Angew. Chem. Int. Ed. Engl. 2012;51:13085–13088. doi: 10.1002/anie.201207171. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Valkevich EM, Guenette RG, Sanchez NA, Chen YC, Ge Y, Strieter ER. J. Am. Chem. Soc. 2012;134:6916–6919. doi: 10.1021/ja300500a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.a Miller HI, Henzel WJ, Ridgway JB, Kuang WJ, Chisholm V, Liu CC. Bio-Technol. 1989;7:698–704. [Google Scholar]; b Johnston SC, Riddle SM, Cohen RE, Hill CP. EMBO J. 1999;18:3877–3887. doi: 10.1093/emboj/18.14.3877. [DOI] [PMC free article] [PubMed] [Google Scholar]; c Larsen CN, Krantz BA, Wilkinson KD. Biochemistry. 1998;37:3358–3368. doi: 10.1021/bi972274d. [DOI] [PubMed] [Google Scholar]; d Pickart CM, Rose IA. J. Biol. Chem. 1985;260:7903–7910. [PubMed] [Google Scholar]; e Luchansky SJ, Lansbury PT, Jr, Stein RL. Biochemistry. 2006;45:14717–14725. doi: 10.1021/bi061406c. [DOI] [PubMed] [Google Scholar]
- 8.a Schellenberger V, Jakubke HD. Angew. Chem. Int. Ed. Engl. 1991;30:1437–1449. [Google Scholar]; b Jakubke HD, Kuhl P, Konnecke A. Angew. Chem. Int. Ed. Engl. 1985;24:85–93. [Google Scholar]; c Bordusa F. Chem. Rev. 2002;102:4817–4867. doi: 10.1021/cr010164d. [DOI] [PubMed] [Google Scholar]
- 9.Komander D, Reyes-Turcu F, Licchesi JD, Odenwaelder P, Wilkinson KD, Barford D. EMBO Rep. 2009;10:466–473. doi: 10.1038/embor.2009.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.a Haj-Yahya M, Fauvet B, Herman-Bachinsky Y, Hejjaoui M, Bavikar SN, Karthikeyan SV, Ciechanover A, Lashuel HA, Brik A. Proc. Natl. Acad. Sci. U.S.A. 2013;110:17726–17731. doi: 10.1073/pnas.1315654110. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Hemantha HP, Bavikar SN, Herman-Bachinsky Y, Haj-Yahya N, Bondalapati S, Ciechanover A, Brik A. J. Am. Chem. Soc. 2014;136:2665–2673. doi: 10.1021/ja412594d. [DOI] [PubMed] [Google Scholar]; c Burchak ON, Jaquinod M, Cottin C, Mugherli L, Iwai K, Chatelain F, Balakirev MY. Chembiochem. 2006;7:1667–1669. doi: 10.1002/cbic.200600283. [DOI] [PubMed] [Google Scholar]
- 11.a Hoskins AA, Friedman LJ, Gallagher SS, Crawford DJ, Anderson EG, Wombacher R, Ramirez N, Cornish VW, Gelles J, Moore MJ. Science. 2011;331:1289–1295. doi: 10.1126/science.1198830. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Selvin PR, Ha T. Single-molecule techniques : a laboratory manual. Cold Spring Harbor, N.Y.: Cold Spring Harbor Laboratory Press; 2008. [Google Scholar]
- 12.Ha T, Rasnik I, Cheng W, Babcock HP, Gauss GH, Lohman TM, Chu S. Nature. 2002;419:638–641. doi: 10.1038/nature01083. [DOI] [PubMed] [Google Scholar]
- 13.a Yildiz A, Forkey JN, McKinney SA, Ha T, Goldman YE, Selvin PR. Science. 2003;300:2061–2065. doi: 10.1126/science.1084398. [DOI] [PubMed] [Google Scholar]; b Crawford DJ, Hoskins AA, Friedman LJ, Gelles J, Moore MJ. RNA. 2008;14:170–179. doi: 10.1261/rna.794808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.a Ziv I, Matiuhin Y, Kirkpatrick DS, Erpapazoglou Z, Leon S, Pantazopoulou M, Kim W, Gygi SP, Haguenauer-Tsapis R, Reis N, Glickman MH, Kleifeld O. Mol. Cell. Proteomics. 2011;10:M111 009753. doi: 10.1074/mcp.M111.009753. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Kim W, Bennett EJ, Huttlin EL, Guo A, Li J, Possemato A, Sowa ME, Rad R, Rush J, Comb MJ, Harper JW, Gygi SP. Mol. Cell. 2011;44:325–340. doi: 10.1016/j.molcel.2011.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]; c Wagner SA, Beli P, Weinert BT, Nielsen ML, Cox J, Mann M, Choudhary C. Mol. Cell. Proteomics. 2011;10:M111 013284. doi: 10.1074/mcp.M111.013284. [DOI] [PMC free article] [PubMed] [Google Scholar]; d Kaiser SE, Riley BE, Shaler TA, Trevino RS, Becker CH, Schulman H, Kopito RR. Nat. Methods. 2011;8:691–696. doi: 10.1038/nmeth.1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.a Chau V, Tobias JW, Bachmair A, Marriott D, Ecker DJ, Gonda DK, Varshavsky A. Science. 1989;243:1576–1583. doi: 10.1126/science.2538923. [DOI] [PubMed] [Google Scholar]; b Finley D, Sadis S, Monia BP, Boucher P, Ecker DJ, Crooke ST, Chau V. Mol. Cell. Biol. 1994;14:5501–5509. doi: 10.1128/mcb.14.8.5501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ambroggio XI, Rees DC, Deshaies RJ. PLoS Biol. 2004;2:E2. doi: 10.1371/journal.pbio.0020002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cooper EM, Cutcliffe C, Kristiansen TZ, Pandey A, Pickart CM, Cohen RE. EMBO J. 2009;28:621–631. doi: 10.1038/emboj.2009.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.a Cooper EM, Boeke JD, Cohen RE. J. Biol. Chem. 2010;285:10344–10352. doi: 10.1074/jbc.M109.059667. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Shao G, Lilli DR, Patterson-Fortin J, Coleman KA, Morrissey DE, Greenberg RA. Proc. Natl. Acad. Sci. U.S.A. 2009;106:3166–3171. doi: 10.1073/pnas.0807485106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McCullough J, Clague MJ, Urbe S. J. Cell Biol. 2004;166:487–492. doi: 10.1083/jcb.200401141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.a Yao T, Cohen RE. Nature. 2002;419:403–407. doi: 10.1038/nature01071. [DOI] [PubMed] [Google Scholar]; b Verma R, Aravind L, Oania R, McDonald WH, Yates JR, 3rd, Koonin EV, Deshaies RJ. Science. 2002;298:611–615. doi: 10.1126/science.1075898. [DOI] [PubMed] [Google Scholar]
- 21.a Sobhian B, Shao G, Lilli DR, Culhane AC, Moreau LA, Xia B, Livingston DM, Greenberg RA. Science. 2007;316:1198–1202. doi: 10.1126/science.1139516. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Wang B, Matsuoka S, Ballif BA, Zhang D, Smogorzewska A, Gygi SP, Elledge SJ. Science. 2007;316:1194–1198. doi: 10.1126/science.1139476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Clague MJ, Urbe S. Trends Cell Biol. 2006;16:551–559. doi: 10.1016/j.tcb.2006.09.002. [DOI] [PubMed] [Google Scholar]
- 23.Rasnik I, McKinney SA, Ha T. Nat. Methods. 2006;3:891–893. doi: 10.1038/nmeth934. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.