Abstract
The transcriptional repressor B-cell lymphoma 6 (BCL6) is required for the development of T helper (Th) follicular cells and it has been shown to suppress Th2 cell differentiation. We demonstrate that BCL6 is a key regulator of Th9 cell development. BCL6 expression is transiently downregulated in polarized Th9 cells and forced expression of BCL6 in Th9 cells impairs Th9 cell differentiation. In contrast, BCL6 knockdown up-regulated interleukin (IL)-9 production in Th9 cells. The function of BCL6 in Th9 cells is under the control of IL-2/Janus kinase 3 (JAK3)/signal transducer and activator of transcription 5 (STAT5) signaling pathway. Using chromatin immunoprecipitation (ChIP), we show that in Th9 cells, BCL6 and STAT5 bind to adjacent motifs in the Il9 promoter. Furthermore, we found that STAT5 binding was associated with the abundance of a permissive histone mark at the Il9 promoter, while under conditions where BCL6 binding was predominant a repressive histone mark was prevalent. The effects of STAT5 and BCL6 on IL-9 transcription were further demonstrated using an IL-9-luciferase reporter assay where BCL6 repressed STAT5-mediated Il9 transactivation. In experimental autoimmune encephalomyelitis (EAE), forced expression of BCL6 in myelin oligodendrocyte glycoprotein (MOG)35–55-specific Th9 cells resulted in decreased IL-9 production and induction of IFNγ causing an exacerbation of the clinical disease. Our findings demonstrate a novel role of BCL6 in the regulation of Th9 cell development and their encephalitogenicity.
INTRODUCTION
Following antigen stimulation, naïve CD4+ T cells differentiate into one of several functional classes of effector cells. In addition to the classical Th1 and Th2 lineages, Th17 cells have been described and extensively characterized. Recently, a new subset of IL-9-producing Th cells induced in vitro by IL-4 and transforming growth factor-β1 has been identified (1, 2). Traditionally associated with the Th2 response, IL-9 is a pleiotropic cytokine that impacts inflammation by exerting broad effects on a variety of cell types such as CD4+ T cells, mast cells and epithelial cells. Recent reports by our group and others demonstrated that IL-9 exerts pro- or anti-inflammatory properties depending on the inflammatory milieu by regulating Th17 and regulatory CD4+FoxP3+ T cells (Tregs) expansion and survival (3–6). Moreover, adoptive transfer of Th9 cells has shown divergent functions from other transferred subsets in models of tumor immunity, autoimmune encephalomyelitis, and allergic airway disease (7–9).
Networks of cytokines and transcription factors are critical for determining CD4+ T cell fates and effector cytokine production. Indeed, each subset utilizes a master regulatory transcription factor and a particular signal transducer and activator of transcription (10). The relationships are as follows: Th2, GATA-binding protein 3 (GATA-3)/STAT5; Th1, T-box transcription factor expressed in T cells (T-bet)/STAT4; Th17, retinoid orphan receptor γt (RORγt)/STAT3; inducible Treg, forkhead box protein 3 (Foxp3)/STAT5. Recent studies suggest that T follicular helper cells may also fit the paradigm with the factors being B-cell lymphoma 6 (Bcl-6)/STAT3. Interestingly, in many instances, the STAT involved also plays a role in the induction of the master transcriptional regulator (reviewed in (11)). The Il9 locus is responsive to multiple factors that bind and induce a conserved non-coding sequence (CNS) in reporter assays including IRF4, PU.1, NF-κB, and Smad/Notch complexes (3, 12–14). Recently, transcription factors of the STAT family, STAT5 and STAT6, were shown to be critical for Th9 cell development (15, 16). The Bcl6 gene, originally identified as an oncogene for B cell lymphoma, encodes a transcriptional repressor protein that regulates T cell differentiation by repressing Th1 and Th2 cell development (17–19). BCL6 knockout (KO) mice exhibit significant growth retardation and invariably die by ten weeks of age (20, 21). BCL6KO mice have multiple immunological defects, including lack of germinal center formation and spontaneous development of severe Th2-type inflammatory disease, particularly affecting the heart and lungs (20, 21). The DNA motifs recognized by BCL6 are highly homologous to the core consensus binding sequence TTC-NNN-GAA (where N is any nucleotide) of STAT5 (20, 22), a positive regulator of Th9 cell development (16), which suggests that BCL6 may play a role in the transcriptional regulation of the Il9 locus and Th9 cell development.
In the present study, we analyzed the role of BCL6 in the regulation of Th9 cell development and encephalitogenicity. We demonstrate that BCL6 controls Th9 cell differentiation by direct binding and regulation of the Il9 locus. Furthermore, BCL6 function in Th9 cells is regulated by the IL-2/JAK3/STAT5 signaling pathway.
MATERIALS AND METHODS
Mice and Reagents
C57BL/6 and Rag2−/− mice were purchased from the Jackson Laboratories and MOG35–55 T cell receptor transgenic mice (2D2) were previously described (23). Mice were housed in the pathogen-free animal facility at Harvard Medical School, New Research Building, in accordance with the guidelines of the Committee of Animal Research at the Harvard Medical School and the National Institutes of Health animal research guidelines as set forth in the Guide for the Care and Use of Laboratory Animals. The following fluorescent conjugated primary mAb were purchased from BD Biosciences (San Jose, CA): anti-mouse IL-9-PE, IL-17-APC, and anti-mouse CD4-Pacific Blue. JAK3 inhibitor VI (420126) and STAT5 inhibitor (573108) were obtained from Calbiochem (San Diego, CA). 7AAD stain for dead cell exclusion was obtained from BD Biosciences.
In vitro T Cell Differentiation, Retroviral Cell Transduction and Cytokine Assay
Naïve CD4+ T cells were purified from C57BL/6 WT mice using anti-CD4 beads (Miltenyi, Auburn, CA) and flow sorted into naïve CD4+CD62Lhi T cells by flow cytometry on a FACSAria T cell sorter (BD Biosciences). CD4+ T cells were stimulated with plate-bound anti-CD3 (4 μg/ml) (145-2C11; BD Biosciences, San Diego, CA) and soluble anti-CD28 (2 μg/ml; BD Biosciences) for 4 days in a serum-free culture medium (X-VIVO-20; Lonza, Hopkinton, MA) supplemented with 50 μM 2-mercaptoethanol, 1 mM sodium pyruvate, non-essential amino acids, L-glutamine and 100 μU/ml penicillin/100 μg/ml streptomycin in the presence of recombinant cytokines. Polarization of Th9 cells was in the presence of human TGF-β1 (3 ng/ml) plus mouse IL-4 (10 ng/ml). Cells were supplemented with recombinant IL-2 (40 ng/ml) where indicated. Rat anti-mouse IL-2 antibody (BD Biosciences) was used for blocking of IL-2 activity in vitro. All recombinant proteins were from R&D Systems. JAK3 (250 μM) and STAT5 (50 μM) inhibitors were added along with the polarization medium at the start of differentiation. For intracellular flow cytometry staining, cells were restimulated with 12-O-tetradecanoylphorbol-13-acetate (20 ng/ml; Sigma, St. Louis, MO), ionomycin (300 ng/ml; Sigma), and 2 mM monensin (GolgiStop, BD Biosciences) for 4 h at 37°C. Cells were washed, stained for surface markers, fixed and permeabilized, and fluochrome-conjugated antibodies were added. For the measurement of cytokines released in the culture supernatants, cell culture supernatants were collected on day 4 after differentiation and the secreted cytokines were determined by fluorescent bead-based Luminex technology (Luminex, Austin, TX) for the indicated cytokines, in accordance with the manufacturers’ instructions. For intracellular cytokine staining of infiltrated T cells, mice were sacrificed and perfused with PBS. Spinal cord tissues were digested with collagenase IV (Sigma-Aldrich) for 30 min at 37°C, resuspended in 30% Percoll, and loaded onto 70% Percoll. After centrifuge at 1300 × g for 20 min, the CNS inflammatory cells were retrieved from the 30/70% Percoll interface. Lymphocytes were prepared for intracellular flow cytometry staining as described above using anti-IL-9-PE and anti-IFNγ-APC conjugated antibodies.
For forced expression of BCL6, naïve CD4+ T cells were transduced with a MSCV-based retroviral vector (24). Briefly, the RV particles were produced by co-transfecting the plasmids encoding for the ecotropic envelope gag-pol with the retroviral vector with Effectene (QIAGEN) into the Phoenix packaging cells. RV supernatant was harvested 48hr after transfection and centrifuged prior to use. CD4+CD62L+ T cells isolated by magnetic-activated cell sorting (MACS) from WT mice were activated with anti-CD3 and anti-CD28 for 24 hr, retroviral supernatant was added to the cells together with polybrene (8 μg/ml), and cells were incubated at 37°C. After incubation, live cells were sorted with the Dead Cell Removal Kit (Miltenyi) and cells were expanded for an additional 4 days under Th9 cell polarization condition.
Expression Analysis by Real-Time PCR
RNA was purified using Stratagene RNA kit and transferred directly into the RT reagent using the Applied Biosystems Taqman reverse transcriptase reagents. Samples were subjected to real-time PCR analysis on PRISM 7000 Sequencer Detection System (Applied Biosystems, Foster City, CA) under standard conditions. Genes analyzed were detected using commercially available assays (Applied Biosystems). Relative mRNA abundance was normalized against GAPDH.
Western blot
Cells were lysed in RIPA buffer (Thermo Scientific) with a protease inhibitor (Roche Diagnostics) and a phosphatase inhibitor mixture (Sigma-Aldrich); 20 μg total protein was loaded into each well of a SDS-PAGE gel for separation by electrophoresis and then transferred on nitrocellulose membrane. The resulting blots were blocked for 1 h with TBS-Tween 20 containing 5% bovine serum albumin and then probed for 3 hrs at room temperature with primary Abs directed against: phospho-STAT5 (Tyr694), phospho-JAK3 (Tyr980/981), STAT5, JAK3, or BCL6 (Cell Signaling Technology). β-actin mouse mAb (Sigma) was used as the loading control. Blots were then washed five times and probed for 1 h with the appropriate HRP-conjugated secondary Ab. Membranes were developed with Immobilon Western Chemiluminescent HRP substrate (Millipore).
RNA interference
To knock down the expression of Stat5a and Bcl6, CD4+ T cells were purified and nucleoporated with small interfering RNA (siRNA) specific for Stat5a or Bcl6 (Santa Cruz Biotechnology). Scrambled siRNA was used as control. Cells were transfected using an Amaxa nucleoporator system. Briefly, 5–10 × 106 CD4+ T cells were resuspended in 100 μl Nucleofector solution and transfected with 100 nM siRNA using Amaxa Nucleofector (Lonza, Basel, Switzerland). After transfection, the cells were incubated for 6 h at 37°C followed by polarization under Th9 conditions.
Passive EAE Model
For the generation of myelin-specific Th9 cells, CD4+CD62L+ cells were prepared from spleens of MOG35–55 T cell receptor transgenic mice (2D2) using MACS beads. CD4+ cells were transduced with MSCV-BCL6-GFP or control empty vector MSCV-GFP followed by stimulation with MOG35–55 peptide (20 μg/ml) in the presence of IL-4 (20 ng/ml) and TGF-β1 (3 ng/ml) for 3 days in the presence of splenic CD11c+ dendritic cells isolated from 2D2 mice. One million cells were transferred into Rag2−/− recipient mice via i.p. injection followed by immunization with the emulsion made of 50 μg MOG35–55 (MEVGWYRSPFSRVVHLYRNGK; New England Peptide) and complete Freund’s adjuvant (CFA). Each animal also received i.p. injections of 69 ng pertussis toxin (PT) on days 1 and 3 after transfer. The EAE clinical score was determined as follows: 0, no disease; 0.5, partial tail paralysis; 1, complete tail paralysis; 2, partial hind limb paralysis; 3, complete hind limb paralysis; 4, complete hind limb and partial front limb paralysis; and 5, moribund or dead animals.
ChIP and qPCR
CD4+ T cells were purified by MACS sorting and were polarized to Th9 phenotype. ChIP was performed according to the protocol described in (25) with the following modifications: Chromatin was sheared using Micrococcal Nuclease (New England Biolabs) and Protein A Magnetic Beads (New England Biolabs) were used. Cell lysates were used for immunoprecipitation with anti-STAT5, anti-BCL6, anti-mono-methyl-Histone H3 lysine 4 (H3K4me1), and anti-pan-methyl-H3K9 (all from Cell Signaling Technology) and were compared to control IgG. One region of the Il9 promoter containing putative STAT5 and BCL6 binding sites as well as a region of the Bcl6 promoter containing putative STAT5 binding sites was amplified by SYBR Green qPCR (Applied Biosystems) and quantified in duplicate with the percentage of input method. The following primers were used: Il9 Promoter: Site 1 Fwd: ACTGAGTTCCAGACTCCCGT, Rev: GCCCAGCACAGAACTGAAGA; Site 2 Fwd: GGATCCTCAAGGCCAATGCT, Rev: ACACCTCTGAGAAGTCGCTC; Site 3 Fwd: ACAGAAGTGTGCTGTCTGGT; Rev: CCCCTTGAGCCACTGGATAC. Bcl6 Promoter: Site 1 Fwd: CTGCGGAGCAATGGTAAAGC, Rev: ATAATCACCTGGTGTCCGGC; Site 2 Fwd: CGAGGAGCCGAGTTTATGGG, Rev: GAGAGTGCGCTTTGCTTTCC; Site 3 Fwd: CGAATGACAGTCCCGACGAT, Rev: GCTTGGGATGCTCCTGTTGT.
Luciferase Reporter Assay
Reporter vector coding for the Firefly Luciferase under the control of the Il9 promoter encompassing nucleotides −1310 to +32 bp was cloned into the promoterless pGL3 Basic luciferase reporter gene vector (Promega). Murine stem cell virus vector expressing wild-type mouse Stat5a (MSCV) was previously described (26). Plasmid encoding mouse Bcl6 (mBCL6/pCMV-SPORT6.1) was purchased from Open Biosystems. Reporter assays were carried out as described previously (3). Briefly, 293T cells were transfected with 0.2 μg of the reporter vector coding for the Firefly Luciferase under the control of the Il9 promoter and with 0.2 μg of the STAT5A or BCL6 encoding plasmids. Cells were cultured for 24 hrs before harvesting and the relative Il9 promoter activity was measured using Promega kit in accordance with the manufacturer’s instructions.
Statistical Analysis
The Mann-Whitney test was used for clinical disease analysis. Data are expressed as mean ± SEM and were compared using the Unpaired Student’s t test for experiments with two groups by Prism software v5. Data were considered statistically significant at P < 0.05.
RESULTS
BCL6 Is a Negative Regulator of IL-9 Expression in Th9 Cells
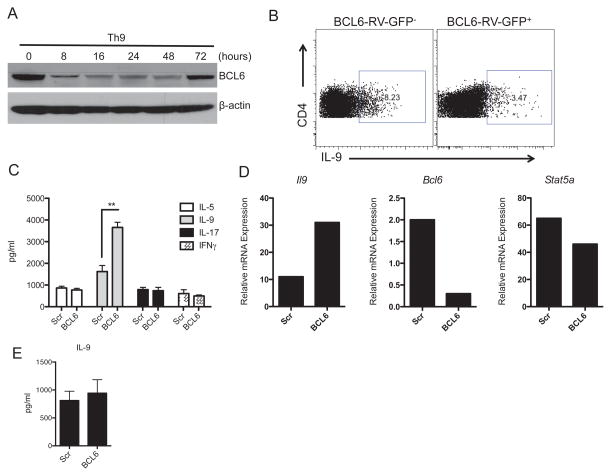
The transcriptional repressor BCL6 has emerged as a multifunctional regulator of lymphocyte differentiation and immune responses. Here, we investigated the possibility whether BCL6 is involved in murine Th9 cell development. We measured BCL6 protein expression in Th9 cells polarized in vitro with IL-4 and TGF-β1 and found it decreased as early as 8 hours following differentiation as shown by Western blot. BCL6 downregulation was transient suggesting an intrinsic mechanism for the control of BCL6 expression in Th9 cells (Figure 1A). To understand the functional relevance of the regulation of BCL6 expression in Th9 cells, we asked whether BCL6 forced expression influences Th9 cell development in vitro. To address this, we transduced naïve CD4+CD4lowCD62Lhi T cells with MSCV-BCL6-GFP retrovirus (BCL6-RV-GFP), cultured the cells under Th9 conditions for 4 days, and measured intracellular cytokine expression using flow cytomtery. We found that the frequency of IL-9-positive cells was altered by ~60% after BCL6 (GFP+) overexpression compared to BCL6-negative (GFP−) Th9 cells suggesting that the transcription factor BCL6 is a negative regulator of Th9 cells (Figure 1B). To ascertain the involvement of BCL6 in Th9 cell generation, we used BCL6-specific siRNA to knockdown BCL6 gene expression in Th9 cells. Naïve CD4+ T cells were nucleoporated with BCL6-specific siRNA and subsequently polarized to the Th9 lineage and harvested 4 days after transfection. Consistent with the data obtained with BCL6 over-expression, BCL6 knockdown resulted in an up-regulation of IL-9 expression at both the protein and gene levels as measured by bead-based Luminex assay and Taqman PCR, respectively (Figure 1C, D). Moreover, to address the functional role of increased BCL6 level at later stage of Th9 differentiation, we treated Th9 cells with BCL6-specific siRNA or scrambled siRNA on day 3 after polarization. Cells were kept for another 3 days under Th9 conditions followed by analysis for IL-9 production by Luminex. We found that BCL6 knockdown did not alter IL-9 production suggesting that BCL6 is primarily involved in the early development of Th9 cells (Figure 1E).
Figure 1. BCL6 Is a Negative Regulator of Th9 Cells.
(A) BCL6 is transiently downregulated in polarized Th9 cells. Immunoblot of BCL6 in Th9 cells polarized for 0–72 hours in the absence of exogenous recombinant IL-2. β-actin was used as internal control for protein loading. (B) BCL6 forced expression reduced IL-9 expression. Naïve CD4+ T cells were prepared from the spleens of WT mice and cultures were transduced with BCL6-RV-GFP encoding for BCL6-GFP. Cells were differentiated under Th9 cell conditions for 4 days followed by intracellular cytokine staining and BCL6-GFP-positive cells were gated on the basis of the GFP expression. (C) BCL6 knockdown in Th9 cells up-regulates IL-9 production. Naïve CD4+ T cells were transfected with siRNA specific for BCL6 or with scrambled (Scr) siRNA using Amaxa followed by polarization under Th9 cell conditions for 4 days. Cytokine analysis of IL-5, IL-9, IL-17 and IFNγ expression in the supernatants was assessed by Luminex. (D) A representative gene expression of Il9, Bcl6 and Stat5a in Th9 cells that were transfected with siRNA specific for BCL6 is shown. Transfection with scrambled siRNA was used as a control. (E) IL-9 production in Th9 cells. Th9 cells were differentiated for 3 days followed by transfection with siRNA specific for BCL6 or with scrambled siRNA. IL-9 release in the culture supernatants was measured by Luminex. Data are representative of three experiments with similar results. **p < 0.005.
Activation of IL-2/JAK3/STAT5 Axis Inversely Correlates With BCL6 Down-regulation in Th9 Cells
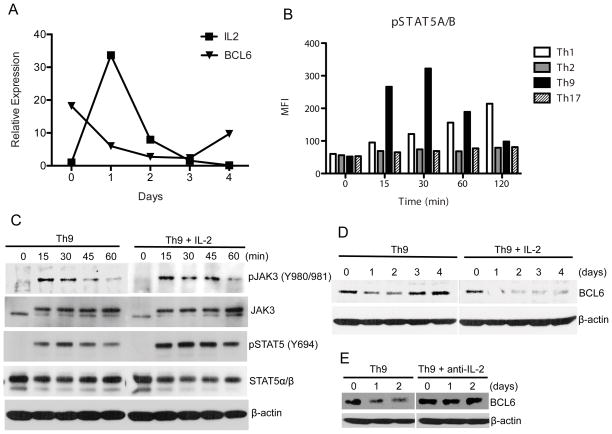
IL-2 is a major growth factor for activated T cells and plays an important role in murine and human Th9 cell expansion and differentiation in vitro (27, 28). Additionally, activation of STAT5 is an important component of IL-2 signaling and has been recently proposed as a positive regulator of Th9 cells (16, 29). To measure the IL-2 signaling pathway downstream components in Th cells, we first extracted gene expression profiling from RNA-sequencing (RNA-Seq) data deposited on Gene Expression Omnibus (GEO) (30). Analysis of IL-2 signaling in murine Th1, Th2, Th9, Th17 and inducible CD4+FoxP3+ regulatory T cells (iTregs) differentiated in vitro according to standard protocols shows that the expression of Il2ra, Stat5a and Stat5b is up-regulated in Th9 cells (Supplementary Figure 1A). Using Taqman PCR to verify the transcriptome sequencing results in different Th subsets, we found that mRNA levels of Il2ra, Stat5a and Stat5b are indeed elevated in Th9 cells compared to other cell phenotypes (Supplementary Figure 1B). Moreover, temporal analysis of IL-2 expression in polarized Th9 cells demonstrates that IL-2 is transiently induced in Th9 cells and this expression inversely correlates with a transient decrease in BCL6 mRNA expression in these cells (Figure 2A).
Figure 2. Activation of IL-2/JAK3/STAT5 Axis Inversely Correlates With BCL6 Down-regulation in Th9 Cells.
(A) Opposite regulation of IL-2 and BCL6 in polarized Th9 cells. Naïve CD4+ T cells were cultured under Th9 conditions for 4 days in the absence of exogenous recombinant IL-2 and BCL6 and IL-2 mRNA expression was assessed by Taqman PCR. (B) Increased STAT5 activation in Th9 cells. Polarized T helper and inducible Tregs were differentiated in vitro for 0–120 minutes (min) and phospho-STAT5 (pSTAT5) was assessed using bead-based Luminex assay. (C) Representative immunoblot of phospho-dependent and -independent JAK3 and STAT5 in naïve CD4+CD62Lhi polarized under Th9 conditions for the indicated time points (0–60 min) with recombinant IL-4 plus TGF-β1 in the presence or absence of exogenous recombinant IL-2. Phospho-epitope of each protein kinase is indicated. β-actin was used as internal control for protein loading. (D) Recombinant IL-2 inhibited BCL6 expression in Th9 cells. Naïve CD4+ T cells were polarized under Th9 cell conditions in the presence or absence of exogenous recombinant IL-2 and BCL6 expression was assessed for up to 4 days. β-actin expression is shown. (E) Anti-IL-2 neutralizing antibody rescued the decrease in BCL6 expression in Th9 cells. Naïve CD4+ T cells were polarized under Th9 cell conditions in the presence or absence of anti-IL-2 neutralizing antibody and BCL6 expression was assessed on days 0, 1 and 2 days. β-actin expression is shown.
Given that IL-2 is required for Th9 cells polarized in vitro and is abundantly produced by these cells as measured by Luminex bead-based assay (Supplementary Figure 1C), and since IL-2/STAT5 pathway has been shown to regulate BCL6 signals in follicular T helper cells (31, 32), we sought to analyze the activity of the key transducers of IL-2 signaling, STAT5 and its upstream kinase JAK3, in polarized Th9 cells. Using bead-based Luminex assay for phosphorylated STAT5αβ, we compared the activity of STAT5 in different T helper subsets differentiated in vitro for several time points between 0–120 minutes according to standard protocols in the absence of exogenous IL-2. We found that STAT5αβ activation is predominantly induced in Th9 cells as early as 15 minutes after polarization (Figure 2B). Furthermore, we confirmed that STAT5 signaling is activated in Th9 cells, by measuring STAT5αβ phosphorylation and its upstream protein kinase, JAK3. Thus, naïve CD4+ T cells were stimulated under Th9 conditions in the presence or absence of recombinant IL-2 (0–60 minutes), and cells were lysed and used for immunoblot analysis using antibodies specific for phosphorylation-dependent and independent JAK3 and STAT5. We found that the phosphorylation of both JAK3 and STAT5 was induced in Th9 cells and this was sustained in the presence of added recombinant IL-2 (Figure 2C). Quantification of phospho-JAK3 and STAT5 by measurements of the optical density of the corresponding bands is shown (Supplementary Figure 1D). Altogether, these data suggest the JAK3/STAT5 signaling cascade is activated in Th9 cells and is further enhanced by IL-2 signaling.
To investigate whether IL-2 is involved in the down-regulation of BCL6 in polarized Th9 cells, we measured BCL6 protein expression in Th9 cells differentiated in vitro for several days in the presence of recombinant IL-2 or anti-IL-2 neutralizing antibody. We found that while BCL6 protein levels were transiently decreased during Th9 cell differentiation, supplementation with recombinant IL-2 induced sustained suppression of BCL6 protein expression (Figure 2D). Interestingly, when Th9 cells were differentiated in the presence of anti-IL-2 neutralizing antibody, BCL6 expression was maintained throughout the polarization suggesting that IL-2 signaling is a negative regulator of BCL6 in Th9 cells (Figure 2E). Quantification of BCL6 levels by densitometry is shown (Supplementary Figure 1D).
IL-2/JAK3/STAT5 Inhibition Up-regulates BCL6 Expression and Suppresses Th9 Cell Differentiation
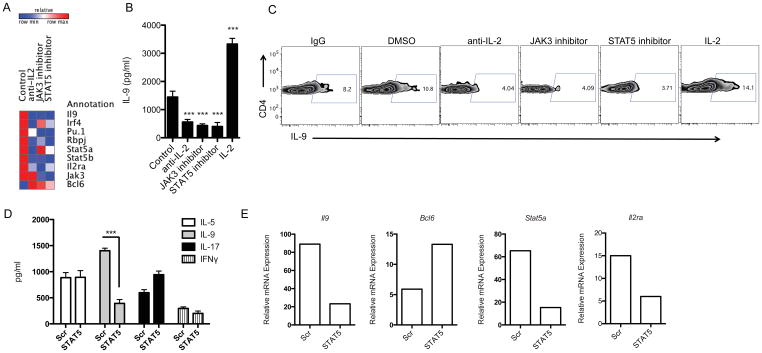
Recent reports demonstrated that STAT5 signaling promotes Th9 cell differentiation and that STAT5 and BCL6 are cross-regulated in Th1 cells (33, 34). Given that JAK3 is an upstream positive regulator of STAT5 (35), we hypothesized that BCL6 may be regulated by STAT5 in Th9 cells. Thus, we analyzed the effects of the inhibition of IL-2/JAK3/STAT5 signaling on the outcome of Th9 cell differentiation and BCL6 expression. CD4+ T cells were differentiated under Th9 conditions in the presence or absence of JAK3 (250 μM) or STAT5 (100 μM) specific inhibitors for 4 days. We found that addition of either inhibitor up-regulated BCL6 mRNA expression as measured by Taqman PCR (Figure 3A). Moreover, JAK3 and STAT5 inhibitors suppressed the production of IL-9 as measured by intracellular cytokine staining as well as by Luminex bead-based assay (Figure 3B, C).
Figure 3. Opposite Role of STAT5A and BCL6 in the Regulation of Th9 Cell Development.
(A, B, C) Inhibition of IL-2/JAK3/STAT5 signaling decreases IL-9 production in Th9 cells. (A) Heatmap of gene regulated in Th9 cells treated with anti-IL-2 neutralizing antibody, and JAK3 and STAT5 inhibitors. Naive CD4+ T cells from WT mice were differentiated into Th9 cells in the presence or absence of the indicated treatment and gene expression was measured by Taqman PCR. (B, C) IL-9 protein expression by bead-based Luminex assay (B) as well as by intracellular flow cytometry (C) in treated Th9 cells is shown. (D, E) STAT5 knockdown reduces IL-9 and up-regulated BCL6 expression in Th9 cells. Naïve CD4+ T cells transfected using Amaxa with siRNA specific for STAT5A or scrambled (Scr) siRNA followed for polarization under Th9 cell conditions for 4 days. (D) Cytokine analysis of IL-5, IL-9, IL-17 and IFNγ expression in the supernatants was assessed by Luminex. (E) A representative gene expression of Il9, Bcl6, Stat5a and Il2ra in Th9 cells that were transfected with siRNA specific for STAT5A, or scrambled siRNA and measured by Taqman PCR. Data are representative of three experiments with similar results. ***p < 0.0001.
Next, we used STAT5A-specific siRNA to inhibit STAT5 signaling at the gene level. Thus, naïve CD4+ T cells were nucleoporated with STAT5A-specific siRNA or with scrambled siRNA and subsequently polarized to the Th9 lineage and harvested 4 days after transfection. Silencing of STAT5A specifically inhibited IL-9 production but did not affect the production of others cytokines, including IL-5, IL-17 and IFNγ (Figure 3D). Quantitative RT-PCR confirmed that STAT5A siRNA reduced the expression of Il9 and Stat5a mRNA (Figure 3E). To check whether the reduction in IL-9 in STAT5A siRNA-treated cells was due to differential cell survival, we performed 7AAD flow staining of nucleoporated cells, which revealed no changes between control and STAT5A siRNA-treated cells (data not shown).
Further analysis of STAT5A-specific siRNA showed that the treatment was associated with an increase in Bcl6 mRNA expression (Figure 3E), in agreement with the elevated Bcl6 expression in Th9 cells treated with IL-2/JAK3/STAT5 inhibitors (Figure 3A) and the decrease in BCL6 protein expression in Th9 cells exposed to IL-2 (Figure 2C). These findings suggest that BCL6, a transcriptional repressor that has been shown to modulate STAT signaling in B cells (36, 37) plays a role in the regulation of Th9 cell differentiation under the control of STAT5 signaling.
Opposite Roles of BCL6 and STAT5 in Binding and Regulating the Il9 Promoter in Th9 Cells
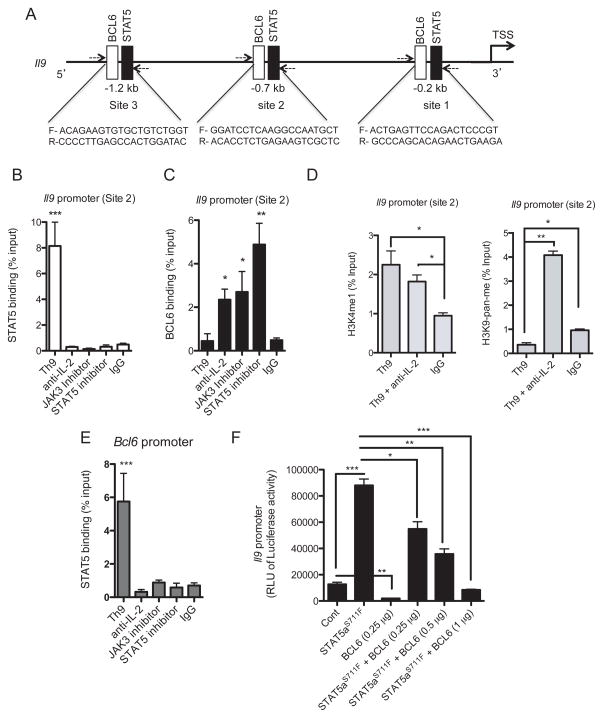
Because IL-2 enhances the differentiation of Th9 cells and exerts opposite effects on STAT5 and BCL6 expression, we hypothesized that these transcription factors may have opposing roles in the regulation of the Il9 promoter in Th9 cells. To investigate this, we searched the Il9 promoter for potential binding sites for STAT5 and BCL6 using Biobase database. We identified three overlapping putative binding sites for STAT5 and BCL6 upstream of the transcription start site (TSS) (Figure 4A). To determine the Il9 promoter occupancies by STAT5 and BCL6, binding motifs were used to design chromatin immunoprecipitation (ChIP) experiments. Primer sets flanking the STAT5 and BCL6 binding at three sites in the Il9 promoter were designed to amplify the immunoprecipitated ChIP DNA by qPCR (Figure 4A). CD4+CD62Lhi T cells were differentiated under Th9 cell polarizing conditions for 1–4 days and then analyzed by ChIP-PCR. We found that STAT5 binds significantly to sites 1, 2 and 3 in the Il9 promoter in Th9 cells 2 days after cell polarization (Figure 4B and Supplementary Figure 2). We next investigated whether STAT5 binding to the Il9 promoter in Th9 cells is modulated by IL-2 signaling. We observed that IL-2 blockade using IL-2 neutralizing antibody abolished STAT5 binding to the Il9 promoter. Similarly, pharmacological inhibition of both JAK3 and STAT5 abolished STAT5 recruitment to the Il9 promoter in Th9 cells which correlates with the suppression of IL-9 expression (Figure 4B and Supplementary Figure 2). We confirmed the specificity of STAT5 binding by amplifying a region of the Il9 promoter that does not contain STAT5 binding sites (data not shown).
Figure 4. IL-2-Mediated Control of STAT5 and BCL6 Differential Binding to the Il9 Promoter.
(A) Three predicted binding sites for BCL6 (Open box) and STAT5 (filled boxes) in the Il9 promoter. PCR primer sets for each site are shown. (B, C) ChIP analysis of BCL6 and STAT5 binding to the Il9 promoter. Regulation of BCL6 and STAT5 binding to site 2 within the Il9 promoter. Naïve CD4+ T cells from WT mice were polarized under Th9 cell conditions for 2 days in the presence of anti-IL-2 neutralizing antibody, JAK3 or STAT5 inhibitor. ChIP-SYBR Green PCR was performed to determine (B) STAT5 and (C) BCL6 binding to the Il9 promoter. Abs used for IP are anti-STAT5, anti-BCL6 and control IgG. Total input DNA before IP was used for normalization of data. (D) ChIP analysis of H3K4 (left panel) and H3K9 (right panel) modifications normalized to input controls at the BCL6 and STAT5 binding site 2 within the Il9 promoter in Th9 cells differentiated in vitro for 2 days. (E) ChIP analysis of STAT5 binding to the Bcl6 promoter in Th9 cells differentiated in vitro for 2 days. The graphs represent quantitative PCR analysis of the ratio of enriched Il9 and Bcl6 promoters with transcription factor binding sites to the input DNA. Data represent mean ± SE of a representative experiment each performed in triplicate. (F) HEK 293T cells were transfected with active mutant of STAT5 (STAT5aS711F), BCL6, or combination of STAT5aS711F and BCL6 together with a constant amount of Il9 promoter-luciferase vector. Cells were lyzed 24 hrs later and luminescence was measured. Representative of three independent experiments. *p < 0.01, **p < 0.005; ***p < 0.0001.
Given that STAT5 and BCL6 have opposite effects on IL-9 expression in Th9 cells, we investigated whether the two transcription factors compete for binding to the same binding motifs in the Il9 promoter. Indeed, we found that BCL6 binding to the Il9 promoter inversely correlates with STAT5 binding to site 2 and to a lesser extent to sites 1 and 3. Strikingly, this process was under the control of IL-2 signaling since inhibition of either IL-2, JAK3 or STAT5 suppressed STAT5 binding while it facilitated BCL6 recruitment to the same motif, particularly to site 2 and to a lesser extent to sites 1 and 3 in the Il9 promoter (Figure 4C and supplementary Figure 2). We next studied whether chromatin modifications at the STAT5 and BCL6 binding motifs within the Il9 promoter could explain the regulation of IL-9 by IL-2 signaling. Analysis of histone 3 (H3) methylation in Th9 cells revealed increased permissive H3 lysine 4 monomethylation (H3K4me1) modifications at site 2 in the Il9 promoter with decreased repressive H3 lysine 9 pan-methylation (H3K9-pan-me) chromatin modifications on day 2 after in vitro polarization. Interestingly, IL-2 neutralization increased the H3K9-pan repressive mark while it has no significant influence on the H3K4me1 permissive mark (Figure 4D). Previously, STAT5 has been shown to bind the Bcl6 promoter and down-regulate its transcription in B-lymphoma cells and other hematopoietic cell lines (38). To investigate a similar outcome in Th9 cells, we analyzed STAT5 recruitment to the Bcl6 promoter in polarized Th9 cells exposed to IL-2/JAK3/STAT5 blocking agents. Interestingly, we found that under basal conditions, STAT5 binds significantly to the Bcl6 promoter and this process was abolished when cells were exposed to anti-IL-2 blocking antibody or to JAK3 and STAT5 inhibitors (Figure 4E), which correlates with the identified role of STAT5 as a positive regulator of Th9 cell development.
To analyze the functional relevance of the binding of STAT5 and BCL6 to their target sequences in the Il9 locus, we investigated the ability of STAT5 and BCL6 to regulate the activity of the Il9 promoter in reporter assays. We used the reporter construct pGL3-Il9 containing the firefly luciferase gene under the control of the Il9 promoter. First, we found that co-transfection of the pGL3-Il9 luciferase reporter construct with a plasmid encoding constitutively active mutant of STAT5 (STAT5aS711F) (26) in 293T cells resulted in a significant increase in Il9 transcription (Figure 4F). In agreement with the function of BCL6 as a negative regulator of Th9 cell development, we found that BCL6 decreased Il9 promoter activity significantly in a luciferase reporter assay. Strikingly, co-transfection of BCL6 with STAT5 resulted in a very significant suppression of STAT5 positive activity (p < 0.0001) suggesting that BCL6 is a dominant negative regulator of the Il9 transcription in Th9 cells (Figure 4F). STAT5A overexpression in 293T cells is comparable in the presence or absence of exogenous BCL6 (Supplementary Figure 3).
BCL6 Overexpression in Myelin-Specific Th9 Cells Regulates their Encephalitogenicity
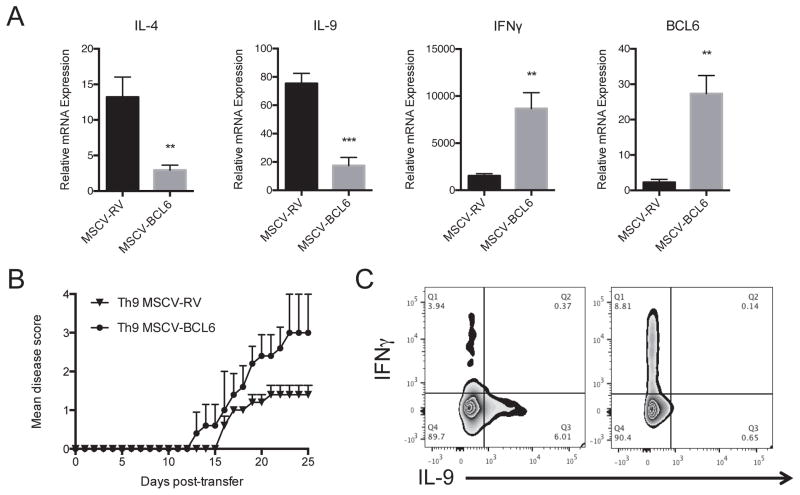
Previous studies demonstrated that adoptive transfer of myelin-reactive Th9 cells induces experimental autoimmune encephalomyelitis (EAE) (7). To investigate whether BCL6-mediated IL-9 repression modulates EAE, we carried out adoptive transfer experiments with MOG35–55-pulsed Th9 cells following BCL6 overexpression. To generate myelin-specific Th9 cells overexpressing BCL6, MOG35–55-TCR transgenic CD4+CD62Lhi naive cells were sorted from 2D2 mice and were transduced with MSCV-BCL6-GFP retrovirus or with control vector and subsequently polarized into Th9 cells for three days in the presence of MOG35–55 (20 μg/ml) peptide and splenic dendritic cells. Analysis of the cytokine profile of BCL6-GFP-positive 2D2 CD4+ T cells demonstrate a decrease in IL-4 and IL-9 expression associated with an increase in IFNγ expression compared to control cells as shown by Taqman PCR (Figure 5A). No changes were detected in IL-17 expression (data not shown). To analyze the effects of BCL6 on the encephalitogenicity of Th9 2D2 cells, cells were transferred into Rag2−/− lymphopenic mice followed by reactivation with MOG35–55/CFA immunization and mice were monitored for clinical disease development. We found that EAE was markedly enhanced in recipients of Th9 MSCV-BCL6 compared with Th9 MSCV-RV control mice (Mean maximal score for Th9 MSCV-BCL6-treated mice 3 ± 0.3 compared with Th9 MSCV-RV-treated mice 1.4 ± 0.2; p < 0.001 by Mann-Whitney test)(Figure 5B). To test whether the cytokine profile of transferred cells is maintained in recipient mice, spinal cord tissues were collected ~10 days post-transfer, 2D2 cells were identified using the surface markers Vα3.2/Vβ11 and cells were analyzed for cytokine expression confirming that BCL6 forced expression converts Th9 cells into Th1-like phenotype as shown by an increase in IFNγ and suppression of IL-9 production (Figure 5C).
Figure 5. BCL6 Regulates Th9 Cell Pathogenicity in EAE mice.
(A) A representative gene expression of IL-4, IL-9, IL-17A, IFNγ and BCL6 in Th9 overexpressing BCL6 and control cells. MOG35–55-specific CD4+ T cells were isolated from 2D2 mice and were transduced with MSCV-BCL6-GFP retrovirus virus (RV) or with control empty vector (MSCV-RV) followed by in vitro polarization under Th9 conditions for 4 days in the presence of MOG35–55 peptide (20 μg/ml) and irradiated antigen-presenting cells. Cells were lyzed and analyzed for gene expression by Taqman. (B) A representative EAE experiment showing clinical scores (means ± s.e.m.) in mice after adoptive transfer of MOG35–55-stimulated 2D2 polarized under Th9 cell conditions with or without BCL6 overexpression (10 mice/group). Rag2−/− mice received MSCV-BCL6-transduced Th9 cells or control cells followed by immunization with MOG35–55 peptide (100 μg/ml). (C) Intracellular staining of transferred 2D2 cells. Infiltrated cells were prepared from the spinal cords of recipient mice (3 mice/group) followed by flow cytometry intracellular staining of IFNγ and IL-9. Data are representative of two experiments with similar results. **p < 0.01; ***p < 0.001.
DISCUSSION
Previous reports uncovered a key role of BCL6 as a transcriptional regulator in various cell types including B cells, T lymphocyte subsets within and outside of the germinal centers, including CD4+ follicular helper T cells (39), T regulatory (40) and T helper cells and in the control of T-cell-dependent inflammation and autoimmunity (17, 20). The role of BCL6 in the regulation of T helper cells including Th1, Th2 and Th17 cells has been reported by several groups (17, 18, 21, 41–43). Here we provide evidence demonstrating that BCL6 is a repressor of Th9 cell development through direct regulation of the Il9 promoter. We report that BCL6 expression is reduced early in polarized Th9 cells under IL-4 plus TGF-β1 conditions in an IL-2-dependent manner. At the transcription level, BCL6 binds and induces Il9 repression, and this process is in competition with the STAT5 signaling, a positive regulator of Th9 cell differentiation. In autoimmunity, silencing of BCL6 in myelin-reactive Th9 cells increases IL-9 and IL-4 expression and decreases IFNγ production leading to a reduction of their encephalitogenicity upon adoptive transfer in lymphopenic hosts.
IL-2, a growth factor linked to human autoimmune diseases (44), is a critical element for the development of Th1 (45) and Th2 (46) cells while inhibiting Th17 (47) and T follicular helper cells (48). The alpha-subunit of the IL-2 receptor (IL-2Rα) plays a pivotal role in the regulation of T cell function. Levels of a soluble form of IL-2Rα (sIL-2Rα) lacking the transmembrane and cytoplasmic domains were shown to be increased in several autoimmune diseases including multiple sclerosis (49). In addition, IL-2Rα genetic variants correlate with the levels of soluble IL-2Rα in subjects with type 1 diabetes and MS (50, 51). Addition of IL-2 to murine and human Th9 cells enhances their differentiation (27, 28, 52). However, the molecular mechanisms of IL-2-mediated amplification of Th9 cell differentiation are still unclear. IL-2 is produced mainly by activated T cells after engagement of the T-cell receptor and the CD28 co-stimulatory molecule (53). Our data demonstrate that in addition to Th1 and Th2 cells, IL-2 is massively produced early after Th9 cell polarization suggesting a role of IL-2 signaling in the regulation of Th9 cell development. Three subunits constitute the IL-2 receptor (IL-2R): the α chain (IL-2Rα, also known as CD25), the β chain (IL-2Rβ, also known as CD122, shared by the IL-15R), and the common cytokine receptor γ chain (γc, also known as CD132, shared by IL-4R, IL-7R, IL-9R, IL-15R and IL-21R). The IL-2/IL-2R complex mediates downstream signaling through IL-2Rβ and γc by inducing the association and phosphorylation of the tyrosine kinases Janus kinase 1 (JAK1) and JAK3 (54, 55), leading to the activation of STAT5 pathway (56). Here we provide novel evidence characterizing a regulatory network where STAT5 functions as an upstream regulator of the expression and activity of BCL6 and promoting the development of Th9 cells. These findings are in agreement with a recent report describing involvement of STAT5 signaling in Th9 cell polarization in animal models of allergic airway inflammation (16, 29).
The molecular dissection of BCL6-mediated Th9 cell differentiation inhibition indicates that BCL6 directly represses the Il9 locus. We have previously reported that Th9 cell differentiation is defective in GATA3-deficient mice suggesting that GATA3 is required for IL-9 transcription (1). Although our present report provides direct evidence demonstrating that BCL6 directly regulates the Il9 promoter, it remains possible that GATA3, a known target of BCL6 (17), may provide an additional mechanism linking BCL6 to the regulation of Th9 cell differentiation.
The BCL6 consensus-binding site resembles the interferon-gamma activated sequence motif recognized by the STAT family of transcription factors, raising speculation that BCL6 may repress some cytokine response genes (57). Our data demonstrate that STAT5 not only binds and activates the Il9 promoter transactivation but also competes with BCL6 binding activity to the Il9 promoter in an IL-2-dependent manner. These findings support previous studies demonstrating that STAT5 and BCL6 bind to distinct but overlapping sequence motifs, supporting the proposal that STAT factors and BCL6 directly compete for DNA binding (20, 58). BCL6 repression could thus involve direct competition for STAT5 binding, as well as epigenetic modifications induced by BCL6-associated factors. Indeed, we observed an increase in the repressive H3K9 pan-methylation mark surrounding BCL6 binding site in Th9 cells cultured with anti-IL-2 neutralizing antibody. It should be noted that the described interplay between STAT5 and BCL6 is likely modulated for the long-term because STAT5 was found to act as a transcriptional repressor on the BCL6 gene itself (38). We also found that in Th9 cells, STAT5 is recruited to the BCL6 promoter and this was regulated by IL-2 signaling.
T helper cells exhibit various degrees of plasticity, namely, the capacity to change their phenotype when exposed to a new inflammatory milieu or upon regulation of transcription factors. An increasing body of evidence demonstrates that the regulation of Th9 cell development is a complex process requiring multiple cis-regulatory elements and specific trans-activating transcription factors including PU.1, IRF4, and RBPJ that are in common with other CD4+ T cell lineages particularly Th2 cells (12, 14, 59, 60). Our identification of BCL6 transcriptional program in Th9 cell differentiation provides additional evidence of the plasticity of Th9 cells where BCL6 forced expression resulted in a down-regulation of Th9/Th1 ratio as shown by a decrease in IL-4/IL-9 expression and an increase in IFNγ production leading to EAE exacerbation following adoptive transfer. It is worth noting that other transcription factors such as STAT1 have been reported to modulate Th9 cell plasticity such that genetic deletion of STAT1 in these cells increased the Th9/Th1 ratio while IL-27 suppressed IL-9 production and up-regulated IFNγ expression in Th9 cells in a STAT1-dependent manner (61).
In conclusion, our work indicates that IL-2/BCL6 pathway orchestrates Il9 transcription and modulates the pathogenicity of Th9 cells in EAE mice. Further studies are therefore warranted to determine the effects of IL-9 on the plasticity of T helper cells in the context of autoimmune responses.
Supplementary Material
Acknowledgments
We would like to thank Dr. K. Bunting (Emory) for providing the STAT5aS711F mutant plasmid.
The research was supported by Awards from the National Institutes of Health (AI071448 to SJK and AI093838 to SJK and WE) and the National Multiple Sclerosis Society (RG3945 to SJK and PP1734 to WE).
References
- 1.Dardalhon V, Awasthi A, Kwon H, Galileos G, Gao W, Sobel RA, Mitsdoerffer M, Strom TB, Elyaman W, Ho IC, Khoury S, Oukka M, Kuchroo VK. IL-4 inhibits TGF-beta-induced Foxp3+ T cells and, together with TGF-beta, generates IL-9+ IL-10+ Foxp3(−) effector T cells. Nature immunology. 2008;9:1347–1355. doi: 10.1038/ni.1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Veldhoen M, Uyttenhove C, van Snick J, Helmby H, Westendorf A, Buer J, Martin B, Wilhelm C, Stockinger B. Transforming growth factor-beta ‘reprograms’ the differentiation of T helper 2 cells and promotes an interleukin 9-producing subset. Nat Immunol. 2008;9:1341–1346. doi: 10.1038/ni.1659. [DOI] [PubMed] [Google Scholar]
- 3.Elyaman W, Bassil R, Bradshaw EM, Orent W, Lahoud Y, Zhu B, Radtke F, Yagita H, Khoury SJ. Notch receptors and Smad3 signaling cooperate in the induction of interleukin-9-producing T cells. Immunity. 2012;36:623–634. doi: 10.1016/j.immuni.2012.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Elyaman W, Bradshaw EM, Uyttenhove C, Dardalhon V, Awasthi A, Imitola J, Bettelli E, Oukka M, van Snick J, Renauld JC, Kuchroo VK, Khoury SJ. IL-9 induces differentiation of TH17 cells and enhances function of FoxP3+ natural regulatory T cells. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:12885–12890. doi: 10.1073/pnas.0812530106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lu LF, Lind EF, Gondek DC, Bennett KA, Gleeson MW, Pino-Lagos K, Scott ZA, Coyle AJ, Reed JL, Van Snick J, Strom TB, Zheng XX, Noelle RJ. Mast cells are essential intermediaries in regulatory T-cell tolerance. Nature. 2006;442:997–1002. doi: 10.1038/nature05010. [DOI] [PubMed] [Google Scholar]
- 6.Li H, Nourbakhsh B, Ciric B, Zhang GX, Rostami A. Neutralization of IL-9 ameliorates experimental autoimmune encephalomyelitis by decreasing the effector T cell population. J Immunol. 2010;185:4095–4100. doi: 10.4049/jimmunol.1000986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jager A, Dardalhon V, Sobel RA, Bettelli E, Kuchroo VK. Th1, Th17, and Th9 effector cells induce experimental autoimmune encephalomyelitis with different pathological phenotypes. J Immunol. 2009;183:7169–7177. doi: 10.4049/jimmunol.0901906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Purwar R, Schlapbach C, Xiao S, Kang HS, Elyaman W, Jiang X, Jetten AM, Khoury SJ, Fuhlbrigge RC, Kuchroo VK, Clark RA, Kupper TS. Robust tumor immunity to melanoma mediated by interleukin-9-producing T cells. Nature medicine. 2012 doi: 10.1038/nm.2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jones CP, Gregory LG, Causton B, Campbell GA, Lloyd CM. Activin A and TGF-beta promote T(H)9 cell-mediated pulmonary allergic pathology. J Allergy Clin Immunol. 2012;129:1000–1010 e1003. doi: 10.1016/j.jaci.2011.12.965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen Y, Lin G, Huo JS, Barney D, Wang Z, Livshiz T, States DJ, Qin ZS, Schwartz J. Computational and functional analysis of growth hormone (GH)-regulated genes identifies the transcriptional repressor B-cell lymphoma 6 (Bc16) as a participant in GH-regulated transcription. Endocrinology. 2009;150:3645–3654. doi: 10.1210/en.2009-0212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhu J, Paul WE. Peripheral CD4+ T-cell differentiation regulated by networks of cytokines and transcription factors. Immunological reviews. 2010;238:247–262. doi: 10.1111/j.1600-065X.2010.00951.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Staudt V, Bothur E, Klein M, Lingnau K, Reuter S, Grebe N, Gerlitzki B, Hoffmann M, Ulges A, Taube C, Dehzad N, Becker M, Stassen M, Steinborn A, Lohoff M, Schild H, Schmitt E, Bopp T. Interferon-regulatory factor 4 is essential for the developmental program of T helper 9 cells. Immunity. 2010;33:192–202. doi: 10.1016/j.immuni.2010.07.014. [DOI] [PubMed] [Google Scholar]
- 13.Jash A, Sahoo A, Kim GC, Chae CS, Hwang JS, Kim JE, Im SH. Nuclear factor of activated T cells 1 (NFAT1)-induced permissive chromatin modification facilitates nuclear factor-kappaB (NF-kappaB)-mediated interleukin-9 (IL-9) transactivation. The Journal of biological chemistry. 2012;287:15445–15457. doi: 10.1074/jbc.M112.340356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chang HC, Sehra S, Goswami R, Yao W, Yu Q, Stritesky GL, Jabeen R, McKinley C, Ahyi AN, Han L, Nguyen ET, Robertson MJ, Perumal NB, Tepper RS, Nutt SL, Kaplan MH. The transcription factor PU.1 is required for the development of IL-9-producing T cells and allergic inflammation. Nature immunology. 2010;11:527–534. doi: 10.1038/ni.1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goswami R, Jabeen R, Yagi R, Pham D, Zhu J, Goenka S, Kaplan MH. STAT6-dependent regulation of Th9 development. J Immunol. 2012;188:968–975. doi: 10.4049/jimmunol.1102840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yao W, Zhang Y, Jabeen R, Nguyen ET, Wilkes DS, Tepper RS, Kaplan MH, Zhou B. Interleukin-9 is required for allergic airway inflammation mediated by the cytokine TSLP. Immunity. 2013;38:360–372. doi: 10.1016/j.immuni.2013.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sawant DV, Sehra S, Nguyen ET, Jadhav R, Englert K, Shinnakasu R, Hangoc G, Broxmeyer HE, Nakayama T, Perumal NB, Kaplan MH, Dent AL. Bcl6 controls the Th2 inflammatory activity of regulatory T cells by repressing Gata3 function. J Immunol. 2012;189:4759–4769. doi: 10.4049/jimmunol.1201794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mondal A, Sawant D, Dent AL. Transcriptional repressor BCL6 controls Th17 responses by controlling gene expression in both T cells and macrophages. J Immunol. 2010;184:4123–4132. doi: 10.4049/jimmunol.0901242. [DOI] [PubMed] [Google Scholar]
- 19.Cimmino L, Martins GA, Liao J, Magnusdottir E, Grunig G, Perez RK, Calame KL. Blimp-1 attenuates Th1 differentiation by repression of ifng, tbx21, and bcl6 gene expression. J Immunol. 2008;181:2338–2347. doi: 10.4049/jimmunol.181.4.2338. [DOI] [PubMed] [Google Scholar]
- 20.Dent AL, Shaffer AL, Yu X, Allman D, Staudt LM. Control of inflammation, cytokine expression, and germinal center formation by BCL-6. Science. 1997;276:589–592. doi: 10.1126/science.276.5312.589. [DOI] [PubMed] [Google Scholar]
- 21.Ye BH, Cattoretti G, Shen Q, Zhang J, Hawe N, de Waard R, Leung C, Nouri-Shirazi M, Orazi A, Chaganti RS, Rothman P, Stall AM, Pandolfi PP, Dalla-Favera R. The BCL-6 proto-oncogene controls germinal-centre formation and Th2-type inflammation. Nature genetics. 1997;16:161–170. doi: 10.1038/ng0697-161. [DOI] [PubMed] [Google Scholar]
- 22.Horvath CM. STAT proteins and transcriptional responses to extracellular signals. Trends in biochemical sciences. 2000;25:496–502. doi: 10.1016/s0968-0004(00)01624-8. [DOI] [PubMed] [Google Scholar]
- 23.Bettelli E, Sullivan B, Szabo SJ, Sobel RA, Glimcher LH, Kuchroo VK. Loss of T-bet, but not STAT1, prevents the development of experimental autoimmune encephalomyelitis. J Exp Med. 2004;200:79–87. doi: 10.1084/jem.20031819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yu RY, Wang X, Pixley FJ, Yu JJ, Dent AL, Broxmeyer HE, Stanley ER, Ye BH. BCL-6 negatively regulates macrophage proliferation by suppressing autocrine IL-6 production. Blood. 2005;105:1777–1784. doi: 10.1182/blood-2004-08-3171. [DOI] [PubMed] [Google Scholar]
- 25.Lin J, Wang X, Dorsky RI. Progenitor expansion in apc mutants is mediated by Jak/Stat signaling. BMC developmental biology. 2011;11:73. doi: 10.1186/1471-213X-11-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li G, Miskimen KL, Wang Z, Xie XY, Tse W, Gouilleux F, Moriggl R, Bunting KD. Effective targeting of STAT5-mediated survival in myeloproliferative neoplasms using ABT-737 combined with rapamycin. Leukemia. 2010;24:1397–1405. doi: 10.1038/leu.2010.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Beriou G, Bradshaw EM, Lozano E, Costantino CM, Hastings WD, Orban T, Elyaman W, Khoury SJ, Kuchroo VK, Baecher-Allan C, Hafler DA. TGF-beta induces IL-9 production from human Th17 cells. J Immunol. 2010;185:46–54. doi: 10.4049/jimmunol.1000356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schmitt E, Germann T, Goedert S, Hoehn P, Huels C, Koelsch S, Kuhn R, Muller W, Palm N, Rude E. IL-9 production of naive CD4+ T cells depends on IL-2, is synergistically enhanced by a combination of TGF-beta and IL-4, and is inhibited by IFN-gamma. J Immunol. 1994;153:3989–3996. [PubMed] [Google Scholar]
- 29.Yang XO, Zhang H, Kim BS, Niu X, Peng J, Chen Y, Kerketta R, Lee YH, Chang SH, Corry DB, Wang D, Watowich SS, Dong C. The signaling suppressor CIS controls proallergic T cell development and allergic airway inflammation. Nature immunology. 2013;14:732–740. doi: 10.1038/ni.2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li P, Spolski R, Liao W, Wang L, Murphy TL, Murphy KM, Leonard WJ. BATF-JUN is critical for IRF4-mediated transcription in T cells. Nature. 2012;490:543–546. doi: 10.1038/nature11530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nurieva RI, Podd A, Chen Y, Alekseev AM, Yu M, Qi X, Huang H, Wen R, Wang J, Li HS, Watowich SS, Qi H, Dong C, Wang D. STAT5 protein negatively regulates T follicular helper (Tfh) cell generation and function. The Journal of biological chemistry. 2012;287:11234–11239. doi: 10.1074/jbc.M111.324046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Johnston RJ, Choi YS, Diamond JA, Yang JA, Crotty S. STAT5 is a potent negative regulator of TFH cell differentiation. J Exp Med. 2012;209:243–250. doi: 10.1084/jem.20111174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pepper M, Pagan AJ, Igyarto BZ, Taylor JJ, Jenkins MK. Opposing signals from the Bcl6 transcription factor and the interleukin-2 receptor generate T helper 1 central and effector memory cells. Immunity. 2011;35:583–595. doi: 10.1016/j.immuni.2011.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Oestreich KJ, Mohn SE, Weinmann AS. Molecular mechanisms that control the expression and activity of Bcl-6 in TH1 cells to regulate flexibility with a TFH-like gene profile. Nature immunology. 2012;13:405–411. doi: 10.1038/ni.2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hebenstreit D, Horejs-Hoeck J, Duschl A. JAK/STAT-dependent gene regulation by cytokines. Drug news & perspectives. 2005;18:243–249. doi: 10.1358/dnp.2005.18.4.908658. [DOI] [PubMed] [Google Scholar]
- 36.Scheeren FA, Naspetti M, Diehl S, Schotte R, Nagasawa M, Wijnands E, Gimeno R, Vyth-Dreese FA, Blom B, Spits H. STAT5 regulates the self-renewal capacity and differentiation of human memory B cells and controls Bcl-6 expression. Nature immunology. 2005;6:303–313. doi: 10.1038/ni1172. [DOI] [PubMed] [Google Scholar]
- 37.Duy C, Yu JJ, Nahar R, Swaminathan S, Kweon SM, Polo JM, Valls E, Klemm L, Shojaee S, Cerchietti L, Schuh W, Jack HM, Hurtz C, Ramezani-Rad P, Herzog S, Jumaa H, Koeffler HP, de Alboran IM, Melnick AM, Ye BH, Muschen M. BCL6 is critical for the development of a diverse primary B cell repertoire. J Exp Med. 2010;207:1209–1221. doi: 10.1084/jem.20091299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Walker SR, Nelson EA, Frank DA. STAT5 represses BCL6 expression by binding to a regulatory region frequently mutated in lymphomas. Oncogene. 2007;26:224–233. doi: 10.1038/sj.onc.1209775. [DOI] [PubMed] [Google Scholar]
- 39.Alinikula J, Lassila O. Gene interaction network regulates plasma cell differentiation. Scandinavian journal of immunology. 2011;73:512–519. doi: 10.1111/j.1365-3083.2011.02556.x. [DOI] [PubMed] [Google Scholar]
- 40.Chung Y, Tanaka S, Chu F, Nurieva RI, Martinez GJ, Rawal S, Wang YH, Lim H, Reynolds JM, Zhou XH, Fan HM, Liu ZM, Neelapu SS, Dong C. Follicular regulatory T cells expressing Foxp3 and Bcl-6 suppress germinal center reactions. Nature medicine. 2011;17:983–988. doi: 10.1038/nm.2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Arima M, Toyama H, Ichii H, Kojima S, Okada S, Hatano M, Cheng G, Kubo M, Fukuda T, Tokuhisa T. A putative silencer element in the IL-5 gene recognized by Bcl6. J Immunol. 2002;169:829–836. doi: 10.4049/jimmunol.169.2.829. [DOI] [PubMed] [Google Scholar]
- 42.Hirata H, Arima M, Fukushima Y, Ishii Y, Tokuhisa T, Fukuda T. Effects of Th2 pulmonary inflammation in mice with bleomycin-induced pulmonary fibrosis. Respirology. 2008;13:788–798. doi: 10.1111/j.1440-1843.2008.01361.x. [DOI] [PubMed] [Google Scholar]
- 43.Nakayamada S, Kanno Y, Takahashi H, Jankovic D, Lu KT, Johnson TA, Sun HW, Vahedi G, Hakim O, Handon R, Schwartzberg PL, Hager GL, O’Shea JJ. Early Th1 cell differentiation is marked by a Tfh cell-like transition. Immunity. 2011;35:919–931. doi: 10.1016/j.immuni.2011.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schimpl A, Berberich I, Kneitz B, Kramer S, Santner-Nanan B, Wagner S, Wolf M, Hunig T. IL-2 and autoimmune disease. Cytokine & growth factor reviews. 2002;13:369–378. doi: 10.1016/s1359-6101(02)00022-9. [DOI] [PubMed] [Google Scholar]
- 45.Liao W, Lin JX, Wang L, Li P, Leonard WJ. Modulation of cytokine receptors by IL-2 broadly regulates differentiation into helper T cell lineages. Nature immunology. 2011;12:551–559. doi: 10.1038/ni.2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liao W, Schones DE, Oh J, Cui Y, Cui K, Roh TY, Zhao K, Leonard WJ. Priming for T helper type 2 differentiation by interleukin 2-mediated induction of interleukin 4 receptor alpha-chain expression. Nature immunology. 2008;9:1288–1296. doi: 10.1038/ni.1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Laurence A, Tato CM, Davidson TS, Kanno Y, Chen Z, Yao Z, Blank RB, Meylan F, Siegel R, Hennighausen L, Shevach EM, O’Shea JJ. Interleukin-2 signaling via STAT5 constrains T helper 17 cell generation. Immunity. 2007;26:371–381. doi: 10.1016/j.immuni.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 48.Ballesteros-Tato A, Leon B, Graf BA, Moquin A, Adams PS, Lund FE, Randall TD. Interleukin-2 inhibits germinal center formation by limiting T follicular helper cell differentiation. Immunity. 2012;36:847–856. doi: 10.1016/j.immuni.2012.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Carrieri PB, Soscia E, Iacovitti B, Pellicano M, D’Antonio A, Provitera V, Perrella O. Soluble interleukin-2 receptor and soluble CD8 molecules in cerebrospinal fluid and serum of patients with multiple sclerosis. European cytokine network. 1992;3:495–498. [PubMed] [Google Scholar]
- 50.Maier LM, Anderson DE, Severson CA, Baecher-Allan C, Healy B, Liu DV, Wittrup KD, De Jager PL, Hafler DA. Soluble IL-2RA levels in multiple sclerosis subjects and the effect of soluble IL-2RA on immune responses. J Immunol. 2009;182:1541–1547. doi: 10.4049/jimmunol.182.3.1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Maier LM, Lowe CE, Cooper J, Downes K, Anderson DE, Severson C, Clark PM, Healy B, Walker N, Aubin C, Oksenberg JR, Hauser SL, Compston A, Sawcer S, De Jager PL, Wicker LS, Todd JA, Hafler DA. IL2RA genetic heterogeneity in multiple sclerosis and type 1 diabetes susceptibility and soluble interleukin-2 receptor production. PLoS genetics. 2009;5:e1000322. doi: 10.1371/journal.pgen.1000322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Houssiau FA, Renauld JC, Fibbe WE, Van Snick J. IL-2 dependence of IL-9 expression in human T lymphocytes. J Immunol. 1992;148:3147–3151. [PubMed] [Google Scholar]
- 53.Yang SY, Denning SM, Mizuno S, Dupont B, Haynes BF. A novel activation pathway for mature thymocytes. Costimulation of CD2 (T,p50) and CD28 (T,p44) induces autocrine interleukin 2/interleukin 2 receptor-mediated cell proliferation. J Exp Med. 1988;168:1457–1468. doi: 10.1084/jem.168.4.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nelson BH, Lord JD, Greenberg PD. Cytoplasmic domains of the interleukin-2 receptor beta and gamma chains mediate the signal for T-cell proliferation. Nature. 1994;369:333–336. doi: 10.1038/369333a0. [DOI] [PubMed] [Google Scholar]
- 55.Russell SM, Johnston JA, Noguchi M, Kawamura M, Bacon CM, Friedmann M, Berg M, McVicar DW, Witthuhn BA, Silvennoinen O, et al. Interaction of IL-2R beta and gamma c chains with Jak1 and Jak3: implications for XSCID and XCID. Science. 1994;266:1042–1045. doi: 10.1126/science.7973658. [DOI] [PubMed] [Google Scholar]
- 56.Gaffen SL. Signaling domains of the interleukin 2 receptor. Cytokine. 2001;14:63–77. doi: 10.1006/cyto.2001.0862. [DOI] [PubMed] [Google Scholar]
- 57.Harris MB, Chang CC, Berton MT, Danial NN, Zhang J, Kuehner D, Ye BH, Kvatyuk M, Pandolfi PP, Cattoretti G, Dalla-Favera R, Rothman PB. Transcriptional repression of Stat6-dependent interleukin-4-induced genes by BCL-6: specific regulation of iepsilon transcription and immunoglobulin E switching. Molecular and cellular biology. 1999;19:7264–7275. doi: 10.1128/mcb.19.10.7264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Diehl SA, Schmidlin H, Nagasawa M, van Haren SD, Kwakkenbos MJ, Yasuda E, Beaumont T, Scheeren FA, Spits H. STAT3-mediated up-regulation of BLIMP1 Is coordinated with BCL6 down-regulation to control human plasma cell differentiation. J Immunol. 2008;180:4805–4815. doi: 10.4049/jimmunol.180.7.4805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Amsen D, Antov A, Jankovic D, Sher A, Radtke F, Souabni A, Busslinger M, McCright B, Gridley T, Flavell RA. Direct regulation of Gata3 expression determines the T helper differentiation potential of Notch. Immunity. 2007;27:89–99. doi: 10.1016/j.immuni.2007.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fang TC, Yashiro-Ohtani Y, Del Bianco C, Knoblock DM, Blacklow SC, Pear WS. Notch directly regulates Gata3 expression during T helper 2 cell differentiation. Immunity. 2007;27:100–110. doi: 10.1016/j.immuni.2007.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Murugaiyan G, Beynon V, Pires Da Cunha A, Joller N, Weiner HL. IFN-gamma limits Th9-mediated autoimmune inflammation through dendritic cell modulation of IL-27. J Immunol. 2012;189:5277–5283. doi: 10.4049/jimmunol.1200808. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.