SUMMARY
The Saccharomyces cereivisiae gene deletion project revealed that approximately 20% of yeast genes are required for viability. The analysis of essential genes traditionally relies on conditional mutants, typically temperature-sensitive (ts) alleles. We developed a systematic approach (termed “diploid shuffle”) useful for generating a ts allele for each essential gene in S. cerevisiae and for improved genetic manipulation of mutant alleles and gene constructs in general. Importantly, each ts allele resides at its normal genomic locus, flanked by specific cognate UPTAG and DNTAG bar codes. A subset of 250 ts mutants, including ts alleles for all uncharacterized essential genes and prioritized for genes with human counterparts, is now ready for distribution. The importance of this collection is demonstrated by biochemical and genetic screens that reveal essential genes involved in RNA processing and maintenance of chromosomal stability.
INTRODUCTION
Deciphering the functional roles of all encoded gene products within a genome is the central challenge in understanding the biology of an organism. The study of mutant phenotypes has proven to be a fundamental approach for obtaining a detailed understanding of gene function. While genetic screens based on random mutagenesis have been seminal in identifying genes involved in biological processes, the random mutagenesis approach rarely achieves saturation, because mutability, resulting in haploid viable mutants, varies widely among genes.
The efficiency of mutant phenotype studies was greatly accelerated in the yeast Saccharomyces cerevisiae following the creation of a comprehensive Saccharomyces gene deletion (YKO) collection by a consortium of laboratories. This collection includes isogenic knockouts for virtually all annotated yeast ORFs, each of which was replaced by a kanMX4 cassette that confers resistance to G418 (Giaever et al., 2002). One of the most powerful features of the YKO collection is the systematic incorporation of two 20-mer molecular bar codes (UPTAG and DNTAG) unique to each YKO strain (Shoemaker et al., 1996; Winzeler et al., 1999). Functional profiling of populations using TAG microarrays has greatly expedited genetic screens and made them more quantitative (Birrell et al., 2001; Lum et al., 2004; Ooi et al., 2001).
The YKO mutants are a valuable resource for genome-wide functional analysis of the 4700 nonessential genes; however, no equivalent systematic bar-coded mutant collection exists for the 1107 essential genes. Such a collection of essential gene haploid mutants would be scientifically valuable, enabling systematic genome-wide genetic screens, the study of individual genes, and a more comprehensive view of genetic interaction networks.
Genetic analysis of essential genes typically relies on conditional mutants carrying point mutations, which retain the function of a specific gene under one set of conditions (permissive), lack that function under a different set of conditions (nonpermissive), and exhibit partial (hypomorphic) function under semipermissive conditions. Conditional alleles can also be generated by the replacement of the native promoter of a gene with one that can be rapidly repressed, such as the tetracycline (tet)-regulatable promoter. This approach has recently been used to generate a collection of promoter-shutoff strains for two-thirds of the essential yeast gene (Mnaimneh et al., 2004). Another approach is a technique termed decreased abundance by mRNA perturbation (DAmP); in this method, the 3′UTR of an essential gene is disrupted by insertion of an antibiotic-resistance marker, destabilizing the target mRNA. The result is a protein potentially under its natural transcriptional regulation but at reduced levels (Schuldiner et al., 2005).
Traditionally, temperature-sensitive (ts) alleles have been used with great success for the genetic analysis of essential proteins, and these still remain a mainstay of genetic analysis (Hartwell et al., 1970; Simchen, 1978). In vitro mutagenesis by the method known as “plasmid shuffling” is widely used to generate ts mutations in individual genes of interest (Sikorski and Boeke, 1991); however, the method is not easily scaled to encompass the whole genome, and the generated alleles are not integrated into the genome and do not capitalize on the bar code strategy. An alternative method for constructing ts alleles involves engineering the chromosomal locus of a gene such that the amino terminus of the encoded protein is fused to a “heat-inducible degron.” The fusion protein is targeted for degradation at 37°C, which modulates the stability of the protein (Kanemaki et al., 2003; Labib et al., 2000). The corresponding mutants, termed td (for temperature-activated degron) (Dohmen and Varshavsky, 2005), exhibit lethality at high temperature for approximately 50% of the essential genes tested.
In this paper, we describe an efficient, scalable method to generate ts alleles for each of the essential genes in S. cerevisiae. The resultant strains are isogenic to the YKO deletion collection. Each of the ts alleles is present at its normal genomic locus and is flanked by specific cognate UPTAG and DNTAG bar codes. In addition, the basic principles of the diploid shuffle method and its corollaries are broadly applicable to yeast molecular genetic analysis. We have used this method to generate ts alleles for 250 essential genes, 45 of which encode proteins with relatively unknown function and the remaining alleles prioritized based on sequence similarity to human proteins. We also describe biochemical and genetic screens in which ts mutants corresponding to the 45 genes of unknown function were used. These screens reveal new mutants that affect RNA processing and chromosome transmission and strongly validate the use of this ordered array as a resource for biological discovery.
RESULTS
Diploid Shuffle—A Systematic Method for Generating ts Alleles for the Essential Genes in S. cereviviae
Our goal was to develop a systematic approach to screen for missense mutations that result in ts alleles in each essential gene at its endogenous chromosomal location and flanked with the appropriate bar codes.
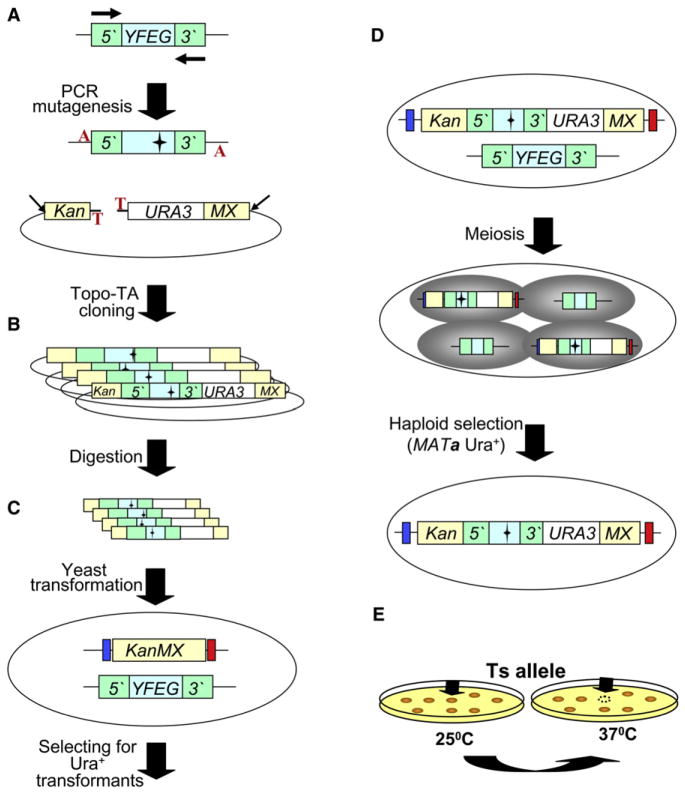
The first step in generating a ts allele for Your Favorite Essential Gene (YFEG) is the use of a genomic DNA as a template to amplify its open reading frame (ORF) including the 5′ promoter and 3′ terminator regions using mutagenic amplification (Figure 1A). The mutagenized PCR product from step 1 is cloned into SB221+Topo-TA. This plasmid contains the URA3 gene flanked by the 5′ and 3′ regions of KanMX. The Topo-TA cloning site (Invitrogen) has been inserted in between the KanMX 5′ end and the URA3 gene. This site allows direct cloning of each of the PCR products without the need for any further modifications. The result of the cloning step is a library of mutagenized YFEG, which is then transformed into E. coli and digested to release linear fragments (following DNA purification). The linear fragments are directly transformed into the corresponding strain from the haploid convertible “heterozygous” diploid YKO collection (Figures 1B and 1C). This collection was made by individually incorporating the modified SGA reporter can1Δ::LEU2-MFA1pr-HIS3 into the full set of heterozygous diploid YKOs originally constructed by the Saccharomyces Genome Deletion project (available from Open Biosystems, J.D.B., S. Sookhai-Mahadeo, X. Wang, C. Tiffany, O. Chen, F.A. Spencer, D.S. Yuan, and P.B. Meluh, unpublished data). The ~700 bp KanMX5′ and KanMX3′ fragments direct the mutagenized YFEG library into the yfegΔ::KanMX genomic locus via homologous recombination (Figure 1C). Ura+ transformants are selected. Pools of Ura+ cells containing the mutant alleles are sporulated (Figure 1D). Spores are spread on haploid-selective medium and incubated at 25°C for 5 days for colony formation (Figure 1D). Only haploid MATa Ura+ spores containing the integrated mutagenized allele of YFEG can grow on this medium. Colonies formed on selective medium are replica plated and incubated at 37°C for 2 days (Figure 1E). Colonies growing at 25°C, but not 37°C, are selected as potential ts alleles and restreaked at both temperatures for retesting. Retesting is done by backcrossing the potential ts allele to a donor strain (see the Experimental Procedures). Following sporulation and tetrad dissection, we confirmed that (1) the ts phenotype segregates in a Mendelian manner (2:2), indicative that it depends on a single mutated gene; (2) it is linked to URA3 and therefore cosegregates with the mutated PCR product; and (3) the mutagenized PCR product was integrated at the correct genomic locus, rendering the cells G418 sensitive (5′kanMX::yfeg-ts-URA3::3′kanMX).
Figure 1. Diploid Shuffle—A Method for Generating ts Alleles for the Essential Genes in Saccharomyces cereviviae.
(A) Genomic DNA containing YFEG and its 5′ and 3′ regions is used as the template for PCR mutagenesis. Two black horizontal arrows represent the gene-specific primers used. The mutagenized PCR product is cloned into the vector SB221+ Topo-TA (mutations are represented by black stars). The Topo-TA cloning site is represented by a red T, and the A overhang protruding from the PCR product is represented by a red A. Left beige bar represents the 5′ half of the KanMX selectable marker (Kan), while the right beige bar represents the other half of the KanMX selectable marker (MX). The NotI restriction sites are indicated by two diagonal black arrows.
(B) The product of the cloning step is a library of a mutagenized YFEG. The library is then transformed into E. coli and digested with NotI to release linear fragments (following DNA purification).
(C) The linearized library is transformed into the corresponding heterozygous diploid strain. Blue and red bars that flank the KanMX knockout represent the two bar codes.
(D) Heterozygous diploid transformants are sporulated (following meiosis), and MATa Ura+ haploids spores are selected on haploid-selective medium at 25°C.
(E) Selection of ts candidates following the replica plating and incubating at 25°C and 37°C. Black arrows identify a potential ts allele.
In summary, the final product of the method that we have described is a confirmed MATa strain from the YKO collection genetic background containing a URA3-marked ts allele of a specific essential gene integrated into its endogenous locus and flanked by both bar codes. In addition, the diploid shuffle method and its variations (see Figures S3 and S4 available online and the Discussion) represent significantly improved methods for generating strains carrying specific alleles, for transferring ts alleles to a variety of strain backgrounds and genetic contexts, and for the introduction of gene constructs such as fusion proteins, heterozygous expression cassettes, etc.
Construction of ts Strains for 45 Essential Genes of Unknown Function
We generated an initial collection of ts strains to provide an early-stage resource of value for community genetic and phenotypic screening. We chose 45 yeast ORFs known at the time that we started to be essential for viability but defined by the S. cerevisiae Genome Data Base (SGD) as uncharacterized ORFs, proteins with unknown function or unknown molecular function. Thirty of these ORFs (67%) have a significant human homolog with an E value that ranges from 3.00E-05 to 1.00E-261 (Table S1).
For each gene, ~15,000 independent PCR-mutagenized clones were screened for potential ts phenotypes. In 42 of 45 cases, we successfully isolated a ts allele in the first round of screening. In the remaining three cases, a second round of mutagenesis was required, resulting in successful isolation of one or more ts mutants in all genes attempted. By sequencing 12 independent ts alleles, we found that the average mutation frequency is 0.54 base substitution per 100 bp, which translates to an average of 0.9 mutations per 100 amino acids (Figure S1). To demonstrate the potential for new discovery, we screened this collection of 45 ts mutants using established biochemical and genetic screens used extensively in the past.
Screen for RNA-Processing Defects Reveals Previously Unidentified Mutants
Production of ribosomal RNA in eukaryotic cells is considerably more complex than in prokaryotes, where a single enzyme, RNase III, makes the needed cleavages and ribosome assembly is cotranscriptional. Although eukaryotic rRNA is organized similarly, in a 35S precursor, the cleavages are carried out by many protein and ribonucleoprotein complexes (including RNase III [Kufel et al., 1999]). In eukaryotes, the molecularly intertwined steps of rRNA processing and ribosome assembly are much slower, and many steps are posttranscriptional. The combined molecular weight of the small nucleolar ribonucloprotein particle (snoRNP) complexes involved in ribosome assembly (not present in prokaryotes) exceeds the molecular weight of the ribosome by >15-fold (Venema and Tollervey, 1999). Production of rRNA in the nucleolus, transport of r proteins into (and then out of) the nucleus, imposes requirements on assembly absent from prokaryotes (Hage and Tollervey, 2004). At least 170 proteins and 100 small RNAs are estimated to participate in eukaryotic rRNA processing and ribosome assembly (Andersen et al., 2002; Fromont-Racine et al., 2003; Venema and Tollervey, 1999) in addition to the 79 proteins and four RNAs that make up the ribosome.
Because of this complexity and because RNA-processing defects are easily identified by simple RNA-blotting techniques (Lindahl et al., 1992; Peng et al., 2003), we hypothesized that some of the unknown essential genes might contribute to RNA processing. We performed specific assays to detect defects in rRNA, mRNA, snoRNA, and snRNA processing in the collection of 45 ts mutants with (relatively) unknown function (Table S1) as well as a positive control nme1 ts mutant (a kind gift of Mark Schmitt). Mutants were grown at permissive temperature and shifted to the nonpermissive temperature for 2 hr, and total RNA was harvested, resolved on appropriate gels, and blotted with a series of hybridization probes that detect the appropriate RNA precursors. Processing defects are readily revealed by the buildup of intermediates larger than the mature RNA species under study (Table S2). Consistent with the large number of known genes affecting rRNA processing, we identified ten new ts mutants with obvious rRNA-processing defects (Figure 4). These were revealed by probing with internal transcribed spacer 1 (ITS1) sequences (Figure 2) and mature rRNA probes (Figure S2, Table 1). No ts mutants severely affecting the pre-mRNA, snoRNA, or snRNA-processing pathways were detected in this set of mutants.
Figure 4. A Summary of ts Mutant Phenotypes, Showing CIN or RNA-Processing Defects.
Central nodes colored in red, green, blue, or light purple represent the CIN (BiM, ALF, CTF) and RNA-processing screens, respectively. Arrows that project from the central nodes point toward the mutants that were positive in a specific screen. Small circles colored in blue represent ts mutants with unknown function that were defective for CIN or RNA processing at semipermissive temperature. The ORFs that were used as controls for the CIN assay were colored in red. SSU components isolated bicohemically while this work was in progress are colored purple.
Figure 2. Ribosomal RNA-Processing Screen.
(A) Scheme of rRNA processing. Ribosomal RNA is encoded as part of ~9 kb tandem repeat in yeast and is processed from a 35S precursor in the multistep pathway outlined here. The cartoon indicates rDNA repeat structure and positions of mature 25S, 5.8S, and 18S rRNAs (black bars in DNA and precursor forms, gray bars represent mature forms) derived from the precursor, as well as the separately transcribed 5S rRNA. 3′ ends are indicated by arrowheads. Intermediates and pathways of rRNA processing have been identified and are shown in the bottom part of the panel. Dotted lines indicate effects of processing; an alternate pathway to the formation of mature 5.8S and 25S rRNAs from the topmost 27S precursor is indicated by square bracket. Note that there are multiple species with mobilities of about 27S and 7S; our experiments do not distinguish among these. The internal transcribed spacer (ITS) regions used as probes to detect precursor forms are indicated in the inset.
(B) Isolation of ribosomal RNA-processing-defective mutants. RNA preparations were resolved on 1% agarose formaldehyde gels and blotted to nylon membranes and probed with an ITS2 probe. Duplicate isolates of the wild-type and identified new mutants are shown. Mutant YAL043 is not part of the set of 45 unknown mutants but was also studied.
Table 1.
Genes Identified through ts Mutants that Affect rRNA Processing
| ORF Name | Gene Name(s) | Description | rRNA Defect | Percent Reduction |
|---|---|---|---|---|
| YAL043C | PTA1, FUN39 | Subunit of holo-CPF, mRNA, snoRNA pre-tRNA processing; binds phospho-CTD of RNAPII | 18Sa | 22.4 |
| YDR196C | Coenzyme A biosynthesis; mitochondrial | 18S | 11.3 | |
| YDR339C | FCF1, UTP24 | SSU processome component | 18S | 11 |
| YGR145W | ENP2 | Nucleolar | 18S | 23.5 |
| YGR277C | Coenzyme A biosynthesis | Not significant | ||
| YJL069C | UTP18 | Possible U3 snoRNP protein | 25S | 5.9 |
| YKL033W | FMP47 | Mitochondrial | 18S | 16.9 |
| YKR022C | NTR2 | Spliceosome disassembly | 18S | 9 |
| YLR132C | Mitochondria and nucleus | Not significant | ||
| YOL022C | Cytoplasmic | 18S | 9.1 | |
| YOR004W | UTP23 | SSU processome component | 18S | 8.1 |
As is apparent from Figure 2 and Table 1, rRNA-processing mutants ranging from nearly as severe as the control (the nme1 mutant) to milder defects were observed as hyperaccumulation of 35S, 33S, and, in some cases, reduced accumulation of 27S precursor rRNA species. Most of the mutants also had a significant deficit in mature 18S or 25S rRNA, suggesting a direct involvement in rRNA processing (Figure S2 and Table S3). The proteins corresponding to two of these mutants, ydr339c and yor004w, were biochemically identified as important components, Utp23p and 24p, of the small-subunit rRNA-processing machinery (Bleichert et al., 2006; Rempola et al., 2006) during the course of these experiments, providing a critical internal validation for the assay and for the approach. While these are clearly integral parts of the rRNA-processing machinery, other mutants may affect rRNA processing less directly. There may be many such mechanisms; half of all yeast’s energy goes into ribosomes. For example, defects in secretion downregulate ribosome biogenesis (Mizuta and Warner, 1994).
Crosstalk among RNA-Processing Pathways
We searched for pathway connections among the newly identified ts mutants. YJL069C (UTP18) was previously predicted to function in processing of U3 snoRNA (Bernstein et al., 2004; Samanta and Liang, 2003). The rRNA-processing defect in yjl069-ts provides genetic evidence supporting that hypothesis, because U3 snoRNA is essential for rRNA processing. Similarly, the YGR145W (ENP2) gene encodes a protein known to interact with the MPP10p component of the SSU processome (SGD); the SSU processome is responsible for processing of the 18S rRNA. The protein encoded by YKR022C (NTR2) is reportedly a component of the Prp43 helicase spliceosome disassembly complex (Tsai et al., 2005); our data suggest that it might also facilitate rRNA processing, in accord with recent studies (Combs et al., 2006; Lebaron et al., 2005; Leeds et al., 2006). Interestingly, both YKR022C and another gene identified here, YLR132C, were predicted pre-mRNA splicing factors by a computational analysis (Hazbun et al., 2003), but we observed no such defects. The YAL043C (PTA1) gene encodes a subunit of the holo-CPF complex involved in mRNA 3′ end formation/polyadenylation (Zhao et al., 1999) and tRNA biogenesis (O’Connor and Peebles, 1992). The discovery of a set of proteins that appear to function in multiple RNA-processing networks is exciting, consistent with prior studies (Burckin et al., 2005; Leeds et al., 2006), and provides new evidence for cross-RNA-processing pathway interactions.
A Connection between Coenzyme A Biosynthesis and rRNA Processing?
Two of the rRNA-processing mutants were ts in genes with predicted roles in CoA biosynthesis (YDR196C and YGR277C), based on homology to prokaryotic CoA biosynthesis proteins. The observed defects are relatively specific for rRNA processing and seem not to represent a general pleiotropic defect. It is likely that a CoA derivative, rather than these proteins per se, affects (directly or indirectly) rRNA processing.
Screens for Chromosome Instability
S. cerevisiae is attractive for identifying and characterizing genes involved in chromosome segregation. Recently, systematic screening of the 4700 nonessential gene deletion mutants has identified more than 200 nonessential genes contributing to faithful chromosome segregation (Yuen et al., 2007), but no systematic screening of essential gene mutants has been possible. We therefore decided to search for novel CIN genes by screening the 45 ts mutant collection for a CIN phenotype. Three different tests were used (Figure 3): the chromosome transmission fidelity (CTF) (Spencer et al., 1990) and a-like faker (ALF) (Gerring et al., 1990) assays in haploids and the diploid bimater test (BiM) (Kouprina et al., 1988) in MATa/MATα diploids. Screening with these three different assays broadens the spectrum of identifiable genes by allowing various defects in chromosome transmission to be detected (Yuen et al., 2007).
Figure 3. Screening for Mutants Defective in Faithful Chromosome Segregation.
(A) A CTF assay: (Aa) A supernumerary linear artificial chromosome fragment (CF) containing the SUP11 gene serves as a sensitive indicator of chromosome stability by its ability to suppress the ade2-101 ochre mutation (resulting in white cells). (Ab) Chromosome loss is indicated by streaking the cells on media non-selective for the artificial chromosome and examination of the appearance of white/red sectored colonies. The sectoring phenotype of dre2 starts to appear at 32°C; at higher temperature (34°C) the phenotype becomes more severe.
(B) An ALF assay. (Ba) ALF is based on the fact that the default mating type in yeast is MATa. If the MATα cells of the ts mutants lose the MATα locus (due to the loss of chromosome III), they mate with a MATα tester as MATa and are thus called “a-like fakers.” (Bb) Three patches of a MATα ts isolate and control strains (WT Alpha and bim1) were replica plated on a lawn of MATα tester strain. Growing colonies are the indication of the ability to mate with the tester strains. As shown, ALF of swi4 appears at 32°C as compared to the controls.
(C) A BiM assay. (Ca) Normally, diploid cells heterozygous at the mating type locus (MATa/MATα) do not mate with either MATa or MATα haploids. However, loss of either the MATa or the MATα mating type locus generates a higher frequency of mating-competent cells within a population of cells, which then exhibits a bimating phenotype. (Cb) Three independent isolates of a homozygous diploid ts isolate plus the control strains (WT homozygous diploid and the homozygous diploid of chl1 deletion) were patched and replica plated to a MATa (top) and MATα (bottom) testers. The ability to mate as a diploid is assessed by the number of colonies on both tester strains. The BiM phenotype of ynl152w appears at 34°C, as compared to the control strains. The mechanisms that can lead to CF loss, ALF, and BiM are shown in (Ac), (Bc), and (Cc), respectively.
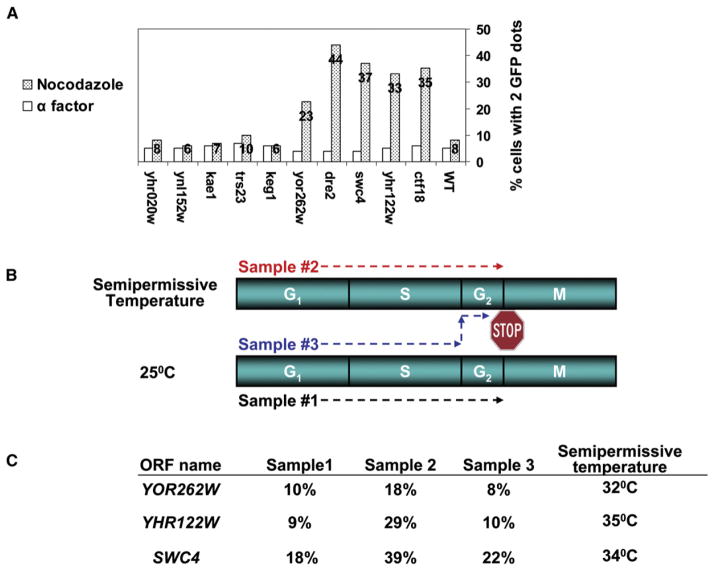
For each of the CIN assays, specific ts alleles were tested in a wide range of semi- and nonpermissive temperatures (25°C, 30°C, 32°C, 34°C, and 37°C). In total, nine of 45 ts mutants (20%) exhibited increased chromosome instability (CIN) and were defined as new CIN genes (Table 2 and Figure 4). Typical results are presented for three of these mutants (Figure 3). The severity of the CIN phenotypes is quantified in Table 2. Six of these mutants (ynl152w, yhr020w, keg1, yhr122w, dre2, and yor262w) define uncharacterized ORFs. Three additional mutants, swc4, trs23, and kae1, were previously identified as coding for components of the Swr1 (Krogan et al., 2003), TRAPP (Sacher et al., 2000), and KEOPS (Bianchi and Shore, 2006) complexes, respectively, but their molecular function is still unknown.
Table 2.
Genes Identified through ts Mutants that Affect CIN, and Quantification of the CIN Phenotype
| ORF Name | Gene Name | Description | ALF | BiM | CTF |
|---|---|---|---|---|---|
| YKR038C | KAE1 | A component of the KEOPS protein complex | 0b | 0c | 3d |
| YHR020W | Protein of unknown function | 32 | 4 | 3 | |
| YOR262W | Cytoplasmic protein of unknown function | 0 | 0 | 3 | |
| YDR246W | TRS23 | One of ten subunits of the transport protein particle (TRAPP) | 6 | 2 | 2 |
| YHR122W | Putative protein of unknown function | 23 | 3 | 3 | |
| YGR002C | SWC4 | Component of the Swr1p complex | 19 | 4 | 2 |
| YFR042W | KEG1 | Protein of unknown function | 34 | 6 | 2 |
| YNL152W | Protein of unknown function | 59 | 6 | 0 | |
| YKR071Ca | DRE2 | Protein of unknown function | 0 | 0 | 3 |
| YJL072Ca | PSF2 | Subunit of the GINS complex | 61 | 0 | 2 |
| YDR013Wa | PSF1 | Subunit of the GINS complex | 52 | 6 | 3 |
| YDR489Wa | SLD5 | Subunit of the GINS complex | 0 | 5 | 3 |
| YDR288Wa | NSE3 | Subunit of the Mms21-Smc5-Smc6 complex | 52 | 3 | 1 |
| YML023Ca | NSE5 | Subunit of the Mms21-Smc5-Smc6 complex | 0 | 6 | 0 |
| YDL105Wa | NSE4 | Subunit of the Mms21-Smc5-Smc6 complex | 18 | 4 | 1 |
These strains were used as a control for the CIN assays; see text for details.
Mutants were scored as positive if they exhibited >4-fold increase over wild-type frequency. For more details, see the Supplemental Experimental Procedures.
Homozygous ts mutants with an average of 2- to >5-fold increase in mating tests with both MATa and MATa testers were identified as bimaters. For more details, see the Supplemental Experimental Procedures.
Colony sectoring phenotypes were scored qualitatively by eye as mild, intermediate, and severe (indicated as 1, 2, and 3, respectively). For more details, see the Supplemental Experimental Procedures.
We wanted to estimate how frequently to expect a ts mutant (isolated via a random mutagenesis procedure) to subsequently reveal a detectable hypomorphic CIN phenotype at reduced (semipermissive) temperature. This addresses the issue of false negatives upon phenotypic screening of hypomorphs. To accomplish this, we generated ts alleles for PSF2 and NSE4 and tested for a CIN phenotype at a range of reduced temperatures. Psf2 represents a known subunit of the GINS complex (Sld5p, Psf1p, Psf2p, Psf3p) (Takayama et al., 2003). Loss of Psf2p function by depletion or deletion has been shown to cause chromosome missegregation in Schizosaccharomyces pombe and in human cells (Huang et al., 2005). Nse4 is known to interact with Nse3 and Nse5 as components of the Mms21-Smc5-Smc6 DNA repair complex. It has been suggested that Nse4 plays a role in maintenance of higher-order chromosome structure (Hu et al., 2005). As shown in Figure 4 and Table 2, newly constructed ts alleles for both psf2 and nse4 show the expected CIN phenotype at semipermissive temperatures. In addition, similar phenotypes were caused by newly constructed ts alleles for the other components of the GINS complex (Psf1 and Sld5) and the Mms21-Smc5-Smc6 complex (Nse3 and Nse5). Thus, we were able to isolate new ts alleles and subsequently detect a CIN phenotype at semipermissive temperatures for all alleles in six genes tested.
Four Previously Unidentified Essential Genes Involved in Sister Chromatid Cohesion
To test for sister chromatid cohesion defects in specific CIN mutants, each mutant was crossed to the tet repressor-GFP/tet operator repeat strain (Michaelis et al., 1997). The markers in this strain allow visualization of a specific chromosomal locus to assess whether sister chromatids are in juxtaposition or are separated. The resulting diploids were sporulated, and haploid strains containing the appropriate markers were identified and analyzed for cohesion defects.
In the wild-type, only 8% of cells arrested in G2/M had two GFP signals, indicating sister chromatids efficiently paired prior to anaphase. As expected (Mayer et al., 2001), in a ctf18 control strain, 35% of cells arrested in G2/M had two GFP signals, indicating a sister chromatid cohesion defect. Of nine new ts mutants exhibiting a CIN phenotype, four (yor262w, dre2, swc4, and yhr122w) were associated with sister chromatid cohesion defects with values ranging from 23% to 44% (Figure 5A). These defects were indeed due to sister chromatid cohesion defects as opposed to aneuploidy; only 4%–7% of the wild-type and ts mutant cells arrested in G1 had two GFP signals.
Figure 5. Identification of Genes Critical for Proper Sister Chromatid Cohesion.
(A) A secondary screen for ts mutants defective in sister chromatid cohesion. Cells were arrested in G1 or in G2/M. The number of GFP signals was scored in wild-type, ctf18 mutant, and the nine CIN mutants. The data shown represent the percentage of cells with two GFP signals. One hundred cells were counted for the G1-arrested cells and 300–400 cells for the G2/M-arrested cells.
(B and C) Sister chromatid cohesion mutants that are required for the establishment of sister chromatids: three samples were released from α factor arrest into nocodazole-containing media to allow arrest at G2/M (labeled with a stop sign). Samples 1 (black arrow), and 2 (red arrow) were released and arrested at 25°C or the semipermissive temperature, respectively. Sample 3 (blue arrow) completed its S phase at 25°C; however, upon the completion of S phase, it was shifted to the semipermissive temperature, to arrest at G2/M. Three hours after the arrest, the percentage of cells with two GFP signals was counted (100 cells). The results are presented in (C). In all cases, the numbers obtained for samples 1 and 3 are similar, indicative of a potential role in sister chromatid establishment.
YHR122W, SWC4, and YOR262W Are Required for Establishment of Sister Chromatid Cohesion
Sister chromatid cohesion is established during S phase and maintained until anaphase onset. Three mutants with sister chromatid cohesion defects (yhr122w, swc4, and yor262w) were used in temperature-shift experiments to assess roles in establishment or maintenance of cohesion. The experimental design is described in Figure 5B. When the gene of interest affects the establishment, the results obtained for sample 3 should resemble sample 1, whereas, for maintenance defects, results for sample 3 should resemble sample 2. For all three ts mutants, the values for the GFP signals in sample 3 are similar to those obtained in sample 1, not in sample 2 (Figure 5C). We conclude that these three essential genes may play a role in the establishment of sister chromatid cohesion.
A Resource of 250 Essential Gene ts Mutants
We expanded the initial collection of 45 ts mutants by an additional 205 essential genes. We decided to focus on only those essential genes for which no ts alleles are available to the yeast community. To assess whether a ts allele was available for each individual ORF, we referred to an updated list of ts alleles collected by the Boone laboratory (C. Boone, personal communication). This collection represents ~40% of the essential gene spectrum corresponding to 450 essential genes. These extant alleles were generated by the yeast community over the past 40 years and are now being transferred to the YKO genetic background in a non-bar-coded format (C. Boone, personal communication). Of the remaining 657 essential genes for which no ts alleles are currently available, we chose 131 genes with homology to a human sequence (E value range of 3.00E-05 to 1.00E-261) and 74 additional genes (chosen randomly) to add to the collection (see Table S4) and generated ts alleles for each of them. In total, 250 ts mutants in an array format are now available to the community upon request. For each ts allele, matched MATa and MATα strains are available.
DISCUSSION
We describe a method for making a comprehensive set of ts conditional mutants in each of the essential genes in Saccharomyces cerevisiae. To demonstrate feasibility and create an initial community resource, we generated a subset of ts alleles for essential genes with unknown function. We used biochemical (RNA processing) and genetic (chromosomal stability and sister chromatid cohesion) screens to identify new genes important for these essential functions. The initial collection of 45 ts mutants was expanded to 250 strains, in which essential genes with significant human homologs were prioritized. The collection is now ready for distribution.
Generating ts alleles for individual genes by conventional methods is tedious and not readily scalable genome-wide. The approach described here exploits features of the “haploid-convertible” heterozygous diploid collection, using the SGA reporter to select for an integrated ts-allele flanked by corresponding bar codes. We name this method the diploid shuffle to distinguish it from the standard plasmid shuffle method. The Topo-TA modified plasmid saves time and effort in the cloning step of the mutagenized library. The approach was validated for generating ts alleles; however, it can be used for other applications. Using the Topo-TA plasmid and protocol described (Figure 1), any extant ts allele can be easily transferred to the deletion collection genetic background (“allele transfer-in”). The result is an integrated allele, marked by URA3, and flanked by the appropriate bar codes (see Figure S3 for more details). Indeed, using the Topo-TA plasmid, any PCR product (mutant allele, fusion protein, heterologous gene expression cassette, etc.) can be introduced at any of the 6000 genomic sites carrying a KanMX replacement cassette as the integration site, depending on the specific heterozygote chosen as the recipient strain. In addition, by using primers external to the primers used in the original mutagenesis, the URA3-marked ts allele or gene construct can be easily transferred from the deletion set genetic background to any other strain of interest (“allele transfer-out,” see Figure S4). Thus, each ts mutation or gene construct can be analyzed for more specific phenotypes of interest in various genetic contexts.
In model organisms, the function of every known gene or predicted ORF can be analyzed by assessing a specific phenotype when the gene or ORF is deleted or inactivated. A most important milestone for yeast functional genomics was the complete set of deletion mutants (Giaever et al., 2002). This collection is widely used for direct screening for recessive phenotypes in haploids and for establishing genetic interactions in haploid double mutants. However, these systematic screens are currently possible only for the ~4700 nonessential genes. For essential genes, haploids carrying conditional or hypomorphic alleles are required. The two screens of the first-release ts allele resource described in this paper demonstrate the value of a systematic ts mutant resource.
A screen for ribosomal RNA processing netted 11 mutants affecting the process relatively severely and specifically. This considerably expands a list of approximately 110 genes previously identified using tet-regulated essential gene constructs and a microarray approach for detecting rRNA precursors and displaying increases in rRNA precursor abundance of at least 10-fold (Peng et al., 2003). In addition to two genes previously identified biochemically as components of the processome, nine additional genes were identified. Several gene products were implicated in other forms of RNA processing, implying crosstalk among central rRNA, pre-mRNA, snoRNA, and tRNA-processing pathways. Many of the identified genes had products localized to the nucleus or nucleolus as expected for proteins directly involved in rRNA processing, and many showed a significant defect in mature rRNA. However, for some of the mutants, the reduction in activity of the affected protein may well affect rRNA processing indirectly, e.g., affect processing of snoRNAs, which themselves direct rRNA processing.
Two mutants predicted to affect CoA biosynthesis strongly affected rRNA processing, suggesting a requirement for a CoA derivative for rRNA processing. Two possible explanations for this are that specific protein acetylation regulates rRNA processing, or, alternatively, free CoA or some other derivative might serve as a signal for the metabolic state of the cell. It would make sense if a decrease in readily usable cellular carbon sources downregulated ribosomes/translation machinery.
In this paper, we also describe CIN screening of the ts mutant collection resource. Over the past 20 years, phenotype-based screens for chromosome missegregation have generated a number of mutant collections (CTF, CHL, MCM, CIN), identifying many genes that functionally conserved in other eukaryotes (Hoyt et al., 1990; Kouprina et al., 1988; Maine et al., 1984; Meeks-Wagner et al., 1986; Ouspenski et al., 1999; Spencer et al., 1990; Yuen et al., 2007). Results of the extended effort to isolate mutants that affect chromosome stability could suggest that the yeast S. cerevisiae is close to saturation in this respect. Using a subset of ts alleles from our new collection, we isolated nine new CIN mutants. The original ctf collection identified ~50 genes necessary for high-fidelity segregation of chromosomes (Spencer et al., 1990). To date, 25 genes represented in the original ctf collection have been cloned and characterized, ten of which encode functions important for sister chromatid cohesion (40%). The connection between sister chromatid cohesion function and CIN was further emphasized in our secondary screen, in which four of nine new CIN mutants were associated with a cohesion defect (Figure 5A). We also found a potential role for three of the cohesion mutants at the establishment stage (Skibbens et al., 1999). Since Ctf7 was shown to be involved in the establishment of sister chromatid cohesion, no other essential genes with such a role have been described. Based on these results, we believe that an early release of a subset of 250 ts mutants will provide a valuable resource to the community and enrich the spectrum of mutants affecting various cellular functions.
The most obvious use of this resource is to study the function of individual genes, including poorly characterized ORFs. In a broader view, the collection can be used directly, with no other modifications, to complete genome-wide studies that used the nonessential gene deletion collection as a resource. Other applications include incorporation of hypomorphic alleles into the synthetic genetic network. The MATa version of the collection contains a uniform genetic background (see the Experimental Procedures), in which each of the strains contains a LEU2-marked SGA reporter. Therefore, the ts collection can be also used directly for synthetic genetic array (SGA) (Tong et al., 2001) or epistatic miniarray profiles (eMAPs) (Schuldiner et al., 2005). In both cases, the MATa strains can be provided in an array form to enrich the deletion set. In addition, each of the individual mutants can be used as a query for SGA analysis. An alternative approach to SGA is SLAM (Ooi et al., 2003). One of the unique features of our collection is that each of the integrated alleles is flanked with two bar codes. Since the growth rates of the ts mutants differ and the nonpermissive temperatures of different alleles vary, we believe that the diploid version of SLAM, dSLAM, is preferable (Pan et al., 2004). In addition, any of the ts mutants can be introduced as a query mutation by transformation to assess its synthetic lethal profile (using the allele transfer-out method, Figure S4).
Several approaches have been developed by the yeast community for the study of essential genes, which resulted in a few collections that are already available to the community: DAmP alleles (Schuldiner et al., 2005), the N-degron (Kanemaki et al., 2003; Labib et al., 2000), and the tet promoter collections (Mnaimneh et al., 2004). The main drawback of our approach is the fact that, following the integration of the mutagenized PCR product, a relatively laborious screening process is needed to isolate a ts allele. In the other methods, the final product is ready to use following one simple step of homologous recombination, avoiding such screening. However, our approach has an important advantage—it promises a high rate of success. If for a certain gene no ts is isolated at the first round, a second round of screening, or changing the mutagenesis conditions, will usually result in a positive isolate. The criterion for defining a mutant as ts is very clear (inability to grow at high temperatures), and most ts mutations isolated are in the gene of interest. For the other approaches, only one step is needed to get the modified gene, but the resultant allele is not always associated with a clear phenotype. For the tet promoter collection, rapid and/or sufficient elimination of protein function is problematic for a large fraction of proteins (Aparicio, 2003). In nearly half of the cases tested, shutting off the promoter did not affect phenotypes or gave only a very slight phenotype (Mnaimneh et al., 2004). When 104 essential genes were fused to the N-degron sequences, nearly 40% of them did not result in inviability at the nonpermissive temperature (Kanemaki et al., 2003), indicating that the rapid protein depletion by N-degron is not uniform across the proteome. Similarly, the DAmP approach works very well for only approximately one-third of essential genes and less well for the majority. For cases in which no effect is observed following the modification, experimental options are limited, and therefore the rate of success for each of these methods is lower than the one we obtained for ts alleles.
Genetic analysis of essential genes typically relies on conditional mutants. In many cases, ts alleles provide a wide spectrum of activity ranging from permissive to nonpermissive temperatures, providing flexibility in phenotype assessment. In the tet promoter strains, the promoter is either in its on or off state, making it difficult to achieve intermediate expression levels (Schuldiner et al., 2005). DAmP alleles are not conditional alleles—reduced expression is constitutive. Finally, in our method, the integrated allele is controlled by its natural promoter and terminator and the protein has a wild-type structure lacking any extensions or tags. For the tet and DAmP strains, the promoter and terminator are modified. In the degron system, the amino terminus of the encoded protein is fused with a 20 KD fragment, which may affect the function of the targeted protein.
Taking these considerations together, a comprehensive collection of ts missense mutations, in an integrated, bar-coded format, represents an excellent resource for analyzing essential gene function and will significantly enhance discovery, genome crossreferencing, and functional analysis.
EXPERIMENTAL PROCEDURES
For more details, see the Supplemental Experimental Procedures.
Yeast Strains
ts strains were derived from the yeast heterozygous gene deletion collection carrying the haploid selection marker can1Δ::LEU2-MFA1pr-HIS3 (Open Biosystems catalogue number YSC4428).
Plasmids
SB221 was modified starting from the plasmid M4758 (Voth et al., 2003) to allow more efficient targeted integration at the KanMX locus in the heterozygous deletion strains. The BamHII site of M4758 was modified with a Topo-TA site for easier recombinant cloning.
RNA-Processing Mutant Screens
Two independent clones from each mutant strain were grown at the permissive and nonpermissive temperature prior to harvesting pellets. RNA was prepared, and a standard northern blot procedure was preformed. RNA blots were probed with biotin-labeled riboprobes. Subsequently, the blots were reprobed with a probe containing one entire unit of yeast rDNA.
Construction of ts Mutants
ts strains were constructed as described in the Results; for detailed procedures, see the Supplemental Experimental Procedures.
CIN Assays
CTF, genomic BiM, and ALF were performed as previously described (Yuen et al., 2007).
Supplementary Material
Acknowledgments
We thank A. Carter, M. Tao, M. Kofoed, B. Le, and L. Tang for technical assistance; M. Kupiec, K. McManus, G. Simchen, and X. Pan for helpful discussions; and J. Woolford for helpful comments on an earlier version of the manuscript. Supported in part by National Institutes of Health (NIH) grant P01 CA16519 to P.H. and J.D.B., Canadian Institute for Health Research (CIHR) grant MOP-38096 to P.H., and NIH roadmap grant U54 RR020839 (to J.D.B.). S.B.-A. was supported by an Human Frontier Science Program (HFSP) long-term fellowship and by a research grant from the Killam trust.
Footnotes
Supplemental Data include Supplemental Experimental Procedures, four figures, and four tables and can be found with this article online at http://www.molecule.org/cgi/content/full/30/2/248/DC1/.
References
- Andersen JS, Lyon CE, Fox AH, Leung AK, Lam YW, Steen H, Mann M, Lamond AI. Directed proteomic analysis of the human nucleolus. Curr Biol. 2002;12:1–11. doi: 10.1016/s0960-9822(01)00650-9. [DOI] [PubMed] [Google Scholar]
- Aparicio OM. Tackling an essential problem in functional proteomics of Saccharomyces cerevisiae. Genome Biol. 2003;4:230. doi: 10.1186/gb-2003-4-10-230. Published online September 24, 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein KA, Gallagher JE, Mitchell BM, Granneman S, Baserga SJ. The small-subunit processome is a ribosome assembly intermediate. Eukaryot Cell. 2004;3:1619–1626. doi: 10.1128/EC.3.6.1619-1626.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianchi A, Shore D. The KEOPS complex: a rosetta stone for telomere regulation? Cell. 2006;124:1125–1128. doi: 10.1016/j.cell.2006.03.008. [DOI] [PubMed] [Google Scholar]
- Birrell GW, Giaever G, Chu AM, Davis RW, Brown JM. A genome-wide screen in Saccharomyces cerevisiae for genes affecting UV radiation sensitivity. Proc Natl Acad Sci USA. 2001;98:12608–12613. doi: 10.1073/pnas.231366398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleichert F, Granneman S, Osheim YN, Beyer AL, Baserga SJ. The PINc domain protein Utp24, a putative nuclease, is required for the early cleavage steps in 18S rRNA maturation. Proc Natl Acad Sci USA. 2006;103:9464–9469. doi: 10.1073/pnas.0603673103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burckin T, Nagel R, Mandel-Gutfreund Y, Shiue L, Clark TA, Chong JL, Chang TH, Squazzo S, Hartzog G, Ares M., Jr Exploring functional relationships between components of the gene expression machinery. Nat Struct Mol Biol. 2005;12:175–182. doi: 10.1038/nsmb891. [DOI] [PubMed] [Google Scholar]
- Combs DJ, Nagel RJ, Ares M, Jr, Stevens SW. Prp43p is a DEAH-box spliceosome disassembly factor essential for ribosome biogenesis. Mol Cell Biol. 2006;26:523–534. doi: 10.1128/MCB.26.2.523-534.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohmen RJ, Varshavsky A. Heat-inducible degron and the making of conditional mutants. Methods Enzymol. 2005;399:799–822. doi: 10.1016/S0076-6879(05)99052-6. [DOI] [PubMed] [Google Scholar]
- Fromont-Racine M, Senger B, Saveanu C, Fasiolo F. Ribosome assembly in eukaryotes. Gene. 2003;313:17–42. doi: 10.1016/s0378-1119(03)00629-2. [DOI] [PubMed] [Google Scholar]
- Gerring SL, Spencer F, Hieter P. The CHL 1 (CTF 1) gene product of Saccharomyces cerevisiae is important for chromosome transmission and normal cell cycle progression in G2/M. EMBO J. 1990;9:4347–4358. doi: 10.1002/j.1460-2075.1990.tb07884.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giaever G, Chu AM, Ni L, Connelly C, Riles L, Veronneau S, Dow S, Lucau-Danila A, Anderson K, Andre B, et al. Functional profiling of the Saccharomyces cerevisiae genome. Nature. 2002;418:387–391. doi: 10.1038/nature00935. [DOI] [PubMed] [Google Scholar]
- Hage AE, Tollervey D. A surfeit of factors: why is ribosome assembly so much more complicated in eukaryotes than bacteria? RNA Biol. 2004;1:10–15. [PubMed] [Google Scholar]
- Hartwell LH, Culotti J, Reid B. Genetic control of the cell-division cycle in yeast. I Detection of mutants. Proc Natl Acad Sci USA. 1970;66:352–359. doi: 10.1073/pnas.66.2.352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazbun TR, Malmstrom L, Anderson S, Graczyk BJ, Fox B, Riffle M, Sundin BA, Aranda JD, McDonald WH, Chiu CH, et al. Assigning function to yeast proteins by integration of technologies. Mol Cell. 2003;12:1353–1365. doi: 10.1016/s1097-2765(03)00476-3. [DOI] [PubMed] [Google Scholar]
- Hoyt MA, Stearns T, Botstein D. Chromosome instability mutants of Saccharomyces cerevisiae that are defective in microtubule-mediated processes. Mol Cell Biol. 1990;10:223–234. doi: 10.1128/mcb.10.1.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu B, Liao C, Millson SH, Mollapour M, Prodromou C, Pearl LH, Piper PW, Panaretou B. Qri2/Nse4, a component of the essential Smc5/6 DNA repair complex. Mol Microbiol. 2005;55:1735–1750. doi: 10.1111/j.1365-2958.2005.04531.x. [DOI] [PubMed] [Google Scholar]
- Huang HK, Bailis JM, Leverson JD, Gomez EB, Forsburg SL, Hunter T. Suppressors of Bir1p (Survivin) identify roles for the chromosomal passenger protein Pic1p (INCENP) and the replication initiation factor Psf2p in chromosome segregation. Mol Cell Biol. 2005;25:9000–9015. doi: 10.1128/MCB.25.20.9000-9015.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanemaki M, Sanchez-Diaz A, Gambus A, Labib K. Functional proteomic identification of DNA replication proteins by induced proteolysis in vivo. Nature. 2003;423:720–724. doi: 10.1038/nature01692. [DOI] [PubMed] [Google Scholar]
- Kouprina N, Pashina OB, Nikolaishwili NT, tsouladze AM, Larionov VL. Genetic control of chromosome stability in the yeast Saccharomyces cerevisiae. Yeast. 1988;4:257–269. doi: 10.1002/yea.320040404. [DOI] [PubMed] [Google Scholar]
- Krogan NJ, Keogh MC, Datta N, Sawa C, Ryan OW, Ding H, Haw RA, Pootoolal J, Tong A, Canadien V, et al. A Snf2 family ATPase complex required for recruitment of the histone H2A variant Htz1. Mol Cell. 2003;12:1565–1576. doi: 10.1016/s1097-2765(03)00497-0. [DOI] [PubMed] [Google Scholar]
- Kufel J, Dichtl B, Tollervey D. Yeast Rnt1p is required for cleavage of the pre-ribosomal RNA in the 3′ ETS but not the 5′ ETS. RNA. 1999;5:909–917. doi: 10.1017/s135583829999026x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labib K, Tercero JA, Diffley JF. Uninterrupted MCM2–7 function required for DNA replication fork progression. Science. 2000;288:1643–1647. doi: 10.1126/science.288.5471.1643. [DOI] [PubMed] [Google Scholar]
- Lebaron S, Froment C, Fromont-Racine M, Rain JC, Monsarrat B, Caizergues-Ferrer M, Henry Y. The splicing ATPase prp43p is a component of multiple preribosomal particles. Mol Cell Biol. 2005;25:9269–9282. doi: 10.1128/MCB.25.21.9269-9282.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leeds NB, Small EC, Hiley SL, Hughes TR, Staley JP. The splicing factor Prp43p, a DEAH box ATPase, functions in ribosome biogenesis. Mol Cell Biol. 2006;26:513–522. doi: 10.1128/MCB.26.2.513-522.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindahl L, Archer RH, Zengel JM. A new rRNA processing mutant of Saccharomyces cerevisiae. Nucleic Acids Res. 1992;20:295–301. doi: 10.1093/nar/20.2.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lum PY, Armour CD, Stepaniants SB, Cavet G, Wolf MK, Butler JS, Hinshaw JC, Garnier P, Prestwich GD, Leonardson A, et al. Discovering modes of action for therapeutic compounds using a genome-wide screen of yeast heterozygotes. Cell. 2004;116:121–137. doi: 10.1016/s0092-8674(03)01035-3. [DOI] [PubMed] [Google Scholar]
- Maine GT, Sinha P, Tye BK. Mutants of S. cerevisiae defective in the maintenance of minichromosomes. Genetics. 1984;106:365–385. doi: 10.1093/genetics/106.3.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer ML, Gygi SP, Aebersold R, Hieter P. Identification of RFC(Ctf18p, Ctf8p, Dcc1p): an alternative RFC complex required for sister chromatid cohesion in S. cerevisiae. Mol Cell. 2001;7:959–970. doi: 10.1016/s1097-2765(01)00254-4. [DOI] [PubMed] [Google Scholar]
- Meeks-Wagner D, Wood JS, Garvik B, Hartwell LH. Isolation of two genes that affect mitotic chromosome transmission in S. cerevisiae. Cell. 1986;44:53–63. doi: 10.1016/0092-8674(86)90484-8. [DOI] [PubMed] [Google Scholar]
- Michaelis C, Ciosk R, Nasmyth K. Cohesins: chromosomal proteins that prevent premature separation of sister chromatids. Cell. 1997;91:35–45. doi: 10.1016/s0092-8674(01)80007-6. [DOI] [PubMed] [Google Scholar]
- Mizuta K, Warner JR. Continued functioning of the secretory pathway is essential for ribosome synthesis. Mol Cell Biol. 1994;14:2493–2502. doi: 10.1128/mcb.14.4.2493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mnaimneh S, Davierwala AP, Haynes J, Moffat J, Peng WT, Zhang W, Yang X, Pootoolal J, Chua G, Lopez A, et al. Exploration of essential gene functions via titratable promoter alleles. Cell. 2004;118:31–44. doi: 10.1016/j.cell.2004.06.013. [DOI] [PubMed] [Google Scholar]
- O’Connor JP, Peebles CL. PTA1, an essential gene of Saccharomyces cerevisiae affecting pre-tRNA processing. Mol Cell Biol. 1992;12:3843–3856. doi: 10.1128/mcb.12.9.3843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ooi SL, Shoemaker DD, Boeke JD. A DNA microarray-based genetic screen for nonhomologous end-joining mutants in Saccharomyces cerevisiae. Science. 2001;294:2552–2556. doi: 10.1126/science.1065672. [DOI] [PubMed] [Google Scholar]
- Ooi SL, Shoemaker DD, Boeke JD. DNA helicase gene interaction network defined using synthetic lethality analyzed by microarray. Nat Genet. 2003;35:277–286. doi: 10.1038/ng1258. [DOI] [PubMed] [Google Scholar]
- Ouspenski II, Elledge SJ, Brinkley BR. New yeast genes important for chromosome integrity and segregation identified by dosage effects on genome stability. Nucleic Acids Res. 1999;27:3001–3008. doi: 10.1093/nar/27.15.3001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan X, Yuan DS, Xiang D, Wang X, Sookhai-Mahadeo S, Bader JS, Hieter P, Spencer F, Boeke JD. A robust toolkit for functional profiling of the yeast genome. Mol Cell. 2004;16:487–496. doi: 10.1016/j.molcel.2004.09.035. [DOI] [PubMed] [Google Scholar]
- Peng WT, Robinson MD, Mnaimneh S, Krogan NJ, Cagney G, Morris Q, Davierwala AP, Grigull J, Yang X, Zhang W, et al. A panoramic view of yeast noncoding RNA processing. Cell. 2003;113:919–933. doi: 10.1016/s0092-8674(03)00466-5. [DOI] [PubMed] [Google Scholar]
- Rempola B, Karkusiewicz I, Piekarska I, Rytka J. Fcf1p and Fcf2p are novel nucleolar Saccharomyces cerevisiae proteins involved in pre-rRNA processing. Biochem Biophys Res Commun. 2006;346:546–554. doi: 10.1016/j.bbrc.2006.05.140. [DOI] [PubMed] [Google Scholar]
- Sacher M, Barrowman J, Schieltz D, Yates JR, 3rd, Ferro-Novick S. Identification and characterization of five new subunits of TRAPP. Eur J Cell Biol. 2000;79:71–80. doi: 10.1078/S0171-9335(04)70009-6. [DOI] [PubMed] [Google Scholar]
- Samanta MP, Liang S. Predicting protein functions from redundancies in large-scale protein interaction networks. Proc Natl Acad Sci USA. 2003;100:12579–12583. doi: 10.1073/pnas.2132527100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuldiner M, Collins SR, Thompson NJ, Denic V, Bhamidipati A, Punna T, Ihmels J, Andrews B, Boone C, Greenblatt JF, et al. Exploration of the function and organization of the yeast early secretory pathway through an epistatic miniarray profile. Cell. 2005;123:507–519. doi: 10.1016/j.cell.2005.08.031. [DOI] [PubMed] [Google Scholar]
- Shoemaker DD, Lashkari DA, Morris D, Mittmann M, Davis RW. Quantitative phenotypic analysis of yeast deletion mutants using a highly parallel molecular bar-coding strategy. Nat Genet. 1996;14:450–456. doi: 10.1038/ng1296-450. [DOI] [PubMed] [Google Scholar]
- Sikorski RS, Boeke JD. In vitro mutagenesis and plasmid shuffling: from cloned gene to mutant yeast. Methods Enzymol. 1991;194:302–318. doi: 10.1016/0076-6879(91)94023-6. [DOI] [PubMed] [Google Scholar]
- Simchen G. Cell cycle mutants. Annu Rev Genet. 1978;12:161–191. doi: 10.1146/annurev.ge.12.120178.001113. [DOI] [PubMed] [Google Scholar]
- Skibbens RV, Corson LB, Koshland D, Hieter P. Ctf7p is essential for sister chromatid cohesion and links mitotic chromosome structure to the DNA replication machinery. Genes Dev. 1999;13:307–319. doi: 10.1101/gad.13.3.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer F, Gerring SL, Connelly C, Hieter P. Mitotic chromosome transmission fidelity mutants in Saccharomyces cerevisiae. Genetics. 1990;124:237–249. doi: 10.1093/genetics/124.2.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takayama Y, Kamimura Y, Okawa M, Muramatsu S, Sugino A, Araki H. GINS, a novel multiprotein complex required for chromosomal DNA replication in budding yeast. Genes Dev. 2003;17:1153–1165. doi: 10.1101/gad.1065903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong AH, Evangelista M, Parsons AB, Xu H, Bader GD, Page N, Robinson M, Raghibizadeh S, Hogue CW, Bussey H, et al. Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science. 2001;294:2364–2368. doi: 10.1126/science.1065810. [DOI] [PubMed] [Google Scholar]
- Tsai RT, Fu RH, Yeh FL, Tseng CK, Lin YC, Huang YH, Cheng SC. Spliceosome disassembly catalyzed by Prp43 and its associated components Ntr1 and Ntr2. Genes Dev. 2005;19:2991–3003. doi: 10.1101/gad.1377405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venema J, Tollervey D. Ribosome synthesis in Saccharomyces cerevisiae. Annu Rev Genet. 1999;33:261–311. doi: 10.1146/annurev.genet.33.1.261. [DOI] [PubMed] [Google Scholar]
- Voth WP, Jiang YW, Stillman DJ. New ‘marker swap’ plasmids for converting selectable markers on budding yeast gene disruptions and plasmids. Yeast. 2003;20:985–993. doi: 10.1002/yea.1018. [DOI] [PubMed] [Google Scholar]
- Winzeler EA, Lee B, McCusker JH, Davis RW. Whole genome genetic-typing in yeast using high-density oligonucleotide arrays. Parasitology (Suppl) 1999;118:S73–S80. doi: 10.1017/s0031182099004047. [DOI] [PubMed] [Google Scholar]
- Yuen KW, Warren CD, Chen O, Kwok T, Hieter P, Spencer FA. Systematic genome instability screens in yeast and their potential relevance to cancer. Proc Natl Acad Sci USA. 2007;104:3925–3930. doi: 10.1073/pnas.0610642104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Kessler M, Helmling S, O’Connor JP, Moore C. Pta1, a component of yeast CF II, is required for both cleavage and poly(A) addition of mRNA precursor. Mol Cell Biol. 1999;19:7733–7740. doi: 10.1128/mcb.19.11.7733. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.