Abstract
Background
Ionizing radiation causes the generation of damaging reactive oxygen species that lead to cellular damage and death. Organisms such as Deinococcus radiodurans have evolved mechanisms for extreme resistance to ionizing radiation, and resistance has been shown to be a consequence of protection of critical proteins from oxidative inactivation.
Objectives
D. radiodurans accumulates high levels of manganese and of small peptides that together are protective. Our aim was to rationally design antioxidant peptides.
Methods
Amino acid analysis was utilized to determine the rates of loss of the 20 amino acids exposed to varying doses of irradiation. The activity of glutamine synthetase and methionine sulfoxide reductase was assayed to follow their inactivation by irradiation.
Results
The ability of an amino acid to protect enzymes from inactivation by ionizing radiation paralleled its sensitivity to ionizing radiation. Based on this observation and the ability of histidine to confer water solubility, we synthesized the hexapeptide His-Met-His-Met-His-Met and found that it provided markedly increased protection against irradiation.
Discussion
Small peptides containing histidine and methionine were readily soluble and provided enzymes with remarkable protection from inactivation by ionizing radiation.
Keywords: Ionizing radiation, Radioprotection, Amino acid oxidation, Methionine, Histidine
Ionizing radiation kills cells and organisms through induction of oxidative stress, produced via direct generation of reactive species or by generation in the cell as a response to the radiation. A delivered dose of ∼200 Gy is sufficient to kill an Escherichia coli1 and 5–10 Gy a human, but some organisms are resistant to very high doses. These radiation-resistant organisms are often resistant to other stresses, such as dessication. This is the case for the freshwater invertebrate bdelloid rotifers2 and the bacterium, Deinococcus radiodurans.3 D. radiodurans can survive radiation doses greater than 10 000 Gy, and the basis of its radioresistance has been the subject of numerous investigations and many reviews.4–8 For over 50 years it has been considered established that ionizing and ultraviolet irradiation kill cells by damaging their DNA, a mechanism most clearly and emphatically stated by Hutchinson in 1966.9 However, the DNA of D. radiodurans is not resistant to ionizing radiation. For any given dose, D. radiodurans incurs about the same number of double strand breaks as do radiation-sensitive bacteria.7,8 What distinguishes D. radiodurans from radiosensitive organisms is its impressive ability to repair the DNA damage through homologous recombination.10 Yet the enzymes that mediate DNA repair in D. radiodurans are not unusual either. For example, mutation of DNA polymerase I renders D. radiodurans very sensitive to irradiation, but resistance is restored by expression of the E. coli DNA polymerase I.11
Recent studies by Daly et al.8 led them to propose that the basis for radioresistance is the ability of D. radiodurans to prevent oxidative inactivation of its proteins. The logic can be put rather simply. Repair of the DNA damage suffered by D. radiodurans can only be effected by enzymes, which of course are proteins. Proteins have long been known to be damaged and functionally disabled by irradiation.12–15 If proteins essential to the DNA repair process are inactivated by irradiation, there can be no DNA repair. Thus, while the DNA of D. radiodurans is fragmented by irradiation, it is repaired because the cell's proteins have been protected from inactivation by irradiation. Subsequent experimental investigations and modeling studies support the proposition that the radioresistance of D. radiodurans is due to protection of its proteins, not its DNA, from oxidative damage.16,17
What molecules provide this protection? Bruce and colleagues18 showed in 1976 that D. radiodurans accumulated large amounts of manganese, and they speculated that the manganese might be important in the organism's resistance to ultraviolet radiation. Later, Daly et al.19 established experimentally that the manganese was important for resistance to ionizing radiation. Working with Daly more recently, we undertook a classical biochemical approach to identify molecules required for radiation resistance.1 A cell-free extract of D. radiodurans protected purified proteins and also protected proteins in an E. coli homogenate from oxidative damage. After several purification steps, the protective activity was localized to a protein-free, low molecular weight fraction. The fraction contained a number of components including manganese and many small peptides derived from a variety of D. radiodurans proteins. A synthetic decapeptide and millimolar manganese were then shown to protect purified enzymes from inactivation by irradiation. The investigations reported in this paper were undertaken to provide a rational basis for the design of peptides with increased protective ability.
Materials and methods
Materials
Phosphate-buffered saline, 10 × , was purchased from KD Medical (Columbia, MD, USA). It contained 9 g NaCl, 1.44 g KH2PO4, and 7.95 g Na2HPO4 per liter. When diluted 10-fold, its pH was 7.4. The decapeptide, DEHGTAVMLK, and its scrambled version, THMVLAKGED, were synthesized by American Peptide Co. (Vista, CA, USA) and by Elim Biopharmaceuticals (Hayward, CA, USA); the Elim peptides were a gift from Heather Pangburn and Thomas Lamkin of the Air Force Research Laboratory, Wright Patterson Air Force Base. American Peptide Co. synthesized HM, HMHM, and HMHMHM.
Amino acids were from Sigma (St Louis, MO, USA).
Glutamine synthetase was purified20 from E. coli pgln6/YMC1021 and assayed with the pH 7.57 triethanolamine-dimethyglutarate buffer system.22 Recombinant mouse methionine sulfoxide reductase A was a gift from Geumsoo Kim of our laboratory, prepared and assayed as described.23
Irradiation
Irradiations were performed in a JL Shepherd and Associates 60Co irradiator, model 484R-2 (San Fernando, CA, USA). The dose rate was calculated at the beginning of each experiment and was in the range of 150 Gy/min. One hundred microliters air-saturated solution was placed in a 1 ml screw cap vial (Agilent Technologies #5182-0715, Santa Clara, CA, USA) fitted with a PTFE/silicone septum cap (Agilent 5190-3156), and irradiated on ice. In experiments in which the dose of radiation was varied, a separate vial was incubated for each dose. Glutamine synthetase was 280 µg/ml in 25 mM K2HPO4, 100 mM KCl, 10 mM MgCl2, pH to 7.4. Methionine sulfoxide reductase A was 125–150 µg/ml in phosphate-buffered saline, pH 7.4.
Amino acid analysis
Analysis of amino acids and of proteins was performed with and without incubation with CNBr in order to quantitate both methionine and methionine sulfoxide. The procedures were performed as described24 except that the column was a Zorbax Eclipse AAA (#966400-902) fitted with a guard column (Agilent #820950-931), and the gradient program was from Agilent.25
Statistical analysis
All results reported in this paper are the average and standard deviation of at least two replicate experiments performed on separate days. Differences were assessed by Student's t-test.
Results
Protection is not dependent on sequence
In a previous study, we synthesized a decapeptide with sequence DEHGTAVMLK to test whether it could protect enzymes from inactivation during irradiation.1 The amino acid content of the peptide was selected to match that of peptides present in the protective fraction prepared from D. radiodurans, but the sequence was chosen arbitrarily. The decapeptide provided the enzymes BamH1 and glutamine synthetase with remarkable protection against inactivation by irradiation, and protection was enhanced by addition of millimolar manganese. We have now compared protection provided by the original decapeptide with that of a decapeptide in which the sequence was scrambled to THMVLAKGED (Fig. 1). The two peptides were equally protective, leading us to conclude that composition and not sequence is of primary importance.
Figure 1.
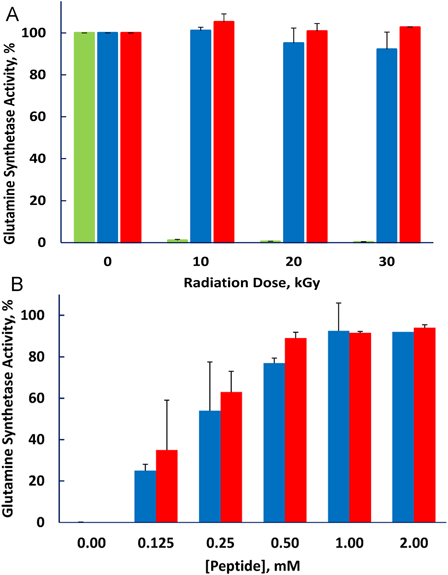
Protection of glutamine synthetase from irradiation is not dependent on the sequence of the decapeptide. 1 mM MnCl2 was present in all samples. (A) Peptide concentration fixed at 3 mM and radiation dose varied. (B) Radiation dose fixed at 10 kGy and peptide concentration varied. No peptide, green bar; Standard sequence DEHGTAVMLK, blue bar; Scrambled peptide, THMVLAKGED, red bar.
Determinants of protection efficiency
The protective efficiency of the various amino acids should correlate with the efficiency of their scavenging of reactive species produced by irradiation. We measured the reactivity of the 20 amino acids typically present in proteins by determining the fraction that was modified by exposure to 5 kGy (Fig. 2). Methionine, the aromatic amino acids histidine, tyrosine, tryptophan, and phenylalanine, along with cysteine were the most reactive while the hydrophilic amino acids were the least reactive. We then investigated the ability of the 20 amino acids to protect glutamine synthetase from inactivation during irradiation (Fig. 3). The aromatic amino acids, histidine, phenylalanine, tryptophan, and tyrosine, and the sulfur containing amino acids, cysteine and methionine, were by far more effective than the other 14. Thus, the protective ability of these individual amino acids paralleled their susceptibility to oxidation. However, the branched chain amino acids, isoleucine, leucine, and valine, were relatively susceptible to irradiation but were poorly protective of glutamine synthetase. We do not have a mechanistic explanation for this dichotomy. Methionine sulfone, whose sulfur cannot be further oxidized, did not protect glutamine synthetase, and methionine sulfoxide was less protective than methionine (Fig. 4A). The notable ability of methionine to protect a protein from oxidative inactivation is consistent with its incorporation into proteins as an antioxidant.26,27
Figure 2.
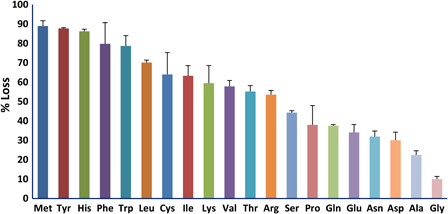
Sensitivity of individual amino acids to irradiation. Both the amino acid and MnCl2 concentrations were 0.5 mM. The irradiation dose was 5 kGy.
Figure 3.
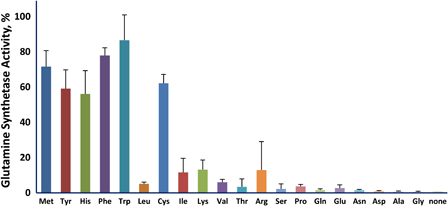
Ability of individual amino acids to protect glutamine synthetase from inactivation. The amino acid concentration was 3.0 mM and that of MnCl2 was 1.0 mM. The irradiation dose was 20 kGy.
Figure 4.
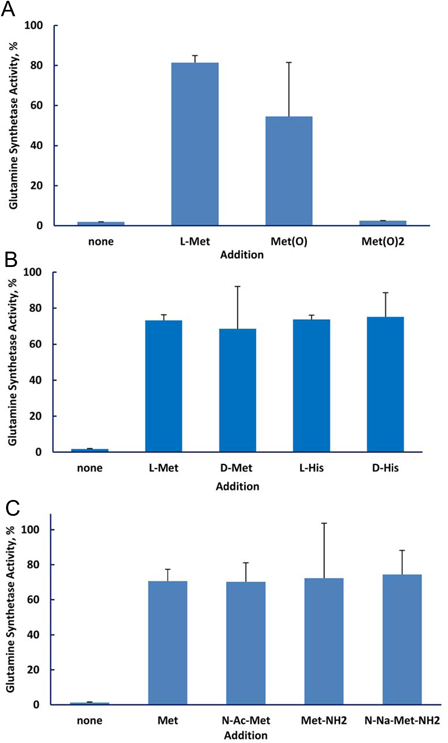
Protection of glutamine synthetase by amino acids. The amino acid concentration was 3.0 mM and that of MnCl2 was 1.0 mM. (A) Methionine and its oxidation products. The irradiation dose was 10 kGy. (B) d and l amino acids. The irradiation dose was 20 kGy. (C) Methionine and its amino or carboxyl modified forms. The irradiation dose was 20 kGy.
We also found that the d and l forms of the amino acid were equally protective (Fig. 4B). Thus, employing the d form might be useful when it is desired to protect cells with a compound that would be poorly taken up by the cells or would be relatively inert metabolically. The status of the amino and carboxyl groups did not affect the ability of methionine to protect glutamine synthetase: There was no difference in protection by methionine, N-acetyl methionine, methionine amide, and N-acetyl methionine amide (Fig. 4C).
Mechanism of manganese effect
Manganese enhances the protective effect of the decapeptide, DEHGTAVMLK, for glutamine synthetase and BamH1 at modest irradiation doses.1 At least three mechanisms for the manganese effect may be operative: (1) Manganese scavenges superoxide, a reaction that is greatly stimulated by bicarbonate/CO2.28,29 (2) Manganese, again in a bicarbonate-dependent reaction, forms a complex with amino acids and peptides that catalytically scavenges hydrogen peroxide.30 (3) Manganese can bind directly to divalent cation binding sites that are common in enzymes, and it typically binds with a higher affinity than does magnesium.31 We previously compared protection by manganese alone or manganese with decapeptide in phosphate buffer or bicarbonate/carbon dioxide buffer under conditions known to produce compounds capable of scavenging reactive oxygen species in a bicarbonate-dependent reaction.30,32 The bicarbonate/carbon dioxide buffer did not enhance protection (Fig. 4E1), arguing against operation of the scavenging mechanisms (1) and (2). To probe whether the direct binding of manganese to the protein was protective, we compared the protective effect of histidine with and without manganese on the inactivation of two enzymes, glutamine synthetase and methionine sulfoxide reductase. Glutamine synthetase has two well defined binding sites for divalent cations while methionine sulfoxide reductase has no known divalent cation binding site.33 If binding of manganese to the protein were the only mechanism by which manganese enhances protection, then we should observe manganese-dependent protection of glutamine synthetase but not of methionine sulfoxide reductase. That is what was observed (Fig. 5): addition of manganese enhanced the radioprotection provided by histidine to glutamine synthetase but not to methionine sulfoxide reductase, consistent with a protective mechanism mediated simply by binding to the cation binding site on glutamine synthetase.
Figure 5.
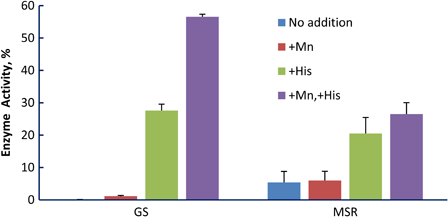
Effect of manganese and/or histidine on protection of enzymes from irradiation damage. When added, manganese was 1 mM and histidine was 3 mM. Enzymes were irradiated to cause ∼75% loss of activity in the presence of histidine alone in order to facilitate detection of a manganese effect in either direction. For glutamine synthetase (GS), the dose was 20 kGy and for methionine sulfoxide reductase (MSR), it was 10 kGy. At these high doses, manganese alone provides no protection.1 In the presence of histidine, manganese significantly increased the protection of glutamine synthetase (P < 0.01) but not of methionine sulfoxide reductase (P = 0.30).
Effect of peptide linkage and design of a peptide with improved protective ability
If the protective effect of the amino acids in a peptide were additive, then a given concentration of the peptide would be as effective as the same concentration of free amino acids. There are situations where that may be advantageous, for example, when one wants to provide protection without exposure to a substantial increase in osmolality. We synthesized peptides containing two of the most protective amino acids, His and Met, with the expectation that inclusion of histidine would increase the solubility of the peptides. The protective ability of the free amino acids and those in peptide linkage was determined by the total concentration of His and Met (Fig. 6). Thus, 0.5 mM of the hexamer, His-Met-His-Met-His-Met, was as protective as 3 mM total free amino acids. Glutamine synthetase has an allosteric site that binds l-histidine,34 but this result and the equal protection provided by d- and l-histidine indicate that binding to the allosteric site is not involved in the protective mechanism.
Figure 6.
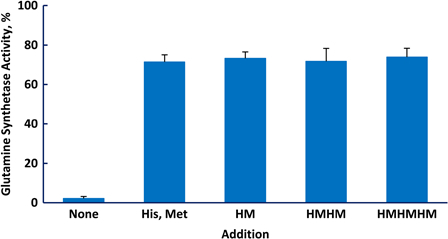
Protection of glutamine synthetase from inactivation by methionine and histidine, either free or in peptide linkage. Each incubation contained 1.5 mM histidine, 1.5 mM methionine, and 1.0 mM MnCl2. The irradiation dose was 20 kGy.
The major reason for performing the studies reported above was to provide a rational basis for the design of peptides with even greater protection than that provided by the original decapeptide.1 We therefore compared the protective effect for glutamine synthetase of 10 mM total amino acids provided by either 1.0 mM decapeptide (DEHGTAVMLK) or 1.6 mM of the hexapeptide HMHMHM (Fig. 7). As expected, the decapeptide was highly protective, increasing the irradiation dosage required for half-inactivation (D50) from 2 to 33 kGy. The His/Met containing hexapeptide was remarkably more effective, increasing the D50 to 112 kGy.
Figure 7.
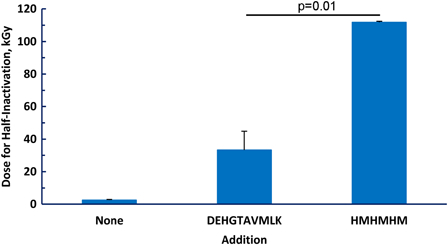
Protection of glutamine synthetase by peptides. Inactivation of the enzyme was measured at doses of irradiation from 0 to 30 kGy. The inactivation curve was fit to a first-order process by regression analysis with Prism for Windows version 5 (GraphPad Software, La Jolla, CA, USA). The fit equation was then used to extrapolate to the irradiation dose required for a 50% loss of activity (half-inactivation). When added, DEHGTAVMLK was 1.0 mM and HMHMHM was 1.7 mM, giving a total of 10 mM of amino acid residues.
Discussion
The partially purified fraction prepared from D. radiodurans that had strong radioprotective properties was rich in peptides, with the sum of the concentrations of methionine, histidine, and tyrosine exceeding 3 mM.1 This finding prompted us to synthesize the decapeptide DEHGTAVMLK. Its amino acid composition was selected to include most of the prevalent amino acids in the protective fraction, but the sequence was chosen arbitrarily. The scrambled peptide tested in this study was equally protective, indicating that the amino acid composition determines protection. We thought it would be possible to improve the efficiency of protection by altering the composition of the peptide to include amino acids most sensitive to irradiation because these would be the best scavengers. There is a vast literature reporting studies on radioprotective compounds, on irradiation of proteins, peptides, and amino acids, and reporting kinetic data for reactive species produced by irradiation – under a variety of experimental conditions.35 We utilized amino acid analysis to provide a simple mechanism for assessing protective efficacy under the specific irradiation conditions used in studies with Deinococcus and other cells.
The two sulfur containing amino acids and the four aromatic amino acids were most protective of glutamine synthetase activity, with or without added Mn2+. These were also the amino acids most degraded by a given irradiation dose. However, the branched chain amino acids, isoleucine, leucine, and valine, were also highly degraded but were poorly protective of glutamine synthetase, suggesting that the particular reactive species that inactivates glutamine synthetase is especially well scavenged by the aromatic and sulfur-containing amino acids. An alternative explanation is that inactivation of glutamine synthetase can be effected by certain products of irradiation of the branched chain amino acids such as hydroperoxides or aldehydes. We tested this possibility by exposing solutions of 1 mM manganese and 3 mM of an amino acid (histidine, methionine, leucine, isoleucine, or valine) to 20 kGy irradiation. Glutamine synthetase was then added to the solutions and allowed to stand for 1 hour at 4 °C before assay. None of the irradiated amino acid solutions affected glutamine synthetase activity.
Manganese alone at 1 mM provided slight protection; addition of manganese to 3 mM amino acid or peptide improved the protective ability of the amino acids or peptides. Comparison of the manganese effects on glutamine synthetase to that on methionine sulfoxide reductase provided evidence that protection by manganese requires binding of manganese to defined divalent cation sites on the proteins. Glutamine synthetase has such sites while methionine sulfoxide reductase does not, and only glutamine synthetase was provided additional protection by manganese. The mechanism of protection is thought to be by prevention of binding of iron to the cation site.36 The binding of metals such as magnesium, zinc, calcium, and manganese to divalent cation binding sites is dynamic, with the fraction of time that the site is occupied by that metal cation determined by the affinity of the site. Thus, some fraction of time the site is temporarily unoccupied. Adventitious iron, which is ubiquitous, may bind to the site during that period. In the presence of oxygen and, especially, reactive oxygen species such as hydrogen peroxide, Fenton chemistry will generate a reactive species. Because the reactive species is generated at the essential cation binding site, the site is damaged and the enzyme is inactivated or the protein loses structural integrity and cannot function properly. Although the oxidation is usually mediated by a free radical, it occurs in the ‘cage’ formed by the cation binding site and is thus site specific.36 We conclude that manganese enhances protection from irradiation by binding to pre-exisiting divalent cation binding sites on proteins. We suggest that this is the likely mechanism by which the high intracellular concentration of manganese in D. radiodurans contributes to the organism's radioresistance because almost all of the enzymes involved in DNA repair have an essential divalent cation binding site at their active sites.
The sulfur-containing and aromatic amino acids provided impressive protection to glutamine synthetase against ionizing radiation. This is consistent with the recognized ability of cysteine in glutathione and proteins and of methionine in proteins to provide antioxidant protection to the cell.26,37 They are superior to the arbitrarily chosen peptide composition used in our earlier studies that itself provided impressive protection.1 Practical applications have already been developed. Gaidamakova et al.38 showed that viruses and bacteria could be sterilized by high-dose irradiation while preserving the antigenic integrity of the organism's proteins, providing a novel approach to vaccine development.
D. radiodurans is resistant to many stresses besides ionizing irradiation. These include UV radiation, dessication, DNA damaging agents, and reactive species such as hydrogen peroxide.7,39 The common feature of all of these stresses is that they are mediated through reactive oxygen species.7 Thus, the sulfur-containing and aromatic amino acids, whether free or in peptide linkage, and whether the d or l forms, may also protect against these other stresses.
Funding
This research was supported by the Intramural Research Program of the National Heart, Lung, and Blood Institute.
References
- 1.Daly MJ, Gaidamakova EK, Matrosova VY, Kiang JG, Fukumoto R, Lee DY,. et al. Small-molecule antioxidant proteome-shields in Deinococcus radiodurans. PLoS One 2010;5(9):e12570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gladyshev E, Meselson M. Extreme resistance of bdelloid rotifers to ionizing radiation. Proc Natl Acad Sci USA 2008;105(13):5139–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anderson AW, Nordon HC, Cain RF, Duggan D. Studies on a radio-resistant micrococcus. I. Isolation, morphology, cultural characteristics, and resistance to gamma radiation. Food Technol 1956;10:575–8. [Google Scholar]
- 4.Cox MM, Battista JR. Deinococcus radiodurans – the consummate survivor. Nat Rev Microbiol 2005;3(11):882–92. [DOI] [PubMed] [Google Scholar]
- 5.Daly MJ. Modulating radiation resistance: insights based on defenses against reactive oxygen species in the radioresistant bacterium Deinococcus radiodurans. Clin Lab Med 2006;26(2):491–504. [DOI] [PubMed] [Google Scholar]
- 6.Minton KW. Repair of ionizing-radiation damage in the radiation resistant bacterium Deinococcus radiodurans. Mutat Res 1996;363(1):1–7. [DOI] [PubMed] [Google Scholar]
- 7.Slade D, Radman M. Oxidative stress resistance in Deinococcus radiodurans. Microbiol Mol Biol Rev 2011;75(1):133–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Daly MJ, Gaidamakova EK, Matrosova VY, Vasilenko A, Zhai M, Leapman RD,. et al. Protein oxidation implicated as the primary determinant of bacterial radioresistance. PLoS Biol 2007;5(4):e92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hutchinson F. The molecular basis for radiation effects on cells. Cancer Res 1966;26(9):2045–52. [PubMed] [Google Scholar]
- 10.Daly MJ, Minton KW. Interchromosomal recombination in the extremely radioresistant bacterium Deinococcus radiodurans. J Bacteriol 1995;177(19):5495–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gutman PD, Fuchs P, Minton KW. Restoration of the DNA damage resistance of Deinococcus radiodurans DNA polymerase mutants by Escherichia coli DNA polymerase I and Klenow fragment. Mutat Res 1994;314(1):87–97. [DOI] [PubMed] [Google Scholar]
- 12.Hardy WB. The action of salts of radium upon globulins. J Physiol 1903;29:xxix–xxx. [Google Scholar]
- 13.Arnow LE. Effects produced by the irradiation of proteins and amino acids. Physiol Rev 1936;16(4):671–85. [Google Scholar]
- 14.Barron ESG, Ambrose J, Johnson P. Studies on the mechanism of action of ionizing radiations XIII. The effect of X-irradiation on some physico-chemical properties of amino acids and proteins. Radiat Res 1955;2(2):145–58. [PubMed] [Google Scholar]
- 15.Garrison WM. Reaction mechanisms in radiolysis of peptides, polypeptides, and proteins. Chem Rev 1987;87:381–98. [Google Scholar]
- 16.Krisko A, Radman M. Protein damage and death by radiation in Escherichia coli and Deinococcus radiodurans. Proc Natl Acad Sci USA 2010;107(32):14373–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shuryak I, Brenner DJ. Mechanistic analysis of the contributions of DNA and protein damage to radiation-induced cell death. Radiat Res 2012;178(1):17–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leibowitz PJ, Schwartzberg LS, Bruce AK. The in vivo association of manganese with the chromosome of Micrococcus radiodurans. Photochem Photobiol 1976;23(1):45–50. [DOI] [PubMed] [Google Scholar]
- 19.Daly MJ, Gaidamakova EK, Matrosova VY, Vasilenko A, Zhai M, Venkateswaran A,. et al. Accumulation of Mn(II) in Deinococcus radiodurans facilitates gamma-radiation resistance. Science 2004;306(5698):1025–8. [DOI] [PubMed] [Google Scholar]
- 20.Miller RE, Shelton E, Stadtman ER. Zinc-induced paracrystalline aggregation of glutamine synthetase. Arch Biochem Biophys 1974;163:155–71. [DOI] [PubMed] [Google Scholar]
- 21.Backman K, Chen YM, Magasanik B. Physical and genetic characterization of the glnA–glnG region of the Escherichia coli chromosome. Proc Natl Acad Sci USA 1981;78(6):3743–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stadtman ER, Smyrniotis PZ, Davis JN, Wittenberger ME. Enzymic procedures for determining the average state of adenylylation of Escherichia coli glutamine synthetase. Anal Biochem 1979;95(1):275–85. [DOI] [PubMed] [Google Scholar]
- 23.Kim G, Cole NB, Lim JC, Zhao H, Levine RL. Dual sites of protein initiation control the localization and myristoylation of methionine sulfoxide reductase A. J Biol Chem 2010;285(23):18085–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Levine RL, Berlett BS, Moskovitz J, Mosoni L, Stadtman ER. Methionine residues may protect proteins from critical oxidative damage. Mech Ageing Dev 1999;107(3):323–32. [DOI] [PubMed] [Google Scholar]
- 25.Henderson J, Ricker R, Bidlingmeyer BA, Woodward C. Rapid, accurate, sensitive, and reproducible HPLC analysis of amino acids. Santa Clara, CA: Agilent Techologies, Part No. 5980-1193E; 2000. [Google Scholar]
- 26.Levine RL, Mosoni L, Berlett BS, Stadtman ER. Methionine residues as endogenous antioxidants in proteins. Proc Natl Acad Sci USA 1996;93:15036–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bender A, Hajieva P, Moosmann B. Adaptive antioxidant methionine accumulation in respiratory chain complexes explains the use of a deviant genetic code in mitochondria. Proc Natl Acad Sci USA 2008;105(43):16496–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Archibald FS, Fridovich I. The scavenging of superoxide radical by manganous complexes: in vitro. Arch Biochem Biophys 1982;214(2):452–63. [DOI] [PubMed] [Google Scholar]
- 29.Barnese K, Gralla EB, Valentine JS, Cabelli DE. Biologically relevant mechanism for catalytic superoxide removal by simple manganese compounds. Proc Natl Acad Sci USA 2012;109(18):6892–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stadtman ER, Berlett BS, Chock PB. Manganese-dependent disproportionation of hydrogen peroxide in bicarbonate buffer. Proc Natl Acad Sci USA 1990;87:384–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hunt JB, Smyrniotis PZ, Ginsburg A, Stadtman ER. Metal ion requirement by glutamine synthetase of Escherichia coli in catalysis of gamma-glutamyl transfer. Arch Biochem Biophys 1975;166:102–24. [DOI] [PubMed] [Google Scholar]
- 32.Berlett BS, Chock PB, Yim MB, Stadtman ER. Manganese(II) catalyzes the bicarbonate-dependent oxidation of amino acids by hydrogen peroxide and the amino acid-facilitated dismutation of hydrogen peroxide. Proc Natl Acad Sci USA 1990;87:389–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lowther WT, Brot N, Weissbach H, Matthews BW. Structure and mechanism of peptide methionine sulfoxide reductase, an “anti-oxidation” enzyme. Biochemistry 2000;39(44):13307–12. [DOI] [PubMed] [Google Scholar]
- 34.Woolfolk CA, Stadtman ER. Regulation of glutamine synthetase. 3. Cumulative feedback inhibition of glutamine synthetase from Escherichia coli. Arch Biochem Biophys 1967;118(3):736–55. [DOI] [PubMed] [Google Scholar]
- 35.Buxton GV, Greenstock CL, Helman WP, Rose AB. Critical review of rate constants for reactions of hydrated electrons, hydrogen atoms and hydroxyl radicals (.OH/.O−) in aqueous solution. J Phys Chem Ref Data 1988;17:513–886. [Google Scholar]
- 36.Stadtman ER, Levine RL. Protein oxidation. Ann NY Acad Sci 2000;899:191–208. [DOI] [PubMed] [Google Scholar]
- 37.Hansen RE, Roth D, Winther JR. Quantifying the global cellular thiol-disulfide status. Proc Natl Acad Sci USA 2009;106(2):422–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gaidamakova EK, Myles IA, McDaniel DP, Fowler CJ, Valdez PA, Naik S,. et al. Preserving immunogenicity of lethally irradiated viral and bacterial vaccine epitopes using a radio-protective Mn2 +-peptide complex from Deinococcus. Cell Host Microbe 2012;12(1):117–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Battista JR. Against all odds: the survival strategies of Deinococcus radiodurans. Annu Rev Microbiol 1997;51:203–24. [DOI] [PubMed] [Google Scholar]


