Abstract
Despite its superiority for evaluating gene expression, real-time quantitative polymerase chain reaction (qPCR) results can be significantly biased by the use of inappropriate reference genes under different experimental conditions. Reaumuria soongorica is a dominant species of desert ecosystems in arid central Asia. Given the increasing interest in ecological engineering and potential genetic resources for arid agronomy, it is important to analyze gene function. However, systematic evaluation of stable reference genes should be performed prior to such analyses. In this study, the stabilities of 10 candidate reference genes were analyzed under 4 kinds of abiotic stresses (drought, salt, dark, and heat) within 4 accessions (HG010, HG020, XGG030, and XGG040) from 2 different habitats using 3 algorithms (geNorm, NormFinder, and BestKeeper). After validation of the ribulose-1,5-bisphosphate carboxylase/oxygenase large unite (rbcL) expression pattern, our data suggested that histone H2A (H2A) and eukaryotic initiation factor 4A-2 (EIF4A2) were the most stable reference genes, cyclophilin (CYCL) was moderate, and elongation factor 1α (EF1α) was the worst choice. This first systematic analysis for stably expressed genes will facilitate future functional analyses and deep mining of genetic resources in R. soongorica and other species of the Reaumuria genus.
Introduction
Gene expression analysis is increasingly important in many fields of biological research. Understanding gene expression patterns is expected to provide insight into complex regulatory networks and will help to identify which genes are relevant to new biological processes. Recently developed methods to measure transcript abundance have been frequently applied in many model biological systems; however, these approaches require the same normalization procedures as traditional mRNA quantification methods. Real-time quantitative polymerase chain reaction (qPCR) is one of the most common technologies used for gene expression and transcriptome analysis and is characterized by high sensitivity and specificity, good reproducibility, a widely dynamic quantification range, and high-throughput capacity for a limited number of target genes [1]–[3]. Despite its superiority over other methods available for evaluating gene expression, qPCR remains underused due in part to conflicting results and weak repeatability. Traditional reference genes, such as actin (ACT), ubiquitin (UBQ), alpha-tubulin (TUA), eukaryotic initiation factor 4A-2 (EIF4A2), 60S ribosomal protein L2 (L2), TIP41-like protein (TIP41), elongation factor 1-alpha (EF1α), cyclophilin (CYCL), histone H2A (H2A) and DNAJ-like protein (DNAJ), are mostly used in model plants and crops [4]–[6] and do not always maintain stable expression levels among different tissues, experimental conditions [7]–[12], or species [7], [11]–[16]. Systematic validations of reference genes have mainly focused on models and important crop species, such as Arabidopsis [17], rice [6], poplar [18], soybean [19], wheat [14], barley [20], tomato [21], grape [22], and potato [5]. However, with the increasing importance of agronomy and ecology, greater numbers of non-model plants are being assessed with molecular functional analyses, and it is crucial to identify proper reference genes for new species. To date, reference gene selection has been performed for some non-model plants like bamboo [23], peach [15], Caragana intermedia [12], and eggplant [24], but seldom in desert plants.
Reaumuria (Tamaricaceae) plants are perennial xeric shrubs widely distributed in arid regions in North Africa, Asia, and southern Europe. R. soongorica (2n = 22, 778 Mb genome size) [25] is a constructive and dominant species of desert ecosystems in central Asia [26]. It has evolved typical phenotypes of desert plants, such as an extremely thick cuticle; hollow stomata; specialized leaf shape; and a root system that reduces the transpiration rate, increases water use efficiency, and maintains stem vigor to survive desiccation [27], [28]. Thus, R. soongorica is a good species to study the molecular mechanisms of drought adaptation. Many studies have attempted to elucidate the mechanisms of the adaptation and tolerance of R. soongorica under the extreme drought conditions [29]–[33], which suggested its more and more importance of ecology. It is in urgent to systematically identify appropriate references genes for R. soongorica, for better understanding of the processes and molecular functions of its potential gene resources since its transcriptome analysis had been released [34].
Three statistical algorithms packages have been developed to assess reference gene stability: geNorm [35], NormFinder [11], and BestKeeper [36]. In this study, the stability of 10 candidate reference genes used in many plants (ACT, UBQ, TIP41, CYCL, EF1α, EIF4A, H2A, L2, DNAJ, and TUA) were assessed based on their expression abundance in two contrasting populations under four stress conditions. To validate the best candidate reference genes, the expression levels of the ribulose-1,5-bisphosphate carboxylase/oxygenase (rbcL) homolog were analyzed in comparison to 10 reference genes. This work will facilitate future studies on R. soongorica gene expression, speed up functional analyses of special genetic resources, and improve our understanding of the molecular mechanisms of drought adaptation.
Materials and Methods
Ethics Statement
R. soongorica is a species widely distributed in Shashichang, Jingtai County, Gansu province and other arid regions, and it is not included on any list of endangered or protected species. Before collecting the samples, oral permission was obtained from the local management of forestry after sending introduction letters from the CAREERI (Cold and Arid Regions Environmental and Engineering Research Institute, Chinese Academy of Sciences).
Plant Materials and Treatments
Seeds of four accessions (HG010, HG020, XGG030, and XGG040) were collected from two different habitats (Honggu [HG] N36°15′20.16″, E103°1′29.82″, annual precipitation [AP] ∼400 mm based on WorldClim database 1950–2000; Xiaogangou [XGG] N36°54′21.60″, E103°53′31.44″, AP ∼150 mm) were sampled in the year 2011. After 2 years in frozen storage at −20°C to completely break physiological dormancy, the seeds were surface sterilized with 0.1% sodium hypochlorite and rinsed with distilled water. The sterilized seeds were then grown in MS-medium with photosynthetically active radiation under long day condition (16/8-hour light-dark cycles); the relative humidity was 75%, and the temperatures were 25°C in the light and 22°C in the dark. After 2 weeks, 10 seedlings from each accession were collected for RNA extraction. For drought stress, seedlings were subjected to 15% PEG (polyethylene glycol) for 1 hour. Heat stress treatment was performed at 38°C. For salt stress, plants were treated with 100 mM NaCl for 3 hours. For dark stress, plants were wrapped with foil paper for 2 hours. Control plants were kept at 25°C during light hours on MS-medium.
Total RNA Isolation and cDNA Synthesis
Total RNA was extracted using an E.Z.N.A. plant RNA kit (Omega Bio-tek), and the remaining DNA was removed by RNase-free DNase (Omega Bio-tek) according to the instruction manual. Total RNA concentration and purity were determined with ratio optical density (OD) 260/280 by NanoDrop1000 (Thermo Scientific). All the samples passed QC with the ratio OD260/280 between 1.9 and 2.2 and ratio OD260/230 <2.0. RNA integrity was verified by 1.5% agarose gel electrophoresis with two clear bands of 28S/18S ribosomal RNA. In each treatment and control, three biological replicates of RNA samples were extracted from at least 10 seedlings of each genotype first, and then mixed equally as genotype representative sample. For each representative sample, 1 µg total RNA was reverse transcribed in a 20-µl volume reaction with oligo dT primers by the RevertAidTM First Strand cDNA Synthesis Kit (Fermentas). In total, we got 20 cDNA samples (four genotypes with four treatments and one control) to be analyzed in this study. The cDNAs were diluted 1∶10 with nuclease-free water prior to the qPCR analyses.
Selection and Validation of Candidate Genes
Candidate reference genes were screened based on the transcriptome de novo assembly sequences [34] with lengths >1 kb and transcription abundance FPKM (fragments per kilobase of exon per million fragments mapped) values >5. Unigenes were annotated by National Center for Biotechnology Information (NCBI) non-redundant protein (Nr) database with E-values less than 1e−28. A reciprocal BLAST hits approach was then conducted against Arabidopsis with a permissive E-value cutoff of 10−3 to obtain hits to confirm the gene homologs in R. soongorica. The final list of 10 candidate reference genes included ACT, EF1α, TUA, TIP41, DNAJ, UBQ, CYCL, EIF4A2, H2A, and L2, which were assessed in later expression analyses (Table 1).
Table 1. Candidate reference gene descriptions.
| Gene Symbol | Length (bp) | Tm (°C) | LinRegPCR Efficiency | 5′-Sequences | 3′-Sequences | GeneID | Gene Length | Nr-Annotation |
| ACT | 145 | 80.57 | 1.834 | AGCAAGGTCAAGACGAAGGA | TGTTGCTATTCAGGCTGTGC | CL448.Contig2_A | 1784 | Actin |
| CYCL | 287 | 83.56 | 1.946 | CCCTGAAGCATGAAGCTAGG | GCCTTGATCTGGTGTTTGGT | Unigene10547_A | 1193 | Cyclophilin |
| DNAJ | 141 | 81.91 | 1.911 | GATTATCTCAGCGCGCCTAC | GATTGGTTTCGGGTCTTTCA | Unigene46962_A | 819 | DnaJ-like protein |
| EF1α | 128 | 81.88 | 1.929 | CCCCAGGTTAATCCTTCCAT | GGAAAGAAGCCCAAGGAATC | Unigene18820_A | 1127 | elongation factor 1 alpha |
| EIF4A2 | 116 | 77.24 | 1.854 | TCAGCAACATTTGCAGGAAG | TTTGGAAGAAAGGGTGTTGC | CL9396.Contig2_A | 1717 | Eukaryotic initiation factor 4A-2 |
| H2A | 164 | 81.54 | 1.875 | GGCGCTGCAAAGAAGTCTAC | AGGACCTCAGCAGCAAGGTA | CL9675.Contig2_A | 818 | Histone H2A |
| L2 | 280 | 82.9 | 1.885 | TCATTGCGAACGAAACTCTG | ACGGTGACAATTCAGGGAAG | CL8967.Contig2_A | 1342 | 60S ribosomal protein L2 |
| TIP41 | 110 | 79.16 | 1.899 | GGTTTCTGCTCTTGCGTTTC | CCGTTTAATGATGGGGAATG | CL7670.Contig1_A | 1134 | TIP41-like protein |
| TUA | 264 | 82.82 | 1.893 | CAACAAGCCGCTAAGGAAAG | GCCTTCGTCCACTGGTATGT | CL12940.Contig1_A | 1916 | alpha-tubulin |
| UBQ | 113 | 80.16 | 1.878 | CCGGCCTTCTGGTAAACATA | TTGAGGAAGGAATGCCCTAA | CL5323.Contig1_A | 471 | Ubiquitin |
| rbcL * | 185 | 80.93 | 1.890 | AGAGCACGTAGGGCTTTGAA | GACAACTGTGTGGACCGATG | Unigene19142_A | 5856 | Ribulose-1,5-bisphosphate carboxylase oxygenase |
Notes:
* used for normalization validation under stress conditions in four accessions.
rbcL gene (ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit, chloroplast) was chosen for the validation of reference genes. We tookUnigene19142_A, a rbcL homolog in the R. soongorica de novo transcriptome sequences (from Shi et.al. 2013) to predict its peptide sequence and gene structure with the GENSCAN Web Server at MIT (http://genes.mit.edu/GENSCAN.html). The protein sequence was then blasted against Nr database in NCBI and got the best hit ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit (chloroplast) in Eucalyptus umbra (YP_008575886.1, with total score of 978, coverage of 100% and identity of 99%). The protein sequences of the rbcL homolog was well aligned with those from other species in File S3.pdf, which suggested the high homologous of rbcL genes to each other. The annotation information of the ten reference genes and one validation gene were listed in the Table S1.
qPCR Primer Design and Verification of Amplification
Primers of each reference gene were designed by the platform OligoPerfect Designer (Invitrogen), with optimal melting temperatures 60°C, optimal GC content 50%, optimal primer size 20 bp, and product size was between 100 and 300 bp (Table 1, and Figure S1). Each primer showed good specificity and efficiency (File S1 and S2). PCR efficiencies per amplicon were calculated by LinRegPCR [37]–[39], and the mean PCR efficiency between 1.8 and 2.0 from three technology replicates was regarded as the efficiency of one sample. The final primer efficiency was obtained from the average of all sample amplified with each pair of primer.
qPCR amplification products of the 10 candidate reference from one genotype (XGG030) were sequenced with ABi 3730 automated DNA Sequencer. Both the product sizes and sequences of amplificiaton product were consistent with the unigenes from de novo sequencing (File S4).
qPCR
qPCR experiments were conducted in the Stratagene Mx3000P QPCR system (Agilent Technologies), in which combined gene amplification, detection and data analysis steps with FAM, HEX, ROX, and TET filters. A 20-µl reaction system volume was used for amplification. Each reaction contained 10 µl DyNAmo Flash SYBR Green qPCR Kit Master Mix (Thermo Scientific), 0.5 µl each of forward and reverse primer, 0.2 µl F-402 buffer, 2 µl cDNA synthesized from total RNA. The PCR program contained an initial denaturation step of 5 min at 95°C, followed by denaturation for 15 s at 95°C, annealing for 30 s at 60°C, and extension for 30 s at 72°C for 40 cycles. Three technical replicates were used in qPCR experiments, and the average Cq values were used for quantification. The amplification curves and dissociation curves were shown in File S1 and S2. No-template controls were used as negative control for each pair of primers. The PCR products were checked by 1.5% agarose gel electrophoresis to verify the specificity and expected sizes of amplicons. The amplification curve was analyzed by LinRegPCR for efficiency correction and baseline subtraction [37]–[39].
Statistical Analysis
Three different types of Microsoft Excel-based software – geNorm, NormFinder, and BestKeeper – were used to rank the expression stabilities of reference genes across all of the experimental sets. The candidate reference genes were ranked by geNorm based on the expression stability value M, which was calculated for all genes investigated (the lower the M value, the higher the gene's expression stability) [35]. NormFinder program is a Visual Basic application tool for Microsoft Excel used to determine the expression stabilities of reference genes that ranks all reference gene candidates based on intra- and inter-group variations and combines both results into a stability value for each candidate reference gene [11]. BestKeeper identifies the most stably expressed genes based on the coefficient of correlation to the BestKeeper Index, which is the geometric mean of the candidate reference gene Cq values. BestKeeper also calculates the standard deviation (SD) and the coefficient of variation (CV) based on the Cq values of all candidate reference genes [36]. Genes with SD >1 are considered unacceptable [10]. Thus, with BestKeeper package the best reference genes should be the most stable ones, which should exhibit the lowest CV ± SD [40]. This analysis was done using the Cq values after efficiency correction. Following qPCR data collection, Cq values corrected after efficiency correction by LinRegPCR were converted to relative quantities using the formula: 2−ΔCq, in which ΔCq equals each corresponding Cq value minus the minimum Cq value. The sample with the maximum expression level (the minimum Cq value) was used as a calibrator and was set to a value of 1. Relative quantities were used for geNorm and NormFinder, while BestKeeper analysis was based on untransformed Cq values.
Results
Expression levels of candidate reference genes across all samples
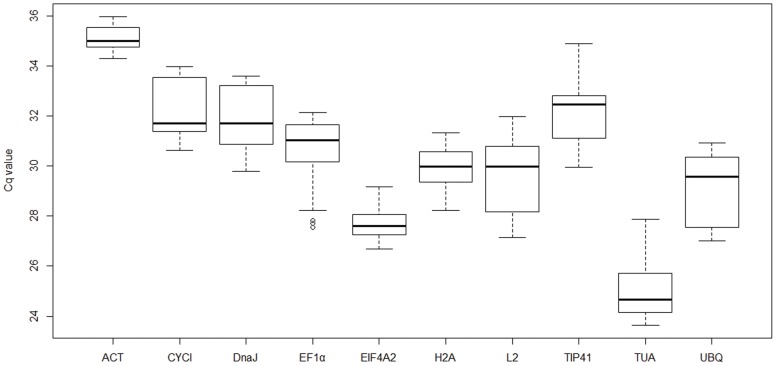
A total of 10 candidate reference genes of R. soongorica were chosen based on pBlast similarity against the nr database (Table 1), and their transcript levels were quantified under different stress conditions. The relative expression levels of the candidate reference genes were determined as Cq values, and the transcripts of these genes showed different levels of abundance in pooled samples (as Total), and each subset of treatments in all four genotypes (PEG, Heat, Dark, and Salt) (Figure 1). The mean Cq values of the genes ranged from 22–40, with most between 28 and 31 across all tested samples. Among all the candidate genes, TUA had the lowest Cq value (24.8), indicating its highest level of expression; EIF4A2, H2A, and L2 were moderately expressed; and ACT was expressed at the lowest level (35.1 of Cq value). Stability was regarded as the most important characteristic for reference genes, and ACT, EIF4A2 and H2A showed the least gene expression variation (CV of 1.42%, 2.32% and 3.01% respectively), while EF1α (4.82%) and TUA (5.20%) were the most variable within the total group.
Figure 1. Expression levels of candidate reference genes across all samples.
Lines across the boxes depict the medians, boxes indicate the interquartile range, whiskers represent 95% confidence intervals, and open circles represent outliers.
Expression Stability of Candidate Reference Genes under different treatments geNorm analysis
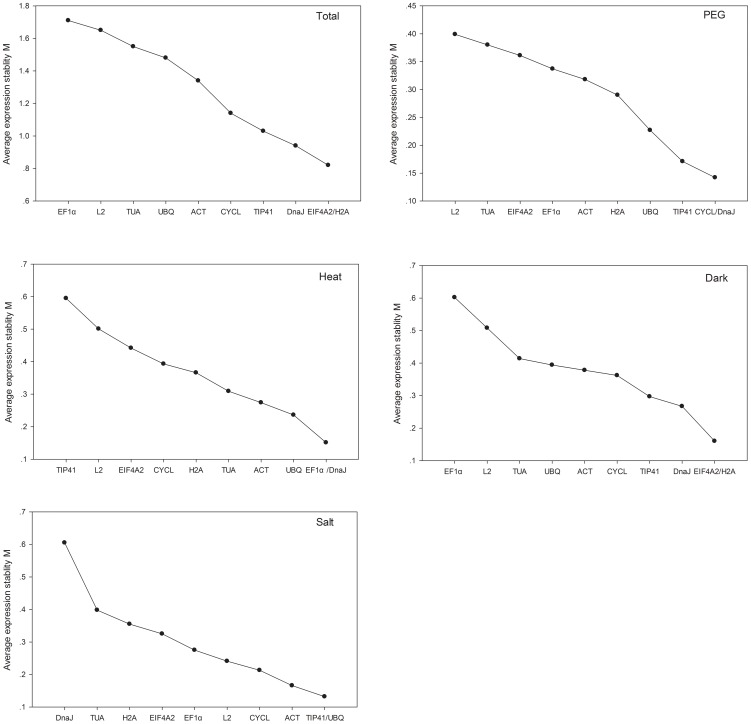
The expression stabilities of the 10 reference genes were assessed using the geNorm software [35] in the pooled and separate treatments (PEG, heat, dark, and salt) across four accessions from two contrasting habitats. Figure 2 shows the ranking of the tested genes on the basis of their expression stability (M). From the pooled data, the order from the most stable gene to the least was: H2A/EIF4A2, DNAJ, TIP41, CYCL, ACT, UBQ, TUA, L2, and EF1α (Figure 2A). Their calculated “M” values were 0.82, 0.94, 1.03, 1.14, 1.34, 1.48, 1.55, 1.65, and 1.71, respectively. Successive elimination of the least stable genes led to the identification of H2A and EIF4A2 as the most stable genes. However, we found that CYCL and DNAJ were the most stable genes in PEG treatment (Figure 2B), EF1α and DNAJ in heat (Figure 2C), H2A and EIF4A2 in dark (Figure 2D), and TIP41 and UBQ in salt treatment (Figure 2E). EIF4A2 was the moderate stable gene in individual PEG, heat, and salt treatments, which is inconsistent with the CV analysis of the pooled data.
Figure 2. Gene expression stability and ranking of 10 reference genes calculated by geNorm.
Mean expression stability (M) was calculated following stepwise exclusion of the least stable gene across all the treatment groups. The least stable genes are on the left, and the most stable ones are on the right.
geNorm also performs stepwise calculations of the pair-wise variation (Vn/Vn+1) between sequential normalization factors (NFn and NFn+1) to determine the optimal number of reference genes required for accurate normalization. A large variation means that the added gene had a significant effect and should preferably be included to calculate a reliable normalization factor [35]. The cut-off value of 0.15 was proposed by the geNorm program [35]; below this cut-off value there is no need of inclusion of an additional control gene. As shown in Figure 3, when all the samples were taken into account, the pairwise variation V2/3 was 0.311, higher than 0.15, indicating that the addition of the third reference gene was necessary to normalize gene expression. H2A and EIF4A2 were recommended as the best reference genes based on their M value (Figure 2A). However, for individual treatments, V2/3 values were 0.092, 0.058, 0.057, and 0.104 in Heat, PEG, Salt, and Dark treatments, respectively. It is apparent that the inclusion of a third gene did not have a significant effect in separate treatments.
Figure 3. Determination of the optimal number of reference genes required for effective normalization.
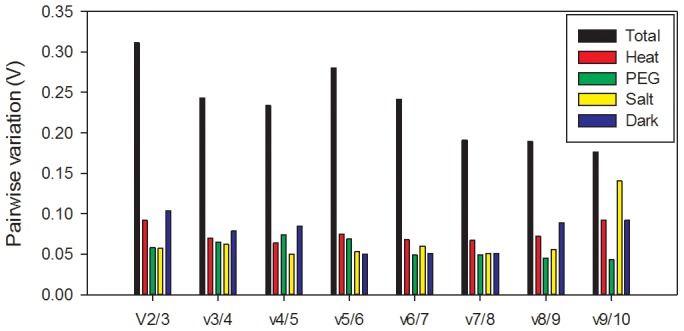
Pairwise variation (Vn/Vn+1) analysis between the normalization factors (NFn and NFn+1) was performed by the geNorm program to determine the optimal number of reference genes and was carried out for qPCR data normalization in various sample pools.
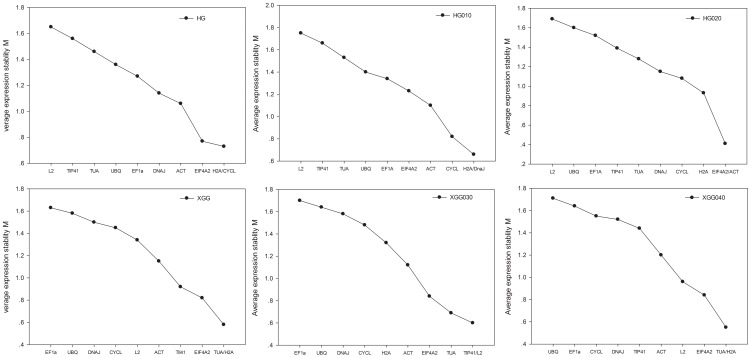
The expression stabilities of the 10 reference genes were also assessed among genotypes (Figure 4). In the HG genotypes (HG010 and HG020), the CYCL, H2A and EIF4A2 were most stable; while in XGG genotypes (XGG030 and XGG040) were EIF4A2, H2A and TUA. Among individual genotypes, H2A and EIF4A2 were frequently observed in the top of the most sable genes. However, compared to H2A and EIF4A2, ACT and UBQ which were usually used as reference gene for qPCR were not a good choice.
Figure 4. Expression stability and ranking of housekeeping genes by geNorm analysis within and among populations.
Mean expression stability (M) was calculated following stepwise exclusion of the least stable gene.
NormFinder analysis
NormFinder analysis results (shown in Table 2) revealed that the expression of three candidate reference genes, EIF4A2, ACT, and H2A had lower stability values (the lower, the better) across all the samples pooled together. However, the stability ranking of genes in individual treatments were significantly different; for example, the best reference genes were found as UBQ in PEG, EF1α and DNAJ in Heat, DNAJ in Dark, and UBQ and CYCL in Salt, and all the genes harbored the stable values <0.1. Interestingly, according to NormFinder analysis, ACT was suggested as a good reference gene among the four treatments, which is inconsistent with the conclusion from the geNorm analysis. However, the results from the NormFinder and geNorm analyses were consistent, suggesting that EIF4A2 and H2A are stable reference genes.
Table 2. Expression stability of the reference genes calculated by NormFinder software.
| Total | SV | PEG | SV | Heat | SV | Dark | SV | Salt | SV |
| EIF4A2 | 0.368 | UBQ | 0.073 | EF1α | 0.053 | DNAJ | 0.093 | UBQ | 0.043 |
| ACT | 0.4 | CYCL | 0.144 | DNAJ | 0.062 | TIP41 | 0.147 | CYCL | 0.063 |
| H2A | 0.573 | DNAJ | 0.186 | ACT | 0.203 | ACT | 0.173 | TIP41 | 0.11 |
| DNAJ | 0.677 | H2A | 0.19 | TUA | 0.216 | EIF4A2 | 0.198 | TUA | 0.203 |
| CYCL | 0.874 | TIP41 | 0.212 | UBQ | 0.238 | CYCL | 0.215 | L2 | 0.209 |
| TUA | 1.017 | EF1α | 0.212 | CYCL | 0.244 | UBQ | 0.254 | EF1α | 0.21 |
| UBQ | 1.046 | ACT | 0.231 | H2A | 0.253 | TUA | 0.302 | ACT | 0.235 |
| TIP41 | 1.119 | EIF4A2 | 0.234 | EIF4A2 | 0.382 | H2A | 0.311 | H2A | 0.32 |
| L2 | 1.157 | TUA | 0.287 | L2 | 0.451 | L2 | 0.515 | EIF4A2 | 0.329 |
| EF1α | 1.185 | L2 | 0.291 | TIP41 | 0.632 | EF1α | 0.628 | DNAJ | 0.978 |
Note: SV, stability value.
BestKeeper analysis
The results of the BestKeeper analysis are shown in Table 3. Four candidate reference genes (EIF4A2, ACT, H2A, and DNAJ) in the Total group showed an SD value <1, which qualified as reference genes. When taking into account the separate treatments, all the candidate genes showed SD values <1 in individual groups. Here, only the top two genes were listed, as UBQ and CYCL in PEG treatment, EF1α and DNAJ in heat treatment, DNAJ and TIP41 in dark treatment, and UBQ, CYCL in salt treatment. Further data processing using Pearson correlation coefficient and regression analysis (r) showed that ACT exhibited the lowest correlation with other candidate genes (data not shown, r = −0.676, p-value = 0.001), suggesting its higher independence. DNAJ was the most correlated to the other nine genes in pool (r = 0.68, p-value = 0.001). Pairwise correlation revealed that TIP41 and TUA (r = 0.804, p-value = 0.001) were highly correlated. As these two genes were not stable in pooled data, we did not take these two genes as good reference. Although EIF4A2 and H2A was the second most correlated gene pair (r = 0.773, p-value = 0.001), considering their higher scores in geNorm and NormFinder, EIF4A2 and H2A could be selected as two likely reference genes with good correlation.
Table 3. Expression stability of the reference genes calculated by BestKeeper software.
| Total | SD | CV | PEG | SD | CV | Heat | SD | CV | Dark | SD | CV | Salt | SD | CV |
| ACT | 0.43 | 1.22 | ACT | 0.09 | 0.26 | EF1α | 0.12 | 0.39 | ACT | 0.19 | 0.54 | L2 | 0.08 | 0.26 |
| EIF4A2 | 0.49 | 1.75 | H2A | 0.12 | 0.4 | DNAJ | 0.12 | 0.39 | DNAJ | 0.2 | 0.63 | UBQ | 0.12 | 0.4 |
| H2A | 0.74 | 2.47 | UBQ | 0.19 | 0.69 | ACT | 0.18 | 0.51 | TIP41 | 0.23 | 0.76 | EF1α | 0.14 | 0.45 |
| DNAJ | 0.98 | 3.07 | EF1α | 0.2 | 0.73 | H2A | 0.21 | 0.71 | TUA | 0.23 | 0.94 | TIP41 | 0.16 | 0.48 |
| TUA | 1 | 4.01 | L2 | 0.26 | 0.84 | CYCL | 0.22 | 0.71 | EIF4A2 | 0.24 | 0.87 | CYCL | 0.16 | 0.52 |
| TIP41 | 1.07 | 3.32 | TIP41 | 0.28 | 0.8 | UBQ | 0.24 | 0.78 | CYCL | 0.25 | 0.73 | ACT | 0.22 | 0.62 |
| CYCL | 1.08 | 3.33 | CYCL | 0.33 | 1.05 | TUA | 0.25 | 1.01 | H2A | 0.34 | 1.11 | EIF4A2 | 0.3 | 1.08 |
| EF1α | 1.16 | 3.78 | DNAJ | 0.37 | 1.22 | EIF4A2 | 0.38 | 1.39 | UBQ | 0.35 | 1.27 | H2A | 0.39 | 1.28 |
| UBQ | 1.26 | 4.33 | EIF4A2 | 0.51 | 1.79 | L2 | 0.45 | 1.62 | EF1α | 0.56 | 1.79 | TUA | 0.4 | 1.61 |
| L2 | 1.33 | 4.5 | TUA | 0.58 | 2.13 | TIP41 | 0.74 | 2.32 | L2 | 0.69 | 2.44 | DNAJ | 1.24 | 3.86 |
Note: Reference genes were identified as the most stable genes, i.e. those with the lowest coefficient of variance (CV) with standard deviation (SD).
Reference Gene Validation
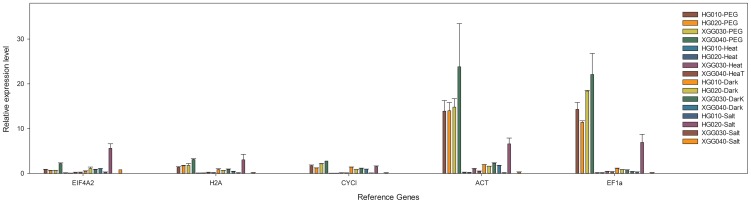
To validate candidate reference gene selection, the expression pattern of rbcL was analyzed against all 10 candidate reference genes (Figure 5). RuBisCO is an enzyme involved in the first major step of carbon fixation, a process by which atmospheric carbon dioxide is converted by plants to energy-rich molecules, such as glucose. In chemical terms, it catalyzes the carboxylation of ribulose-1,5-bisphosphate (also known as RuBP). It is probably the most abundant protein on Earth [41], [42]. It was obviously down-regulated due to its light-dependent characteristic, and rbcL has also been shown to be down-regulated in soybean under stress [43], [44]. In our results, rbcL was down-regulated against all genes except PEG treatments in three accessions (HG010, XGG030, and XGG040). However, in PEG treatments, rbcL was slightly down-regulated against EF1α, CYCL, H2A, DNAJ, and UBQ, which were the most stable genes in PEG. It was strongly up-regulated compared to TUA, ACT, EIF4A2, and L2, which were the most unstable genes in PEG treatment. In accession HG020, It was obvious that its expression level was down-regulated against most candidate reference genes, except in PEG and salt treatments. This could be due to the divergent genetic backgrounds between this genotype and the other three genotypes. Actually, the huge genetic differentiation within populations was supported by a phylogeography study based on chloroplast genes (unpublished data), and different genotypes may exhibit different physiological responses under drought conditions. Thus, the high expression level of rbcL in HG020 accession may be related to local environment adaptation. Except this, the validation results against the most stable genes were all consistent with the results from soybean [43], [44], suggesting that the DNAJ and H2A genes could be used as practical reference genes in multiple treatments.
Figure 5. Relative quantification of rbcL expression using 10 reference genes for normalization in the four accessions.
The results are represented as mean fold changes in relative expression compared to untreated samples.
Discussion
qPCR has significantly improved the detection and quantification of expression profiles of genes in distinct biological samples. The main advantages of this technique are based on its high sensitivity, high specificity, and broad quantification range [45] [46]. For these reasons, qPCR is the first choice for detecting and quantifying gene expression profiles. An appropriate internal control gene is required for valid qPCR analyses. The ideal control gene should have relatively stable expression in distinct biological samples, which can be from different cell types, developmental stages, and samples exposed to different experimental conditions, even for different genotypes.
R. soongorica is regarded as a plant that is tolerant to extremely drought conditions [26]–[28]. Moreover, R. soongorica is a typical recretohalophyte, and its molecular mechanism of salt resistance remains unclear [47]. Only a few studies have focused on the molecular mechanisms of adaption to extreme environments [27]. Since his shrub species is prevalent in ecological engineering and restoration to mitigate further desertification of the arid land in northwest China, with its numerous novel genetic resources (around 40% of genes were newly found in its transcriptome sequences [34]),, it is convenient and feasible to focus on molecular functional analyses of the expression of unique genes. However, it is first necessary to identify stable reference genes.
In this study, the four stresses of drought, salt, dark, and heat were investigated in different R. soongorica genotypes to identify stable reference genes. Three different statistical approaches (geNorm, NormFinder, and BestKeeper) were used to evaluate the stability of the 10 most commonly used candidate reference genes. The most prominent observation was that each type of software produced a different set of top-ranked reference genes. This is not unexpected because the three programs employed different algorithms and analytical procedures. The top three stable reference genes were EIF4A2, H2A, and DNAJ using geNorm. EIF4A2, ACT, and H2A had lower stability values across all samples using NormFinder, which was consistent with the BestKeeper results. We used a previously described method [48] to determine the best reference gene based on all three algorithms (Table S2). According to the frequencies of the reference genes in the rank crossing the difference treatments (PEG, Salt, Dark, Heat and Total), H2A and EIF4A2 were the best reference genes, CYCL was moderate stable, and EF1α was the fourth choice. This was supported in the subsequent validation analysis with rbcL (File S4). All the 10 genes are normally expressed under all the treatments in the 4 genotypes (Figure S2), indicating that normalization of total RNA was sufficient. In PEG treatments, rbcL was slightly or down-regulated against EF1α, CYCL, H2A, DNAJ, and UBQ, which were the most stable genes in accordance with results in Soybean [43], [44]. Once we identify practical internal reference gene, it is easy to characterize the true expression level for a candidate gene.
ACT has been considered as one of the best reference genes for assessing gene expression in many plant tissues and under different experimental conditions [49]–[51]. In present study, GeNorm, NormFinder, and Bestkeeper analyses demonstrated that ACT was not that stable and exhibited a lower expression level compared to other good reference genes, which suggested that it is not a good reference gene for R. soongorica under such conditions. Actually, the instablity of ACT homolog genes was also confirmed in two other plant species [12], [15]. The ACT homolog gene exhibited similar low expression levels in Caragana intermedia and R. soongorica, which are widely spread throughout the arid zone in China. Even though C. intermedia and R. soongorica belong to different families, it is likely that both of them harbor similar pathways involved in strong drought- and salt-resistance ability.
The other two most commonly used reference genes are UBQ and TUA. UBQ showed highly stable expression in Arabidopsis [52] and tomato [9], but the UBQ gene was ranked differently across different treatments in the present study. It was listed on the top under PEG with NormFinder; however, it was in a moderate stable position with geNorm and BestKeeper under other treatments. It was suggested to be an inappropriate internal control for qPCR studies at different developmental stages in rice [53] and soybean [54]. TUA has also been widely used as a reference gene in studies in water lily and soybean [16], [19]. However, some studies revealed that TUA did not satisfy certain basic requirements for use as an internal control [18]. In our results in R. soongorica, TUA was once of the least stable genes according to geNorm, BestKeeper, and NormFinder analyses together with rbcL expression level. Consequently, ACT, UBQ, and TUA should be used with caution as an internal control in R. soongorica.
Conclusions
To our knowledge, this study is the first to systematically analyze reference genes for qPCR in R. soongorica under different abiotic stress conditions (drought, salt, dark, and heat) among four accessions. Analysis of expression stability using geNorm, NormFinder, and BestKeeper revealed that EIF4A2 and H2A were appropriate reference genes under different abiotic stress conditions, whereas EF1α showed relatively low expression stability. In contrast, the most commonly used reference genes ACT, UBQ, and TUA were inappropriate. This work will benefit future gene expression studies assessing different abiotic stress conditions in R. soongorica and other Reaumuria species.
Supporting Information
Agarose gel electrophoresis for PCR products of 10 candidate reference genes and rbcL .
(TIFF)
Expression levels of candidate reference genes across all the four accessions.
(TIFF)
Annotation information of 10 reference genes and the one validation gene from the de novo transcriptome sequences.
(XLS)
Ranking of the best reference gene based on the all three algorithms.
(DOC)
Contains 11 figures from Figure S3 to S13 representing the qPCR amplification curves of 10 candidate reference genes ( ACT, CYCL, DNAJ, EF1, EIF4A2, H2A, L2, TIP41, TUA and UBQ ) and one validation gene rbcL , respectively.
(ZIP)
Contains 11 figures from Figure S14 to S24 representing the melting curves of qPCR products of the 10 candidate reference genes ( ACT, CYCL, DNAJ, EF1, EIF4A2, H2A, L2, TIP41, TUA and UBQ ) and one validation gene rbcL , respectively.
(ZIP)
Alignment of the protein sequences of rbcL homologs.
(PDF)
Contains 11 sequence alignment files from Fasta S1 to S11,for verification of the homologs of 10 reference genes and one validation gene rbcL .
(ZIP)
Acknowledgments
We thank for the instructions on the data analysis from Jianfeng Zhu, and appreciate the constructive and helpful suggestions from Dr. Jinping Li and the two anonymous reviewers.
Funding Statement
This work was supported by the “One Hundred Talents” Project of the Chinese Academy of Sciences (Grant No. 29Y127E71) and National Natural Science Foundation of China (No. 91125025). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Fink L, Seeger W, Ermert L, Hanze J, Stahl U, et al. (1998) Real-time quantitative RT-PCR after laser-assisted cell picking. Nat Med 4: 1329–1333. [DOI] [PubMed] [Google Scholar]
- 2. Heid CA, Stevens J, Livak KJ, Williams PM (1996) Real time quantitative PCR. Genome Res 6: 986–994. [DOI] [PubMed] [Google Scholar]
- 3. Higuchi R, Fockler C, Dollinger G, Watson R (1993) Kinetic PCR analysis: real-time monitoring of DNA amplification reactions. Biotechnology (NY) 11: 1026–1030. [DOI] [PubMed] [Google Scholar]
- 4. Brunner AM, Yakovlev IA, Strauss SH (2004) Validating internal controls for quantitative plant gene expression studies. BMC Plant Biol 4: 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Nicot N, Hausman J, Hoffmann L, Evers D (2005) Housekeeping gene selection for real-time RT-PCR normalization in potato during biotic and abiotic stress. J Exp Bot 56: 2907–2914. [DOI] [PubMed] [Google Scholar]
- 6. Jain M, Nijhawan A, Tyagi AK, Khurana JP (2006) Validation of housekeeping genes as internal control for studying gene expression in rice by quantitative real-time PCR. Biochem Biophys Res Commun 345: 646–651. [DOI] [PubMed] [Google Scholar]
- 7. Die J, Roman B, Nadal S, Gonzalez-Verdejo C (2010) Evaluation of candidate reference genes for expression studies in Pisum sativum under different experimental conditions. Planta 232: 145–153. [DOI] [PubMed] [Google Scholar]
- 8. Gimenez C, Mitchell VJ, Lawlor DW (1992) Regulation of photosynthetic rate of 2 sunflower hybrids under water-stress. Plant Physiol 98: 516–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lovdal T, Lillo C (2009) Reference gene selection for quantitative real-time PCR normalization in tomato subjected to nitrogen, cold, and light stress. Anal Biochem 387: 238–242. [DOI] [PubMed] [Google Scholar]
- 10. Migocka M, Papierniak A (2011) Identification of suitable reference genes for studying gene expression in cucumber plants subjected to abiotic stress and growth regulators. Mol Breeding 28: 343–357. [Google Scholar]
- 11. Zhong HY, Chen JW, Li CQ, Chen L, Wu JY, et al. (2011) Selection of reliable reference genes for expression studies by reverse transcription quantitative real-time PCR in litchi under different experimental conditions. Plant Cell Rep 30: 641–653. [DOI] [PubMed] [Google Scholar]
- 12. Zhu J, Zhang L, Li W, Han S, Yang W, et al. (2013) Reference gene selection for quantitative real-time PCR normalization in Caragana intermedia under different abiotic stress conditions. PloS one 8: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Saito T, Matsukura C, Ban Y, Shoji K, Sugiyama M, et al. (2008) Salinity stress affects assimilate metabolism at the gene-expression level during fruit development and improves fruit quality in tomato (Solanum lycopersicum L.). J Jpn Soc Hort Sci 77: 61–68. [Google Scholar]
- 14. Paolacci A, Tanzarella O, Porceddu E, Ciaffi M (2009) Identification and validation of reference genes for quantitative RT-PCR normalization in wheat. BMC Molecular Biology 10: 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Tong Z, Gao Z, Wang F, Zhou J, Zhang Z (2009) Selection of reliable reference genes for gene expression studies in peach using real-time PCR. BMC Molecular Biology 10: 71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Luo H, Chen S, Wan H, Chen F, Gu C, et al. (2010) Candidate reference genes for gene expression studies in water lily. Anal Biochem 404: 100–102. [DOI] [PubMed] [Google Scholar]
- 17. Czechowski T, Stitt M, Altmann T, Udvardi M, Scheible W-R (2005) Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis . Plant Physiol 139: 5–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Xu M, Zhang B, Su X, Zhang S, Huang M (2011) Reference gene selection for quantitative real-time polymerase chain reaction in Populus . Anal Biochem 408: 337–339. [DOI] [PubMed] [Google Scholar]
- 19. Hu R, Fan C, Li H, Zhang Q, Fu YF (2009) Evaluation of putative reference genes for gene expression normalization in soybean by quantitative real-time RT-PCR. BMC Mol Biol 10: 93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Janská A, Hodek J, Svoboda P, Zámečník J, Prášil IT, et al. (2013) The choice of reference gene set for assessing gene expression in barley (Hordeum vulgare L.) under low temperature and drought stress. Molecular genetics and genomics 288: 639–649. [DOI] [PubMed] [Google Scholar]
- 21. Exposito-Rodriguez M, Borges A, Borges-Perez A, Perez J (2008) Selection of internal control genes for quantitative real-time RT-PCR studies during tomato development process. BMC Plant Biology 8: 131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Dubrovina AS, Kiselev KV, Khristenko VS (2013) Expression of calcium-dependent protein kinase (CDPK) genes under abiotic stress conditions in wild-growing grapevine Vitis amurensis . J Plant Physiol 170: 1491–1500. [DOI] [PubMed] [Google Scholar]
- 23. Fan C, Ma J, Guo Q, Li X, Wang H, et al. (2013) Selection of reference genes for quantitative real-time PCR in bamboo (Phyllostachys edulis). PLoS One 8: e56573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gantasala NP, Papolu PK, Thakur PK, Kamaraju D, Sreevathsa R, et al. (2013) Selection and validation of reference genes for quantitative gene expression studies by real-time PCR in eggplant (Solanum melongena L). BMC Res Notes 6: 312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wang X, Zhang T, Wen Z, Xiao H, Yang Z, et al. (2011) The chromosome number, karyotype and genome size of the desert plant diploid Reaumuria soongorica (Pall.) Maxim. Plant Cell Rep 30: 955–964. [DOI] [PubMed] [Google Scholar]
- 26. Qian Z-Q, Xu L, Wang Y-L, Yang J, Zhao G-F (2008) Ecological genetics of Reaumuria soongorica (Pall.) Maxim. population in the oasis–desert ecotone in Fukang, Xinjiang, and its implications for molecular evolution. Biochemical Systematics and Ecology 36: 593–601. [Google Scholar]
- 27. Liu Y, Zhang T, Li X, Wang G (2007) Protective mechanism of desiccation tolerance in Reaumuria soongorica: leaf abscission and sucrose accumulation in the stem. Sci China C Life Sci 50: 15–21. [DOI] [PubMed] [Google Scholar]
- 28. Liu JQ, Qiu MX, Pu JC, Lu ZM (1982) The typical extreme xerophyte-Reaumuria soongorica in the desert of China. Act bot sinica 24: 485–488. [Google Scholar]
- 29. Bai J, Gong CM, Chen K, Kang HM, Wang G (2009) Examination of antioxidative system's responses in the different phases of drought stress and during recovery in desert plant Reaumuria soongorica (Pall.) Maxim. Journal of Plant Biology 52: 417–425. [Google Scholar]
- 30. Liu YB, Wang G, Liu J, Zhao X, Tan HJ, et al. (2007) Anatomical, morphological and metabolic acclimation in the resurrection plant Reaumuria soongorica during dehydration and rehydration. Journal of Arid Environments 70: 183–194. [Google Scholar]
- 31. Liu Y, Zhang T, Li X, Wang G (2007) Protective mechanism of desiccation tolerance in Reaumuria soongorica: leaf abscission and sucrose accumulation in the stem. Sci China C Life Sci 50: 15–21. [DOI] [PubMed] [Google Scholar]
- 32. Ma J, Chen F, Zhang H, Xia D (2007) Spatial distribution characteristics of stable carbon isotope compositions in desert plant Reaumuria soongorica (Pall.) Maxim. Frontiers of Earth Science in China 1: 150–156. [Google Scholar]
- 33. Qian Z-Q, Xu L, Wang Y-L, Yang J, Zhao G-F (2008) Ecological genetics of Reaumuria soongorica (Pall.) Maxim. population in the oasis–desert ecotone in Fukang, Xinjiang, and its implications for molecular evolution. Biochemical Systematics and Ecology 36: 593–601. [Google Scholar]
- 34. Shi Y, Yan X, Zhao P, Yin H, Zhao X, et al. (2013) Transcriptomic Analysis of a Tertiary Relict Plant, Extreme Xerophyte Reaumuria soongorica to Identify Genes Related to Drought Adaptation. PLoS ONE 8: e63993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, et al. (2002) Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome biology 3: RESEARCH0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Pfaffl MW, Tichopad A, Prgomet C, Neuvians TP (2004) Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper–Excel-based tool using pair-wise correlations. Biotechnol Lett 26: 509–515. [DOI] [PubMed] [Google Scholar]
- 37. Ramakers C, Ruijter JM, Deprez RH, Moorman AF (2003) Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neuroscience letters 339: 62–66. [DOI] [PubMed] [Google Scholar]
- 38. Ruijter JM, Ramakers C, Hoogaars WM, Karlen Y, Bakker O, et al. (2009) Amplification efficiency: linking baseline and bias in the analysis of quantitative PCR data. Nucleic Acids Res 37: e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Khodadadi A, Hamidi- Nia M, Ghaforian M, Mohammadi- Asl J (2012) Evaluation of real-time PCR efficiency by the use of two strategies: standard curve and linear regression. Jundishapur Sci Med J 11 1: 85–95. [Google Scholar]
- 40. Chang E, Shi S, Liu J, Cheng T, Xue L, et al. (2012) Selection of reference genes for quantitative gene expression studies in Platycladus orientalis (Cupressaceae) Using Real-Time PCR. PLoS ONE 7: e33278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Dhingra A, Portis AR Jr, Daniell H (2004) Enhanced translation of a chloroplast-expressed RbcS gene restores small subunit levels and photosynthesis in nuclear RbcS antisense plants. Proc Natl Acad Sci USA 101: 6315–6320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Salvucci ME, Portis AR, Ogren WL (1986) Light and CO(2) response of ribulose-1,5-bisphosphate carboxylase/oxygenase activation in Arabidopsis leaves. Plant Physiol 80: 655–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Vu JC, Allen LH, Bowes G (1987) Drought stress and elevated CO(2) effects on soybean ribulose bisphosphate carboxylase activity and canopy photosynthetic rates. Plant Physiol 83: 573–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Carmo-Silva AE, Keys AJ, Andralojc PJ, Powers SJ, Arrabaca MC, et al. (2010) rbcL activities, properties, and regulation in three different C4 grasses under drought. J Exp Bot 61: 2355–2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ginzinger DG (2002) Gene quantification using real-time quantitative PCR: an emerging technology hits the mainstream. Experimental hematology 30: 503–512. [DOI] [PubMed] [Google Scholar]
- 46. Bustin SA, Benes V, Nolan T, Pfaffl MW (2005) Quantitative real-time RT-PCR–a perspective. Journal of Molecular Endocrinology 34: 597–601. [DOI] [PubMed] [Google Scholar]
- 47. Dang ZH, Zheng LL, Wang J, Gao Z, Wu SB, et al. (2013) Transcriptomic profiling of the salt-stress response in the wild recretohalophyte Reaumuria trigyna . BMC Genomics 14: 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Klie M, Debener T (2011) Identification of superior reference genes for data normalisation of expression studies via quantitative PCR in hybrid roses (Rosa hybrida). BMC Research Notes 4: 518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Rai V, Dey N (2012) Identification of programmed cell death related genes in bamboo. Gene 497: 243–248. [DOI] [PubMed] [Google Scholar]
- 50. Rai V, Ghosh JS, Pal A, Dey N (2011) Identification of genes involved in bamboo fiber development. Gene 478: 19–27. [DOI] [PubMed] [Google Scholar]
- 51. Xu H, Chen LJ, Qu LJ, Gu HY, Li DZ (2010) Functional conservation of the plant embryonic flower2 gene between bamboo and Arabidopsis . Biotechnol Lett 32: 1961–1968. [DOI] [PubMed] [Google Scholar]
- 52. Czechowski T, Stitt M, Altmann T, Udvardi M, Scheible W-R (2005) Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis . Plant Physiol 139: 5–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Jain M, Nijhawan A, Tyagi AK, Khurana JP (2006) Validation of housekeeping genes as internal control for studying gene expression in rice by quantitative real-time PCR. Biochem Biophys Res Commun 345: 646–651. [DOI] [PubMed] [Google Scholar]
- 54. Jian B, Liu B, Bi Y, Hou W, Wu C, et al. (2008) Validation of internal control for gene expression study in soybean by quantitative real-time PCR. BMC Mol Biol 9: 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Agarose gel electrophoresis for PCR products of 10 candidate reference genes and rbcL .
(TIFF)
Expression levels of candidate reference genes across all the four accessions.
(TIFF)
Annotation information of 10 reference genes and the one validation gene from the de novo transcriptome sequences.
(XLS)
Ranking of the best reference gene based on the all three algorithms.
(DOC)
Contains 11 figures from Figure S3 to S13 representing the qPCR amplification curves of 10 candidate reference genes ( ACT, CYCL, DNAJ, EF1, EIF4A2, H2A, L2, TIP41, TUA and UBQ ) and one validation gene rbcL , respectively.
(ZIP)
Contains 11 figures from Figure S14 to S24 representing the melting curves of qPCR products of the 10 candidate reference genes ( ACT, CYCL, DNAJ, EF1, EIF4A2, H2A, L2, TIP41, TUA and UBQ ) and one validation gene rbcL , respectively.
(ZIP)
Alignment of the protein sequences of rbcL homologs.
(PDF)
Contains 11 sequence alignment files from Fasta S1 to S11,for verification of the homologs of 10 reference genes and one validation gene rbcL .
(ZIP)