Abstract
Engineered nanoparticles are widely used in commercial products, and yet due to the paucity of safety information, there are concerns surrounding potential adverse health effects, especially from inhaled nanoparticles and their putative contribution to allergic airway disease. The objective of this study was to investigate whether size or surface chemistry of engineered nanoparticles can influence the immune enhancing properties of these agents on antigen-specific T cell responses. Ovalbumin (OVA)-derived peptides were presented to T cells by either spleen-derived endogenous antigen presenting cells or a mouse dendritic cell (DC) line, DC2.4. In all models, interferon (IFN)-γ and interleukin (IL)-2 production by CD8+ or CD4+ T cells in response to peptide OVA257–264 or OVA323–339, respectively, was measured by flow cytometry. To address the study objective, silica nanoparticles (SNPs) were modified with alkyne-terminated surfaces and appended with polyethylene glycol chains via “click” chemistry. These modified SNPs were resistant to agglomerate in in vitro culture media, suggesting that their modulation of T cell responses is the result of true nanoscale-mediated effects. Under conditions of suboptimal T-cell activation, modified SNPs (up to 10 μg/ml) enhanced the proportion of CD8+, but not CD4+, T cells producing IFN-γ and IL-2. Various functional groups (–COOH, –NH2 and –OH) on modified SNPs enhanced IFN-γ and IL-2 production to different levels, with –COOH SNPs being the most effective. Furthermore, 51 nm –COOH SNPs exhibited a greater enhancing effect on the CD8+ T cell response than other sized particles. Collectively, our results show that modified SNPs can enhance antigen-specific CD8+ T cell responses, suggesting that certain modified SNPs exhibit potential adjuvant-like properties.
Keywords: Silica nanoparticles, cytokines, T cells, ovalbumin antigens, adjuvant
Introduction
Nanoparticles can occur naturally in the environment or can be artificially engineered for a wide range of applications in industry, such as electronics, medicine, food, clothing and cosmetics (Stern & McNeil, 2008). Nanoparticles have potentially beneficial effects; for example, their therapeutic use in vaccine development (Demento et al., 2009). By contrast, the same mechanisms that account for their beneficial attributes in vaccine development can contribute to putative adverse effects on human health (Colvin, 2003; Nel et al., 2006). Some of these concerns have arisen from animal studies, which have demonstrated that nanoparticles can exhibit adjuvant-like properties and can also contribute to allergic pulmonary disease through increased granulocyte infiltration, production of proinflammatory cytokines and allergen-specific antibodies (Inoue et al., 2006; Li et al., 2009). Little is presently known concerning the consequences resulting from interactions of engineered nanoparticles with the immune system, especially at the cellular level.
In this investigation, silica nanoparticles (SNPs) were synthesized, which were modified with alkyne-terminated surfaces and appended with polyethylene glycol (PEG)-azides via “click” chemistry. Pegylated SNPs are dispersed in water and can readily interact with, as well as enter, cells (Lin & Haynes, 2009; Luhmann et al., 2008). Previous in vivo studies using bronchoalveolar lavage fluid (BALF) and lymph node (LN)-derived cells from ovalbumin (OVA)/SNP-treated mice have demonstrated the immune enhancing effects of these SNPs on cell counts and activation of lymphocytes as well as other cell populations (Brandenberger et al., 2013). However, since we were limited by the quantity of materials available and the number of T cells that could be obtained from BALF and LN samples, in vitro models were employed to delineate the effect of SNPs on T-cell function directly. In addition, these in vitro models were easily manipulated allowing for the study of mechanisms associated with SNP-mediated immune enhancing effects.
In this study, antigen-specific T cell responses were induced in vitro. T cells were obtained from spleens of Ova-T-cell receptor-specific (TCR) transgenic mice, which have been engineered to express TCRs specific for OVA-derived peptides OVA257–264 (SIINFEKL) or OVA323–339 (ISQAVHAAHAEINEAGR), respectively (Clarke et al., 2000; Sette et al., 1987). In the OT-I model, OVA257–264 peptides that bind to major histocompatibility complex (MHC)-I molecules were presented by either endogenous antigen presenting cells (APCs) from the splenic population or a mouse dendritic cell (DC) line, DC2.4, to activate OT-I CD8+ T cells specifically. By using DC2.4 as APCs, OVA peptides were directly loaded onto DC2.4 cells, which were then co-cultured with splenic cells. In the OT-II model, OVA323–339 peptides that bind to MHC II molecules were presented by endogenous APCs from the splenic population to OT-II CD4+ T cells specifically. Production of cytokines, including interferon (IFN)-γ and interleukin (IL)-2, were used as a measure of T cell function.
The physical and chemical properties of the engineered nanoparticles determine their uptake by cells and therefore their activity in the cells (Dobrovolskaia & McNeil, 2007; Dobrovolskaia et al., 2008; Wilhelm et al., 2003). Therefore, SNPs were synthesized in different sizes and with modifications to surface chemistry through the addition of functional groups, including –COOH, –NH2 and –OH, which were resistant to agglomeration in in vitro culture media by performing dynamic light scattering (DLS) analysis. Using the above-mentioned antigen specific models, the objective of this study was to investigate whether size or surface chemistry of engineered nanoparticles can influence the immune enhancing properties of these agents on antigen-specific T cell responses.
Methods
SNPs
LUDOX® TM-40 colloidal silica (LTM40, 40wt% suspension in H2O), 3-(triethoxysilyl)propyl isocyanate (IPTEOS), propargyl alcohol, sodium ascorbate and sodium azide were purchased from Sigma-Aldrich (St. Louis, MO) and used as received. Colloidal silica SNOWTEX XS (SNTXS, 4–6 nm, 20wt% suspension in H2O), SNOWTEX 20 L (SNT20L, 40–50 nm, 20wt% suspension in H2O) and SNOWTEX ZL (SNTZL, 71 nm, 40wt% suspension in H2O) were gifts from Nissan Chemical Industries, Ltd. (Pasadena, TX). 1-azido-2-(2-(2-(2-methoxyethoxy)ethoxy) ethoxyethane (N34PEG) was synthesized according to a literature procedure (Kitto et al., 2008). Dialysis membranes (Spectra/Por® Biotech Cellulose ester, molecular weight cut off (MWCO): 300 K, 12–14 K) were purchased from Spectrum Laboratories Inc. (Rancho Dominguez, CA). Ion exchange resin (Amberlite® IRC-748) was purchased from Alfa Aesar (Ward Hill, MA).
Instruments and SNP characterization
1H nuclear magnetic resonance (NMR) and 13C NMR spectra were recorded in CDCl3 on a Varian 300 MHz or VXR-500 MHz instrument (Agilent Technologies, Santa Clara, CA). The CDCl3 resonance was used as the internal standard for 13C NMR (δ = 77.0 ppm) and residual CHCl3 for 1H NMR (δ = 7.24 ppm). Fourier transform infrared spectroscopy (FTIR) spectra were recorded on a Mattson Galaxy series FTIR 3000 (Mattson Instruments, Inc., Madison, WI). Thermogravimetric analyses (TGA) were obtained in air from a Perkin-Elmer TGA 7 (Perkin Elmer Corporation, Norwalk, CT). Samples were held at 120 °C for 30 min to remove adsorbed water from the particle surfaces, and then heated to 850 °C at a rate of 10 °C/min. All FTIR and TGA samples were dried under vacuum at room temperature for 24 h.
DLS data were obtained with a Malvern Nano ZS ZetaSizer (Malvern Instruments Inc., Westborough, MA) with 178° back-scattering detection. Intensity and average diameters were calculated from the autocorrelation function using Malvern’s Zetasizer Software 6.12 (Malvern Instruments, Inc., Westborough, MA, US). Samples for DLS analyses were sonicated at 60 °C prior to measuring particle sizes at 25 °C (FS20H sonicator, Fisher Scientific, Pittsburgh, PA). For nanoparticle stability studies, particles were added to fresh in vitro culture media (see below), dispersed by vortex mixing and characterized by DLS at 0, 4, 8, 21, 42, 68 h at 25 °C. Particles only and media only were included as experimental controls.
Synthesis of reagents for modifying nanoparticles and click chemistry
Synthesis of ((2-propynylcarbamate)propyl)triethoxysilane
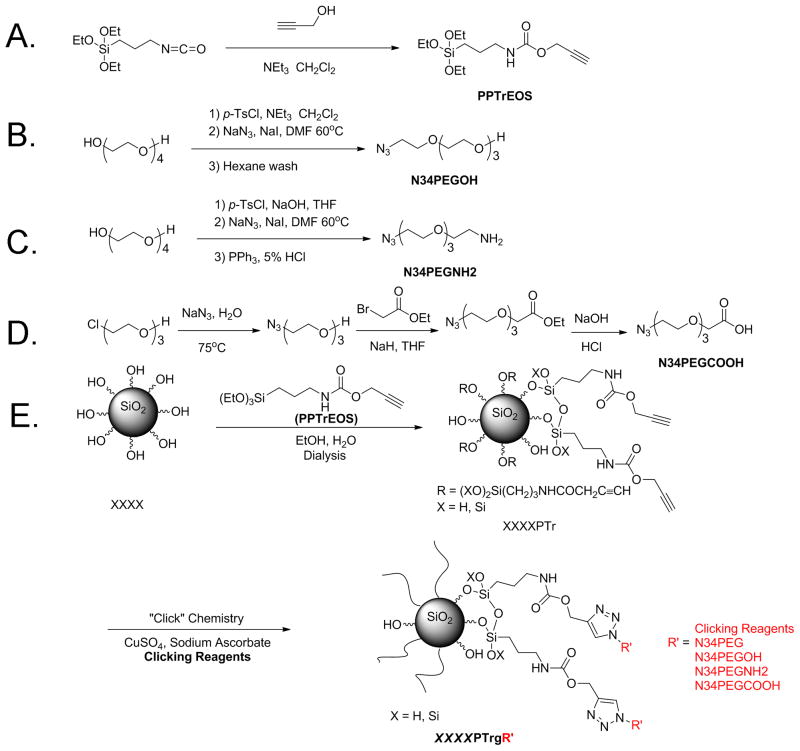
Propargyl alcohol (12 g, 0.21 mol) and triethylamine (19 g, 0.19 mol) were dissolved in 120 mL of dry CH2Cl2 (Figure 1A). The mixture was cooled in an ice bath, and under nitrogen, a solution of IPTEOS (46 g, 0.18 mol) in 60 mL of dry CH2Cl2, was added dropwise to the flask. The reaction mixture was stirred for 24 h at room temperature and then filtered. CH2Cl2, excess propargyl alcohol and NEt3 were evaporated under reduced pressure to afford 50 g of ((2-propynylcarbamate) propyl)triethoxysilane (PPTrEOS) as pale-yellow oil. 1H NMR (500 MHz, CDCl3) 0.52 (t, 2H, J = 8 Hz), 1.11 (t, 9H, J = 8 Hz), 1.52 (m, 2H), 2.37 (s, 1H), 3.08 (m, 2H), 3.71 (q, 6H, J = 7Hz) and 4.55 (s, 2H).
Figure 1.
Synthesis of different SNPs. (A–D) Synthesis steps are illustrated for PPTrEOS (A), N34PEGOH (B), N34PEGNH2(C) and N34PEGCOOH (D). (E) Particle synthesis procedure is illustrated for modified SNPs. XXXX represents different commercial SNPs, including LTM40, SNTXS, SNT20L and SNTZL. PTr represents particles modified with alkyne functional groups. R′ represents different clicking reagents, including N34PEG, N34PEGOH, N34PEGNH2 and N34PEGCOOH, which are PEG chains of four repeating units terminated with –OMe, –OH, –NH2 and –COOH functional groups, respectively. g represents that the clicking reagents were grafted to the particles via “click” chemistry.
Synthesis of 2-[2-[2-[2-azidoethoxy]ethoxy]ethoxy]ethanol
A solution of TsCl (14 g, 0.071 mol) in 250 mL of tetrahydrofuran (THF) was added dropwise to a solution of tetraethylene glycol (34g, 0.18mol), triethylamine (14 mL) and THF (250 mL) (Figure 1B). After stirring the resulting solution for 36 h at room temperature, THF was removed by rotary evaporation, and the crude product was dissolved in 200 mL of CH2Cl2. The organic layer was washed with water (3 × 150 mL), aqueous NaHCO3 (200 mL) and brine (200 mL). After drying over Na2SO4, the solution was concentrated and dried under vacuum to give 18 g of a light yellow liquid comprised of a mixture of mono-tosylate (85%) and di-tosylate (15%), which corresponded to 14.3 g of mono-tosylate (58% yield). The product was used in the next step without further purification. 1H NMR (500 MHz, CDCl3) δ 7.77 (m, 2H), 7.32 (m, 2H), 4.29 (m, 2H), 3.69–3.50 (m, 14H), 2.5 (br, 1H) and 2.42 (s, 3H). The tosylates were dissolved in dimethylformamide (DMF) (210 mL), treated with NaN3 (10.4 g, 15 mmol) and then heated to 60 °C for 24 h. Then the mixture was concentrated, re-dissolved in ethyl acetate, filtered to remove the solid sodium tosylate and concentrated under reduced pressure. The resulting oily product was dissolved in water and washed with hexane until α,ω-diazido tetraethylene glycol was not detected in the aqueous layer. Drying under vacuum gave the product as a light yellow liquid (8.0 g, 72%). 1H NMR (500 MHz, CDCl3) δ 3.69 (m, 2H), 3.64 (m, 10H), 3.58 (m, 2H), 3.36 (t, 2H, J = 5.5 Hz), 2.8 (br, 1H); 13C NMR (125 MHz, CDCl3) 72.41, 70.55, 70.51, 70.43, 70.18, 69.90, 61.56 and 50.54.
Synthesis of 2-[2-[2-[2-azidoethoxy]ethoxy]ethoxy]ethyl amine (N34PEGNH2)
A solution of TsCl (39.9 g, 0.21 mol) in THF (150 mL) was added dropwise into an ice-cold mixture of tetraethylene glycol (13.5 g, 0.07 mol), aqueous KOH (100 g, 47%, w/w) and THF (100 mL) (Figure 1C). After stirring the solution for 36 h at room temperature, THF was removed by rotary evaporation, and then CH2Cl2 (200 mL) was added to the residue. The organic layer was washed with water (3 × 150 mL), aqueous NaHCO3 (200 mL) and brine (200 mL). After drying the organic layer over Na2SO4, the solution was concentrated and dried under vacuum to give 32 g of a light yellow liquid (91%), which was used in the next step without further purification. 1H NMR (300 MHz, CDCl3) δ 7.81 (m, 4H), 7.36 (m, 4H), 4.20 (m, 4H), 3.70–3.65 (m, 12H) and 2.47 (s, 6H). The di-tosylate of tetraethylene glycol was treated with NaN3 (11.7 g, 18 mmol) in DMF (300 mL) at 90 °C for 24 h. The mixture was concentrated, dissolved in water and extracted with CH2Cl2 (4 × 100 mL). The solution was dried over anhydrous Na2SO4 and concentrated under reduced pressure to give 12 g of a yellow liquid. Aqueous HCl (5%, 100 mL) was added to the yellow liquid, and under vigorous stirring at room temperature, a solution of PPh3 (11.4 g, 0.041 mol) in 75 mL of ether was added dropwise. After stirring for 24 h, the ether was removed by rotary evaporation and the aqueous layer was extracted with CH2Cl2 until triphenylphosphine oxide was not detected in the aqueous layer. The aqueous layer was adjusted to pH = 12, and amine was extracted from the aqueous layer with CH2Cl2 (6 × 50 mL). Drying under vacuum gave the product as a light yellow liquid (9.0 g 68%). 1H NMR (500 MHz, CDCl3) δ 3.72–3.58 (m, 10H), 3.48 (t, 2H, J = 5 Hz), 3.35 (t, 2H, J = 5 Hz), 2.84 (t, 2H, J = 5 Hz), 2.24 (s, 2H); 13C NMR (125 MHz, CDCl3) 72.79, 70.50, 70.47, 70.43, 70.07, 69.85, 50.50 and 41.44.
Synthesis of 2-[2-[2-[2-azidoethoxy]ethoxy]ethoxy]acetic acid (N34PEGCOOH)
A solution of [2-[2-[2-chloroethoxy]ethoxy]ethanol (16.8 g, 0.1 mol) and NaN3 (13 g, 0.2 mol) in 75 mL of water was heated at 75 °C with stirring for 24 h (Figure 1D). Water was removed by rotary evaporation, and the crude product was extracted from the salts with 200 mL of CH2Cl2. Concentration and drying under vacuum gave the product as a light yellow liquid (17 g, 97%), which was used without further purification. 1H NMR (500 MHz, CDCl3) δ 3.72–3.62 (m, 8H), 3.58 (m, 2H) and 3.37 (t, 2H, J = 5 Hz). The light yellow liquid product (14 g, 0.08 mol), prepared above, was added to NaH (0.1 mol) in 350 mL of dry THF cooled in an ice bath. After stirring for 1 h at room temperature, ethyl bromoacetate (17.7 mL, 0.16 mol) was added dropwise to the solution and then stirred for 48 h at room temperature. The THF was removed by rotary evaporation, and then 150 mL of water was added. The aqueous layer was extracted with CH2Cl2 (3 × 50 mL), dried over anhydrous Na2SO4 and concentrated under reduced pressure at 55 °C to give an amber liquid (26 g), which was used without further purification. 1H NMR (500 MHz, CDCl3) δ 4.24 (q, 2H, J = 10Hz), 4.17 (s, 2H), 3.77–3.65 (m, 10H) and 3.41 (t, 2H, J = 5 Hz). The ester was hydrolyzed with 53 mL of 3M NaOH at room temperature for 18 h. After extracting the aqueous layer with CH2Cl2 (10 × 30 mL) to remove mineral oil and other impurities, the aqueous layer was acidified to pH = 1 and extracted with ethyl acetate (EtOAc) (1 × 80 mL, 7 × 30 mL). After drying over anhydrous Na2SO4, the residue was concentrated to a pale red liquid (14g, 75%). 1H NMR (500 MHz, CDCl3) δ 4.18 (s, 2H), 3.78–3.67 (m, 10H), 3.41 (t, 2H, J = 5 Hz); 13C NMR (125 MHz, CDCl3) 177.88, 76.71, 75.95, 75.69, 75.48, 75.31, 74.02 and 55.9.
Surface modification of SNPs
Synthesis of XXXXPTr: one-step alkyne modification of SNPs (Figure 1E)
XXXX represented different commercial SNPs, including LTM40, SNTXS, SNT20L and SNTZL (9g, 40% suspension in water), which were diluted with 75 mL of 1:1 ethanol (EtOH)–H2O and sonicated. Particle size was measured by DLS prior to modification. pH of the silica suspension was adjusted to ∼9 by adding drops of aqueous ammonia. At room temperature, a solution of PPTrEOS (3 g, 10 mmol) in 10 mL of EtOH was added dropwise to the stirred silica suspension. After stirring for 3 days, the suspension was diluted with EtOH until it was homogeneous. The modified particles were purified by dialysis in EtOH and water (9:1) using dialysis tubing (MWCO: 300 K). The surface coverage of the alkyne functional group was evaluated by TGA. The particle concentration (mg/g) was determined by evaporating 2–4 mL of purified particle solutions.
Synthesis of XXXXPTrgR’ with different functional groups: “Click” chemistry of SNPs
Click chemistry between alkyne-modified SNPs and clicking reagents (R′, including N34PEG, 2-[2-[2-[2-azidoethoxy]ethoxy] ethoxy]ethanol, N34PEGNH2 and N34PEGCOOH) is described using the synthesis of LTM40PTrg4PEG as an example (Figure 1E). LTM40PTr (100 mg, 0.036 mmol of alkyne functional groups) and N34PEG (41.9 mg, 0.18 mmol, 5 equiv.) were added to 13.8 g of EtOH and water (9:1). Then, this mixture was diluted in 3:1 DMF/water until the solution was clear. After adding sodium ascorbate (14.3 mg, 2 equiv.) to the solution, trace O2 was removed by three freeze–pump–thaw cycles. Aqueous CuSO4 (72 μL, 0.1 M, 0.2 equiv.) was added to the solution followed by a final degassing process, and the solution changed to light brown–yellow. After stirring at room temperature for 36 h, 1 mL of aqueous ethylenediaminetetraacetic acid (EDTA) (0.1 M) was added and the solution was stirred for another 6 h. The resulting particles were dialyzed against (MWCO:12–14 K) EtOH/water (9:1, 5 × 2L) and against distilled water to afford purified surface-modified SNPs with different surface chemistry. For the synthesis of LTM40PTrg4PEGNH2, EDTA was not added after click chemistry, but was dialyzed directly as stated above, and then the solution of LTM40PTrg4PEGNH2 was sonicated to be homogeneous. Ion exchange resin (2 g, Amberlite® IRC-748) was added to remove the Cu2+, and it was removed via filtration after 8 h. The modified SNPs, including LTM40PTrg4PEG, LTM40PTrg4PEGOH, LTM40PTrg4PEGNH2, LTM40PTrg4PE GCOOH, SNTXSPTrg4PEGCOOH, SNT20LPTrg4PEGCOOH and SNTZLPTrg4PEGCOOH, were kept in distilled water until further characterization. The surface coverage of surface functional groups after each step modification was evaluated by TGA. g indicates that the clicking reagents were grafted to the particles via “click” chemistry. DLS samples were prepared by diluting or concentrating solutions to 0.1 or 1 mg/mL. The particle concentration (mg/g) was determined by evaporating 2–4 mL of purified particle solutions.
Mice
Female OT-I and OT-II mice were purchased from Jackson Laboratory (Bar Harbor, ME) and used at 8–12 weeks of age. Mice were randomized at no more than five animals per cage and provided rodent chow (Harlan Teklad, Madison, WI) and water ad libitum. Rooms were kept on a 12-hr light/dark cycle at 21–24 °C and 40–60% humidity. All experiments were performed following the guidelines set forth by Michigan State University Institutional Animal Care and Use Committee.
Cell culture conditions
DC2.4 cells (a kind gift from Dr. Kenneth Rock at University of Massachusetts) were maintained in complete Roswell Park Memorial Institute (RPMI) media (10% bovine calf serum [BCS; HyClone, Logan, UT], 100 units/ml penicillin and 100 μg/ml streptomycin [pen/strep]).
Activation of OT-I or OT-II T cells with OVA peptides in vitro
Splenocytes were isolated aseptically from OT-I or OT-II mouse spleens and made into single-cell suspensions. Splenocytes (8 × 105 cells/well) were treated with various concentrations (10−8 to 10−2 μg/ml) of OVA257–264 peptide SIINFEKL or OVA323–339 peptide ISQAVHAAHAEINEAGR (Anaspec, San Jose, CA) in 48-well plates for 2 days in 2% BCS Roswell Park Memorial Institute (RPMI) media (2% BCS, pen/strep, and 50 μM 2-mercaptoethanol). When DC2.4 cells were used as APCs, they were plated at 2 × 106 cell/well in six-well pates and pretreated with 1 μg/ml lipopolysaccharide (LPS) (Salmonella typhosa, Sigma-Aldrich) in 2% BCS RPMI media overnight. DC2.4 cells were then incubated with various concentrations of SIINFEKL (10−2 to 10 μg/ml) for 3–4 h, x-ray irradiated at 35 gray to prevent their proliferation, washed three times to remove the extra SIINFEKL and plated 2 × 105 cell/well in 96 U-Bottom plates with 1 × 106 cells/well OT-I splenocytes for 4 days in 5% BCS RPMI media (5% BCS, pen/strep and 50 μM 2-mercaptoethanol). Peptide concentration for suboptimal stimulation of T cell response was determined and used to further study the influence of modified SNPs on T cell function.
Modified SNPs treatment in cell culture
Once the suboptimal stimulation condition was determined, splenocytes were treated with modified SNPs with or without different surface functional groups or in different sizes, including LTM40PTrg4PEG, LTM40PTrg4PEGOH, LTM40PTrg4PEGNH2, LTM40PTrg4PEGCOOH, SNTXSPTrg4PEGCOOH, SNT20LP Trg4PEGCOOH and SNTZLPTrg4PEGCOOH. These modified SNPs were sonicated at 60 °C for 35 min and then added to the splenocyte suspension at various concentrations (10−2 to 10 μg/ml) prior to the OVA peptide stimulation as describe above. As reviewed by Gangwal et al. (2011), a 10 μg/ml concentration of SNP falls into the range of concentrations (∼30 to 400 μg/ml) of nanoparticles used for in vitro assay testing based on estimates of lung deposition after occupational exposure.
Flow cytometry analysis
On day 2 post the initiation of peptide stimulation, brefeldin A (Biolegend, San Diego, CA) was added to the culture during the last 6 h of incubation to prevent cytokine release and allow for the identification of cytokine producing T cells. Cells were washed with 1 × Hank’s Balanced Salt Solution (HBSS) buffer (Invitrogen, Carlsbad, CA) and incubated with LIVE/DEAD Fixable Near-IR Dead Cell Stain Kit (Invitrogen) to assess cell viability according to manufacturer’s instructions. For surface molecule staining, cells were blocked for Fc receptors with purified rat anti-mouse CD16/CD32 (“Fc block”, BD Pharmingen, San Diego, CA), then incubated with PE/Cy7-conjugated rat anti-mouse antibodies against CD8α (clone 53–6.7, Biolegend,) or CD4 (clone RM4–5, Biolegend) and fixed with Cytofix (BD Pharmingen). On the day of flow cytometric analysis, cells were permeabilized with 1 × Perm/Wash (BD Pharmingen) and stained with PE-conjugated anti-mouse IFN-γ antibody (clone XMG1.2, Biolegend) and APC-conjugated anti-mouse IL-2 antibody (clone JES6-5H4, Biolegend). Single stain controls were included in all experiments to compensate for fluorescence interference between detectors. Cells were washed with 1 × Perm/Wash, resuspended in fluorescence activated cell sorting (FACS) buffer (1 × HBSS containing 1% bovine serum albumin (BSA) and 0.1% sodium azide), assessed on a FACS Canto II flow cytometer (BD Biosciences, San Jose, CA) and analyzed with FlowJo v8.8.6 (Tree Star, Ashland, OR) or Kaluza 1.1 (Beckman Coulter, Miami, FL) software. In the experiments where DC2.4 cells were used for elicitation, on day 4 post the initiation of the co-culture, cells were restimulated with SIINFEKL (10 μg/ml) in the presence of brefeldin A for 6 h in 2% BCS RPMI media. Cells were then stained for viability, surface CD8 expression and intracellular cytokine production as described above.
Statistical analysis
Graphing and statistical analyses were performed using GraphPad Prism v4.0 (Graphpad Software, San Diego, CA). The mean ± SE is presented in all bar graphs. To determine the statistically significant changes between peptide stimulated and unstimulated cells, two-way analysis of variance (ANOVA) for parametric data was performed. Bonferroni post-test was used to compare samples to their control group. Differences between means from each treatment group were determined using one-way ANOVA. When significant differences were detected, treatment groups were compared to the appropriate control using Dunnett’s test.
Results
Surface chemistry of modified LTM40PTrg4PEG SNPs
PPTrEOS was synthesized as a pure material with a high yield. Characterization by 1H NMR and 13C NMR indicated that further purification was not required (Choualeb et al., 2003; Lu et al., 2009). Commercial SNPs were condensed with PPTrEOS in basic EtOH–H2O mixture for 3 days, as outlined in Figure 1(E). After condensation, alkyne-modified SNPs were purified via dialysis, using tubing with MWCO values appropriate for the sizes of the particles. Purification continued until no organic chemicals were detected in the dialysate residue by 1H NMR analysis, usually lasting between 1.5 and 8 days. After dialysis was completed, particles were recovered by evaporating the dialysis solvent for further characterization. “Click” chemistry on SNP surfaces was conducted to generate SNPs with different functional group moieties. These modified SNPs were purified by dialysis and used for further surface characterization, as well as in in vitro culture studies.
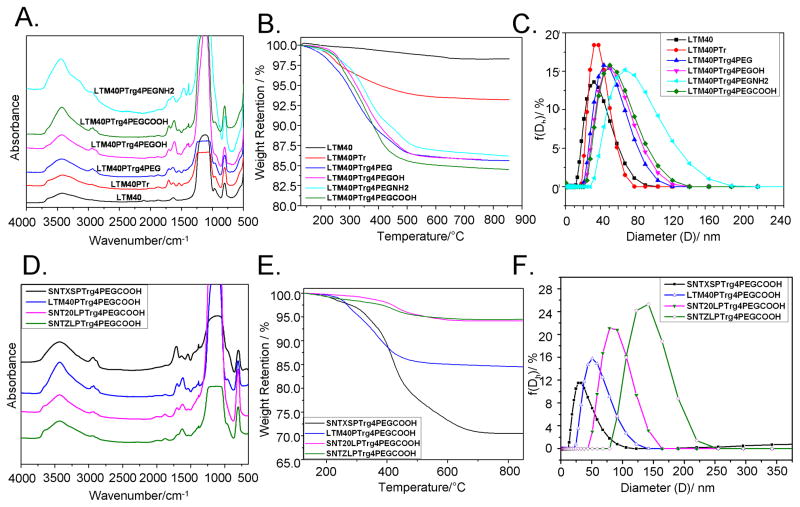
Changes in nanoparticle surface chemistry were tracked by FTIR. Figure 2(A) shows normalized FTIR spectra of LTM40 and its modified SNPs as described in “Methods” section. The band at 806 cm−1, characteristic of bulk silica, allowed normalization of the data and semi-quantitative comparison of the nanoparticles. For example, LTM40PTr showed new absorption at 2900–3300 cm−1, 2123 cm−1, 1700 cm−1 and 1430 cm−1, which corresponded to the alkyl group, triple bond, carbonyl group and another alkyl group, respectively, and confirmed the functionalization of particle surface with alkynes. After “click” modification of the particles, bands at 2930 and 1640 cm−1 increased and were consistent with adding methylene groups from PEG units and the triazole ring (top four traces; Figure 2A). FTIR was limited for characterizing pegylation, since the characteristic C–O bands of PEG were buried under intense FTIR bands from the SNPs. However, C–H bending and stretching absorption bands were consistent with the PEG chain in LTM40PTrg4PEG (–OMe terminated) particles, as was seen in all other pegylated SNPs (–OH, –NH2 and –COOH terminated particles).
Figure 2.
Surface chemistry of modified SNPs with different functional groups and at different sizes. (A and D) FTIR spectra were obtained by scanning the pellets of different SNPs and KBr from 500 to 4000 cm−1, and the absorbance was recorded. FTIR spectra were normalized using the band intensity at 806 cm−1 for semi-quantitative comparison of the SNPs. (B and E) TGA traces of different SNPs were measured by burning the SNPs in air at a rate of 10 °C/min, and the weight retention of SNPs at different temperatures (120–850°C) was recorded. (C and F) Different SNPs (100 μg/ml) were sonicated, and DLS data (size distribution by intensity) were measured at 25 °C.
The TGA data presented in Figure 2(B) and Table 1 confirmed the step-by-step modification of SNPs possessing different surface functional groups. The grafting density on particle surfaces was calculated from TGA mass loss in air, which corresponded to oxidation of the organic layer on the nanoparticle surfaces. Table 1 showed a 5.1% weight loss for alkyne-modified nanoparticles (LTM40PTr), which was calculated to a grafting density of 1.54 alkyne groups/nm2. After clicking pegylated functional groups to LTM40PTr, TGA analyses showed 7.5–8.7% weight loss for all SNPs. The mass losses corresponded to a grafting density of ∼1.5 chain/nm2 and >90% high click conversion, indicating successful synthesis of water-dispersed nanoparticles.
Table 1.
Grafting amount and size of SNPs.
| Sample | Weight loss (%)a | Grafting amount (mmol/g)a | Grafting density (group/nm2)a | Click conversiona (%) | Size (nm)b |
|---|---|---|---|---|---|
| LTM40 | – | – | – | – | 30 |
| LTM40PTr | 5.1 | 0.36 | 1.5 | – | 37c |
| LTM40PTrg4PEG | 7.6 | 0.33 | 1.4 | 92 | 56 |
| LTM40PTrg4PEGOH | 7.5 | 0.34 | 1.4 | 94 | 50 |
| LTM40PTrg4PEGNH2 | 7.2 | 0.33 | 1.4 | 90 | 66 |
| LTM40PTrg4PEGCOOH | 8.7 | 0.37 | 1.5 | 97 | 51 |
Weight loss, grafting amount, density and click conversion were determined after each modification step.
Size measured by DLS after sonication for 30 min in H2O.
Size measured in EtOH.
DLS measurements were conducted with LTM40, LTM40PTr and the four click products, LTM40PTrg4PEG (–OMe), LTM40PTrg4PEGOH (–OH), LTM40PTrg4PEGNH2 (–NH2) and LTM40PTrg4PEGCOOH (–COOH) (Table 1 and Figure 2C). All surface-modified nanoparticles had monomodal distributions. After modifying the surface of LTM40 with alkynyl groups to make LTM40PTr, its size increased from 30 to 37 nm with a zeta potential of −35 mV measured in zwitterionic buffer (100 mM KC1/10 mM HEPES). The sizes of the four products obtained from LTM40PTr using click chemistry increased to 56, 50, 66 and 51 nm for LTM40PTrg4PEG, LTM40PTrg4PEGOH, LTM40PTrg4PEGNH2 and LTM40PTrg4PEGCOOH, respectively, and zeta potentials were changed to −10 mV, −13 mV, −7 mV and −17 mV, respectively. The DLS size of the click products slightly depended on the surface chemistry and their steric stabilization.
Surface chemistry of modified –COOH SNPs at different sizes
Grafting data from different sized –COOH SNPs are summarized in Table 2. The initial particle diameters ranged from 20 to 120 nm. Figure 2(D) showed normalized FTIR spectra for SNPs terminated with –COOH groups, including LTM40PTrg4 PEGCOOH, SNTXSPTrg4PEGCOOH, SNT20LPTrg4PEG COOH and SNTZLPTrg4PEGCOOH. Again, the FTIR band at 806 cm−1 (silica) was used to normalize the spectra and allowed semi-quantitative comparisons of the nanoparticles. All spectra showed FTIR absorption bands at 2900–3300 cm−1 and 1700 cm−1, which corresponded to C–H stretching and a carbonyl group, respectively, confirming successful modification of the particle surface. Since the specific surface areas of small SNPs are higher than large particles, small particles appear to have higher grafted layers than larger particles. Therefore, the FTIR bands in Figure 2(D) decreased in intensity from top to bottom (smallest particle to largest).
Table 2.
Grafting data from different sized SNPs terminated with carboxylic acids.
| Sample | Weight loss (%)a | Grafting amount (mmol/g)a | Grafting density (group/nm2)a | Click conversiona (%) | Size (nm)b |
|---|---|---|---|---|---|
| SNTXS | – | – | – | – | 20b |
| SNTXSPTr | 10.4 | 0.74 | 0.8 | – | 19c |
| SNTXSPTrg4PEGCOOH | 15.9 | 0.68 | 0.7 | 91.9 | 33c |
| LTM40 | – | – | – | – | 30b |
| LTM40PTr | 5.1 | 0.36 | 1.5 | – | 37c |
| LTM40PTrg4PEGCOOH | 8.7 | 0.37 | 1.5 | 100 | 51b |
| SNT20L | – | – | – | – | 80b |
| SNT20LPTr | 1.9 | 0.14 | 1.4 | – | 90c |
| SNT20LPTrg4PEGCOOH | 2.8 | 0.12 | 1.2 | 84.3 | 82b |
| SNTZL | – | – | – | – | 150b |
| SNTZLPTr | 1.6 | 0.12 | 2.1 | – | 117b |
| SNTZLPTrg4PEGCOOH | 2.2 | 0.10 | 1.7 | 79.2 | 137b |
Weight loss, grafting amount, density and click conversion were determined after each modification step.
Size measured in H2O by DLS after sonication.
Size measured in EtOH.
Table 2 and Figure 2(E) showed TGA results for –COOH– terminated SNPs. With increasing particle sizes (33 to 137 nm), TGA weight losses decreased from 15.9% to 2.2%, as expected due to the decreasing surface to mass ratios. The click conversion calculated from TGA data showed high conversion for all particles (>80%).
The DLS traces (Figure 2F) and the data from SNPs modified with –COOH groups are presented in Table 2. All modified nanoparticles had monomodal distributions (intensity distribution), a result of the steric stabilization of surface modified particles.
Aqueous media dispersion stability of modified SNPs
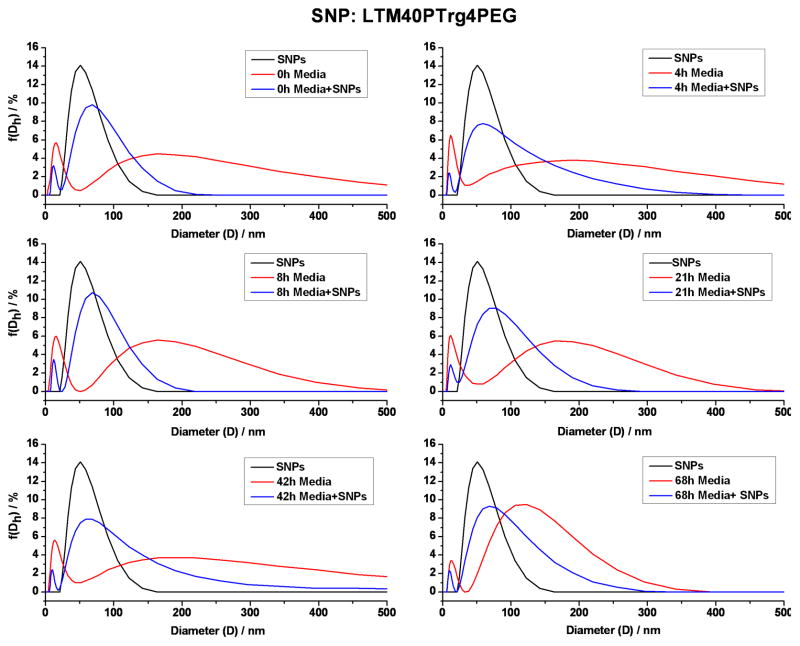
DLS was used to determine whether the modified SNPs remained dispersed in cell culture media (RPMI containing 2% BCS) as well as their stability over time. Four SNPs with different surface chemistry (–OMe, –OH, –NH2 and –COOH) were measured by DLS, and all were stabile for 2–3 days in the in vitro culture media without aggregation. For example, Figure 3 showed data for LTM40PTrg4PEG (–OMe) in in vitro media. At 0 h in water, the distribution of the nanoparticles was monomodal with a hydrodynamic size of ∼60nm (black curve). The results from in vitro culture media showed two peaks, a small narrow peak at ∼10 nm, and a broad particle population from >50 nm to <500 nm (red curve). After combining the particles and culture media (blue curve), the small peak at ∼10 nm, seen in the culture media, shifted slightly to lower diameters (<10 nm) and the combined scattering from the nanoparticle and the media formed a single peak, which broadened toward higher particle sizes. On the high size of the distribution, the scattering was comprised of the sum of the particles and the media. Importantly, the onset for scattering from the particles, the near-constant slope of the low side of the distribution and the peak of the distribution did not change as a function of time. We concluded that the pegylated SNPs in in vitro culture media showed no significant aggregation, and the main effect of the media was to broaden the distribution. The other three SNPs showed similar DLS behavior when added to in vitro culture media as presented in Supplemental Figure S1–S3.
Figure 3.
Aqueous dispersion stability of LTM40PTrg4PEG SNPs in in vitro culture media. LTM40PTrg4PEG SNPs were sonicated, and dispersed in culture media by vortexing. DLS data (size distribution by intensity) from LTM40PTrg4PEG SNPs alone (100 μg/ml, black curve), culture media alone (red curve) and LTM40PTrg4PEG SNPs in culture media (blue curve) at different time points (indicated on the graphs) were recorded.
OVA257–264 (SIINFEKL)-stimulated cytokine production by CD8+ T cells
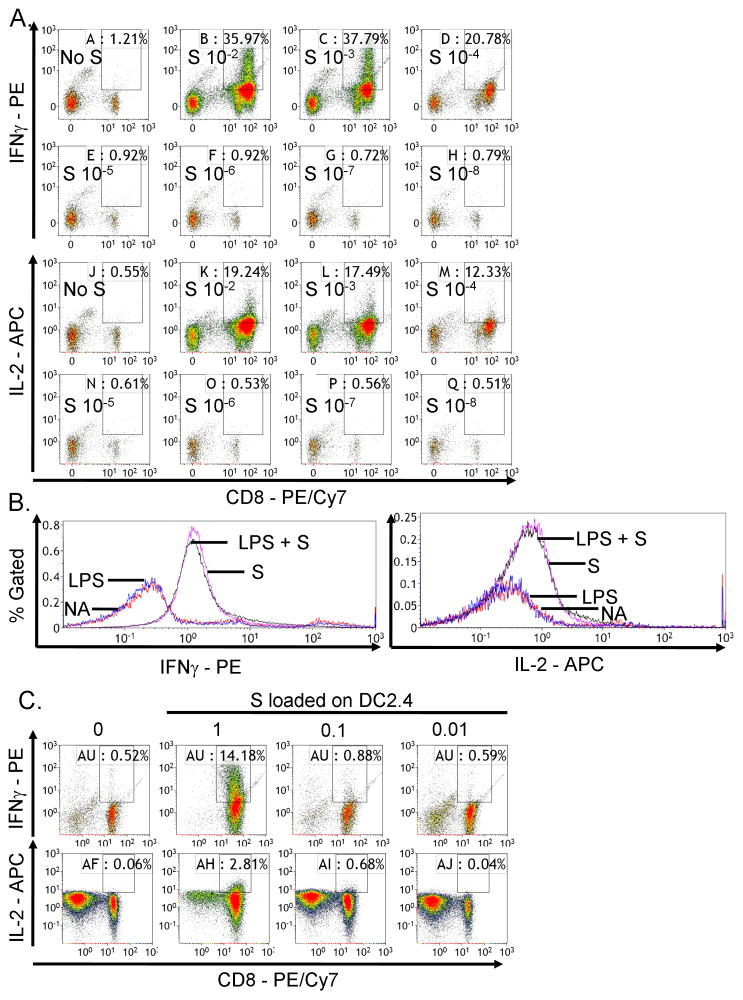
Using the OT-I model in which SIINFEKL was presented by endogenous APCs to CD8+ T cells, the percent of viable CD8+ T cells that produced IFNγ (Figure 4A, upper panel) or IL-2 (Figure 4A, bottom panel) increased with increasing concentrations of SIINFEKL (S; 10−8 to 10−2 μg/ml). There was little background cytokine production in the absence of SIINFEKL stimulation (No S). In addition, the fluorescence intensity of IFNγ was higher than IL-2 (Figure 4A). SIINFEKL (10−4 μg/ml) produced suboptimal, but measurable, stimulation of the CD8+ T cell response and was used in subsequent experiments.
Figure 4.
SIINFEKL concentration-dependent activation of CD8+ T cell responses. (A) OT-I splenocytes (CD8+ T cells, 8 × 105 cells/well) were incubated with various concentrations of SIINFEKL (S) peptide (10−8 to 10−2 μg/ml) for 2 days. Brefeldin A was added during the last 6 h of incubation. Splenocytes were stained for viability, surface CD8 and intracellular IFNγ or IL-2 expression. The percent of IFNγ+ or IL-2+ cells within CD8+ T cells is presented in the quadrants of dot plots, with X axis and Y axis representing the fluorescence intensity for CD8 and IFNγ or IL-2, respectively. (B) DC2.4 cells were pretreated with LPS (1 μg/ml) overnight, loaded with SIINFEKL peptide (1 μg/ml) for 3–4 h, irradiated and plated at 2 × 105 cells/well with 1 × 106 cells/well OT-I splenocytes for 4 days. Splenocytes were restimulated with SIINFEKL (10 μg/ml) in the presence of brefeldin A for 6 h. Splenocytes were stained for viability, surface CD8 and intracellular IFNγ or IL-2 expression. The percent of IFNγ+ or IL-2+ cells within CD8+ T cells is presented in histograms, with x axis representing fluorescence intensity for IFNγ or IL-2 and Y axis representing percent of CD8+ T cells. (C) DC2.4 cells were not pretreated with LPS and incubated with various concentrations of SIINFEKL (10−2 to 1 μg/ml), co-cultured with OT-I splenocytes as described in (B). The dot plots are presented as described in (A). (A–C) Three replicates were concatenated for each treatment in the flow cytometric analysis. Data are representative of at least two separate experiments.
Cytokine production was also measured using the model in which DC2.4 cells were used as APCs to present SIINFEKL to OT-I CD8+ T cells. DC maturation can be induced by various stimuli, including LPS (Banchereau & Steinman, 1998). However, in this model under current conditions, LPS pretreatment of DC2.4 cells did not further increase IFNγ (Figure 4B, left panel) or IL-2 (Figure 4B, right panel) production by SIINFEKL-activated viable CD8+ T cells. Thus, LPS pretreatment was not utilized in subsequent experiments. Production of IFNγ (Figure 4C, upper panel) or IL-2 (Figure 4C, bottom panel) by viable CD8+ T cells was increased with increasing concentrations of SIINFEKL (0.01–1 μg/ml) loaded onto MHC I of DC2.4 cells. Furthermore, there was more IFNγ production than IL-2, which was consistent with the trend observed in Figure 4(A). These studies showed that the concentration of SIINFEKL for MHC I loading on DC2.4, in order to achieve suboptimal activation of CD8+ T cells, was 0.1 μg/ml.
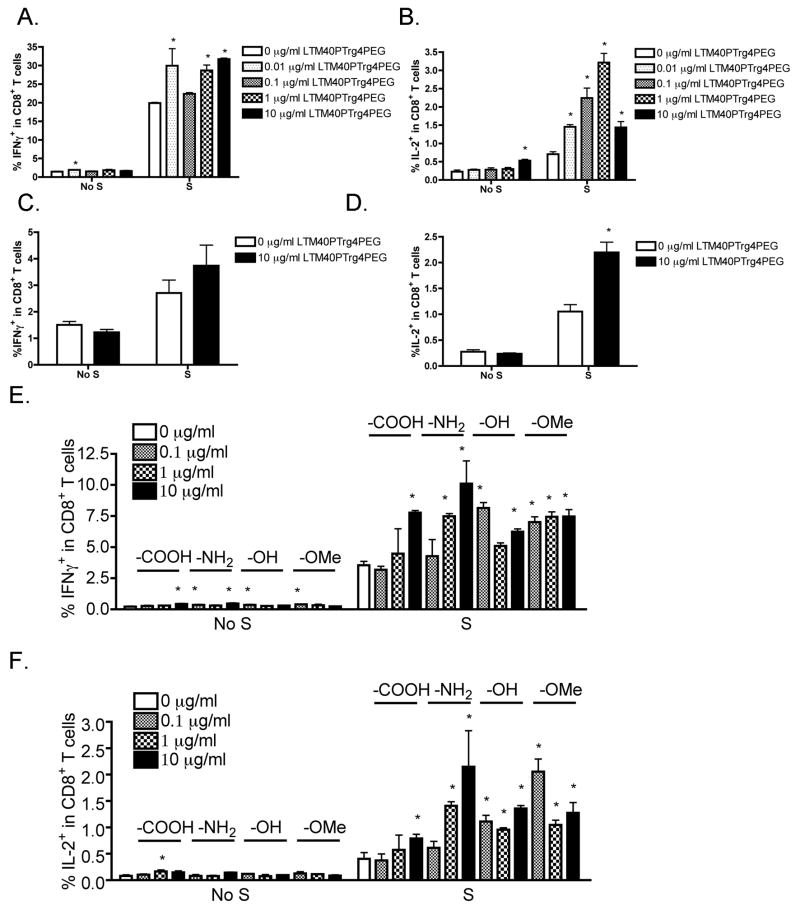
Modified SNPs enhanced SIINFEKL-activated CD8+ T cell responses
LTM40PTrg4PEG SNPs alone showed minimal effects on IFNγ and IL-2 production by viable CD8+ T cells; whereas LTM40PTrg4PEG enhanced the production of both cytokines by CD8+ T cells that were suboptimally activated using SIINFEKL peptide directly (Figure 5A and B). The effect was deemed statistically significant at multiple LTM40PTrg4PEG concentrations, including 0.01, 1 and 10 μg/ml for IFNγ, and all concentrations for IL-2. In the co-culture OT-I model in which irradiated DC2.4 cells were used as APCs, LTM40PTrg4PEG SNPs showed a similar trend of effect on CD8+ T cell responses. LTM40PTrg4PEG had no effect in the absence of SIINFEKL activation; whereas in the presence of a suboptimal concentration of SIINFEKL loaded onto DC2.4 cells, LTM40PTrg4PEG at 10 μg/ml further enhanced IFNγ (Figure 5C) and IL-2 (Figure 5D) production by viable CD8+ T cells, with the latter being significantly different from the no particle treatment control. This suggests that SNP-mediated enhancement is likely to act directly on CD8+ T cell responses.
Figure 5.
Enhancement by modified LTM40PTrg4PEG on CD8+ T cell responses induced by a suboptimal concentration of SIINFEKL. (A and B) LTM40PTrg4PEG SNPs were sonicated and added to OT-I splenocytes (CD8+ T cells, 8 × 105 cells/well) at various concentrations (10−2 to 10 μg/ml) prior to SIINFEKL (S) stimulation (10−4 μg/ml) for 2 days. (C and D) SIINFEKL (S, 0.1 μg/ml)-loaded DC2.4 cells (2 × 106 cells/well) were irradiated and plated at 2 × 105 cells/well. OT-I splenocytes (1 × 106 cells/well) were treated with LTM40PTrg4PEG SNPs (10 μg/ml) and then co-cultured with SIINFEKL-loaded DC2.4 cells for 4 days. Splenocytes were restimulated with SIINFEKL (10 μg/ml) in the presence of brefeldin A. (E and F) SNPs with different functional groups, including LTM40PTrg4PEG (–OMe), LTM40PTrg4PEGOH (–OH), LTM40PTrg4PEGCOOH (–COOH) and LTM40PTrg4PEGNH2 (–NH2), were sonicated and then added to OT-I splenocytes (8 × 105 cells/well) at various concentrations (10−1 to 10 μg/ml) prior to SIINFEKL (S) stimulation (10−4 μg/ml) for 2 days. (A–F) Brefeldin A was added during the last 6 h of incubation. Splenocytes were stained for viability, surface CD8 and intracellular IFNγ or IL-2 expression. The percent of IFNγ+ or IL-2+ cells within CD8+ T cells is presented as the mean % ± SE of triplicate cultures. *p < 0.05 as compared to respective no SNP treatment (0 μg/ml) control. Data are representative of at least two separate experiments.
Different functional groups on SNPs influenced the immune enhancing effect to different levels
The effect of engineered SNPs on T cell responses was compared between particles with different surface functional groups, including LTM40PTrg4PEG (–OMe), LTM40PTrg4PEGOH (–OH), LTM40PTrg4PEGCOOH (–COOH) and LTM40PTrg4 PEGNH2 (–NH2). Similar to the above observations, in the absence of SIINFEKL stimulation, SNPs produced minimal effects on the cytokine response, especially for IL-2 (Figure 5E and F). When OT-I CD8+ T cells were suboptimally activated with 10−4 μg/ml of SIINFEKL, all tested SNPs showed significant enhancement of IFNγ (Figure 5E) and IL-2 (Figure 5F) production by CD8+ T cells. In particular, –COOH and –NH2 particles exhibited concentration-dependent enhancement on cytokine production.
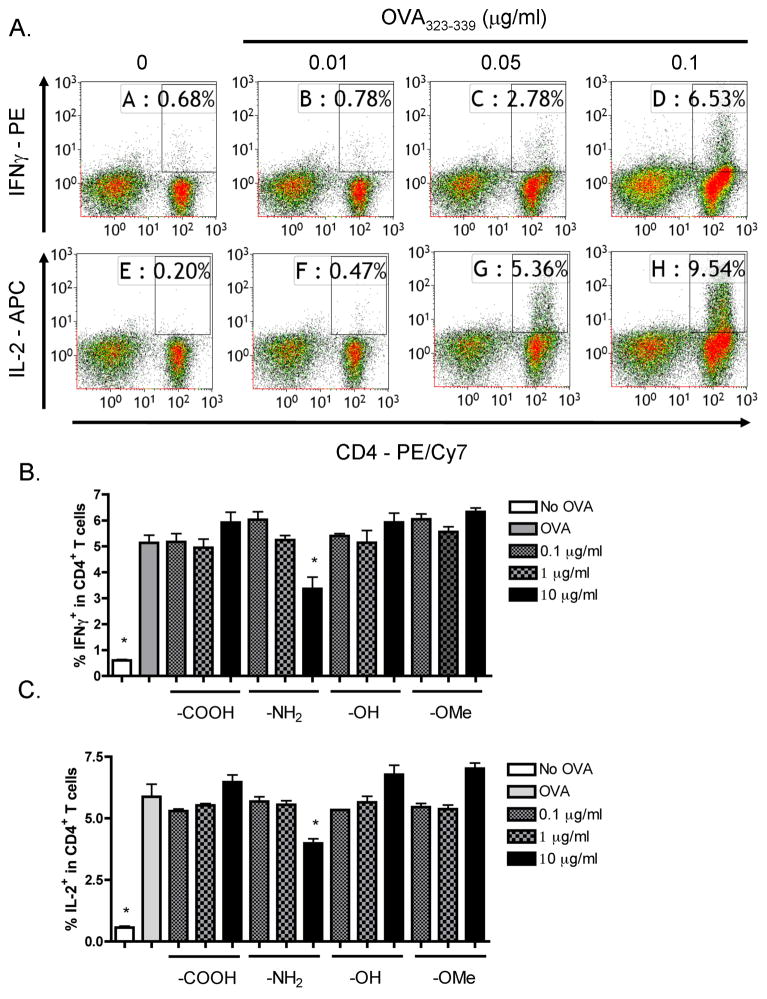
Absence of immune enhancement by SNPs on OVA323–339-induced CD4+ T cell responses
In the OT-II model in which OVA323–339 is presented by MHC II molecules on endogenous APCs to OT-II CD4+ T cells, production of IFNγ and IL-2 was minimal in the absence of OVA323–339 peptide activation and increased with increasing concentrations of OVA323–339 (0.01–0.1 μg/ml) (Figure 6A). In contrast to the OT-I model, there was more IL-2 production than IFNγ in the OT-II model. The OVA323–339 concentration for suboptimal CD4+ T cell activation was determined to be 0.05 μg/ml. Next, effects of SNPs with different functional groups on CD4+ T cell responses were characterized. In contrast to their enhancing effects on CD8+ T cell responses, none of the particles showed enhancing activity on IFNγ or IL-2 production by CD4+ T cells at any of the tested concentrations (Figure 6B and C).
Figure 6.
Absence of enhancement by SNPs with different functional groups on CD4+ T cell responses under suboptimal OVA323–339 stimulation. (A) OT-II splenocytes (CD4+ T cells, 8 × 105 cells/well) were incubated with various concentrations of OVA323–339 peptide (10−2 to 0.1 μg/ml) for 2 days. (B and C) SNPs with different functional groups, including LTM40PTrg4PEG (–OMe), LTM40PTrg4PEGOH (–OH), LTM40PTrg4PEGCOOH (–COOH) and LTM40PTrg4PEGNH2 (–NH2), were sonicated and added to OT-II splenocytes at various concentrations (10−1 to 10 μg/ml) prior to OVA323–339 peptide stimulation (0.05 μg/ml) for 2 days. (A–C) Brefeldin A was added during the last 6 h of incubation. Splenocytes were stained for viability, surface CD4 and intracellular IFNγ or IL-2 expression. The percent of IFNγ+ or IL-2+ cells within CD4+ T cells is presented in the quadrants in dot plots (A), with X axis and Y axis representing fluorescence intensity for CD4 and IFNγ or IL-2, respectively. Three replicates were concatenated for each treatment in the flow cytometry analysis (A). The percent of IFNγ+ (B) or IL-2+ (C) cells within CD4+ T cells is presented as the mean % ± SE of triplicate cultures. *p < 0.05 as compared to respective no SNP treatment (0 μg/ml) control. Data are representative of at least two separate experiments.
Enhancing effect of SNPs on CD8+ T cell responses is size-dependent
Next, we investigated the role of particle size on the enhancement of CD8+ T cell responses. Particles with the –COOH functional group at different sizes, including SNTXSPTrg4PEGCOOH (33 nm), LTM40PTrg4PEGCOOH (51 nm), SNT20LPTrg4 PEGCOOH (82 nm) and SNTZLPTrg4PEGCOOH (137 nm), were compared. Interestingly, only the 51 nm –COOH particles at 10 μg/ml produced a significantly greater enhancement of IFNγ and IL-2 production by CD8+ T cells activated with a suboptimal concentration of SIINFEKL. SNPs either smaller or larger in size than 51 nm particles lacked immune enhancing activity as assessed by IL-2 and IFNγ production (Figure 7A and B).
Figure 7.
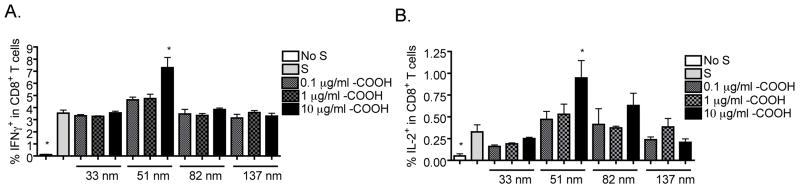
Effect of –COOH SNPs at different sizes on CD8+ T cell responses under suboptimal induction. (A and B) SNPs with –COOH functional group at different sizes, including SNTXSPTrg4PEGCOOH (33 nm), LTM40PTrg4PEGCOOH (51 nm), SNT20LPTrg4PEGCOOH (82 nm) and SNTZLPTrg4PEGCOOH (137 nm) were sonicated and then added to OT-I splenocytes (8 × 105 cells/well) at various concentrations (10−1 to 10 μg/ml) prior to SIINFEKL (S) stimulation (10−4 μg/ml) for 2 days. Brefeldin A was added during the last 6 h of incubation. Splenocytes were stained for viability, surface CD8 and intracellular IFNγ or IL-2 expression. The percent of IFNγ+ (A) or IL-2+ (B) cells within CD8+ T cells is presented as the mean % ± SE of triplicate cultures. *p < 0.05 as compared to respective no SNP treatment (0 μg/ml) control. Data are representative of at least two separate experiments.
Discussion
As the commercial use of engineered nanoparticles has grown, their potential for causing adverse health issues has also received increasing attention. To date, most investigations focusing on the potential toxicity of engineered nanoparticles have been conducted in laboratory animals. Some of these studies have demonstrated that certain engineered nanoparticles possess adjuvant-like properties, especially on pulmonary inflammation and lung disease. Although inhalation is one of the principal routes of particle exposure (Colvin, 2003; Inoue et al., 2006; Li et al., 2008, 2009; Nel et al., 2006), little is still known concerning the effect of engineered nanoparticles on immune responses in airways or on the immune system in general. In this study, we utilized unique in vitro mouse models in order to investigate the effects of size and surface chemistry of engineered nanoparticles on antigen-specific CD8+ (OT-I) and CD4+ (OT-II) T cell responses. It is worth mentioning that two related but distinct OT-I models possessing either endogenous splenic APCs containing multiple cellular populations or the cell line, DC2.4, for antigen presentation of SIINFEKL showed similar trends, as evidenced by enhancement of antigen-specific CD8+ T cell responses by engineered nanoparticles. As reviewed by Banchereau and co-workers, DCs are highly efficient APCs (Banchereau & Steinman, 1998). DC2.4 cells were irradiated before co-culturing with splenocytes to arrest their proliferation, so they would not deplete the media of nutrients and/or outgrow naïve T cells; however, their ability to activate naïve T cells in the context of antigen-MHC complexes is not affected by irradiation (He et al., 2007; Ni et al., 2001). By using DC2.4 as the only APC population in the second OT-I model, OVA peptides were directly loaded onto DC2.4; therefore, the effects of nanoparticles on antigen processing via APCs as a potential confounder were eliminated, allowing the direct assessment of their potential effects on T cell responses. With two OT-I models, we demonstrated that SNPs produced their immune enhancing effects most likely by affecting T-cell function directly.
The aqueous-dispersed engineered nanoparticles used in this study underwent chemical modification by varying the surface functional group moieties and overall particle sizes. An important as well as rare physico-chemical property of the engineered nanoparticles employed in this study was that these intermediate-sized particles were resistant to aggregating even in the culture media, which allowed them to be used in in vitro assay systems to investigate their influence on leukocyte function in their true nanoscale context. Although the size distribution peak for the –NH2 SNPs in culture media appeared to shift to the right, around 100 nm, distinct from the peaks for other particles, which appeared close to 50 nm, we believe they are unlikely to agglomerate in the culture media. If modest changes in the size of –NH2 SNPs led to significant shifts between no agglomeration and agglomeration, one would expect to observe significant peaks at markedly larger sizes than 100 nm (i.e. which would be representative of association between two particles). Alternatively, the change in size could reflect an interaction between SNPs and serum proteins as previously suggested (Cohen et al., 2013; Jiang et al., 2009). However, if the nanoparticles interacted with protein components in the serum, the particle size would increase dramatically due to formation of agglomerates and keep increasing over time as reported in the literature (Katas et al., 2013), which was not detected by DLS in this study. In addition, a relatively low serum concentration in the RPMI media was maintained in our studies; therefore, keeping the zeta potentials relatively similar to the no-serum condition. It is also noteworthy that the average hydrodynamic diameters of the bare SNPs, measured by DLS in water, were larger than the sizes claimed by manufacturers, which is likely due to the differences in methodologies for particle size measurement, such as using transmission electron microscopy or scanning electron microscopy images.
Since surface chemistry of nanoparticles directly influences cellular uptake and thereby their activity as well as interaction with cellular compartments in the cells (Wilhelm et al., 2003), three surface functional groups, including –COOH, –NH2 and –OH, were engineered to modify the pegylated SNPs individually. The zeta potential of all coated SNPs displayed higher values than the bare SNP, indicating that the silanol groups were covered by PEG layers. The –NH2-modified SNPs showed the highest zeta potential, which indicates the zeta potential of bare SNP was compensated by both the PEG layer and the NH2 (+) surface charge. The –COOH-modified SNPs with the lowest zeta potential, which was higher than bare SNP, is due to the PEG layer and carboxylate group in the buffer. PEG and –OH-modified SNPs showed medium zeta potential, which is due to the neutral surface function and PEG layers. It has been reported recently that surface modified SNPs have a negative zeta potential for neutral surface layers and +0.6 mV only for amine modified SNPs in PBS buffer (Kardys et al., 2013; Yildirim et al., 2013). Our negative zeta potential is most likely due, at least in part, to the remaining negatively charged silanol groups. Among these three functional groups, –COOH SNPs were chosen for further characterization in part because no agglomeration in culture media was observed with –COOH particles. It has been suggested that –COOH particles help to facilitate cellular uptake and transport into intracellular compartments with less aggregation inside the cells (Thevenot et al., 2008). Furthermore, since –COOH, –NH2, and –OH particles have different surface charges at physiological pH, cationic charge of the cell membrane due to interaction with –NH2 particles may cause excessive proton transport and accumulate inside the cells, therefore generating reactive oxygen species, which may injury mitochondria and eventually disrupt the cellular function (Wang et al., 2007; Xia et al., 2006).
In this assay system, cytokine production by CD8+, but not CD4+, T cells was significantly increased by modified SNPs, suggesting that the particles selectively target CD8+ T cell responses. It has been suggested that certain physico-chemical properties of modified SNPs may induce upregulation of MHC I expression on DCs by increasing phosphorylation of mitogen-activated protein kinases, such as p38 kinase, c-Jun N-terminal kinases and extracellular signal-regulated kinases, which are involved in DC activation signaling pathways, and thereby indirectly enhance CD8+ T cell responses (Uto et al., 2009). However, because DC2.4 cells were irradiated for experiments employing the co-culture model, MHC I expression on these cells should not be further regulated by SNPs, hence suggesting that SNPs did not enhance CD8+ T cell responses via regulating APCs. Therefore, mechanisms underlining the observed enhancement on CD8+ T cell responses remain to be elucidated. It is possible that engineered nanoparticles in our models specifically modulate CD8+ T cell receptor signaling or directly interact with surface receptors on CD8+ T cells, e.g. low-density lipoprotein receptor (Yeh et al., 1993), and thereby increase particle cellular uptake to enhance CD8+ T cell responses, but further investigations will be needed. In addition, there was no direct cytotoxicity from SNPs on the T cells as assessed using viability staining.
We have demonstrated that only particles at a size of 51 nm significantly enhanced CD8+ T cell responses, suggesting that the nanoparticle-mediated enhancement of CD8+ T cell responses is size-dependent. Similar nanoparticle size-dependent effects have been demonstrated previously, indicating that within the size range of 2–100 nm, 50 nm gold nanoparticles showed the greatest effect on membrane receptor internalization, which in turn affected the downstream cellular proliferation and survival responses (Jiang et al., 2008). As suggested by others using gold nanoparticles in cells or theoretical models, 50 nm particles might fall into the critical size range for receptor-mediated internalization. Smaller particles might dissociate from the receptors on the membrane before being endocytosed due to their small avidity; whereas large particles might cause decreased membrane wrapping efficiency, which would directly impair the nanoparticle internalization process (Chithrani et al., 2006, Gao et al., 2005, Jiang et al., 2008).
Collectively, we demonstrated immune enhancing properties by engineered SNPs on DC-induced antigen-specific CD8+ T cell responses using an OT-I model in vitro, suggesting the potential adjuvant-like effects of these engineered SNPs on immune function at the cellular level. We also show that by modifying the chemical and physical properties of the particles, such as the surface chemistry and size, the immunomodulatory properties of these particles can be differentially affected. Further studies at the organismal, cellular and subcellular level with special emphasis on engineered nanoparticle-membrane interactions are needed to provide more in-depth understanding of the mechanisms responsible for immune modulation by SNPs.
Conclusions
In summary, our results demonstrated that the engineered SNPs were successfully modified with different functional group moieties on the surface and in different sizes, which allowed them to be used to characterize the effect of diverse physical and chemical properties of SNPs on T cell function. In addition, modified SNPs, except for –NH2 SNP, which might interact with proteins in the media, did not agglomerate under the in vitro culture conditions, which was critical for them to maintain their aqueous distribution at the nanoscale and was an essential prerequisite for studying their immunological effects (Figure 3 and S1–S3). Conditions for suboptimal stimulation of T cell responses were determined using OT-I and OT-II models (Figure 4 and 6A) in order to measure the immune enhancing effects of the SNPs. Modified SNPs (up to 10 μg/ml) enhanced OVA-specific IFNγ and IL-2 production by CD8+, but not CD4+, T cells, suggesting a cell-type selective effect by engineered SNPs on immune function, in particular on antigen-specific CD8+ T cell responses. Enhancement of CD8+ T responses by SNPs was differentially affected by the surface functional groups placed on the SNPs, which is consistent with findings of others (Thevenot et al., 2008; Wilhelm et al., 2003). SNPs with –COOH moieties were chosen for subsequent studies, since they exhibited concentration-dependent enhancement of CD8+ T responses in the current assay system as well as advantages over SNPs with other functional groups as suggested in the literature (Thevenot et al., 2008; Wang et al., 2007; Xia et al., 2006), such as enhanced cellular uptake, less agglomeration and limited cellular toxicity. Finally, 51 nm sized SNPs showed greater enhancement of CD8+ T cell responses than the other sized SNPs, which might be dictated by the nanoparticle size-dependent internalization processes as suggested by others (Chithrani et al., 2006; Gao et al., 2005; Jiang et al., 2008). Our findings provide strong evidence that nanoparticles have potential adjuvant-like properties by enhancing immune function, which will cause adverse effects on allergic airway disease in humans. In addition, our studies have significant implications in understanding the role of particle size and surface chemistry on regulating the biological responses and providing guidelines for the design of nanoproducts, with great potential for the development of novel biomedical applications in diagnostics and therapeutics.
Supplementary Material
Acknowledgments
The authors would like to thank Mr Robert Crawford for flow cytometric analysis.
Footnotes
Authors’ contributions
W. C. designed and performed experiments, analyzed the data and wrote the manuscript. Q. Z. synthesized and characterized the particles and contributed to writing the associated sections. B. L. F. K. and G. L. B. helped design the study and reviewed the manuscript. N. E. K. designed the study, oversaw all experimental work and edited the manuscript. All of the authors have read and approved the final manuscript.
Declaration of Interest
The authors declare that they have no competing interests. This work is funded by National Institutes of Health Grant RC2 ES018756.
References
- Banchereau J, Steinman RM. Dendritic cells and the control of immunity. Nature. 1998;392:245–52. doi: 10.1038/32588. [DOI] [PubMed] [Google Scholar]
- Brandenberger C, Rowley NL, Jackson-Humbles DN, Zhang Q, Bramble LA, Lewandowski RP, et al. Engineered silica nanoparticles act as adjuvants to enhance allergic airway disease in mice. Part Fibre Toxicol. 2013;10:26. doi: 10.1186/1743-8977-10-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chithrani BD, Ghazani AA, Chan WC. Determining the size and shape dependence of gold nanoparticle uptake into mammalian cells. Nano Lett. 2006;6:662–8. doi: 10.1021/nl052396o. [DOI] [PubMed] [Google Scholar]
- Choualeb A, Rosé J, Braunstein P, Welter R. Routes to ruthenium-cobalt clusters and dicobalt complexes with new alkoxysilyl- or sulfur-functionalized alkynes. X-ray structures of [NEt4][RuCo3(CO) 10{μ4-η2-HC2(CH2)2OC(O)NH(CH2)3Si(OEt)3}] and [Co2(CO)6 {μ2-η2-HC2CH2NHC(O)NH(CH2)3Si(OEt)3}] Organometallics. 2003;22:2688–93. [Google Scholar]
- Clarke SR, Barnden M, Kurts C, Carbone FR, Miller JF, Heath WR. Characterization of the ovalbumin-specific TCR transgenic line OT-I: MHC elements for positive and negative selection. Immunol Cell Biol. 2000;78:110–7. doi: 10.1046/j.1440-1711.2000.00889.x. [DOI] [PubMed] [Google Scholar]
- Cohen J, Deloid G, Pyrgiotakis G, Demokritou P. Interactions of engineered nanomaterials in physiological media and implications for in vitro dosimetry. Nanotoxicology. 2013;7:417–31. doi: 10.3109/17435390.2012.666576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colvin VL. The potential environmental impact of engineered nanomaterials. Nat Biotech. 2003;21:1166–70. doi: 10.1038/nbt875. [DOI] [PubMed] [Google Scholar]
- Demento SL, Eisenbarth SC, Foellmer HG, Platt C, Caplan MJ, Mark Saltzman W, et al. Inflammasome-activating nanoparticles as modular systems for optimizing vaccine efficacy. Vaccine. 2009;27:3013–21. doi: 10.1016/j.vaccine.2009.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobrovolskaia MA, Aggarwal P, Hall JB, Mcneil SE. Preclinical studies to understand nanoparticle interaction with the immune system and its potential effects on nanoparticle biodistribution. Mol Pharm. 2008;5:487–95. doi: 10.1021/mp800032f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobrovolskaia MA, Mcneil SE. Immunological properties of engineered nanomaterials. Nat Nanotechnol. 2007;2:469–78. doi: 10.1038/nnano.2007.223. [DOI] [PubMed] [Google Scholar]
- Gangwal S, Brown JS, Wang A, Houck KA, Dix DJ, Kavlock RJ, Hubal EA. Informing selection of nanomaterial concentrations for ToxCast in vitro testing based on occupational exposure potential. Environ Health Perspect. 2011;119:1539–46. doi: 10.1289/ehp.1103750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao H, Shi W, Freund LB. Mechanics of receptor-mediated endocytosis. Proc Natl Acad Sci USA. 2005;102:9469–74. doi: 10.1073/pnas.0503879102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He T, Tang C, Xu S, Moyana T, Xiang J. Interferon gamma stimulates cellular maturation of dendritic cell line DC2.4 leading to induction of efficient cytotoxic T cell responses and antitumor immunity. Cell Mol Immunol. 2007;4:105–11. [PubMed] [Google Scholar]
- Inoue K-I, Takano H, Yanagisawa R, Hirano S, Sakurai M, Shimada A, Yoshikawa T. Effects of airway exposure to nanoparticles on lung inflammation induced by bacterial endotoxin in mice. Environ Health Perspect. 2006;114:1325–30. doi: 10.1289/ehp.8903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J, Oberdorster GN, Biswas P. Characterization of size, surface charge, and agglomeration state of nanoparticle dispersions for toxicological studies. J Nanoparticle Res. 2009;11:77–89. [Google Scholar]
- Jiang W, Kimbetty YS, Rutka JT, Chanwarren CW. Nanoparticle-mediated cellular response is size-dependent. Nat Nano. 2008;3:145–50. doi: 10.1038/nnano.2008.30. [DOI] [PubMed] [Google Scholar]
- Kardys AY, Bharali DJ, Mousa SAR. Amino-functionalized silica nanoparticles: in vitro evaluation for targeted delivery and therapy of pancreatic cancer. J Nanotechnol. 2013;2013:8. [Google Scholar]
- Katas H, Hussain Z, Awang SAR. Bovine serum albumin-loaded chitosan/dextran nanoparticles: preparation and evaluation of ex vivo colloidal stability in serum. J Nanomat. 2013;2013:9. [Google Scholar]
- Kitto HJ, Schwartz E, Nijemeisland M, Koepf M, Cornelissen JJLM, Rowan AE, Nolte RJM. Post-modification of helical dipeptido polyisocyanides using the “click” reaction. J Mat Chem. 2008;18:5615–24. [Google Scholar]
- Li N, Wang M, Bramble LA, Schmitz DA, Schauer JJ, Sioutas C, et al. The adjuvant effect of ambient particulate matter is closely reflected by the particulate oxidant potential. Environ Health Perspect. 2009;117:1116–23. doi: 10.1289/ehp.0800319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N, Xia T, Nel AE. The role of oxidative stress in ambient particulate matter-induced lung diseases and its implications in the toxicity of engineered nanoparticles. Free Radic Biol Med. 2008;44:1689–99. doi: 10.1016/j.freeradbiomed.2008.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y-S, Haynes CL. Synthesis and characterization of biocompatible and size-tunable multifunctional porous silica nanoparticles. Chem Mat. 2009;21:3979–86. [Google Scholar]
- Lu X, Sun F, Wang J, Zhong J, Dong Q. A facile route to prepare organic/inorganic hybrid nanomaterials by ‘Click Chemistry’. Macromol Rapid Commun. 2009;30:2116–20. doi: 10.1002/marc.200900356. [DOI] [PubMed] [Google Scholar]
- Luhmann T, Rimann M, Bittermann AG, Hall H. Cellular uptake and intracellular pathways of PLL-g-PEG-DNA nanoparticles. Bioconjug Chem. 2008;19:1907–16. doi: 10.1021/bc800206r. [DOI] [PubMed] [Google Scholar]
- Nel A, Xia T, Mädler L, Li N. Toxic potential of materials at the nanolevel. Science. 2006;311:622–7. doi: 10.1126/science.1114397. [DOI] [PubMed] [Google Scholar]
- Ni HT, Spellman SR, Jean WC, Hall WA, Low WC. Immunization with dendritic cells pulsed with tumor extract increases survival of mice bearing intracranial gliomas. J Neurooncol. 2001;51:1–9. doi: 10.1023/a:1006452726391. [DOI] [PubMed] [Google Scholar]
- Sette A, Buus S, Colon S, Smith JA, Miles C, Grey HM. Structural characteristics of an antigen required for its interaction with Ia and recognition by T cells. Nature. 1987;328:395–9. doi: 10.1038/328395a0. [DOI] [PubMed] [Google Scholar]
- Stern ST, Mcneil SE. Nanotechnology safety concerns revisited. Toxicol Sci. 2008;101:4–21. doi: 10.1093/toxsci/kfm169. [DOI] [PubMed] [Google Scholar]
- Thevenot P, Cho J, Wavhal D, Timmons RB, Tang L. Surface chemistry influences cancer killing effect of TiO2 nanoparticles. Nanomedicine. 2008;4:226–36. doi: 10.1016/j.nano.2008.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uto T, Akagi T, Hamasaki T, Akashi M, Baba M. Modulation of innate and adaptive immunity by biodegradable nanoparticles. Immunol Lett. 2009;125:46–52. doi: 10.1016/j.imlet.2009.05.008. [DOI] [PubMed] [Google Scholar]
- Wang CC, Lu JN, Young TH. The alteration of cell membrane charge after cultured on polymer membranes. Biomaterials. 2007;28:625–31. doi: 10.1016/j.biomaterials.2006.09.025. [DOI] [PubMed] [Google Scholar]
- Wilhelm C, Billotey C, Roger J, Pons JN, Bacri JC, Gazeau F. Intracellular uptake of anionic superparamagnetic nanoparticles as a function of their surface coating. Biomaterials. 2003;24:1001–11. doi: 10.1016/s0142-9612(02)00440-4. [DOI] [PubMed] [Google Scholar]
- Xia T, Kovochich M, Brant J, Hotze M, Sempf J, Oberley T, et al. Comparison of the abilities of ambient and manufactured nanoparticles to induce cellular toxicity according to an oxidative stress paradigm. Nano Lett. 2006;6:1794–807. doi: 10.1021/nl061025k. [DOI] [PubMed] [Google Scholar]
- Yeh T-C, Zhang W, Ildstad ST, Ho C. Intracellular labeling of T-cells with superparamagnetic contrast agents. Magn Reson Med. 1993;30:617–25. doi: 10.1002/mrm.1910300513. [DOI] [PubMed] [Google Scholar]
- Yildirim A, Ozgur E, Bayindir M. Impact of mesoporous silica nanoparticle surface functionality on hemolytic activity, thrombogenicity and non-specific protein adsorption. J Mater Chem B. 2013;1:1909–20. doi: 10.1039/c3tb20139b. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.