Abstract
Traditional combinatorial peptidyl substrate library approaches generally utilize natural amino acids, limiting the usefulness of this tool in generating selective substrates for proteases that share similar substrate specificity profiles. To address this limitation, we synthesized a Hybrid Combinatorial Substrate Library (HyCoSuL) with the general formula of Ac-P4-P3-P2-Asp-ACC, testing the approach on a family of closely related proteases – the human caspases. The power of this library for caspase discrimination extends far beyond traditional PS-SCL approach, as in addition to 19 natural amino acids we also used 110 diverse unnatural amino acids that can more extensively explore the chemical space represented by caspase-active sites. Using this approach we identified and employed peptide-based substrates that provided excellent discrimination between individual caspases, allowing us to simultaneously resolve the individual contribution of the apical caspase-9 and the executioner caspase-3 and caspase-7 in the development of cytochrome-c-dependent apoptosis for the first time.
Apoptosis, the most well-understood form of programmed cell death, is a highly regulated process controlled and executed by proteolytic enzymes called caspases. The apoptotic process is somewhat hierarchical and caspases can be assigned as initiators (2, 8, 9, and 10) and executioners (3, 6, and 7).1, 2, 3 Apoptosis can be triggered extrinsically via ligation of a death receptor by its cognate ligands, leading to the activation of caspases 8 and 10, or intrinsically following the release of cytochrome c from mitochondria with formation of a caspase 9 activation complex known as the apoptosome.3, 4, 5 Mechanistically, caspases display a near absolute preference for aspartate at the P1 position of their substrates. In addition, they require a minimum substrate length of four amino acids N-terminal of the scissile bond. Thornberry et al.6, 7 used a combinatorial library of fluorogenic substrates to profile nine human caspases at the P4–P2 region, demonstrating that the caspases tended to have specificity profiles that enabled grouping based on substrate preferences.6, 7 This work provided a great insight into caspase recognition patterns and opened the door for others to pursue small molecule probes for caspase investigations.
To date, various types of substrates and inhibitors have been developed and biologically evaluated against caspases.2, 8, 9, 10, 11 Unfortunately, most of them lack selectivity and cannot be used for selectively targeting or analyzing particular enzymes in complex biological environments.12, 13, 14, 15 This is entirely because of the overlapping specificities of the caspases on their preferred natural amino acid sequences. To address this problem we designed and synthesized a Hybrid Combinatorial Substrate Library (HyCoSuL) containing 19 natural amino acids (omitting cysteine) and 110 unnatural amino acids. We propose that such a large and varied set of chemical structures provides an excellent tool to investigate caspases and distinguish between them. In this work we dissected the kinetic profiles of six human apoptotic recombinant caspases through HyCoSuL screening. We then designed and synthesized new caspase substrates with the ability to discriminate these enzymes within a group. To further test the specificity and utility of the designed hybrid substrates, we performed a series of experiments in a cell-free model of apoptosis where multiple caspases are activated.
Results
HyCoSuL synthesis strategy and its application
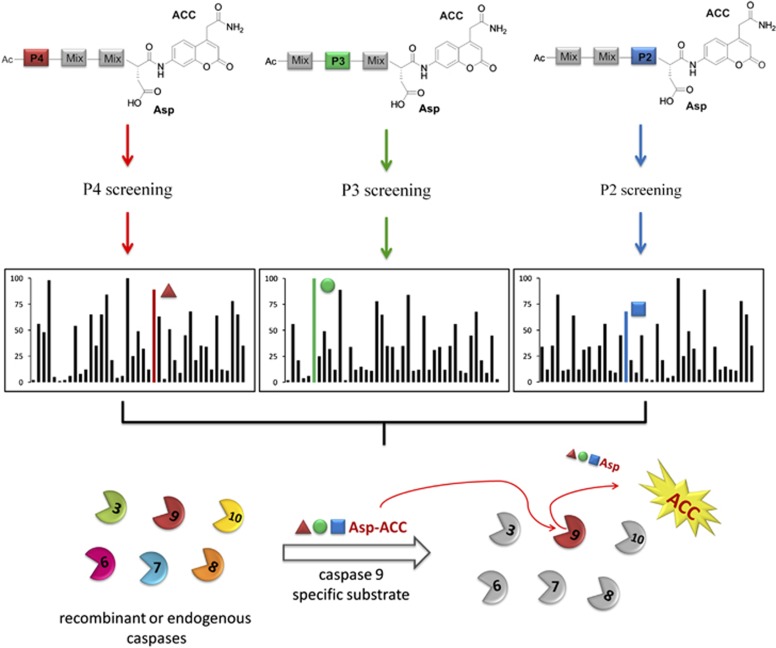
Our objective was to exploit a chemical tool that could be used to develop highly selective and sensitive short peptide-based substrates for caspases. We hypothesized that the use of a large number of unnatural amino acids would allow us to explore chemical space encompassed by the caspase-active site cleft, and to this end we synthesized a hybrid combinatorial library with the general formula of Ac-P4-P3-P2-Asp-ACC (Figure 1). Based on this structural scaffold we fixed aspartic acid at the P1 position (an essential requirement for caspases6, 16, 17) and synthesized three sublibraries (P4, P3, and P2) for dissecting caspase preferences at the S4–S2 region. As a reporter group, an ACC (7-amino-4-carbamoylmethyl coumarin) fluorophore moiety was selected for convenience in solid-phase synthesis.18 Each sublibrary contained one of 129 amino acids fixed at either the P4, P3, or P2 positions. These amino acids consisted of 19 amino acids (18 natural, cysteine was omitted and methionine was replaced with norleucine) and 110 unnatural amino acids, where we define unnatural amino acids as those that are not encoded in proteins. Each pool contained a total of 129 × 19 × 19=46 569 possible tetrapeptide sequences so that the total number of possible tetrapeptide combinations that can be made using this library is 129 × 129 × 129=2 146 689, and this is significantly greater than in previously published approach (20 × 20 × 20=8000).6
Figure 1.
General scheme for HyCoSuL design and its use for the discovery of selective and sensitive substrates for caspases. HyCoSuL is composed of three sublibraries containing 129 individual natural and unnatural amino acids. Screening results reveal the identity of residues that contribute to activity at a specific position for each caspase. In the example in the colored bars represent the most selective residue when compared with other caspases (not shown in this illustration). Sometimes, but not always, the most selective residue is also the most active. Thus, HyCoSuL is useful not only for the design and synthesis of sensitive caspase substrates, but also for the discovery of caspase substrates with high selectivity
We selected commercially available unnatural amino acids with diverse physicochemical attributes to cover a broad range of possible substrate–enzyme interactions: acidic and basic, small and large, hydrophilic and hydrophobic, with defined stereochemistry (L or D) or lacking stereochemistry (dehydro amino acids). After the screening of six human recombinant caspases, we were armed with data that allowed us to design and synthesize new substrates with a very high selectivity index. The general architecture of HyCoSuL and its use for discovery of selective and sensitive caspase substrates is presented in Figure 1. The most selective substrates can be used in cell-based assays as well as serve as the scaffold for selective inhibitor design. On the other hand, most active substrates (the highest kcat/Km values) can be used in regular kinetic studies on recombinant caspases.
Substrate specificity determination
Each of the P4, P3, and P2 sublibraries was screened at 50 μM final total substrate mixture, with caspase concentrations in the range of 10–200 nM. The concentration of each caspase in the assay was selected in preliminary experiments to provide an optimal fluorescence readout (see Materials and Methods). Initial rates of hydrolysis were recorded as relative fluorescence units (RFUs) over time and normalized by inclusion of at least three references substrate mixtures. Each caspase was screened at least three times (n=3–5) and the average value of RFU/s (expressed as % normalized to the best substrate) is presented as a specificity matrix. All substrate specificity profiles for each tested caspase can be found in the Supplementary Data section; however we have distilled their essence into Table 1.
Table 1. The aggregated substrate specificity profiles of six recombinant human caspases.
| Casp-3 | Casp-6 | Casp-7 | Casp-8 | Casp-9 | Casp-10 | |
|---|---|---|---|---|---|---|
| P4 position | ||||||
| Asp | ••••• | • | ••••• | •• | • | • |
| Asp esters | • | • | ••• | • | • | • |
| Ile, Leu, Val | • | ••••• | • | ••••• | ••••• | ••••• |
| Abu, hLeu, Nva, Tle | • | •••• | • | •••• | ••• | •••• |
| Hyp, Thz, Hyp(Bzl), Oic | • | • | •• | ••• | •••• | • |
| D-hPhe, D-Phg | • | • | • | ••• | • | • |
| dhLeu, dhAbu | • | • | • | ••• | •• | • |
| Phe(2-Cl) | • | •• | • | • | • | ••• |
| Others | • | • | • | • | • | • |
| P3 position | ||||||
| Glu | ••••• | ••••• | ••••• | ••••• | •••• | ••••• |
| Glu esters | ••• | ••• | •••• | •• | •• | •••• |
| hGlu | ••• | •• | •••• | ••••• | ••• | •••• |
| Val, Tle | •• | • | ••••• | •• | ••••• | ••• |
| D-amino acids | • | • | • | • | • | • |
| Phe-derivatives | ••• | • | •• | • | • | • |
| Bulky amino acids | •• | •• | ••• | •• | •• | •• |
| Basic amino acids | •• | • | ••• | • | •• | •• |
| Small, hydrophobic | •• | • | •• | •• | •• | •• |
| P2 position | ||||||
| Thr(Bzl) | ••••• | ••••• | ••••• | ••••• | • | •• |
| Val, Ile | ••• | •• | •• | •• | • | •• |
| Oic, Pip, Tic | •• | • | •• | •• | •• | •••• |
| His, His(Bzl) | • | ••••• | • | •• | ••••• | •••• |
| Ser(Bzl), hPhe, hTyr | • | ••• | • | •• | • | ••••• |
| D-amino acids | • | • | • | • | • | • |
| Others | • | • | • | • | • | ••• |
We present only some of most important information extracted from the full HyCoSuL screen. The code used for substrate specificity description is as follows: •••••, very high activity (81–100%); ••••, high activity (61–80%); •••, medium activity (41–60%); ••, weak activity (21–40%); •, very weak or no activity (0–20%)
The selectivity overview presented in Table 1 confirms that some caspases share common preferences at a particular subsite, and that these preferences are substantially extended in several cases by incorporating unnatural amino acids into the peptidyl substrates. However, along with the expected preferences come some unexpected ones, and we have summarized these below. We propose that understanding the amino acid preferences at each position, and perhaps more importantly the restricted amino acids at each position, would allow for the design of highly selective substrates.
The P4 position
The P4 position is the most distinguishing characteristic discriminating substrate selectivity in the caspase family.6, 19 In general, our observed P4 preferences followed established rules, with caspases 3 and 7 favoring Asp, and caspases 6, 8, 9, and 10 favoring aliphatic residues.6, 19 Caspase 3 and 7 are very closely related enzymes sharing 57% sequence identity and very similar substrate specificity.20 At the P4 position, caspase 3 favored Asp ∼10-fold over other amino acids but caspase 7 was more catholic, preferring Asp but also tolerating aspartic acid methyl, cyclohexyl and benzyl esters, and also histdine benzyl ester, thioproline. The broader tolerance of caspase 7, also seen at the P3 position (see below), is the key distinguishing feature between the closely related paralogs caspases 3 and 7. Distinctions between caspases at the P4 position can also be found via negative selection. Caspases 3 and 7 accept very few amino acids from our library, except Asp and a few others mentioned previously. Caspases 6 and 10 also have strict preferences at the P4 position. The only group that is not tolerated by caspase 8 at the P4 position is basic amino acids. The most promiscuous is caspase 9 that recognizes almost all chemical structures at P4. We detected only a few amino acids that were completely ignored by this enzyme.
The P3 position
The P3 position provides the least discrimination between caspases. This position is intolerant of D-amino acids and secondary amino acids, and generally prefers Glu. Interestingly, Glu-esters were also well tolerated at this position, particularly for caspases 7 and 10. In available crystal structures, the γ-carboxylate of P3 Glu generally forms an ion pair with Arg-341 that is absolutely conserved in all human caspases,21 and one might therefore expect that the ion pair was necessary for substrate recognition. However, the tolerance for Glu-esters, as well as aliphatic residues by caspases 7 and 9, suggest that this ion pair is not essential for optimal P3 occupancy. Most likely, Arg-341 conservation is required for interaction with P1 Asp, to which it also makes an ion pair. The substrate specificity of caspase 9 at P3 was unusual, with tert-Leucine and valine dominating over glutamic acid.
The P2 position
The P2 position was not previously thought to provide a high degree of discrimination,6 yet analysis of the P2 HyCoSuL sublibrary revealed some unexpected preferences. We found that the best recognized amino acid at this position for caspases 3, 6 ,and 7 was threonine benzyl ester, revealing that the S2 pocket is large enough to accommodate the bulky benzyl group. Other amino acids that were well tolerated at P2 position were proline derivatives (octahydroindole-2-carboxylic acid (Oic), pipecolinic acid (Pip), and 1,2,3,4-tetrahydroisoquinoline-3-carboxylic acid (Tic)), and this is also quite unexpected based on canonical natural amino acid preferences.6 Subsite preferences suggested that the caspase 6 S2 subsite is large enough to accept even bulkier moieties than the other executioner caspases. Caspase 9 demonstrated essentially exclusive selectivity for His, N(im)-benzyl-histidine (His(Bzl)), and Tic. Caspase 10 recognizes large and hydrophobic amino acids (serine benzyl ester (Ser(Bzl)), homophenylalanine (hPhe), homocyclohexylalanine (hCha)) as well as small and basic amino acids. This preference distinguishes caspase 10 from its close paralog caspase 8. Caspases 3, 6, and 7 do not accommodate basic amino acids and caspase 8 has a very similar ‘negative pattern', but it can accommodate basic amino acids. Caspase 9 is ultra-selective at P2 and >95% of amino acids tested are prohibited at this position. Of all the caspases tested, only caspase 10 demonstrated a wide tolerance at P2, failing to accept only D- and dehydro-amino acids.
Design of the most sensitive caspases substrates through HyCoSuL screening
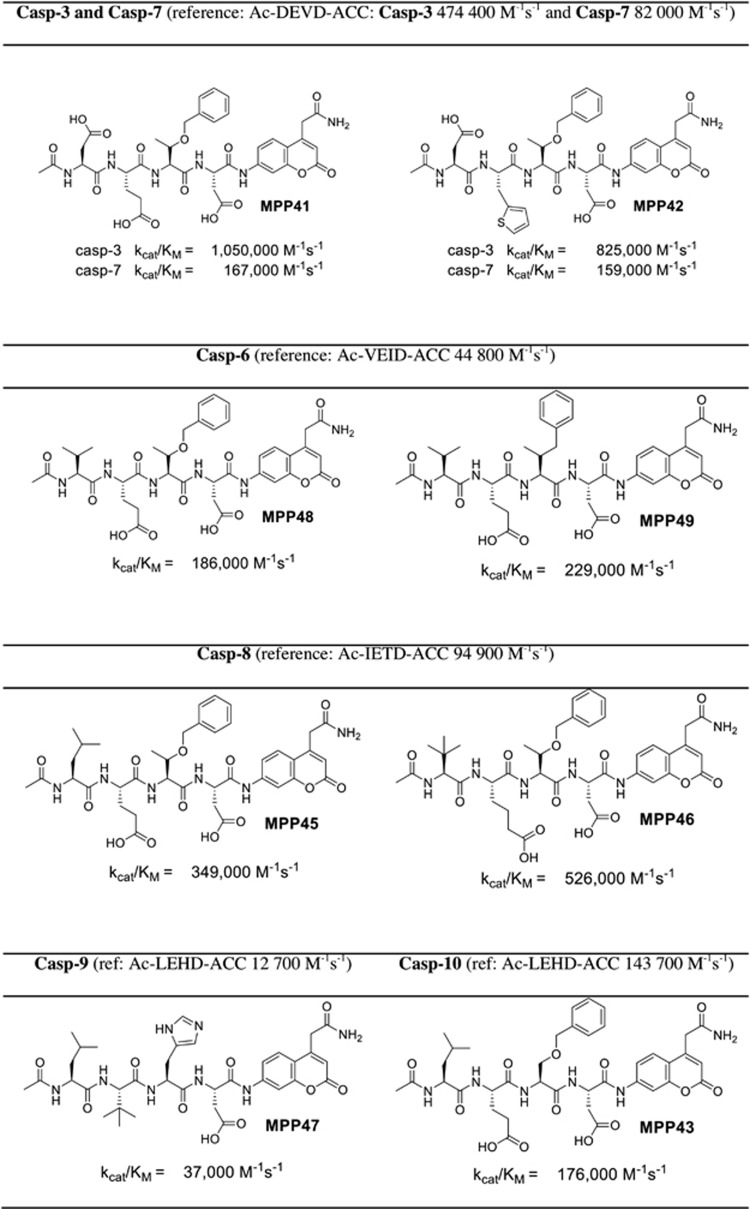
Armed with substrate specificity profiles of six human caspases, we proceeded to synthesize new substrates displaying enhanced sensitivity over existing substrates. The best substrates for caspase 3 and 7 (MPP41 and MPP42) contain threonine O-benzyl ester (Thr(Bzl)) at the P2 position that seemed to enhance exploration of the S2 chemical space. We also noticed that glutamic acid at P3 can be successfully replaced by many other amino acids (like 2-thienyl Ala) with only slight decreases in overall activity. The best substrates for caspases 3 and 7 are twofold better that those based on DEVD sequence (Figure 2). For caspase 6 we found that Val at P4 and Glu at P3 are obligatory for substrates; however, at P2 a few bulky amino acids are tolerated (like Thr(Bzl) or hPhe). These substrates (MPP48 and MPP49) are 4–5-fold better than the reference one (Ac-VEID-ACC). An analysis of caspase 8 substrate specificity revealed that the most active substrate (MPP46) contains three unnatural amino acids: tert-leucine (Tle) at P4, homoglutamic acid (2-aminohexanedioic acid (Aad)) at P3, and threonine benzyl ester (Thr(Bzl)) at the P2 position. The kcat/KM for this substrate is ∼5.5-fold higher than for the reference Ac-IETD-ACC substrate. For caspase 9 we discovered that replacing Glu at P3 with the unnatural tert-leucine (Trt) resulted in a threefold increase in activity (MPP47). Caspase 10 is the only enzyme for which we did not obtain improved activity. The replacement of histdine at P2 for serine benzyl ester (Ser(Bzl)) resulted in a slight increase of activity (∼20%) (MPP43). Therefore, overall, although we were able to enhance catalytic rates of peptide hydrolysis by incorporating unnatural amino acids, the degree of enhancement could be considered moderate.
Figure 2.
Some examples of novel highly sensitive caspase ACC substrates containing unnatural amino acids
Design of caspase-selective substrates
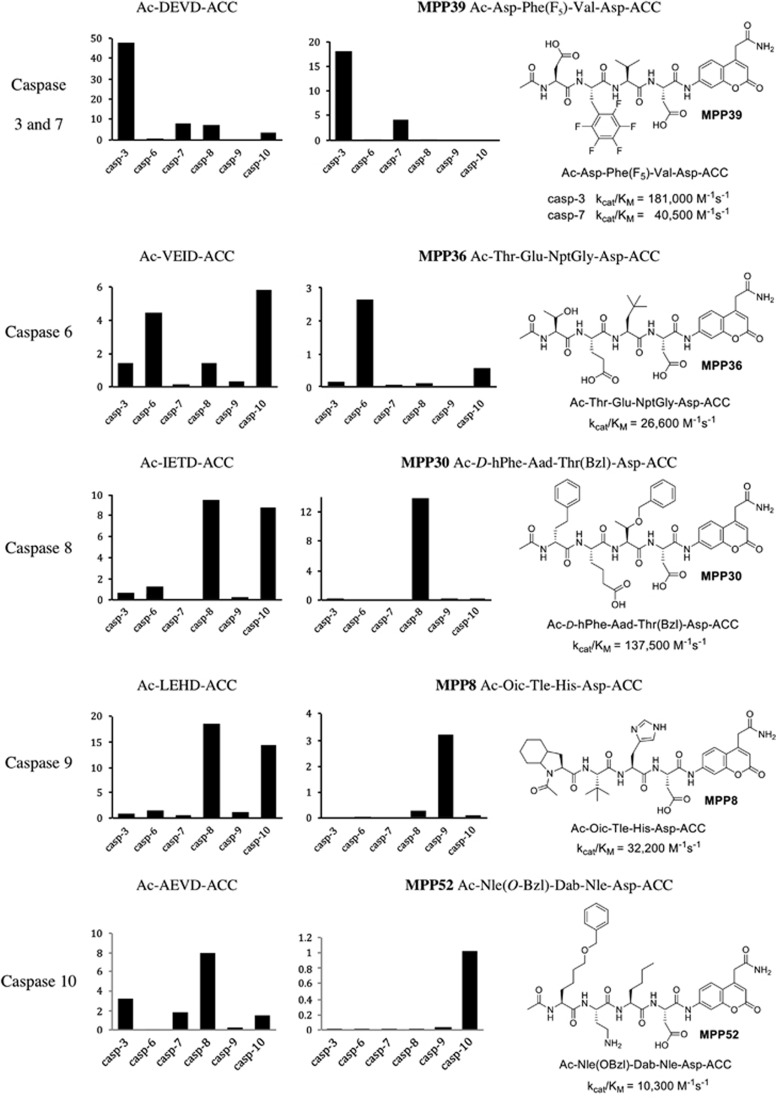
The commonly used caspase substrates containing only natural amino acids lack selectivity (Figure 3). To overcome these limitations, we decided to explore whether a HyCoSuL approach, which extends far beyond traditional positional scanning-substrate combinatorial library (PS-SCL), could deliver enhanced selectivity – that is, could we generate individual substrates that showed dramatically enhanced selectivity for individual caspases. Our initial goal was to discover new fluorogenic substrates with the ability to discriminate caspases in a virtual experiment where all caspases are present at the same concentration. Clearly, it would not be possible to achieve absolute selectivity, and hence we aimed at providing the best possible selectivity, displayed as relative catalytic efficiencies. This ‘proof of concept' experiment demonstrated that for almost all caspases our expectations were met. We could not produce a substrate that efficiently discriminated caspases 3 and 7, but we found that replacing Glu from the DEVD sequence with 2,3,4,5,6-pentafluorophenylalanine, Phe(F5) (compound MPP39) showed excellent discrimination of caspases 3 and 7 compared with other caspases. The most selective caspase 6 substrate is MPP36 that contains threonine at P4, glutamic acid at P3, and neo-pentylglycine (NptGly) at P2. MPP30, the most selective caspase 8 substrate, contains three unnatural amino acids, with the D-homophenylalanine (D-hPhe) at P4, Aad at P3, and Thr(Bzl) at P2. To distinguish caspase 9 from others enzyme, we synthesized MPP8 that possesses Oic (a cyclohexyl proline derivative) at P4, Tle at P3, and His at P2. Finally, for caspase 10 we found several selective substrates, among which MPP52 displayed the highest kcat/KM selectivity index among all tested substrates. This substrate contains 2-amino-6-benzyloxyhexanoic acid (Nle(O-Bzl)) at P4, 2,4-diaminobutyric acid (Dab) at P3, and norleucine at P2. In Figure 3 we present our best hits, and a detailed kinetic analysis for these (and several other) substrates can be found in the Supplementary Data section. We were delighted to find substrates that would distinguish each of the apical apoptotic caspases (8, 9, and 10) from each other and from other caspases.
Figure 3.
Comparison of commonly used and HyCoSuL-derived caspases substrates. Six ACC substrates with commercially available sequences were tested toward six recombinant human caspases (left panels). For each substrate, all three kinetic parameters were determined (see Supplementary Data section). Here we present only kcat/KM values ( × 104 M−1s−1). Comparison with the most selective substrates derived from HyCoSuL screening reveals the utility of this approach in developing highly selective tetrapeptide substrates
Determination of caspase-selective substrates for cell-based assays
One of the main driving principles of this study was to develop substrates that could be used to assess the role of individual caspases in complex mixtures, a goal that has previously remained elusive.12 The HyCoSuL strategy allowed us to design new, very selective caspase substrates, measured by the specificity constant kcat/KM. However, in many biochemical experiments to obtain a measurable fluorescent readout, substrates are often used at a concentration significantly above their KM values, and thus the rate of their hydrolysis depends only on the kcat parameter. Such an example is caspase detection in cytosolic extracts. To date, there are no commercial substrates that can demonstrate selectivity in this kind of assay, as all described substrates exhibit off target reactivity, especially with caspase 3. Unfortunately, we were not able to measure kcat values for all substrates for all caspases as some of these substrates, being highly selective of one caspase, were very poorly recognized by other caspases. Second, for some substrates, their KM parameters were above their solubility index. However, as we thought that this analysis would be of significant value, we decided to evaluate our most selective substrates by screening them at a final concentration of 100 μM, well above KM values for the best substrates, and compared them with commercial substrates under the same conditions (Supplementary Table 5). By this experiment we demonstrate that almost all of our substrates with the best kcat/KM selectivity index can also be used for assays running in ‘high substrate concentration' conditions, where an individual substrate would exhibit the lowest degree of selectivity for different caspases. Thus, reducing substrate concentrations would result in even more selectivity, but at the price of lower fluorescence yields. Moreover, apart from the substrates described in the previous section, we found some other structures that are promising in this experimental paradigm (see Supplementary Data section).
Simultaneous detection of caspases 3 and 9 in cell-free apoptosis
To validate our results and assess the utility of the selective substrates, we tested individual substrates in a complex system where several caspases are active and where they operate in what approaches a natural environment. Because of the uncertainty of cellular penetration, we avoided intact cells, and opted instead for a model of cytochrome c programmed in vitro apoptosis that has frequently been used in the literature.22, 23, 24 In this model, cytochrome c is added to cytosolic extracts of human cells where it triggers the formation of a soluble activation platform known as the apoptosome, driving caspase 9 activation.25 Active caspase 9 then activates caspases 3 and 7 that are responsible for apoptosis execution. Caspase 9 shows little activity on previously reported peptide-based synthetic substrates, making it impossible to infallibly measure its activity in complex systems where several caspases are active (especially caspase 3 that is known to hydrolyze existing caspase 9 substrates).
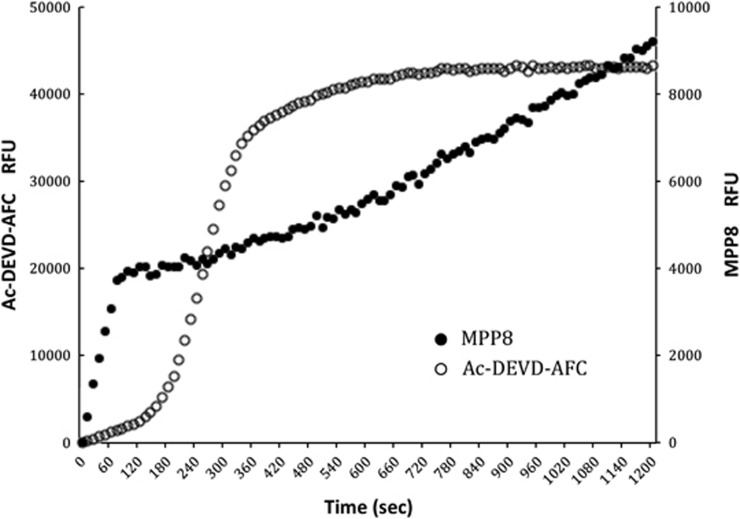
We first compared the cleavage of the caspase 3 reference substrate Ac-DEVD-AFC with the most selective caspase 9 substrate MPP8. A portion of cytosolic extracts pre-equilibrated with 100 μM of both substrates were activated by adding 10 μM cytochrome c and 1 mM dATP at 37°C. To avoid unwanted proteolysis disturbing our assay, we also added MG132 (a proteasome inhibitor) to the cytosolic extract. As MPP8 contains the ACC fluorophore, we were able to assay the independent cleavage of each substrate, simultaneously, in the same reaction by leveraging the distinct excitation and emission spectra of ACC (355 and 460 nm) and 7-amino-4-trifluoromethylcoumarin (AFC; 400 and 505 nm; Figure 4).
Figure 4.
Caspase activation time course. Cytosolic extracts were treated with cytochrome c and caspase activity was monitored simultaneously at different wavelengths by the caspase-9-selective substrate MPP8 that initiates after ∼1 min of addition of cytochrome c, whereas activity reported by Ac-DEVD-AFC exhibits a lag of ∼2–3 min before full activity is obtained. Eventually, Ac-DEVD-AFC substrate is depleted (∼6 min) and the curve relaxes to full substrate depletion. Because MPP8 is cleaved more slowly than Ac-DEVD-AFC, substrate is not depleted within the time course. The ‘burst' in the first 40–60 s is because of the interaction of the ACC substrate with cytochrome c through a mechanism that we do not yet understand, but is independent of any proteolytic activity, as it is observed in controls and is not observed for the AFC substrate. All substrates were used at 250 μM, and we could detect activity at 50 μM with the spectrofluorimeter employed in this study
Activity on Ac-DEVD-AFC exhibited a typical lag phase previously reported,26 but activity against the optimized caspase 9 substrate MPP8 appeared earlier. This is consistent with the requirement of caspase 9 to activate caspase 3 in a hierarchical manner. Although these data are consistent with selectivity of MPP8 for caspase 9, it is possible that the different sensitivities of the substrates somehow masked nonspecificity. To test this, we assayed caspase activity in the cytochrome c activated cytosolic extracts in the presence of highly specific protein inhibitors of the caspases. We chose the BIR2 (baculovirus inhibitor of apoptosis protein repeat) domain of human X-linked inhibitor of apoptosis protein (XIAP), which is stringently selective for caspases 3 and 7, and the BIR3 domain of XIAP, which is stringently selective for caspase 9.21 We also employed the commercially available LEHD sequence that is sold as a caspase 9 substrate in addition to MPP8 and Ac-DEVD-ACC. Cytosolic extracts were pre-activated by adding 10 μM cytochrome c and 1 mM dATP at 37°C for 10 min before addition of substrates and BIR domains as appropriate (Figure 5).
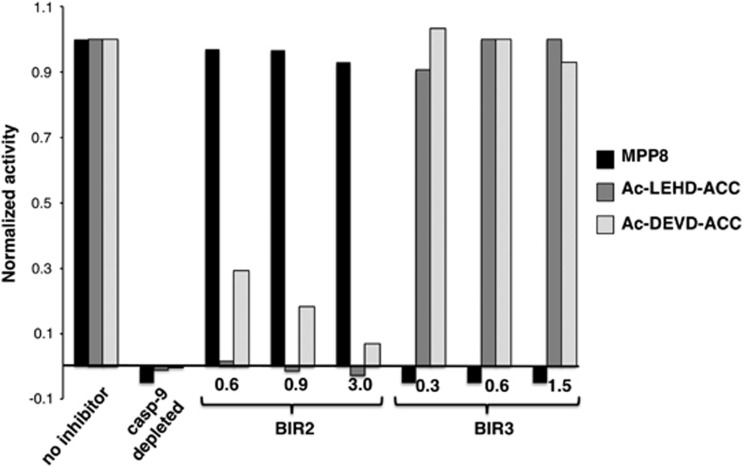
Figure 5.
Detection of caspase 3 and caspase 9 in cytosolic extracts using synthetic substrates. Cytosolic extracts were incubated with cytochrome c for 10 min at 37°C before measuring activity with MPP8, Ac-LEHD-ACC, or Ac-DEVD-ACC. Activated cytosolic extracts were either left untreated (no inhibitor) or treated with the indicated concentrations of BIR2 or BIR3 (μM). As a control, caspase 9 was immunodepleted from extracts before addition of cytochrome c
The kinetic analysis showed that both Ac-LEHD-ACC and Ac-DEVD-ACC substrates are processed extremely fast, whereas MPP8 is cleaved poorly. This is evidence that LEHD and MPP8 are preferentially cleaved by different enzymes in activated cytosolic extracts as both substrates have comparable kinetic values for recombinant caspase 9. Importantly, MPP8 activity was abrogated by BIR3, and not by BIR2, consistent with an exquisite selectivity of this substrate for caspase 9. In contrast, both activity on Ac-DEVD-ACC and Ac-LEHD-ACC was inhibited by BIR2 and not BIR3. This is strong evidence that the activity against Ac-DEVD-ACC is because of caspases 3 and 7, as expected, and that the activity observed with the supposed caspase 9 substrate Ac-LEHD-ACC in cytosolic extracts is mainly because of caspases 3 and 7, and not caspase 9. The caspase 9 immunodepletion control confirms that activity on the substrates is caspase dependent, because only caspases are expected to be activated in this system – see Supplementary Figure 13B for validation of caspase 9 depletion. Addition of recombinant caspase 9 as well as its cleavage site mutant D330A recapitulated activity on MPP8 (Supplementary Figures 13–15) validating the in vitro caspase activation paradigm.
Discussion
Caspases are key regulatory enzymes in apoptosis and inflammation. The development of selective and sensitive biochemical tools for the investigation of their activation and regulation in vivo are obligatory for understanding their biological functions in both normal cells and pathophysiological states. The synthesis and biochemical evaluation of the first combinatorial substrate library targeting caspases over 15 years ago had a great impact on the understanding of their substrate preferences and predicted functions. Unfortunately, almost all caspase substrates lack specificity and thus cannot be used for monitoring the participation of individual caspases in a complex system. We demonstrate that HyCoSuL is a powerful tool for the discovery of new, more selective and more sensitive caspase substrates. The use of a wide range of unnatural amino acids allowed us to design selective substrates that are able to distinguish between all the apoptotic caspases tested, with the notable exceptions of the closely related executioner caspases 3 and 7. We note a caveat to our approach, because the direct conversion of information obtained through HyCoSuL screening (as well as from traditional PS-SCL) into the design of specific substrates is not straightforward. This is because caspases, akin to many other proteases, likely exhibit subsite cooperativity. Binding of particular amino acid to its cognate subsite is not always an independent process and can be influenced (assisted or disturbed) by neighboring amino acids.27 Taking this into account, results from both approaches (HyCoSuL and PS-SCL) must be validated by resynthesis and kinetic analysis. Consequently, a selection of best hits from the HyCoSuL screens were used to inform the synthesis of a selection of substrates, and from these individual substrates we determined the optimal and most selective substrates for each caspase.
The biological utility of this approach extends to the ability, for the first time, to observe the activity of individual caspases during cytochrome-c-triggered apoptosis. We have not yet attempted to observe the value of the selective substrates in intact cells, mainly because of cell penetration concerns, but this is clearly a venue for future exploration. Moreover, we propose that caspase-selective substrates found through HyCoSuL screening provide an excellent scaffold for the synthesis of highly selective activity-based probes to study caspase activity and activation during apoptosis. We predict that this novel approach can secure a much more thorough understanding of the apoptotic caspase cascade regulation and possibly guide the design of new compounds with enhanced specificity for therapeutic utility.
Materials and Methods
Reagents
Chemicals were obtained from commercial suppliers and used without further purification. To libraries and individual substrates synthesis, Rink amide RA resin (particle size 200–300 mesh, loading 0.48 mmol/g, Iris Biotech GmbH, Marktredwitz, Germany), Fmoc-protected amino acids (purity>99% Iris Biotech GmbH, Bachem (Torrance, CA, USA), Creosalus (Louisville, KY, USA)), N-hydroxybenzotriazole (HOBt monohydrate purity>98%, Creosalus), diisopropylcarbodiimide (DICI, peptide grade, Iris Biotech GmbH), N,N-diisopropylethylamine (DIPEA, peptide grade, VWR International, Gdansk, Poland), O-benzotriazole-N,N,N',N'-tetramethyl-uronium-hexafluoro-phosphate (HBTU, peptide grade, Iris Biotech GmbH), 2-(1-H-7-azabenzotriazol-1-yl)-1,1,3,3-tetramethyl uranium hexafluorophosphatemethanaminium (HATU, peptide grade, Iris Biotech GmbH), 2,4,6-trimethylpyridine (collidine, peptide grade, Sigma-Aldrich Sp. z o.o., Poznan, Poland), N,N'-dimethylformamide (DMF, peptide grade, Avantor, Center Valley, PA, USA), dichloromethane (DCM, pure for analysis, POCh, Gliwice, Poland), methanol (MeOH, pure for analysis, POCh), acetonitrile (ACN, HPLC gradient grade, Avantor), diethyl ether (Et2O, pure for analysis, POCh), piperidine (PIP, purity>99%, Sigma-Aldrich Sp. z o.o.), trifluoroacetic acid (TFA, purity 99%, Iris Biotech GmbH), triisopropylsilane (TIPS, purity 99%, Sigma-Aldrich Sp. z o.o.), acetic acid (AcOH, purity >98%, POCh), and phosphorus pentoxide (P2O5, purity 98%, POCh) were used. Individual substrates were purified by HPLC on a Waters M600 solvent delivery module with a Waters M2489 detector system using a semi-preparative Waters Spherisorb S10ODS2 column Waters Sp. z o.o. (Warszawa, Polska). The solvent composition was as follows: phase A (water/0.1% TFA) and phase B (ACN/H2O 80/20 (%, v/v) with 0.1% of TFA). The purity of each individual compound was confirmed by analytical HPLC using a Waters Spherisorb S5ODS2 column. Finally, the molecular weight of each substrate was confirmed by high-resolution mass spectrometry on High Resolution Mass Spectrometer WATERS LCT Premier XE with Electrospray ionization (ESI) and Time of Flight (TOF). Data files for these studies can be found in the Supplementary Data section.
Synthesis of NH2-ACC-Rink Amide resin
Preparation of ACC was carried out as described previously according to Maly et al.18 To glass reaction vessel, 1 eq (6.24 mmol, 13 g) of Rink AM resin was added and stirred gently once per 10 min in DCM for 1 h, and then filtered and washed 3 times with DMF. Fmoc-protecting group was removed using 20% piperidine in DMF (5, 5, and 25 min), filtered each time and washed with DMF (six times). Next, 2.50 eq of Fmoc-ACC-OH (15.6 mmol, 6.9 g) was preactivated with 2.50 eq HOBt monohydrate (15.6 mmol, 2.34 g) and 2.50 eq DICI (15.6 mmol, 2.0 ml) in DMF and mixture was added to the resin. Reaction was stirred gently for 24 h at room temperature. Resin was washed four times with DMF and reaction was repeated using 1.5 eq of above reagents to improve yield of ACC coupling to the resin. After reaction, resin was washed with DMF and Fmoc group was removed using 20% piperidine in DMF (5, 5, and 25 min), filtered and washed with DMF (six times).
Synthesis of NH2-Asp(t-Bu)-ACC-Rink Amide resin
Next, 2.5 eq Fmoc-Asp(t-Bu)-OH (15.6 mmol, 6.42 g) with 2.5 eq HATU (15.6 mmol, 5.93 g), 2.5 eq collidine (15.6 mmol, 2.03 ml) in DMF were activated for 2 min and added to filter cannula with 1 eq (6.24 mmol) NH2-ACC-resin and reaction was carried out for 24 h. Next, resin was washed four times with DMF and reaction was repeated using 1.5 eq of above reagents. After washing with DMF, Fmoc-protecting group was removed using 20% piperidine in DMF (5, 5, and 25 min). Resin was additional washed with DCM (3 times) and MeOH (3 times) and dried over P2O5.
Synthesis of P2, P3, and P4 sublibrary exemplified in detail with the P2 sublibrary
Each library consisted of 129 sublibraries in which each of the natural amino acids (minus cysteine) and a pool of unnatural amino acids were used at a defined position and an isokinetic mixture of 19 amino acids (without cysteine and with norleucine mimicking methionine) was coupled. Equivalents ratios of amino acids in the isokinetic mixture were created based on their reported coupling rates. A fivefold excess (over resin load) of mixture was used. For fixed positions, a 2.5 eq of single amino acid was used. Reactions were carried out with coupling reagent DICI and HOBt. For P2 coupling, the synthesis of the library was performed using a MultiChem 48-wells synthesis apparatus (FlexChem from SciGene, Sunnyvale, CA, USA). To wells of the reaction apparatus, 1 eq of dry NH2-Asp(t-Bu)-ACC-resin (0.04 mmol, 80 mg) was added and stirred gently for 1 h in DCM, and then washed with DMF (four times). In separated vials, 2.5 eq (0.1 mmol) Fmoc-P2-OH was preactivated with 2.5 eq HOBt (0.1 mmol, 15 mg) and 2.5 eq DICI (0.1 mmol, 14 μl) in DMF. Next, preactivated amino acids were added to wells of the apparatus containing NH2-Asp(t-Bu)-ACC-resin, followed by 3 h agitation. Then, reaction mixture was filtered, washed with DMF (4 times), and a ninhydrin test was carried out. Subsequently Fmoc-protecting groups were removed using 20% piperidine in DMF (5, 5, and 25 min). For P3 and P4 positions coupling, an isokinetic mixture for 48 portions was prepared of 18 Fmoc-protected natural amino acids (omitting cysteine) and with norleucine mimicking methionine (19 amino acids in total). Then, 5 eq of isokinetic mixture, 5 eq HOBt (9.22 mmol, 1.4 g), and 5 eq DICI (9.22 mmol, 1.2 ml) were diluted in DMF and preactivated for 3 min. To each of 48 wells containing 1 eq of NH2-P2-Asp(t-Bu)-ACC-resin, the activated isokinetic mixture was added. After 3 h of agitation, the slurry was filtered and washed with DMF (4 times). A ninhydrin test was carried out and Fmoc-protecting group was removed using 20% piperidine in DMF (5, 5, and 25 min). The same procedure was applied for the remaining substrates. In the same manner the isokinetic mixture was added to sample the P4 position. For N-terminus acetylation, to a vial 5 eq AcOH (9.22 mmol, 527 μl), 5 eq HBTU (9.22 mmol, 3.5 g), and 5 eq DIPEA (9.22 mmol, 1.60 ml) in DMF were added. After stirring for 1 min, the mixture was added to each well in the reaction apparatus, containing NH2-X-X-P2-Asp(t-Bu)-ACC-resin, followed by gentle agitation for 30 min. Next, the resin was washed with DMF (5 times), DCM (3 times), and MeOH (3 times) and dried over P2O5. For cleavage from the resin, after completing the synthesis, peptides were cleaved from the resin by treating for 2 h (shaking once per 15 min) with a mixture of TFA:TIPS:H2O (%, v/v/v 95 : 2.5 : 2.5; 2 ml/well). The solution from each well was collected separately and the resin was washed once by a portion of cleavage solution (1 ml), followed by addition of diethyl ether (Et2O, 14 ml). After precipitation (0.5 h in −20°C) the mixture was centrifuged and washed again with Et2O (4 ml). After centrifugation, a white precipitate was dissolved in ACN.H2O (v/v, 1 : 1.4 ml) and lyophilized. The final products were dissolved in DMSO to a concentration of 20 mM and used without further purification. The synthesis of P3 and P4 sublibraries was in the same manner as described above, and P3 and P4 sublibraries were synthesized by coupling fixed amino acid residues to P3 (isokinetic mixture to P2 and P4) and respectively P4 position (isokinetic mixture to P2 and P3).
Preparation of recombinant caspases
The detailed description of caspases expression and purification can be found elsewhere.28
Enzymatic kinetic studies
These were performed using a fMax fluorimeter (Molecular Devices, Sunnyvale, CA, USA) operating in the kinetic mode in 96-well Corning (Corning, NY, USA) plates. The excitation wavelength for ACC-based substrates was 355 nm, and the emission wavelength was 460 nm with a cutoff of 455 nm. For HyCoSuL screening the assay buffer was 10% w/v sucrose, 20 mM Pipes, 10 mM NaCl, 1 mM EDTA, 10 mM DTT, pH=7.2–7.4 (the buffer for caspases 8, 9, and 10 was supplemented with 1.0 M sodium citrate to ensure maximal activity29, 30). Buffers were prepared at 23°C, screening of the libraries and assays were performed at 37°C. Caspases 3, 6, and 7 were preincubated for 15 min and caspases 8, 9, and 10 were preincubated for 30 min at 37°C before adding to the wells containing substrate.
Characterization of caspases specificity using HyCoSuL
Standard enzyme assay conditions for P2, P3, and P4 positions were as follows: reaction volume 100 μl; total final substrate mixture concentration 50 μM; enzyme concentration caspase 3: 10–20 nM, caspase 6: 100–200 nM, caspase 7: 50–100 nM, caspase 8: 50–100 nM, caspase 9: 100–150 nM, and caspase 10: 50–100 nM. The total assay time was 30 min, and the linear portion of the progress curve (generally 15–30 min) was used to calculate velocity. All experiments were repeated at least three times and the results presented are means. The difference between individual values was <10% in every case. Analysis of the results was based on total RFUs for each sublibrary, setting the highest value to 100% and adjusting the other results accordingly.
Synthesis of individual optimized substrates
The 2.5 eq Fmoc-P2-OH was preactivated with 2.5 eq HOBt and 2.5 eq DICI in DMF and added to cartridge with 1 eq NH2-Asp(t-Bu)-ACC-resin (all substrates contained Asp at P1 position) and followed by gentle agitation for 3 h. Then, it was filtered and washed with DMF (six times). Fmoc-protecting group was removed using 20% piperidine in DMF (5, 5, and 25 min). Ninhydrin test was carried out each time after coupling and deprotection. A solution of 2.5 eq Fmoc-P3-OH, 2.5 eq HOBt, and 2.5 eq DICI in DMF was added to the resin and the slurry was agitated for 3 h. After removal of the solution, the resin was washed with DMF (six times), and coupling and deprotection of Fmoc-P4-OH was carried in identical conditions like P2 position. N-terminus was protected with acetyl group using 5 eq AcOH, 5 eq HBTU, and 5 eq DIPEA in DMF as previous described. After solvent removal, the resin was washed with DMF (six times), DCM (three times), and MeOH (three times) dried over P2O5 and cleaved from the resin with a mixture of TFA/TIPS/H2O (%, v/v/v 95 : 2.5 : 2.5). The crude product was purified by HPLC and lyophilized. Its purity was confirmed by analytical HPLC. Each optimized substrate was analyzed using HRMS. Optimized substrates were dissolved in peptide grade DMSO to 20 mM concentration and stored at −80°C until use.
Comparison of kinetic parameters (kcat, KM, and kcat/KM) for individual substrates
Substrates were screened against human recombinant caspases at 37°C in the above assay buffers. Buffers were prepared at 23°C. Caspase 3, 6, and 7 solutions in buffer were preincubated for 15 min (caspases 8, 9, and 10 were preincubated for 30 min) at 37°C before adding to the substrate in the wells of a 96-well plate reader operating in the kinetic mode. Total assay time was 10–20 min. ACC concentrations were calculated by the hydrolysis of eight independent ACC-coupled substrate solutions at known concentration, and an average value was determined. Enzyme kinetic assay conditions were as follows: 100 μl total reaction volume, eight different substrate concentrations, and enzymes at appropriate concentrations. Release of ACC fluorophore was monitored as above. Each experiment was repeated at least three times, and the results are presented as an average with the error bars describing the S.D. Final substrate concentrations for kcat/KM determination ranged from 0.05 to 500 μM. Concentration of DMSO in the assay was <2% (v/v). Calculation of kinetic parameters was carried out using GraphPad Prism software (La Jolla, CA, USA).
FreeStyle 293-F cell cytosolic extracts assay
Hypotonic lysis of FreeStyle 293-F (Life Technologies, Grand Island, NY, USA) cells was performed, and cytosols collected as describedpreviously.24 Each 100 μl portion of FreeStyle 293-F cytosolic extract was activated by addition of 1 μl of dATP (12 mM in water), and 2 μl of horse cytochrome c (1 mM in water) in the presence of the proteasome inhibitor MG132 (10 mM in DMSO), 2 μl. The total reaction volume was 80 μl. The proteolytic activities of caspase 3 and caspase 9 were measured using four short peptide-based fluorogenic substrates (MPP8, Ac-LEHD-ACC, Ac-DEVD-ACC, or Ac-DEVD-AFC) at the final concentration of 250 μM (the excitation wavelength for AFC-based DEVD substrate was 400 nm, and the emission wavelength was 505 nm). Release of ACC or AFC fluorophore was monitored as above. Each experiment was repeated at least three times, and the results are presented as an average RFU/s value. The S.D. for each value was calculated from at least three independent measurements.
Caspase inhibition by XIAP-BIR2 and XIAP-BIR3
Caspase 3/7 activity was inhibited in cellular extracts by using XIAP-BIR2 domain (concentration range: 0.6–3.0 μM), and caspase 9 was inhibited by addition of XIAP-BIR3 domain (concentration range: 0.3–1.5 μM). Cytosolic extracts were preincubated with BIR2 or BIR3 domains for 30 min at 37°C before cytochrome c stimulation. The XIAP fragment BIR2 (124-237) and BIR3 (262-348) were cloned into a modified pET15b (EMD Millipore, Billerica, MA, USA) vector containing 8 × His residues at the N-terminus and expressed in BL21 (DE3) E. coli by induction with 0.2 mM IPTG at 30°C for 4 h.
Immunoblotting and caspase 9 immunodepletion
To monitor caspase 9 processing after cytochrome c activation, cytosolic extracts were stimulated and fractions were collected at the corresponding time points. Each fraction was mixed with SDS loading buffer, boiled, subjected to Laemmli SDS-Page (12% gel), and transferred to a nitrocellulose membrane for western blotting. Primary antibodies were detected by fluorophore-coupled secondary antibodies (LiCOR, Lincoln, NB, USA), and signal acquired using the Odyssey system. For caspase 9 immunodepletion, 100 μl of Protein A/G-Plus agarose resin (Santa Cruz Biotechnology, Paso Robles, CA, USA) was mixed with 50 μl of anti-caspase 9 antibody in PBS and incubated overnight at 4°C to couple the antibody on the beads. Resin was extensively washed with PBS, and subsequently mixed with 1 ml of cytosolic extracts and incubated for 5 h at 4°C. Immunodepletion efficiency was evaluated by western blotting, and the immunodepleted extracts were subsequently used for kinetic studies. The following antibodies were used for immunoblots: anti-caspase 9 rabbit antisera (a gift from Dr. Douglas R Green, Department of Immunology, St. Jude Children's Research Hospital, Memphis, TN, USA), anti-Caspase 3 rabbit polyclonal (Cell Signaling Technology, Beverly, MA, USA), anti-cleaved caspase 3 (D175) rabbit polyclonal (Cell Signaling Technology), and anti-HSP90 mouse monoclonal (BD Transduction Laboratories, San Diego, CA, USA).
Acknowledgments
This work was supported by the National Science Centre grant 2011/03/B/ST5/01048 in Poland to MD and the Foundation for Polish Science (START to MP and FOCUS to MD) and NIH grants GM09040, CA163743, and 5P30 CA030199 to GSS. The work was also supported by a statutory activity subsidy from the Polish Ministry of Science and Higher Education for the Faculty of Chemistry at Wroclaw University of Technology. We thank Artur Krezel for help with caspase analysis.
Glossary
- HyCoSuL
Hybrid Combinatorial Substrate Library
- ACC
7-amino-4-carbamoylmethylcoumarin
- RFU
relative fluorescence unit
- PS-SCL
positional scanning-substrate combinatorial library
- Dab
2,4-diaminobutyric acid
- D-hPhe
D-homophenylalanine
- Ser(Bzl)
serine benzyl ester
- hCha
homocyclohexylalanine
- AFC
7-amino-4-trifluoromethylcoumarin
- XIAP
X-linked inhibitor of apoptosis protein
- BIR domain
baculovirus inhibitor of apoptosis protein repeat
- Phe(2-Cl)
2-chlorophenylalanine
- Pip
pipecolinic acid
- hTyr
homotyrosine
- Oic
octahydroindole-2-carboxylic acid
- Tic
1,2,3,4-tetrahydroisoquinoline-3-carboxylic acid
- His(Bzl)
N(im)-benzyl-histidine
- hPhe
homophenylalanine
- Thr(Bzl)
threonine O-benzyl ester
- Phe(F5)
2,3,4,5,6-pentafluorophenylalanine
- Aad
2-Aminohexanedioic Acid
- Nle(O-Bzl)
2-amino-6-benzyloxyhexanoic acid
- NptGly
neo-pentylglycine
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Cell Death and Differentiation website (http://www.nature.com/cdd)
Edited by D Vaux
Supplementary Material
References
- McIlwain DR, Berger T, Mak TW. Caspase functions in cell death and disease. Cold Spring Harb Perspect Biol. 2013;5:a008656. doi: 10.1101/cshperspect.a008656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford ED, Wells JA. Caspase substrates and cellular remodeling. Annu Rev Biochem. 2011;80:1055–1087. doi: 10.1146/annurev-biochem-061809-121639. [DOI] [PubMed] [Google Scholar]
- Pop C, Salvesen GS. Human caspases: activation, specificity, and regulation. J Biol Chem. 2009;284:21777–21781. doi: 10.1074/jbc.R800084200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y. Mechanical aspects of apoptosome assembly. Curr Opin Cell Biol. 2006;18:677–684. doi: 10.1016/j.ceb.2006.09.006. [DOI] [PubMed] [Google Scholar]
- van Raam BJ, Salvesen GS. Proliferative versus apoptotic functions of caspase-8 Hetero or homo: the caspase-8 dimer controls cell fate. Biochim Biophys Acta. 2012;1824:113–122. doi: 10.1016/j.bbapap.2011.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornberry NA, Rano TA, Peterson EP, Rasper DM, Timkey T, Garcia-Calvo M, et al. A combinatorial approach defines specificities of members of the caspase family and granzyme B. Functional relationships established for key mediators of apoptosis. J Biol Chem. 1997;272:17907–17911. doi: 10.1074/jbc.272.29.17907. [DOI] [PubMed] [Google Scholar]
- Rano TA, Timkey T, Peterson EP, Rotonda J, Nicholson DW, Becker JW, et al. A combinatorial approach for determining protease specificities: application to interleukin-1beta converting enzyme (ICE) Chem Biol. 1997;4:149–155. doi: 10.1016/s1074-5521(97)90258-1. [DOI] [PubMed] [Google Scholar]
- Poreba M, Strozyk A, Salvesen GS, Drag M. Caspase substrates and inhibitors. Cold Spring Harb Perspect Biol. 2013;5:a008680. doi: 10.1101/cshperspect.a008680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linton SD. Caspase inhibitors: a pharmaceutical industry perspective. Curr Top Med Chem. 2005;5:1697–1717. doi: 10.2174/156802605775009720. [DOI] [PubMed] [Google Scholar]
- MacKenzie SH, Schipper JL, Clark AC. The potential for caspases in drug discovery. Curr Opin Drug Discov Dev. 2010;13:568–576. [PMC free article] [PubMed] [Google Scholar]
- Callus BA, Vaux DL. Caspase inhibitors: viral, cellular and chemical. Cell Death Differ. 2007;14:73–78. doi: 10.1038/sj.cdd.4402034. [DOI] [PubMed] [Google Scholar]
- McStay GP, Salvesen GS, Green DR. Overlapping cleavage motif selectivity of caspases: implications for analysis of apoptotic pathways. Cell Death Differ. 2008;15:322–331. doi: 10.1038/sj.cdd.4402260. [DOI] [PubMed] [Google Scholar]
- Benkova B, Lozanov V, Ivanov IP, Mitev V. Evaluation of recombinant caspase specificity by competitive substrates. Anal Biochem. 2009;394:68–74. doi: 10.1016/j.ab.2009.07.012. [DOI] [PubMed] [Google Scholar]
- Pereira NA, Song Z. Some commonly used caspase substrates and inhibitors lack the specificity required to monitor individual caspase activity. Biochem Biophys Res Commun. 2008;377:873–877. doi: 10.1016/j.bbrc.2008.10.101. [DOI] [PubMed] [Google Scholar]
- Berger AB, Sexton KB, Bogyo M. Commonly used caspase inhibitors designed based on substrate specificity profiles lack selectivity. Cell Res. 2006;16:961–963. doi: 10.1038/sj.cr.7310112. [DOI] [PubMed] [Google Scholar]
- Sleath PR, Hendrickson RC, Kronheim SR, March CJ, Black RA. Substrate specificity of the protease that processes human interleukin-1 beta. J Biol Chem. 1990;265:14526–14528. [PubMed] [Google Scholar]
- Howard AD, Kostura MJ, Thornberry N, Ding GJ, Limjuco G, Weidner J, et al. IL-1-converting enzyme requires aspartic acid residues for processing of the IL-1 beta precursor at two distinct sites and does not cleave 31-kDa IL-1 alpha. J Immunol. 1991;147:2964–2969. [PubMed] [Google Scholar]
- Maly DJ, Leonetti F, Backes BJ, Dauber DS, Harris JL, Craik CS, et al. Expedient solid-phase synthesis of fluorogenic protease substrates using the 7-amino-4-carbamoylmethylcoumarin (ACC) fluorophore. J Org Chem. 2002;67:910–915. doi: 10.1021/jo016140o. [DOI] [PubMed] [Google Scholar]
- Talanian RV, Quinlan C, Trautz S, Hackett MC, Mankovich JA, Banach D, et al. Substrate specificities of caspase family proteases. J Biol Chem. 1997;272:9677–9682. doi: 10.1074/jbc.272.15.9677. [DOI] [PubMed] [Google Scholar]
- Boucher D, Blais V, Denault JB. Caspase-7 uses an exosite to promote poly(ADP ribose) polymerase 1 proteolysis. Proc Natl Acad Sci USA. 2012;109:5669–5674. doi: 10.1073/pnas.1200934109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuentes-Prior P, Salvesen GS. The protein structures that shape caspase activity, specificity, activation and inhibition. Biochem J. 2004;384 (Pt 2:201–232. doi: 10.1042/BJ20041142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P, Nijhawan D, Budihardjo I, Srinivasula SM, Ahmad M, Alnemri ES, et al. Cytochrome c and dATP-dependent formation of Apaf-1/caspase-9 complex initiates an apoptotic protease cascade. Cell. 1997;91:479–489. doi: 10.1016/s0092-8674(00)80434-1. [DOI] [PubMed] [Google Scholar]
- Jiang X, Wang X. Cytochrome c promotes caspase-9 activation by inducing nucleotide binding to Apaf-1. J Biol Chem. 2000;275:31199–31203. doi: 10.1074/jbc.C000405200. [DOI] [PubMed] [Google Scholar]
- Denault JB, Eckelman BP, Shin H, Pop C, Salvesen GS. Caspase 3 attenuates XIAP (X-linked inhibitor of apoptosis protein)-mediated inhibition of caspase 9. Biochem J. 2007;405:11–19. doi: 10.1042/BJ20070288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riedl SJ, Salvesen GS. The apoptosome: signalling platform of cell death. Nat Rev Mol Cell Biol. 2007;8:405–413. doi: 10.1038/nrm2153. [DOI] [PubMed] [Google Scholar]
- Stennicke HR, Deveraux QL, Humke EW, Reed JC, Dixit VM, Salvesen GS. Caspase-9 can be activated without proteolytic processing. J Biol Chem. 1999;274:8359–8362. doi: 10.1074/jbc.274.13.8359. [DOI] [PubMed] [Google Scholar]
- Ng NM, Pike RN, Boyd SE. Subsite cooperativity in protease specificity. Biol Chem. 2009;390:401–407. doi: 10.1515/BC.2009.065. [DOI] [PubMed] [Google Scholar]
- Stennicke HR, Salvesen GS. Caspases: preparation and characterization. Methods. 1999;17:313–319. doi: 10.1006/meth.1999.0745. [DOI] [PubMed] [Google Scholar]
- Boatright KM, Deis C, Denault JB, Sutherlin DP, Salvesen GS. Activation of caspases 8 and 10 by FLIP L. Biochem J. 2004;382 (Pt 2:651–657. doi: 10.1042/BJ20040809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boatright KM, Renatus M, Scott FL, Sperandio S, Shin H, Pedersen IM, et al. A unified model for apical caspase activation. Mol Cell. 2003;11:529–541. doi: 10.1016/s1097-2765(03)00051-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.