Abstract
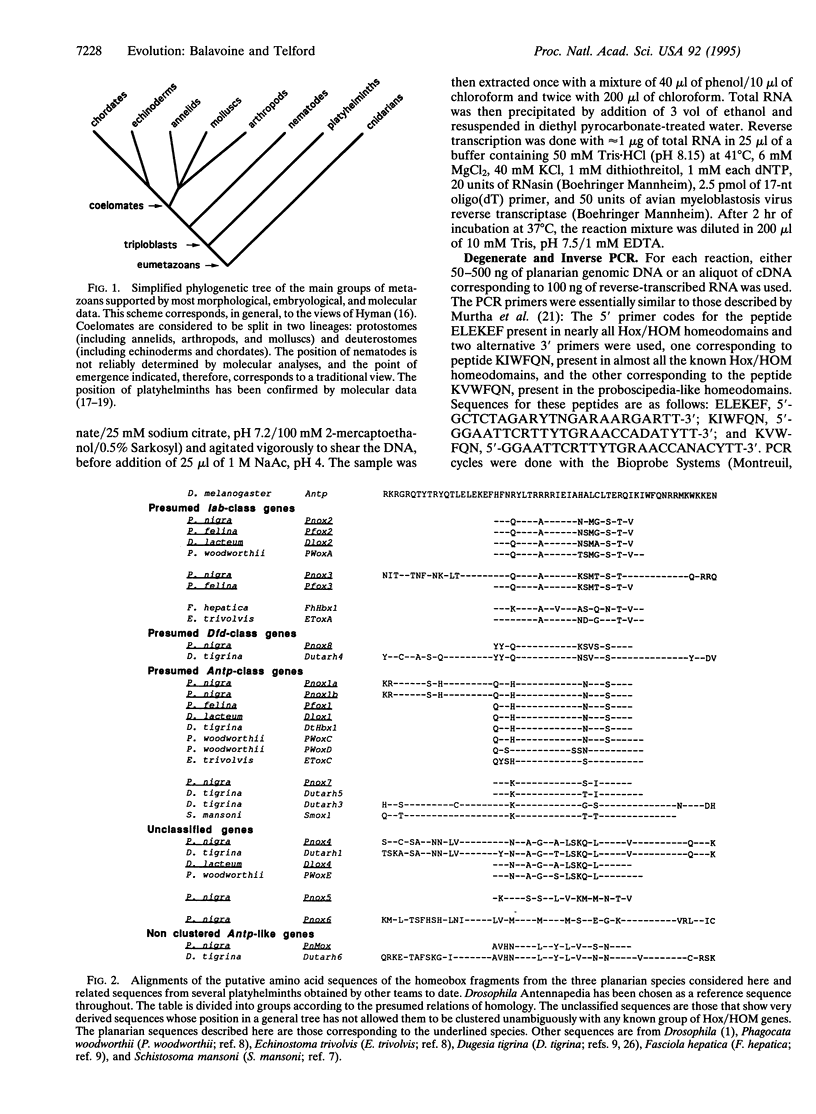
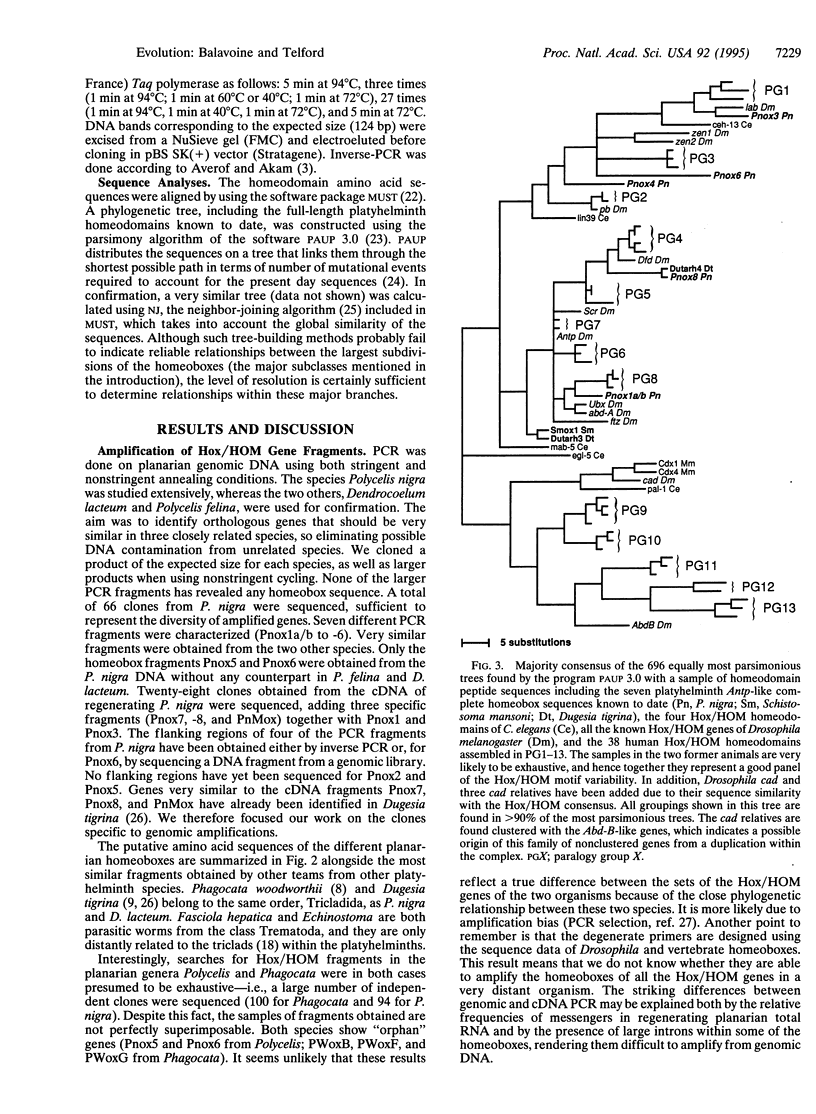
The homeotic gene complex (HOM-C) is a cluster of genes involved in the anteroposterior axial patterning of animal embryos. It is composed of homeobox genes belonging to the Hox/HOM superclass. Originally discovered in Drosophila, Hox/HOM genes have been identified in organisms as distantly related as arthropods, vertebrates, nematodes, and cnidarians. Data obtained in parallel from the organization of the complex, the domains of gene expression during embryogenesis, and phylogenetic relationships allow the subdivision of the Hox/HOM superclass into five classes (lab, pb/Hox3, Dfd, Antp, and Abd-B) that appeared early during metazoan evolution. We describe a search for homologues of these genes in platyhelminths, triploblast metazoans emerging as an outgroup to the great coelomate ensemble. A degenerate PCR screening for Hox/HOM homeoboxes in three species of triclad planarians has revealed 10 types of Antennapedia-like genes. The homeobox-containing sequences of these PCR fragments allowed the amplification of the homeobox-coding exons for five of these genes in the species Polycelis nigra. A phylogenetic analysis shows that two genes are clear orthologues of Drosophila labial, four others are members of a Dfd/Antp superclass, and a seventh gene, although more difficult to classify with certainty, may be related to the pb/Hox3 class. Together with previously identified Hox/HOM genes in other flatworms, our analyses demonstrate the existence of an elaborate family of Hox/HOM genes in the ancestor of all triploblast animals.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adoutte A., Philippe H. The major lines of metazoan evolution: summary of traditional evidence and lessons from ribosomal RNA sequence analysis. EXS. 1993;63:1–30. doi: 10.1007/978-3-0348-7265-2_1. [DOI] [PubMed] [Google Scholar]
- Averof M., Akam M. HOM/Hox genes of Artemia: implications for the origin of insect and crustacean body plans. Curr Biol. 1993 Feb;3(2):73–78. doi: 10.1016/0960-9822(93)90158-k. [DOI] [PubMed] [Google Scholar]
- Bartels J. L., Murtha M. T., Ruddle F. H. Multiple Hox/HOM-class homeoboxes in Platyhelminthes. Mol Phylogenet Evol. 1993 Jun;2(2):143–151. doi: 10.1006/mpev.1993.1014. [DOI] [PubMed] [Google Scholar]
- Bürglin T. R., Ruvkun G. The Caenorhabditis elegans homeobox gene cluster. Curr Opin Genet Dev. 1993 Aug;3(4):615–620. doi: 10.1016/0959-437x(93)90097-9. [DOI] [PubMed] [Google Scholar]
- Candia A. F., Hu J., Crosby J., Lalley P. A., Noden D., Nadeau J. H., Wright C. V. Mox-1 and Mox-2 define a novel homeobox gene subfamily and are differentially expressed during early mesodermal patterning in mouse embryos. Development. 1992 Dec;116(4):1123–1136. doi: 10.1242/dev.116.4.1123. [DOI] [PubMed] [Google Scholar]
- Cartwright P., Dick M., Buss L. W. HOM/Hox type homeoboxes in the chelicerate Limulus polyphemus. Mol Phylogenet Evol. 1993 Sep;2(3):185–192. doi: 10.1006/mpev.1993.1019. [DOI] [PubMed] [Google Scholar]
- Chomczynski P., Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987 Apr;162(1):156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- Kenyon C. If birds can fly, why can't we? Homeotic genes and evolution. Cell. 1994 Jul 29;78(2):175–180. doi: 10.1016/0092-8674(94)90288-7. [DOI] [PubMed] [Google Scholar]
- Krumlauf R. Hox genes in vertebrate development. Cell. 1994 Jul 29;78(2):191–201. doi: 10.1016/0092-8674(94)90290-9. [DOI] [PubMed] [Google Scholar]
- McGinnis W., Krumlauf R. Homeobox genes and axial patterning. Cell. 1992 Jan 24;68(2):283–302. doi: 10.1016/0092-8674(92)90471-n. [DOI] [PubMed] [Google Scholar]
- Murtha M. T., Leckman J. F., Ruddle F. H. Detection of homeobox genes in development and evolution. Proc Natl Acad Sci U S A. 1991 Dec 1;88(23):10711–10715. doi: 10.1073/pnas.88.23.10711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naito M., Ishiguro H., Fujisawa T., Kurosawa Y. Presence of eight distinct homeobox-containing genes in cnidarians. FEBS Lett. 1993 Nov 1;333(3):271–274. doi: 10.1016/0014-5793(93)80668-k. [DOI] [PubMed] [Google Scholar]
- Oliver G., Vispo M., Mailhos A., Martínez C., Sosa-Pineda B., Fielitz W., Ehrlich R. Homeoboxes in flatworms. Gene. 1992 Nov 16;121(2):337–342. doi: 10.1016/0378-1119(92)90140-k. [DOI] [PubMed] [Google Scholar]
- Pendleton J. W., Nagai B. K., Murtha M. T., Ruddle F. H. Expansion of the Hox gene family and the evolution of chordates. Proc Natl Acad Sci U S A. 1993 Jul 1;90(13):6300–6304. doi: 10.1073/pnas.90.13.6300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippe H. MUST, a computer package of Management Utilities for Sequences and Trees. Nucleic Acids Res. 1993 Nov 11;21(22):5264–5272. doi: 10.1093/nar/21.22.5264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou N., Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987 Jul;4(4):406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Schierwater B., Murtha M., Dick M., Ruddle F. H., Buss L. W. Homeoboxes in cnidarians. J Exp Zool. 1991 Dec;260(3):413–416. doi: 10.1002/jez.1402600316. [DOI] [PubMed] [Google Scholar]
- Schubert F. R., Nieselt-Struwe K., Gruss P. The Antennapedia-type homeobox genes have evolved from three precursors separated early in metazoan evolution. Proc Natl Acad Sci U S A. 1993 Jan 1;90(1):143–147. doi: 10.1073/pnas.90.1.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schummer M., Scheurlen I., Schaller C., Galliot B. HOM/HOX homeobox genes are present in hydra (Chlorohydra viridissima) and are differentially expressed during regeneration. EMBO J. 1992 May;11(5):1815–1823. doi: 10.1002/j.1460-2075.1992.tb05233.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shankland M. Leech segmentation: a molecular perspective. Bioessays. 1994 Nov;16(11):801–808. doi: 10.1002/bies.950161106. [DOI] [PubMed] [Google Scholar]
- Slack J. M., Holland P. W., Graham C. F. The zootype and the phylotypic stage. Nature. 1993 Feb 11;361(6412):490–492. doi: 10.1038/361490a0. [DOI] [PubMed] [Google Scholar]
- Tarabykin V. S., Lukyanov K. A., Potapov V. K., Lukyanov S. A. Detection of planarian Antennapedia-like homeobox genes expressed during regeneration. Gene. 1995 Jun 9;158(2):197–202. doi: 10.1016/0378-1119(95)00006-r. [DOI] [PubMed] [Google Scholar]
- Webster P. J., Mansour T. E. Conserved classes of homeodomains in Schistosoma mansoni, an early bilateral metazoan. Mech Dev. 1992 Jul;38(1):25–32. doi: 10.1016/0925-4773(92)90035-i. [DOI] [PubMed] [Google Scholar]



