Abstract
In the preparation of synthetic conotoxins containing multiple disulfide bonds, oxidative folding can produce numerous permutations of disulfide bond connectivities. Establishing the native disulfide connectivities thus presents a significant challenge when the venom-derived peptide is not available, as is increasingly the case when conotoxins are identified from cDNA sequences. Here, we investigate the disulfide connectivity of μ-conotoxin KIIIA, which was predicted originally to have a [C1-C9,C2-C15,C4-C16] disulfide pattern based on homology with closely-related μ-conotoxins. The two major isomers of synthetic μ-KIIIA formed during oxidative folding were purified and their disulfide connectivities mapped by direct mass spectrometric CID fragmentation of the disulfide-bonded polypeptides. Our results show that the major oxidative folding product adopts a [C1-C15,C2-C9,C4-C16] disulfide connectivity, while the minor product adopts a [C1-C16,C2-C9,C4-C15] connectivity. Both of these peptides were potent blockers of NaV1.2 (Kd 5 and 230 nM, respectively). The solution structure for μ-KIIIA based on NMR data was recalculated with the [C1-C15,C2-C9,C4-C16] disulfide pattern; its structure was very similar to the μ-KIIIA structure calculated with the incorrect [C1-C9,C2-C15,C4-C16] disulfide pattern, with an α-helix spanning residues 7–12. In addition, the major folding isomers of μ-KIIIB, an N-terminally extended isoform of μ-KIIIA identified from its cDNA sequence, were isolated. These folding products had the same disulfide connectivities as for μ-KIIIA, and both blocked NaV1.2 (Kd 470 and 26 nM, respectively). Our results establish that the preferred disulfide pattern of synthetic μ-KIIIA/μ-KIIIB folded in vitro is 1-5/2-4/3-6 but that other disulfide isomers are also potent sodium channel blockers. These findings raise questions about the disulfide pattern(s) of μ-KIIIA in the venom of Conus kinoshitai; indeed, the presence of multiple disulfide isomers in the venom could provide a means to further expand the snail's repertoire of active peptides.
Disulfide bonds are a common feature of peptides and proteins that function outside the cell and are generally regarded as being essential for both stability and maintenance of the native structure.1,2 There is evidence, however, that they can be reduced and scrambled in vivo by agents such as glutathione, serum albumin or redox enzymes,3–5 with possible losses of native structure and biological activity. Several strategies have been implemented in order to replace disulfide bridges with more stable linkages, including diselenides,6,7 thioethers such as lanthionine, in which one of the sulfur atoms of the disulfide bond is eliminated,8 cystathionine, in which one of the sulfur atoms is replaced by a methylene group,9,10 or dicarba bridges, in which the disulfide is replaced with a carbon-carbon bridge.11–13
In peptides with multiple disulfide bonds, establishing the native disulfide connectivities can be challenging. Disulfide mapping methods have generally relied on selective reduction and alkylation of bonded thiol pairs, followed by sequencing using traditional Edman sequencing methods or mass spectrometry.14–16 This approach is limited by difficulties of selective reduction in the case of multiple disulfide bonded peptides.17 Conventional two-dimensional 1H NMR spectroscopy can also identify these connectivities in principle,16 but spectral overlap of resonances from multiple Cys spin systems often makes it difficult or impossible to decipher the native connectivities in peptides with multiple disulfide bonds.18,19 As such, alternative NMR-based strategies have been introduced in order to overcome this problem.20,21
Recently, an alternative approach was proposed by Poppe et al.17 in which the disulfide connectivity was obtained by applying Bayesian rules of inference to the local topology of cysteine residues. This method was then applied to experimental NMR data for three exemplar peptides with complex disulfide connectivities: hepcidin, kalata-B1, and μ-conotoxin KIIIA. In the case of μ-KIIIA, the inferred connectivity [C1-C15,C2-C9,C4-C16] differed from the previously described pattern for this conotoxin [C1-C9,C2-C15,C4-C16].22 The [C1-C9,C2-C15,C4-C16] pattern was consistent with the original NMR data set although not uniquely defined by it (for the reasons alluded to above), and the choice of that pattern was dictated predominantly by the ample precedent in the literature for the 1-4/2-5/3-6 disulfide pattern in numerous other μ-conotoxins.21,23–27 It was further supported by the observation that two-disulfide analogues of μ-KIIIA containing just the [C2-C15] and [C4-C16] bridges showed a similar structure and similar activity profile to those of μ-KIIIA itself.22,28,29
In this paper, we have examined the two major isomers of synthetic μ-KIIIA (named μ-KIIIA–P1 and μ-KIIIA-P2) formed during oxidative folding. Their disulfide connectivities have been mapped by a newly developed MS procedure which is based on the interpretation of the product ions produced upon direct gas phase fragmentation of the native, disulfide bonded intact peptides.15,30 NMR spectra of both the folding isoforms have also been analysed. The results show that the more abundant product from oxidative folding has the [C1-C15,C2-C9,C4-C16] disulfide pattern observed by Poppe et al.17 while the minor product has a [C1-C16,C2-C9,C4-C15] pattern. Remarkably, both isomers exhibited blockade of sodium channels, raising questions about which, if either, is the `native' disulfide pattern in the venom of Conus kinoshitai, the Conus species from which the cDNA sequence encoding this peptide was first isolated.31 Upon further examination of the cDNA sequence of KIIIA, it was determined that the mature peptide sequence of μ-KIIIA produced in the venom of Conus kinoshitai possesses an additional two residues preceding the N-terminus (Asn1 and Gly2). The N-terminally extended isoform of μ-KIIIA is named μ-KIIIB. The major oxidative isomers of this analogue (μ-KIIIB-P1 and μ-KIIIB-P2) have also been synthesized and their disulfide connectivities and sodium channel blocking activities compared with that of μ-KIIIA.
MATERIALS AND METHODS
Cloning of μ-KIIIB
μ-KIIIB was identified from a cDNA library prepared as described previously.32 Briefly, a cDNA library was created from the venom duct of an individual Conus kinoshitai specimen. Total RNA was isolated from the venom duct with TRIzol reagent (TRIzol Total RNA Isolation - Life Technologies/Gibco BRL, Grand Island, NY), and cDNA prepared using the SMART PCR cDNA Synthesis Kit (Clontech Laboratories, Palo Alto CA). The cDNA was cloned into pNEB206A vector (New England BioLabs, Inc., Bever1y, MA) and sequences of individual clones were determined by standard automated sequencing.
Synthesis and oxidative folding
Peptides were synthesized at 30 μmol scale on pre-loaded Fmoc-Cys(Trt)-Rink Amide-MBHA resin (substitution = 0.32 mmol/g) using standard N-(9-fluorenyl)methoxycarbonyl (Fmoc) chemistry. The peptides were cleaved from the resin by 3–4 h treatment with reagent K (TFA/water/ethanedithiol/phenol/thioanisole; 82.5/5/2.5/5/5 by volume). The cleaved peptides were filtered, precipitated with cold methyl tert-butyl ether (MTBE) and washed several times with cold MTBE. The reduced peptides were purified by reversed-phase HPLC using a semi-preparative C18 Vydac column (218TP510, 250 mm × 10 mm, 5 μm particle size) and eluted with a linear gradient from 5 to 35% solvent B in 35 min, where solvent A was 0.1% (v/v) TFA in water, and B was 0.1% (v/v) TFA in 90% aqueous acetonitrile. The flow rate was 4 mL/min, and absorbance was monitored at 220 nm. Purified peptides were quantified by UV absorbance at 280 nm.
Oxidative folding of synthetic μ-KIIIA was accomplished by 2 h glutathione-assisted folding at room temperature under the following conditions: 20 μM linear peptide, 0.1 M Tris-HCl, pH 7.5, 1 mM EDTA, 1:1 mM reduced and oxidized glutathione. Folding was quenched by acidification through addition of 8% (v/v) formic acid. Folded peptides were purified by reversed-phase HPLC using a semi-preparative C18 column (#218TP510, 250 mm × 10 mm) over a linear gradient ranging from 5 to 35% solvent B in 35 min. Purities of the folded peptides were assessed by analytical HPLC using a linear gradient ranging from 10 to 40% solvent B in 30 min. Quantities of the minor folding product were obtained by re-purification using an analytical C18 column (#218TP54, 250 mm × 4.6 mm, 5 μm particle size) using identical HPLC conditions. Peptide amounts were quantified by UV absorbance at 280 nm. Purified peptides were finally dried by lyophilisation and peptide masses were confirmed by MALDI-ToF mass spectrometry.
Mass spectrometry
The mass spectrometric experiments were performed on a HCT Ultra ETDII ion trap mass spectrometer (Bruker Daltonics, Bremen, Germany). All experiments were performed through LC-MS analysis of the samples by coupling the ion trap mass spectrometer with an Agilent 1100 HPLC system. The peptide samples were subjected to LC-MS using a reversed-phase C18 analytical column, with H2O/ acetonitrile (with 0.1% formic acid) as the solvent system, at a flow rate of 0.2 ml/min. The CID experiment was performed by selecting the precursor ion and subsequently fragmenting it through collision with He gas. The fragmentation amplitude (Vp−p) was kept between 1 and 3. The spectra were averaged over four scans.
NMR spectroscopy
NMR spectra were recorded on μ-KIIIA-P1 (86 μM) and of μ-KIIIA-P2 (43 μM) in 95% H2O / 5% 2H2O at pH 4.8 and 5 °C on a Bruker DRX-600 spectrometer. Spectra were processed using TOPSPIN (Version 1.3, Bruker Biospin). Spectra recorded as described in Khoo et al. 22 were analysed using XEASY (Version 1.3.13).33
Structure calculations
Structure calculations were run using the original NOE and dihedral restraint list as described in Khoo et al.22 (BMRB: 20048) but with the disulfide connectivities [C1-C15,C2-C9,C4-C16]. Additional structural calculations were run with six additional NOE distance restraints that were consistent with the [C1-C15,C2-C9,C4-C16] disulfide connectivity but could not be unambiguously determined previously due to peak overlap. Structures were recalculated in XPLOR-NIH34 using the simulated annealing script. Lowest energy structures were subjected to energy minimization in water. Final families of 20 lowest-energy structures were chosen for analysis using PROCHECK-NMR35 and MOLMOL.36 Final structures had no experimental distance violations greater than 0.2 Ǻ or dihedral angle violations greater than 5°. Structural figures were prepared using the programs MOLMOL36 and PyMOL (Delano, W.L. The PyMOL Molecular Graphics System (2002) Delano Scientific, San Carlos, CA, USA. http://www.pymol.org). Final structures have been deposited in the PDB with accession code 2LXG.
Electrophysiology of rat clone of NaV1.2 expressed in Xenopus oocytes
Oocytes expressing NaV1.2 α-subunits were prepared and two-electrode voltage clamped essentially as described previously.37 Briefly, oocytes were placed in a 30 μL chamber containing ND96 and two-electrode voltage clamped at a holding potential of −80 mV. To activate VGSCs, the membrane potential was stepped to −10 mV for a 50 ms period every 20 s. To apply toxin, the perfusion was halted, 3 μL of toxin solution (at ten times the final concentration) was applied to the 30 μL bath, and the bath manually stirred for about 5 s by gently aspirating and expelling a few μL of the bath fluid several times with a pipettor. Toxin exposures were in static baths to conserve material. On-rate constants were obtained assuming the equation kobs = kon•[peptide] + koff,38 where kobs was determined from the single-exponential fit of the time course of block by a fixed concentration of 10 μM peptide, and koff estimated from the level of recovery from block 20 min following toxin washout and assuming recovery followed a single-exponential time course; this procedure to obtain koff was adopted because recovery from block was too slow to measure by single-exponential fits.37 All recordings were made at room temperature (~21 °C).
RESULTS
Cloning of μ-KIIIB
The amino acid sequence of μ-KIIIB was deduced from cDNA derived from venom ducts of Conus kinoshitai, as described previously.32 μ-KIIIB shared an identical amino acid sequence with μ-KIIIA, but included an additional two residues at the N-terminus as follows: Asn-Gly.
Synthesis and oxidative folding
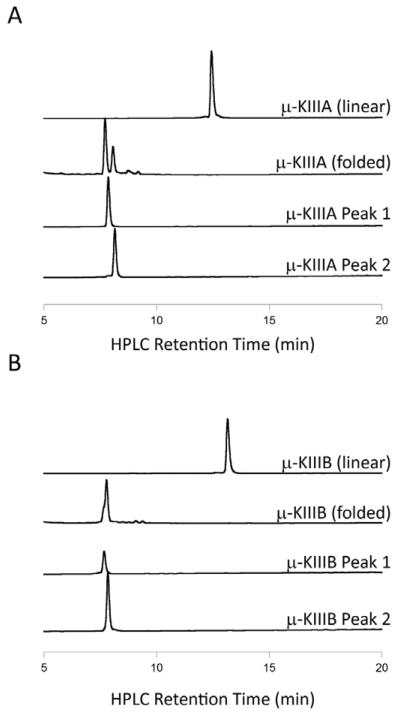
μ-KIIIA and μ-KIIIB were synthesized chemically using the Fmoc protocols described elsewhere.31 After oxidation of the synthetic linear peptides, two distinct HPLC peaks for both μ-KIIIA and μ-KIIIB were obtained that yielded identical mass values (1882.6 Da for μ-KIIIA, 2055.4 Da for μ-KIIIB), corresponding to the respective native sequences with three disulfide bonds. Representative reversed-phase HPLC chromatograms of the oxidative folding of μ-KIIIA and μ-KIIIB are shown in Figure 1. The major isomer (μ-KIIIA-P1) elutes earlier than the minor isomer (μ-KIIIA-P2) for μ-KIIIA while the major isomer (μ-KIIIB-P2) elutes later than the minor isomer (μ-KIIIB-P1) for μ-KIIIB.
Figure 1.
Representative reversed-phase HPLC chromatograms of the oxidative folding of (A) μ-KIIIA and (B) μ-KIIIB. μ-KIIIA Peaks 1 and 2 represent the [C1-C15,C2-C9,C4-C16] and [C1-C16,C2-C9,C4-C15] connectivities, respectively (see text). Folding was carried out in buffered solution (pH 7.5) containing a 1:1 mM mixture of oxidized and reduced glutathione for 2 h at room temperature.
Mass spectrometric determination of disulfide connectivities
Two distinct HPLC peaks of μ-KIIIA, obtained through the oxidation of the synthetic linear peptide, yielded identical mass values (1882.6 Da) corresponding to the native sequence with three disulfide bonds. In principle, 15 disulfide foldamers corresponding to distinct disulfide linkage patterns are possible (Figure S1). Direct MS fragmentation of intact disulfide-bonded peptides or proteolyticaly nicked peptides provides a route to de novo determination of disulfide connectivity in natural and synthetic peptides. This procedure relies on interpretation of different modes of disulfide bond cleavage under ion trap mass spectrometric conditions. The structures of the ions are determined through a program, DisConnect, developed to analyse the CID MS/MS data of the native disulfide bonded molecule. The key steps in establishing disulfide connectivity for the two μ-KIIIA and μ-KIIIB isomers (P1 and P2) are described below.
μ-KIIIA-P1
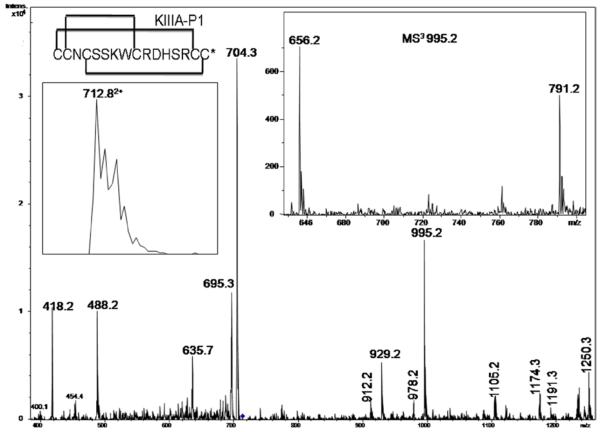
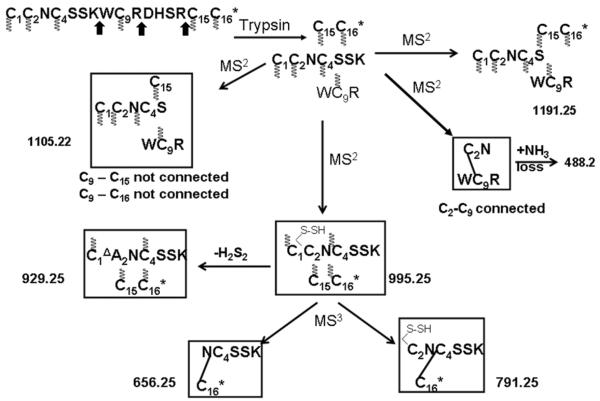
Upon trypsin digestion the fraction shows a mass of 1423.6Da (inset to Figure 2) that corresponds to a peptide in which the central tetrapeptide segment (DHSR) has been excised out by trypsin cleavage at the R-D and R-C15 peptide bonds. In addition, a proteolytic cleavage at the K-W bond is also observed. This means that the tryptic peptide consists of three individual peptide chains that are held together by three disulfide bonds. Foldamers F1, 4 and 7 (Figure S1) cannot yield the observed tryptic peptide. In these cases, due to the C15-C16 connectivity, Arg14 is not inside a disulfide loop and thus would cause separation of C15-C16 from the rest of the peptide. Hence, 12 possible foldamers (Figure S2) need to be considered. Figure 2 shows the CID MS/MS spectra of the tryptic peptide. The structures of the major fragment ions are determined through DisConnect. The chemical structures of the key ions, derived through the fragmentation of the disulfide-bonded molecule are shown in Figure 3. The structures shown correspond uniquely to the observed m/z values. A key ion is observed at m/z 488.2 that arises through the loss of NH3 from an initial product ion, which necessarily must have C2 disulfide bonded to C9. This leaves only two probable foldamers, F11 and F14 to be considered further. A final distinction between these two foldamers is achieved by a subsequent MS3 fragmentation of the ion 995.2 (inset to Figure 2). The probable structures of the fragment ions from 995.2 are shown in Figure 3. The ions at 656.1 and 791.2, establish the C4-C16 connectivity unambiguously. The overall connectivity pattern is therefore that of foldamer F11 (1-5/2-4/3-6).
Figure 2.
CID MS2 spectrum of the peptide derived upon trypsin digestion of μ-KIIIA-P1 (712.8, (M+2H)2+). Inset shows the MS spectrum of the precursor ion and MS3 spectra of 995.2.
Figure 3.
Assignments of the key MSn fragment ions of tryptic μ-KIIIA-P1. The m/z values of each of the ions are indicated against the respective structures and they correspond to the singly charged values, unless otherwise specified. For every structure, Cys residues with indeterminate connectivity are indicated with the wavy lines. Subsequently, in the structures from which a particular Cys connectivity is evident, the connected Cys residues are joined through a dashed line. The arrows indicate the site of proteolysis. The 2–4 connectivity is established by the product ion 488.2, while the ions 656.2 and 791.2 confirm the 3–6 connectivity.
μ-KIIIA-P2
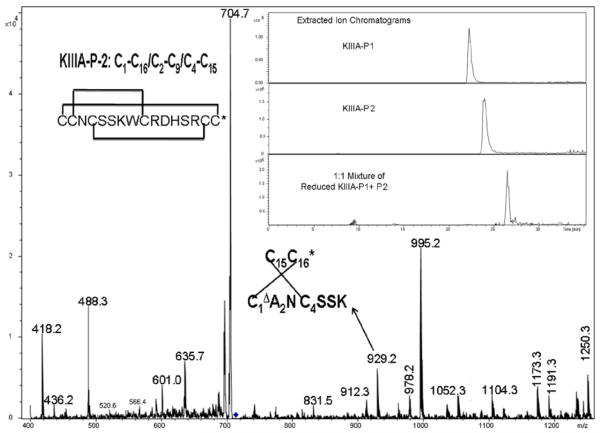
For μ-KIIIA-P2, an identical mass was obtained upon trypsin digestion. Fragmentation of the doubly-charged tryptic peptide yielded an identical MS/MS spectrum (Figure 4) which again leaves F11 and F14 as the two probable foldamers (as described previously for μ-KIIIA-P1). Unfortunately, the MS3 fragmentation of 995.2 did not yield product ions of measurable intensities. However, assignment of the disulfide connectivity in μ-KIIIA-P2 to foldamer F14 may be made through an alternative approach. As described above, the observation of the ion 995.2 is only compatible with foldamers F11 and F14. μ-KIIIA-P1 has already been assigned unambiguously, through mass spectral fragmentation, as F11. HPLC analysis of μ-KIIIA-P1 and P2 reveal two distinct retention times, suggesting that they are indeed two distinct foldamers (inset to Figure 4). Upon reduction, the linearized product from the two foldamers must be identical. This is demonstrated by HPLC co-elution of reduced products of both the μ-KIIIA foldamers (inset to Figure 4). Hence the connectivity of μ-KIIIA-P2 is that of F14 (1-6/2-4/3-5) (Figure 4).
Figure 4.
CID MS2 spectrum of the peptide derived upon trypsin digestion of μ-KIIIA-P2 (712.8, (M+2H)2+). Inset shows the extracted ion chromatogram from the LC-MS analysis of μ-KIIIA-P1, P2 and a reduced equimolar mixture of both fractions.
μ-KIIIB-P1 and -P2
The peptide μ-KIIIB, as mentioned previously, differs from μ-KIIIA by an addition of N-terminal Asn-Gly. In a similar way, described in the Supplementary Material (Figure S3–S5, Supplementary Text), the disulfide connectivity of the μ-KIIIB isomers were determined. The disulfide connectivity of the major isomer, μ-KIIIB-P2, was determined as 1-5/2-4/3-6, while that of μ-KIIIB-P1 was 1-6/2-4/3-5.
NMR of μ-KIIIA
Comparison of 1D spectra at 5 °C of μ-KIIIA-P1 with that published originally for μ-KIIIA22 (Figure S6) indicates that the original sample studied was identical to μ-KIIIA-P1, the major isomer obtained from oxidative folding. Spectra of μ-KIIIA-P2, on the other hand, displayed significant differences, indicating a different fold as a result of a different disulfide connectivity (Figure S6).
Recalculated solution structure for μ-KIIIA-P1
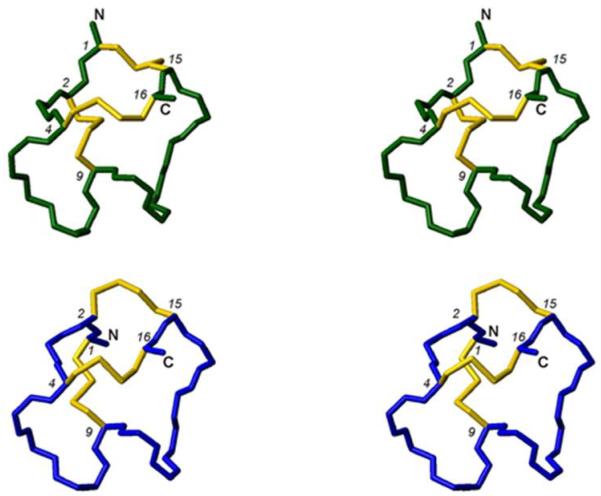
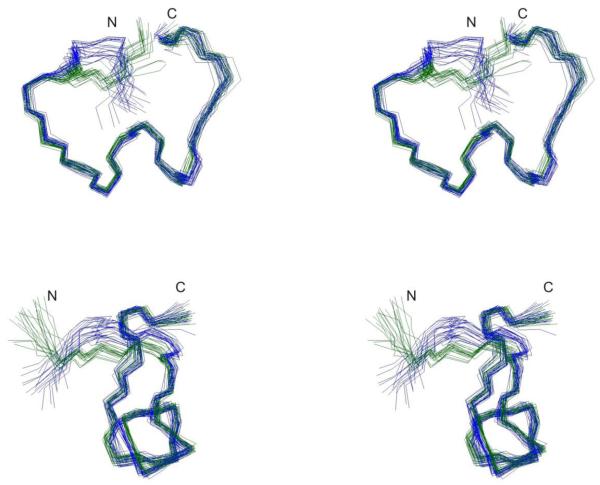
The [C1-C15,C2-C9,C4-C16] disulfide connectivity was consistent with the original NOE and dihedral data set,22 with no major violations observed during calculations, although there was a slight increase in NOE energy function for the final structures calculated. A summary of experimental constraints and structural statistics for μ-KIIIA[C1-C15,C2-C9,C4-C16] (ie. μ-KIIIA-P1) compared with the previously calculated structure for μ-KIIIA assuming the [C1-C9,C2-C15,C4-C16] disulfide connectivity is given in Table 1. The angular order parameters for φ and ψ in the final ensemble of 20 structures for μ-KIIIA[C1-C15,C2-C9,C4-C16] were both > 0.8 over all residues (Figure S8), indicating that the backbone dihedral angles are well defined across the family of structures. The mean pairwise RMS difference over the backbone heavy atoms over all residues for the family of structures of μ-KIIIA[C1-C15,C2-C9,C4-C16] was 0.51 Å. Comparisons of the calculated families of structures as well as closest-to-average structures of μ-KIIIA[C1-C9,C2-C15,C4-C16] and μ-KIIIA[C1-C15,C2-C9,C4-C16] are shown in Figures 5 and 6. Similar to the findings of Poppe et al.,17 structures recalculated with the [C1-C15,C2-C9,C4-C16] disulfide connectivity had an overall topology resembling the previously published structure,22 with an α-helix spanning residues 7-12 in the closest-to-average structure. The backbones generally align well with global RMSD of 0.57 ± 0.14 Å over residues 4-16. The main difference lies in the orientation of the N-terminus, with Cys2 now pulled closer to the α-helix and Cys1 oriented towards the C-terminal tail as a result of the [C2-C9] and [C1-C15] disulfide bonds, respectively. With the [C1-C15,C2-C9,C4-C16] disulfide connectivity, backbone φ and ψ angles of Cys2 correspond to the β-sheet region of the Ramachandran plot instead of the α-helix region, as observed with the [C1-C9,C2-C15,C4-C16] disulfide connectivity (Figure S9).
Table 1.
Structural statistics for μ-KIIIA[C1-C15,C2-C9,C4-C16] and μ-KIIIA[C1-C9,C2-C15,C4-C16]
| μ-KIIIA[C1-C15,C2-C9,C4-C16] | μ-KIIIA[C1-C9,C2-C15,C4-C16]e | |
|---|---|---|
| Distance restraints | 231 | 225 |
| Intra (i = j) | 97 | 97 |
| Sequential (| i – j | =1) | 70 | 70 |
| Short (l < | i – j | < 6 ) | 45 | 45 |
| Long | 19 | 13 |
| Dihedral restraints | 8 | 8 |
| Energies (kcal mol−1)a | ||
| ENOE | 1.7 ± 0.4 | 1.6 ± 0.3 |
| Deviations from ideal geometryb | ||
| Bonds (Å) | 0.0015 ± 0.0001 | 0.0017 ± 0.0002 |
| Angles (°) | 0.503 ± 0.005 | 0.503 ± 0.013 |
| Impropers (°) | 0.379 ± 0.009 | 0.371 ± 0.009 |
| Mean global RMSD (Å)c | ||
| Backbone heavy atoms | 0.51 ± 0.12 | 0.58 ± 0.11 |
| All heavy atoms | 1.39 ± 0.26 | 1.42 ± 0.28 |
|
| ||
| Ramachandran plotd | ||
| Most favoured (%) | 81.1 | 78.9 |
| Allowed (%) | 18.9 | 21.1 |
| Additionally allowed (%) | 0 | 0 |
| Disallowed (%) | 0 | 0 |
The values for ENOE are calculated from a square well potential with force constants of 50 kcal mol−1 Å2.
The values for the bonds, angles, and impropers show the deviations from ideal values based on perfect stereochemistry.
The pairwise RMSD over the indicated residues calculated in MOLMOL.
As determined by the program PROCHECK-NMR for all residues except Gly and Pro.
Structural statistics for μ-KIIIA[C1-C9,C2-C15,C4-C16] from previously published data22 for comparison
Figure 5.
Stereo views of backbone and closest-to-average structures of μ-KIIIA[C1-C15,C2-C9,C4-C16] (green) and μ-KIIIA[C1-C9,C2-C15,C4-C16]22 (blue). Disulfide bonds are displayed in yellow.
Figure 6.
Stereo views of overlay of ensemble of 20 NMR structures of μ-KIIIA[C1-C15,C2-C9,C4-C16] (green) and μ-KIIIA[C1-C9,C2-C15,C4-C16]22 (blue), superimposed over backbone heavy atoms of residues 4–16. Top and bottom panel views are related by a 90° anticlockwise rotation about the vertical-axis.
Electrophysiology assays
The ability of the minor oxidative folding isomer of μ-KIIIA (μ-KIIIA-P2) and the two major oxidative folding isomers of μ-KIIIB (μ-KIIIB-P1 and μ-KIIIB-P2) to block NaV1.2 expressed in oocytes was assessed by the voltage-clamp protocol described in Materials and Methods. Table 2 summarizes the activity of 10 μM μ-KIIIA-P2, μ-KIIIB-P1 and μ-KIIIB-P2 on NaV1.2 in comparison with that of the major oxidative folding isomer of μ-KIIIA (μ-KIIIA-P1) reported previously.37 Representative sodium-current traces of the block by μ-KIIIB-P1 and μ-KIIIB-P2 of NaV1.2 are shown in Figure S10. Both μ-KIIIB isomers had slow off-rates comparable to that of μ-KIIIA, indicating their almost irreversible binding to the NaV1.2 channel. Of the two μ-KIIIB isomers, the major oxidative folding isomer, μ-KIIIB-P2, displayed a greater affinity (Kd 0.026 μM) for NaV1.2 than μ-KIIIB-P1 (Kd 0.47 μM). Thus, μ-KIIIB-P2 was about five times less potent than μ-KIIIA-P1 (Kd 0.005 μM). Comparing the two μ-KIIIA isomers, the major oxidative folding isomer, μ-KIIIA-P1, was more potent than the minor isomer μ-KIIIA-P2, which had a Kd of 0.23 μM.
Table 2.
Block of Nav1.2 by μ-KIIIA and μ-KIIIBa
| Toxin | koff (min−1) | kon (μM•min)−1 | Kdb (μM) |
|---|---|---|---|
| μ-KIIIA-P1c | 0.0016 ± 0.0016 | 0.30 ± 0.03 | 0.005 ± 0.005 |
| μ-KIIIA-P2 | 0.0044 ± 0.0023 | 0.019 ± 0.002d | 0.23 ± 0.12 |
| μ-KIIIB-P1 | 0.0052 ± 0.0017 | 0.011 ± 0.003d | 0.47 ± 0.20 |
| μ-KIIIB-P2 | 0.0034 ± 0.0018 | 0.13 ± 0.01d | 0.026 ± 0.014 |
Values (mean ± S.D, n ≥ 3 oocytes) were obtained by two-electrode voltage clamp of Xenopus oocytes expressing rat NaV1.2 channels as described in Methods and Materials
From koff/kon.
Values for μ-KIIIA as determined previously.22
From [kobd – koff] / 10μM.
The minor isomer μ-KIIIA-P2 was also tested against the NaV1.4 and NaV1.7 subtypes, displaying Kd values of 0.83 and 1.57 μM, respectively (Table S4). Amongst these three subtypes, therefore, the selectivity profile of μ-KIIIA-P2 remained the same as for the major μ-KIIIA isomer, namely, NaV1.2>NaV1.4>NaV1.7.39
DISCUSSION
μ-conotoxins belonging to the M-superfamily of conotoxins have the characteristic Cys framework (-CC-C-C-CC-) of this superfamily,40 and have been subdivided previously into five branches (M1–M5) based on the number of residues between the fourth and fifth half-cystine residues. Interestingly, among known structures, different disulfide patterns can be found in the M1 (1-5/2-4/3-6), M2 (1-6/2-4/3-5) and M4 (1-4/2-5/3-6) branch conotoxins, suggesting that the number of residues in the last cysteine loop might determines the disulfide connectivity in this family.41,42 The first class of μ-conotoxins to be characterized (sequences in Table S1) belonged to the M4 branch, which included μ-GIIIA, μ-GIIIB, μ-PIIIA and μ-TIIIA, and the first of these to be characterized was μ-GIIIA from Conus geographus.43 Typically, early characterization involved isolation and sequencing of the venom-derived peptide,43 following which the peptide was synthesized chemically with the disulfide bonds formed by oxidative-reductive folding with glutathione.44 HPLC co-elution with the venom-derived toxin was then employed to confirm that the synthetic peptide had an identical fold to the native peptide and therefore possessed the same disulfide connectivity.23,44 Mass spectrometric methods have also been employed to elucidate disulfide connectivities.14,26,45 Disulfide mapping was previously carried out on synthetic μ-GIIIA, establishing the disulfide pattern as 1-4/2-5/3-6.45 Disulfide mapping with other M4 branch μ-conotoxins μ-PIIIA and μ-SxIIIA also showed that the disulfide connectivity for this class of μ-conotoxins was consistent with that of μ-GIIIA.21,26
Specific disulfide mapping for the newer class of μ-conotoxins belonging to the M5 branch, which includes μ-KIIIA (Table S1), has, however, never been conducted. Disulfide connectivities were instead based on sequence alignment with the closely related μ-conotoxins in the M4 branch. Solution structures of μ-SmIIIA,24 μ-SIIIA,27 μ-KIIIA22 and most recently μ-CnIIIC46 were solved assuming those canonical disulfide connectivities. The results of our study on μ-KIIIA now confirm that the thermodynamically favored product of oxidative folding in vitro has a disulfide connectivity pattern differing from the canonical pattern established for the M4 μ-conotoxins. The 1-5/2-4/3-6 disulfide connectivity pattern17 adopted by the major folding isomer, μ-KIIIA-P1, is similar to that observed in the M1 branch of conotoxins, while the 1-6/2-4/3-5 pattern of the minor isomer, μ-KIIIA-P2, is similar to that of the M2 branch. Similar studies on the other M5 μ-conotoxins will need to be conducted to determine whether all conotoxins in this class also adopt the 1-5/2-4/3-6 disulfide pattern, although we note that μ-KIIIA and μ-KIIIB have a shorter first loop compared with other M5 μ-conotoxins, with only one amino acid residue between the second and third cysteine residues (Table S1), and this may also influence folding.
In addition to the two major disulfide isomers characterized in this paper, the biological activity and structure of the disulfide-deficient analog μ-KIIIA[C1A,C9A], which has the [2–15,4–16] disulfide connectivity, were characterized previously.22 Intercysteine NOEs for μ-KIIIA[C1A,C9A]22 (Table S3) were better resolved as only two disulfide bonds were present, and confirmed its expected disulfide connectivity. It is intriguing that μ-KIIIA[C1A,C9A] showed potent blockade of NaV1.2 (Kd of 8 nM, comparable to that of 5 nM for μ-KIIIA) and NaV1.4 (240 nM compared with 50 nM)22 even though it contained [C2-C15] and [C4-C16] disulfides rather than the [C2-C9] and [C4-C16] disulfides in the major product of in vitro folding of μ-KIIIA. Clearly, several distinct disulfide connectivities are compatible with a μ-KIIIA structure capable of potent sodium channel blockade.
The biological activities and structures of the major isomers of synthetic μ-PIIIA, an M4 branch μ-conotoxin, formed during oxidative folding were investigated recently.47 Disulfide connectivity analysis by a combination of intercysteine NOE analysis and MALDI-TOF MS/MS revealed different disulfide connectivities for each of the three isomers investigated, with one adopting the canonical 1-4/2-5/3-6 connectivity. In keeping with our findings for μ-KIIIA, all three disulfide isomers of μ-PIIIA were able to block NaV1.4 with considerable potency.47 In contrast to μ-KIIIA, however, the different isomers adopted different overall folds, although the two most potent isomers did retain a common helical region between Phe7 and Arg12. Intriguingly, the disulfide isomer with a 1-5/2-6/3-4 connectivity was more than twice as potent as the 1-4/2-5/3-6 disulfide isomer,47 which is the major form of this peptide in the venom gland.26
These studies, together with our current results, highlight the different disulfide connectivities that can arise as a result of in vitro oxidative folding. The question this then poses is whether the disulfide connectivity in the major product of in vitro oxidative folding corresponds to the native disulfide pattern in the venom of the cone shell Conus kinoshitai. Indeed, the mechanisms of oxidative folding in vivo that lead to formation of the native toxin in the venom are still not well understood. In the biosynthesis of a conotoxin within the venom gland, several factors such as the propeptide precursor sequence, folding enzymes (protein disulfide isomerases) and molecular chaperones can play a role in directing folding and disulfide formation, allowing the efficient synthesis of the native disulfide pattern.48 Recent work by Safavi-Hemami et al.49 provided evidence for the presence of both the globular and ribbon forms of the α-conotoxin ImI in the venom of Conus imperialis. Non-native disulfide isomers have also been reported for other α-conotoxins such as α-GI and α-AuIB.50,51 In these α-conotoxins, two additional non-native disulfide bond isomers could be formed during oxidative folding, the `ribbon' isomer (1–4/2–3 disulfide connectivity) and the `bead' isomer (1–2/3–4). Each of these two isomers adopted a different fold from the native isomer (1–3/2–4), which had a globular fold. Pharmacological studies have traditionally focused on the activities of the globular form, assuming this form to be the native disulfide connectivity. However, in numerous cases (e.g. α-AuIB), it is actually the ribbon form that exhibits the greatest affinity/potency for the molecular target.50 Taken together, these results suggest that multiple isoforms may have evolved, yielding even greater toxin diversity within Conus venoms. Although these studies emphasized the presence of multiple folding isoforms of α-conotoxins, it is conceivable that multiple “misfolded” isomers of μ-conotoxins are also present in the venom and should be explored for their biological activities.
In this study we have also addressed the ambiguity regarding the mature peptide sequence of μ-KIIIA which was derived from the cDNA clone.31 Similar to μ-SmIIIA,38 two putative proteolytic processing sites were identified, and to date the shorter sequence lacking the Asn and Gly residues at the N-terminus has been taken to be the mature peptide sequence of μ-KIIIA. However, upon further examination of the cDNA sequence of μ-KIIIA, it was determined that, if cleavage occurs immediately after the putative `KR' proteolytic site in the propeptide sequence, as observed in several other μ-conotoxins (Table S2), then the mature peptide sequence of μ-KIIIA produced in the venoms of Conus kinoshitai would include the two additional residues preceding the N-terminus. The N-terminally extended isomer has been named μ-KIIIB, and its major folding isomer μ-KIIIB-P2 has been shown to block NaV1.2 with a Kd five-fold higher than that of μ-KIIIA.
Our study shows that, for μ-KIIIA, both the [C1-C9,C2-C15,C4-C16] and [C1-C15,C2-C9,C4-C16] disulfide connnectivities were consistent with the NMR data set, and interchanging between the two patterns altered the structure minimally, with the α-helix bearing the key residues for sodium channel blockade being preserved. This observation validates efforts to miniaturize μ-KIIIA29 and mimic the pharmacophore in truncated, lactam-stabilized analogues of μ-KIIIA.52 Indeed, the uncertainty surrounding disulfide bond connectivities in synthetic conotoxins provides a compelling rationale for replacing them with more stable isosteres or removing them completely with the aim of designing more stable compounds for therapeutic use.
Supplementary Material
ACKNOWLEDGMENTS
We thank Alan A. Goldin for the NaV1.2 clone and Layla Azam for preparing cRNA from this clone. We also thank Joanna Gajewiak and Konkallu H. Gowd for helpful discussions and suggestions.
Funding This work was supported in part by grants to R.S.N., G.B. and T.O. from the Australian Research Council (DP1094212) and National Institutes of Health grant GM 48677 to GB, BMO and DY. K.G. acknowledges CSIR, Government of India, for a senior research fellowship. The work at Bangalore (IISc, India) is supported by grants from Department of Biotechnology, Government of India. R.S.N. acknowledges fellowship support from the National Health and Medical Research Council of Australia.
ABBREVIATIONS
- CID
Collision induced dissociation
- μ-GIIIA μ-GIIIB
μ-conotoxins GIIIA and GIIIB, respectively, from Conus geographus
- μ-KIIIA, μ-KIIIB
μ-conotoxin KIIIA and KIIIB, respectively, from C. kinoshitai
- μ-KIIIA[C1A,C9A]
μ-KIIIA with Cys1 and 9 replaced by Ala
- MTBE
methyl tert-butyl ether
- NaV1.2
the α-subunit of the voltage-gated sodium channel subtype 1.2
- μ-PIIIA
μ-conotoxin PIIIA from C. purpurascens
- μ-SIIIA
μ-conotoxin SIIIA from C. striatus
- μ-SmIIIA
μ-conotoxin SmIIIA from C. stercusmuscarum
- RMSD
root mean square deviation
Footnotes
‡ Chemical shift assignments and the family of structures for μ-KIIIA-P1 have been deposited in the BioMagResBank and Protein Data Bank with accession number 20048 and PDB ID 2LXG, respectively.
Supporting Information Ten figures and four tables. This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interest.
REFERENCES
- (1).Khoo KK, Norton RS. Role of disulfide bonds in peptide and protein conformation. In: Hughes AB, editor. Amino Acids, Peptides and Proteins in Organic Chemistry. Wiley-VCH, Weinheim; Germany: 2012. pp. 395–417. [Google Scholar]
- (2).Trivedi MV, Laurence JS, Siahaan TJ. The role of thiols and disulfides on protein stability. Curr Protein Pept Sci. 2009;10:614–625. doi: 10.2174/138920309789630534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Gilbert HF. Thiol/disulfide exchange equilibria and disulfide bond stability. Methods Enzymol. 1995;251:8–28. doi: 10.1016/0076-6879(95)51107-5. [DOI] [PubMed] [Google Scholar]
- (4).Holmgren A, Bjornstedt M. Thioredoxin and thioredoxin reductase. Methods Enzymol. 1995;252:199–208. doi: 10.1016/0076-6879(95)52023-6. [DOI] [PubMed] [Google Scholar]
- (5).Buczek O, Green BR, Bulaj G. Albumin is a redox-active crowding agent that promotes oxidative folding of cysteine-rich peptides. Biopolymers. 2007;88:8–19. doi: 10.1002/bip.20615. [DOI] [PubMed] [Google Scholar]
- (6).Armishaw CJ, Daly NL, Nevin ST, Adams DJ, Craik DJ, Alewood PF. Alpha-selenoconotoxins, a new class of potent α7 neuronal nicotinic receptor antagonists. J Biol Chem. 2006;281:14136–14143. doi: 10.1074/jbc.M512419200. [DOI] [PubMed] [Google Scholar]
- (7).Walewska A, Zhang MM, Skalicky JJ, Yoshikami D, Olivera BM, Bulaj G. Integrated oxidative folding of cysteine/selenocysteine containing peptides: improving chemical synthesis of conotoxins. Angew Chem Int Ed Engl. 2009;48:2221–2224. doi: 10.1002/anie.200806085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Paul M, Donk W. A. v. d. Chemical and enzymatic synthesis of lanthionines. Min. Rev. Org. Chem. 2005;2:23–37. [Google Scholar]
- (9).Knerr PJ, Tzekou A, Ricklin D, Qu H, Chen H, van der Donk WA, Lambris JD. Synthesis and activity of thioether-containing analogues of the complement inhibitor compstatin. ACS Chem Biol. 2011;6:753–760. doi: 10.1021/cb2000378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Muttenthaler M, Andersson A, de Araujo AD, Dekan Z, Lewis RJ, Alewood PF. Modulating oxytocin activity and plasma stability by disulfide bond engineering. J Med Chem. 2010;53:8585–8596. doi: 10.1021/jm100989w. [DOI] [PubMed] [Google Scholar]
- (11).Platt RJ, Han TS, Green BR, Smith MD, Skalicky J, Gruszczynski P, White HS, Olivera B, Bulaj G, Gajewiak J. Stapling mimics noncovalent interactions of γ-carboxyglutamates in conantokins, Peptidic antagonists of N-Methyl-D-aspartic acid receptors. J Biol Chem. 2012;287:20727–20736. doi: 10.1074/jbc.M112.350462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).MacRaild CA, Illesinghe J, van Lierop BJ, Townsend AL, Chebib M, Livett BG, Robinson AJ, Norton RS. Structure and activity of (2,8)-dicarba-(3,12)-cystino α-ImI, an α-conotoxin containing a nonreducible cystine analogue. J Med Chem. 2009;52:755–762. doi: 10.1021/jm8011504. [DOI] [PubMed] [Google Scholar]
- (13).Robinson AJ, van Lierop BJ, Garland RD, Teoh E, Elaridi J, Illesinghe JP, Jackson WR. Regioselective formation of interlocked dicarba bridges in naturally occurring cyclic peptide toxins using olefin metathesis. Chem Commun (Camb) 2009:4293–4295. doi: 10.1039/b909056h. [DOI] [PubMed] [Google Scholar]
- (14).Gray WR. Disulfide structures of highly bridged peptides: a new strategy for analysis. Protein Sci. 1993;2:1732–1748. doi: 10.1002/pro.5560021017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Gupta K, Kumar M, Balaram P. Disulfide bond assignments by mass spectrometry of native natural peptides: cysteine pairing in disulfide bonded conotoxins. Anal Chem. 2010;82:8313–8319. doi: 10.1021/ac101867e. [DOI] [PubMed] [Google Scholar]
- (16).Ye M, Khoo KK, Xu S, Zhou M, Boonyalai N, Perugini MA, Shao X, Chi C, Galea CA, Wang C, Norton RS. A helical conotoxin from Conus imperialis has a novel cysteine framework and defines a new superfamily. J Biol Chem. 2012;287:14973–14983. doi: 10.1074/jbc.M111.334615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Poppe L, Hui JO, Ligutti J, Murray JK, Schnier PD. PADLOC: a powerful tool to assign disulfide bond connectivities in peptides and proteins by NMR spectroscopy. Anal Chem. 2012;84:262–266. doi: 10.1021/ac203078x. [DOI] [PubMed] [Google Scholar]
- (18).Jordan JB, Poppe L, Haniu M, Arvedson T, Syed R, Li V, Kohno H, Kim H, Schnier PD, Harvey TS, Miranda LP, Cheetham J, Sasu BJ. Hepcidin revisited, disulfide connectivity, dynamics, and structure. J Biol Chem. 2009;284:24155–24167. doi: 10.1074/jbc.M109.017764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Lauth X, Babon JJ, Stannard JA, Singh S, Nizet V, Carlberg JM, Ostland VE, Pennington MW, Norton RS, Westerman ME. Bass hepcidin synthesis, solution structure, antimicrobial activities and synergism, and in vivo hepatic response to bacterial infections. J Biol Chem. 2005;280:9272–9282. doi: 10.1074/jbc.M411154200. [DOI] [PubMed] [Google Scholar]
- (20).Mobli M, King GF. NMR methods for determining disulfide-bond connectivities. Toxicon. 2010;56:849–854. doi: 10.1016/j.toxicon.2010.06.018. [DOI] [PubMed] [Google Scholar]
- (21).Walewska A, Skalicky JJ, Davis DR, Zhang MM, Lopez-Vera E, Watkins M, Han TS, Yoshikami D, Olivera BM, Bulaj G. NMR-based mapping of disulfide bridges in cysteine-rich peptides: application to the μ-conotoxin SxIIIA. J Am Chem Soc. 2008;130:14280–14286. doi: 10.1021/ja804303p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Khoo KK, Feng ZP, Smith BJ, Zhang MM, Yoshikami D, Olivera BM, Bulaj G, Norton RS. Structure of the analgesic μ-conotoxin KIIIA and effects on the structure and function of disulfide deletion. Biochemistry. 2009;48:1210–1219. doi: 10.1021/bi801998a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Hill JM, Alewood PF, Craik DJ. Three-dimensional solution structure of μ-conotoxin GIIIB, a specific blocker of skeletal muscle sodium channels. Biochemistry. 1996;35:8824–8835. doi: 10.1021/bi960073o. [DOI] [PubMed] [Google Scholar]
- (24).Keizer DW, West PJ, Lee EF, Yoshikami D, Olivera BM, Bulaj G, Norton RS. Structural basis for tetrodotoxin-resistant sodium channel binding by μ-conotoxin SmIIIA. J Biol Chem. 2003;278:46805–46813. doi: 10.1074/jbc.M309222200. [DOI] [PubMed] [Google Scholar]
- (25).Lancelin JM, Kohda D, Tate S, Yanagawa Y, Abe T, Satake M, Inagaki F. Tertiary structure of conotoxin GIIIA in aqueous solution. Biochemistry. 1991;30:6908–6916. doi: 10.1021/bi00242a014. [DOI] [PubMed] [Google Scholar]
- (26).Shon KJ, Olivera BM, Watkins M, Jacobsen RB, Gray WR, Floresca CZ, Cruz LJ, Hillyard DR, Brink A, Terlau H, Yoshikami D. μ-Conotoxin PIIIA, a new peptide for discriminating among tetrodotoxin-sensitive Na channel subtypes. J Neurosci. 1998;18:4473–4481. doi: 10.1523/JNEUROSCI.18-12-04473.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Yao S, Zhang MM, Yoshikami D, Azam L, Olivera BM, Bulaj G, Norton RS. Structure, dynamics, and selectivity of the sodium channel blocker μ-conotoxin SIIIA. Biochemistry. 2008;47:10940–10949. doi: 10.1021/bi801010u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Han TS, Zhang MM, Walewska A, Gruszczynski P, Robertson CR, Cheatham TE, 3rd, Yoshikami D, Olivera BM, Bulaj G. Structurally minimized μ-conotoxin analogues as sodium channel blockers: implications for designing conopeptide-based therapeutics. ChemMedChem. 2009;4:406–414. doi: 10.1002/cmdc.200800292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Stevens M, Peigneur S, Dyubankova N, Lescrinier E, Herdewijn P, Tytgat J. Design of bioactive peptides from naturally occurring μ-conotoxin structures. J Biol Chem. 2012;287:31382–31392. doi: 10.1074/jbc.M112.375733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Gupta K, Bhattacharyya M, Gowd KH, Balaram P. Rapid mass spectrometric determination of disulfide folds in peptides and proteins: A robust solution for a long standing problem in protein chemistry. J. Am. Soc. Mass. Spectrom. Submitted. 2012 [Google Scholar]
- (31).Bulaj G, West PJ, Garrett JE, Watkins M, Zhang MM, Norton RS, Smith BJ, Yoshikami D, Olivera BM. Novel conotoxins from Conus striatus and Conus kinoshitai selectively block TTX-resistant sodium channels. Biochemistry. 2005;44:7259–7265. doi: 10.1021/bi0473408. [DOI] [PubMed] [Google Scholar]
- (32).Biggs JS, Watkins M, Puillandre N, Ownby JP, Lopez-Vera E, Christensen S, Moreno KJ, Bernaldez J, Licea-Navarro A, Corneli PS, Olivera BM. Evolution of Conus peptide toxins: analysis of Conus californicus Reeve, 1844. Mol Phylogenet Evol. 2010;56:1–12. doi: 10.1016/j.ympev.2010.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Bartels C, Xia TH, Billeter M, Güntert P, Wüthrich K. The program XEASY for computer-supported NMR spectral-analysis of biological macromolecules. J Biomol NMR. 1995;6:1–10. doi: 10.1007/BF00417486. [DOI] [PubMed] [Google Scholar]
- (34).Schwieters CD, Kuszewski JJ, Tjandra N, Clore GM. The Xplor-NIH NMR molecular structure determination package. J Magn Reson. 2003;160:65–73. doi: 10.1016/s1090-7807(02)00014-9. [DOI] [PubMed] [Google Scholar]
- (35).Laskowski RA, Rullmannn JA, MacArthur MW, Kaptein R, Thornton JM. AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J Biomol NMR. 1996;8:477–486. doi: 10.1007/BF00228148. [DOI] [PubMed] [Google Scholar]
- (36).Koradi R, Billeter M, Wuthrich K. MOLMOL: a program for display and analysis of macromolecular structures. J Mol Graph. 1996;14:51–55. 29–32. doi: 10.1016/0263-7855(96)00009-4. [DOI] [PubMed] [Google Scholar]
- (37).Zhang MM, Green BR, Catlin P, Fiedler B, Azam L, Chadwick A, Terlau H, McArthur JR, French RJ, Gulyas J, Rivier JE, Smith BJ, Norton RS, Olivera BM, Yoshikami D, Bulaj G. Structure/function characterization of μ-conotoxin KIIIA, an analgesic, nearly irreversible blocker of mammalian neuronal sodium channels. J Biol Chem. 2007;282:30699–30706. doi: 10.1074/jbc.M704616200. [DOI] [PubMed] [Google Scholar]
- (38).West PJ, Bulaj G, Garrett JE, Olivera BM, Yoshikami D. μ-conotoxin SmIIIA, a potent inhibitor of tetrodotoxin-resistant sodium channels in amphibian sympathetic and sensory neurons. Biochemistry. 2002;41:15388–15393. doi: 10.1021/bi0265628. [DOI] [PubMed] [Google Scholar]
- (39).Wilson MJ, Yoshikami D, Azam L, Gajewiak J, Olivera BM, Bulaj G, Zhang MM. μ-Conotoxins that differentially block sodium channels NaV1.1 through 1.8 identify those responsible for action potentials in sciatic nerve. Proc Natl Acad Sci U S A. 2011;108:10302–10307. doi: 10.1073/pnas.1107027108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Norton RS. μ-conotoxins as leads in the development of new analgesics. Molecules. 2010;15:2825–2844. doi: 10.3390/molecules15042825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Corpuz GP, Jacobsen RB, Jimenez EC, Watkins M, Walker C, Colledge C, Garrett JE, McDougal O, Li W, Gray WR, Hillyard DR, Rivier J, McIntosh JM, Cruz LJ, Olivera BM. Definition of the M-conotoxin superfamily: characterization of novel peptides from molluscivorous Conus venoms. Biochemistry. 2005;44:8176–8186. doi: 10.1021/bi047541b. [DOI] [PubMed] [Google Scholar]
- (42).Jacob RB, McDougal OM. The M-superfamily of conotoxins: a review. Cell Mol Life Sci. 2010;67:17–27. doi: 10.1007/s00018-009-0125-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Yanagawa Y, Abe T, Satake M, Odani S, Suzuki J, Ishikawa K. A novel sodium channel inhibitor from Conus geographus: purification, structure, and pharmacological properties. Biochemistry. 1988;27:6256–6262. doi: 10.1021/bi00417a009. [DOI] [PubMed] [Google Scholar]
- (44).Cruz LJ, Kupryszewski G, LeCheminant GW, Gray WR, Olivera BM, Rivier J. μ-conotoxin GIIIA, a peptide ligand for muscle sodium channels: chemical synthesis, radiolabeling, and receptor characterization. Biochemistry. 1989;28:3437–3442. doi: 10.1021/bi00434a043. [DOI] [PubMed] [Google Scholar]
- (45).Hidaka Y, Sato K, Nakamura H, Kobayashi J, Ohizumi Y, Shimonishi Y. Disulfide pairings in geographutoxin I, a peptide neurotoxin from Conus geographus. FEBS Lett. 1990;264:29–32. doi: 10.1016/0014-5793(90)80756-9. [DOI] [PubMed] [Google Scholar]
- (46).Favreau P, Benoit E, Hocking HG, Carlier L, D DH, Leipold E, Markgraf R, Schlumberger S, Cordova MA, Gaertner H, Paolini-Bertrand M, Hartley O, Tytgat J, Heinemann SH, Bertrand D, Boelens R, Stocklin R, Molgo J. A novel μ-conopeptide, CnIIIC, exerts potent and preferential inhibition of NaV 1.2/1.4 channels and blocks neuronal nicotinic acetylcholine receptors. Br J Pharmacol. 2012;166:1654–1668. doi: 10.1111/j.1476-5381.2012.01837.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Tietze AA, Tietze D, Ohlenschlager O, Leipold E, Ullrich F, Kuhl T, Mischo A, Buntkowsky G, Gorlach M, Heinemann SH, Imhof D. Structurally diverse μ-conotoxin PIIIA isomers block sodium channel NaV1.4. Angew Chem Int Ed Engl. 2012;51:4058–4061. doi: 10.1002/anie.201107011. [DOI] [PubMed] [Google Scholar]
- (48).Bulaj G, Olivera BM. Folding of conotoxins: formation of the native disulfide bridges during chemical synthesis and biosynthesis of Conus peptides. Antioxid Redox Signal. 2008;10:141–155. doi: 10.1089/ars.2007.1856. [DOI] [PubMed] [Google Scholar]
- (49).Safavi-Hemami H, Gorasia DG, Steiner AM, Williamson NA, Karas JA, Gajewiak J, Olivera BM, Bulaj G, Purcell AW. Modulation of conotoxin structure and function is achieved through a multienzyme complex in the venom glands of cone snails. J Biol Chem. 2012;287:34288–34303. doi: 10.1074/jbc.M112.366781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Dutton JL, Bansal PS, Hogg RC, Adams DJ, Alewood PF, Craik DJ. A new level of conotoxin diversity, a non-native disulfide bond connectivity in α-conotoxin AuIB reduces structural definition but increases biological activity. J Biol Chem. 2002;277:48849–48857. doi: 10.1074/jbc.M208842200. [DOI] [PubMed] [Google Scholar]
- (51).Gehrmann J, Alewood PF, Craik DJ. Structure determination of the three disulfide bond isomers of α-conotoxin GI: a model for the role of disulfide bonds in structural stability. J Mol Biol. 1998;278:401–415. doi: 10.1006/jmbi.1998.1701. [DOI] [PubMed] [Google Scholar]
- (52).Khoo KK, Wilson MJ, Smith BJ, Zhang MM, Gulyas J, Yoshikami D, Rivier JE, Bulaj G, Norton RS. Lactam-stabilized helical analogues of the analgesic μ-conotoxin KIIIA. J Med Chem. 2011;54:7558–7566. doi: 10.1021/jm200839a. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.