Abstract
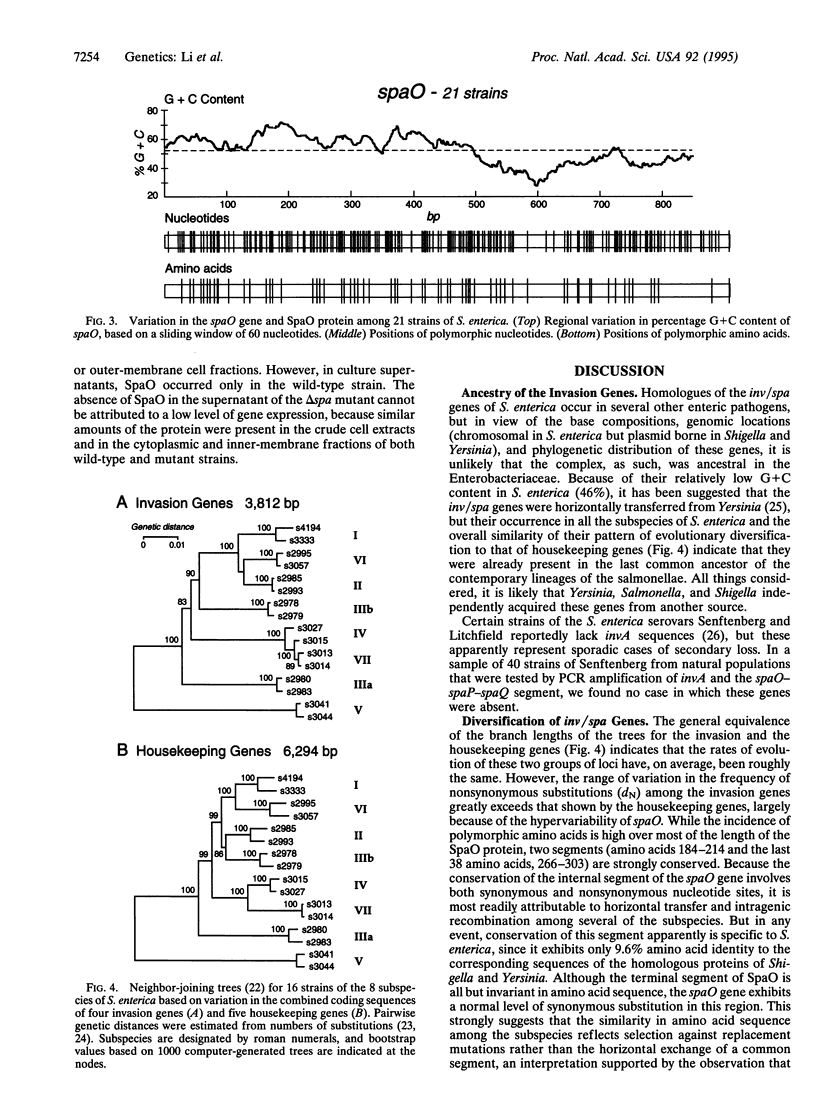
For 21 strains of Salmonella enterica, nucleotide sequences were obtained for three invasion genes, spaO, spaP, and spaQ, of the chromosomal inv/spa complex, the products of which form a protein export system required for entry of the bacteria into nonphagocytic host cells. These genes are present in all eight subspecies of the salmonellae, and homologues occur in a variety of other bacteria, including the enteric pathogens Shigella and Yersinia, in which they are plasmid borne. Evolutionary diversification of the invasion genes among the subspecies of S. enterica has been generally similar in pattern and average rate to that of housekeeping genes. However, the range of variation in evolutionary rate among the invasion genes is unusually large, and there is a relationship between the evolutionary rate and cellular location of the invasion proteins, possibly reflecting diversifying selection on exported proteins in adaptation to variable host factors in extracellular environments. The SpaO protein, which is hypervariable in S. enterica and exhibits only 24% sequence identity with its homologues in Shigella and Yersinia, is secreted. In contrast, the membrane-associated proteins SpaP, SpaQ, and InvA are weakly polymorphic and have > 60% sequence identity with the corresponding proteins of other enteric bacteria. Acquisition of the inv/spa genes may have been a key event in the evolution of the salmonellae as pathogens, following which the invention of flagellar phase shifting facilitated niche expansion to include warm-blooded vertebrates.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Altmeyer R. M., McNern J. K., Bossio J. C., Rosenshine I., Finlay B. B., Galán J. E. Cloning and molecular characterization of a gene involved in Salmonella adherence and invasion of cultured epithelial cells. Mol Microbiol. 1993 Jan;7(1):89–98. doi: 10.1111/j.1365-2958.1993.tb01100.x. [DOI] [PubMed] [Google Scholar]
- Bergman T., Erickson K., Galyov E., Persson C., Wolf-Watz H. The lcrB (yscN/U) gene cluster of Yersinia pseudotuberculosis is involved in Yop secretion and shows high homology to the spa gene clusters of Shigella flexneri and Salmonella typhimurium. J Bacteriol. 1994 May;176(9):2619–2626. doi: 10.1128/jb.176.9.2619-2626.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd E. F., Nelson K., Wang F. S., Whittam T. S., Selander R. K. Molecular genetic basis of allelic polymorphism in malate dehydrogenase (mdh) in natural populations of Escherichia coli and Salmonella enterica. Proc Natl Acad Sci U S A. 1994 Feb 15;91(4):1280–1284. doi: 10.1073/pnas.91.4.1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collazo C. M., Zierler M. K., Galán J. E. Functional analysis of the Salmonella typhimurium invasion genes invl and invJ and identification of a target of the protein secretion apparatus encoded in the inv locus. Mol Microbiol. 1995 Jan;15(1):25–38. doi: 10.1111/j.1365-2958.1995.tb02218.x. [DOI] [PubMed] [Google Scholar]
- Galán J. E., Curtiss R., 3rd Cloning and molecular characterization of genes whose products allow Salmonella typhimurium to penetrate tissue culture cells. Proc Natl Acad Sci U S A. 1989 Aug;86(16):6383–6387. doi: 10.1073/pnas.86.16.6383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galán J. E., Ginocchio C., Costeas P. Molecular and functional characterization of the Salmonella invasion gene invA: homology of InvA to members of a new protein family. J Bacteriol. 1992 Jul;174(13):4338–4349. doi: 10.1128/jb.174.13.4338-4349.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galán J. E., Pace J., Hayman M. J. Involvement of the epidermal growth factor receptor in the invasion of cultured mammalian cells by Salmonella typhimurium. Nature. 1992 Jun 18;357(6379):588–589. doi: 10.1038/357588a0. [DOI] [PubMed] [Google Scholar]
- Ginocchio C. C., Galán J. E. Functional conservation among members of the Salmonella typhimurium InvA family of proteins. Infect Immun. 1995 Feb;63(2):729–732. doi: 10.1128/iai.63.2.729-732.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginocchio C. C., Olmsted S. B., Wells C. L., Galán J. E. Contact with epithelial cells induces the formation of surface appendages on Salmonella typhimurium. Cell. 1994 Feb 25;76(4):717–724. doi: 10.1016/0092-8674(94)90510-x. [DOI] [PubMed] [Google Scholar]
- Groisman E. A., Ochman H. Cognate gene clusters govern invasion of host epithelial cells by Salmonella typhimurium and Shigella flexneri. EMBO J. 1993 Oct;12(10):3779–3787. doi: 10.1002/j.1460-2075.1993.tb06056.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaniga K., Bossio J. C., Galán J. E. The Salmonella typhimurium invasion genes invF and invG encode homologues of the AraC and PulD family of proteins. Mol Microbiol. 1994 Aug;13(4):555–568. doi: 10.1111/j.1365-2958.1994.tb00450.x. [DOI] [PubMed] [Google Scholar]
- Karaolis D. K., Lan R., Reeves P. R. Sequence variation in Shigella sonnei (Sonnei), a pathogenic clone of Escherichia coli, over four continents and 41 years. J Clin Microbiol. 1994 Mar;32(3):796–802. doi: 10.1128/jcm.32.3.796-802.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyte J., Doolittle R. F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982 May 5;157(1):105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- Le Minor L., Popoff M. Y., Laurent B., Hermant D. Individualisation d'une septième sous-espèce de Salmonella: S. choleraesuis subsp. indica subsp. nov. Ann Inst Pasteur Microbiol. 1986 Sep-Oct;137B(2):211–217. [PubMed] [Google Scholar]
- Le Minor L., Véron M., Popoff M. Taxonomie des Salmonella. Ann Microbiol (Paris) 1982 Sep-Oct;133(2):223–243. [PubMed] [Google Scholar]
- Li J., Nelson K., McWhorter A. C., Whittam T. S., Selander R. K. Recombinational basis of serovar diversity in Salmonella enterica. Proc Natl Acad Sci U S A. 1994 Mar 29;91(7):2552–2556. doi: 10.1073/pnas.91.7.2552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills D. M., Bajaj V., Lee C. A. A 40 kb chromosomal fragment encoding Salmonella typhimurium invasion genes is absent from the corresponding region of the Escherichia coli K-12 chromosome. Mol Microbiol. 1995 Feb;15(4):749–759. doi: 10.1111/j.1365-2958.1995.tb02382.x. [DOI] [PubMed] [Google Scholar]
- Mills J. A., Buysse J. M., Oaks E. V. Shigella flexneri invasion plasmid antigens B and C: epitope location and characterization with monoclonal antibodies. Infect Immun. 1988 Nov;56(11):2933–2941. doi: 10.1128/iai.56.11.2933-2941.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ménard R., Sansonetti P. J., Parsot C. Nonpolar mutagenesis of the ipa genes defines IpaB, IpaC, and IpaD as effectors of Shigella flexneri entry into epithelial cells. J Bacteriol. 1993 Sep;175(18):5899–5906. doi: 10.1128/jb.175.18.5899-5906.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M., Gojobori T. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol Biol Evol. 1986 Sep;3(5):418–426. doi: 10.1093/oxfordjournals.molbev.a040410. [DOI] [PubMed] [Google Scholar]
- Nelson K., Selander R. K. Analysis of genetic variation by polymerase chain reaction-based nucleotide sequencing. Methods Enzymol. 1994;235:174–183. doi: 10.1016/0076-6879(94)35139-2. [DOI] [PubMed] [Google Scholar]
- Nelson K., Selander R. K. Evolutionary genetics of the proline permease gene (putP) and the control region of the proline utilization operon in populations of Salmonella and Escherichia coli. J Bacteriol. 1992 Nov;174(21):6886–6895. doi: 10.1128/jb.174.21.6886-6895.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson K., Selander R. K. Intergeneric transfer and recombination of the 6-phosphogluconate dehydrogenase gene (gnd) in enteric bacteria. Proc Natl Acad Sci U S A. 1994 Oct 11;91(21):10227–10231. doi: 10.1073/pnas.91.21.10227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson K., Whittam T. S., Selander R. K. Nucleotide polymorphism and evolution in the glyceraldehyde-3-phosphate dehydrogenase gene (gapA) in natural populations of Salmonella and Escherichia coli. Proc Natl Acad Sci U S A. 1991 Aug 1;88(15):6667–6671. doi: 10.1073/pnas.88.15.6667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochman H., Whittam T. S., Caugant D. A., Selander R. K. Enzyme polymorphism and genetic population structure in Escherichia coli and Shigella. J Gen Microbiol. 1983 Sep;129(9):2715–2726. doi: 10.1099/00221287-129-9-2715. [DOI] [PubMed] [Google Scholar]
- Ochman H., Wilson A. C. Evolution in bacteria: evidence for a universal substitution rate in cellular genomes. J Mol Evol. 1987;26(1-2):74–86. doi: 10.1007/BF02111283. [DOI] [PubMed] [Google Scholar]
- Parra-Lopez C., Lin R., Aspedon A., Groisman E. A. A Salmonella protein that is required for resistance to antimicrobial peptides and transport of potassium. EMBO J. 1994 Sep 1;13(17):3964–3972. doi: 10.1002/j.1460-2075.1994.tb06712.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahn K., De Grandis S. A., Clarke R. C., McEwen S. A., Galán J. E., Ginocchio C., Curtiss R., 3rd, Gyles C. L. Amplification of an invA gene sequence of Salmonella typhimurium by polymerase chain reaction as a specific method of detection of Salmonella. Mol Cell Probes. 1992 Aug;6(4):271–279. doi: 10.1016/0890-8508(92)90002-f. [DOI] [PubMed] [Google Scholar]
- Reeves P. Evolution of Salmonella O antigen variation by interspecific gene transfer on a large scale. Trends Genet. 1993 Jan;9(1):17–22. doi: 10.1016/0168-9525(93)90067-R. [DOI] [PubMed] [Google Scholar]
- Saitou N., Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987 Jul;4(4):406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Sasakawa C., Komatsu K., Tobe T., Suzuki T., Yoshikawa M. Eight genes in region 5 that form an operon are essential for invasion of epithelial cells by Shigella flexneri 2a. J Bacteriol. 1993 Apr;175(8):2334–2346. doi: 10.1128/jb.175.8.2334-2346.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevenson G., Neal B., Liu D., Hobbs M., Packer N. H., Batley M., Redmond J. W., Lindquist L., Reeves P. Structure of the O antigen of Escherichia coli K-12 and the sequence of its rfb gene cluster. J Bacteriol. 1994 Jul;176(13):4144–4156. doi: 10.1128/jb.176.13.4144-4156.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Gijsegem F., Genin S., Boucher C. Conservation of secretion pathways for pathogenicity determinants of plant and animal bacteria. Trends Microbiol. 1993 Aug;1(5):175–180. doi: 10.1016/0966-842x(93)90087-8. [DOI] [PubMed] [Google Scholar]
- Van Gijsegem F., Gough C., Zischek C., Niqueux E., Arlat M., Genin S., Barberis P., German S., Castello P., Boucher C. The hrp gene locus of Pseudomonas solanacearum, which controls the production of a type III secretion system, encodes eight proteins related to components of the bacterial flagellar biogenesis complex. Mol Microbiol. 1995 Mar;15(6):1095–1114. doi: 10.1111/j.1365-2958.1995.tb02284.x. [DOI] [PubMed] [Google Scholar]
- Venkatesan M. M., Buysse J. M., Kopecko D. J. Characterization of invasion plasmid antigen genes (ipaBCD) from Shigella flexneri. Proc Natl Acad Sci U S A. 1988 Dec;85(23):9317–9321. doi: 10.1073/pnas.85.23.9317. [DOI] [PMC free article] [PubMed] [Google Scholar]




