Abstract
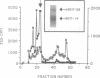
The peptide somatostatin exists as two different molecular species. In addition to the most common form, somatostatin-14, there is also a fourteen amino acid N-terminally extended form of the tetradecapeptide, somatostatin-28. Both peptides are synthesized as larger precursors containing paired basic and monobasic amino acids at their processing sites, which upon cleavage generate either somatostatin-14 or -28, respectively. In some species of fish two distinct, but homologous, precursors (prosomatostatin-I and -II) give rise to somatostatin-14 and -28, respectively. Whereas anglerfish prosomatostatin-II was previously shown to release exclusively somatostatin-28, the yeast Saccharomyces cerevisiae proteolytically matures the homologous prosomatostatin-I precursor to somatostatin-28 and -14 as well as to a lysine-extended form of somatostatin-14. The Kex2 endoprotease appears to be essential for the formation of lysine somatostatin-14 and is involved either directly or indirectly in the release of mature somatostatin-14. The isolation of yeast mutants defective in somatostatin-28 expression (sex mutant) allowed the cloning of a non-essential gene, which encodes an aspartyl protease, whose disruption severely affects the cleavage of mature somatostatin-28 from both somatostatin precursors. We conclude that two distinct endoproteases, which demonstrate some cross specificity in vivo, are involved in the proteolytic maturation of prosomatostatin at mono- and dibasic processing sites in yeast.
Full text
PDF
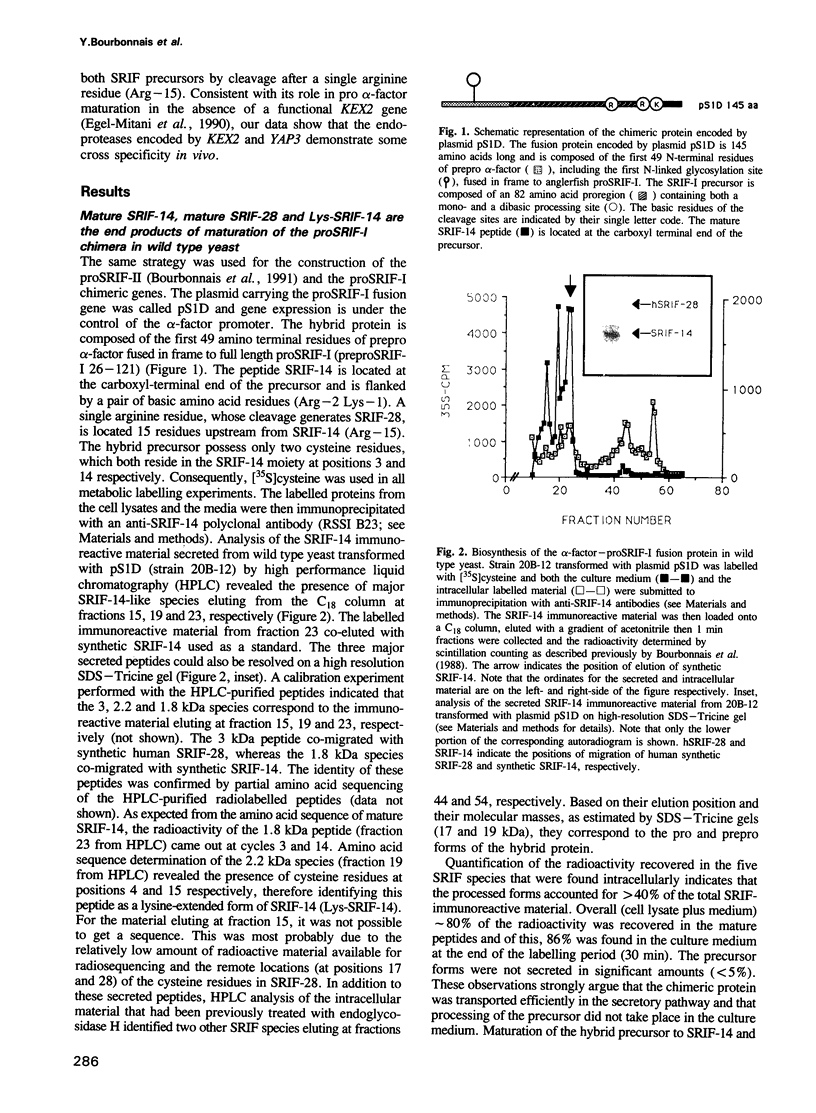


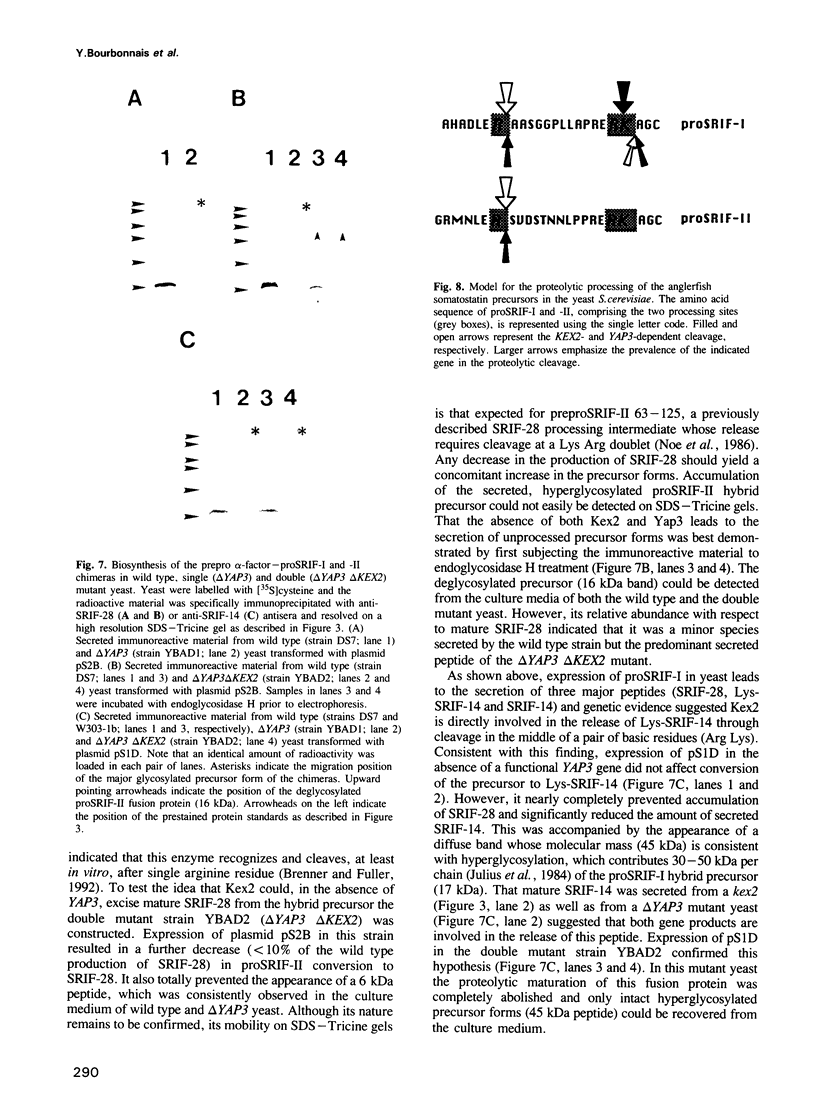




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Beinfeld M. C., Bourdais J., Kuks P., Morel A., Cohen P. Characterization of an endoprotease from rat small intestinal mucosal secretory granules which generates somatostatin-28 from prosomatostatin by cleavage after a single arginine residue. J Biol Chem. 1989 Mar 15;264(8):4460–4465. [PubMed] [Google Scholar]
- Bourbonnais Y., Bolin D., Shields D. Secretion of somatostatin by Saccharomyces cerevisiae. Correct proteolytic processing of pro-alpha-factor-somatostatin hybrids requires the products of the KEX2 and STE13 genes. J Biol Chem. 1988 Oct 25;263(30):15342–15347. [PubMed] [Google Scholar]
- Bourbonnais Y., Danoff A., Thomas D. Y., Shields D. Heterologous expression of peptide hormone precursors in the yeast Saccharomyces cerevisiae. Evidence for a novel prohormone endoprotease with specificity for monobasic amino acids. J Biol Chem. 1991 Jul 15;266(20):13203–13209. [PubMed] [Google Scholar]
- Brenner C., Fuller R. S. Structural and enzymatic characterization of a purified prohormone-processing enzyme: secreted, soluble Kex2 protease. Proc Natl Acad Sci U S A. 1992 Feb 1;89(3):922–926. doi: 10.1073/pnas.89.3.922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danoff A., Cutler D. F., Shields D. Heterologous expression of preprosomatostatin. Intracellular degradation of prosomatostatin-II. J Biol Chem. 1991 May 25;266(15):10004–10010. [PubMed] [Google Scholar]
- Devi L. Consensus sequence for processing of peptide precursors at monobasic sites. FEBS Lett. 1991 Mar 25;280(2):189–194. doi: 10.1016/0014-5793(91)80290-j. [DOI] [PubMed] [Google Scholar]
- Dignard D., Whiteway M., Germain D., Tessier D., Thomas D. Y. Expression in yeast of a cDNA copy of the K2 killer toxin gene. Mol Gen Genet. 1991 May;227(1):127–136. doi: 10.1007/BF00260717. [DOI] [PubMed] [Google Scholar]
- Dmochowska A., Dignard D., Henning D., Thomas D. Y., Bussey H. Yeast KEX1 gene encodes a putative protease with a carboxypeptidase B-like function involved in killer toxin and alpha-factor precursor processing. Cell. 1987 Aug 14;50(4):573–584. doi: 10.1016/0092-8674(87)90030-4. [DOI] [PubMed] [Google Scholar]
- Douglass J., Civelli O., Herbert E. Polyprotein gene expression: generation of diversity of neuroendocrine peptides. Annu Rev Biochem. 1984;53:665–715. doi: 10.1146/annurev.bi.53.070184.003313. [DOI] [PubMed] [Google Scholar]
- Egel-Mitani M., Flygenring H. P., Hansen M. T. A novel aspartyl protease allowing KEX2-independent MF alpha propheromone processing in yeast. Yeast. 1990 Mar-Apr;6(2):127–137. doi: 10.1002/yea.320060206. [DOI] [PubMed] [Google Scholar]
- Fricker L. D. Carboxypeptidase E. Annu Rev Physiol. 1988;50:309–321. doi: 10.1146/annurev.ph.50.030188.001521. [DOI] [PubMed] [Google Scholar]
- Germain D., Zollinger L., Racine C., Gossard F., Dignard D., Thomas D. Y., Crine P., Boileau G. The yeast KEX-2-processing endoprotease is active in the Golgi apparatus of transfected NIH 3T3 fibroblasts. Mol Endocrinol. 1990 Oct;4(10):1572–1579. doi: 10.1210/mend-4-10-1572. [DOI] [PubMed] [Google Scholar]
- Gluschankof P., Gomez S., Morel A., Cohen P. Enzymes that process somatostatin precursors. A novel endoprotease that cleaves before the arginine-lysine doublet is involved in somatostatin-28 convertase activity of rat brain cortex. J Biol Chem. 1987 Jul 15;262(20):9615–9620. [PubMed] [Google Scholar]
- Gomez S., Gluschankof P., Lepage A., Cohen P. Relationship between endo- and exopeptidases in a processing enzyme system: activation of an endoprotease by the aminopeptidase B-like activity in somatostatin-28 convertase. Proc Natl Acad Sci U S A. 1988 Aug;85(15):5468–5472. doi: 10.1073/pnas.85.15.5468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green R., Schaber M. D., Shields D., Kramer R. Secretion of somatostatin by Saccharomyces cerevisiae. Correct processing of an alpha-factor-somatostatin hybrid. J Biol Chem. 1986 Jun 5;261(16):7558–7565. [PubMed] [Google Scholar]
- Ito H., Fukuda Y., Murata K., Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983 Jan;153(1):163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones J. S., Prakash L. Yeast Saccharomyces cerevisiae selectable markers in pUC18 polylinkers. Yeast. 1990 Sep-Oct;6(5):363–366. doi: 10.1002/yea.320060502. [DOI] [PubMed] [Google Scholar]
- Julius D., Brake A., Blair L., Kunisawa R., Thorner J. Isolation of the putative structural gene for the lysine-arginine-cleaving endopeptidase required for processing of yeast prepro-alpha-factor. Cell. 1984 Jul;37(3):1075–1089. doi: 10.1016/0092-8674(84)90442-2. [DOI] [PubMed] [Google Scholar]
- Kunkel T. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc Natl Acad Sci U S A. 1985 Jan;82(2):488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackin R. B., Noe B. D. Direct evidence for two distinct prosomatostatin converting enzymes. Detection using a rapid, sensitive, and specific assay for propeptide converting enzymes. J Biol Chem. 1987 May 15;262(14):6453–6456. [PubMed] [Google Scholar]
- Mackin R. B., Noe B. D., Spiess J. Identification of a somatostatin-14-generating propeptide converting enzyme as a member of the kex2/furin/PC family. Endocrinology. 1991 Oct;129(4):2263–2265. doi: 10.1210/endo-129-4-2263. [DOI] [PubMed] [Google Scholar]
- Mackin R. B., Noe B. D., Spiess J. The anglerfish somatostatin-28-generating propeptide converting enzyme is an aspartyl protease. Endocrinology. 1991 Oct;129(4):1951–1957. doi: 10.1210/endo-129-4-1951. [DOI] [PubMed] [Google Scholar]
- McDonald J. K., Greiner F., Bauer G. E., Elde R. P., Noe B. D. Separate cell types that express two different forms of somatostatin in anglerfish islets can be immunohistochemically differentiated. J Histochem Cytochem. 1987 Feb;35(2):155–162. doi: 10.1177/35.2.2878951. [DOI] [PubMed] [Google Scholar]
- Nakayama K., Kim W. S., Torii S., Hosaka M., Nakagawa T., Ikemizu J., Baba T., Murakami K. Identification of the fourth member of the mammalian endoprotease family homologous to the yeast Kex2 protease. Its testis-specific expression. J Biol Chem. 1992 Mar 25;267(9):5897–5900. [PubMed] [Google Scholar]
- Noe B. D., Andrews P. C., Dixon J. E., Spiess J. Cotranslational and posttranslational proteolytic processing of preprosomatostatin-I in intact islet tissue. J Cell Biol. 1986 Oct;103(4):1205–1211. doi: 10.1083/jcb.103.4.1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noe B. D., Spiess J. Evidence fore biosynthesis and differential post-translational proteolytic processing of different (pre)prosomatostatins in pancreatic islets. J Biol Chem. 1983 Jan 25;258(2):1121–1128. [PubMed] [Google Scholar]
- Orr-Weaver T. L., Szostak J. W., Rothstein R. J. Yeast transformation: a model system for the study of recombination. Proc Natl Acad Sci U S A. 1981 Oct;78(10):6354–6358. doi: 10.1073/pnas.78.10.6354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel Y. C., Wheatley T., Ning C. Multiple forms of immunoreactive somatostatin: comparison of distribution in neural and nonneural tissues and portal plasma of the rat. Endocrinology. 1981 Dec;109(6):1943–1949. doi: 10.1210/endo-109-6-1943. [DOI] [PubMed] [Google Scholar]
- Redding K., Holcomb C., Fuller R. S. Immunolocalization of Kex2 protease identifies a putative late Golgi compartment in the yeast Saccharomyces cerevisiae. J Cell Biol. 1991 May;113(3):527–538. doi: 10.1083/jcb.113.3.527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose M. D., Broach J. R. Cloning genes by complementation in yeast. Methods Enzymol. 1991;194:195–230. doi: 10.1016/0076-6879(91)94017-7. [DOI] [PubMed] [Google Scholar]
- Rothstein R. J. One-step gene disruption in yeast. Methods Enzymol. 1983;101:202–211. doi: 10.1016/0076-6879(83)01015-0. [DOI] [PubMed] [Google Scholar]
- Schwartz T. W. The processing of peptide precursors. 'Proline-directed arginyl cleavage' and other monobasic processing mechanisms. FEBS Lett. 1986 May 5;200(1):1–10. doi: 10.1016/0014-5793(86)80500-2. [DOI] [PubMed] [Google Scholar]
- Seidah N. G., Gaspar L., Mion P., Marcinkiewicz M., Mbikay M., Chrétien M. cDNA sequence of two distinct pituitary proteins homologous to Kex2 and furin gene products: tissue-specific mRNAs encoding candidates for pro-hormone processing proteinases. DNA Cell Biol. 1990 Jul-Aug;9(6):415–424. doi: 10.1089/dna.1990.9.415. [DOI] [PubMed] [Google Scholar]
- Smeekens S. P., Avruch A. S., LaMendola J., Chan S. J., Steiner D. F. Identification of a cDNA encoding a second putative prohormone convertase related to PC2 in AtT20 cells and islets of Langerhans. Proc Natl Acad Sci U S A. 1991 Jan 15;88(2):340–344. doi: 10.1073/pnas.88.2.340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smeekens S. P., Steiner D. F. Identification of a human insulinoma cDNA encoding a novel mammalian protein structurally related to the yeast dibasic processing protease Kex2. J Biol Chem. 1990 Feb 25;265(6):2997–3000. [PubMed] [Google Scholar]
- Thomas G., Thorne B. A., Thomas L., Allen R. G., Hruby D. E., Fuller R., Thorner J. Yeast KEX2 endopeptidase correctly cleaves a neuroendocrine prohormone in mammalian cells. Science. 1988 Jul 8;241(4862):226–230. doi: 10.1126/science.3291117. [DOI] [PubMed] [Google Scholar]
- Whiteway M., Hougan L., Thomas D. Y. Expression of MF alpha 1 in MATa cells supersensitive to alpha-factor leads to self-arrest. Mol Gen Genet. 1988 Sep;214(1):85–88. doi: 10.1007/BF00340184. [DOI] [PubMed] [Google Scholar]
- Zhu Y. S., Zhang X. Y., Cartwright C. P., Tipper D. J. Kex2-dependent processing of yeast K1 killer preprotoxin includes cleavage at ProArg-44. Mol Microbiol. 1992 Feb;6(4):511–520. doi: 10.1111/j.1365-2958.1992.tb01496.x. [DOI] [PubMed] [Google Scholar]
- Zollinger L., Racine C., Crine P., Boileau G., Germain D., Thomas D. Y., Gossard F. Intracellular proteolytic processing of proopiomelanocortin in heterologous COS-1 cells by the yeast KEX2 endoprotease. Biochem Cell Biol. 1990 Mar;68(3):635–640. doi: 10.1139/o90-090. [DOI] [PubMed] [Google Scholar]