Abstract
Cellular ribonucleic acid (RNA) plays a crucial role in the initial conversion of cellular prion protein PrPC to infectious PrPSc or scrapie. The nature of this RNA remains elusive. Previously, RNA aptamers against PrPC have been isolated and found to form G-quadruplexes (G4s). PrPC binding to G4 RNAs destabilizes its structure and is thought to trigger its conversion to PrPSc. Here it is shown that PrP messenger RNA (mRNA) itself contains several G4 motifs, located in the octarepeat region. Investigation of the RNA structure in one of these repeats by circular dichroism, nuclear magnetic resonance and ultraviolet melting studies shows evidence of G4 formation. In vitro translation of full-length PrP mRNA, naturally harboring five consecutive G4 motifs, was specifically affected by G4-binding ligands, lending support to G4 formation in PrP mRNA. A possible role of PrP binding to its own mRNA and the role of anti-prion drugs, many of which are G4-binding ligands, in prion disease are discussed.
INTRODUCTION
Prion diseases such as Creutzfeldt–Jakob disease (CJD) and scrapie are characterized by the deposition of aggregates of the misfolded form (PrPSc) of the cellular prion protein (PrPC) in the central nervous system (1). Once formed, the misfolded PrPSc conformation can be transmitted to native PrPC molecules via a poorly understood process. The resulting chain reaction leads to accumulation of largely protease resistant aggregates, which can be infectious to other mammals via oral transmission (2).
How the initial misfolding event takes place is still a mystery. It is assumed that next to PrP itself other factors are required to induce the fatal conformational change. Among the factors investigated are polysulphates, polyanions, lipids and nucleic acids (3,4). Currently, ribonucleic acid (RNA) is a major suspect as a co-factor (5). In protein cyclic misfolding assays (PCMAs) using recombinant PrP, RNA isolated from hamster or mouse brain cells was found to strongly enhance conversion to PrPSc in contrast to RNAs isolated from invertebrate species (6). Using bacterially expressed PrP, it has been shown in PCMAs that PrP conversion can be triggered by a combination of the synthetic phospholipid POPG (1-palmitoyl-2-oleoylphosphatidylglycerol) and RNA from brain cells (4) or synthetic polyA RNA (7). Synthetic polyG RNA was also reported to stimulate conversion but like polyA RNA it is less efficient than brain RNA (8). Deleault et al. (9) proposed that phosphatidylethanolamine is sufficient for conversion, however, this could not be confirmed by others (8). Up till now the identity of the natural RNA has not been elucidated and even about its size there exists no consensus; according to Kellings et al. (10) it is smaller than 250 nucleotides (nts), whereas Deleault et al. (6) suggest a size of more than 300 nts.
Since 1997 several RNA aptamers have been selected that bind with high affinity to PrPc (11). Many of these aptamers can form a G-quadruplex [G4 (12,13)] and although capable of preventing the conversion to PrPSc, binding of PrPC to G4 RNA may actually lower the free energy barrier between PrPC and PrPSc and thereby trigger conversion (14). This finding raises the possibility that the enigmatic RNA co-factor also contains G4 motifs. Another possibility that has not been investigated is the role of PrP messenger RNA (mRNA) itself in this process.
MATERIALS AND METHODS
Circular dichroism, nuclear magnetic resonance and ultraviolet melting studies
Circular dichroism (CD) spectra were recorded on a Jasco J-810 spectropolarimeter, equipped with a Peltier temperature controller, using 10-mm path length quartz cuvettes and a scanning speed of 50 nm/min. Concentration of RNAs was 1–2 μM. The average of eight CD scans was calculated and the spectrum of buffer alone was substracted. The ellipticity at 320 nm was set at zero. Molar ellipticity was calculated from θobs using the formula: [θ] = 100 × θobs/(C × l), where C is the concentration in mol/l and l is the cell path length in cm.
1H nuclear magnetic resonance (NMR) experiments were recorded on a Bruker 600-MHz spectrometer equipped with a cryoprobe using Watergate suppression. Samples were dissolved in 10-mM Na2HPO4/NaH2PO4 buffer (pH 6.7) containing 10% D2O.
Thermal difference spectra (15) were recorded in the 220–350-nm range, with a scan speed of 50 nm/min and a data interval of 0.1 nm. Spectra recorded at 20°C were substracted from those recorded at 90°C and normalized to the highest absorbance. Absorbance values at 260 nm were ∼0.6 OD.
Ultraviolet (UV) melting experiments were performed on a Varian Cary300 spectrophotometer in 10-mm quartz cuvettes. Absorption data were collected at 1°C intervals between 20°C and 90°C during repeated heating and cooling cycles at a rate of 0.5°C/min. Samples were kept at 90°C for 2 min. Melting temperatures (Tm) were calculated from two heating and cooling runs according to van der Werf et al. (16).
Preparation of RNA
High-performance liquid chromatography-purified RNA oligonucleotides were purchased from Sigma-Aldrich (0.2-mmole scale). The various prion mRNAs were synthesized on polymerase chain reaction (PCR) generated deoxyribonucleic acid (DNA) templates using the T7 RiboMax Large Scale RNA production system (Promega). A DNA template encoding the full-length prion open reading frame [ORF (Prp1-253)] was generated by PCR using primers PRI1 and PRI2 and prion cDNA [17; a gift from A. Bossers, Wageningen University, The Netherlands). PRI1: 5′GTAATACGACTCACTATAGGATTCCATATTATGGCGAACCTTGGCTGCTGG, (T7 promoter sequence in bold, start codon underlined). PRI2: 5′T40CCTCTAGACTCATCCCACTATCAGGAAG (stop codon underlined). A truncated mRNA (Prp89-253) was synthesized using primers PRI7 (GTAATACGACTCACTATAGGATTCCTTATAATGGGTCAAGGAGGTGGCACCCACAGTCAGT) and PRI2. RLuc, RLuc2, Pri-RLuc and Mut-RLuc plasmid DNAs were constructed by inserting pairs of complementary DNA oligonucleotides, containing also a T7 promoter sequence, directly upstream of the Renilla luciferase start codon of plasmid pMRL (18). After linearization with XhoI, transcription was carried out as described above. 5′ untranslated region (UTR) sequence of RLuc: GGCUAGUUAACUAUAUAACAUCGCGGUACAAACGGCACCAUG; 5′UTR sequence of RLuc2: GGCUAGUUAAGAUAUAACAUUGUGGUGUGGAAUCCAAACCAUG; 5′UTR of Pri-RLuc: GGCUAGUUAACUAUAUAACAUCAGGGCGGUGGUGGCUGGGGGCAGCCUCUUGGUGGUGGCUGGGGGCAGCCUAAUCCAAACCAUG, an upstream AUG codon was removed by substituting A by U (underlined). 5′UTR sequence of Mut-RLuc: GGCUAGUUAACUAUAUAACAUCAGGGCAGUGGUAGCUGAGGGCAGCCUCUUGGUAGUGGCUGAGGGCAGCCUAAUCCAAACCAUG, A to G mutations are underlined.
In vitro translation
Translation mixtures (5 μl) contained 2.5 nM of mRNA, 2.5 μl of nuclease-treated rabbit reticulocyte lysate (RRL; Promega), 0.5 μl of 1-mM amino acids mix without methionine, 0.125 μl of 35S-methionine [>1000 Ci (37.0TBq)/mmol, EasyTag, Perkin Elmer], and indicated amounts of PhenDC3 or KCl, and were incubated at 30°C for 1 h. Reactions were processed as described previously (19). Firefly luciferase (FLuc) control mRNA (Promega) was used as a control in some assays. For non-radioactive assays amino acid mixes without leucine and cysteine were added. After incubation, the reaction was stopped by addition of 45 μl of Passive Lysis Buffer (Promega). Twenty-five microliter was transferred to a 96-well microplate and luciferase activity was measured after addition of 2.5 μl of diluted substrate (Stop&Glo reagent, Promega) in a GloMax multi luminometer.
RESULTS
The mature PrP consists of 208 amino acids (aa) that make up a loosely structured N-terminal domain (aa 23-123) containing two RNA-binding motifs, and a C-terminal domain (aa 124-230) that is involved in prion formation (Figure 1A). Although the N-terminal domain is dispensable for prion propagation, it has been suggested to affect folding and stability of the C-terminal domain. The RNA-binding motifs are flanking the so-called octarepeat (OR) domain, which in most humans is composed of five repeats of the octapeptide sequence P(H/Q)GGGWGQ (Figure 1A). Interestingly, the underlying mRNA sequence harbors five putative G4 motifs (Figure 1B). The highly conserved nature of the GGGWG amino acid motif and thus also the GGNGGNGGNUGGGGN RNA sequence in all eutherian mammals (20) indicates the possibility of G4 formation in all these species. The G4 motifs are present within a region for which previously several RNA structures have been proposed (21–24). None of these structures are currently supported by experimental data or phylogenetic comparisons. As this region is translated by ribosomes, which melt RNA structures, the structure will not be static and will likely assume different conformations, potentially including G4s. To investigate this possibility, four RNA constructs corresponding to different sections of one OR were designed (Figure 1C).
Figure 1.
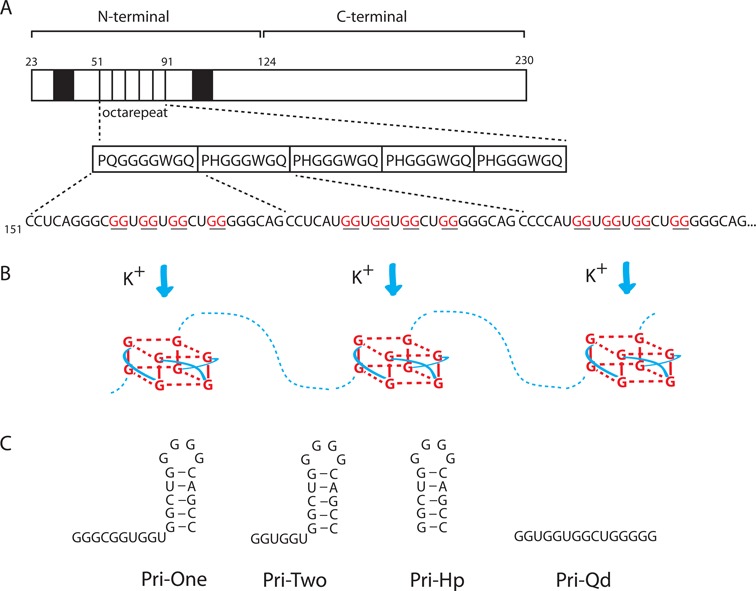
Putative G-quadruplex structures in human PrP mRNA. (A) Schematic representation of the mature PrP 23-230. Black boxes represent RNA-binding motifs. The peptide sequence of the five ORs is shown with the nucleotide sequence of the first three repeats depicted below. Guanines capable of forming G-quadruplexes are shown underlined/in red. (B) Cartoon showing the formation of G-quadruplexes structures in the presence of potassium ions. Hydrogen bonds between guanines are shown as dashed red lines. Connecting loops within G-quadruplexes are indicated by solid blue lines. (C) RNA constructs corresponding to different regions of the first OR which were used in this study.
Biophysical analysis of synthetic PrP RNA oligonucleotides
CD was used to investigate the secondary structures of the four RNAs. In the presence of MgCl2, Pri-One, Pri-Two and Pri-Hp RNAs displayed the typical CD pattern of an A-form RNA helix, with a maximum at 265 nm and minimum at 210 nm (Figure 2A–C, red curves). The spectrum of Pri-Qd RNA was in accordance with the formation of a parallel G4 with a minimum at 240 nm either in the presence of MgCl2 or KCl (Figure 2D). KCl had no effect on the structure of Pri-Hp (Figure 2C, blue dashed line), which remained in the hairpin conformation, but KCl did induce structural changes in Pri-One and Pri-Two RNAs (Figure 2A and B, blue lines) indicating the formation of a G4 in these RNAs similar to Pri-Qd (Figure 2D).
Figure 2.
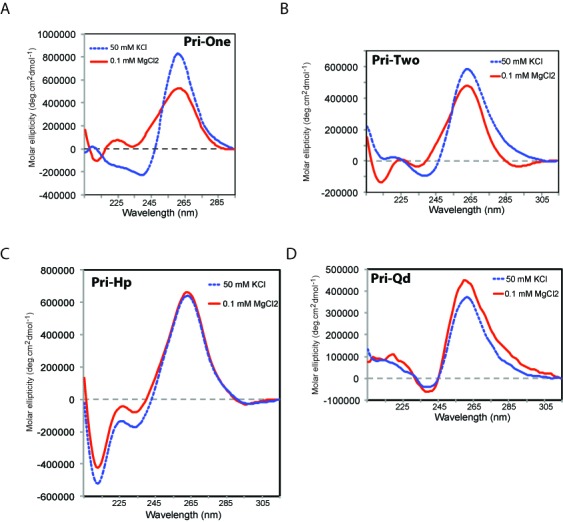
CD spectra of the four synthetic RNAs. RNAs were dissolved in 10-mM sodium phosphate buffer (pH 6.8) containing MgCl2 or KCl, and their spectra recorded at 20°C. (A) Pri-One RNA. (B) Pri-Two RNA. (C) Pri-Hp RNA. (D) Pri-Qd RNA..
Pri-Two and Pri-Hp RNAs were obtained in sufficient quantities to perform NMR as well. In 10-mM sodium phosphate buffer (pH 7.0), the spectrum of Pri-Hp was in agreement with a stem–loop structure with typical resonances at 13.5 ppm and one near 12.2 ppm corresponding to regular Watson–Crick (WC) base pairs (Figure 3C). The spectrum of Pri-Two in the presence of 0.1-mM MgCl2 showed in addition to WC peaks at 13.5 ppm also a number of other peaks and in particular a broad resonance in the region of non-WC base pairs around 11 ppm (Figure 3A). In the presence of 100-mM KCl, most resonances that corresponded to WC base pairs disappeared and peaks in the 11-ppm region were strongly enhanced (Figure 3B). This behavior is typical of G4 formation involving multiple non-WC base pairs (25). The spectrum of Pri-Hp RNA was hardly affected by the addition of KCl (Figure 3D).
Figure 3.
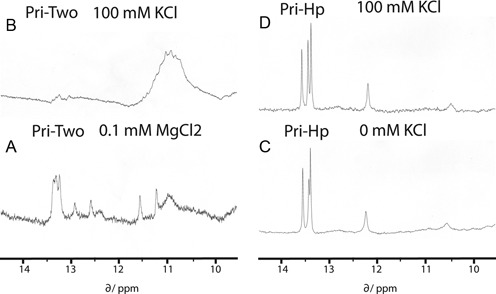
1D proton NMR spectroscopy analysis of two synthetic RNAs. Spectra were recorded at 5°C. (A) Pri-Two RNA (60 μM) in the presence of 0.1 mM MgCl2. (B). Pri-Two RNA (60 μM) in the presence of 100 mM KCl. (C) Pri-Hp RNA (250 μM). (D) Pri-Hp RNA (250 μM) in the presence of 100 mM KCl.
Next, UV absorbance profiles were measured as this also allows one to differentiate between quadruplex and hairpin forming nucleic acids. First, a thermal difference spectrum (TDS) was recorded in the range 220–320 nm for Pri-One in sodium phosphate buffer without or with KCl. Figure 4A shows the normalized absorbance spectra of Pri-One in the absence (red curve) or presence of 50-mM KCl (blue dashed curve). The shape of the red curve is typical for a DNA or RNA duplex involving WC base pairs while the blue curve with a negative absorbance peak at 300 nm is typical for Hoogsteen base pairs such as present in G4 structures (15). Thus, the TDS signature of Pri-One is also in agreement with a hairpin to G4 transition by potassium ions.
Figure 4.
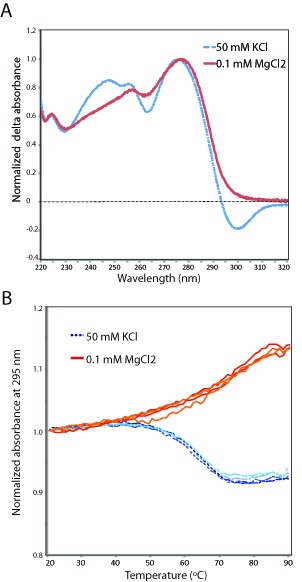
Thermal difference spectra and absorption profiles of Pri-One RNA in the absence and presence of KCl. (A) Normalized thermal difference spectra of Pri-One RNA in 10-mM sodium phosphate buffer (pH 6.7) with (blue dashed curve) or without (red curve) 50-mM KCl. (B) Normalized UV absorption profiles at 295 nm of Pri-One RNA in 10-mM sodium phosphate buffer (pH 6.7) with (blue dashed curves) or without (red curves) 50-mM KCl.
To obtain information on the stability of the G4 and hairpin conformation, UV melting profiles were recorded at 295 nm. At this wavelength, the TDS signature of the G4 conformation of Pri-One is negative while that of the hairpin conformation is still measurably positive (Figure 4A) meaning that upon melting the absorbance will either decrease or increase depending on the conformation of Pri-One. Figure 4B shows that the absorbance of Pri-One in 50-mM KCl decreased upon heating and increased again along the same curve upon cooling down (Figure 4B, blue dashed lines). This corresponds to the melting (and refolding) of a G4 (26) with a Tm of 62.8 ± 0.77°C. The opposite melting profile was observed in the absence of KCl (Figure 4B, red lines) which indicates that under these conditions Pri-One folded and unfolded as a regular RNA hairpin with a Tm of 71.5 ± 0.61°C. The absorbance profile at 295 nm of the reference RNA hairpin Pri-Hp (in the presence of KCl) was similar to that of Pri-One (in the absence of KCl) and was in agreement with conventional melting of an RNA hairpin (Supplementary Figure S1). The UV melting curve at 295 nm of the reference RNA Pri-Qd showed the typical absorbance profile of a G4 (Supplementary Figure S2). In conclusion, the UV melting studies are also indicative of a potassium-induced conformational switch from hairpin to G4 in Pri-One RNA.
In vitro translation of PrP mRNA
To investigate whether G4s can form in full-length PrP mRNA, a different approach was followed. First PrP mRNA was synthesized by T7 RNA polymerase-directed transcription of a PCR-generated template corresponding to the PrP ORF. This mRNA was used in cell-free translation assays in the presence of 35S-labelled methionine. In RRL, the T7-transcript directed the production of the full-length PrP protein of 27 kD (Figure 5A, lane 1). In addition, a number of higher molecular weight (HMW) products were observed, which probably resulted from glycosylation activity in RRL. HMW products were absent when translation was carried out with a bacterial lysate, which is lacking glycosylation activity (data not shown). When PrP mRNA translation was performed in the presence of potassium chloride, the yield of PrP dropped more than 4-fold at 100-mM KCl (50 mM from RRL plus 50 mM added KCl; Figure 5A, lane 3) while the yield of a firefly control mRNA remained essentially constant at 75- and 100-mM KCl (lanes 5 and 6). These data fit with a potassium-mediated stabilization of G4 structures in PrP mRNA. To provide additional support for this, translation was performed in the presence of a G4-stabilizing ligand; the bis-quinolinium compound PhenDC3 (27). Addition of PhenDC3 at 1 μM reduced PrP translation about 2-fold (Figure 5B, lanes 3 and 4). In contrast, PhenDC3 had no inhibiting effect on the translation of PrP89-253 mRNA, which lacks the five ORs capable of forming the G4 structures (Figure 5B, lanes 5 and 6) or on translation of FLuc mRNA (Figure 5B, lanes 1 and 2). This result strongly suggests that G4s can form in full-length PrP mRNA and that they can actually interfere with translation elongation. Although PrP89-253 mRNA contains two putative G4 forming sequences (266-GGGGUCAAGGAGGUGG-282 and 355-GGGGCAGUGGUGGGGGG-371), these are apparently not sufficiently stable on their own or not stabilized by PhenDC3 to impede ribosome movement. G4 stability is very sensitive to the length of the connecting loops (28) and in these two sequences at least one of the loops consists of four bases (see underlined sequence above).
Figure 5.
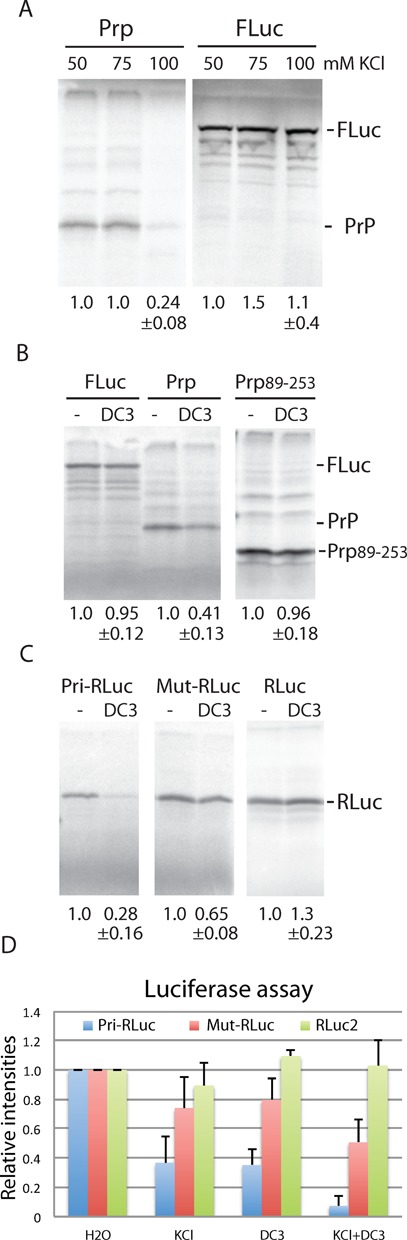
Effect of potassium and G4 ligand on translation of PrP mRNA in vitro (A)–(C). SDS-PAGE of 35S-methionine-labeled products from in vitro translation reactions using RRL. (A) PrP mRNA and FLuc mRNA were preincubated with or without KCl. As undiluted RRL contains ∼100-mM KCl, the final concentrations are 50, 75 and 100 mM. The relative yields of the bands corresponding to full-length PrP protein or FLuc protein are indicated. Standard deviation at 100-mM KCl was determined from four (PrP) and two (Fluc) experiments. (B) FLuc, PrP and PrP89-253 mRNA were preincubated for 1 h with or without PhenDC3 (final concentration 1 μM). Positions of the expected products are indicated at the right. Standard deviations were determined from two (FLuc and PrP89-253) and four (PrP) experiments. (C) Preincubation as under (B). Standard deviations were determined from five (Pri-RLuc) and four (RLuc and Mut-RLuc) experiments. (D) Final concentrations of KCl and PhenDC3 were 100 mM and 1 μM, respectively. The Y-axis shows the relative light units normalized to the reference samples without KCl or PhenDC3. Error bars represent the standard deviations of nine (Pri-RLuc), six (Mut-Rluc) and five (RLuc2) experiments.
In vitro translation of OR luciferase hybrid mRNAs
To demonstrate that the OR regions were indeed responsible for the inhibitory effect of PhenDC3 on translation, two copies of the OR repeat were cloned into the 5′UTR of a Renilla luciferase reporter mRNA. Addition of PhenDC3 reduced the yield of Renilla luciferase in Pri-RLuc to 28% while there was a slight enhancing effect on the translation of the RLuc control mRNA (Figure 5C), indicating that the formation of G4s in the 5′UTR was interfering with ribosomal scanning. In Mut-RLuc, the putative G4s were destabilized by substitution of several G's by A's (see the Materials and Methods section). As a consequence, the overall translation was enhanced and also the inhibiting effect of PhenDC3 was strongly reduced (Figure 5C, lanes 3 and 4). In an independent assay, the luciferase activity of Pri-RLuc and Mut-RLuc was measured. Pre-incubation of Pri-RLuc mRNA with either KCl or PhenDC3 resulted in an almost 3-fold decrease in luciferase activity while in the presence of both KCl and PhenDC3 luciferase level dropped more than 13-fold (Figure 5D, blue bars). Under the latter conditions, the translation of Mut-RLuc mRNA was inhibited only 2-fold (Figure 5D, red bars). The luciferase activity of a control RLuc mRNA (RLuc2) was only marginally affected by KCl and/or PhenDC3 (Figure 5D, green bars). This suggests that the remaining G-residues in Mut-RLuc are capable of adopting a weak G4 structure in the presence of KCl and PhenDC3. Although not visible in Figure 5D, the level of luciferase activity of Mut-RLuc in the absence of KCl or PhenDC3 was at least 3-fold higher than that of Pri-RLuc, indicating that sequences derived from the OR region can already form G4 structures under standard in vitro conditions.
DISCUSSION
The above data demonstrate that sequences within PrP mRNA have the propensity to switch between hairpin structures and G-quadruplexes depending on environmental conditions. Fluctuations in potassium levels are common for cells including those in the central nervous system in which PrP is highly expressed (29). Combined with the observations that G4s are strong binders of PrP itself a view is emerging in which binding of PrP to its own mRNA maybe involved in the regulation of the co-translational folding of PrP. Disturbances in the RNA binding of PrP could trigger the conversion of PrPC to PrPSc. Although PrPC is normally exported into the Endoplasmic Reticulum, there are several reports describing accumulation of PrP in cytosolic (30,31) and even nuclear fractions (32). These endocytosed or mis-translocated PrPs can potentially bind to PrP mRNA G4s and interfere with translation elongation and/or the protein folding activity of the ribosome [PFAR (33)] thereby somehow triggering the formation of the first misfolded PrPSc.
Recently, it was shown that G4 motifs can halt translation elongation (34) and even lead to ribosomal frameshifting (19,35). Moreover, the presence of G4 motifs in the human estrogen receptor alpha (hERalpha) mRNA has been demonstrated to affect the correct folding of hERalpha and its proteolysiss (36). Certain anti-prion drugs have been described that inhibit PFAR and diminish prion misfolding (33). Thus, events occurring during mRNA translation, possibly mediated by G4 motifs, may play a key role in prion folding and misfolding.
Many small molecules that are able to inhibit PrPSc accumulation like bis-acridines (37), porphyrins, phthalocyanines (38,39) and fumaridines (40) are also known to bind and stabilize DNA and RNA G4s. Possibly these drugs exert their anti-aggregation effect by binding to PrP mRNA rather than to the prion protein itself, and it is this RNA–drug complex that prevents the initial conversion of PrPC to PrPSc. In this respect, it is worth noting that many anti-prion drugs are only active in cells and do not bind the purified PrPC ex vivo (41). It has been suggested that these drugs require an auxiliary factor that is present only in cells. Quite likely this factor is PrP mRNA. This would also explain why RNA from brain cells, which have high levels of PrP, is more efficient in the conversion of PrPC to PrPSc than RNA from other sources. To investigate the role of PrP mRNA in PrPSc formation it may be included during PCMAs. In such assays also the effect of potassium and G4-ligands could be tested. Another way to address the role of G4s is to introduce mutants of this motif into a transgenic mouse (42) and determine if mutations alter susceptibility to (interspecies) prion transmission. However, it is difficult to destroy the G4 motifs without interfering with the amino acid sequence of PrP.
In conclusion, the presence of G4 forming motifs in PrP mRNA may provide the missing link in the initial conversion of PrPC to PrPSc. Understanding how mRNA structures are involved in the (mis-)folding of PrPC and possibly many other RNA-binding proteins with prion-like properties is of prime importance for the development of better treatments of CJD and related diseases.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Acknowledgments
I thank Alex Bossers for H. sapiens prion cDNA, Marie-Paule Teulade-Fichou for PhenDC3, Fons Lefeber, Karthick Sai Sankar Gupta and Kees Erkelens for excellent NMR assistance, Hans Heus for his help with UV melting experiments and the members of the former RNA Genetics group Leiden for stimulating discussions.
FUNDING
Leiden Institute of Chemistry, Leiden University, Leiden, The Netherlands. Funding for open access charge: Leiden Institute of Chemistry.
Conflict of interest statement. None declared.
REFERENCES
- 1.Colby D.W., Prusiner S.B. Prions. Cold Spring Harb. Perspect. Biol. 2011;3:a006833. doi: 10.1101/cshperspect.a006833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kraus A., Groveman B.R., Caughey B. Prions and the potential transmissibility of protein misfolding diseases. Annu. Rev. Microbiol. 2013;67:543–564. doi: 10.1146/annurev-micro-092412-155735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Deleault N.R., Harris B.T., Rees J.R., Supattapone S. Formation of native prions from minimal components in vitro. Proc. Natl. Acad. Sci. U.S.A. 2007;104:9741–9746. doi: 10.1073/pnas.0702662104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang F., Wang X., Yuan C.G., Ma J. Generating a prion with bacterially expressed recombinant prion protein. Science. 2010;327:1132–1135. doi: 10.1126/science.1183748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gomes M.P.B., Vieira T.C.R.G., Cordeiro Y., Silva J.L. The role of RNA in mammalian prion protein conversion. Wiley Interdiscip. Rev. RNA. 2012;3:415–428. doi: 10.1002/wrna.118. [DOI] [PubMed] [Google Scholar]
- 6.Deleault N.R, Lucassen R.W., Supattapone S. RNA molecules stimulate prion protein conversion. Nature. 2003;425:717–720. doi: 10.1038/nature01979. [DOI] [PubMed] [Google Scholar]
- 7.Wang F., Zhang Z., Wang X., Li J., Zha L., Yuan C.G., Weissmann C., Ma J. Genetic informational RNA is not required for recombinant prion infectivity. J. Virol. 2012;86:1874–1876. doi: 10.1128/JVI.06216-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Saá P., Sferrazza G.F., Ottenberg G., Oelschlegel A.M., Dorsey K., Lasmézas C.I. Strain-specific role of RNAs in prion replication. J. Virol. 2012;86:10494–10504. doi: 10.1128/JVI.01286-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Deleault N.R., Piro J.R., Walsh D.J., Wang F., Ma J., Geoghegan J.C., Supattapone S. Isolation of phosphatidylethanolamine as a solitary cofactor for prion formation in the absence of nucleic acids. Proc. Natl. Acad. Sci. U.S.A. 2012;109:8546–8551. doi: 10.1073/pnas.1204498109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kellings K., Meyer N., Mirenda C., Prusiner S., Riesner D. Further analysis of nucleic acids in purified scrapie prion preparations by improved return refocusing gel electrophoresis. J. Gen. Virol. 1992;73:1025–1029. doi: 10.1099/0022-1317-73-4-1025. [DOI] [PubMed] [Google Scholar]
- 11.Gilch S., Schätzl H.M. Aptamers against prion proteins and prions. Cell. Mol. Life Sci. 2009;66:2445–2455. doi: 10.1007/s00018-009-0031-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Proske D., Gilch S., Wopfner F., Schätzl H.M., Winnacker E.L., Famulok M. Prion-protein-specific aptamer reduces PrPSc formation. Chembiochem. 2003;3:717–725. doi: 10.1002/1439-7633(20020802)3:8<717::AID-CBIC717>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 13.Mashima T., Nishikawa F., Kamatari Y.O., Fujiwara H., Saimura M., Nagata T., Kodaki T., Nishikawa S., Kuwata K., Katahira M. Anti-prion activity of an RNA aptamer and its structural basis. Nucleic Acids Res. 2013;41:1355–1362. doi: 10.1093/nar/gks1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cavaliere P., Pagano B., Granata V., Prigent S., Rezaei H., Giancola C., Zagari A. Cross-talk between prion protein and quadruplex-forming nucleic acids: a dynamic complex formation. Nucleic Acids Res. 2013;41:327–339. doi: 10.1093/nar/gks970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Merngy J.-L., Li J., Lacroix L., Amrane S., Chaires J.B. Thermal difference spectra: a specific signature for nucleic acid structures. Nucleic Acids Res. 2005;33:e138. doi: 10.1093/nar/gni134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.van der Werf R., Wijmenga S.S., Heus H.A., Olsthoorn R.C.L. Structural and thermodynamic signatures that define pseudotriloop RNA hairpins. RNA. 2013;19:1833–1839. doi: 10.1261/rna.039636.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bossers A., Belt P.B.G.M., Raymond G.J., Caughey B., De Vries R., Smits M.A. Scrapie susceptibility-linked polymorphisms modulate the in vitro conversion of sheep prion protein to protease-resistant forms. Proc. Natl. Acad. Sci. U.S.A. 1997;94:4931–4936. doi: 10.1073/pnas.94.10.4931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Girard G., Gultyaev A.P., Olsthoorn R.C.L. Upstream start codon in segment 4 of North American H2 avian influenza A viruses. Infect. Genet. Evol. 2011;11:489–495. doi: 10.1016/j.meegid.2010.12.014. [DOI] [PubMed] [Google Scholar]
- 19.Yu C.-H., Teulade-Fichou M.P., Olsthoorn R.C.L. Stimulation of ribosomal frameshifting by RNA G-quadruplex structures. Nucleic Acids Res. 2014;42:1887–1892. doi: 10.1093/nar/gkt1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Premzl M., Delbridge M., Gready J.E., Wilson P., Johnson M., Davis J., Kuczek E., Marshall Graves J.A. The prion protein gene: identifying regulatory signals using marsupial sequence. Gene. 2005;349:121–134. doi: 10.1016/j.gene.2004.11.049. [DOI] [PubMed] [Google Scholar]
- 21.Wills P.R., Hughes A.L. Stem loops in HIV and prion protein mRNAs. J. Acquir. Immune Defic. Syndr. 1990;3:95–97. [PubMed] [Google Scholar]
- 22.Wills P.R. Potential pseudoknots in the PrP-encoding mRNA. J. Theor. Biol. 1992;159:523–527. doi: 10.1016/s0022-5193(05)80696-4. [DOI] [PubMed] [Google Scholar]
- 23.Lück R., Steger G., Riesner D. Thermodynamic prediction of conserved secondary structure application to the RRE element of HIV, the tRNA element of CMV and the mRNA of prion protein. J. Mol. Biol. 1996;258:813–826. doi: 10.1006/jmbi.1996.0289. [DOI] [PubMed] [Google Scholar]
- 24.Barrette I., Poisson G., Gendron P., Major F. Pseudoknots in prion protein mRNAs confirmed by comparative sequence analysis and pattern searching. Nucleic Acids Res. 2001;29:753–758. doi: 10.1093/nar/29.3.753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bugaut A., Murat P., Balasubramanian S. An RNA hairpin to G-quadruplex conformational transition. J. Am. Chem. Soc. 2012;134:19953–19956. doi: 10.1021/ja308665g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mergny J.L., Phan A.T., Lacroix L. Following G-quartet formation by UV-spectroscopy. FEBS Lett. 1998;435:74–78. doi: 10.1016/s0014-5793(98)01043-6. [DOI] [PubMed] [Google Scholar]
- 27.De Cian A., Delemos E., Mergny J.-L., Teulade-Fichou M.-P., Monchaud D. Highly efficient G-quadruplex recognition by bisquinolinium compounds. J. Am. Chem. Soc. 2007;129:1856–1857. doi: 10.1021/ja067352b. [DOI] [PubMed] [Google Scholar]
- 28.Zhang A.Y.Q., Bugaut A., Balasubramanian S. A sequence-independent analysis of the loop length dependence of intramolecular RNA G-quadruplex stability and topology. Biochemistry. 2011;50:7251–7258. doi: 10.1021/bi200805j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brown D.R., Schmidt B., Kretzschmar H.A. Role of microglia and host prion protein in neurotoxicity of a prion protein fragment. Nature. 1996;380:345–347. doi: 10.1038/380345a0. [DOI] [PubMed] [Google Scholar]
- 30.Rane N.S., Yonkovich J.L., Hegde R.S. Protection from cytosolic prion protein toxicity by modulation of protein translocation. EMBO J. 2004;23:4550–4559. doi: 10.1038/sj.emboj.7600462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ma J., Lindquist S. Conversion of PrP to a self-perpetuating PrPSc-like conformation in the cytosol. Science. 2002;298:1785–1788. doi: 10.1126/science.1073619. [DOI] [PubMed] [Google Scholar]
- 32.Crozet C., Vezilier J., Delfieu V., Nishimura T., Onodera T., Casanova D., Lehmann S., Beranger F. The truncated 23–230 form of the prion protein localizes to the nuclei of inducible cell lines independently of its nuclear localization signals and is not cytotoxic. Mol. Cell. Neurosci. 2006;32:315–323. doi: 10.1016/j.mcn.2006.05.004. [DOI] [PubMed] [Google Scholar]
- 33.Pang Y., Kurella S., Voisset C., Samanta D., Banerjee D., Schabe A., Das Gupta C., Galons H., Blondel M., Sanyal S. The antiprion compound 6-aminophenanthridine inhibits the protein folding activity of the ribosome by direct competition. J. Biol. Chem. 2013;288:19081–19089. doi: 10.1074/jbc.M113.466748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Endoh T., Kawasaki Y., Sugimoto N. Suppression of gene expression by G-quadruplexes in open reading frames depends on G-quadruplex stability. Angew. Chem. Int. Ed. Engl. 2013;52:5522–5526. doi: 10.1002/anie.201300058. [DOI] [PubMed] [Google Scholar]
- 35.Endoh T., Sugimoto N. Unusual -1 ribosomal frameshift caused by stable RNA G-quadruplex in open reading frame. Anal. Chem. 2013;85:11435–11439. doi: 10.1021/ac402497x. [DOI] [PubMed] [Google Scholar]
- 36.Endoh T., Kawasaki Y., Sugimoto N. Stability of RNA quadruplex in open reading frame determines proteolysis of human estrogen receptor α. Nucleic Acids Res. 2013;41:6222–6231. doi: 10.1093/nar/gkt286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.May B.C., Fafarman A.T., Hong S.B., Rogers M., Deady L.W., Prusiner S.B., Cohen F.E. Potent inhibition of scrapie prion replication in cultured cells by bis-acridines. Proc. Natl. Acad. Sci. U.S.A. 2003;100:3416–3421. doi: 10.1073/pnas.2627988100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Caughey W.S., Raymond L.D., Horiuchi M., Caughey B. Inhibition of protease-resistant prion protein formation by porphyrins and phthalocyanines. Proc. Natl. Acad. Sci. U.S.A. 1998;95:12117–12122. doi: 10.1073/pnas.95.21.12117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Priola S.A., Raines A., Caughey W.S. Porphyrin and phthalocyanine antiscrapie compounds. Science. 2000;287:1503–1506. doi: 10.1126/science.287.5457.1503. [DOI] [PubMed] [Google Scholar]
- 40.Stanton J.B., Schneider D.A., Dinkel K.D., Balmer B.F., Baszler T.V., Mathison B.A., Boykin D.W., Kumar A. Discovery of a novel, monocationic, small-molecule inhibitor of Scrapie prion accumulation in cultured sheep microglia and Rov cells. Plos ONE. 2012;7:e51173. doi: 10.1371/journal.pone.0051173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kocisko D.A., Baron G.S., Rubenstein R., Chen J., Kuizon S., Caughey B. New inhibitors of scrapie-associated prion protein formation in a library of 2000 drugs and natural products. J. Virol. 2003;77:10288–10294. doi: 10.1128/JVI.77.19.10288-10294.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Watts J.C., Giles K., Patel S., Oehler A., Dearmond S.J., Prusiner S.B. Evidence that bank vole PrP is a universal acceptor for prions. PLoS Pathog. 2014;10:e1003990. doi: 10.1371/journal.ppat.1003990. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


