Abstract
The mechanism for generating double minutes chromosomes (dmin) and homogeneously staining regions (hsr) in cancer is still poorly understood. Through an integrated approach combining next-generation sequencing, single nucleotide polymorphism array, fluorescent in situ hybridization and polymerase chain reaction-based techniques, we inferred the fine structure of MYC-containing dmin/hsr amplicons harboring sequences from several different chromosomes in seven tumor cell lines, and characterized an unprecedented number of hsr insertion sites. Local chromosome shattering involving a single-step catastrophic event (chromothripsis) was recently proposed to explain clustered chromosomal rearrangements and genomic amplifications in cancer. Our bioinformatics analyses based on the listed criteria to define chromothripsis led us to exclude it as the driving force underlying amplicon genesis in our samples. Instead, the finding of coexisting heterogeneous amplicons, differing in their complexity and chromosome content, in cell lines derived from the same tumor indicated the occurrence of a multi-step evolutionary process in the genesis of dmin/hsr. Our integrated approach allowed us to gather a complete view of the complex chromosome rearrangements occurring within MYC amplicons, suggesting that more than one model may be invoked to explain the origin of dmin/hsr in cancer. Finally, we identified PVT1 as a target of fusion events, confirming its role as breakpoint hotspot in MYC amplification.
INTRODUCTION
Despite the high frequency of homogeneously staining regions (hsr) and double minute chromosomes (dmin) in human cancer (1), the mechanisms underlying their genesis remain unclear. The breakage-fusion-bridge model postulates the ‘head-to-head’ amplification of regions close to double strand breaks (DSBs) (2), while the episome model proposes the excision of a DNA segment followed by its circularization and ‘head-to-tail’ amplification to generate dmin or hsr (3). We provided evidence in favor of the episome model in acute myeloid leukemia (AML) cases carrying MYC (8q24.21) amplifications (4), and in neuroblastoma and small cell lung cancer (SCLC) cell lines showing MYCN (2p24) amplifications (5). Intriguingly, we described dmin and hsr carrying identical amplicons, thus sharing the same molecular origin (5). The episome model has been proposed also for other tumor types (6). A model postulating a one-step early catastrophic genomic event, named chromothripsis, was recently invoked to explain complex chromosome rearrangements in cancer (7). This model was first introduced to explain the genesis of 8q24 MYC-containing dmin in a SCLC cell line, showing a heavily rearranged derivative chromosome 8 (8). Afterward, the telomeric fusion of two chromosomes subsequently shattered by chromothripsis was proposed to explain the origin of complex dmin (9). However, the mechanism behind the formation of dmin involving more than two chromosomes is still unknown. To clarify the origin of MYC-containing dmin or hsr, we investigated these structures in seven tumor cell lines including SCLC, AML and colon carcinoma (CC) samples. We finely characterized their dmin and hsr amplicons through a combined approach including next-generation sequencing (NGS), fluorescent in situ hybridization (FISH), single nucleotide polymorphism (SNP) array and polymerase chain reaction (PCR). Our study took advantage from the investigation of cell lines deriving from the same patient, with amplifications differing in their inner chromosome content and organization. This heterogeneity suggested the involvement of a stepwise process, starting from single-chromosome ancestral episomes, for the genesis of complex dmin and hsr, rather than chromothripsis. Finally, we mapped an unprecedented number of hsr insertion sites and investigated the mechanisms driving the hsr seeding and evolution. We evaluated the expression level of genes involved in the amplifications or mapped at hsr insertion sites, and identified novel fusion genes originated at junctions on dmin/hsr.
MATERIALS AND METHODS
Tumor cell lines
For this study, we selected seven tumor cell lines carrying MYC amplifications in form of dmin and/or hsr (Table 1). Cell lines were preferred to primary tumors due to material availability reasons and to avoid contamination from non-neoplastic or stromal tissue. SCLC (GLC1DM/HSR, GLC2 and GLC3) and the AML cell line (HL-60) were obtained as previously described (10,11,12). GLC1DM/HSR cell lines, as well as the commercially available COLO320DM/HSR cell lines (13,14), derived from the same patient.
Table 1. Tumor cell lines included in the study.
| Cell line | Tumor type | Amplification type |
|---|---|---|
| GLC1DM | SCLC | dmin/hsr |
| GLC1HSR | SCLC | hsr |
| GLC2 | SCLC | dmin/hsr |
| GLC3 | SCLC | hsr |
| COLO320DM | CC | dmin/hsr |
| COLO320HSR | CC | hsr |
| HL-60 | AML | hsr |
FISH and PCR-based assays
FISH, Multicolor-FISH, long-range PCR, Vectorette PCR, Sanger sequencing and genomic real-time quantitative-PCR (qPCR) were performed as already described (4).
SNP array
All cell lines were analyzed by Affymetrix Genome Wide Human SNP Array 6.0 platform (Affymetrix, Santa Clara, CA, USA), as previously described (5). SNP array data are available in the ArrayExpress database (www.ebi.ac.uk/arrayexpress) under accession number E-MTAB-2248.
Whole-genome NGS
Genomic DNA was extracted, quantified and purified using standard methods. Whole-genome NGS DNA libraries were prepared using the Illumina TruSeq protocol (Illumina, San Diego, CA, USA). Briefly, 1 ug DNA was sheared with a Bioruptor sonicator (Diagenode, Denville, NJ, USA) in 50 μl TE for two cycles of 30”on/30”off on high power. DNA fragments were end-repaired, A-tailed and ligated to Illumina adaptors, and then fragments between 350 and 500 bp were selected by gel-cut on a 1% agarose gel. The library was amplified by PCR with adaptor-specific primers, and sequenced on one lane of an Illumina HiSeq 2000 instrument, in a paired-end 100-cycle run, at the Oregon Health and Science University Massively Parallel Sequencing Shared Resource (Portland, OR, USA), obtaining up to ∼150 million reads per sample (Supplementary Table S1).
NGS data analysis
All positions refer to the GRCh37/hg19 human genome assembly, February 2009 release. Reads were aligned to the human reference genome using Novoalign (v.2.0.7), run with parameters ‘-k -r Random -a’ (http://www.novocraft.com). PCR duplicates were removed using Picard (v.1.72) (http://picard.sourceforge.net/). Candidate structural variations (SVs) and breakpoints were identified using Delly software (15), with parameters ‘-q 20’ and ‘-p’. A further analysis by BreakDancer software (v.1.1_2011_02_21) (16), with parameters ‘-q 10 -r 2’, was performed in all cell lines. The detections by Delly and BreakDancer were merged to obtain the final SV call set. For GLC1 cell lines, a further analysis of candidate SVs was made by using Hydra (v. 0.5.3) (17) and GASV (v. 2.0) (18). GASV was also employed in COLO320 cell lines. Hydra analysis was performed as recommended by its authors: we first realigned all discordant and unmapped reads using Novoalign (setting ‘-t 210 -a -r Ex 1100’) and the quality calibration file from the initial alignment; then, discordant reads were de-duplicated using the provided script; finally, Hydra was executed with parameters ‘-li -use all’. GASV analysis was performed using the default parameters. Finally, we identified the breakpoints (BPs) shared by the cell lines, analyzing the alignments of the supporting reads for each candidate with BEDTools (v 2.16.2) (19). Anonymous insertions were confirmed by local assemblies of NGS reads around the BPs. Finally, we verified the presence of longer (up to ∼250 bp) non-template insertions, through local pair-wise re-assemblies of reads having an unmapped mate or only a partial match on the reference genome (see Supplementary Materials).
SV filtering for validation
We discarded recurrent SVs mapping outside the amplified regions or within repeated elements, and filtered SVs also found on a normal human genomic DNA by BreakDancer (3X coverage). For GLC1DM/HSR, as well as for COLO320DM/HSR, we selected the shared SVs to focus on early occurring events. All the identified fusion sequences were also checked by Integrative Genomics Viewer (IGV) visual inspection. Validated SV sequences were submitted to GenBank repository (http://www.ncbi.nlm.nih.gov/genbank/), under accession numbers KF857235–KF857259 and KF898873–KF898919 (Supplementary Table S2). Finally, FISH experiments were carried out to validate SVs involving regions not previously defined as amplified, evaluating their co-localization with MYC-containing amplicons. Real-time PCR, Long Range- and Vectorette-PCR were performed to facilitate the full amplicon assembly.
Chromothripsis analysis
To infer chromothripsis, we evaluated the criteria described by Korbel and Campbell (7). We could not investigate the criterion: ‘prevalence of rearrangements affecting a specific haplotype’, as germline DNA was not available. We used data from the chromosome 15 of the SNU-C1 cell line as a positive control (7,8), and all chromosomes of a normal human genome HG00096 sequenced by the 1K Genomes Project (20) as negative controls. We focused the analysis on chromosomes involved in the amplification process.
Bioinformatics analysis for sequence motifs searching at hsr insertion sites
We finely mapped the insertion sites of the recurrent hsr by FISH on metaphases and stretched chromosomes. A detailed sequence analysis by NGS focused on the intervals delimited by FISH allowed us to finely map the insertion sites of five hsr. We investigated the genomic regions 20 Kb around each hsr insertion site identified at nucleotide level, searching for enrichment of features and motifs listed in Supplementary Table S3, using a permutation testing strategy. We performed this analysis by Fuzznuc (http://emboss.bioinformatics.nl/cgi-bin/emboss/fuzznuc). To test the significance of these enrichments (P < 0.05), we also tested a control dataset of 10 000 randomly permuted 20 Kb genomic regions, obtained through the shuffleBed tool from the BEDTools suite (19), excluding overlaps with assembly gaps (hg19 UCSC ‘gaps’ track). We compared the target and control dataset using a two-sample Kolmogorov–Smirnov test. We repeated the analysis narrowing the tested regions to 1 Kb in size.
Gene expression quantification
Reverse transcription quantitative PCR assays were performed as previously described (4). We tested 10 housekeeping genes (ACTB, B2M, GAPDH, HMBS, HPRT1, RPL13A, SDHA, RNA28S, UBC and YWHAZ) choosing the three best-performing ones (YWHAZ, HMBS and UBC) by geNorm analysis (21). The geometric mean of chosen housekeeping genes was used as normalization factor. Normal colon RNA (Clontech Laboratories, Inc., Palo Alto CA, USA), three pooled normal lung RNAs and commercial bone marrow (BM) RNA (Clontech, Jesi, Italy) were used as calibrators for CC, SCLC and AML cell lines, respectively. We included MYC-amplification negative cell lines as additional controls: GLC8 and GLC14 for SCLC, HCT-116 and LOVO for CC and the BM from three pooled AML patients for HL-60. Finally, statistical analyses were performed using the Relative expression software tool (22).
Detection of fusion transcripts
To confirm the presence of putative fusion transcripts detected by NGS, we performed RT-PCR assays. Primers specific for exons 1 and 2 of PVT1 (accession number M34428) were tested in combination with those specific for exon 2 of AKT3 (NM_005465) in GLC1DM/HSR cell lines and exon 3 of EYA1 (NM_172058.2) in GLC3 cell line. Sanger sequenced fusion products were analyzed by the NCBI BLAST tool (http://blast.ncbi.nlm.nih.gov). Fusion transcript sequences are deposited in GenBank repository (http://www.ncbi.nlm.nih.gov/genbank/) under accession numbers KJ452453–KJ452457. In silico translation of the full-length chimeric transcripts was performed by NCBI ORFfinder tool.
RESULTS
Detailed characterization of amplicons
SNP array data disclosed a high-level amplification of multiple chromosome regions in all the analyzed cell lines, confirming the shared amplification of the MYC domain. HL-60, GLC2 and GLC3 cell lines showed amplifications derived all from chromosome 8, co-localizing with MYC on dmin/hsr (as shown by FISH) (Supplementary Figure S1). Both GLC1 cell lines shared the amplification of sequences from chromosomes 1q24.2, 1q44 and 21q22.12, all co-localizing with MYC on dmin and hsr (Supplementary Figure S2A). COLO320DM and HSR showed amplification of sequences from chromosomes 6p25.3, 13q12.2 and 16p11.2, all co-localizing with MYC on dmin and hsr (Supplementary Figure S1). To unravel the inner complexity of dmin/hsr amplicons in each cell line, we performed NGS analysis and identified a total number of 17 473 SVs. Combining FISH and SNP array results, we selected 1263 SVs involved in amplified regions, validating a subset of them by Sanger sequencing (Supplementary Table S2). Finally, we found 68 validated SVs crucial for the circular amplicon structure assembly (Supplementary Table S2). We observed the presence of 33 microhomologies sequences, 29 anonymous insertions and 11 blunt-end joining events at the fusion junctions (Supplementary Table S2).
Amplicon structure in HL-60/GLC2/GLC3 cell lines
In these cell lines, we found amplicons composed of multiple, non-contiguous, non-collinear genomic segments repeated head-to-tail (Figure 1). HL-60 showed a 1.9 Mb amplicon consisting of 10 sub-segments, amplified in a single hsr within the pericentromeric region of chromosome 9 [hsr(9)] (Figure 1A). Interestingly, by IGV visual inspection, we identified a region showing an amplification level two times higher than the other co-amplified segments, thus disclosing two nested sub-amplicons (E6/E7 within E8/E9; Figure 1A). GLC2 and GLC3 showed the additional amplification of a region containing EYA1 at 8q13.3 (Figure 1B and C, and Supplementary Figure S1). The former cell line showed a 2.8 Mb amplicon consisting of five sub-segments, mapped on dmin and on a single hsr on chromosome 12 [hsr(12)], while the latter disclosed a 4.6 Mb amplicon, made of 12 sub-segments amplified on a single hsr located on chromosome 5 [hsr(5)] (Figure 1, Supplementary Figure S1).
Figure 1.
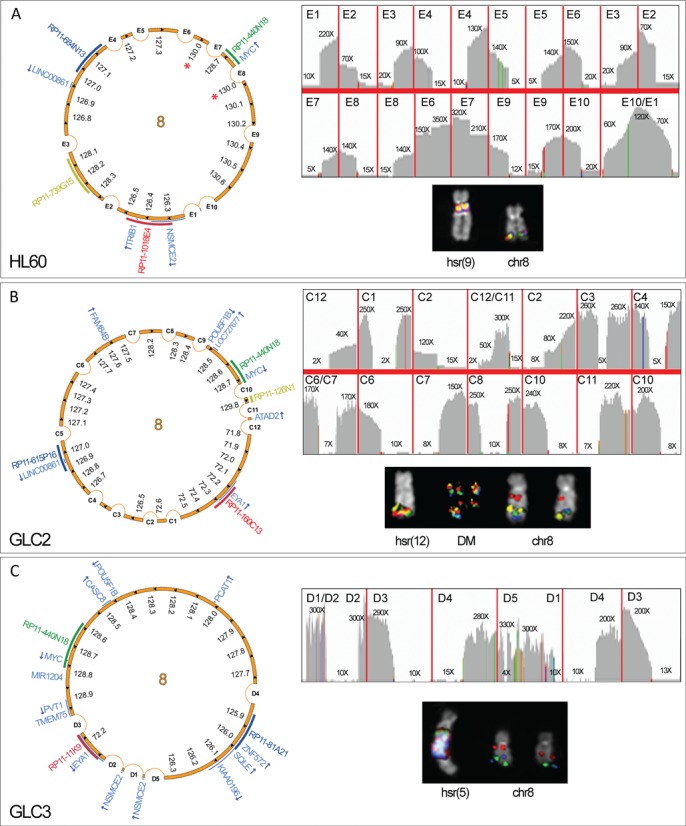
Circular amplicon structures in HL-60, GLC2 and GLC3 cell lines. Image showing HL-60 (A), GLC2 (B) and GLC3 (C) amplicon structure. Asterisks represent nested amplicons. For each cell line, top right panel: IGV plot of NGS read depth for recurrent amplicon SVs; left panel: scaled Circos plots showing the circular structure of the overall amplicons, with internal arrows indicating amplicon orientation, and genes in pale blue (arrows represent the transcriptional orientation); bottom right panel: FISH pseudocolor images showing co-localizing amplified probes, consistently colored as in the Circos plot.
Amplicon structure in GLC1DM/HSR cell lines
These two cell lines shared the amplification of the MYC domain and multiple non-contiguous segments derived from 1q24.2 (amplifying the POU2F1 3’-end, CD247 and CREG1), 1q43–q44 (containing SDCCAG8 3’-end and AKT3) and 21q22.12 (harboring the CLIC6 3’-end and RUNX1). Some amplicons also contained centromeric alpha-satellite DNA from chromosome 1 (Supplementary Figure S1D), however not co-localizing with the centromeric CENP-C protein (data not shown). Interestingly, FISH experiments disclosed a highly heterogeneous dmin/hsr composition (Figure 2). In GLC1DM, for instance, we detected five dmin subpopulations (DM), occurring within the same cells (Figure 2A, and Supplementary Figure S2). Likewise, GLC1HSR displayed a very heterogeneous hsr composition (Figure 2B). Combining data from NGS, SNP array and FISH, we detected some differences in these two sister cell lines. Particularly, GLC1HSR showed shorter distal borders of 1q24.2 and 1q43–q44 amplicons on hsr(1), hsr(8)b and hsr(20). Furthermore, both proximal and distal borders of 8q24 amplicons differed between GLC1DM (DM1) and GLC1HSR [hsr(2)], while 21q22 amplicons showed an internal gap in GLC1HSR [hsr(8)b and hsr(1)] but not in GLC1DM (DM5) (Supplementary Figure S2). SVs shared by the two cell lines were considered as ‘ancestral’ in reconstructing the evolutionary path leading to their differentiation (Figure 3, and Supplementary Figure S3). We thus identified head-to-tail amplifications harboring material only from chromosome 1 in DM2 and hsr(16), 21 in DM3 and 8 in hsr(2) (Figure 3A–C). These three single-chromosome amplicons were detected in both cell lines by NGS and PCR, but not by FISH (due to its lower resolution). Conversely, we inferred a shared head-to-tail 2.4 Mb amplicon on hsr(8), harboring sequences only from chromosomes 1 and 8 (Figure 3D). Finally, we detected a 2 Mb amplicon involving chromosomes 1 and 8 sequences in GLC1DM (DM4) and a 2.6 Mb array of amplified sequences from chromosomes 1, 8 and 21 in GLC1HSR [hsr(8)b and hsr(1)] (Figure 3E and F) and in GLC1DM (DM5) (Figure 2).
Figure 2.
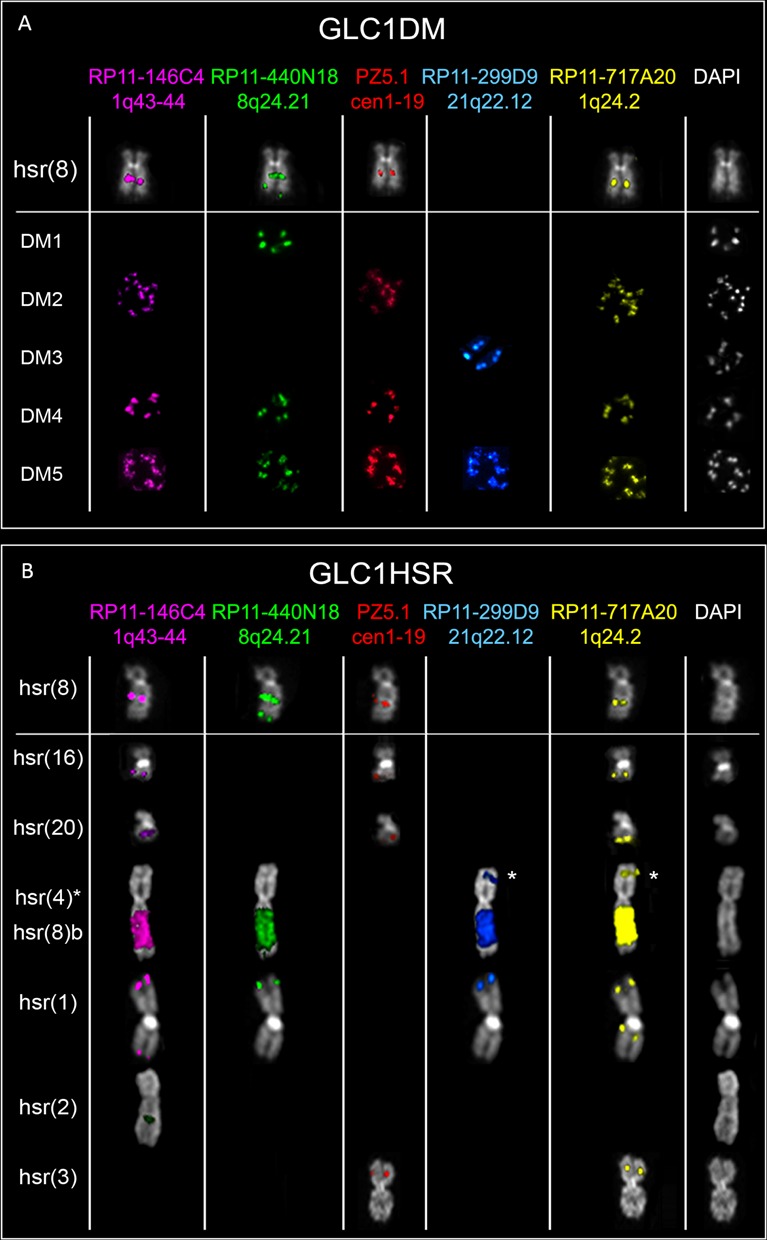
Heterogeneous dmin/hsr structure in GLC1DM/HSR cell lines. Partial metaphases showing FISH results obtained using the probes listed on the top. Each row refers to an amplicon population in GLC1DM (A) and GLC1HSR (B). The asterisk indicates hsr(4), mapping within a chromosome 4p joined to the 8q harboring hsr(8)b. (A) hsr at 8q21 [hsr(8)], shared by the two lines, showing the co-localization of all chromosomes 1q and 8q24 amplicons; DM1 containing only 8q24 amplifications; DM2 showing co-amplification of all 1q amplicons; DM3 with amplification of only 21q22.12 sequences; DM4 displaying the co-amplification of sequences found in DM1–2 dmin subgroups; DM5 with the co-localization of DM1–3 dmin amplified sequences together; (B) hsr(8) (see above in A); two hsr on 16q and 20q [hsr(16) and hsr(20), respectively], both positive for chromosome 1 sequences only (including alphoid DNA); a hsr within the long arm of one chromosome 4 [hsr(4)] (labeled with an asterisk), translocated, at its short arm, to a segment of chromosome 8q, harboring a second hsr [hsr(8)b]; a hsr within the short arm of one chromosome 1 [hsr(1)], showing co-localization of 1q (without alpha-satellite), 8q and 21q sequences; a hsr in 2q [hsr(2)], positive only for 8q24 probes; a hsr within the short arm of chromosome 3 [hsr(3)], with co-amplification of 1q24.2 and alphoid sequences.
Figure 3.
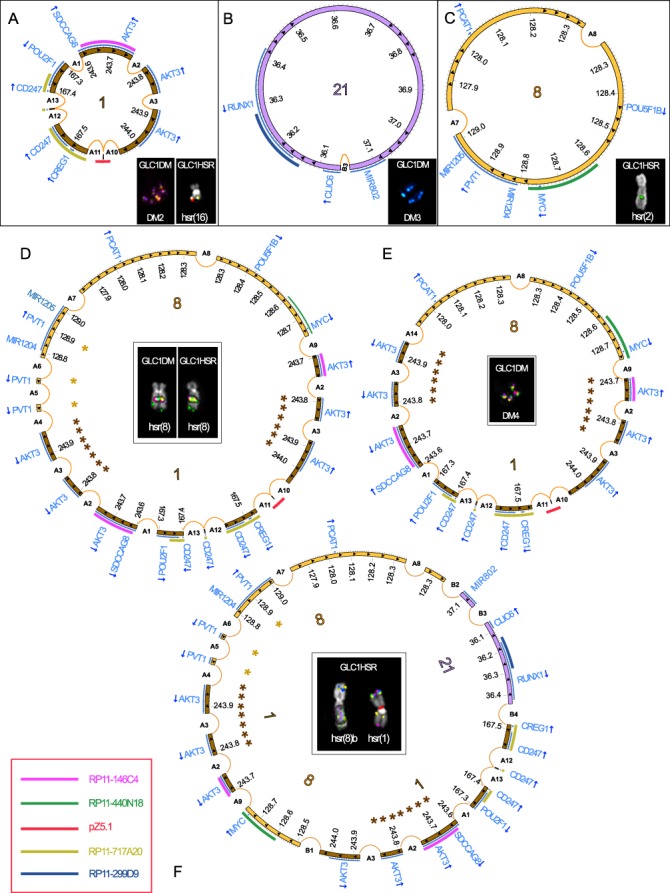
Circular amplicon structures in GLC1DM/HSR cell lines. Scaled Circos plots representing the amplicon structure of dmin/hsr in GLC1DM/HSR. Chromosomes 1, 8 and 21 fragments are shown by brown, yellow and purple innermost circular panels, respectively. Internal arrows indicate the amplicon orientation. Consistently colored asterisks represent nested amplicons. Genes are in pale blue, with arrows corresponding to their transcriptional orientation. Each pseudocolor FISH image shows the co-localizing amplified probes listed in the legend (bottom-left), consistently colored in the outermost semicircular panels of Circos plots. (A–C) Tandemly amplified segments originating from single chromosomes (1, 21 and 8, respectively). (D, E) Circular arrays of co-amplified chromosomes 1 and 8 segments [hsr(8) and DM4, respectively, in d and e]. (F) Co-amplification of chromosomes 1, 8 and 21 regions, inferred as a head-to-tail amplicon, in hsr(8)b and hsr(1) in GLC1HSR.
Amplicon structure in COLO320DM/HSR cell lines
COLO320DM and HSR carried a large number of hsr, only a few of them recurrent; dmin were peculiar for COLO320DM. These cell lines shared several amplified regions carrying relevant genes, such as MYC at 8q24, DUSP22 at 6p25.3, PDX1, ATP5EP2 and CDX2 at 13q12.2 and pseudogenes like LOC390705, SLC6A10P and TP53TG3C at 16p11.2, all co-localized on dmin and hsr as showed by FISH (Figure 4, Supplementary Figure S1). These cell lines shared the majority of the SVs, but showed some differences in their copy number switch profiles, disclosing the presence of multiple sub-clones carrying different SVs (Supplementary Figure S3, and Supplementary Table S2). This was particularly evident for F7, a SV previously described as peculiar of a sub-population of 8q24 amplicons (23). Furthermore, we found some SVs joining 8q24 and 16p11.2 exclusively in COLO320HSR (Supplementary Table S2). The inner complexity of dmin and hsr in these cell lines hampered the full assembly of the amplicon circular structure. Unfortunately, few SVs remained unsolved due to the presence of repetitive DNA. Among them, there was Hx, the SV linking the telomeric border of 16p11.2 amplicon to the other co-amplified regions (Figure 4 and Supplementary Figure S3). Nevertheless, we were able to infer the head-to-tail connection of the amplicons by taking into account the shared SVs and detected several nested amplifications (Figure 4). Despite the amplicon heterogeneity, both COLO320 lines showed an almost identical copy number switch profile corresponding to the F1 and F2 SVs at 8q24 (Supplementary Figure S3). These SVs likely fused two ‘ancestral’ chromosome 8 sub-amplicons in an inverted sequence orientation. The shared inter-chromosomal SVs joining 8q24 to 6p25.3 (H5) and to 13q12.2 (H6) could also be considered as ancestral.
Figure 4.
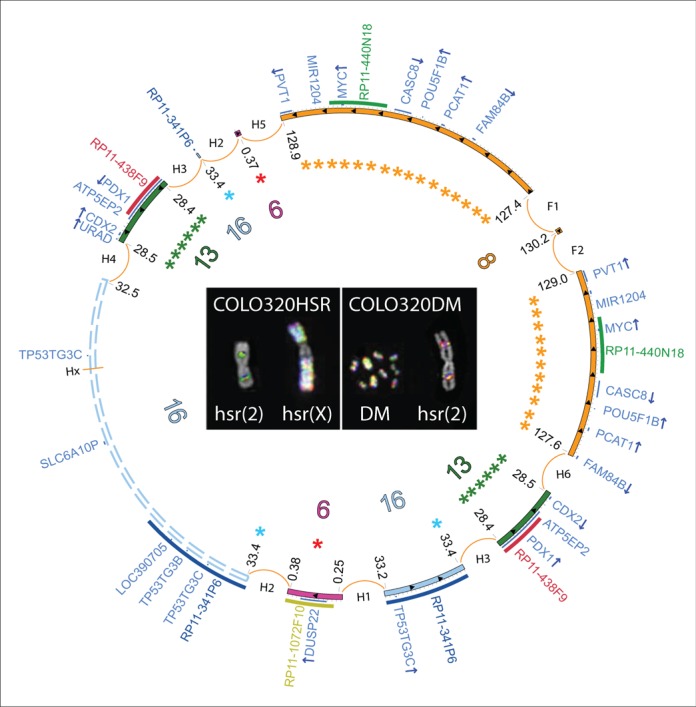
Circular amplicon structures shared by COLO320DM/HSR cell lines. Unscaled Circos plot representing the overall amplicon structure shared by COLO320DM/HSR. Chromosomes 6, 8, 13 and 16 fragments are shown as purple, yellow, green and light blue inner circular segments, respectively. Internal arrows indicate amplicon orientation. Asterisks indicate nested amplicons. Dashed lines represent the ambiguous structure of the 16p11.2 amplicon. Genes are in dark blue, with arrows corresponding to their transcriptional orientation. FISH images show co-localizing amplified probes, consistently colored in the outermost semicircular panels of the Circos plots.
Mapping of hsr insertion sites
According to FISH results, hsr can be classified as Low Copy Number (LCN) or High Copy Number (HCN) (5). It was previously hypothesized that the genome position could influence the amplification level of the inserted sequences (24). Multicolor (M)-FISH and FISH data indicated that the majority of hsr structures were inserted into chromosomes other than 8, with the exception of hsr(8) and hsr(8)b in GLC1 cell lines (Supplementary Figure S4). FISH analysis carried out on metaphases and stretched chromosomes did not detect any deletion flanking the insertion sites (Supplementary Figure S4 and Supplementary Table S4). A detailed sequence analysis focused on the intervals delimited by FISH allowed us to finely map the insertion sites of five hsr, then confirmed by Sanger sequencing [GLC1 LCN hsr(8)/LCN hsr(4), GLC2 LCN hsr(12), GLC3 HCN hsr(5) and COLO320DM hsr(2)] (Figure 5 and Supplementary Table S4). Interestingly, the insertion of three LCN hsr interrupted the coding genes CALB1 and LRBA for GLC1, and RILPL1 for GLC2, although not affecting their expression levels (Supplementary Table S5). The bioinformatics analysis for sequence motifs at the insertion sites, performed to identify features that could discriminate LCN and HCN hsr, disclosed enrichment for pyrimidine traits, putative triple helices and Chi-like sequences, even if not reaching the statistical significance.
Figure 5.
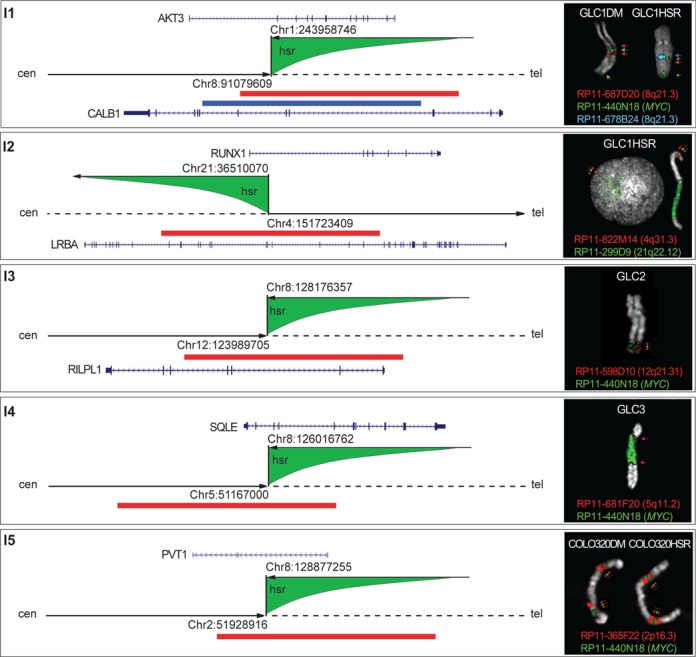
Schematic representation of the hsr insertion sites mapped at the nucleotide level. Each corresponding SV is top-left reported. Arrowhead and dashed dark lines indicate the distal and proximal side of each insertion site along the hosting chromosome, respectively; green shapes represent the inserted hsr, and the upper arrows indicate sequence orientation. All insertion sites show sequences joined in opposite direction. The right boxes show FISH results with splitting probes (red and blue) surrounding the hsr (green) in each cell line. FISH probes are represented as colored bars along each inserted chromosome. Target genes are in blue; arrows indicate their transcriptional orientation.
Chromothripsis analysis
We performed the point-by-point analysis of the criteria established to infer chromothripsis, and the results are shown in Supplementary Figure S5. The first analyzed criterion was the ‘Regularity of Oscillating Copy-Number States’. In chromothripsis, the copy number value should typically oscillate between two or occasionally three states; conversely, all our cell lines showed variations of copy number exceeding three states (Supplementary Figure S5A). The second criterion was the presence of ‘Interspersed regions with loss and retention of heterozygosity’, which means that the segments in the highest copy number state should retain heterozygosity; we found at least one not-deleted region showing loss of heterozygosity along the chromosome in each cell line (Supplementary Figure S5B). The third criterion was the ‘Clustering of breakpoints’; in case of chromothripsis, 5–10 breaks should be observed within 50 Kb genomic intervals. In our cell lines, we never observed this level of breakpoint clustering (Supplementary Figure S5C). The fourth criterion was the ‘Ability to walk the derivative chromosome’, which excludes the possibility of nested rearrangements; the majority of our cell lines were found to ‘reuse’ some segments in dmin/hsr amplifications (indicated as asterisks in Figures 1A, 3D–F and 4; Supplementary Figure S5D). The fifth criterion was the ‘Randomness of DNA segments order and fragment joins’; as chromosome fragments are randomly joined after chromothripsis, the breakpoint order should be not conserved on the resulting derivative chromosome, and an equal distribution of tail-to-head (–+), head-to-tail (+–), head-to-head (++) and tail-to-tail (–) rearrangements is expected. On the contrary, we found a highly conserved order of breakpoints and a prevalence (>28%) of deletions, even representing a subclass of the +– rearrangements (Supplementary Figure S5E). Since at least two of the six criteria should be fulfilled to invoke chromothripsis, we should discard it as a possible mechanism involved in the formation of dmin and hsr in the cell lines under study.
Fusion genes generated by junctions
Three fusion genes, all involving PVT1 5’-end (accession number M34428), were generated by the amplicon rearrangements on dmin/hsr. The first detected fusion transcript was 5’PVT1/3’MYC (generated by the SV F7), already described in COLO320 cell lines (23). The second chimeric transcript fused alternative exons of the PVT1 5’-end, to exon 3 of EYA1 (NM_172058) in GLC3, generating three distinct isoforms by the SV D3 (Supplementary Figure S6). In silico translation revealed the possible production of an EYA1 truncated protein [546 aminoacids (aa)] retaining only the C-terminus of the wild-type form (NP_742055, 592 aa) (Supplementary Figure S6). The third fusion transcript was found to join exon 2 of AKT3 to alternative exons of PVT1 (exon 1 or 2, transcript M34430) as consequence of the SVs A4/A5/A6 in GLC1 lines (Supplementary Figure S6). Both these transcript isoforms encoded shorter forms of the AKT3 protein (NP_005456; 478 and 418 aa), retaining the serine/threonine kinase domain but carrying a truncated pleckstrin homology (PH) domain.
Expression analysis of amplified genes
MYC, as well as some other 8q24 amplified genes, was significantly up-regulated (Supplementary Table S5). However, other genes in the same amplicon did not show an exclusive direct correlation between amplification and over-expression (i.e. NSMCE2, LINC00861, CASC8). EYA1 was found to be significantly over-expressed in GLC2 and GLC3. In both GLC1 cell lines, we observed a significant up-regulation of the amplified genes AKT3 and RUNX1; CREG1 showed up-regulation in GLC1DM but not in GLC1HSR, most likely due to the lower copy number state of the 1q24 region in that cell line (B4 in Supplementary Figure S3). POU2F1, conversely, was up-regulated even if not amplified in all SCLC cell lines. In both COLO320 cell lines, we found a significant up-regulation of the amplified genes PDX1, CDX2, SLC6A10P and LOC390705. Other genes (ATP5EP2, TP53TG3B, TP53TG3C), although amplified, did not show transcriptional activation. Surprisingly, DUSP22, the only amplified gene within the 6p25.3 amplicon in both COLO320 lines, appeared to be significantly down-regulated.
DISCUSSION
MYC amplifications as dmin/hsr are frequently observed in hematological malignancies and solid tumors, although their origin and functional impact are still under debate. So far, several mechanisms have been proposed to explain their genesis. In particular, the episome model was suggested in AML cases, and solid tumors cell lines, showing MYC and MYCN amplifications, respectively (4,5). The same model was also proposed for EGFR amplification in glioblastoma (25). Recently, chromothripsis was invoked to explain the origin of 8q24 MYC-containing dmin in a SCLC cell line showing a heavily rearranged derivative chromosome 8 (8). The genesis of complex dmin in medulloblastoma and SCLC was explained by the telomeric fusion of chromosomes shattered by chromothripsis (8,26,27). However, the mechanism behind the formation of dmin involving more than two chromosomes is still unclear. To unravel the mechanisms underlying the genesis of MYC-containing amplicons, we finely characterized the dmin and hsr structures in seven tumor cell lines including SCLC, AML and CC samples, through a combined approach integrating NGS, FISH, SNP array and PCR techniques. Our combined approach proved to be crucial to overcome the shortcomings of NGS in SV detection analysis, mainly due to the presence of long (>100 bp) non-template sequence insertions and repetitive DNA.
The sequencing of the amplicon junctions disclosed microhomologies and anonymous insertions, typical of non-homologous end joining (NHEJ) (28) or microhomology-mediated break-induced replication (29), in agreement with previous data on dmin/hsr origin (4–6,25,30–33).
In reconstructing amplicon structures, we found dmin and hsr sharing segments originated from single chromosomes (i.e. chr8), and others from at least two chromosomes. Complex amplicons, i.e. generated by the assembly of segments derived from different chromosomes, have been already described in several tumor types (5,6,9,30). In particular, GLC1 and COLO320 cell lines carried amplicons derived from chromosomes 1, 8 and 21 and from chromosomes 6, 8, 13 and 16, respectively. These domains harbor genes potentially involved in tumorigenesis.
The hypothesis of a telomeric fusion and subsequent shattering of the derivative chromosomes to generate dmin and hsr is not applicable to these cell lines, as confirmed by our analysis of the criteria to infer chromothripsis. The variety of shared SVs detected in GLC1DM and HSR, as well as in COLO320DM and HSR cell lines, strongly supported the evidence of a common origin for dmin and hsr, as previously suggested (5,31). On the contrary, the presence of multiple amplicon subpopulations, some consisting in single-chromosome amplified episomes, other carrying amplifications as combinations of different chromosomes (Figures 2 and 3), suggested the involvement of a stepwise process for the genesis of these heterogeneous dmin/hsr structures. The characterization of this amplicon heterogeneity allowed us to hypothesize a ‘multi-step’ evolutionary path to explain the genesis and evolution of the complex dmin/hsr observed in GLC1 cell lines (Figure 6). The common hsr(8) found in both GLC1 cell lines, and likely present in the original tumor sample, carried amplicons derived from the recombination of chromosomes 1 and 8 ‘ancestral’ episomes, actually present on hsr(2), hsr(16) and DM2 subpopulations. This event was accompanied by large duplications of sequences flanking the recombination site. Subsequent deletion and/or recombination (i.e. the recruitment of sequences from an ancestral chromosome 21-specific episome) could explain the observed heterogeneity (Figure 6). Of note, in support of our model, recombination between an episomally amplified plasmid, bearing a mammalian replication initiation region and a matrix attachment region, and pre-existing dmin structures was already observed in COLO320DM (34). The discrete mutation events (recombinations, deletions, duplications) observed at each step of the proposed ‘multi-step’ model in amplicon genesis substantially differ from the massive shattering of chromosomes described as occurring in chromothripsis. In COLO320 cell lines, amplicon heterogeneity was previously described for the 8q24 amplicons (23). In addition, we identified a further source of heterogeneity since we found amplified segments of chromosomes 8 and 16 fused in COLO320HSR, but not in COLO320DM. Even though COLO320 lines displayed co-localization of all the amplified regions on every observed dmin or hsr, we detected differences in the copy number switch profiles of the two cell lines, revealing the presence of sub-clones with dmin or hsr differing in their inner organization, but not in their content. For this reason it was not possible for us to trace an evolutionary path, even if a complex multi-step evolutionary process in which intermediate amplicons were lost cannot be excluded. Evidence of this hypothesis can be found in the presence of the F1 and F2 SVs, showing an identical copy number switch profile between the two cell lines, and so likely fusing two ancestral chromosome 8 sub-amplicons in an inverted orientation (Supplementary Figure S3). This ‘ancestral’ amplicon might have subsequently undergone secondary rearrangements leading either to the connections with chromosomes 6, 13 and 16 (Figure 4). None of the studied cell lines showed a deletion corresponding to the amplified regions, thus indicating that the excision–circularization–amplification of the ancestral episomes could have occurred as a post-replicative mechanism (4,5,25). Conversely, studies on a glioma tumor showing five amplicons with the alternating amplification of four chromosome loci (1q32.1, 7p11, 5p15 and 9p22) disclosed the deletion of a portion of the amplified segments (1q32.1 and 7p11) from their original location (6). These authors suggested a possible role for replication stress at breakpoint regions able to induce the simultaneous genesis of multiple episomes at different chromosomal sites. According to this hypothesis, the subsequent fusion of some of these fragments by NHEJ, possibly driven by their close proximity in the nucleus, could have generated the initial complex extrachromosomal circular DNA molecule.
Figure 6.

Evolutionary path for amplicons genesis in GLC1DM/HSR cell lines. Green lightning indicates breakage events; red dashed semicircles and curved arrows represent sequence deletions and duplications, respectively. Orange cross lines indicate homologous recombination (HR) events. Pink semicircle lines indicate the creation of new fusion junctions, labeled by appropriate SV codes. If undetermined, they are defined by a question mark. (A) The ancestral single-chromosome episomes, excised from their original location at chromosomes 1, 8 and 21, are amplified in the form of hsr(2), DM2/hsr(16) and DM3, respectively. The subsequent non-HR between chromosomes 1 and 8 episomes originated the shared hsr(8) amplicon structure, being associated with the duplication of sequences flanking the breakpoint regions on both ancestral episomes. The newly originated amplicon underwent three independent rearrangement types, indicated in (B) as enclosed in a gray dashed rectangle. Pink and pale blue squares (C, D) encompass the evolutionary paths observed in GLC1DM and GLC1HSR, respectively. (C) A single deletion event (SV A14) (B, left) generated the amplicon found at DM4. A further internal rearrangement (B, center) or alternatively its recombination with the chromosome 21 amplicon (B, right) originated the DM1 and DM5 amplicons, respectively. (D) The excision of a sub-region of 1q24.1 from the ancestral hsr(8) amplicon generated the amplified segment at hsr(3). A recombination-deletion event involving the hsr(8) and DM3 amplicons, followed by an inversion (B1) and HR, gave rise to the amplicon at hsr(8)b and hsr(1) in GLC1HSR. Finally, the subsequent excision of a sub-region encompassing 21q22.12 and 1q24 originated the hsr(4) amplicon.
Additional data in the literature support the view that DSBs induced at stalled replication forks, particularly within common fragile sites (CFSs), could be a driving force toward genome amplifications in tumors (35–37). Interestingly, two fragile sites mapping within 8q24 [FRA8C (8q24.1) and FRA8D (8q24.3)] were shown to be prone to breakage when cell lines harboring MYC-containing hsr were exposed to aphidicolin (31).
A comparison of our amplicon breakpoints with mapped fragile sites (38) showed that, in addition to the CFS at 8q24, other CFSs [FRA1S (1q43), FRA1I (1q44), FRA8F (8q13), FRA16F (16p11/16q11) and FRA21B (21q22.1)] may overlap with DSBs involving amplified regions in our cell lines. Their mapping, however, was established only at cytogenetic level. Replication stress could also be implicated in hsr insertion of amplicons, although assessing its actual implication in hsr integration would need further elucidations.
In this study, we report the highest number of hsr insertion sites sequenced in human cancer so far. This enabled us to draw some relevant conclusions about these sites. In particular, micro-homologies and anonymous insertions found at the integration sites indicate that hsr seeding is likely to be mediated by NHEJ (33). Nevertheless, we did not detect any particular sequence motif enrichment or genomic feature indicative of genome fragility or instability that could explain why some regions undergo tandemly repeated amplifications more than others. Similarly, we could not identify any genomic property that might differentiate the insertion sites of LCN and HCN hsr. Genes interrupted by hsr seeding surprisingly did not show altered expression patterns. MYC is considered the target gene of amplification in many cancers, including SCLC (26), CC (39) and AML (4,40). Here we found that other genes (KIAA0196, SQLE, ZNF572, TRIB1, FAM84B, PCAT1, POU5F1B, PVT1) found at 8q24 in addition to MYC were highly expressed when amplified, disclosing their crucial role in tumorigenesis (41). For the first time, we report the amplification and significant up-regulation of EYA1, AKT3 and RUNX1 in SCLC. These genes were already described as involved in leukemia and other solid tumors (42–44). Of note, amplified NSMCE2, LINC00861, and CASC8 genes did not show an exclusive direct correlation between amplification and over-expression. In addition, we detected a down-regulation of the amplified DUSP22, a tumor suppressor gene already described in lymphoproliferative disorders (45), breast (46) and lung cancer (47). Some epigenetic mechanisms might act to down-regulate this amplified gene, but the functional significance of this novel 6p25.3 amplification in SCLC remains unclear. Moreover, we detected three fusion genes, all involving the PVT1 gene, as consequence of the juxtaposition of amplified regions. PVT1, a non-coding RNA gene, was already reported as a frequent fusion gene partner in SCLC (26,48) and other tumor types, such as multiple myeloma (49), medulloblastoma (27) and gastric cancer (50). Our data highlight the role of PVT1 as a hotspot for chromosomal breaks during MYC amplification. PVT1 amplification and rearrangements have been correlated with poor prognosis (51) and resistance to therapy (52), although the functional role of its normal and chimeric transcripts remains unclear. The novel PVT1 fusion genes described here give rise to EYA1 and AKT3 chimeric proteins lacking their N-terminus, which harbor a transactivation (53) and a PH domain responsible for membrane translocation and activation, respectively (54).
All the experiments and results described here were obtained using established cell lines (rather than fresh tumor material) as biological model of the SVs participating in dmin/hsr formation. Although some potential interpretation problems might occur, literature data minimize these complications, demonstrating that cell lines may provide genetic proxies for primary tumors in many cancer types (55,56). Concerning MYC/MYCN amplifications, their persistence in culture has been documented (57–59) and, indeed, many relevant results, like the discovery of the chromothripsis phenomenon, were achieved using established cell lines (8). Moreover, the MYCN amplicon structure we documented in neuroblastoma and SCLC cell lines (5) turned out to be perfectly similar to what reported by other groups on fresh cancer material using SNP array (60,61). It is also worth noting that the only two available pieces of information on MYC or MYCN hsr insertion sites were achieved on cell lines (31,33). Thus, we think that the dmin/hsr described here have not been altered by the culture conditions, and these cell lines, for what concerns the amplicon structure, exactly resemble the primary tumor tissue they derive from.
CONCLUDING REMARKS
Thanks to an integrated approach combining NGS, SNP array, FISH and PCR-based techniques, we were able to overcome some of the inherent limitations of NGS in disentangle amplicon heterogeneity, gathering a complete view of the chromosomal rearrangements occurring in the amplicons of the seven tumor cell lines under investigation. Our results exclude chromothripsis as the driving force behind the genesis of dmin and hsr carrying heterogeneous amplicons. Therefore, we believe that a multi-step evolutionary process starting from single-chromosome ancestral amplicons could better explain the involvement of more than two chromosomes in the amplification process. In this view, other already known mechanisms, such as the episome model, should be reconsidered as still valid to describe amplicon emergence in the genesis of dmin and hsr. Furthermore, we confirmed the role of PVT1 as a breakpoint hotspot in MYC amplification, and reported EYA1, AKT3 and RUNX1 as novel target genes of amplification in SCLC.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Acknowledgments
We thank Dr Giulia Daniele, Ms Santina Venuto, Ms Grazia Marsico and Ms. Raffaella Stallone for technical assistance.
Footnotes
The authors wish it to be known that, in their opinion, the first two authors should be regarded as joint First Authors.
FUNDING
Italian Association on Cancer Research [My First AIRC Grant number 11405 to C.T.S]; National Institutes of Health [P30 AA019355 and P51 RR000163 to L.C.].
Conflict of interest statement. None declared.
REFERENCES
- 1.Mitelman F., Johansson B., Mertens F. 2014. Mitelman Database of Chromosome Aberrations and Gene Fusions in Cancer. http://cgap.nci.nih.gov/Chromosomes/Mitelman. [Google Scholar]
- 2.Vukovic B., Beheshti B., Park P., Lim G., Bayani J., Zielenska M., Squire J.A. Correlating breakage-fusion-bridge events with the overall chromosomal instability and in vitro karyotype evolution in prostate cancer. Cytogenet. Genome Res. 2007;116:1–11. doi: 10.1159/000097411. [DOI] [PubMed] [Google Scholar]
- 3.Carroll S.M., DeRose M.L., Gaudray P., Moore C.M., Needham-Vandevanter D.R., Von Hoff D.D., Wahl G.M. Double minute chromosomes can be produced from precursors derived from a chromosomal deletion. Mol. Cell. Biol. 1988;8:1525–1533. doi: 10.1128/mcb.8.4.1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Storlazzi C.T., Fioretos T., Surace C., Lonoce A., Mastrorilli A., Strombeck B., D'Addabbo P., Iacovelli F., Minervini C., Aventin A., et al. MYC-containing double minutes in hematologic malignancies: evidence in favor of the episome model and exclusion of MYC as the target gene. Hum. Mol. Genet. 2006;15:933–942. doi: 10.1093/hmg/ddl010. [DOI] [PubMed] [Google Scholar]
- 5.Storlazzi C.T., Lonoce A., Guastadisegni M.C., Trombetta D., D'Addabbo P., Daniele G., L'Abbate A., Macchia G., Surace C., Kok K., et al. Gene amplification as double minutes or homogeneously staining regions in solid tumors: origin and structure. Genome Res. 2010;20:1198–1206. doi: 10.1101/gr.106252.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gibaud A., Vogt N., Hadj-Hamou N.S., Meyniel J.P., Hupe P., Debatisse M., Malfoy B. Extrachromosomal amplification mechanisms in a glioma with amplified sequences from multiple chromosome loci. Hum. Mol. Genet. 2010;19:1276–1285. doi: 10.1093/hmg/ddq004. [DOI] [PubMed] [Google Scholar]
- 7.Korbel J.O., Campbell P.J. Criteria for inference of chromothripsis in cancer genomes. Cell. 2013;152:1226–1236. doi: 10.1016/j.cell.2013.02.023. [DOI] [PubMed] [Google Scholar]
- 8.Stephens P.J., Greenman C.D., Fu B., Yang F., Bignell G.R., Mudie L.J., Pleasance E.D., Lau K.W., Beare D., Stebbings L.A., et al. Massive genomic rearrangement acquired in a single catastrophic event during cancer development. Cell. 2011;144:27–40. doi: 10.1016/j.cell.2010.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rausch T., Jones D.T., Zapatka M., Stutz A.M., Zichner T., Weischenfeldt J., Jager N., Remke M., Shih D., Northcott P.A., et al. Genome sequencing of pediatric medulloblastoma links catastrophic DNA rearrangements with TP53 mutations. Cell. 2012;148:59–71. doi: 10.1016/j.cell.2011.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Leij L., Postmus P.E., Buys C.H., Elema J.D., Ramaekers F., Poppema S., Brouwer M., van der Veen A.Y., Mesander G., The T.H. Characterization of three new variant type cell lines derived from small cell carcinoma of the lung. Cancer Res. 1985;45:6024–6033. [PubMed] [Google Scholar]
- 11.Zijlstra J.G., de Vries E.G., Mulder N.H. Multifactorial drug resistance in an adriamycin-resistant human small cell lung carcinoma cell line. Cancer Res. 1987;47:1780–1784. [PubMed] [Google Scholar]
- 12.Gallagher R., Collins S., Trujillo J., McCredie K., Ahearn M., Tsai S., Metzgar R., Aulakh G., Ting R., Ruscetti F., et al. Characterization of the continuous, differentiating myeloid cell line (HL-60) from a patient with acute promyelocytic leukemia. Blood. 1979;54:713–733. [PubMed] [Google Scholar]
- 13.Quinn L.A., Moore G.E., Morgan R.T., Woods L.K. Cell lines from human colon carcinoma with unusual cell products, double minutes, and homogeneously staining regions. Cancer Res. 1979;39:4914–4924. [PubMed] [Google Scholar]
- 14.Lin C.C., Alitalo K., Schwab M., George D., Varmus H.E., Bishop J.M. Evolution of karyotypic abnormalities and C-MYC oncogene amplification in human colonic carcinoma cell lines. Chromosoma. 1985;92:11–15. doi: 10.1007/BF00327240. [DOI] [PubMed] [Google Scholar]
- 15.Rausch T., Zichner T., Schlattl A., Stutz A.M., Benes V., Korbel J.O. DELLY: structural variant discovery by integrated paired-end and split-read analysis. Bioinformatics. 2012;28:i333–i339. doi: 10.1093/bioinformatics/bts378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen K., Wallis J.W., McLellan M.D., Larson D.E., Kalicki J.M., Pohl C.S., McGrath S.D., Wendl M.C., Zhang Q., Locke D.P., et al. BreakDancer: an algorithm for high-resolution mapping of genomic structural variation. Nat. Methods. 2009;6:677–681. doi: 10.1038/nmeth.1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Quinlan A.R., Clark R.A., Sokolova S., Leibowitz M.L., Zhang Y., Hurles M.E., Mell J.C., Hall I.M. Genome-wide mapping and assembly of structural variant breakpoints in the mouse genome. Genome Res. 2010;20:623–635. doi: 10.1101/gr.102970.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sindi S., Helman E., Bashir A., Raphael B.J. A geometric approach for classification and comparison of structural variants. Bioinformatics. 2009;25:i222–i230. doi: 10.1093/bioinformatics/btp208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Quinlan A.R., Hall I.M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Abecasis G.R., Auton A., Brooks L.D., DePristo M.A., Durbin R.M., Handsaker R.E., Kang H.M., Marth G.T., McVean G.A. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vandesompele J., De Preter K., Pattyn F., Poppe B., Van Roy N., De Paepe A., Speleman F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-7-research0034. RESEARCH0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pfaffl M.W., Horgan G.W., Dempfle L. Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 2002;30:e36. doi: 10.1093/nar/30.9.e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schwab M., Klempnauer K.H., Alitalo K., Varmus H., Bishop M. Rearrangement at the 5’ end of amplified c-myc in human COLO 320 cells is associated with abnormal transcription. Mol. Cell. Biol. 1986;6:2752–2755. doi: 10.1128/mcb.6.7.2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gajduskova P., Snijders A.M., Kwek S., Roydasgupta R., Fridlyand J., Tokuyasu T., Pinkel D., Albertson D.G. Genome position and gene amplification. Genome Biol. 2007;8:R120. doi: 10.1186/gb-2007-8-6-r120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vogt N., Lefevre S.H., Apiou F., Dutrillaux A.M., Cor A., Leuraud P., Poupon M.F., Dutrillaux B., Debatisse M., Malfoy B. Molecular structure of double-minute chromosomes bearing amplified copies of the epidermal growth factor receptor gene in gliomas. Proc. Natl Acad. Sci. U.S.A. 2004;101:11368–11373. doi: 10.1073/pnas.0402979101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Iwakawa R., Takenaka M., Kohno T., Shimada Y., Totoki Y., Shibata T., Tsuta K., Nishikawa R., Noguchi M., Sato-Otsubo A., et al. Genome-wide identification of genes with amplification and/or fusion in small cell lung cancer. Genes Chromosomes Cancer. 2013;52:802–816. doi: 10.1002/gcc.22076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Northcott P.A., Shih D.J., Peacock J., Garzia L., Morrissy A.S., Zichner T., Stutz A.M., Korshunov A., Reimand J., Schumacher S.E., et al. Subgroup-specific structural variation across 1,000 medulloblastoma genomes. Nature. 2012;488:49–56. doi: 10.1038/nature11327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moore J.K., Haber J.E. Cell cycle and genetic requirements of two pathways of nonhomologous end-joining repair of double-strand breaks in Saccharomyces cerevisiae. Mol. Cell. Biol. 1996;16:2164–2173. doi: 10.1128/mcb.16.5.2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hastings P.J., Ira G., Lupski J.R. A microhomology-mediated break-induced replication model for the origin of human copy number variation. PLoS Genet. 2009;5:e1000327. doi: 10.1371/journal.pgen.1000327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sanborn J.Z., Salama S.R., Grifford M., Brennan C.W., Mikkelsen T., Jhanwar S., Katzman S., Chin L., Haussler D. Double minute chromosomes in glioblastoma multiforme are revealed by precise reconstruction of oncogenic amplicons. Cancer Res. 2013;73:6036–6045. doi: 10.1158/0008-5472.CAN-13-0186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gibaud A., Vogt N., Brison O., Debatisse M., Malfoy B. Characterization at nucleotide resolution of the homogeneously staining region sites of insertion in two cancer cell lines. Nucleic Acids Res. 2013;41:8210–8219. doi: 10.1093/nar/gkt566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Harada S., Sekiguchi N., Shimizu N. Amplification of a plasmid bearing a mammalian replication initiation region in chromosomal and extrachromosomal contexts. Nucleic Acids Res. 2011;39:958–969. doi: 10.1093/nar/gkq882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Campbell P.J., Stephens P.J., Pleasance E.D., O'Meara S., Li H., Santarius T., Stebbings L.A., Leroy C., Edkins S., Hardy C., et al. Identification of somatically acquired rearrangements in cancer using genome-wide massively parallel paired-end sequencing. Nat. Genet. 2008;40:722–729. doi: 10.1038/ng.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shimizu N., Shingaki K., Kaneko-Sasaguri Y., Hashizume T., Kanda T. When, where and how the bridge breaks: anaphase bridge breakage plays a crucial role in gene amplification and HSR generation. Exp. Cell Res. 2005;302:233–243. doi: 10.1016/j.yexcr.2004.09.001. [DOI] [PubMed] [Google Scholar]
- 35.Arlt M.F., Mulle J.G., Schaibley V.M., Ragland R.L., Durkin S.G., Warren S.T., Glover T.W. Replication stress induces genome-wide copy number changes in human cells that resemble polymorphic and pathogenic variants. Am. J. Hum. Genet. 2009;84:339–350. doi: 10.1016/j.ajhg.2009.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mizuno K., Miyabe I., Schalbetter S.A., Carr A.M., Murray J.M. Recombination-restarted replication makes inverted chromosome fusions at inverted repeats. Nature. 2013;493:246–249. doi: 10.1038/nature11676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Blumrich A., Zapatka M., Brueckner L.M., Zheglo D., Schwab M., Savelyeva L. The FRA2C common fragile site maps to the borders of MYCN amplicons in neuroblastoma and is associated with gross chromosomal rearrangements in different cancers. Hum. Mol. Genet. 2011;20:1488–1501. doi: 10.1093/hmg/ddr027. [DOI] [PubMed] [Google Scholar]
- 38.Mrasek K., Schoder C., Teichmann A.C., Behr K., Franze B., Wilhelm K., Blaurock N., Claussen U., Liehr T., Weise A. Global screening and extended nomenclature for 230 aphidicolin-inducible fragile sites, including 61 yet unreported ones. Int. J. Oncol. 2010;36:929–940. doi: 10.3892/ijo_00000572. [DOI] [PubMed] [Google Scholar]
- 39.Network., C.G.A. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Storlazzi C.T., Fioretos T., Paulsson K., Strombeck B., Lassen C., Ahlgren T., Juliusson G., Mitelman F., Rocchi M., Johansson B. Identification of a commonly amplified 4.3 Mb region with overexpression of C8FW, but not MYC in MYC-containing double minutes in myeloid malignancies. Hum. Mol. Genet. 2004;13:1479–1485. doi: 10.1093/hmg/ddh164. [DOI] [PubMed] [Google Scholar]
- 41.Ji W., Bian Z., Yu Y., Yuan C., Liu Y., Yu L., Li C., Zhu J., Jia X., Guan R., et al. Expulsion of micronuclei containing amplified genes contributes to a decrease in double minute chromosomes from malignant tumor cells. Int. J. Cancer. 2014;134:1279–1288. doi: 10.1002/ijc.28467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Blyth K., Cameron E.R., Neil J.C. The RUNX genes: gain or loss of function in cancer. Nat. Rev. Cancer. 2005;5:376–387. doi: 10.1038/nrc1607. [DOI] [PubMed] [Google Scholar]
- 43.Sun J., Karoulia Z., Wong E.Y., Ahmed M., Itoh K., Xu P.X. The phosphatase-transcription activator EYA1 is targeted by anaphase-promoting complex/Cdh1 for degradation at M-to-G1 transition. Mol. Cell. Biol. 2013;33:927–936. doi: 10.1128/MCB.01516-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Davies M.A. Regulation, role, and targeting of Akt in cancer. J. Clin. Oncol. 2011;29:4715–4717. doi: 10.1200/JCO.2011.37.4751. [DOI] [PubMed] [Google Scholar]
- 45.Feldman A.L., Dogan A., Smith D.I., Law M.E., Ansell S.M., Johnson S.H., Porcher J.C., Ozsan N., Wieben E.D., Eckloff B.W., et al. Discovery of recurrent t(6;7)(p25.3;q32.3) translocations in ALK-negative anaplastic large cell lymphomas by massively parallel genomic sequencing. Blood. 2011;117:915–919. doi: 10.1182/blood-2010-08-303305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bernard-Pierrot I., Gruel N., Stransky N., Vincent-Salomon A., Reyal F., Raynal V., Vallot C., Pierron G., Radvanyi F., Delattre O. Characterization of the recurrent 8p11–12 amplicon identifies PPAPDC1B, a phosphatase protein, as a new therapeutic target in breast cancer. Cancer Res. 2008;68:7165–7175. doi: 10.1158/0008-5472.CAN-08-1360. [DOI] [PubMed] [Google Scholar]
- 47.Lee W., Jiang Z., Liu J., Haverty P.M., Guan Y., Stinson J., Yue P., Zhang Y., Pant K.P., Bhatt D., et al. The mutation spectrum revealed by paired genome sequences from a lung cancer patient. Nature. 2010;465:473–477. doi: 10.1038/nature09004. [DOI] [PubMed] [Google Scholar]
- 48.Pleasance E.D., Stephens P.J., O'Meara S., McBride D.J., Meynert A., Jones D., Lin M.L., Beare D., Lau K.W., Greenman C., et al. A small-cell lung cancer genome with complex signatures of tobacco exposure. Nature. 2010;463:184–190. doi: 10.1038/nature08629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nagoshi H., Taki T., Hanamura I., Nitta M., Otsuki T., Nishida K., Okuda K., Sakamoto N., Kobayashi S., Yamamoto-Sugitani M., et al. Frequent PVT1 rearrangement and novel chimeric genes PVT1-NBEA and PVT1-WWOX occur in multiple myeloma with 8q24 abnormality. Cancer Res. 2012;72:4954–4962. doi: 10.1158/0008-5472.CAN-12-0213. [DOI] [PubMed] [Google Scholar]
- 50.Kim H.P., Cho G.A., Han S.W., Shin J.Y., Jeong E.G., Song S.H., Lee W.C., Lee K.H., Bang D., Seo J.S. Novel fusion transcripts in human gastric cancer revealed by transcriptome analysis. Oncogene. 2013 doi: 10.1038/onc.2013.490. doi:10.1038/onc.2013.490. [DOI] [PubMed] [Google Scholar]
- 51.Borg A., Baldetorp B., Ferno M., Olsson H., Sigurdsson H. c-myc amplification is an independent prognostic factor in postmenopausal breast cancer. Int. J. Cancer. 1992;51:687–691. doi: 10.1002/ijc.2910510504. [DOI] [PubMed] [Google Scholar]
- 52.You L., Chang D., Du H.Z., Zhao Y.P. Genome-wide screen identifies PVT1 as a regulator of Gemcitabine sensitivity in human pancreatic cancer cells. Biochem. Biophys. Res. Commun. 2011;407:1–6. doi: 10.1016/j.bbrc.2011.02.027. [DOI] [PubMed] [Google Scholar]
- 53.Xu P.X., Cheng J., Epstein J.A., Maas R.L. Mouse Eya genes are expressed during limb tendon development and encode a transcriptional activation function. Proc. Natl Acad. Sci. U.S.A. 1997;94:11974–11979. doi: 10.1073/pnas.94.22.11974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kim D., Sun M., He L., Zhou Q.H., Chen J., Sun X.M., Bepler G., Sebti S.M., Cheng J.Q. A small molecule inhibits Akt through direct binding to Akt and preventing Akt membrane translocation. J. Biol. Chem. 2010;285:8383–8394. doi: 10.1074/jbc.M109.094060. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 55.Wilding J.L., Bodmer W.F. Cancer cell lines for drug discovery and development. Cancer Res. 2014;74:2377–2384. doi: 10.1158/0008-5472.CAN-13-2971. [DOI] [PubMed] [Google Scholar]
- 56.Barretina J., Caponigro G., Stransky N., Venkatesan K., Margolin A.A., Kim S., Wilson C.J., Lehar J., Kryukov G.V., Sonkin D., et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483:603–607. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Leal M.F., Calcagno D.Q., Borges da Costa Jde F., Silva T.C., Khayat A.S., Chen E.S., Assumpcao P.P., de Arruda Cardoso Smith M., Burbano R.R. MYC, TP53, and chromosome 17 copy-number alterations in multiple gastric cancer cell lines and in their parental primary tumors. J. Biomed. Biotechnol. 2011 doi: 10.1155/2011/631268. doi:10.1155/2011/631268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sos M.L., Dietlein F., Peifer M., Schottle J., Balke-Want H., Muller C., Koker M., Richters A., Heynck S., Malchers F., et al. A framework for identification of actionable cancer genome dependencies in small cell lung cancer. Proc. Natl Acad. Sci. U.S.A. 2012;109:17034–17039. doi: 10.1073/pnas.1207310109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Solomon D.A., Kim J.S., Ressom H.W., Sibenaller Z., Ryken T., Jean W., Bigner D., Yan H., Waldman T. Sample type bias in the analysis of cancer genomes. Cancer Res. 2009;69:5630–5633. doi: 10.1158/0008-5472.CAN-09-1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Caren H., Erichsen J., Olsson L., Enerback C., Sjoberg R.M., Abrahamsson J., Kogner P., Martinsson T. High-resolution array copy number analyses for detection of deletion, gain, amplification and copy-neutral LOH in primary neuroblastoma tumors: four cases of homozygous deletions of the CDKN2A gene. BMC Genomics. 2008;9:353. doi: 10.1186/1471-2164-9-353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fix A., Lucchesi C., Ribeiro A., Lequin D., Pierron G., Schleiermacher G., Delattre O., Janoueix-Lerosey I. Characterization of amplicons in neuroblastoma: high-resolution mapping using DNA microarrays, relationship with outcome, and identification of overexpressed genes. Genes Chromosomes Cancer. 2008;47:819–834. doi: 10.1002/gcc.20583. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


