Abstract
The transition between initiation and productive elongation during RNA Polymerase II (Pol II) transcription is a well-appreciated point of regulation across many eukaryotes. Elongating Pol II is modified by phosphorylation of serine 2 (Ser2) on its carboxy terminal domain (CTD) by two kinases, Bur1/Ctk1 in yeast and Cdk9/Cdk12 in metazoans. Here, we discuss the roles and regulation of these kinases and their relationship to Pol II elongation control, and focus on recent data from work in C. elegans that point out gaps in our current understand of transcription elongation.
Keywords: C. elegans, Bur1, Cdk12, Cdk9, Ctk1, P-TEFb, RNA Polymerase II, Serine 2, elongation, transcription
Introduction
The eukaryote RNA polymerase II (Pol II) holoenzyme is a 12-subunit complex that transcribes protein coding genes and many non-coding RNA (ncRNA) genes. Pol II activity is tightly regulated at three distinct phases: (1) initiation, which includes transcription start site selection, formation of the open complex, and production of the first few phosphodiester bonds in the RNA transcript; (2) elongation, or progression of RNA polymerase through a locus as it lengthens the RNA transcript; and (3) termination, or the release of polymerase when it reaches the end of the gene being transcribed. Recruitment of Pol II to a locus is perhaps the most conceptually straightforward mode of regulating gene expression, and thus Pol II initiation was thought to be the rate-limiting step in gene expression for many years. Indeed, today we understand the regulation of Pol II initiation by the general transcription factors (GTFs) at a detailed molecular level.1,2
During the past several years, biochemical and genomic analyses have revealed that Pol II recruitment is not the only point of transcriptional regulation. Rather, the transition between initiation and elongation is a multi-step process where any point between recruitment and productive elongation can potentially be regulated for proper gene expression in eukaryotes.3,4 Accompanying this transition is the phosphorylation of specific residues on the extended C-terminal domain (CTD) of Pol II. In this review, we will discuss the relationship between Pol II elongation and phosphorylation of one of the CTD residues, Serine 2 (Ser2), across several model systems. Although it is appreciated that this modification correlates with elongation, how Ser2 phosphorylation is regulated in a multi-cellular context and its requirement during elongation is an emerging field of interest. Recent results from studies on tissue-specific regulation of this modification in C. elegans have bearing on these questions, and are informative for how Ser2 phosphorylation, and the kinases that add this modification, are linked to Pol II elongation.
The Pol II CTD
The large subunit of Pol II, Rbp1, contains a repeated motif on the C-terminal end, called the “C terminal domain,” or Pol II CTD.5,6 A highly conserved feature of the CTD is that it is composed of dozens of repeats of the heptapeptide sequence Y1-S2-P3-T4-S5-P6-S7 (for a review see refs. 7 and 8). All residues within the CTD heptad repeat can be modified either by phosphorylation (tyrosine, threonine, serine) or isomerization (proline).9 However, serine 5 and serine 2 phosphorylation (Ser5-P and Ser2-P) are the best studied and appear to be the most conserved general marks of transcription.10 Importantly, it was recognized 14 years ago that these modifications correlate with different steps of the transcription cycle. In 2000, Buratowski and colleagues showed that Pol II phosphorylated on Ser5 is found near the 5′ end of genes and Pol II accumulates phosphorylated Ser2 as it progresses through a gene.11 This has been shown to be a general phenomenon using genome-wide analysis in yeast12 and mammalian cells.13 Thus, the current model is that for the vast majority of protein coding genes, Pol II is recruited to a gene with a hypophosphorylated CTD; the CTD then becomes phosphorylated on Ser5 during initiation, and subsequently phosphorylated on Ser2 during productive elongation (for a review see refs. 14 and 15). This differential phosphorylation of Ser2 and Ser5 functions to recruit transcription-associated proteins at the appropriate phase of the transcription cycle (for a review see refs. 9 and 14), thus acting as an important “docking station” that can be regulated for gene expression.
More recently, additional CTD residues (Tyr1, Thr4, and Ser7) have also been shown to be phosphorylated in vivo.16-20 However, these phosphorylation events either are not generally required for transcription of protein coding genes, or their specific role in transcription is not as yet understood. In particular, Ser7 phosphorylation can be found throughout gene bodies in yeast.12,21-23 However, despite its presence on protein-coding genes, Ser7-P is particularly important for proper transcription of snRNA genes16 and may or may not be required for transcription or processing of mRNA genes.16,17 Thus, because Ser2 phosphorylation has the tightest association with transcription elongation regulation, we focus our discussion on the kinases that carry out this modification. More complete reviews of the additional CTD modifications are available elsewhere.9,19
Before discussing Ser2 kinases, several caveats of current experimental approaches should be taken into consideration. A large majority of the studies on Pol II CTD kinases have been performed using antibodies thought to be specific for distinct CTD modifications. These studies are limited by the same caveats that plague other antibody studies: variations in specificity under different situations and epitope masking. For example, antibodies thought to recognize specific phosphoepitopes can be significantly influenced by the status of phosphorylation on neighboring residues.17,24 This can produce dramatic differences in the conclusions drawn about the status of specific CTD phosphoepitopes within a gene.21 Additionally, as both immunofluorescence and immunoprecipitation experiments often require fixation of samples, it is possible that crosslinking between the CTD and CTD-binding proteins may mask the accessibility of the CTD to antibodies.12 Thus, low signal from antibody studies may not arise from the lack of a CTD modification, but from masking of the epitope by a cross-linked protein that itself associates with the epitope in the cell. Finally, studies of the Pol II CTD are particularly challenging to interpret because of the repetitive nature of the CTD. Studies using antibodies against CTD modifications cannot inform how many heptad repeats are modified or which repeats on the CTD are modified. Thus, increases in signal could indicate that more Pol II molecules are phosphorylated, that more repeats of the CTD on one Pol II molecule are modified, or simply that the epitope is more accessible. Additionally, it should be noted that not all of the repeats on the CTD from any eukaryote have the conserved heptad sequence, and many eukaryotes harbor Pol II CTDs that vary quite dramatically from the consensus repeat.17 As an illustration, many of the yeast CTD heptads match the consensus,8 while only two of the Drosophila heptads perfectly match the consensus25 (for a review, see refs. 8 and 10). The functional differences between these distinct repeats are not well understood; however, it should be noted these different heptads can also influence antibody studies.
Positive Elongation Factors: Pol II CTD Serine 2 Kinases
Eukaryotes have two kinases that phosphorylate Ser2: Bur1/CDK9 and Ctk1/Lsk1/CDK12 (Table 1). Although initially thought to have distinct functions in yeast and mammals, recent studies have uncovered a common mode of Ser2 phosphorylation regulation across eukaryotes by these kinases (described below, Fig. 1B and C).
Table 1. RNA Polymerase II elongation factor homologs.
| S. cerevisae | S. pombe | C. elegans | Drosophila | Mammals | ||
|---|---|---|---|---|---|---|
| 5′ Ser2 | kinase | Bur1 | Cdk9 | CDK-9 | CDK9 | CDK9/PITARE |
| cyclin(s) | Bur2 | Pch1 | CIT-1.1 | CYCT | CCNT1 | |
| CIT-1.2 | CCNT2a/b | |||||
| 3′ Ser2 | kinase(s) | Ctk1 | Lsk1 | CDK-12 | CDK12 | CDK12/ CrkRS |
| CDK13/CDC2L5 | ||||||
| cyclin(s) | Ctk2 | Lsc1 | CCNK-1 | CYCK | CCNK | |
| Ctk3 | Lsg1 | * | * | * | ||
| DSIF | Spt4 | Spt4 | SPT-4 | SPT4/p160 | SPT4/p160 | |
| Spt5 | Spt5 | SPT-5 | SPT5/p14 | SPT5/p14 | ||
| NELF | * | * | * | NELF-A | NELFA/WHSC2 | |
| * | * | * | NELF-B | NELFB/COBRA1 | ||
| * | * | * | NELF-D/TH1 | NELFC/NELFD/ TH1/HSPC130 | ||
| * | * | * | NELF-E | NELFE/RD† | ||
*no significant homolog detected. †homologs detected in lower eukaryotes contain an RRM similar to that found in NELFE but do not appear to be functional homologs
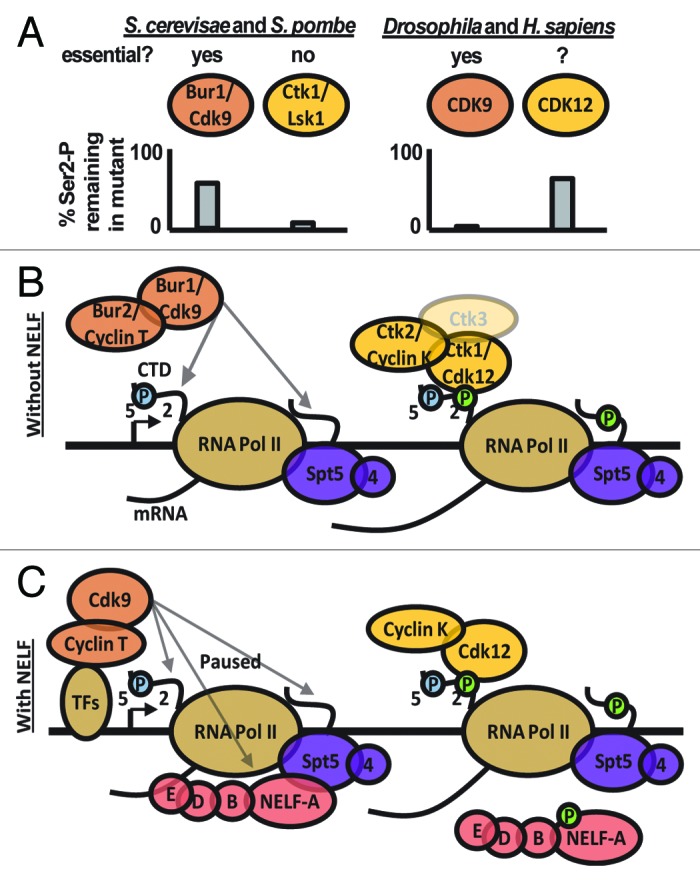
Figure 1. Ser2 kinases in eukaryotic transcriptional elongation regulation. (A) Loss of Bur1/CDK9 or Ctk1/Lsk1/CDK12 activity results in different effects on Ser2-P levels in yeast vs. metazoans. While Ctk1/Lsk1 are responsible for the majority of Ser2-P in yeast S. cerevisae and S. pombe, both CDK9 and CDK12 significantly contribute to Ser2-P levels in Drosophila and huamans. Importantly, loss of Cdk9 in metazoans results in complete loss of Ser2-P suggesting it is required upstream of CDK12 activity. Finally, while Bur1 and CDK9 are largely essential proteins (S. cerevisae bur1 mutants are extremely slow growing), Ctk1/Lsk1 is not essential in yeast. (B) Transcription elongation is not a rate-limiting step of gene expression in organisms without NELF homologs, including yeast. In these organisms, the kinases Bur1/CDK9 and Ctk1/CDK12 are recruited through general mechanisms to phosphorylate Ser2 of the Pol II CTD and SPT5. These phosphorylation marks enhance recruitment of other Pol II elongation associated factors such as RNA processing factors and chromatin remodelers. (C) In organisms that contain the NELF complex, such as Drosophila and mammals, the transition from Pol II initiation to elongation is a regulated step of gene expression. Here, DSIF (SPT4/SPT5) mediates NELF binding to the Pol II complex, “pausing” Pol II and preventing productive elongation. Regulated recruitment of CDK9 and Cyclin T results in the release of this pause. CDK9 phosphorylates the Pol II CTD on Ser2, SPT5, and NELF. Downstream of this regulated step, the CDK12 complex further phosphorylates the Pol II CTD.
The yeast Ser2 kinases: Ctk1/Lsk1 and Bur1/Cdk9
The first CTD Ser2 kinase was identified in biochemical studies in budding yeast.26 When cloned, this kinase showed homology to the cyclin dependent kinases and was named carboxy terminal kinase 1, Ctk1.27 Ctk1 forms a complex with its cyclin partner, Ctk2, and Ctk3, which is important for stability of the complex.28,29 The other budding yeast Ser2 kinase complex components, Bur1 and Bur2, were identified in a screen for factors that could bypass the need for upstream activating sequences (UAS) of a reporter gene (BUR, bypass UAS requirement30). These proteins were also found to comprise a cyclin dependent kinase complex in which Bur1 is the kinase and Bur2 is the cyclin.31 While genetic studies linked Bur1 activity with phosphorylation of the CTD,32 it was not shown to be a direct CTD kinase in vivo until several years later.33,34
Interestingly, although Ctk1 is considered the major Ser2 kinase in S. cerevisiae, it is not an essential protein. Null mutants for Ctk1 complex components are viable and healthy at normal temperatures, but show cold sensitive phenotypes.28 In contrast, Bur1 is important for normal growth as both bur1 and bur2 null mutants are extremely slow growing.31,35 Similarly, in fission yeast, mutants of the Ctk1 homolog, Lsk1, are also viable under normal growth conditions,36 while mutants of the Bur1 homolog, Cdk9, are lethal37 (Fig. 1A). This suggests that the activity of Bur1 is more important for yeast growth under normal conditions than the major Ser2 kinase, Ctk1. This could be due to its role in phosphorylating other sites on the CTD such as Ser538,39 and Ser722 or other proteins besides Pol II including the elongation factor Spt5.39,40
Bur1/Cdk9 and Ctk1/Lsk1 regulate yeast Ser2 phosphorylation together
Recent studies using analog sensitive mutants of Ctk1 and Bur1 established an epistatic relationship between these factors.33 This work showed that loss of Bur1 activity results in dramatic decreases in Ser2 phosphorylation.33 Additionally, the authors showed that the trace amounts (~10%) of Ser2-P detected in a ctk1 deletion strain are lost with Bur1 inhibition.33 Thus, while Ctk1 is responsible for the majority of Ser2-P, loss of Bur1 activity also shows significant decreases in Ser2-P. Significantly, a ctk1 mutant shows more dramatic decreases in Ser2-P at the 3′ end of a gene than the 5′ end.33 Thus, the authors suggested that Bur1 acts on Pol II at the 5′ end of genes, and this phosphorylation enhances phosphorylation by Ctk1 further downstream in the gene (Fig. 1A and B).
Fission yeast regulate Ser2-P in a similar fashion. While an overwhelming majority of detectable Ser2-P is absent with loss of activity of the Ctk1 homolog, Lsk1,36,41 recent data using analog sensitive mutants has also shown that fission yeast Cdk9 is responsible for some level of Ser2-P.41-43
Regulation of yeast Ser2 kinases
Mirroring their roles in Ser2 phosphorylation, both Ctk1 and Bur1 are associated with elongating Pol II by chromatin immunoprecipitation (ChIP),32,44,45 and this has been shown genome wide for Ctk1.46 In yeast, these kinases appear to be recruited to loci through general mechanisms, rather than gene-specific factors. Specifically, Bur1 contains a phospho-CTD interaction domain, which interacts with Ser5-P to enhance Bur1 recruitment to genes; Ctk1 is proposed to subsequently interact with Pol II through the Bur1-phosphorylated CTD.33 Because the factors involved in Ser2 kinase activity identified thus far in yeast are not gene-specific regulators, recruitment of Ser2 kinases may not play a role in the regulation of Pol II elongation, unlike in higher eukaryotes (discussed below). Instead, these kinases seem to act largely as general factors that mark elongating Pol II for the recruitment of other factors at the proper stage of transcription. Therefore, regulation of this process at individual genes may have evolved as a process in multi-cellular organisms to control spatial and temporal expression during development.
The metazoan Ser2 kinases: CDK9 and CDK12
Metazoans have well-conserved homologs of Bur1 (CDK9) and Ctk1 (CDK12). Until recently,47 CDK9 was thought to be the sole Ser2 kinase in metazoans. When in a complex with its cyclin partner, Cyclin T, CDK9 is referred to as positive transcription elongation factor b, or P-TEFb, as it was first identified as having an ability to enhance Pol II elongation in vitro.48 CDK9 is a well-studied protein, in part because of early associations of CDK9 function with HIV infection (for a review see ref. 49) and more recently because of the role of CDK9 in the regulation of Pol II elongation (discussed below). However, the role of the metazoan Ctk1 homolog, CDK12, in Ser2 phosphorylation has been more recently appreciated.
Identification of CDK9
The P-TEFb complex, CDK9/Cyclin T, was identified as a factor that positively regulates transcriptional activity50 and phosphorylates the CTD48,51 in Drosophila and mammalian systems (for a review see ref. 49). Although these early studies established the CTD as the major target of CDK9 within Pol II,52,53 there have been conflicting reports about the specific residues targeted by CDK9. In vitro studies suggest CDK9 may phosphorylate Ser5, Ser2, and Ser7.52,54-57 In contrast, in vivo studies that have shown that Ser2 phosphorylation is most affected when CDK9 is inactivated.58-62
CDK9 activity is tightly regulated, and in higher eukaryotes it is largely contained within multi-protein regulatory complexes that have been extensively characterized in Drosophila, mouse, and human. These complexes have been discussed in previous reviews (see below). Briefly, P-TEFb is negatively regulated by the 7SK/HEXIM complex (for a review see refs. 63,64). Additionally, P-TEFb on active genes is found in a large complex of other elongation factors, called the “Super Elongation Complex,” or SEC65-70 (for a review see refs. 64, 71, and 72). Current models suggest that for P-TEFb to enhance elongation, it must be actively recruited to loci by other factors such as the Mediator proteins, CDK8,73,74 Med26,75 the bromodomain protein, Brd4,76,77 splicing factors,78 or by transcription factors, deemed “P-TEFb facilitators” (for a review see refs. 79 and 80). Some of these factors not only influence P-TEFb activity but also help release P-TEFb from the inhibitory 7SK/HEXIM complex (for a review see ref. 81).
It should be noted that Brd4 has also been identified as a Ser2 kinase in human cells;82 however, its role in the transcription cycle is unclear10 as it also regulates CDK9 activity.83 Furthermore, Brd4 is not present in lower eukaryotes.
CDK12/Cyclin K: the recently appreciated CTD kinase complex
The role of CDK12 in CTD phosphorylation was demonstrated in 2010 when Greenleaf and colleagues showed that it phosphorylates Ser2 in vitro and in vivo.47 ChIP studies revealed that CDK12 is enriched toward the 3′ end of genes, which correlates with the 3′ enrichment of Ser2-P. In addition, the authors showed that a S. cerevisiae Bur1 chimera with the CDK9 kinase domain rescues a bur1 deletion while a Ctk1/CDK12 kinase domain chimera rescues a ctk1 mutant, illustrating conservation of function.47 Based on these observations, as well as the fact that inhibition of CDK9 activity frequently reduces all detectable Ser2-P, Greenleaf and colleagues proposed a model for metazoan Ser2-P regulation that more closely matches Ser2-P regulation in yeast: CDK9 phosphorylates Ser2-P at the 5′ end of genes and this activity is upstream of and required for the CDK12-mediated Ser2-P toward the 3′ end of genes84 (discussed below, Fig. 1A and C). Therefore in both yeast and metazoans, Bur1/CDK9 acts upstream, both physically within the ORF and epistatically, of Ctk1/CDK12 to regulate Ser2 phosphorylation.
This model of Ser2 phosphorylation in higher eukaryotes is particularly attractive as it places a Ser2 kinase, CDK12, in the location of where the majority of Ser2-P is located on a gene, at the 3′ end. In many studies, CDK9 is largely located toward the 5′ end of genes85-87 while the majority of Ser2-P is found to increase toward the 3′ end of genes. While this low Ser2-P signal at CDK9 binding sites could be due to antibody detection issues discussed above, or the activity of a the Ser2 phosphatase FCP1,88 the identification of CDK12 as a Ser2 kinase helps to resolve this apparent paradox.
Recent studies have showed that CDK12/Cyclin K is important for the expression of long genes with a large number of exons.89 In particular, DNA damage response genes are down-regulated in cells with reduced CDK12 and cells lacking CDK12/Cyclin K accumulate spontaneous DNA damage.89 Regulation of CDK12/Cyclin K levels may also be important for differentiation. While Cyclin K levels are high in pluripotent embryonic stem cells (ESCs), they decrease through differentiation and experimentally decreasing CDK12 levels in ESCs leads to spurious differentiation.90 Thus, CDK12/Cyclin K is an important transcriptional regulator that needs to be more extensively characterized.
Summary of kinase requirements
It now appears that most if not all eukaryotes probably regulate Ser2 phosphorylation in a similar manner: Bur1/Cdk9 phosphorylates Ser2 toward the 5′ end of genes, and this enhances further phosphorylation by Ctk1/Cdk12 (Fig. 1B and C). Although it is satisfying that recent results have resolved two seemingly disparate modes of Ser2 regulation into one conserved model, there is one important difference between yeast and metazoans. In yeast, while Bur1 is important for Ser2 phosphorylation levels, it is not required for Ctk1-mediated phosphorylation (Fig. 1A and B). In contrast, CDK9 is essential for CDK12-mediated CTD phosphorylation in Drosophila and mammalian systems (Fig. 1A and C). The reason for this dissimilarity is due to differences in the regulation of transcriptional elongation between these systems.
Pol II Elongation Control: A CDK-9-Mediated Transition in Metazoans
Pol II pausing is pervasive in some higher eukaryotes
As mentioned in the introduction, the transitions between Pol II recruitment and productive elongation are now recognized as possible points of transcription regulation in many eukaryotes.91 One of the most general and well-documented examples of post-recruitment regulation is the transition between initiation and productive elongation.
Because many early studies of transcription were performed in yeast, where Pol II activity is largely regulated at the level of recruitment (discussed below), this was thought to be the major point of transcription regulation. However, studies in Drosophila and human cells reveal that higher eukaryotes exhibit substantial post-initiation regulation of transcription. Genome-wide analyses showed that at many genes, Pol II localization is highest 25–60 bp downstream of transcription start sites where Pol II “pauses” between early elongation and productive elongation.92-98 This abundance of binding is thought to reflect the fact that Pol II quickly transitions from the pre-initiation complex (PIC) to the paused state at most genes,99 and can be stably positioned at paused sites for several minutes prior to productive elongation.96,98-102 Thus, as opposed to yeast systems, the transition between transcription initiation and productive elongation is regulated in higher metazoans. While there are many ideas about the purpose of Pol II pausing, it is clear that regulating polymerase elongation allows fine-tuning of gene expression (for a discussion about proposed functions of pausing see refs. 4, 80, and 103).
NELF is the established regulator of polymerase pausing
Although the widespread regulation of Pol II elongation was not appreciated until recently, beautiful biochemistry by the Handa and Price labs established a framework for understanding the regulation of early transcription elongation. Specifically, this work uncovered factors that are required to mediate promoter proximal pausing: the DSIF complex (SPT4/SPT5) and the NELF complexes. DSIF interacts directly with Pol II,104 bridging contact between NELF and Pol II.105 Although the molecular details of how NELF negatively regulates Pol II is poorly understood (for a review see refs. 4 and 80), it is clear that NELF is a major mediator of pausing in systems studied to date106-108 (discussed below). Elongation repression is alleviated by P-TEFb,109,110 which, in addition to phosphorylating the Pol II CTD, also phosphorylates SPT5 of the DSIF complex111 and the NELF complex.112 Following this, NELF leaves the elongating Pol II complex while DSIF remains associated.113,114 After dissociation of NELF, DSIF acts to enhance transcription elongation111 (details below). Thus, DSIF may repress elongation simply by acting as a facilitator of NELF-mediated repression (Fig. 1C).
While the NELF complex members are only conserved in higher eukaryotes, including Drosophila and mammals,107 both DSIF components, SPT4 and SPT5, are highly conserved proteins (for a review see refs. 115 and 116). Studies performed in yeast, which lack NELF, suggest that DSIF acts only as a positive elongation factor,117-119 supporting the model that DSIF functions as a negative elongation factor in higher eukaryotes by binding NELF. In yeast, Spt5 helps to recruit transcription-associated factors such as RNA processing factors and chromatin remodelers (for a review see refs. 115 and 120-122). In addition, structural studies have determined that DSIF likely directly enhances Pol II processivity.123-125 Thus, this role of Spt5 in transcriptional regulation likely evolved early, with successive layers of regulation added throughout evolution.
Studies in other organisms also support the correlation between presence of NELF homologs and regulation of Pol II elongation. Pol II pausing has only been extensively studied in Drosophila and mammalian systems; however, NELF homologs are also present many other eukaryotic model systems such as Xenopus and Zebrafish. Genome-wide studies of Pol II in these systems both show a prominent 5′ accumulation of Pol II in metagene analyses.126,127 These studies suggest that elongation is a regulated event in these systems and is likely to be mediated by NELF. In contrast, Arabidopsis does not have obvious NELF homologs, and promoter proximal Pol II pausing has been undetected in this organism.128 In summary, evolutionary conservation of NELF correlates with widespread Pol II pausing in many eukaryotes.64
Pol II elongation is a general step of transcription regulation in higher eukaryotes
The roles of DSIF, NELF, and P-TEFb in pausing were first characterized using biochemical studies on naked DNA templates. This, along with the fact that these factors appear to have consistent effects on the genes tested in vitro and in vivo, led to the model that the transition of Pol II from initiation to elongation is a general point of regulation for all genes, and that these elongation factors are basal transcription factors (Fig. 1C). Genome-wide ChIP studies corroborated this conclusion as these factors are located on a surprisingly large number of genes, both active and inactive.129,130 Indeed, genes bound by DSIF and NELF significantly overlap (> 90%) with those bound by Pol II.97,131,132 Finally, functional data supports this model, as incubation of HeLa cells with flavopiridol, an inhibitor of P-TEFb, reduces the level of transcription to a level strikingly similar to incubation with α−amanitin, a well-studied Pol II inhibitor.133 Thus, P-TEFb-mediated DSIF/NELF release is a generally required and regulated step of transcription in higher eukaryotes. This explains why higher eukaryotes require CDK9 upstream of CDK12: CDK9 is required for the release from NELF-mediated pausing and subsequent elongation-associated phosphorylation of Ser2 by CDK12.
Promoter proximal escape is not generally regulated in yeast
Early studies in yeast established initiation as the major point of transcription regulation,134,135 and this has been confirmed for the vast majority of genes in genome-wide analyses. Pol II is fairly evenly distributed across gene bodies in budding yeast during log phase growth, suggesting there are no major rate-limiting steps in the transition between recruitment and elongation12,136 (for a review see ref. 137). This is also the case for fission yeast, as they also lack a Pol II peak near the TSS in metagene analyses.138
Several other studies have provided more direct evidence for initiation as the major point of transcription regulation in yeast. Although recent genome-wide polymerase run-on sequencing (GRO-seq) studies in budding yeast have shown that the elongation rate can vary across a gene,119 this is not analogous to promoter proximal escape. Instead, this variation may reflect effects on Pol II processivity by the underlying DNA sequence or nucleosome positioning.139 Furthermore, changes in Pol II accumulation on a gene following a stimulus largely reflect changes in mRNA.140 Finally, genes that have initiated transcription (i.e., have Ser5 phosphorylation) highly correlate with genes where elongating Pol II is present (i.e., have Ser2 phosphorylation).12 Thus, the transition between initiation and elongation is not a common barrier to gene expression in yeast. During normal growth, genes that have initiated transcription will generally produce a full-length mRNA product.
The lack of promoter proximal pausing in yeast also correlates with Pol II Ser2 kinase requirements: Pol II elongation does not require Bur1/Cdk9 in yeast. Reducing Bur1 activity does not increase promoter proximal Pol II levels.141 Instead, there is a reduction in both initiated and elongating Pol II.119,141,142 As in budding yeast, the Bur1 homolog, Cdk9 is not required for elongation in fission yeast.143 Thus, while Bur1/Cdk9 phosphorylate the CTD toward the 5′ end of genes, is not required for the transition of Pol II into productive elongation in yeast. This is likely why Bur1/Cdk9 is not required for Ctk1/Lsk1-mediated Ser2 phosphorylation (Fig. 1A and B).
Transcriptional Elongation Regulation in C. elegans
A new facet of Ser2-P regulation and transcription elongation is now emerging in the C. elegans model system. C. elegans has been a unique model for understanding transcription regulation because it is one of the few well-studied models that has trans-splicing of Pol II transcripts. Trans-splicing, which occurs on ~70% of C. elegans genes,144,145 involves the replacement of the 5′ end of transcripts with a short, 21–22 nucleotide RNA sequence, called a splice leader.146 Because trans-splicing occurs on the majority of genes, the TSS for many Pol II genes cannot be mapped from the 5′ sequence of mature mRNA as is commonly done in other organisms.147,148 Thus, although the TSS has been mapped for individual genes in C. elegans,149 genome-wide approaches for TSS identification have only recently been performed.145,147,148,150 These recent results have, for the first time, allowed scientists to analyze the regulation of early steps of Pol II transcription on a global scale. CTD phosphorylation patterns in C. elegans are similar to other metazoans. Genome-wide studies show that Ser5-P shows stronger signal toward the promoter and Ser2 phosphorylation within the body of the genes151,152 (for a review see ref. 153). However, recent studies have identified tissue-specific regulation of Ser2 phosphorylation in C. elegans (for a review of C. elegans germline biology see refs. 154 and 155).
C. elegans Somatic Ser2-P Requires CDK-9 and CDK-12
In C. elegans embryo somatic tissues, Ser2 phosphorylation is regulated similar to other metazoans, i.e., inhibition of CDK-9 activity results in near total loss of detectable Ser2 phosphorylation while inhibition of CDK-12 reduces Ser2 phosphorylation levels by about 60%.58,59 Additionally, CDK-9 is required for bulk transcription in C. elegans somatic tissues as loss of CDK-9 activity causes phenotypes similar to those caused by loss of embyronic RNA Pol II activity, e.g., embryonic death and defective gastrulation.59,156 Furthermore, CDK-9 activity is required for expression of somatic developmental genes in C. elegans embryos.58 Thus, in embryonic somatic tissues, as in other metazoans, it appears that CDK-9 acts upstream of CDK-12 in Ser2 phosphorylation and is required for productive transcription (Fig. 2A).

Figure 2. Ser2 kinases in C. elegans transcriptional elongation regulation. (A) Loss of CDK-9 or CDK-12 activity results in different effects on Ser2-P levels in the C. elegans germline vs. soma (compare with Fig. 1A). While CDK-12 is responsible for the bulk of Ser2-P in the germline, both CDK-9 and CDK-12 regulate Ser2-P levels in the soma. Importantly, loss of CDK-9 in the soma results in complete loss of Ser2-P, suggesting it is required upstream of CDK-12 activity. Finally, while both CDK-9 and CDK-12 are essential for somatic development, only CDK-9 is essential for germline development. (B) As an analogy to eukaryotic organisms that do not have a NELF homolog, transcription elongation may not be a rate-limiting step of gene expression in the germline. Because CDK-9 is essential in the germline, it may target SPT-5 or other factors, though it is not required for phosphorylation of Ser2 on the Pol II CTD. (C) In the soma, the transition from Pol II initiation to elongation can be a regulated step of gene expression. Here, DSIF (SPT4/SPT5) alone or in combination with an additional pausing factor (PF) may regulate a 5′ check point prior to productive Pol II elongation. Regulated recruitment of CDK9 and Cyclin T through transcription factors and possibly phosphorylation of pausing factors may release Pol II from this check point. At the same time, CDK9 phosphorylates the Pol II CTD on Ser2 and may phosphorylate the SPT5 CTR. Downstream of this regulated step, the CDK12 complex further phosphorylates the Pol II CTD.
While regulation of Ser2-P in the C. elegans soma by CDK-9 parallels observations in other metazoans, there are likely distinct differences in the regulation of CDK-9 activity. For example, in higher eukaryotes, CDK9 activity is regulated by large protein complexes that include the inhibitory 7SK/HEXIM and stimulatory SEC complex. C. elegans does not contain obvious homologs for the 7SK/HEXIM components, and thus likely does not sequester CDK-9 in an analogous inhibitory complex. While C. elegans does contain homologs for a subset of the SEC components, specifically ELL1 (Y24D9A.1), AFF9 (Y55B1BR.1 and Y55B1BR.2), and EAF (D1007.16), these factors do not appear to play an essential role in transcription.157 Furthermore, knock down of ELL and AFF9 does not significantly affect either Ser2-P levels or viability (E.A. Bowman, W.G. Kelly, unpublished). Finally, whereas CDK9 activity is regulated by Brd4 and Med26 in higher eukaryotes, there is not a clear Brd4 homolog in C. elegans and Med26 does not affect bulk Ser2-P levels in somatic tissues (EA Bowman, W Kelly, unpublished).
C. elegans elongation regulation
Does C. elegans regulate Pol II promoter proximal escape?
The requirement of CDK-9 for Ser2-P in C. elegans soma suggests that CDK-9 may play a pivotal role in transcriptional elongation in this tissue. As described above, Cdk9 is required for transcription elongation in Drosophila and mammalian systems to counteract Pol II pausing mediated by NELF and DSIF. C. elegans does not have an obvious NELF complex (Table 1); however, the fact that (1) CDK-9 appears to be required for somatic transcription,58,59 (2) cells lacking CDK-9 retain a marker of initiated Pol II, Ser5-P, in the absence of the elongation marker, Ser2-P,59 and (3) Ser2-P in C. elegans somatic tissues is regulated similarly to higher metazoans suggests that Pol II elongation is still critically regulated by CDK-9 in these tissues. Is transcription elongation regulated in C. elegans and does CDK-9/P-TEFb regulate promoter proximal escape of Pol II?
Genome-wide studies show conflicting evidence for elongation regulation in C. elegans. Data from the modENCODE project using C. elegans embryos and larvae suggested that Pol II binds to genes that have low and even undetectable levels of expression.158 This result suggests that Pol II binding at these genes may reflect a paused or stalled polymerase that is regulated for expression later in development.158 A separate analysis of Pol II ChIP data showed a 5′ peak of Pol II binding at both spliced and unspliced genes,150 although this peak can be further downstream of the TSS (100–170 bp) than paused Pol II in Drosophila and mammals (25–60 bp). However, using the current gold standard for identification of Pol II pausing, genome-wide polymerase run-on sequencing (GRO-seq), Meyer and colleagues did not observe evidence of wide-spread Pol II pausing in worms under optimal growth conditions.147
Despite the inconsistent evidence for Pol II pausing in genome-wide analysis and averaging, some reports have suggested that pausing indeed exists at individual genes. Snyder and colleagues identified enriched Pol II promoter proximal binding at 2% of genes analyzed in normally growing embryos and larvae by ChIP-seq (“enriched” defined as 4-fold higher binding near the TSS than within the gene body; about 250 genes).159 Additionally, Meyer and colleagues identified several genes with a Pol II “pausing” profile in embryos and fed larvae by GRO-seq (0.38% in embryos, 15 of 3975 genes; 2.0% in larvae; “pausing” defined as TSS:body signal ratio ≥ 2).147
The strongest evidence that Pol II elongation is regulated in C. elegans comes from studies in starved larvae. Under these conditions, Baugh and colleagues identified an accumulation of Pol II at the 5′ end of a substantial fraction of genes160 (for a review of larval starvation arrest see ref. 161). Most recently, two studies have confirmed that some of these genes with 5′ Pol II accumulation do indeed harbor paused Pol II. As described above, promoter proximal paused Pol II as described in Drosophila is transcriptionally competent rather than arrested or backtracked,96 and remains associated with short capped RNAs (scRNA) instead of terminating.162 Indeed, recent GRO-seq analysis from starved larvae showed that 7.7% of genes analyzed have a 5′ accumulation of Pol II and are transcriptionally competent.147 Furthermore, Baugh and colleagues showed that genes with this 5′ GRO-seq signal also produce scRNAs (short capped RNAs), which extend approximately 30–65 bp downstream of TSSs.163 Furthermore, these scRNAs are longer in worms mutant for TFIIS.163 This lengthening of scRNAs is consistent with Pol II pausing and backtracking seen in Drosophila,162 and makes the case for Pol II pausing in C. elegans even more compelling. Thus, Pol II elongation is indeed regulated for a substantial number of genes in starved C. elegans larvae.
While the release of Pol II from initiation into elongation may not be a ubiquitous rate-limiting step during normal growth in C. elegans, there is strong evidence that a fraction of genes are regulated at this step and are thus “paused.” At this point, it is unclear if Pol II release requires CDK-9 as is the case in Drosophila and mammals; however, as mentioned above CDK-9 is generally required for transcription in somatic tissues (see above). Thus, it seems likely that CDK-9 mediates promoter proximal escape at paused genes and it is attractive to consider that CDK-9 may be a general regulator of transcriptional regulation in C. elegans.
Possible mechanisms for C. elegans promoter proximal escape
Pol II elongation can be regulated during C. elegans transcription, and whether this is a general mechanism at all genes as in Drosophila and mammals, or only important for a small subset of genes, this regulation appears to occur by a mechanism that does not involve NELF. One of the biggest reasons that elongation regulation has been controversial in C. elegans is because it lacks homologs to the NELF complex. Thus, Pol II elongation could be regulated by DSIF activity alone or via a novel, unidentified pausing factor.147 Importantly, C. elegans does not have clear homologs of the recently identified pausing factors Gdown1,164,165 GAGA factor,166 or M1BP.167
Can pausing be mediated by DSIF alone? DSIF is commonly referred to as a negative regulator of elongation; however, it is generally thought to perform this function in concert with NELF (discussed above). In support of a possible role for DSIF-mediated pausing, an early study showed that a heat shock gene, which requires CDK-9 for expression, is re-expressed when both CDK-9 and DSIF are knocked down in the C. elegans embryo.58 However, this may be unique to heat-shock regulation, or there may be additional elongation factors regulating other genes, as the authors did not observe re-expression of developmental genes when both CDK-9 and DSIF were knocked down.58 In summary, if CDK-9 does act to release Pol II for productive elongation in the soma, there is likely to be an additional factor that mediates this step, which may or may not work in combination with DSIF in a mechanism analogous to NELF in higher eukaryotes (Fig. 2C).
Recently, Grishok and colleagues have identified the ZFP-1 (AF10 homolog)/DOT-1 complex as a negative regulator of Pol II elongation in C. elegans.168 Specifically, ZFP-1 mutants cause a decrease in Pol II abundance at the promoter of several C. elegans genes. ZFP-1/DOT-1 regulates elongation by reducing histone ubiquitination that normally promotes transcription. It is unclear if this complex plays a role analogous to the NELF complex in Pol II pausing but it does suggest an interesting mode of elongation regulation through chromatin.4,168 Finally it should be noted that the ZFP-1/DOT-1 complex is not found at all genes that have a predominant 5′ Pol II accumulation, so additional factors are likely involved in this step of Pol II elongation.
Unique regulation of Ser2-P in the C. elegans germline suggests tissue-specific Pol II elongation regulation
Recent studies have uncovered tissue-specific regulation of Ser2-P in C. elegans (for a review of C. elegans germline biology see refs. 154 and 155). In contrast to its essential role in somatic Ser2-P, CDK-9 is not required for the bulk of Ser2 phosphorylation in the germline, which instead requires CDK-12.59,169 While it is unexpected that C. elegans germline Ser2-P does not require CDK-9, this pattern of Ser2-P regulation is surprisingly similar to that seen in yeast (Fig. 1A). As in yeast, Ser2 phosphorylation does not require upstream activity of the Bur1/CDK-9 kinase, but instead is largely dependent on Ctk1/CDK-12 (Fig. 2A). Additionally, in both systems, Bur1/CDK-9 is not required for bulk Ser2 phosphorylation although they are important for normal growth in each cell type. Finally, as in yeast, the Ser2 kinase, CDK-12, is also not essential for germline development in standard laboratory conditions (Fig. 2A).
Overall, Ser2-P regulation in germline tissues matches the regulation seen in yeast while Ser2-P regulation soma matches what is seen in Drosophila and mammalian systems. What is the mechanism for the differential regulation of Ser2 phosphorylation between somatic and germline tissues? As Ser2-P is most tightly associated with transcription elongation, it is appealing to think that tissue-specific Ser2-P regulation reflects differences in transcription elongation control between these two basic tissue types. While it is clear that transcription elongation can be a regulated step in C. elegans transcription (see above), it is enticing to speculate that this regulation is limited to the soma and not the germline. Perhaps somatic tissues require CDK-9 for Ser2-P because CDK-9 activity is required for elongation as in higher metazoans (Fig. 2C). In contrast, germline Ser2-P may not require CDK-9 because (as in yeast) CDK-9 is not required for transcription elongation in the germline (Fig. 2B). While speculative, it is exciting to consider that transcription elongation may be differentially regulated in a tissue-specific manner in some metazoan systems.
Perspective
After an exciting 20 years of research on Pol II CTD phosphorylation and transcription, we are developing a clearer picture of the relationship between Ser2-P and Pol II elongation. As we have uncovered species-specific differences in Pol II regulation, it is also becoming clear that differences in Ser2-P can exist within a species as tissue-specific regulation. Thus, it is important that we begin to dissect mechanisms of Pol II elongation in distinct tissues.
Future tissue-specific studies are likely to be aided by new methods that isolate cell-type specific nuclei. Importantly many previous genome-wide studies of Pol II elongation regulation in metazoans have been performed using mixed cell-types. Based on the recent data suggesting tissue-specific regulation, it is possible that a mixture of cell types masks details of tissue-specific Pol II regulation. Isolation of tissue-specific material, either through flow cytometry, cell-type specific tagging,170 or the recently developed INTACT (isolation of nuclei tagged in specific cell types) method171,172 is likely to have a major impact on the ability to understand tissue-specific phenomenon. This method has already been used to isolate C. elegans muscle nuclei172 and it seems likely that this could be easily adapted to additional somatic tissues or the germline. As we move toward the development of methods that can dissect these events in multicellular organisms, it is likely that we will find unique exceptions to well-established models of transcription. Powerful genetic model systems such as C. elegans will continue to provide this unique insight.
Disclosure of Potential Conflicts of Interest
No potential conflict of interest was disclosed.
Acknowledgments
We would like to thank Dr Karen Adelman, Dr Ryan Baugh, Rebecca Adams, and reviewers for thoughtful comments and helpful discussions during the preparation of this manuscript.
Glossary
Abbreviations:
- Pol II
RNA Polymerase II
- CTD
carboxy terminal domain, referring to the Pol II CTD
- Ser
serine
- Ser2-P
Pol II CTD serine 2 phosphorylation
- Ctk
carboxy terminal kinase, Ctk1/2/3 is a Ser2 kinase complex
- Bur
bypasses upstream requirement, Bur1/2 is a Ser2 kinase complex
- Lsk
latrunculin sensitive kinase, Lsk1 is a Ser2 kinase
- Cdk
cyclin dependent kinase, Cdk9, 12, and 13 are Ser2 kinases
- Spt
suppressor of Ty’s, Spt4/5 makes up the DSIF transcription elongation complex
- DSIF
DRB sensitive inhibitory factor
- DRB
5,6-dichloro-1-β-D-ribofuranosylbenzimidazole, transcription elongation inhibitor
- NELF
negative elongation factor
- P-TEF
positive transcription elongation factor, P-TEFb is composed of Cdk9 and Cyclin T
References
- 1.He Y, Fang J, Taatjes DJ, Nogales E. Structural visualization of key steps in human transcription initiation. Nature. 2013;495:481–6. doi: 10.1038/nature11991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Murakami K, Elmlund H, Kalisman N, Bushnell DA, Adams CM, Azubel M, Elmlund D, Levi-Kalisman Y, Liu X, Gibbons BJ, et al. Architecture of an RNA polymerase II transcription pre-initiation complex. Science. 2013;342:1238724. doi: 10.1126/science.1238724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Michel M, Cramer P. Transitions for regulating early transcription. Cell. 2013;153:943–4. doi: 10.1016/j.cell.2013.04.050. [DOI] [PubMed] [Google Scholar]
- 4.Adelman K, Lis JT. Promoter-proximal pausing of RNA polymerase II: emerging roles in metazoans. Nat Rev Genet. 2012;13:720–31. doi: 10.1038/nrg3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cadena DL, Dahmus ME. Messenger RNA synthesis in mammalian cells is catalyzed by the phosphorylated form of RNA polymerase II. J Biol Chem. 1987;262:12468–74. [PubMed] [Google Scholar]
- 6.Allison LA, Moyle M, Shales M, Ingles CJ. Extensive homology among the largest subunits of eukaryotic and prokaryotic RNA polymerases. Cell. 1985;42:599–610. doi: 10.1016/0092-8674(85)90117-5. [DOI] [PubMed] [Google Scholar]
- 7.Stump AD, Ostrozhynska K. Selective constraint and the evolution of the RNA polymerase II C-Terminal Domain. Transcription. 2013;4:77–86. doi: 10.4161/trns.23305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eick D, Geyer M. The RNA polymerase II carboxy-terminal domain (CTD) code. Chem Rev. 2013;113:8456–90. doi: 10.1021/cr400071f. [DOI] [PubMed] [Google Scholar]
- 9.Heidemann M, Hintermair C, Voß K, Eick D. Dynamic phosphorylation patterns of RNA polymerase II CTD during transcription. Biochim Biophys Acta. 2013;1829:55–62. doi: 10.1016/j.bbagrm.2012.08.013. [DOI] [PubMed] [Google Scholar]
- 10.Jeronimo C, Bataille AR, Robert F. The writers, readers, and functions of the RNA polymerase II C-terminal domain code. Chem Rev. 2013;113:8491–522. doi: 10.1021/cr4001397. [DOI] [PubMed] [Google Scholar]
- 11.Komarnitsky P, Cho EJ, Buratowski S. Different phosphorylated forms of RNA polymerase II and associated mRNA processing factors during transcription. Genes Dev. 2000;14:2452–60. doi: 10.1101/gad.824700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mayer A, Lidschreiber M, Siebert M, Leike K, Söding J, Cramer P. Uniform transitions of the general RNA polymerase II transcription complex. Nat Struct Mol Biol. 2010;17:1272–8. doi: 10.1038/nsmb.1903. [DOI] [PubMed] [Google Scholar]
- 13.Odawara J, Harada A, Yoshimi T, Maehara K, Tachibana T, Okada S, Akashi K, Ohkawa Y. The classification of mRNA expression levels by the phosphorylation state of RNAPII CTD based on a combined genome-wide approach. BMC Genomics. 2011;12:516. doi: 10.1186/1471-2164-12-516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Egloff S, Murphy S. Cracking the RNA polymerase II CTD code. Trends Genet. 2008;24:280–8. doi: 10.1016/j.tig.2008.03.008. [DOI] [PubMed] [Google Scholar]
- 15.Zhou Q, Li T, Price DH. RNA polymerase II elongation control. Annu Rev Biochem. 2012;81:119–43. doi: 10.1146/annurev-biochem-052610-095910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Egloff S, O’Reilly D, Chapman RD, Taylor A, Tanzhaus K, Pitts L, Eick D, Murphy S. Serine-7 of the RNA polymerase II CTD is specifically required for snRNA gene expression. Science. 2007;318:1777–9. doi: 10.1126/science.1145989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chapman RD, Heidemann M, Albert TK, Mailhammer R, Flatley A, Meisterernst M, Kremmer E, Eick D. Transcribing RNA polymerase II is phosphorylated at CTD residue serine-7. Science. 2007;318:1780–2. doi: 10.1126/science.1145977. [DOI] [PubMed] [Google Scholar]
- 18.Hintermair C, Heidemann M, Koch F, Descostes N, Gut M, Gut I, Fenouil R, Ferrier P, Flatley A, Kremmer E, et al. Threonine-4 of mammalian RNA polymerase II CTD is targeted by Polo-like kinase 3 and required for transcriptional elongation. EMBO J. 2012;31:2784–97. doi: 10.1038/emboj.2012.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hsin JP, Manley JL. The RNA polymerase II CTD coordinates transcription and RNA processing. Genes Dev. 2012;26:2119–37. doi: 10.1101/gad.200303.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mayer A, Heidemann M, Lidschreiber M, Schreieck A, Sun M, Hintermair C, Kremmer E, Eick D, Cramer P. CTD tyrosine phosphorylation impairs termination factor recruitment to RNA polymerase II. Science. 2012;336:1723–5. doi: 10.1126/science.1219651. [DOI] [PubMed] [Google Scholar]
- 21.Bataille AR, Jeronimo C, Jacques PE, Laramée L, Fortin ME, Forest A, Bergeron M, Hanes SD, Robert F. A universal RNA polymerase II CTD cycle is orchestrated by complex interplays between kinase, phosphatase, and isomerase enzymes along genes. Mol Cell. 2012;45:158–70. doi: 10.1016/j.molcel.2011.11.024. [DOI] [PubMed] [Google Scholar]
- 22.Tietjen JR, Zhang DW, Rodríguez-Molina JB, White BE, Akhtar MS, Heidemann M, Li X, Chapman RD, Shokat K, Keles S, et al. Chemical-genomic dissection of the CTD code. Nat Struct Mol Biol. 2010;17:1154–61. doi: 10.1038/nsmb.1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim H, Erickson B, Luo W, Seward D, Graber JH, Pollock DD, Megee PC, Bentley DL. Gene-specific RNA polymerase II phosphorylation and the CTD code. Nat Struct Mol Biol. 2010;17:1279–86. doi: 10.1038/nsmb.1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jones JC, Phatnani HP, Haystead TA, MacDonald JA, Alam SM, Greenleaf AL. C-terminal repeat domain kinase I phosphorylates Ser2 and Ser5 of RNA polymerase II C-terminal domain repeats. J Biol Chem. 2004;279:24957–64. doi: 10.1074/jbc.M402218200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zehring WA, Lee JM, Weeks JR, Jokerst RS, Greenleaf AL. The C-terminal repeat domain of RNA polymerase II largest subunit is essential in vivo but is not required for accurate transcription initiation in vitro. Proc Natl Acad Sci U S A. 1988;85:3698–702. doi: 10.1073/pnas.85.11.3698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee JM, Greenleaf AL. A protein kinase that phosphorylates the C-terminal repeat domain of the largest subunit of RNA polymerase II. Proc Natl Acad Sci U S A. 1989;86:3624–8. doi: 10.1073/pnas.86.10.3624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee JM, Greenleaf AL. CTD kinase large subunit is encoded by CTK1, a gene required for normal growth of Saccharomyces cerevisiae. Gene Expr. 1991;1:149–67. [PMC free article] [PubMed] [Google Scholar]
- 28.Sterner DE, Lee JM, Hardin SE, Greenleaf AL. The yeast carboxyl-terminal repeat domain kinase CTDK-I is a divergent cyclin-cyclin-dependent kinase complex. Mol Cell Biol. 1995;15:5716–24. doi: 10.1128/mcb.15.10.5716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hautbergue G, Goguel V. Activation of the cyclin-dependent kinase CTDK-I requires the heterodimerization of two unstable subunits. J Biol Chem. 2001;276:8005–13. doi: 10.1074/jbc.M010162200. [DOI] [PubMed] [Google Scholar]
- 30.Prelich G, Winston F. Mutations that suppress the deletion of an upstream activating sequence in yeast: involvement of a protein kinase and histone H3 in repressing transcription in vivo. Genetics. 1993;135:665–76. doi: 10.1093/genetics/135.3.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yao S, Neiman A, Prelich G. BUR1 and BUR2 encode a divergent cyclin-dependent kinase-cyclin complex important for transcription in vivo. Mol Cell Biol. 2000;20:7080–7. doi: 10.1128/MCB.20.19.7080-7087.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Keogh MC, Podolny V, Buratowski S. Bur1 kinase is required for efficient transcription elongation by RNA polymerase II. Mol Cell Biol. 2003;23:7005–18. doi: 10.1128/MCB.23.19.7005-7018.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Qiu H, Hu C, Hinnebusch AG. Phosphorylation of the Pol II CTD by KIN28 enhances BUR1/BUR2 recruitment and Ser2 CTD phosphorylation near promoters. Mol Cell. 2009;33:752–62. doi: 10.1016/j.molcel.2009.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu Y, Warfield L, Zhang C, Luo J, Allen J, Lang WH, Ranish J, Shokat KM, Hahn S. Phosphorylation of the transcription elongation factor Spt5 by yeast Bur1 kinase stimulates recruitment of the PAF complex. Mol Cell Biol. 2009;29:4852–63. doi: 10.1128/MCB.00609-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chu Y, Sutton A, Sternglanz R, Prelich G. The BUR1 cyclin-dependent protein kinase is required for the normal pattern of histone methylation by SET2. Mol Cell Biol. 2006;26:3029–38. doi: 10.1128/MCB.26.8.3029-3038.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Karagiannis J, Balasubramanian MK. A cyclin-dependent kinase that promotes cytokinesis through modulating phosphorylation of the carboxy terminal domain of the RNA Pol II Rpb1p sub-unit. PLoS One. 2007;2:e433. doi: 10.1371/journal.pone.0000433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pei Y, Shuman S. Characterization of the Schizosaccharomyces pombe Cdk9/Pch1 protein kinase: Spt5 phosphorylation, autophosphorylation, and mutational analysis. J Biol Chem. 2003;278:43346–56. doi: 10.1074/jbc.M307319200. [DOI] [PubMed] [Google Scholar]
- 38.Murray S, Udupa R, Yao S, Hartzog G, Prelich G. Phosphorylation of the RNA polymerase II carboxy-terminal domain by the Bur1 cyclin-dependent kinase. Mol Cell Biol. 2001;21:4089–96. doi: 10.1128/MCB.21.13.4089-4096.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Keogh MC, Podolny V, Buratowski S. Bur1 kinase is required for efficient transcription elongation by RNA polymerase II. Mol Cell Biol. 2003;23:7005–18. doi: 10.1128/MCB.23.19.7005-7018.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhou K, Kuo WH, Fillingham J, Greenblatt JF. Control of transcriptional elongation and cotranscriptional histone modification by the yeast BUR kinase substrate Spt5. Proc Natl Acad Sci U S A. 2009;106:6956–61. doi: 10.1073/pnas.0806302106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Coudreuse D, van Bakel H, Dewez M, Soutourina J, Parnell T, Vandenhaute J, Cairns B, Werner M, Hermand D. A gene-specific requirement of RNA polymerase II CTD phosphorylation for sexual differentiation in S. pombe. Curr Biol. 2010;20:1053–64. doi: 10.1016/j.cub.2010.04.054. [DOI] [PubMed] [Google Scholar]
- 42.Viladevall L, St Amour CV, Rosebrock A, Schneider S, Zhang C, Allen JJ, Shokat KM, Schwer B, Leatherwood JK, Fisher RP. TFIIH and P-TEFb coordinate transcription with capping enzyme recruitment at specific genes in fission yeast. Mol Cell. 2009;33:738–51. doi: 10.1016/j.molcel.2009.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Guiguen A, Soutourina J, Dewez M, Tafforeau L, Dieu M, Raes M, Vandenhaute J, Werner M, Hermand D. Recruitment of P-TEFb (Cdk9-Pch1) to chromatin by the cap-methyl transferase Pcm1 in fission yeast. EMBO J. 2007;26:1552–9. doi: 10.1038/sj.emboj.7601627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cho EJ, Kobor MS, Kim M, Greenblatt J, Buratowski S. Opposing effects of Ctk1 kinase and Fcp1 phosphatase at Ser 2 of the RNA polymerase II C-terminal domain. Genes Dev. 2001;15:3319–29. doi: 10.1101/gad.935901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kim M, Ahn SH, Krogan NJ, Greenblatt JF, Buratowski S. Transitions in RNA polymerase II elongation complexes at the 3′ ends of genes. EMBO J. 2004;23:354–64. doi: 10.1038/sj.emboj.7600053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mayer A, Lidschreiber M, Siebert M, Leike K, Söding J, Cramer P. Uniform transitions of the general RNA polymerase II transcription complex. Nat Struct Mol Biol. 2010;17:1272–8. doi: 10.1038/nsmb.1903. [DOI] [PubMed] [Google Scholar]
- 47.Bartkowiak B, Liu P, Phatnani HP, Fuda NJ, Cooper JJ, Price DH, Adelman K, Lis JT, Greenleaf AL. CDK12 is a transcription elongation-associated CTD kinase, the metazoan ortholog of yeast Ctk1. Genes Dev. 2010;24:2303–16. doi: 10.1101/gad.1968210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Marshall NF, Peng J, Xie Z, Price DH. Control of RNA polymerase II elongation potential by a novel carboxyl-terminal domain kinase. J Biol Chem. 1996;271:27176–83. doi: 10.1074/jbc.271.43.27176. [DOI] [PubMed] [Google Scholar]
- 49.Taube R, Peterlin M. Lost in transcription: molecular mechanisms that control HIV latency. Viruses. 2013;5:902–27. doi: 10.3390/v5030902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Marshall NF, Price DH. Control of formation of two distinct classes of RNA polymerase II elongation complexes. Mol Cell Biol. 1992;12:2078–90. doi: 10.1128/mcb.12.5.2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yang X, Herrmann CH, Rice AP. The human immunodeficiency virus Tat proteins specifically associate with TAK in vivo and require the carboxyl-terminal domain of RNA polymerase II for function. J Virol. 1996;70:4576–84. doi: 10.1128/jvi.70.7.4576-4584.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhou M, Halanski MA, Radonovich MF, Kashanchi F, Peng J, Price DH, Brady JN. Tat modifies the activity of CDK9 to phosphorylate serine 5 of the RNA polymerase II carboxyl-terminal domain during human immunodeficiency virus type 1 transcription. Mol Cell Biol. 2000;20:5077–86. doi: 10.1128/MCB.20.14.5077-5086.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Napolitano G, Majello B, Licciardo P, Giordano A, Lania L. Transcriptional activity of positive transcription elongation factor b kinase in vivo requires the C-terminal domain of RNA polymerase II. Gene. 2000;254:139–45. doi: 10.1016/S0378-1119(00)00278-X. [DOI] [PubMed] [Google Scholar]
- 54.Ramanathan Y, Rajpara SM, Reza SM, Lees E, Shuman S, Mathews MB, Pe’ery T. Three RNA polymerase II carboxyl-terminal domain kinases display distinct substrate preferences. J Biol Chem. 2001;276:10913–20. doi: 10.1074/jbc.M010975200. [DOI] [PubMed] [Google Scholar]
- 55.Ramanathan Y, Reza SM, Young TM, Mathews MB, Pe’ery T. Human and rodent transcription elongation factor P-TEFb: interactions with human immunodeficiency virus type 1 tat and carboxy-terminal domain substrate. J Virol. 1999;73:5448–58. doi: 10.1128/jvi.73.7.5448-5458.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pinhero R, Liaw P, Bertens K, Yankulov K. Three cyclin-dependent kinases preferentially phosphorylate different parts of the C-terminal domain of the large subunit of RNA polymerase II. Eur J Biochem. 2004;271:1004–14. doi: 10.1111/j.1432-1033.2004.04002.x. [DOI] [PubMed] [Google Scholar]
- 57.Glover-Cutter K, Larochelle S, Erickson B, Zhang C, Shokat K, Fisher RP, Bentley DL. TFIIH-associated Cdk7 kinase functions in phosphorylation of C-terminal domain Ser7 residues, promoter-proximal pausing, and termination by RNA polymerase II. Mol Cell Biol. 2009;29:5455–64. doi: 10.1128/MCB.00637-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shim EY, Walker AK, Shi Y, Blackwell TK. CDK-9/cyclin T (P-TEFb) is required in two postinitiation pathways for transcription in the C. elegans embryo. Genes Dev. 2002;16:2135–46. doi: 10.1101/gad.999002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bowman EA, Bowman CR, Ahn JH, Kelly WG. Phosphorylation of RNA polymerase II is independent of P-TEFb in the C. elegans germline. Development. 2013;140:3703–13. doi: 10.1242/dev.095778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Boehm AK, Saunders A, Werner J, Lis JT. Transcription factor and polymerase recruitment, modification, and movement on dhsp70 in vivo in the minutes following heat shock. Mol Cell Biol. 2003;23:7628–37. doi: 10.1128/MCB.23.21.7628-7637.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ni Z, Schwartz BE, Werner J, Suarez JR, Lis JT. Coordination of transcription, RNA processing, and surveillance by P-TEFb kinase on heat shock genes. Mol Cell. 2004;13:55–65. doi: 10.1016/S1097-2765(03)00526-4. [DOI] [PubMed] [Google Scholar]
- 62.Eissenberg JC, Shilatifard A, Dorokhov N, Michener DE. Cdk9 is an essential kinase in Drosophila that is required for heat shock gene expression, histone methylation and elongation factor recruitment. Mol Genet Genomics. 2007;277:101–14. doi: 10.1007/s00438-006-0164-2. [DOI] [PubMed] [Google Scholar]
- 63.Peterlin BM, Brogie JE, Price DH. 7SK snRNA: a noncoding RNA that plays a major role in regulating eukaryotic transcription. Wiley Interdiscip Rev RNA. 2012;3:92–103. doi: 10.1002/wrna.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Guo J, Price DH. RNA polymerase II transcription elongation control. Chem Rev. 2013;113:8583–603. doi: 10.1021/cr400105n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lin C, Smith ER, Takahashi H, Lai KC, Martin-Brown S, Florens L, Washburn MP, Conaway JW, Conaway RC, Shilatifard A. AFF4, a component of the ELL/P-TEFb elongation complex and a shared subunit of MLL chimeras, can link transcription elongation to leukemia. Mol Cell. 2010;37:429–37. doi: 10.1016/j.molcel.2010.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yokoyama A, Lin M, Naresh A, Kitabayashi I, Cleary ML. A higher-order complex containing AF4 and ENL family proteins with P-TEFb facilitates oncogenic and physiologic MLL-dependent transcription. Cancer Cell. 2010;17:198–212. doi: 10.1016/j.ccr.2009.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Biswas D, Milne TA, Basrur V, Kim J, Elenitoba-Johnson KS, Allis CD, Roeder RG. Function of leukemogenic mixed lineage leukemia 1 (MLL) fusion proteins through distinct partner protein complexes. Proc Natl Acad Sci U S A. 2011;108:15751–6. doi: 10.1073/pnas.1111498108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.He N, Chan CK, Sobhian B, Chou S, Xue Y, Liu M, Alber T, Benkirane M, Zhou Q. Human Polymerase-Associated Factor complex (PAFc) connects the Super Elongation Complex (SEC) to RNA polymerase II on chromatin. Proc Natl Acad Sci U S A. 2011;108:E636–45. doi: 10.1073/pnas.1107107108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sobhian B, Laguette N, Yatim A, Nakamura M, Levy Y, Kiernan R, Benkirane M. HIV-1 Tat assembles a multifunctional transcription elongation complex and stably associates with the 7SK snRNP. Mol Cell. 2010;38:439–51. doi: 10.1016/j.molcel.2010.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mueller D, García-Cuéllar MP, Bach C, Buhl S, Maethner E, Slany RK. Misguided transcriptional elongation causes mixed lineage leukemia. PLoS Biol. 2009;7:e1000249. doi: 10.1371/journal.pbio.1000249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Smith E, Lin C, Shilatifard A. The super elongation complex (SEC) and MLL in development and disease. Genes Dev. 2011;25:661–72. doi: 10.1101/gad.2015411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Luo Z, Lin C, Shilatifard A. The super elongation complex (SEC) family in transcriptional control. Nat Rev Mol Cell Biol. 2012;13:543–7. doi: 10.1038/nrm3417. [DOI] [PubMed] [Google Scholar]
- 73.Galbraith MD, Donner AJ, Espinosa JM. CDK8: a positive regulator of transcription. Transcription. 2010;1:4–12. doi: 10.4161/trns.1.1.12373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Donner AJ, Ebmeier CC, Taatjes DJ, Espinosa JM. CDK8 is a positive regulator of transcriptional elongation within the serum response network. Nat Struct Mol Biol. 2010;17:194–201. doi: 10.1038/nsmb.1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Takahashi H, Parmely TJ, Sato S, Tomomori-Sato C, Banks CA, Kong SE, Szutorisz H, Swanson SK, Martin-Brown S, Washburn MP, et al. Human mediator subunit MED26 functions as a docking site for transcription elongation factors. Cell. 2011;146:92–104. doi: 10.1016/j.cell.2011.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yang Z, Yik JH, Chen R, He N, Jang MK, Ozato K, Zhou Q. Recruitment of P-TEFb for stimulation of transcriptional elongation by the bromodomain protein Brd4. Mol Cell. 2005;19:535–45. doi: 10.1016/j.molcel.2005.06.029. [DOI] [PubMed] [Google Scholar]
- 77.Jang MK, Mochizuki K, Zhou M, Jeong HS, Brady JN, Ozato K. The bromodomain protein Brd4 is a positive regulatory component of P-TEFb and stimulates RNA polymerase II-dependent transcription. Mol Cell. 2005;19:523–34. doi: 10.1016/j.molcel.2005.06.027. [DOI] [PubMed] [Google Scholar]
- 78.Ji X, Zhou Y, Pandit S, Huang J, Li H, Lin CY, Xiao R, Burge CB, Fu XD. SR proteins collaborate with 7SK and promoter-associated nascent RNA to release paused polymerase. Cell. 2013;153:855–68. doi: 10.1016/j.cell.2013.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Cheng B, Price DH. Properties of RNA polymerase II elongation complexes before and after the P-TEFb-mediated transition into productive elongation. J Biol Chem. 2007;282:21901–12. doi: 10.1074/jbc.M702936200. [DOI] [PubMed] [Google Scholar]
- 80.Kwak H, Lis JT. Control of transcriptional elongation. Annu Rev Genet. 2013;47:483–508. doi: 10.1146/annurev-genet-110711-155440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Peterlin BM, Price DH. Controlling the elongation phase of transcription with P-TEFb. Mol Cell. 2006;23:297–305. doi: 10.1016/j.molcel.2006.06.014. [DOI] [PubMed] [Google Scholar]
- 82.Devaiah BN, Lewis BA, Cherman N, Hewitt MC, Albrecht BK, Robey PG, Ozato K, Sims RJ, 3rd, Singer DS. BRD4 is an atypical kinase that phosphorylates serine2 of the RNA polymerase II carboxy-terminal domain. Proc Natl Acad Sci U S A. 2012;109:6927–32. doi: 10.1073/pnas.1120422109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Devaiah BN, Singer DS. Cross-talk among RNA polymerase II kinases modulates C-terminal domain phosphorylation. J Biol Chem. 2012;287:38755–66. doi: 10.1074/jbc.M112.412015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bartkowiak B, Greenleaf AL. Phosphorylation of RNAPII: To P-TEFb or not to P-TEFb? Transcription. 2011;2:115–9. doi: 10.4161/trns.2.3.15004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Larochelle S, Amat R, Glover-Cutter K, Sansó M, Zhang C, Allen JJ, Shokat KM, Bentley DL, Fisher RP. Cyclin-dependent kinase control of the initiation-to-elongation switch of RNA polymerase II. Nat Struct Mol Biol. 2012;19:1108–15. doi: 10.1038/nsmb.2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lin C, Garrett AS, De Kumar B, Smith ER, Gogol M, Seidel C, Krumlauf R, Shilatifard A. Dynamic transcriptional events in embryonic stem cells mediated by the super elongation complex (SEC) Genes Dev. 2011;25:1486–98. doi: 10.1101/gad.2059211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ghamari A, van de Corput MP, Thongjuea S, van Cappellen WA, van Ijcken W, van Haren J, Soler E, Eick D, Lenhard B, Grosveld FG. In vivo live imaging of RNA polymerase II transcription factories in primary cells. Genes Dev. 2013;27:767–77. doi: 10.1101/gad.216200.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Cho EJ, Kobor MS, Kim M, Greenblatt J, Buratowski S. Opposing effects of Ctk1 kinase and Fcp1 phosphatase at Ser 2 of the RNA polymerase II C-terminal domain. Genes Dev. 2001;15:3319–29. doi: 10.1101/gad.935901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Blazek D, Kohoutek J, Bartholomeeusen K, Johansen E, Hulinkova P, Luo Z, Cimermancic P, Ule J, Peterlin BMCINCCM. The Cyclin K/Cdk12 complex maintains genomic stability via regulation of expression of DNA damage response genes. Genes Dev. 2011;25:2158–72. doi: 10.1101/gad.16962311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Dai Q, Lei T, Zhao C, Zhong J, Tang YZ, Chen B, Yang J, Li C, Wang S, Song X, et al. Cyclin K-containing kinase complexes maintain self-renewal in murine embryonic stem cells. J Biol Chem. 2012;287:25344–52. doi: 10.1074/jbc.M111.321760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Fuda NJ, Ardehali MB, Lis JT. Defining mechanisms that regulate RNA polymerase II transcription in vivo. Nature. 2009;461:186–92. doi: 10.1038/nature08449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kim TH, Barrera LO, Zheng M, Qu C, Singer MA, Richmond TA, Wu Y, Green RD, Ren B. A high-resolution map of active promoters in the human genome. Nature. 2005;436:876–80. doi: 10.1038/nature03877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Guenther MG, Levine SS, Boyer LA, Jaenisch R, Young RA. A chromatin landmark and transcription initiation at most promoters in human cells. Cell. 2007;130:77–88. doi: 10.1016/j.cell.2007.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Muse GW, Gilchrist DA, Nechaev S, Shah R, Parker JS, Grissom SF, Zeitlinger J, Adelman K. RNA polymerase is poised for activation across the genome. Nat Genet. 2007;39:1507–11. doi: 10.1038/ng.2007.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zeitlinger J, Stark A, Kellis M, Hong JW, Nechaev S, Adelman K, Levine M, Young RA. RNA polymerase stalling at developmental control genes in the Drosophila melanogaster embryo. Nat Genet. 2007;39:1512–6. doi: 10.1038/ng.2007.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Core LJ, Waterfall JJ, Lis JT. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science. 2008;322:1845–8. doi: 10.1126/science.1162228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Rahl PB, Lin CY, Seila AC, Flynn RA, McCuine S, Burge CB, Sharp PA, Young RA. c-Myc regulates transcriptional pause release. Cell. 2010;141:432–45. doi: 10.1016/j.cell.2010.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Min IM, Waterfall JJ, Core LJ, Munroe RJ, Schimenti J, Lis JT. Regulating RNA polymerase pausing and transcription elongation in embryonic stem cells. Genes Dev. 2011;25:742–54. doi: 10.1101/gad.2005511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Li J, Liu Y, Rhee HS, Ghosh SK, Bai L, Pugh BF, Gilmour DS. Kinetic competition between elongation rate and binding of NELF controls promoter-proximal pausing. Mol Cell. 2013;50:711–22. doi: 10.1016/j.molcel.2013.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Core LJ, Waterfall JJ, Gilchrist DA, Fargo DC, Kwak H, Adelman K, Lis JT. Defining the status of RNA polymerase at promoters. Cell Rep. 2012;2:1025–35. doi: 10.1016/j.celrep.2012.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Rhee HS, Pugh BF. Genome-wide structure and organization of eukaryotic pre-initiation complexes. Nature. 2012;483:295–301. doi: 10.1038/nature10799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Henriques T, Gilchrist DA, Nechaev S, Bern M, Muse GW, Burkholder A, Fargo DC, Adelman K. Stable pausing by RNA polymerase II provides an opportunity to target and integrate regulatory signals. Mol Cell. 2013;52:517–28. doi: 10.1016/j.molcel.2013.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Saunders A, Ashe HL. Taking a pause to reflect on morphogenesis. Cell. 2013;153:941–3. doi: 10.1016/j.cell.2013.05.009. [DOI] [PubMed] [Google Scholar]
- 104.Wada T, Takagi T, Yamaguchi Y, Ferdous A, Imai T, Hirose S, Sugimoto S, Yano K, Hartzog GA, Winston F, et al. DSIF, a novel transcription elongation factor that regulates RNA polymerase II processivity, is composed of human Spt4 and Spt5 homologs. Genes Dev. 1998;12:343–56. doi: 10.1101/gad.12.3.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Yamaguchi Y, Wada T, Watanabe D, Takagi T, Hasegawa J, Handa H. Structure and function of the human transcription elongation factor DSIF. J Biol Chem. 1999;274:8085–92. doi: 10.1074/jbc.274.12.8085. [DOI] [PubMed] [Google Scholar]
- 106.Missra A, Gilmour DS. Interactions between DSIF (DRB sensitivity inducing factor), NELF (negative elongation factor), and the Drosophila RNA polymerase II transcription elongation complex. Proc Natl Acad Sci U S A. 2010;107:11301–6. doi: 10.1073/pnas.1000681107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Narita T, Yamaguchi Y, Yano K, Sugimoto S, Chanarat S, Wada T, Kim DK, Hasegawa J, Omori M, Inukai N, et al. Human transcription elongation factor NELF: identification of novel subunits and reconstitution of the functionally active complex. Mol Cell Biol. 2003;23:1863–73. doi: 10.1128/MCB.23.6.1863-1873.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Yamaguchi Y, Takagi T, Wada T, Yano K, Furuya A, Sugimoto S, Hasegawa J, Handa H. NELF, a multisubunit complex containing RD, cooperates with DSIF to repress RNA polymerase II elongation. Cell. 1999;97:41–51. doi: 10.1016/S0092-8674(00)80713-8. [DOI] [PubMed] [Google Scholar]
- 109.Wada T, Takagi T, Yamaguchi Y, Watanabe D, Handa H. Evidence that P-TEFb alleviates the negative effect of DSIF on RNA polymerase II-dependent transcription in vitro. EMBO J. 1998;17:7395–403. doi: 10.1093/emboj/17.24.7395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Renner DB, Yamaguchi Y, Wada T, Handa H, Price DH. A highly purified RNA polymerase II elongation control system. J Biol Chem. 2001;276:42601–9. doi: 10.1074/jbc.M104967200. [DOI] [PubMed] [Google Scholar]
- 111.Ivanov D, Kwak YT, Guo J, Gaynor RB. Domains in the SPT5 protein that modulate its transcriptional regulatory properties. Mol Cell Biol. 2000;20:2970–83. doi: 10.1128/MCB.20.9.2970-2983.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Fujinaga K, Irwin D, Huang Y, Taube R, Kurosu T, Peterlin BM. Dynamics of human immunodeficiency virus transcription: P-TEFb phosphorylates RD and dissociates negative effectors from the transactivation response element. Mol Cell Biol. 2004;24:787–95. doi: 10.1128/MCB.24.2.787-795.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Yamada T, Yamaguchi Y, Inukai N, Okamoto S, Mura T, Handa H. P-TEFb-mediated phosphorylation of hSpt5 C-terminal repeats is critical for processive transcription elongation. Mol Cell. 2006;21:227–37. doi: 10.1016/j.molcel.2005.11.024. [DOI] [PubMed] [Google Scholar]
- 114.Ping YH, Rana TM. DSIF and NELF interact with RNA polymerase II elongation complex and HIV-1 Tat stimulates P-TEFb-mediated phosphorylation of RNA polymerase II and DSIF during transcription elongation. J Biol Chem. 2001;276:12951–8. doi: 10.1074/jbc.M006130200. [DOI] [PubMed] [Google Scholar]
- 115.Werner F. A nexus for gene expression-molecular mechanisms of Spt5 and NusG in the three domains of life. J Mol Biol. 2012;417:13–27. doi: 10.1016/j.jmb.2012.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Tomar SK, Artsimovitch I. NusG-Spt5 proteins-Universal tools for transcription modification and communication. Chem Rev. 2013;113:8604–19. doi: 10.1021/cr400064k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Hartzog GA, Wada T, Handa H, Winston F. Evidence that Spt4, Spt5, and Spt6 control transcription elongation by RNA polymerase II in Saccharomyces cerevisiae. Genes Dev. 1998;12:357–69. doi: 10.1101/gad.12.3.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Schneider S, Pei Y, Shuman S, Schwer B. Separable functions of the fission yeast Spt5 carboxyl-terminal domain (CTD) in capping enzyme binding and transcription elongation overlap with those of the RNA polymerase II CTD. Mol Cell Biol. 2010;30:2353–64. doi: 10.1128/MCB.00116-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Rodríguez-Gil A, García-Martínez J, Pelechano V, Muñoz-Centeno MdeL, Geli V, Pérez-Ortín JE, Chávez S. The distribution of active RNA polymerase II along the transcribed region is gene-specific and controlled by elongation factors. Nucleic Acids Res. 2010;38:4651–64. doi: 10.1093/nar/gkq215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Svetlov V, Nudler E. Clamping the clamp of RNA polymerase. EMBO J. 2011;30:1190–1. doi: 10.1038/emboj.2011.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Hartzog GA, Kaplan CD. Competing for the clamp: promoting RNA polymerase processivity and managing the transition from initiation to elongation. Mol Cell. 2011;43:161–3. doi: 10.1016/j.molcel.2011.07.002. [DOI] [PubMed] [Google Scholar]
- 122.Hartzog GA, Fu J. The Spt4-Spt5 complex: a multi-faceted regulator of transcription elongation. Biochim Biophys Acta. 2013;1829:105–15. doi: 10.1016/j.bbagrm.2012.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Hirtreiter A, Damsma GE, Cheung AC, Klose D, Grohmann D, Vojnic E, Martin AC, Cramer P, Werner F. Spt4/5 stimulates transcription elongation through the RNA polymerase clamp coiled-coil motif. Nucleic Acids Res. 2010;38:4040–51. doi: 10.1093/nar/gkq135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Klein BJ, Bose D, Baker KJ, Yusoff ZM, Zhang X, Murakami KS. RNA polymerase and transcription elongation factor Spt4/5 complex structure. Proc Natl Acad Sci U S A. 2011;108:546–50. doi: 10.1073/pnas.1013828108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Martinez-Rucobo FW, Sainsbury S, Cheung AC, Cramer P. Architecture of the RNA polymerase-Spt4/5 complex and basis of universal transcription processivity. EMBO J. 2011;30:1302–10. doi: 10.1038/emboj.2011.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Vastenhouw NL, Zhang Y, Woods IG, Imam F, Regev A, Liu XS, Rinn J, Schier AF. Chromatin signature of embryonic pluripotency is established during genome activation. Nature. 2010;464:922–6. doi: 10.1038/nature08866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Akkers RC, van Heeringen SJ, Jacobi UG, Janssen-Megens EM, Françoijs KJ, Stunnenberg HG, Veenstra GJ. A hierarchy of H3K4me3 and H3K27me3 acquisition in spatial gene regulation in Xenopus embryos. Dev Cell. 2009;17:425–34. doi: 10.1016/j.devcel.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Hajheidari M, Koncz C, Eick D. Emerging roles for RNA polymerase II CTD in Arabidopsis. Trends Plant Sci. 2013;18:633–43. doi: 10.1016/j.tplants.2013.07.001. [DOI] [PubMed] [Google Scholar]
- 129.Gilchrist DA, Nechaev S, Lee C, Ghosh SK, Collins JB, Li L, Gilmour DS, Adelman K. NELF-mediated stalling of Pol II can enhance gene expression by blocking promoter-proximal nucleosome assembly. Genes Dev. 2008;22:1921–33. doi: 10.1101/gad.1643208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Yung TM, Narita T, Komori T, Yamaguchi Y, Handa H. Cellular dynamics of the negative transcription elongation factor NELF. Exp Cell Res. 2009;315:1693–705. doi: 10.1016/j.yexcr.2009.02.013. [DOI] [PubMed] [Google Scholar]
- 131.Pavri R, Gazumyan A, Jankovic M, Di Virgilio M, Klein I, Ansarah-Sobrinho C, Resch W, Yamane A, Reina San-Martin B, Barreto V, et al. Activation-induced cytidine deaminase targets DNA at sites of RNA polymerase II stalling by interaction with Spt5. Cell. 2010;143:122–33. doi: 10.1016/j.cell.2010.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Gilchrist DA, Dos Santos G, Fargo DC, Xie B, Gao Y, Li L, Adelman K. Pausing of RNA polymerase II disrupts DNA-specified nucleosome organization to enable precise gene regulation. Cell. 2010;143:540–51. doi: 10.1016/j.cell.2010.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Chao SH, Price DH. Flavopiridol inactivates P-TEFb and blocks most RNA polymerase II transcription in vivo. J Biol Chem. 2001;276:31793–9. doi: 10.1074/jbc.M102306200. [DOI] [PubMed] [Google Scholar]
- 134.Ptashne M, Gann A. Transcriptional activation by recruitment. Nature. 1997;386:569–77. doi: 10.1038/386569a0. [DOI] [PubMed] [Google Scholar]
- 135.Stargell LA, Struhl K. Mechanisms of transcriptional activation in vivo: two steps forward. Trends Genet. 1996;12:311–5. doi: 10.1016/0168-9525(96)10028-7. [DOI] [PubMed] [Google Scholar]
- 136.Steinmetz EJ, Warren CL, Kuehner JN, Panbehi B, Ansari AZ, Brow DA. Genome-wide distribution of yeast RNA polymerase II and its control by Sen1 helicase. Mol Cell. 2006;24:735–46. doi: 10.1016/j.molcel.2006.10.023. [DOI] [PubMed] [Google Scholar]
- 137.Wade JT, Struhl K. The transition from transcriptional initiation to elongation. Curr Opin Genet Dev. 2008;18:130–6. doi: 10.1016/j.gde.2007.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Wilhelm BT, Marguerat S, Aligianni S, Codlin S, Watt S, Bähler J. Differential patterns of intronic and exonic DNA regions with respect to RNA polymerase II occupancy, nucleosome density and H3K36me3 marking in fission yeast. Genome Biol. 2011;12:R82. doi: 10.1186/gb-2011-12-8-r82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Churchman LS, Weissman JS. Nascent transcript sequencing visualizes transcription at nucleotide resolution. Nature. 2011;469:368–73. doi: 10.1038/nature09652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Kim TS, Liu CL, Yassour M, Holik J, Friedman N, Buratowski S, Rando OJ. RNA polymerase mapping during stress responses reveals widespread nonproductive transcription in yeast. Genome Biol. 2010;11:R75. doi: 10.1186/gb-2010-11-7-r75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Lidschreiber M, Leike K, Cramer P. Cap completion and C-terminal repeat domain kinase recruitment underlie the initiation-elongation transition of RNA polymerase II. Mol Cell Biol. 2013;33:3805–16. doi: 10.1128/MCB.00361-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Hossain MA, Chung C, Pradhan SK, Johnson TL. The yeast cap binding complex modulates transcription factor recruitment and establishes proper histone H3K36 trimethylation during active transcription. Mol Cell Biol. 2013;33:785–99. doi: 10.1128/MCB.00947-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Sansó M, Lee KM, Viladevall L, Jacques PE, Pagé V, Nagy S, Racine A, St Amour CV, Zhang C, Shokat KM, et al. A positive feedback loop links opposing functions of P-TEFb/Cdk9 and histone H2B ubiquitylation to regulate transcript elongation in fission yeast. PLoS Genet. 2012;8:e1002822. doi: 10.1371/journal.pgen.1002822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Allen MA, Hillier LW, Waterston RH, Blumenthal T. A global analysis of C. elegans trans-splicing. Genome Res. 2011;21:255–64. doi: 10.1101/gr.113811.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Gu W, Lee HC, Chaves D, Youngman EM, Pazour GJ, Conte D, Jr., Mello CC. CapSeq and CIP-TAP identify Pol II start sites and reveal capped small RNAs as C. elegans piRNA precursors. Cell. 2012;151:1488–500. doi: 10.1016/j.cell.2012.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Blumenthal T. Trans-splicing and operons in C. elegans. WormBook. 2012:1–11. doi: 10.1895/wormbook.1.5.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Kruesi WS, Core LJ, Waters CT, Lis JT, Meyer BJ. Condensin controls recruitment of RNA polymerase II to achieve nematode X-chromosome dosage compensation. Elife. 2013;2:e00808. doi: 10.7554/eLife.00808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Chen RA, Down TA, Stempor P, Chen QB, Egelhofer TA, Hillier LW, Jeffers TE, Ahringer J. The landscape of RNA polymerase II transcription initiation in C. elegans reveals promoter and enhancer architectures. Genome Res. 2013;23:1339–47. doi: 10.1101/gr.153668.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Morton JJ, Blumenthal T. Identification of transcription start sites of trans-spliced genes: uncovering unusual operon arrangements. RNA. 2011;17:327–37. doi: 10.1261/rna.2447111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Saito TL, Hashimoto S, Gu SG, Morton JJ, Stadler M, Blumenthal T, Fire A, Morishita S. The transcription start site landscape of C. elegans. Genome Res. 2013;23:1348–61. doi: 10.1101/gr.151571.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Pferdehirt RR, Kruesi WS, Meyer BJ. An MLL/COMPASS subunit functions in the C. elegans dosage compensation complex to target X chromosomes for transcriptional regulation of gene expression. Genes Dev. 2011;25:499–515. doi: 10.1101/gad.2016011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Garrido-Lecca A, Blumenthal T. RNA polymerase II C-terminal domain phosphorylation patterns in Caenorhabditis elegans operons, polycistronic gene clusters with only one promoter. Mol Cell Biol. 2010;30:3887–93. doi: 10.1128/MCB.00325-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Reinke V, Krause M, Okkema P. Transcriptional regulation of gene expression in C. elegans. WormBook. 2013:1–34. doi: 10.1895/wormbook.1.45.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Zanetti S, Puoti A. Sex determination in the Caenorhabditis elegans germline. Adv Exp Med Biol. 2013;757:41–69. doi: 10.1007/978-1-4614-4015-4_3. [DOI] [PubMed] [Google Scholar]
- 155.Pazdernik N, Schedl T. Introduction to germ cell development in Caenorhabditis elegans. Adv Exp Med Biol. 2013;757:1–16. doi: 10.1007/978-1-4614-4015-4_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Powell-Coffman JA, Knight J, Wood WB. Onset of C. elegans gastrulation is blocked by inhibition of embryonic transcription with an RNA polymerase antisense RNA. Dev Biol. 1996;178:472–83. doi: 10.1006/dbio.1996.0232. [DOI] [PubMed] [Google Scholar]
- 157.Cai L, Phong BL, Fisher AL, Wang Z. Regulation of fertility, survival, and cuticle collagen function by the Caenorhabditis elegans eaf-1 and ell-1 genes. J Biol Chem. 2011;286:35915–21. doi: 10.1074/jbc.M111.270454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Gerstein MB, Lu ZJ, Van Nostrand EL, Cheng C, Arshinoff BI, Liu T, Yip KY, Robilotto R, Rechtsteiner A, Ikegami K, et al. modENCODE Consortium Integrative analysis of the Caenorhabditis elegans genome by the modENCODE project. Science. 2010;330:1775–87. doi: 10.1126/science.1196914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Zhong M, Niu W, Lu ZJ, Sarov M, Murray JI, Janette J, Raha D, Sheaffer KL, Lam HY, Preston E, et al. Genome-wide identification of binding sites defines distinct functions for Caenorhabditis elegans PHA-4/FOXA in development and environmental response. PLoS Genet. 2010;6:e1000848. doi: 10.1371/journal.pgen.1000848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Baugh LR, Demodena J, Sternberg PW. RNA Pol II accumulates at promoters of growth genes during developmental arrest. Science. 2009;324:92–4. doi: 10.1126/science.1169628. [DOI] [PubMed] [Google Scholar]
- 161.Baugh LR. To grow or not to grow: nutritional control of development during Caenorhabditis elegans L1 arrest. Genetics. 2013;194:539–55. doi: 10.1534/genetics.113.150847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Nechaev S, Fargo DC, dos Santos G, Liu L, Gao Y, Adelman K. Global analysis of short RNAs reveals widespread promoter-proximal stalling and arrest of Pol II in Drosophila. Science. 2010;327:335–8. doi: 10.1126/science.1181421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Maxwell CS, Kruesi WS, Core LJ, Kurhanewicz N, Waters CT, Lewarch CL, Antoshechkin I, Lis JT, Meyer BJ, Baugh LR. Pol II docking and pausing at growth and stress genes in C. elegans. Cell Rep. 2014;6:455–66. doi: 10.1016/j.celrep.2014.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Cheng B, Li T, Rahl PB, Adamson TE, Loudas NB, Guo J, Varzavand K, Cooper JJ, Hu X, Gnatt A, et al. Functional association of Gdown1 with RNA polymerase II poised on human genes. Mol Cell. 2012;45:38–50. doi: 10.1016/j.molcel.2011.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Jishage M, Malik S, Wagner U, Uberheide B, Ishihama Y, Hu X, Chait BT, Gnatt A, Ren B, Roeder RG. Transcriptional regulation by Pol II(G) involving mediator and competitive interactions of Gdown1 and TFIIF with Pol II. Mol Cell. 2012;45:51–63. doi: 10.1016/j.molcel.2011.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Lee C, Li X, Hechmer A, Eisen M, Biggin MD, Venters BJ, Jiang C, Li J, Pugh BF, Gilmour DS. NELF and GAGA factor are linked to promoter-proximal pausing at many genes in Drosophila. Mol Cell Biol. 2008;28:3290–300. doi: 10.1128/MCB.02224-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Li J, Gilmour DS. Distinct mechanisms of transcriptional pausing orchestrated by GAGA factor and M1BP, a novel transcription factor. EMBO J. 2013;32:1829–41. doi: 10.1038/emboj.2013.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Cecere G, Hoersch S, Jensen MB, Dixit S, Grishok A. The ZFP-1(AF10)/DOT-1 complex opposes H2B ubiquitination to reduce Pol II transcription. Mol Cell. 2013;50:894–907. doi: 10.1016/j.molcel.2013.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Furuhashi H, Takasaki T, Rechtsteiner A, Li T, Kimura H, Checchi PM, Strome S, Kelly WG. Trans-generational epigenetic regulation of C. elegans primordial germ cells. Epigenetics Chromatin. 2010;3:15. doi: 10.1186/1756-8935-3-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Spencer WC, Zeller G, Watson JD, Henz SR, Watkins KL, McWhirter RD, Petersen S, Sreedharan VT, Widmer C, Jo J, et al. A spatial and temporal map of C. elegans gene expression. Genome Res. 2011;21:325–41. doi: 10.1101/gr.114595.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Deal RB, Henikoff S. The INTACT method for cell type-specific gene expression and chromatin profiling in Arabidopsis thaliana. Nat Protoc. 2011;6:56–68. doi: 10.1038/nprot.2010.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Steiner FA, Talbert PB, Kasinathan S, Deal RB, Henikoff S. Cell-type-specific nuclei purification from whole animals for genome-wide expression and chromatin profiling. Genome Res. 2012;22:766–77. doi: 10.1101/gr.131748.111. [DOI] [PMC free article] [PubMed] [Google Scholar]


