Abstract
Chromatin structure can affect the organization and maintenance of chromosomes. Recent discoveries in several filamentous fungi suggest mechanisms for the clustering and co-regulation of secondary metabolite genes or pathogenicity islands. An extreme case of this may be fungal “accessory”, “conditionally dispensable”, or “supernumerary” chromosomes that often confer beneficial traits. Fungal supernumerary chromosomes may be derived by similar mechanisms as animal or plant B chromosomes, and we thus propose that this term should be reconsidered to capture the wide variety of fungal accessory chromosomes. In some fungi, both the “ends” of chromosomes and these “odd” B chromosomes are enriched with a silencing histone modification, H3 lysine 27 trimethylation (H3K27me3), suggesting parallel mechanisms in evolving subtelomeric or B-chromosomal pathogenicity islands and secondary metabolite clusters (SMCs).
Keywords: chromatin, histone modification, B chromosome, supernumerary chromosome, accessory chromosome, H3K27 methylation, H3K9 methylation, fungi, secondary metabolite
Introduction
Genome organization of fungal plant and animal pathogens plays important roles in their pathogenicity. Here we discuss two aspects of this organization in filamentous ascomycetes, first reviewing recent work on the physical linkage of secondary metabolite pathways into gene clusters, their distribution across the genome, and their co-regulation as affected by chromatin structure (the “ends”). We then turn to B chromosomes (“supernumerary”, “accessory” or “lineage-specific” chromosomes - the “odds”) and their importance for pathogenicity. Both features have important consequences for the lifecycle of fungal pathogens and both have been under intense scrutiny in the past few years, driven largely by the development of new technologies. We focus on these phenomena because we feel that they offer unique opportunities for molecular mycologists to contribute to answering fundamental questions in chromosome biology.
Secondary metabolite clusters (SMCs)
Fungi produce a large variety of secondary metabolites – compounds not essential for normal growth but nonetheless important in certain environments or developmental stages [1]. Most fungal secondary metabolites fall into four classes defined by the keystone enzyme in their biosynthetic pathway: isoprenoids by prenyltransferases (PTs), nonribosomal peptides by non-ribosomal peptide synthetases (NRPSs), terpenoids by terpene cyclases (TCs) and polyketides by polyketide synthases (PKSs) [2]. Compounds from SMCs play key roles in fungal pathogenicity of humans, animals and crops. The genus Fusarium, for example, harbors major pathogens of numerous crops, especially cereals, causing diseases that reduce crop yields and may render the remaining harvest unusable as food or feed due to the presence of mycotoxins. The terpenoid deoxynivalenol (DON) inhibits protein synthesis, causing cellular stress and has both acute and chronic effects on humans and animals [3,4]. DON is produced from farnesyl pyrophosphate by the action of 12 enzymes. In Fusarium graminearum, nine of these are clustered at one locus along with two regulatory genes while the remaining three biosynthetic enzymes are at two separate loci [5]. Such clustering of genes within a secondary metabolite pathway is a general rule in fungi [6], and other well-known examples include the 54 kb, 23 gene sterigmatocystin cluster in Aspergillus nidulans [7], the 68 kb, 23 gene sirodesmin cluster in Leptosphaeria maculans [8], the 64 kb, 17 gene lovastatin cluster of Aspergillus terreus [9] and the 75 kb, 15 gene fumonisin cluster of the Gibberella moniliformis species complex [10]. Along with biosynthesis genes, SMCs commonly contain transporter genes that can confer resistance to toxic secondary metabolites. Transcription factors that control expression of the cluster are often, but not always present in or near the SMC. Only about a quarter of all easily predictable SMCs have been assigned final products, and developing methods to coordinately control their expression to enable compound identification is a major goal of current studies.
Regulation of secondary metabolite clusters
Analyses of the many almost complete fungal genome sequences have shed light on the location of SMCs on chromosomes. Overwhelmingly, SMCs are localized closer to the ends of chromosomes, in what can be broadly defined as “subtelomeric regions”, and they are often flanked by repetitive elements [2,11–13]. These findings suggested potential mechanisms of coordinated regulation of multiple clusters by shared transcription factors or chromatin modifications [2,14]. Genes within fungal SMCs are often coordinately regulated by a hierarchy of control systems. Many SMCs encode Zn(II)2Cys6 transcription factors (TFs) that activate the cluster. The best studied example is Aspergillus AflR, which activates sterigmatocystin biosynthesis genes and production of aflatoxin by binding to a preferred consensus sequence, TCG(G/C)(A/T)NN(G/C)CG(A/G), present in the promoters of these genes [15,16]. Positive global regulators, like the fungal-specific putative protein methyltransferase LaeA [17], may control larger regions that are activated by several TFs, for example FapR for fumagillin and pseurotin and an unknown non-SMC TF for the neighboring fumitremorgin cluster [18].
Expression of activating TFs is often not sufficient for cluster expression, as SMCs can be embedded within transcriptionally silent heterochromatin that must be remodeled before expression is possible. It has been proposed that nucleosomes of the Aspergillus aflatoxin cluster are trimethylated on lysine 9 of histone H3 (H3K9me3), which is bound by the chromo domain of Heterochromatin Protein-1 (HP1, in Aspergillus HepA), though genome-wide histone modification maps have not been produced yet. H3K9me3 and HP1 binding results in gene silencing in other systems, which in Aspergillus may somehow be relieved by the action of LaeA [19]. In L. maculans, both HP1 and the H3K9 methyltransferase KMT1 (DIM-5/ClrD/SUVAR39) affect the expression of pathogenicity factors [20] and we predict that SMCs will be shown to be affected once genome-wide ChIP-seq is conducted.
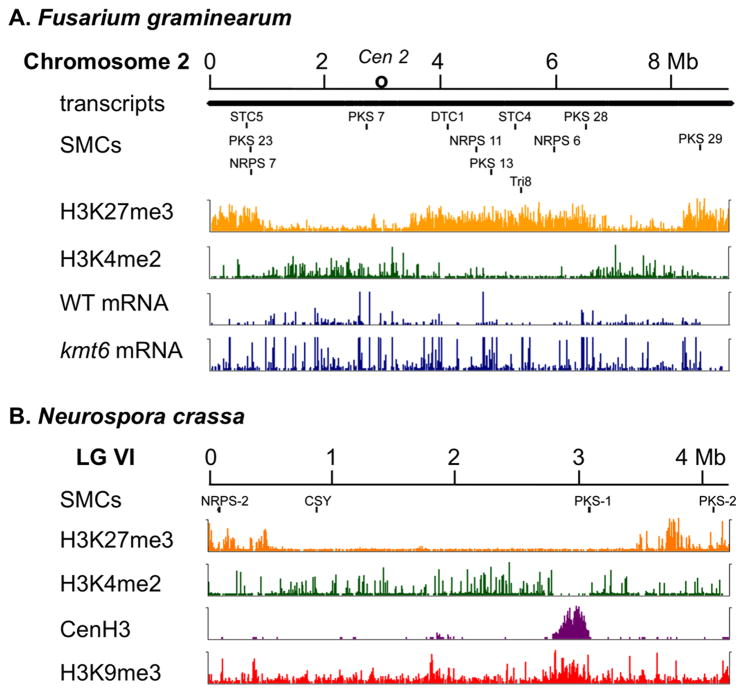
In contrast, in Fusarium graminearum most SMCs are associated with a different repressive chromatin mark, H3K27me3, especially when grown in rich medium with high nitrogen levels (Fig. 1A). Upon deletion of the H3K27 methyltransferase gene, kmt6, more than half of these SMCs are expressed under normally repressive conditions [13]. In Neurospora crassa, deletion of the kmt6 homologue, set-7, also resulted in complete loss of H3K27me3 [21] but in this genus SMCs are mostly not associated with H3K27me3 or H3K9me3 (Fig. 1B). Two SMCs (eas and ltm) in the alkaloid-producing fungus Epichloe festucae show enrichment of H3K9me3 and H3K27me3, in a life stage-dependent manner; enrichment was increased in axenic cultures when compared to symbiotic growth in plant tissue [22]. This study also revealed interesting interactions between H3K9 and H3K27 methylation, as there appears to be cooperativity between the two histone marks in gene silencing. In accord with the currently available data, our unpublished results suggest that similar patterns hold within a given genus (e.g. Fusarium and Zymoseptoria), but that different fungal clades use slightly different versions of chromatin silencing. Overall there does not seem to be a single, universal solution to coordinated regulation of SMCs. Future work will refine the emerging picture and likely also contribute to a basic understanding of silencing pathways in all eukaryotes that make use of H3K27 methylation.
Figure 1. Comparison of chromatin features of Fusarium and Neurospora chromosomes with SMCs.
A. Fusarium graminearum Chromosome 2 contains twelve SMCs that can be identified by keystone enzymes (from left to right: STC5, FGSG_08181; PKS23, FGSG_08208; NRPS7, FGSG_08209; PKS7, FGSG_08795; DTC1, FGSG_03066; NRPS11, FGSG_03245; PKS13, FGSG_03340; STC4, FGSG_03494; Tri8, FGSG_03532; NRPS6, FGSG_03747; PKS28, FGSG_03964; PKS29, FGSG_04588; nomenclature for SMCs according to [2]. Except for PKS7, all SMCs are in silent regions enriched with H3K27me3 (orange). None are in regions enriched with H3K4me2 (green), a modification most often found in the promoters of active genes. This suggested that none of these SMCs are expressed in wild type (WT), which was validated by RNA-seq (WT mRNA). In contrast, all SMCs are expressed or overexpressed in an H3K27me3 mutant, kmt6 (kmt6 mRNA), except for STC5 and PKS7 that either require activating factors (STC5) or are not subject to gene silencing by H3K27me3 (PKS7). The thousands of transcripts generated or predicted from Chromosome 2 are indicated, at this resolution as solid bar (transcripts). The centromeric region (Cen2) is indicated by a circle. B. Neurospora crassa Linkage Group (LG) VI has four SMCs (from left to right: NRPS-2, NCU07119; CSY, NCU04801; PKS-1, NCU06013, PKS-2, NCU05011). Of the four clusters, only NRPS-2 is enriched for H3K27me3 (orange). CSY and PKS-1 are enriched in H3K4me2 (green) and expressed at low levels under standard growth conditions (Vogel’s minimal medium at 32C), while PKS-2 is not enriched for any of the tested histone modifications and not expressed under standard laboratory conditions. The pks-1 gene resides near the right edge of Cen VI (CenH3 localization in purple), directly next to a tract of H3K9me3 (red). Histones and their modifications were assayed byCh IP-seq of chromatin precipitated with antibodies against GFP-tagged CenH3, H3K4me2, H3K9me3 and H3K27me3 as described [13].
Cluster maintenance in fungal lineages
The pressures resulting in mechanisms for clustering of secondary metabolite genes remain unclear. In some cases clustering may be a byproduct from horizontal transfer of entire clusters from a fungus or bacterium into a naïve species. For example, a high degree of sequence and syntenic conservation supports the wholesale transfer of the Aspergillus sterigmatocystin cluster to Podospora anserina [23], and comparative phylogenetics supports the interkingdom transfer of a 6-methylsalicylic acid PKS from actinobacteria to the progenitor of the Ascomycetes [24]. Alternatively, and not mutually exclusive, the need to efficiently regulate SMCs may drive gene clustering. Evidence for this exists in the trichothecene pathway gene distributions within the genus Fusarium. In F. graminearum and F. sporotrichioides the pathway is fragmented across three loci, whereas in F. equiseti it is condensed into two. The ancestral pathway most likely existed across three loci as in F. graminearum and F. sporotrichioides and became consolidated within F. equiseti[10]. Furthermore, the F. equiseti cluster contains a Zn(II)2Cys6 transcription factor that is absent from F. graminearum and F. sporotrichioides. This is consistent with a distinct –if not more efficient– regulatory pathway. Strict coordination of gene expression may be particularly important during the biosynthesis of secondary metabolites as intermediate compounds are potentially toxic. Indeed, a recent analysis of the simplest possible gene clusters, gene pairs, uncovered a strong bias for the pairing of enzymes that share a toxic intermediate [25]. Moreover, many of these gene pairs are divergently oriented around a single promoter, an arrangement that favors tight co-regulation.
Unlinked secondary metabolite pathways genes are at great risk of “disassembly” during meiotic recombination unless mechanisms exist to reduce recombination rates within these regions. Recombination is not evenly distributed across fungal genomes. Notably, the centromeres of Schizosaccharomyces pombe seem to have very low recombination rates in part because they are heterochromatic [26,27]. Reduced recombination rates within heterochromatin may relate to its condensed nature, which makes it refractory to programmed double-strand break formation during meiosis [28]. As double strand breaks are central to meiotic recombination and correlated to high recombination rates [29,30] heterochromatic regions have lower recombination rates than highly expressed regions. Thus secondary metabolite pathways may be clustered within large domains of facultative heterochromatin to limit recombination and thus retain intact clusters. However, recombination profiles of F. graminerarum [31,32] suggest that the facultative heterochromatin marked by H3K27 methylation [13] carries the most variable DNA sequence. Solving this paradox will be a focus of investigations in coming years.
B chromosomes (Bs)
Many eukaryotes, in fact perhaps as many as 15% of all plant species, carry additional (B) chromosomes in excess of the normal haploid or diploid set of A chromosomes (for a review see a chapter in [33]. By definition, Bs are not strictly necessary chromosomes, they are “conditionally dispensable” (CD), “supernumerary” or “accessory” to the core genome contained on the A chromosomes [34–36]. In many plant species and in some fungi, Bs are found only in specific lineages of closely related species, so they are sometimes referred to as “lineage-specific” chromosomes [37]. This is a somewhat biased term as it depends on the current state of knowledge covering specific taxa – what is currently “lineage-specific” may become “genus-specific” by discovery of the proper line or strain. Over the past 20 years, mycologists have roundly rejected “B chromosome” to describe the more specialized supernumerary chromosomes in fungi, though we propose that this term still applies to fungi. Here we outline why.
What do B chromosomes do? Mostly they are harmful to the host plant or animal - at best they are neutral elements. In fungi, however, we find an overwhelming majority of beneficial Bs, perhaps one reason why mycologists prefer specific terms rather than “B chromosome”. Often the benefit to the fungus lies in the ability to use genes encoded on Bs to colonize plant material or detoxify plant defense compounds, perhaps another reason to separate them from the overall gene-poor Bs of plants and animals, explaining why they have been called “pathogenicity” chromosomes [37,38]. Selective advantage via pathogenicity determinants does not seem to hold for all fungal Bs, however [2,39].
Overall, there are more commonalities than differences among Bs from different kingdoms, as revealed by the voluminous literature [33], even though there are differences in sensu stricto definitions [40,41]. In most cases B chromosomes use various forms of meiotic drive and self-accumulation mechanisms to propagate – many of the inheritance patterns are non-Mendelian. Of course this is difficult to study in fungi without extant sexual stage [41]. Nevertheless, general correlations can be derived: (a) higher numbers of Bs in one nucleus, and growth under harsher environmental conditions make negative effects of Bs more pronounced; (b) the behavior of B chromosomes can affect recombination of A chromosomes; (c) in plants, inbreeding promotes the accumulation and spread of beneficial Bs; (d) presence of Bs is positively correlated with low ploidy, low chromosome numbers and large genomes (i.e. larger domains of repetitive DNA); (e) Bs are more prevalent in genomes with acrocentric chromosomes, and smaller Bs are mitotically less stable than large Bs [33,40,42]. Some of these correlations apply to fungal Bs, thus it seems advantageous to mycologists to use the same term as researchers who work with animal and plant species. The other correlations need to be investigated.
How big are B chromosomes? The simple rule in fungi is “rarely larger than the smallest A chromosome”. In most eukaryotes, Bs are indeed small, containing sometimes only the elements required for propagation, i.e. telomeres and some centromeric DNA for kinetochore and spindle attachment. The variety is staggering, however! There are so many different types of Bs in plants and animals that there are no phylogenomic rules discernable for true classification by size and DNA content. This begs the question: What classes of genes or elements are located on Bs? Overall Bs are depleted for genes and enriched for repeated elements, either satellite repeats or active and mutated transposons, often of different types than on the As [35,37]. Few genes have been found on Bs in plants or animals; the most gene-rich Bs are those of various filamentous fungi. Thus, it appears that increase of gene density on Bs is positively correlated with benefit to the host organism. A special case can be made for Bs that contain rDNA clusters. These are much more frequently found among plant and animal Bs and this finding, combined with data showing that rDNA segregates late during division, suggest that rDNA is causally involved in increased non-disjunction and thus mitotic or meiotic drive of Bs [33].
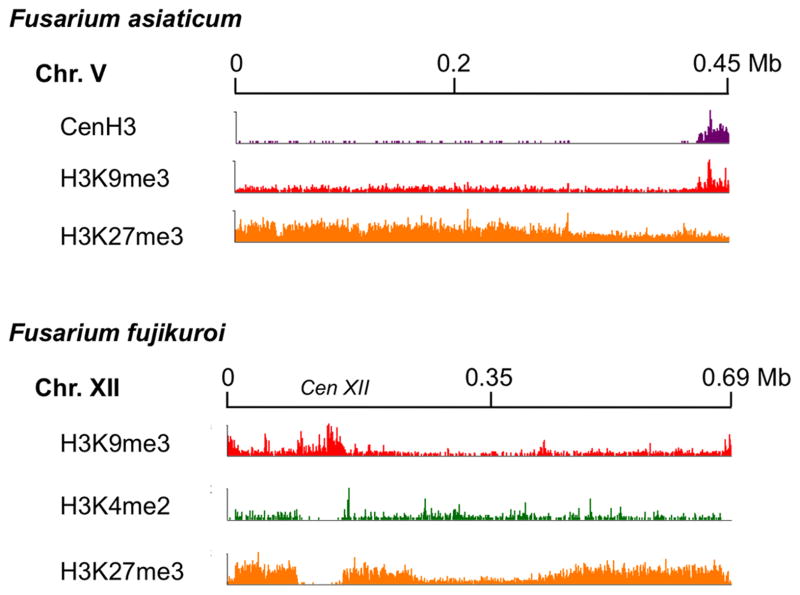
Matching the finding of few active genes, cytological data suggest that Bs are largely heterochromatic. This idea requires further study in fungi, however. Even in plants only half of all Bs are composed of truly constitutive heterochromatin. Long stretches of euchromatin and facultative heterochromatin can be found, though these are matching mostly transposable elements. In preliminary studies, we determined some chromatin features of B chromosomes in several genera of fungi, and found that while they are heterochromatic, they are mostly associated with H3K27me3, a hallmark of facultative heterochromatin that can be activated under the appropriate conditions (Fig. 2). We feel that the genetically tractable fungi, for which many biochemical and cytological methods exist, will become important models to unravel the varied chromatin structures of B chromosomes.
Figure 2. Putative B chromosome of F. asiaticum and F. fujikuroi.
ChIP-seq of chromatin precipitated with antibodies against GFP-tagged CenH3 (purple), H3K4me2 (green), H3K9me3 (red) and H3K27me3 (orange) suggests that H3K27me3 acts as the major gene silencing modification on Fusarium B chromosomes (Sung-Hwan Yun, Lena Studt, Bettina Tudzynski, Lanelle Connolly, Kristina Smith and Michael Freitag, preliminary data). Note the predicted acrocentric centromeres in both species, either determind by Cen H3 ChIP (purple in F. fujikuroi) or predicted by large region of H3K9me3 (red, F. fujikuroi).
Two of the most obvious and vexing questions remain unanswered. Where do Bs originate? And how are they maintained or lost? Clearly Bs are either generated within the host by aberrant chromosome segregation during division, or they are acquired from closely or distantly related taxa. Horizontal Chromosome Transfer (HCT) should be inefficient and rare, as otherwise there would be a large collection of fungal Bs available for study, all actively degenerating from the previous donor A chromosomes to avoid pairing with As during meiosis. Of course, donors may provide already degenerated Bs to the new host, which may explain the very different transposon or repeat content and codon bias observed on some fungal Bs [35,37,43]. While some interspecific hybridization may occur within many genera, particularly in the fungi, endogenous sources seem more obvious. One mechanism would be wholesale duplication of A chromosomes followed by degeneration and accumulation of transposons to generate large Bs that shrink only over time. Alternatively, missegregation or Robertsonian chromosome fusions may result in duplication of short regions from As, presumably containing centromeric or “proto-centromeric” DNA to generate very small Bs that “grow from the centromere out” [33]. Based on what we know about the behavior of centromeric chromatin, the generation and maintenance of heterochromatin, and the inheritance of minichromosomes this latter scenario seems much more likely. There is evidence for both pathways from different organisms [33], but no clear and decisive answers have emerged. Fungi offer opportunities to test both possibilities by experiment, rather than deduction from the age-old evidence generated by natural evolution.
Recent studies in fungi showed that the two pathways are not exclusive, perhaps even for a single species. Horizontal chromosomes transfer (HCT) can occur in F. oxysporum f. sp. lycopersici, at least under strong selection pressure [37]. The Bs of this taxon are enriched for transposable elements and contain genes with distinct evolutionary profiles that are required for pathogenicity on specific host plants. Genes on the Bs are fungal in origin but appear only poorly related to the host’s genes [37]. This study demonstrated transfer of B chromosomes experimentally, converting a non-pathogenic strain into a pathogenic strain. The authors favor the idea that HCT between isolated strains “explain the polyphyletic origin of host specificity and the emergence of new pathogenic lineages”. Interestingly, this study also showed that mixed A and B chromosomes exist in F. oxysporum f. sp. lycopersici, as chromosomes 1 and 2 show repeat content and other sequence features that revealed As with translocated B sequences. B chromosomes 3 and 6 are also larger than the smallest A. This suggests that either translocations after HCT can occur or that endogenous mechanisms may be involved that shuffle chromosomes after acquisition.
The endogenous generation of Bs by chromosome fusion followed by degenerative breakage, as proposed by Barbara McClintock’s model of “breakage-fusion-bridge” (BFB) cycles [44,45], may be the origin of at least one of the eight Bs in the reference strain of Zymoseptoria tritici [36]. This model suggests that repetitive DNA or perhaps specific chromatin structure may be required for non-allelic homologous recombination between repeats to generate a dicentric and acentric chromosome. The dicentric chromosome (here Chromosome 17) may have undergone BFB cycles [36], while the acentric chromosome was simply lost in subsequent divisions. This testable hypothesis will result in renewed efforts to observe how exactly one extra centromere is deleted or degenerated and how acentric, potentially small chromosomes are either lost or may acquire a new centric region, or “neocentromere”. Zymoseptoria is quickly developing into a model genus, combining excellent genome resources [46–48] with opportunities for biochemical and cytological analyses. What is currently lacking is a genetic system under laboratory conditions, though controlled crosses have been carried out in the field [39,49]. Making genetics a routine feature in species with Bs will also aid in addressing how these chromosomes are maintained (or quite frequently lost) during meiosis [39]. We predict that Zymoseptoria and certain Fusarium species will become facile models to answer these very basic questions about the biology of B chromosomes. That these questions are of general interest was illustrated in a recent review on mammalian and yeast genome destabilization [50].
Concluding remarks
The intriguing connection between the “ends”, large subtelomeric blocks of mostly non-syntenic DNA, and the “odds”, supernumerary or B chromosomes, seems to be chromatin structure that is characterized by the presence of nucleosomes with trimethylated H3K27. The F. graminearum species complex presents an interesting case. Many determinants for pathogenicity and most SMCs reside not only in the subtelomeric regions but also within the four chromosomes, which were predicted to result from chromosome fusions of the ancestral 11 or 12 chromosomes [31]. Recombination profiles [31,32] and chromatin structure analyses [13] suggest that the epigenetically defined and usually silent or “cryptic” regions of this genome maintain diversity but paradoxically also have been maintained in this form over millennia. Thus, future work will address how H3K27me3 and other chromatin marks are involved in the formation and maintenance of facultative heterochromatic domains on A versus B chromosomes, with special emphasis on DNA recombination and repair.
Highlights.
Histone H3K27me3 generates facultative heterochromatin in some fungi.
Fungal core genomes are contained on “A chromosomes”.
Supernumerary chromosomes of fungi are specialized “B chromosomes”.
Some fungal B chromosomes are “pathogenicity” chromosomes.
Subtelomeric regions and B chromosomes share widespread H3K27me3.
Acknowledgments
We thank Kristina Smith and Lanelle Connolly for fruitful discussions. Work in our laboratory is supported by funds from the U.S. National Institutes of Health (R01GM097637 and P01GM068087). We are indebted to the Neurospora Functional Genomics Project and the Fungal Genetics Stock Center for strains that greatly enhance our research.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
* of special interest
** of outstanding interest
- 1.Brakhage AA. Regulation of fungal secondary metabolism. Nat Rev Microbiol. 2013;11:21–32. doi: 10.1038/nrmicro2916. [DOI] [PubMed] [Google Scholar]
- 2**.Wiemann P, Sieber CM, von Bargen KW, Studt L, Niehaus EM, Espino JJ, Huss K, Michielse CB, Albermann S, Wagner D, et al. Deciphering the Cryptic Genome: Genome-wide Analyses of the Rice Pathogen Fusarium fujikuroi Reveal Complex Regulation of Secondary Metabolism and Novel Metabolites. PLoS Pathog. 2013;9:e1003475. doi: 10.1371/journal.ppat.1003475. This comprehensive study describes features of the F. fujikuroi genome and delivers a compendium of its SMCs. The authors combine analyses of mRNA by microarrays with ChIP-seq and fluorescent microscopy of a few selected chromatin modifications to determine differential expression under low and high nitrogen conditions and chromatin territories. Importantly they use biochemical and chemical anlyses to describe newly discovered SMCs and the compounds produced. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Goswami RS, Kistler HC. Heading for disaster: Fusarium graminearum on cereal crops. Mol Plant Pathol. 2004;5:515–525. doi: 10.1111/j.1364-3703.2004.00252.x. [DOI] [PubMed] [Google Scholar]
- 4.Pestka JJ. Deoxynivalenol: mechanisms of action, human exposure, and toxicological relevance. Archives of toxicology. 2010;84:663–679. doi: 10.1007/s00204-010-0579-8. [DOI] [PubMed] [Google Scholar]
- 5.Merhej J, Richard-Forget F, Barreau C. Regulation of trichothecene biosynthesis in Fusarium: recent advances and new insights. Appl Microbiol Biotechnol. 2011;91:519–528. doi: 10.1007/s00253-011-3397-x. [DOI] [PubMed] [Google Scholar]
- 6.Hoffmeister D, Keller NP. Natural products of filamentous fungi: enzymes, genes, and their regulation. Natural product reports. 2007;24:393–416. doi: 10.1039/b603084j. [DOI] [PubMed] [Google Scholar]
- 7.Brown DW, Yu JH, Kelkar HS, Fernandes M, Nesbitt TC, Keller NP, Adams TH, Leonard TJ. Twenty-five coregulated transcripts define a sterigmatocystin gene cluster in Aspergillus nidulans. Proceedings of the National Academy of Sciences of the United States of America. 1996;93:1418–1422. doi: 10.1073/pnas.93.4.1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gardiner DM, Cozijnsen AJ, Wilson LM, Pedras MSC, Howlett BJ. The sirodesmin biosynthetic gene cluster of the plant pathogenic fungus Leptosphaeria maculans. Molecular Microbiology. 2004;53:1307–1318. doi: 10.1111/j.1365-2958.2004.04215.x. [DOI] [PubMed] [Google Scholar]
- 9.Kennedy J, Auclair K, Kendrew SG, Park C, Vederas JC, Hutchinson CR. Modulation of polyketide synthase activity by accessory proteins during lovastatin biosynthesis. Science (New York, NY) 1999;284:1368–1372. doi: 10.1126/science.284.5418.1368. [DOI] [PubMed] [Google Scholar]
- 10.Proctor RH, McCormick SP, Alexander NJ, Desjardins AE. Evidence that a secondary metabolic biosynthetic gene cluster has grown by gene relocation during evolution of the filamentous fungus Fusarium. Molecular Microbiology. 2009;74:1128–1142. doi: 10.1111/j.1365-2958.2009.06927.x. [DOI] [PubMed] [Google Scholar]
- 11.Perrin RM, Fedorova ND, Bok JW, Cramer RA, Wortman JR, Kim HS, Nierman WC, Keller NP. Transcriptional regulation of chemical diversity in Aspergillus fumigatus by LaeA. PLoS Pathog. 2007;3:e50. doi: 10.1371/journal.ppat.0030050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shaaban M, Palmer JM, El-Naggar WA, El-Sokkary MA, Habib el SE, Keller NP. Involvement of transposon-like elements in penicillin gene cluster regulation. Fungal Genet Biol. 2010;47:423–432. doi: 10.1016/j.fgb.2010.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13**.Connolly LR, Smith KM, Freitag M. The Fusarium graminearum Histone H3 K27 Methyltransferase KMT6 Regulates Development and Expression of Secondary Metabolite Gene Clusters. PLoS Genet. 2013;9:e1003916. doi: 10.1371/journal.pgen.1003916. Removal of H3K27 methylation in Fusarium results in genome-wide overexpression of ~2,000 genes, many in SMCs. The H3K27 methyltransferase, KMT6, is thus the first truly global repressor of SMCs identified. H3K27me3 overlaps with the highly variable regions identified by recombination profiling of the F. graminearum genome, yet the usually subtelomeric mark has been stably inherited since ancestral chromosomes underwent telomere-telomere fusions to generate the four extant F. graminearum chromosomes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bayram O, Krappmann S, Ni M, Bok JW, Helmstaedt K, Valerius O, Braus-Stromeyer S, Kwon NJ, Keller NP, Yu JH, et al. VelB/VeA/LaeA complex coordinates light signal with fungal development and secondary metabolism. Science. 2008;320:1504–1506. doi: 10.1126/science.1155888. [DOI] [PubMed] [Google Scholar]
- 15.Chang PK, Ehrlich KC, Yu J, Bhatnagar D, Cleveland TE. Increased expression of Aspergillus parasiticus aflR, encoding a sequence-specific DNA-binding protein, relieves nitrate inhibition of aflatoxin biosynthesis. Applied and Environmental Microbiology. 1995;61:2372–2377. doi: 10.1128/aem.61.6.2372-2377.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ehrlich KC, Montalbano BG, Cary JW. Binding of the C6-zinc cluster protein, AFLR, to the promoters of aflatoxin pathway biosynthesis genes in Aspergillus parasiticus. Gene. 1999;230:249–257. doi: 10.1016/s0378-1119(99)00075-x. [DOI] [PubMed] [Google Scholar]
- 17.Bok JW, Keller NP. LaeA, a regulator of secondary metabolism in Aspergillus spp. Eukaryot Cell. 2004;3:527–535. doi: 10.1128/EC.3.2.527-535.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18*.Wiemann P, Guo CJ, Palmer JM, Sekonyela R, Wang CC, Keller NP. Prototype of an intertwined secondary-metabolite supercluster. Proc Natl Acad Sci U S A. 2013;110:17065–17070. doi: 10.1073/pnas.1313258110. Three adjacent SMCs are controlled by LaeA but only two clusters seem to share an activating transcription factor. The authors propose the concept of “superclusters” that are presumably regulated by shared chromatin structure. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reyes-Dominguez Y, Bok JW, Berger H, Shwab EK, Basheer A, Gallmetzer A, Scazzocchio C, Keller N, Strauss J. Heterochromatic marks are associated with the repression of secondary metabolism clusters in Aspergillus nidulans. Mol Microbiol. 2010;76:1376–1386. doi: 10.1111/j.1365-2958.2010.07051.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20*.Soyer JL, El Ghalid M, Glaser N, Ollivier B, Linglin J, Grandaubert J, Balesdent MH, Connolly LR, Freitag M, Rouxel T, et al. Epigenetic Control of Effector Gene Expression in the Plant Pathogenic Fungus Leptosphaeria maculans. PLoS Genet. 2014;10:e1004227. doi: 10.1371/journal.pgen.1004227. The authors show that pathogencity genes, specifically small secreted proteins (SSP) may be controlled by chromatin structure, at least in vitro. To do this they managed to silence both the H3K9 methyltransferase, DIM-5, and HP1 by RNAi. Almost 400 genes were upregulated in silenced strains, and most of these genes were localized in or near AT-rich “isochores” that are subject to silencing by H3K9me3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21**.Jamieson K, Rountree MR, Lewis ZA, Stajich JE, Selker EU. Regional control of histone H3 lysine 27 methylation in Neurospora. Proc Natl Acad Sci U S A. 2013;110:6027–6032. doi: 10.1073/pnas.1303750110. This is the first in-depth paper on H3K27 methylation in fungi; this histone mark was first found in Neurospora (or any fungus) in the same laboratory in 2008. The authors show that H3K27me3 occurs in non-syntenic regions in three Neurospora species. Based on genetic experiments the existence of two different Polycomb Repressive Complexes (PRCs) with SET-7 (KMT6) as catalytic subunit is proposed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22*.Chujo T, Scott B. Histone H3K9 and H3K27 Methylation Regulates Fungal Alkaloid Biosynthesis in a Fungal Endophyte-Plant Symbiosis. Mol Microbiol. 2014 doi: 10.1111/mmi.12567. Epichloe festucae is a well-studied producer of alkaloids. In this study the authors show that expression of bioprotective compounds, lolitrems and ergot alkaloids, is at least partially regulated by H3K9 and H3K27 methylation, and that chromatin states are dependent on growth in axenic culture vs. growth as endophyte on plants. Deletions of the KMT1 (Dim-5/ClrD) and KMT6 (Set-7/EZH2) homologs resulted in decreased growth in culture and on plants, respectively. [DOI] [PubMed] [Google Scholar]
- 23.Slot JC, Rokas A. Horizontal transfer of a large and highly toxic secondary metabolic gene cluster between fungi. Curr Biol. 2011;21:134–139. doi: 10.1016/j.cub.2010.12.020. [DOI] [PubMed] [Google Scholar]
- 24.Schmitt I, Lumbsch HT. Ancient horizontal gene transfer from bacteria enhances biosynthetic capabilities of fungi. PLoS ONE. 2009;4:e4437. doi: 10.1371/journal.pone.0004437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25**.McGary KL, Slot JC, Rokas A. Physical linkage of metabolic genes in fungi is an adaptation against the accumulation of toxic intermediate compounds. Proceedings of the National Academy of Sciences. 2013;110:11481–11486. doi: 10.1073/pnas.1304461110. The authors examined 100 fungal genomes for gene pairs that are metabolic and chromosomal neighbors, so-called “double neighbor gene pairs” (DNGPs). DNGPs are enriched for intermediates that are toxic or have reactive functional groups, and they are more likely divergently oriented, and thus likely transcribed from one promoter. The authors propose that synteny in metabolic pathways is a signature of selection for protection against toxic intermediates. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mancera E, Bourgon R, Brozzi A, Huber W, Steinmetz LM. High-resolution mapping of meiotic crossovers and non-crossovers in yeast. Nature. 2008;454:479–485. doi: 10.1038/nature07135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ellermeier C, Higuchi EC, Phadnis N, Holm L, Geelhood JL, Thon G, Smith GR. RNAi and heterochromatin repress centromeric meiotic recombination. Proc Natl Acad Sci U S A. 2010;107:8701–8705. doi: 10.1073/pnas.0914160107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu S, Hu Y-C, Liu H, Shi Y. Loss of YY1 impacts the heterochromatic state and meiotic double-strand breaks during mouse spermatogenesis. Molecular and Cellular Biology. 2009;29:6245–6256. doi: 10.1128/MCB.00679-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu TC, Lichten M. Meiosis-induced double-strand break sites determined by yeast chromatin structure. Science (New York, NY) 1994;263:515–518. doi: 10.1126/science.8290959. [DOI] [PubMed] [Google Scholar]
- 30.Baudat F, Nicolas A. Clustering of meiotic double-strand breaks on yeast chromosome III. Proceedings of the National Academy of Sciences of the United States of America. 1997;94:5213–5218. doi: 10.1073/pnas.94.10.5213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cuomo CA, Guldener U, Xu JR, Trail F, Turgeon BG, Di Pietro A, Walton JD, Ma LJ, Baker SE, Rep M, et al. The Fusarium graminearum genome reveals a link between localized polymorphism and pathogen specialization. Science. 2007;317:1400–1402. doi: 10.1126/science.1143708. [DOI] [PubMed] [Google Scholar]
- 32.Gale LR, Bryant JD, Calvo S, Giese H, Katan T, O’Donnell K, Suga H, Taga M, Usgaard TR, Ward TJ, et al. Chromosome complement of the fungal plant pathogen Fusarium graminearum based on genetic and physical mapping and cytological observations. Genetics. 2005;171:985–1001. doi: 10.1534/genetics.105.044842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Burt A, Trivers R. Genes in Conflict. Cambridge, MA, U.S.A: Belknap Press of Harvard University; 2006. [Google Scholar]
- 34.Miao VP, Covert SF, VanEtten HD. A fungal gene for antibiotic resistance on a dispensable (“B”) chromosome. Science. 1991;254:1773–1776. doi: 10.1126/science.1763326. [DOI] [PubMed] [Google Scholar]
- 35.Coleman JJ, Rounsley SD, Rodriguez-Carres M, Kuo A, Wasmann CC, Grimwood J, Schmutz J, Taga M, White GJ, Zhou S, et al. The genome of Nectria haematococca: contribution of supernumerary chromosomes to gene expansion. PLoS Genet. 2009;5:e1000618. doi: 10.1371/journal.pgen.1000618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36**.Croll D, Zala M, McDonald BA. Breakage-fusion-bridge Cycles and Large Insertions Contribute to the Rapid Evolution of Accessory Chromosomes in a Fungal Pathogen. PLoS Genet. 2013;9:e1003567. doi: 10.1371/journal.pgen.1003567. The authors suggest mechanisms for chromosomal rearrangements in Zymoseptoria tritici by combining population genomics and “graphical genotyping” with electrophoretic karyotyping and resequencing of parents and progeny from crosses. This revealed diverse complements of B chromosomes that fall into geographic classes when sorted by number and length of B chromosomes. Precise breakpoints for numerous insertions/deletions were shown. Meiotic nondisjunction may have resulted in chromosome loss in progeny of three crosses. The authors provide evidence for the emergence of a novel B chromosome in a few progeny, initiated by fusion between sister chromatids and followed by breakage-fusion-bridge (BFB) cycles to rapidly degenerate chromosomal structure. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ma LJ, van der Does HC, Borkovich KA, Coleman JJ, Daboussi MJ, Di Pietro A, Dufresne M, Freitag M, Grabherr M, Henrissat B, et al. Comparative genomics reveals mobile pathogenicity chromosomes in Fusarium. Nature. 2010;464:367–373. doi: 10.1038/nature08850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rep M, Kistler HC. The genomic organization of plant pathogenicity in Fusarium species. Curr Opin Plant Biol. 2010;13:420–426. doi: 10.1016/j.pbi.2010.04.004. [DOI] [PubMed] [Google Scholar]
- 39.Wittenberg AH, van der Lee TA, Ben M’barek S, Ware SB, Goodwin SB, Kilian A, Visser RG, Kema GH, Schouten HJ. Meiosis drives extraordinary genome plasticity in the haploid fungal plant pathogen Mycosphaerella graminicola. PLoS One. 2009;4:e5863. doi: 10.1371/journal.pone.0005863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jones RN. B chromosomes in plants. New Phytologist. 1995;131:411–434. doi: 10.1111/j.1469-8137.1995.tb03079.x. [DOI] [PubMed] [Google Scholar]
- 41.Covert SF. Supernumerary chromosomes in filamentous fungi. Curr Genet. 1998;33:311–319. doi: 10.1007/s002940050342. [DOI] [PubMed] [Google Scholar]
- 42.Camacho JPM. B chromosomes. In: Gregory TR, editor. The evolution of the genome. Elsevier; 2005. pp. 223–286. [Google Scholar]
- 43*.Hu J, Chen C, Peever T, Dang H, Lawrence C, Mitchell T. Genomic characterization of the conditionally dispensable chromosome in Alternaria arborescens provides evidence for horizontal gene transfer. BMC Genomics. 2012;13:171. doi: 10.1186/1471-2164-13-171. The authors sequenced the genome of A. arborescens to find molecular mechanisms for virulence and the evolution of pathogenicity. They present an assembly of a B chromosome (“conditonally dispensable chromosome”, CDC). As in N. haematococca (F. solani MPVI) and F. oxysporum, differences in codon usage were found between A and B chromosomes genes. PKS and NRPS SMCs cluster on the CDC. In combination, the authors provide sequence-based evidence supporting the origin of this CDC by horizontal transfer from an unrelated fungus. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McClintock B. The Production of Homozygous Deficient Tissues with Mutant Characteristics by Means of the Aberrant Mitotic Behavior of Ring-Shaped Chromosomes. Genetics. 1938;23:315–376. doi: 10.1093/genetics/23.4.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McClintock B. The Stability of Broken Ends of Chromosomes in Zea mays. Genetics. 1941;26:234–282. doi: 10.1093/genetics/26.2.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Goodwin SB, M’Barek SB, Dhillon B, Wittenberg AH, Crane CF, Hane JK, Foster AJ, Van der Lee TA, Grimwood J, Aerts A, et al. Finished genome of the fungal wheat pathogen Mycosphaerella graminicola reveals dispensome structure, chromosome plasticity, and stealth pathogenesis. PLoS Genet. 2011;7:e1002070. doi: 10.1371/journal.pgen.1002070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stukenbrock EH, Dutheil JY. Comparing fungal genomes: insight into functional and evolutionary processes. Methods Mol Biol. 2012;835:531–548. doi: 10.1007/978-1-61779-501-5_33. [DOI] [PubMed] [Google Scholar]
- 48.Stukenbrock EH, Quaedvlieg W, Javan-Nikhah M, Zala M, Crous PW, McDonald BA. Zymoseptoria ardabiliae and Z. pseudotritici, two progenitor species of the septoria tritici leaf blotch fungus Z. tritici (synonym: Mycosphaerella graminicola) Mycologia. 2012;104:1397–1407. doi: 10.3852/11-374. [DOI] [PubMed] [Google Scholar]
- 49.Kema GH, Goodwin SB, Hamza S, Verstappen EC, Cavaletto JR, Van der Lee TA, de Weerdt M, Bonants PJ, Waalwijk C. A combined amplified fragment length polymorphism and randomly amplified polymorphism DNA genetic kinkage map of Mycosphaerella graminicola, the septoria tritici leaf blotch pathogen of wheat. Genetics. 2002;161:1497–1505. doi: 10.1093/genetics/161.4.1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sasaki M, Lange J, Keeney S. Genome destabilization by homologous recombination in the germ line. Nat Rev Mol Cell Biol. 2010;11:182–195. doi: 10.1038/nrm2849. [DOI] [PMC free article] [PubMed] [Google Scholar]