Abstract
An organism’s ability to adapt to its environment depends on its ability to regulate and maintain tissue specific, temporal patterns of gene transcription in response to specific environmental cues. Epigenetic mechanisms are responsible for many of the intricacies of a gene’s regulation that alter expression patterns without affecting the genetic sequence. In particular, DNA methylation has been shown to have an important role in regulating early development and in some human diseases. Within these domains, DNA methylation has been extensively characterized over the past 60 years, but the discovery of its role in regulating behavioral outcomes has led to renewed interest in its potential roles in animal behavior and phenotypic plasticity. The conservation of DNA methylation across the animal kingdom suggests a possible role in the plasticity of genomic responses to environmental cues in natural environments. Here, we review the historical context for the study of DNA methylation, its function and mechanisms, and provide examples of gene/environment interactions in response to social and seasonal cues. Finally, we discuss useful tools to interrogate and dissect the function of DNA methylation in non-model organisms.
Introduction
Epigenetics and DNA methylation
Over the past century, epigenetic mechanisms have been identified with their roles in regulating gene function within cell lineages and/or in the germ line understood. Mechanisms including the covalent modification of DNA (Greenblatt et al. 1994; Elliott et al. 2010), histone modifications, microRNAs (Chuang and Jones 2007), long non-coding RNAs (Mercer et al. 2009), and long-range folding of chromatin (Woodcock and Ghosh 2010) are a few of the mechanisms. Among these, DNA methylation which is the covalent modification of the 5′-cytosine rings via methylation has been widely studied. DNA methylation can provide a dynamic response to environmental cues, and here we review examples of its dynamic responses to the environment.
DNA methylation: the first half century
DNA methylation, discovered >60 years ago, has been described in many taxa of animals (bivalves, echinoderms, amphibians, and mammals) (Bird and Taggart 1980; Tweedie et al. 1997). Historically, however, the study of genetics in tractable model systems overshadowed interest in investigating epigenetic mechanisms such as DNA methylation (Van Speybroeck 2002). This was largely a consequence of DNA methylation being reported as absent in three model systems central to molecular biological analysis, Caenorhabditis elegans (Simpson et al. 1986), Drosophila melanogaster (Rae and Steele 1979; Bird and Taggart 1980; Urieli-Shoval et al. 1982; Patel and Gopinathan 1987), and Saccharomyces cerevisiae (Proffitt et al. 1984). As a consequence, understanding of DNA methylation was technically and conceptually limited. However, the absence of evidence is not evidence of absence since more recent reports found DNA methylation both in some strains of yeast (Tang et al. 2012) and in fruit flies (Lyko et al. 2000).
Interest in the role of DNA methylation in development arose in parallel with discoveries that it was integral to differentiated states of cells and to a regulator of gene function. This led to the suggestion that it was potentially an important mechanism in the fields of cancer and developmental biology (Riggs and Jones 1983). This idea arose from the discovery that a cytidine analog inhibitor of DNA methyltransferases (DNAMTs), the enzyme that catalyzes the DNA methylation reaction, could induce pluripotency and differentiation in 3T3 cell lines (Taylor and Jones 1979).
In cancer biology, aberrant DNA methylation is largely reported at gene-specific loci across the genome leading to the programmed hypomethylation of pro-oncogenic genes (Feinberg and Vogelstein 1983a, 1983b) and to hypermethylation of tumor suppressors genes (Greger et al. 1989; Herman et al. 1995). Aberrant DNA methylation was shown to be regulated by nodal oncogenic (MacLeod and Szyf 1995) and tumor-suppressing pathways (Slack et al. 1999). Inhibition of DNAMT enzymes in turn inhibited tumorigenesis (Ramchandani et al. 1997) and is now being tested in clinical trials in humans as an attractive anticancer target. Metastasis, on the other hand, was found to be driven by demethylation of DNA (Pakneshan et al. 2004) and inhibition of DNA demethylation inhibits cellular invasion and metastasis in vivo (Shukeir et al. 2006). Additionally, global loss of DNA methylation has an effect as well on genomic stability and could be involved in the chromosomal aberrations observed in cancer (Gama-Sosa et al. 1983; Eden et al. 2003). Further, it has been proposed that 5-methylcytosine increases mutability rate of C->T transitions and may serve as an endogenous substrate for oncogenic mutations (Greenblatt et al. 1994). Thus, DNA methylation has become an important factor within the context of a diseased state. In addition, during development, changes in genomic DNA methylation were reported in the primordial germ cells and in pre-implantation embryos in mice. In both instances, genomic erasure of DNA methylation situates cells as pluripotent substrates for genomic methylation and cell differentiation (Mayer et al. 2000; Hajkova et al. 2002; Inoue and Zhang 2011).
In 2004, Weaver et al. showed that maternal behavior of rats could differentially methylate the glucocorticoid receptor (GR) gene of their pups, thus reducing the receptor’s ability to transcribe GR, and ultimately lowering stress responsiveness behavioral outcomes. This seminal study showed that DNA methylation could be induced by behavioral interactions and, in turn, causes subtle alterations in the nervous system and hence behavioral responses. Within a few years of this finding, many investigators showed a role for DNA methylation in a variety of behaviors, including memory (Miller et al. 2010), some pathological states of mental health (Sweatt 2013), and human diseases in addition to cancer (Robertson 2005). These reports revealed the dynamic nature of DNA methylation that matched neural plasticity over a range of time scales.
Functions of DNA methylation
The DNA methylation process has been hypothesized to be evolved to silence evolutionarily accumulated selfish genes (Yoder and Bestor 1998) and transposable elements (Zemach et al. 2010), and to regulate the transcription of genes (Razin and Riggs 1980). In addition, it has been posited to regulate gene splicing (Shukla et al. 2011), X-inactivation, and parental genomic imprinting (Li et al. 1993). Considering the variability of DNA methylation across different species, it is likely to assume various roles. For example, in invertebrate genomes, DNA methylation has been lost to varying degrees and is largely isolated to exonic gene-body elements (Glastad et al. 2011; Sarda et al. 2012).
In mammals, CpG dinucleotides serve as the substrate to which a methyl group is added. Since CpGs are relatively rare in the genome and uniquely arranged, they are thought to be important for transcriptional regulation (Bird 1986). In the genome, regions of higher CpG density comprise CpG islands (CGI) defined as 200 bp windows with a >50% GC content and an observed (within a given sequence) to expected (within the genome) CpG ratio >60% (Fatemi et al. 2005). Mammalian genomes contain CGI islands that can vary in size between 300 and 3000 bp and are found upstream of 40% of transcriptional start-sites. Interestingly, the inverse relationship between a gene’s methylation and its expression has been linked to proximal areas of CGI (defined as CGI shores) (Irizarry et al. 2009) and not the CGI itself. DNA methylation also occurs within a non-CpG context in embryonic stem cells but is mostly absent in somatic tissues (Ramsahoye et al. 2000; Ziller et al. 2011). It should be noted that the mammalian genome is largely methylated in most tissues (70–90%) (Ehrlich et al. 1982).
Mechanistically, DNA methylation has been suggested to cause steric hindrance to transcriptional activators within the major groove of DNA thus blocking expression of a particular gene. In addition, it has been shown to bind several transcriptional repressors such as members of the methyl-binding domain (MBD) family. These act to silence transcription through the recruitment of histone-modifying complexes to block transcription by condensing DNA into heterochromatic complexes (Razin and Riggs 1980; Choy et al. 2010).
DNA methylation toolkit
The methylation of DNA is catalyzed by a family of DNAMTs with highly conserved domains that add methyl moieties from the methyl donor, S-adenosyl-methionine. DNMT1, the first characterized methyltransferase (Bestor 1988), was characterized as a maintenance methyltransferase that targets hemi-methylated daughter strands of newly replicated DNA (Flynn et al. 1996; Bacolla et al. 1999; Fatemi et al. 2001). DNMT3A and DNMT3B catalyze the de novo methylation of DNA (Okano et al. 1999). Both were initially discovered to be upregulated during embryogenesis and have been implicated in generating patterns of de novo DNA methylation during cell differentiation.
The discovery of MBD proteins MBD1-4 and MeCP1 and 2 (Meehan et al. 1989; Nan et al. 1993) provided further understanding of DNA methylation. This family of proteins, characterized with a MBD domain, was shown to have various roles mitigating the function of methylcytosine. Subsequently, several groups have defined their function with roles in gene repression (Jones et al. 1998; Nan et al. 1998) and activation (Collins et al. 2004; Bienvenu and Chelly 2006). For example, MBD2 has been characterized as a transcriptional repressor capable of interacting with histone machinery to form heterochromatin (Ng et al. 1999; Zhang et al. 1999) and as a demethylase capable of inducing transcriptional activation (Bhattacharya et al. 1999; Alvarado et al. 2013).
How is DNA methylation removed? Demethylation through cytosine excision repair has been suggested to occur through growth arrest and DNA-damage-inducible protein 45 (Gadd45a/b) (Wu and Zhang 2014). Similarly, the recently-discovered function of the 10- to 11-translocation (TET) protein family has revealed that hydroxylation of methyl cytosine may be an oxidative intermediate of demethylation. Considering its mechanism, 5-hydroxymethylcytosine has been reported to possibly regulate transcriptional activation as seen in embryonic stem cells (Tahiliani et al. 2009) and the central nervous system (Kriaucionis and Heintz 2009; Lister et al. 2009). Other reports have suggested that, following TET-mediated hydroxylation, either activation induced deaminase (AID/APOBEC) and/or thymine DNA glycosylase (TDG) + MBD4 mediate a demethylation reaction on methylcytosine (Wu and Zhang 2014).
Seasonal regulation and DNA methylation
Many seasonal cues such as temperature and light (Epperson and Martin 2002; Ruby et al. 2002) can have profound effects on the regulation of transcriptional mechanisms in natural environments. Seasons provide predictable environmental changes to which an animal can evolve plastic transcriptional programs producing adaptations to metabolism and reproduction. While the exact role DNA methylation plays in these scenarios has yet to be fully explored, evidence shows that DNA methylation regulates many seasonally related candidate gene pathways (Figure 1).
Fig. 1.
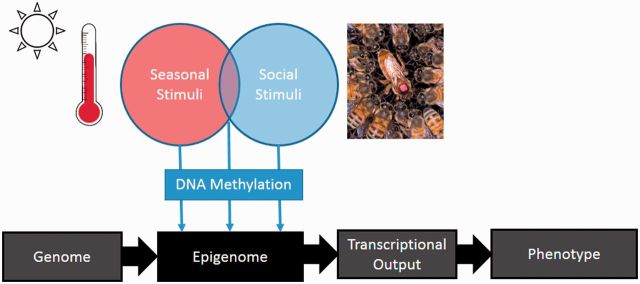
Summary of how social and seasonal stimuli can affect phenotype through transcriptional outputs where gene transcription defines a specific trait.
Hibernation requires seasonally regulated metabolism and mammals conserve energy in the winter by undergoing prolonged bouts of torpor interspersed with brief arousals back to euthermia. During torpor, energy expenditures drop to as low as 1–5% of euthermic rates (Carey et al. 2003) and are accompanied by a suite of transcriptional changes (Srere et al. 1992). Since DNA methylation is largely conserved among vertebrate mammals, we believe that such a mechanism might be central for the differential gene expression that accompanies seasonal change. For example, the hibernation-specific protein-27 (HP-27), known to be upregulated in the blood of hibernating chipmunks (Kondo and Kondo 1992), is regulated by DNA methylation in a tissue-specific manner under the transcriptional control of upstream stimulatory factor-1 (USF1) (Fujii et al. 2006). Considering the mechanisms involved, DNMT3B, a de novo methyltransferase, is regulated by BmaL-regulated circadian rhythms in the liver (Maekawa et al. 2012) that underlie metabolic depression during hibernation in ground squirrels (Ruby et al. 2002). In examining circadian rhythms of the superchiasmatic nuclei during natural diurnal cycles, DNA methylation is shown to be remarkably plastic (Azzi et al. 2014). Considering the importance of these rhythms in response to seasonal change, we speculate their role may be integral for adapting to altered photoperiods.
Since most genomic repertoires of seasonal hibernators are strikingly similar to non-hibernators, we propose that the hibernating phenotype does not necessarily require novel genes but may result from unique epigenetic reconfiguration of its genome. Harris et al. (2000) has proposed a hypothesis of “hibernator as neonate” suggesting that the genetic predisposition for heterothermy is expressed to some extent in all neonates. Further, this hypothesis suggests that heterothermy in hibernation results from the continued expression in the requisite genes. Given the widespread conservation of heterothermy and epigenetic processes, we propose that a mechanism such as DNA methylation could complement an ancestral mammalian capacity for heterothermy (Geiser 1998).
In many vertebrates, seasonally regulated photoperiod changes regulate the development of discrete reproductive phenotypes (Dawson et al. 2001; Goldman 2001; Stevenson and Ball 2011). These distinct phenotypes may be a consequence of epigenetic changes though this is not known. However, in hamsters, changes in photoperiod induce thyroid-hormone-dependent induction of discrete reproductive phenotypes. DNA methylation has been shown to regulate this process through control of type III deiodinase gene in the hypothalamus (Stevenson and Prendergast 2013). Similarly, in humans, the season of conception among rural Gambians can result in aberrant DNA methylation of metastable epialleles (Waterland et al. 2010). This study showed that nutritionally challenged parents that conceived during the rainy season generated offspring with a unique signature of methylation when compared with others conceived out of the rainy season.
Seasonal variation is a broad term to describe a predictable change in environment that elicits an adaptive response that is not solely limited to metabolic regulation or reproduction. For example, the seasonal plasticity of coat color that produces crypsis to avoid predation in the case of deer mice and patterning that results from mutations in the agouti gene (Linnen et al. 2009, 2013). Interestingly, Waterland and Jirtle (2003) showed that the agouti gene promoter could be hypermethylated through dietary folate, a methyl donor, and capable of modulating coat coloring. While there is no direct link between DNA methylation of the agouti gene and seasonal plasticity of crypsis, we propose that DNA methylation may provide plasticity to such genes with important ecological and evolutionary consequences.
Role of social environment
Changes in behavior enable animals to face the challenges of feeding, evading predators, and breeding, but the specific cellular and molecular mechanisms that mediate such behaviors are largely unknown. While genetically set behavior patterns provide a functional basis for behavior, they do not account for the plasticity and dynamic nature of behavior (Figure 1). Since social interactions can regulate context-specific behaviors, they can shape the development and physiology of the brain through neuroanatomical and molecular changes in gene expression (Davis and Fernald 1990; Burmeister et al. 2005; Robinson et al. 2008; Maruska and Fernald 2011; Fernald 2012).
Early social interactions can have lasting effects on gene transcription through DNA methylation. As previously mentioned, maternal licking and grooming can alter DNA methylation and stress responsiveness through the GR (Weaver et al. 2004). These interactions have been described in models of maternal separation (Murgatroyd et al. 2009) and abuse (Roth et al. 2009) modifying the genes expressing arginine vasopressin and brain-derived neurotropic factor, respectively. While deprived social environments can induce diseased states of DNA methylation, social interactions can also protect changes in DNA methylation. For example, neuropathy-induced genomic CpG demethylation in the prefrontal cortex of mice can be protected by enrichment through social interactions with littermates (Tajerian et al. 2013).
Do epigenetic marks influence behavior, particularly social status? DNA methylation at gene-specific promoters in the brain has revealed an interesting signature unique to social status in animals. For example, in mammals, paradigms of social defeat and environmental enrichment both have stable effects on DNA methylation. Resilience in mice treated in a social-defeat model is accompanied by hypermethylation of the corticotrophin releasing factor gene and influences behavioral outcomes (Elliott et al. 2010). Furthermore, in humans, signatures of DNA methylation assayed from many different tissues can serve as biomarkers that categorize components of social behavior or status. For example, genomic signatures of DNA methylation assayed from blood are capable of categorizing socioeconomic status in children (Borghol et al. 2012) and aggression in adults (Guillemin et al. 2014). Similarly, peer rearing in rhesus macaque monkeys provides aggressive phenotypes with distinguishing signatures of methylation between mother-reared and isolated controls (Provencal et al. 2013).
There are also examples of DNA methylation responding to social and nutritional cues in insects. However, differences in DNA methylation in insects occur primarily in exonic areas of the genome (Lyko et al. 2010; Bonasio et al. 2012) suggesting a possible role in alternative splicing (Shukla et al. 2011; Foret et al. 2012). In bees, nutritional cues were shown to regulate patterns of DNA methylation between worker and queen castes (Kucharski et al. 2008). Furthermore, within worker castes, reports also show that complex social tasks result in unique task-specific signatures of gene methylation that may alternatively regulate genes through splicing (Lockett et al. 2012). Furthermore, in bees, DNA methylation can reversibly mark behavioral subcastes of workers that forage and nurse, respectively (Herb et al. 2012).
Molecular investigation of DNA methylation in non-model systems
To discover whether DNA methylation plays any role in social or seasonal phenotypes alone or in response to other exogenous factors, it must be localized and its effects tested. Specifically, in a given tissue, a genome will likely have many genes that are methylated during development and remain so throughout life. To identify these, a paradigm needs to be identified to compare animals in at least two distinct, semi-natural states. If a fully-annotated and sequenced genome does not exist, there are many possibilities for discovering whether DNA methylation might be implicated in different phenotypes. Here, we describe the use of restriction enzymes to assay genomic methylation and identify DNA methylation at specific loci and pharmacology targeted toward DNA methylation machinery. With the cost of sequencing dropping dramatically, sequencing of non-model organisms may become more common.
Methylation sensitive restriction digestion
To study DNA methylation, it is essential to be able to identify genomic loci that show differential methylation or, if possible to assay methylation across the genome. The use of restriction enzymes has been particularly useful in this endeavor as there are many isoschizomer pairs that can be methylation sensitive and insensitive thus capable of generating different patterns of digestion, dependent on DNA methylation. For example, the restriction enzymes MspI and HpaII have been particularly useful in assaying DNA methylation within a CpG context (Waalwijk and Flavell 1978). These enzymes identify methylated loci for sequencing, using methylation-sensitive, amplified fragment-length polymorphisms (MS-AFLP) (Xiong et al. 1999) and are widely applicable to ecological epigenetics in plants and animals (Schrey et al. 2013). Furthermore, the specific fragments identified from MS-AFLP screen can be sequenced at a low cost and situated within certain genomic loci given closely related and sequenced reference genomes. In the same vein, HpaII and MspI generate sticky ends that can be pyrosequenced to generate genomic indices of DNA methylation with the luminometric methylation assay (LUMA) (Karimi et al. 2006). These approaches have been particularly useful in identifying toxicological impacts of mercury on DNA methylation in polar bears, mink, yellow perch, and chickens (Pilsner et al. 2010; Basu et al. 2013; Head et al. 2014). Taken together, a technique such as LUMA can identify genomic changes in methylation and MS-AFLP can identify specific loci worthy of follow-up studies.
Pharmacology of DNA methylation machinery
To find causal link to DNA methylation, cancer biologists have produced a suite of drugs capable of affecting DNA methylation. Specifically the cytidine analogs, 5-aza-cytidine and 5-aza-decytidine are well-characterized nucleoside analogs that incorporate into DNA during replication and halt DNMT1 maintenance methylation, thus demethylating the genome (Ghoshal et al. 2005). In non-dividing tissues, 5-aza-decytidine may have limited effects due to their replication-dependent mechanism; other drugs that target de novo methyltransferases would be better. For example, RG108 targets the conserved C-terminal domain of DNMTs, and can be employed to inhibit de novo methylation (Schirrmacher et al. 2006). In order to hypermethylate the genome, approaches are limited to providing methyl-donors, such as S-adenosyl-methionine and S-adenosyl-homocysteine, to the methylation reaction (Charles et al. 2012). Unfortunately, methylation in general is a ubiquitous process and its substrates may have confounding effects on other subcellular processes relevant to RNA, protein, and lipids. Thus, such pharmacological approaches should be used to complement one another to identify mechanisms central to DNA methylation.
Concluding remarks
Within the past decade, the study of DNA methylation has expanded into many fields. Given its dynamic nature, it may function widely to modulate genomic responses to a given environment. However, the intrinsic regulation of such a mechanism still begs many questions about gene regulation. For example, at what temporal scales (minutes to years) are DNA methylation signatures laid down and removed? Is the methylated state of a gene a permanent molecular memory or the maintenance equilibrium between DNA methylases and demethylases? Does DNA methylation initiate a transition to seasonally/socially-related transcriptional phenotypes, or is it a downstream signature of these environmental changes? The phenotypic variability seen throughout the animal kingdom suggests that adopting new models for DNA methylation studies may reveal how these epigenetic modifications can lend plasticity to a static genome by environmental interactions.
Funding
SA and RDF were funded by an NIH NINDS (NS34950) and SA was supported by a gift from the Sackler Foundation. KBS was funded by an NSERC discovery Grant, MS funded by a grant from the Canadian Institute of Health (MOP42411).
References
- Alvarado S, Wyglinski J, Suderman M, Andrews SA, Szyf M. Methylated DNA binding domain protein 2 (MBD2) coordinately silences gene expression through activation of the microRNA hsa-mir-496 promoter in breast cancer cell line. PLoS One. 2013;8:e74009. doi: 10.1371/journal.pone.0074009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azzi A, Dallmann R, Casserly A, Rehrauer H, Patrignani A, Maier B, Kramer A, Brown SA. Circadian behavior is light-reprogrammed by plastic DNA methylation. Nat Neurosci. 2014;17:377–82. doi: 10.1038/nn.3651. [DOI] [PubMed] [Google Scholar]
- Bacolla A, Pradhan S, Roberts RJ, Wells RD. Recombinant human DNA (cytosine-5) methyltransferase. II. Steady-state kinetics reveal allosteric activation by methylated DNA. J Biol Chem. 1999;274:33011–9. doi: 10.1074/jbc.274.46.33011. [DOI] [PubMed] [Google Scholar]
- Basu N, Head J, Nam DH, Pilsner JR, Carvan MJ, Chan HM, Goetz FW, Murphy CA, Rouvinen-Watt K, Scheuhammer AM. Effects of methylmercury on epigenetic markers in three model species: mink, chicken and yellow perch. Comp Biochem Physiol C Toxicol Pharmacol. 2013;157:322–7. doi: 10.1016/j.cbpc.2013.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bestor TH. Cloning of a mammalian DNA methyltransferase. Gene. 1988;74:9–12. doi: 10.1016/0378-1119(88)90238-7. [DOI] [PubMed] [Google Scholar]
- Bhattacharya SK, Ramchandani S, Cervoni N, Szyf M. A mammalian protein with specific demethylase activity for mCpG DNA. Nature. 1999;397:579–83. doi: 10.1038/17533. [DOI] [PubMed] [Google Scholar]
- Bienvenu T, Chelly J. Molecular genetics of Rett syndrome: when DNA methylation goes unrecognized. Nat Rev Genet. 2006;7:415–26. doi: 10.1038/nrg1878. [DOI] [PubMed] [Google Scholar]
- Bird AP. CpG-rich islands and the function of DNA methylation. Nature. 1986;321:209–13. doi: 10.1038/321209a0. [DOI] [PubMed] [Google Scholar]
- Bird AP, Taggart MH. Variable patterns of total DNA and rDNA methylation in animals. Nucleic Acids Res. 1980;8:1485–97. doi: 10.1093/nar/8.7.1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonasio R, Li Q, Lian J, Mutti NS, Jin L, Zhao H, Zhang P, Wen P, Xiang H, Ding Y, et al. Genome-wide and caste-specific DNA methylomes of the ants Camponotus floridanus and Harpegnathos saltator. Curr Biol. 2012;22:1755–64. doi: 10.1016/j.cub.2012.07.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borghol N, Suderman M, McArdle W, Racine A, Hallett M, Pembrey M, Hertzman C, Power C, Szyf M. Associations with early-life socio-economic position in adult DNA methylation. Int J Epidemiol. 2012;41:62–74. doi: 10.1093/ije/dyr147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burmeister SS, Jarvis ED, Fernald RD. PLoS Biology. 2005 doi: 10.1371/journal.pbio.0030363. 3, art. no. e363: 1996–2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey HV, Andrews MT, Martin SL. Mammalian hibernation: cellular and molecular responses to depressed metabolism and low temperature. Physiol Rev. 2003;83:1153–81. doi: 10.1152/physrev.00008.2003. [DOI] [PubMed] [Google Scholar]
- Charles MA, Johnson IT, Belshaw NJ. Supra-physiological folic acid concentrations induce aberrant DNA methylation in normal human cells in vitro. Epigenetics. 2012;7:689–94. doi: 10.4161/epi.20461. [DOI] [PubMed] [Google Scholar]
- Choy JS, Wei S, Lee JY, Tan S, Chu S, Lee TH. DNA methylation increases nucleosome compaction and rigidity. J Am Chem Soc. 2010;132:1782–3. doi: 10.1021/ja910264z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang JC, Jones PA. Epigenetics and microRNAs. Pediatr Res. 2007;61:24R–29R. doi: 10.1203/pdr.0b013e3180457684. [DOI] [PubMed] [Google Scholar]
- Collins AL, Levenson JM, Vilaythong AP, Richman R, Armstrong DL, Noebels JL, David Sweatt J, Zoghbi HY. Mild overexpression of MeCP2 causes a progressive neurological disorder in mice. Hum Mol Genet. 2004;13:2679–89. doi: 10.1093/hmg/ddh282. [DOI] [PubMed] [Google Scholar]
- Davis MR, Fernald RD. Social control of neuronal soma size. J Neurobiol. 1990;21:1180–8. doi: 10.1002/neu.480210804. [DOI] [PubMed] [Google Scholar]
- Dawson A, King VM, Bentley GE, Ball GF. Photoperiodic control of seasonality in birds. J Biol Rhythms. 2001;16:365–80. doi: 10.1177/074873001129002079. [DOI] [PubMed] [Google Scholar]
- Eden A, Gaudet F, Waghmare A, Jaenisch R. Chromosomal instability and tumors promoted by DNA hypomethylation. Science. 2003;300:455. doi: 10.1126/science.1083557. [DOI] [PubMed] [Google Scholar]
- Ehrlich M, Gama-Sosa MA, Huang LH, Midgett RM, Kuo KC, McCune RA, Gehrke C. Amount and distribution of 5-methylcytosine in human DNA from different types of tissues of cells. Nucleic Acids Res. 1982;10:2709–21. doi: 10.1093/nar/10.8.2709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott E, Ezra-Nevo G, Regev L, Neufeld-Cohen A, Chen A. Resilience to social stress coincides with functional DNA methylation of the Crf gene in adult mice. Nat Neurosci. 2010;13:1351–3. doi: 10.1038/nn.2642. [DOI] [PubMed] [Google Scholar]
- Epperson LE, Martin SL. Quantitative assessment of ground squirrel mRNA levels in multiple stages of hibernation. Physiol Genomics. 2002;10:93–102. doi: 10.1152/physiolgenomics.00004.2002. [DOI] [PubMed] [Google Scholar]
- Fatemi M, Hermann A, Pradhan S, Jeltsch A. The activity of the murine DNA methyltransferase Dnmt1 is controlled by interaction of the catalytic domain with the N-terminal part of the enzyme leading to an allosteric activation of the enzyme after binding to methylated DNA. J Mol Biol. 2001;309:1189–99. doi: 10.1006/jmbi.2001.4709. [DOI] [PubMed] [Google Scholar]
- Fatemi M, Pao MM, Jeong S, Gal-Yam EN, Egger G, Weisenberger DJ, Jones PA. Footprinting of mammalian promoters: use of a CpG DNA methyltransferase revealing nucleosome positions at a single molecule level. Nucleic acids Res. 2005;33:e176. doi: 10.1093/nar/gni180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg AP, Vogelstein B. Hypomethylation of ras oncogenes in primary human cancers. Biochem Biophys Res Commun. 1983a;111:47–54. doi: 10.1016/s0006-291x(83)80115-6. [DOI] [PubMed] [Google Scholar]
- Feinberg AP, Vogelstein B. Hypomethylation distinguishes genes of some human cancers from their normal counterparts. Nature. 1983b;301:89–92. doi: 10.1038/301089a0. [DOI] [PubMed] [Google Scholar]
- Fernald RD, Maruska KP. Social information changes the brain. Proc Natl Acad Sci U S A. 2012;107:21176–80. doi: 10.1073/pnas.1202552109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynn J, Glickman JF, Reich NO. Murine DNA cytosine-C5 methyltransferase: pre-steady- and steady-state kinetic analysis with regulatory DNA sequences. Biochemistry. 1996;35:7308–15. doi: 10.1021/bi9600512. [DOI] [PubMed] [Google Scholar]
- Foret S, Kucharski R, Pellegrini M, Feng S, Jacobsen SE, Robinson GE, Maleszka R. DNA methylation dynamics, metabolic fluxes, gene splicing, and alternative phenotypes in honey bees. Proc Natl Acad Sci U S A. 2012;109:4968–73. doi: 10.1073/pnas.1202392109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii G, Nakamura Y, Tsukamoto D, Ito M, Shiba T, Takamatsu N. CpG methylation at the USF-binding site is important for the liver-specific transcription of the chipmunk HP-27 gene. Biochem J. 2006;395:203–9. doi: 10.1042/BJ20051802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gama-Sosa MA, Slagel VA, Trewyn RW, Oxenhandler R, Kuo KC, Gehrke CW, Ehrlich M. The 5-methylcytosine content of DNA from human tumors. Nucleic Acids Res. 1983;11:6883–94. doi: 10.1093/nar/11.19.6883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiser F. Evolution of daily torpor and hibernation in birds and mammals: importance of body size. Clin Exp Pharmacol Physiol. 1998;25:736–9. doi: 10.1111/j.1440-1681.1998.tb02287.x. [DOI] [PubMed] [Google Scholar]
- Ghoshal K, Datta J, Majumder S, Bai S, Kutay H, Motiwala T, Jacob ST. 5-Aza-deoxycytidine induces selective degradation of DNA methyltransferase 1 by a proteasomal pathway that requires the KEN box, bromo-adjacent homology domain, and nuclear localization signal. Mol Cell Biol. 2005;25:4727–41. doi: 10.1128/MCB.25.11.4727-4741.2005. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Glastad KM, Hunt BG, Yi SV, Goodisman MA. DNA methylation in insects: on the brink of the epigenomic era. Insect Mol Biol. 2011;20:553–65. doi: 10.1111/j.1365-2583.2011.01092.x. [DOI] [PubMed] [Google Scholar]
- Goldman BD. Mammalian photoperiodic system: formal properties and neuroendocrine mechanisms of photoperiodic time measurement. J Biol Rhythms. 2001;16:283–301. doi: 10.1177/074873001129001980. [DOI] [PubMed] [Google Scholar]
- Greenblatt MS, Bennett WP, Hollstein M, Harris CC. Mutations in the p53 tumor suppressor gene: clues to cancer etiology and molecular pathogenesis. Cancer Res. 1994;54:4855–78. [PubMed] [Google Scholar]
- Greger V, Passarge E, Hopping W, Messmer E, Horsthemke B. Epigenetic changes may contribute to the formation and spontaneous regression of retinoblastoma. Hum Genet. 1989;83:155–8. doi: 10.1007/BF00286709. [DOI] [PubMed] [Google Scholar]
- Guillemin C, Provencal N, Suderman M, Cote SM, Vitaro F, Hallett M, Tremblay RE, Szyf M. DNA methylation signature of childhood chronic physical aggression in T cells of both men and women. PLoS One. 2014;9:e86822. doi: 10.1371/journal.pone.0086822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajkova P, Erhardt S, Lane N, Haaf T, El-Maarri O, Reik W, Walter J, Surani MA. Epigenetic reprogramming in mouse primordial germ cells. Mech Dev. 2002;117:15–23. doi: 10.1016/s0925-4773(02)00181-8. [DOI] [PubMed] [Google Scholar]
- Harris MB, Olson LE, Milsom WK. The origin of mammalian heterothermy: a case for perpetual youth? In: Barnes BM, Carey HV, editors. Life in the cold: evolution, mechanisms, adaptation, and application. Vol. 12. 2000. p. 44. [Google Scholar]
- Head JA, Mittal K, Basu N. Application of the LUminometric Methylation Assay (LUMA) to ecological species; tissue quality requirements and a survey of DNA methylation levels in animals. Mol Ecol Resour published online (doi: 10.1111/1755-0998.12244) 2014 doi: 10.1111/1755-0998.12244. [DOI] [PubMed] [Google Scholar]
- Herb BR, Wolschin F, Hansen KD, Aryee MJ, Langmead B, Irizarry R, Amdam GV, Feinberg AP. Reversible switching between epigenetic states in honeybee behavioral subcastes. Nat Neurosci. 2012;15:1371–3. doi: 10.1038/nn.3218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman JG, Merlo A, Mao L, Lapidus RG, Issa JP, Davidson NE, Sidransky D, Baylin SB. Inactivation of the CDKN2/p16/MTS1 gene is frequently associated with aberrant DNA methylation in all common human cancers. Cancer Res. 1995;55:4525–30. [PubMed] [Google Scholar]
- Inoue A, Zhang Y. Replication-dependent loss of 5-hydroxymethylcytosine in mouse preimplantation embryos. Science. 2011;334:194. doi: 10.1126/science.1212483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry RA, Ladd-Acosta C, Wen B, Wu Z, Montano C, Onyango P, Cui H, Gabo K, Rongione M, Webster M, et al. The human colon cancer methylome shows similar hypo- and hypermethylation at conserved tissue-specific CpG island shores. Nat Genet. 2009;41:178–86. doi: 10.1038/ng.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones PL, Veenstra GJ, Wade PA, Vermaak D, Kass SU, Landsberger N, Strouboulis J, Wolffe AP. Methylated DNA and MeCP2 recruit histone deacetylase to repress transcription. Nat Genet. 1998;19:187–91. doi: 10.1038/561. [DOI] [PubMed] [Google Scholar]
- Karimi M, Johansson S, Ekstrom TJ. Using LUMA: a luminometric-based assay for global DNA-methylation. Epigenetics. 2006;1:45–8. doi: 10.4161/epi.1.1.2587. [DOI] [PubMed] [Google Scholar]
- Kondo N, Kondo J. Identification of novel blood proteins specific for mammalian hibernation. J Biol Chem. 1992;267:473–8. [PubMed] [Google Scholar]
- Kriaucionis S, Heintz N. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science. 2009;324:929–30. doi: 10.1126/science.1169786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kucharski R, Maleszka J, Foret S, Maleszka R. Nutritional control of reproductive status in honeybees via DNA methylation. Science. 2008;319:1827–30. doi: 10.1126/science.1153069. [DOI] [PubMed] [Google Scholar]
- Li E, Beard C, Jaenisch R. Role for DNA methylation in genomic imprinting. Nature. 1993;366:362–5. doi: 10.1038/366362a0. [DOI] [PubMed] [Google Scholar]
- Linnen CR, Kingsley EP, Jensen JD, Hoekstra HE. On the origin and spread of an adaptive allele in deer mice. Science. 2009;325:1095–8. doi: 10.1126/science.1175826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linnen CR, Poh YP, Peterson BK, Barrett RD, Larson JG, Jensen JD, Hoekstra HE. Adaptive evolution of multiple traits through multiple mutations at a single gene. Science. 2013;339:1312–6. doi: 10.1126/science.1233213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J, Nery JR, Lee L, Ye Z, Ngo QM, et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462:315–22. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lockett GA, Kucharski R, Maleszka R. DNA methylation changes elicited by social stimuli in the brains of worker honey bees. Genes Brain Behav. 2012;11:235–42. doi: 10.1111/j.1601-183X.2011.00751.x. [DOI] [PubMed] [Google Scholar]
- Lyko F, Ramsahoye BH, Jaenisch R. DNA methylation in Drosophila melanogaster. Nature. 2000;408:538–40. doi: 10.1038/35046205. [DOI] [PubMed] [Google Scholar]
- Lyko F, Foret S, Kucharski R, Wolf S, Falckenhayn C, Maleszka R. The honey bee epigenomes: differential methylation of brain DNA in queens and workers. PLoS Biol. 2010;8:e1000506. doi: 10.1371/journal.pbio.1000506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLeod AR, Szyf M. Expression of antisense to DNA methyltransferase mRNA induces DNA demethylation and inhibits tumorigenesis. J Biol Chem. 1995;270:8037–43. doi: 10.1074/jbc.270.14.8037. [DOI] [PubMed] [Google Scholar]
- Maekawa F, Shimba S, Takumi S, Sano T, Suzuki T, Bao J, Ohwada M, Ehara T, Ogawa Y, Nohara K. Diurnal expression of Dnmt3b mRNA in mouse liver is regulated by feeding and hepatic clockwork. Epigenetics. 2012;7:1046–56. doi: 10.4161/epi.21539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruska KP, Fernald RD. Plasticity of the reproductive axis caused by social status change in an african cichlid fish: II. testicular gene expression and spermatogenesis. Endocrinology. 2011;152:291–302. doi: 10.1210/en.2010-0876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer W, Niveleau A, Walter J, Fundele R, Haaf T. Demethylation of the zygotic paternal genome. Nature. 2000;403:501–2. doi: 10.1038/35000656. [DOI] [PubMed] [Google Scholar]
- Meehan RR, Lewis JD, McKay S, Kleiner EL, Bird AP. Identification of a mammalian protein that binds specifically to DNA containing methylated CpGs. Cell. 1989;58:499–507. doi: 10.1016/0092-8674(89)90430-3. [DOI] [PubMed] [Google Scholar]
- Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10:155–9. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- Miller CA, Gavin CF, White JA, Parrish RR, Honasoge A, Yancey CR, Rivera IM, Rubio MD, Rumbaugh G, Sweatt JD. Cortical DNA methylation maintains remote memory. Nat Neurosci. 2010;13:664–6. doi: 10.1038/nn.2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murgatroyd C, Patchev AV, Wu Y, Micale V, Bockmuhl Y, Fischer D, Holsboer F, Wotjak CT, Almeida OF, Spengler D. Dynamic DNA methylation programs persistent adverse effects of early-life stress. Nat Neurosci. 2009;12:1559–66. doi: 10.1038/nn.2436. [DOI] [PubMed] [Google Scholar]
- Nan X, Meehan RR, Bird A. Dissection of the methyl-CpG binding domain from the chromosomal protein MeCP2. Nucleic Acids Res. 1993;21:4886–92. doi: 10.1093/nar/21.21.4886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nan X, Ng HH, Johnson CA, Laherty CD, Turner BM, Eisenman RN, Bird A. Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature. 1998;393:386–9. doi: 10.1038/30764. [DOI] [PubMed] [Google Scholar]
- Ng HH, Zhang Y, Hendrich B, Johnson CA, Turner BM, Erdjument-Bromage H, Tempst P, Reinberg D, Bird A. MBD2 is a transcriptional repressor belonging to the MeCP1 histone deacetylase complex. Nat Genet. 1999;23:58–61. doi: 10.1038/12659. [DOI] [PubMed] [Google Scholar]
- Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247–57. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- Pakneshan P, Szyf M, Farias-Eisner R, Rabbani SA. Reversal of the hypomethylation status of urokinase (uPA) promoter blocks breast cancer growth and metastasis. J Biol Chem. 2004;279:31735–44. doi: 10.1074/jbc.M401669200. [DOI] [PubMed] [Google Scholar]
- Patel CV, Gopinathan KP. Determination of trace amounts of 5-methylcytosine in DNA by reverse-phase high-performance liquid chromatography. Anal Biochem. 1987;164:164–9. doi: 10.1016/0003-2697(87)90381-2. [DOI] [PubMed] [Google Scholar]
- Pilsner JR, Lazarus AL, Nam DH, Letcher RJ, Sonne C, Dietz R, Basu N. Mercury-associated DNA hypomethylation in polar bear brains via the LUminometric Methylation Assay: a sensitive method to study epigenetics in wildlife. Mol Ecol. 2010;19:307–14. doi: 10.1111/j.1365-294X.2009.04452.x. [DOI] [PubMed] [Google Scholar]
- Proffitt JH, Davie JR, Swinton D, Hattman S. 5-Methylcytosine is not detectable in Saccharomyces cerevisiae DNA. Mol Cell Biol. 1984;4:985–8. doi: 10.1128/mcb.4.5.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Provencal N, Suderman MJ, Caramaschi D, Wang D, Hallett M, Vitaro F, Tremblay RE, Szyf M. Differential DNA methylation regions in cytokine and transcription factor genomic loci associate with childhood physical aggression. PLoS One. 2013;8:e71691. doi: 10.1371/journal.pone.0071691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rae PM, Steele RE. Absence of cytosine methylation at C-C-G-G and G-C-G-C sites in the rDNA coding regions and intervening sequences of Drosophila and the rDNA of other insects. Nucleic Acids Res. 1979;6:2987–95. doi: 10.1093/nar/6.9.2987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramchandani S, MacLeod AR, Pinard M, von Hofe E, Szyf M. Inhibition of tumorigenesis by a cytosine-DNA, methyltransferase, antisense oligodeoxynucleotide. Proc Natl Acad Sci U S A. 1997;94:684–9. doi: 10.1073/pnas.94.2.684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsahoye BH, Biniszkiewicz D, Lyko F, Clark V, Bird AP, Jaenisch R. Non-CpG methylation is prevalent in embryonic stem cells and may be mediated by DNA methyltransferase 3a. Proc Natl Acad Sci U S A. 2000;97:5237–42. doi: 10.1073/pnas.97.10.5237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razin A, Riggs AD. DNA methylation and gene function. Science. 1980;210:604–10. doi: 10.1126/science.6254144. [DOI] [PubMed] [Google Scholar]
- Riggs AD, Jones PA. 5-Methylcytosine, gene regulation, and cancer. Adv Cancer Res. 1983;40:1–30. doi: 10.1016/s0065-230x(08)60678-8. [DOI] [PubMed] [Google Scholar]
- Robertson KD. DNA methylation and human disease. Nat Rev Genet. 2005;6:597–610. doi: 10.1038/nrg1655. [DOI] [PubMed] [Google Scholar]
- Robinson GE, Fernald RD, Clayton DF. Genes and social behavior. Science. 2008;322:896–900. doi: 10.1126/science.1159277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth TL, Lubin FD, Funk AJ, Sweatt JD. Lasting epigenetic influence of early-life adversity on the BDNF gene. Biol Psychiatry. 2009;65:760–9. doi: 10.1016/j.biopsych.2008.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruby NF, Dark J, Burns DE, Heller HC, Zucker I. The suprachiasmatic nucleus is essential for circadian body temperature rhythms in hibernating ground squirrels. J Neurosci. 2002;22:357–64. doi: 10.1523/JNEUROSCI.22-01-00357.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarda S, Zeng J, Hunt BG, Yi SV. The evolution of invertebrate gene body methylation. Mol Biol Evol. 2012;29:1907–16. doi: 10.1093/molbev/mss062. [DOI] [PubMed] [Google Scholar]
- Schirrmacher E, Beck C, Brueckner B, Schmitges F, Siedlecki P, Bartenstein P, Lyko F, Schirrmacher R. Synthesis and in vitro evaluation of biotinylated RG108: a high affinity compound for studying binding interactions with human DNA methyltransferases. Bioconjug Chem. 2006;17:261–6. doi: 10.1021/bc050300b. [DOI] [PubMed] [Google Scholar]
- Schrey AW, Alvarez M, Foust CM, Kilvitis HJ, Lee JD, Liebl AL, Martin LB, Richards CL, Robertson M. Ecological epigenetics: beyond MS-AFLP. Integr Comp Biol. 2013;53:340–50. doi: 10.1093/icb/ict012. [DOI] [PubMed] [Google Scholar]
- Shukeir N, Pakneshan P, Chen G, Szyf M, Rabbani SA. Alteration of the methylation status of tumor-promoting genes decreases prostate cancer cell invasiveness and tumorigenesis in vitro and in vivo. Cancer Res. 2006;66:9202–10. doi: 10.1158/0008-5472.CAN-06-1954. [DOI] [PubMed] [Google Scholar]
- Shukla S, Kavak E, Gregory M, Imashimizu M, Shutinoski B, Kashlev M, Oberdoerffer P, Sandberg R, Oberdoerffer S. CTCF-promoted RNA polymerase II pausing links DNA methylation to splicing. Nature. 2011;479:74–9. doi: 10.1038/nature10442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson VJ, Johnson TE, Hammen RF. Caenorhabditis elegans DNA does not contain 5-methylcytosine at any time during development or aging. Nucleic Acids Res. 1986;14:6711–9. doi: 10.1093/nar/14.16.6711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slack A, Cervoni N, Pinard M, Szyf M. DNA methyltransferase is a downstream effector of cellular transformation triggered by simian virus 40 large T antigen. J Biol Chem. 1999;274:10105–12. doi: 10.1074/jbc.274.15.10105. [DOI] [PubMed] [Google Scholar]
- Srere HK, Wang LC, Martin SL. Central role for differential gene expression in mammalian hibernation. Proc Natl Acad Sci U S A. 1992;89:7119–23. doi: 10.1073/pnas.89.15.7119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevenson TJ, Ball GF. Information theory and the neuropeptidergic regulation of seasonal reproduction in mammals and birds. Proc R Soc Lond B Biol Sci. 2011;278:2477–85. doi: 10.1098/rspb.2010.2181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevenson TJ, Prendergast BJ. Reversible DNA methylation regulates seasonal photoperiodic time measurement. Proc Natl Acad Sci U S A. 2013;110:16651–6. doi: 10.1073/pnas.1310643110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweatt JD. The emerging field of neuroepigenetics. Neuron. 2013;80:624–32. doi: 10.1016/j.neuron.2013.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–5. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajerian M, Alvarado S, Millecamps M, Vachon P, Crosby C, Bushnell MC, Szyf M, Stone LS. Peripheral nerve injury is associated with chronic, reversible changes in global DNA methylation in the mouse prefrontal cortex. PLoS One. 2013;8:e55259. doi: 10.1371/journal.pone.0055259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y, Gao XD, Wang Y, Yuan BF, Feng YQ. Widespread existence of cytosine methylation in yeast DNA measured by gas chromatography/mass spectrometry. Anal Chem. 2012;84:7249–55. doi: 10.1021/ac301727c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor SM, Jones PA. Multiple new phenotypes induced in 10T1/2 and 3T3 cells treated with 5-azacytidine. Cell. 1979;17:771–9. doi: 10.1016/0092-8674(79)90317-9. [DOI] [PubMed] [Google Scholar]
- Tweedie S, Charlton J, Clark V, Bird A. Methylation of genomes and genes at the invertebrate–vertebrate boundary. Mol Cell Biol. 1997;17:1469–75. doi: 10.1128/mcb.17.3.1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urieli-Shoval S, Gruenbaum Y, Sedat J, Razin A. The absence of detectable methylated bases in Drosophila melanogaster DNA. FEBS Lett. 1982;146:148–52. doi: 10.1016/0014-5793(82)80723-0. [DOI] [PubMed] [Google Scholar]
- Van Speybroeck L. From epigenesis to epigenetics: the case of C. H. Waddington. Ann N Y Acad Sci. 2002;981:61–81. [PubMed] [Google Scholar]
- Waalwijk C, Flavell RA. MspI, an isoschizomer of hpaII which cleaves both unmethylated and methylated hpaII sites. Nucleic Acids Res. 1978;5:3231–6. doi: 10.1093/nar/5.9.3231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterland RA, Jirtle RL. Transposable elements: targets for early nutritional effects on epigenetic gene regulation. Mol Cell Biol. 2003;23:5293–300. doi: 10.1128/MCB.23.15.5293-5300.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterland RA, Kellermayer R, Laritsky E, Rayco-Solon P, Harris RA, Travisano M, Zhang W, Torskaya MS, Zhang J, Shen L, et al. Season of conception in rural gambia affects DNA methylation at putative human metastable epialleles. PLoS Genet. 2010;6:e1001252. doi: 10.1371/journal.pgen.1001252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver IC, Cervoni N, Champagne FA, D’Alessio AC, Sharma S, Seckl JR, Dymov S, Szyf M, Meaney MJ. Epigenetic programming by maternal behavior. Nat Neurosci. 2004;7:847–54. doi: 10.1038/nn1276. [DOI] [PubMed] [Google Scholar]
- Woodcock CL, Ghosh RP. Chromatin higher-order structure and dynamics. Cold Spring Harb Perspect Biol. 2010;2:a000596. doi: 10.1101/cshperspect.a000596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Zhang Y. Reversing DNA methylation: mechanisms, genomics, and biological functions. Cell. 2014;156:45–68. doi: 10.1016/j.cell.2013.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong LZ, Xu CG, Saghai Maroof MA, Zhang Q. Patterns of cytosine methylation in an elite rice hybrid and its parental lines, detected by a methylation-sensitive amplification polymorphism technique. Mol Gen Genet. 1999;261:439–46. doi: 10.1007/s004380050986. [DOI] [PubMed] [Google Scholar]
- Yoder JA, Bestor TH. A candidate mammalian DNA methyltransferase related to pmt1p of fission yeast. Hum Mol Genet. 1998;7:279–84. doi: 10.1093/hmg/7.2.279. [DOI] [PubMed] [Google Scholar]
- Zemach A, McDaniel IE, Silva P, Zilberman D. Genome-wide evolutionary analysis of eukaryotic DNA methylation. Science. 2010;328:916–9. doi: 10.1126/science.1186366. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Ng HH, Erdjument-Bromage H, Tempst P, Bird A, Reinberg D. Analysis of the NuRD subunits reveals a histone deacetylase core complex and a connection with DNA methylation. Genes Dev. 1999;13:1924–35. doi: 10.1101/gad.13.15.1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziller MJ, Muller F, Liao J, Zhang Y, Gu H, Bock C, Boyle P, Epstein CB, Bernstein BE, Lengauer T, et al. Genomic distribution and inter-sample variation of non-CpG methylation across human cell types. PLoS Genet. 2011;7:e1002389. doi: 10.1371/journal.pgen.1002389. [DOI] [PMC free article] [PubMed] [Google Scholar]


