Abstract
The breast cancer susceptibility gene BRCA1 encodes a nuclear protein, which functions as a tumor suppressor and is involved in gene transcription and DNA repair processes. Many families with inherited breast and ovarian cancers have mutations in the BRCA1 gene. However, only a few studies have reported on the mechanism underlying the regulation of BRCA1 expression in humans. In this study, we investigated the transcriptional regulation of BRCA1 in HeLa cells treated with etoposide. We found that three Egr-1-binding sequences (EBSs) were located at −1031, −1005, and −385 within the enhancer region of the BRCA1 gene. Forced expression of Egr-1 stimulated the BRCA1 promoter activity. EMSA data showed that Egr-1 bound directly to the EBS within the BRCA1 gene. Knockdown of Egr-1 through the expression of a small hairpin RNA (shRNA) attenuated etoposide-induced BRCA1 promoter activity. We conclude that Egr-1 targets the BRCA1 gene in HeLa cells exposed to etoposide. [BMB Reports 2013; 46(2): 92-96]
Keywords: BRCA1, DNA damage, Egr-1, Etoposide, Promoter activation
INTRODUCTION
The BRCA1 gene is a nuclear phosphoprotein of 1863 amino acids that contains an N-terminal Cys3-His-Cys4 zinc finger domain and a C-terminal acidic domain (1). BRCA1 interacts with several proteins, including Rad51 p53, RNA polymerase II holoenzyme, RNA helicase A, CtBP-interacting protein, CBP/p300 and c-Myc; it plays important roles in DNA damage repair, cell cycle check-point control and apoptosis (2-7). Germline mutations in the BRCA1 gene are closely linked to an increased risk for the development of breast cancer, ovarian cancer and other malignancies (1,8,9), suggesting a tumor suppressor role for BRCA gene.
Several lines of evidence suggest that BRCA1 is associated with the transcriptional regulation of diverse genes, including the upregulation of p21Waf1/Cip1, GADD45, 14-3-3δ, p27Kip1, XPC and TNFα, as well as with the downregulation of cyclin B1, estrogen receptor α-responsive genes and insulin growth factor 1 (7). In MCF7 cells, BRCA1 mRNA expression was increased in response to gamma-irradiation and etoposide, as a response to DNA damage sensing (10). In other studies, BRCA1 was found to be down-regulated at the mRNA level (11) and specifically cleaved and activated by caspase-3 (12) during UVC-rradiation. Although BRCA1 expression is known to be controlled by DNA-damaging agents in diverse cell types, little information is available regarding the regulation of BRCA1 gene expression.
In the present study, we investigated whether BRCA1 expression in HeLa cervix carcinoma cells is regulated at the transcriptional level by etoposide, which is a DNA topoisomerase II inhibitor that induces DNA strand breakage. We show that the transcription factor Egr-1 bound directly to the enhancer region of the BRCA1 gene and that etoposide-induced BRCA1 promoter activity is mediated through Egr-1 activation. These results identify a functional linkage between the DNA damage response and the immediate-early response gene Egr-1 in the regulation of DNA repair and/or induction of apoptosis.
RESULTS AND DISCUSSION
To determine whether etoposide induces the expression of BRCA1 in HeLa cells, Western blot analysis was performed on cells that were exposed to 100 μM etoposide for different time periods. The p53 and p21 proteins were used as positive controls for etoposide stimulation (13). Following exposure of the cells to etoposide, the level of BRCA1 protein increased in a time-dependent manner (Fig. 1A). In addition, the BRCA1 protein displayed retarded electrophoretic migration, probably reflecting phosphorylation of the protein after DNA damage (3,12,14). During UV-induced apoptosis, BRCA1 is cleaved to a ∼90-kDa C-terminal fragment by caspase-3 and plays an important role in the induction of apoptosis (12). We also observed BRCA1 fragments of ∼90 kDa after etoposide treatment of HeLa cells.
Fig. 1. Effect of etoposide on the induction of BRCA1 expression. (A) HeLa cells were treated with 100 μM etoposide for different time periods. Whole cell extracts were prepared and subjected to Western blotting with antibodies directed against BRCA1, p53 and p21. The ∼220-kDa full length and ∼90-kDa fragment of BRCA1 are indicated by an arrow and an arrowhead, respectively. The same blot was reprobed with anti-GAPDH antibody as an internal control. The blots shown are representative of the results obtained from three independent experiments. (B) Total RNA was isolated and the levels of BRCA1 mRNA were measured by QRT-PCR. Relative levels are normalized to the level of gapdh mRNA. The data shown represent the mean ± SD of three independent experiments. *P < 0.05; **P < 0.01, compared with the untreated control cells. (C) HeLa cells grown in 12-well plates were transfected with 0.5 μg of the BRCA1 promoter reporter plasmid, pBRCA1-Luc(–1066/+135), along with 50 ng of the pRL-null vector. After 24 h, the cells were either untreated or treated with 50 μM or 100 μM etoposide for 8 h. The firefly luciferase activity was normalized to the Renilla activity. The data shown represent the mean ± SD of three independent experiments performed in triplicate. *P < 0.05; **P < 0.01, compared with the untreated control cells.
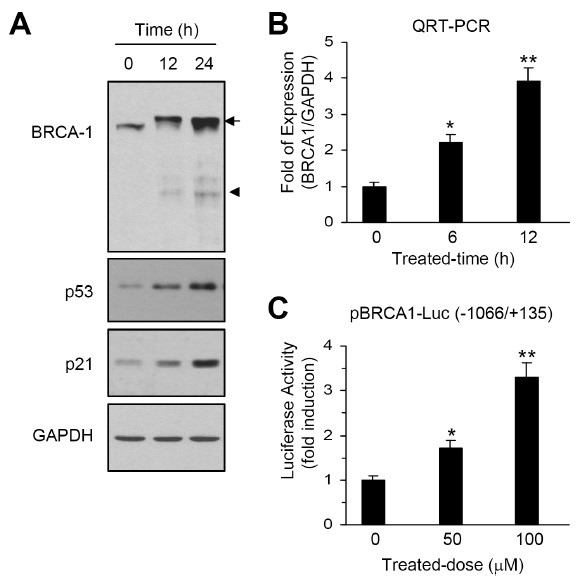
As the cleavage of BRCA1 is an irreversible reaction, we hypothesized that BRCA1 expression is upregulated by etoposide treatment. To test this theory, we examined whether the BRCA1 gene is activated by etoposide. Quantitative Real-Time PCR (QRT-PCR) analysis revealed an approximately 4-fold increase in the level of BRCA1 mRNA after 6 h of treatment with 100 μM etoposide (Fig. 1B). To determine whether etoposide stimulates BRCA1 expression at the transcriptional level, we isolated the 5'-end regulatory region of the human BRCA1 gene, located within 1,066 bp upstream of the transcriptional start site (+1), and subcloned this region into the pGL3-Luc luciferase reporter vector in order to yield pBRCA1-Luc(−1066/+135). This construct was transfected into HeLa cells, and the luciferase activity was measured. Treatment with etoposide resulted in a dose-dependent increase in luciferase reporter activity (Fig. 1C). Approximately a 3.3-fold increase in reporter activity was observed after treatment with 100 μM etoposide (P < 0.01, compared to the mock-treated control). Thus, BRCA1 mRNA expression in HeLa cells is upregulated following etoposide treatment.
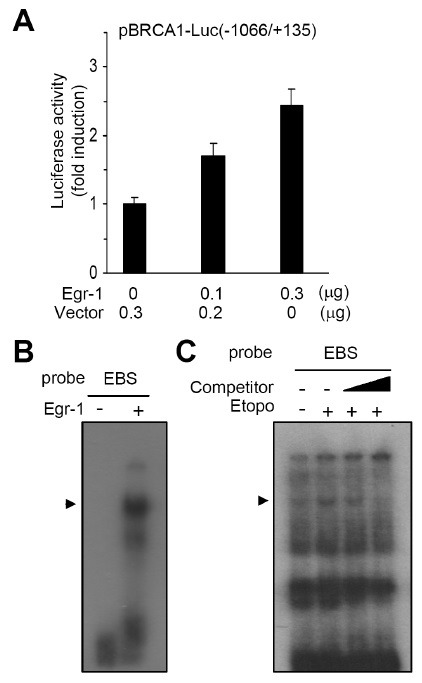
We found that three Egr-1-binding sequences (EBSs) were located at −1031, −1005 and −385 within the enhancer region of the BRCA1 gene. Egr-1 is an immediate-early response gene that is induced by multiple stimuli, including stress, injury, mitogens and DNA damaging agents (15-17). Egr-1 target genes, which include p21, p53, PTEN, TGFβ1, fibronectin and Gadd45, are involved in the regulation of cellular growth, DNA repair and apoptosis (16,18-21). To evaluate whether Egr-1 transactivates the BRCA1 promoter, HeLa cells were transfected with the promoter reporter pBRCA1-Luc(−1066/+135) together with increasing concentrations of the Egr-1 expression plasmid. Forced expression of Egr-1 resulted in the stimulation of pBRCA1-Luc(–1066/+135) reporter activity in a plasmid concentration-dependent manner (Fig. 2A). To investigate whether Egr-1 binds directly to the putative Egr-1-binding sequences (EBS) within the BRCA1 gene, EMSA was performed using the purified recombinant Egr-1 protein. Recombinant Egr-1 bound to the 32P-labeled oligodeoxynucleotides that contained the consensus EBS (Fig. 2B). To determine whether Egr-1 binds to the EBS in response to etoposide treatment, nuclear extracts of HeLa cells were prepared and incubated with the radiolabeled Egr-1-binding probe. Treatment with etoposide increased the level of DNA-protein complex (Fig. 2C). The specificity of Egr-1 binding was confirmed by the loss of the DNA-protein complex when an excess of unlabelled Egr-1-binding oligodeoxynucleotides was added to the reaction. These data demonstrate that Egr-1 binds directly to the EBS site within the BRCA1 promoter region.
Fig. 2. Egr-1 transactivates the BRCA1 promoter through direct binding to the EBS. (A) HeLa cells were co-transfected with 0.5 μg wild-type (WT) pBRCA1-Luc(–1066/+135) and different concentrations of the empty vector (pcDNA3.1zeo) or Egr-1 expression plasmid (pcDNA3.1zeo/Egr1), as indicated. After 48 h, the cells were collected and analyzed for luciferase activity. The firefly luciferase activity was normalized to the Renilla activity. The data shown represent the mean ± SD of three independent experiments performed in triplicate. (B and C) Purified recombinant Egr-1 protein (B) and nuclear extracts from HeLa cells treated with 100 μM etoposide for 1 h (C) were incubated with 32P-labeled oligodeoxynucleotide probes that contain EBS. For competition, unlabeled oligodeoxynucleotides (Competitor) were added at 10-fold or 100-fold excess. Arrowheads indicate DNA-Egr-1 complexes.
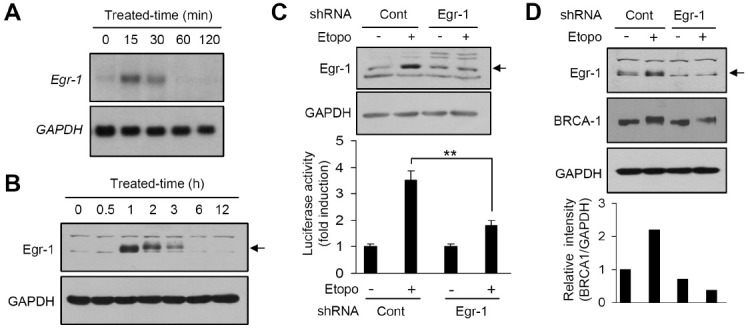
To investigate whether Egr-1 expression is regulated by etoposide, Egr-1 expression was analyzed by Northern and Western blot analyses. The amounts of Egr-1 mRNA (Fig. 3A) and Egr-1 protein (Fig. 3B) were increased by etoposide treatment in a time-dependent manner. The level of Egr-1 mRNA peaked 15 min after etoposide treatment, while the level of Egr-1 protein peaked 1 h after etoposide treatment and gradually decreased thereafter. To confirm that Egr-1 is required for etoposide-induced BRCA1 promoter activity, we used the RNA interference approach. Egr-1 knockdown was verified after etoposide treatment of serum-starved cells. The ability of etoposide to activate the BRCA1 promoter was substantially attenuated by the introduction of Egr-1 siRNA (Fig. 3C). Western blot analysis demonstrated that both basal and etoposide-induced BRCA1 protein levels were substantially reduced in HeLa cells expressing Egr-1 siRNA (Fig. 3D). These results demonstrate that Egr-1 expression is necessary for the etoposide-induced upregulation of BRCA1.
Fig. 3. Role of Egr-1 in etoposide-induced BRCA1 expression. (A) Serum-starved HeLa cells were treated with 100 μM etoposide for different time periods. Total RNA was isolated and Egr-1 mRNA expression was assessed by Northern blotting with 32P-labeled Egr-1 cDNA. The same blot was re-probed with 32P-labeled GAPDH cDNA as an internal control. (B) Serum-starved HeLa cells were treated with 100 μM etoposide for different time periods. Total cell lysates were prepared and subjected to Western blot analysis with rabbit anti-Egr-1 antibody. The same blot was reprobed with anti-GAPDH antibody as an internal control. (C) HeLa cells were transiently co-transfected with 0.5 μg pBRCA1-Luc(–1066/+135) and an shRNA plasmid, pSilencer/scrambled (control siRNA; Cont) or pSilencer/siEgr1 (Egr-1), along with 50 ng of the pRL-null vector plasmid. After 24 h, the cells were left untreated or treated with 100 μM etoposide for 8 h, and the luciferase activity was measured. Egr-1 is indicated by an arrow. The knockdown of Egr-1 expression was verified by Western blot analysis (upper panel). Luciferase activity is shown as the mean ± SD of three independent experiments performed in triplicate (bottom graph). **P < 0.01. (D) HeLa cells were transiently transfected with 0.5 μg shRNA plasmid, pSilencer/scrambled (control siRNA; Cont) or pSilencer/siEgr1 (Egr-1). After 24 h, the cells were left untreated or treated with 100 μM etoposide for 3 h. Whole cell extracts were prepared and subjected to Western blotting with antibodies directed against Egr-1 and BRCA1. Egr-1 is indicated by an arrow. The same blot was reprobed with anti-GAPDH antibody as an internal control. The relative band intensities were measured by quantitative scanning densitometer (bottom graph).
In the present study, we demonstrate that BRCA1 expression is upregulated via Egr-1 following etoposide treatment. Egr-1 binds directly to the EBSs within the BRCA1 promoter region. The essential role of Egr-1 in the activation of BRCA1 transcription is supported by our findings that: (i) Egr-1 binds directly to the EBSs within the BRCA1 enhancer region; (ii) forced expression of Egr-1 transactivates the BRCA1 promoter; and (iii) the expression of Egr-1 siRNA abrogates etoposide-induced activation of the BRCA1 promoter. These results imply that Egr-1 plays a critical role in DNA damage-induced cellular responses.
MATERIALS AND METHODS
Cell culturing and reagents
The human cervix carcinoma cell line HeLa was obtained from the American Type Culture Collection (ATCC, Manassas, VA, USA). Cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM) that was supplemented with 10% fetal bovine serum (FBS; Hyclone, Logan, UT, USA). Etoposide was purchased from Calbiochem (San Diego, CA, USA). Antibodies directed against Egr-1 and GAPDH were obtained from Santa Cruz Biotechnology (Santa Cruz, CA, USA). The firefly and Renilla Dual-GloTM Luciferase Assay System was purchased from Promega (Madison, WI, USA).
Northern blot analysis
Total RNA (10 μg) from each sample was separated by electrophoresis on a formaldehyde/agarose gel and transferred to a Hybond N+ nylon membrane (Amersham Pharmacia Biotech, Piscataway, NJ, USA). Northern blotting was performed using a [α-32P]dCTP-labeled cDNA probe, followed by hybridization with a glyceraldehyde-3-phosphate dehydrogenase (gapdh) cDNA probe, as described previously (22).
Western blot analysis
Cells were lysed in 20 mM HEPES (pH 7.2) that contained 1% Triton X-100, 10% glycerol, 150 mM NaCl, 10 μg/ml leupeptin and 1 mM phenylmethylsulfonyl fluoride (PMSF). The protein samples (15 μg each) were separated by 10% SDS-PAGE, and transferred onto nitrocellulose filters. The blots were incubated with appropriate antibodies and were developed using an enhanced hemiluminescence detection system (Amersham Pharmacia Biotech). The relative band intensities were measured by a quantitative scanning densitometer and image analysis software, Bio-1D version 97.04.
Quantitative Real-Time PCR (QRT-PCR)
Total RNA was extracted using the Trizol RNA extraction kit (Invitrogen, Carlsbad, CA, USA) from cells that were stimulated with etoposide (100 μM) for various periods of time. The first-strand cDNA was synthesized from 500 ng of total RNA using the iScript cDNA synthesis kit (Bio-Rad, Hercules, CA, USA). QRT-PCR was performed with the iCycler iQ Real-Time PCR Detection System (Bio-Rad), using the TaqMan-iQTM Supermix Kit (Bio-Rad), according to the anufacturer's recommendation. The TaqManTM fluorogenic probes and PCR primers for BRCA1 and gapdh were designed by Metabion International AG (Martinsried, Germany). The primer sequences (forward and reverse, respectively) were as follows: for BRCA1, 5'-TGGGAGCCAGCCTTCTAACAG-3 and 5'-GTT AATACTGCTTTTTCTGATGTGCTTTG-3'; and for gapdh, 5'-T CGACAGTCAGCCGCATCTTC-3' and 5'-CGCCCAATACGACC AAATCCG-3'. The TaqManTM fluorogenic probes used were: 5'-FAM-TGACTCTTCTGCCCTTGAGGACCTGCGA-BHQ-1-3' (for BRCA1); and 5'-Yakima YellowTM-CGTCGCCAGCCGAGCC ACATCGC-BHQ-1-3' (for gapdh). The threshold cycle, Ct, which correlates inversely with the target mRNA levels, was calculated as the cycle number at which the reporter fluorescent emission increased above a threshold level. The relative changes in BRCA1 mRNA levels were normalized for gapdh mRNA in the same samples.
Construction of human BRCA1 promoter-reporter construct
A BRCA1 promoter fragment spanning nucleotides (nt) −1066 to +135 was PCR-amplified from human genomic DNA (Promega) using the primers 5'-CACTTGCCCTCAAAACGACC-3' (forward) and 5'-GTTATCTGAGAAACCCCACA-3' (reverse). The amplicons were ligated into the pGL4-basic vector, yielding pBRCA1-Luc(−1066/+135). The resultant construct was verified by DNA sequencing and by restriction enzyme digestion.
Promoter reporter assay
HeLa cells were seeded into 12-well plates and transfected with 0.5 μg of the BRCA1 promoter construct using LipofectamineTM 2000 (Invitrogen), according to the manufacturer’s instructions. To monitor transfection efficiency, 50 ng of the pRL-null plasmid that encodes Renilla luciferase were included in all samples. Where indicated, the empty vector or Egr-1 expression plasmid (pcDNA3.1zeo/Egr1) was included. At 24 h post-transfection, the levels of firefly and Renilla luciferase activity were measured, as described previously (23).
EMSA
Synthetic oligodeoxynucleotides (4 pmol) corresponding to EBS (−385/−360; 5'-AAGTACAAGCGCGCACAGGTCTCC-3') within the BRCA1 enhancer were radioactively labelled by incubation with 10 U of T4 polynucleotide kinase, 5 μl of T4 polynucleotide kinase buffer and 20 μCi of [γ-32P]dATP (Amersham Biosciences) for 30 min at 37℃, followed by inactivation at 65℃ for 10 min. For EMSA, 10 μg of nuclear extract were mixed with the binding buffer (50 mM Tris-HCl [pH 7.5], 5 mM MgCl2, 2.5 mM dithiothreitol, 2.5 mM EDTA, 250 mM NaCl, 20% glycerol) together with 1 μg of poly(dI-dC) (Amersham Biosciences) as a non-specific competitor. The DNA-protein complexes were electrophoresed in non-denaturing 6% polyacrylamide gels and visualised by autoradiography.
Expression of Egr-1 siRNA
The generation of a small hairpin RNA (shRNA) plasmid that targets human Egr-1 mRNA (pSilencer/siEgr1) and its expression were described elsewhere (20).
Statistical analysis
All data are expressed as the means ± SD of at least three independent experiments. The Student’s t-test and ANOVA were used to identify statistically significant differences. P values <0.05 were considered to be statistically significant.
Acknowledgments
This work was supported by a grant of the Korea Healthcare Technology R&D Project, the Ministry for Health, Welfare & Family Affairs (No. A111778). This paper was supported by the SMART Research Professor Program of Konkuk University.
References
- 1.Miki Y., Swensen J., Shattuck-Eidens D., Futreal P. A., Harshman K., Tavtigian S., Liu Q., Cochran C., Bennett L. M., Ding W., Bell R., Rosenthal J., Hussey C., Tran T., McClure M., Frye C., Hattier T., Phelps R., Haugen-Strano A., Katcher H., Yakumo K., Gholami Z., Shaffer D., Stone S., Bayer S., Wray C., Bogden R., Dayananth P., Ward J., Tonin P., Narod S., Bristow P. K., Norris F. H., Helvering L., Morrison P., Rosteck P., Lai M., Barrett J. C., Lewis C., Neuhausen S., Cannon-Albright L., Goldgar D., Wiseman R., Kamb K., Skolnick M. H. A strong candidate for the breast and ovarian cancer susceptibility gene BRCA1. Science. (1994);266:66–71. doi: 10.1126/science.7545954. [DOI] [PubMed] [Google Scholar]
- 2.Zhong Q., Chen C. F., Li S., Chen Y., Wang C. C., Xiao J., Chen P. L., Sharp Z. D., Lee W. H. Association of BRCA1 with the hRad50-hMre11-p95 complex and the DNA damage response. Science. (1999);285:747–750. doi: 10.1126/science.285.5428.747. [DOI] [PubMed] [Google Scholar]
- 3.Scully R., Anderson S. F., Chao D. M., Wei W., Ye L., Young R. A., Livingston D. M., Parvin J. D. BRCA1 is a component of the RNA polymerase II holoenzyme. Proc. Natl. Acad. Sci. U.S.A. (1997);94:5605–5610. doi: 10.1073/pnas.94.11.5605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pao G. M., Janknecht R., Ruffner H., Hunter T., Verma I. M. CBP/p300 interact with and function as transcriptional coactivators of BRCA1. Proc. Natl. Acad. Sci. U.S.A. (2000);97:1020–1025. doi: 10.1073/pnas.97.3.1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Anderson S. F., Schlegel B. P., Nakajima T., Wolpin E. S., Parvin J. D. BRCA1 protein is linked to the RNA polymerase II holoenzyme complex via RNA helicase A. Nat. Genet. (1998);19:254–256. doi: 10.1038/930. [DOI] [PubMed] [Google Scholar]
- 6.Irminger-Finger I., Siegel B. D., Leung W. C. The functions of breast cancer susceptibility gene 1 (BRCA1product and its associated proteins. Biol. Chem. (1999);380:117–128. doi: 10.1515/BC.1999.019. [DOI] [PubMed] [Google Scholar]
- 7.Somasundaram K. Breast cancer gene 1 (BRCA1): role in cell cycle regulation and DNA repair--perhaps through transcription. J. Cell Biochem. (2003);88:1084–1091. doi: 10.1002/jcb.10469. [DOI] [PubMed] [Google Scholar]
- 8.Tavtigian S. V., Simard J., Rommens J., Couch F., Shattuck-Eidens D., Neuhausen S., Merajver S., Thorlacius S., Offit K., Stoppa-Lyonnet D., Belanger C., Bell R., Berry S., Bogden R., Chen Q., Davis T., Dumont M., Frye C., Hattier T., Jammulapati S., Janecki T., Jiang P., Kehrer R., Leblanc J. F., Mitchell J. T., McArthur-Morrison J., Nguyen K., Peng Y., Samson C., Schroeder M., Snyder S. C., Steele L., Stringfellow M., Stroup C., Swedlund B., Swense J., Teng D., Thomas A., Tran T., Tranchant M., Weaver-Feldhaus J., Wong A. K., Shizuya H., Eyfjord J. E., Cannon-Albright L., Labrie F., Skolnick M. H., Weber B., Kamb A., Goldgar D. E. The complete BRCA2 gene and mutations in chromosome 13q-linked kindreds. Nat. Genet. (1996);12:333–337. doi: 10.1038/ng0396-333. [DOI] [PubMed] [Google Scholar]
- 9.Welcsh P. L., King M. C. BRCA1 and BRCA2 and the genetics of breast and ovarian cancer. Hum. Mol. Genet. (2001);10:705–713. doi: 10.1093/hmg/10.7.705. [DOI] [PubMed] [Google Scholar]
- 10.Kroupis C., Stathopoulou A., Zygalaki E., Ferekidou L., Talieri M., Lianidou E. S. Development and applications of a real-time quantitative RT-PCR method (QRT-PCR) for BRCA1 mRNA. Clin. Biochem. (2005);38:50–57. doi: 10.1016/j.clinbiochem.2004.09.012. [DOI] [PubMed] [Google Scholar]
- 11.Fan S., Twu N. F., Wang J. A., Yuan R. Q., Andres J., Goldberg I. D., Rosen E. M. Down-regulation of BRCA1 and BRCA2 in human ovarian cancer cells exposed to adriamycin and ultraviolet radiation. Int. J. Cancer. (1998);77:600–609. doi: 10.1002/(sici)1097-0215(19980812)77:4<600::aid-ijc21>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- 12.Zhan Q., Jin S., Ng B., Plisket J., Shangary S., Rathi A., Brown K. D., Baskaran R. Caspase-3 mediated cleavage of BRCA1 during UV-induced apoptosis. Oncogene. (2002);21:5335–5345. doi: 10.1038/sj.onc.1205665. [DOI] [PubMed] [Google Scholar]
- 13.Ha S. A., Kim H. K., Yoo J., Kim S., Shin S. M., Lee Y. S., Hur S. Y., Kim Y. W., Kim T. E., Chung Y. J., Jeun S. S., Kim D. W., Park Y. G., Kim J., Shin S. Y., Lee Y. H., Kim J. W. Transdifferentiation-inducing HCCR-1 oncogene. BMC Cell Biol. (2010);11:49. doi: 10.1186/1471-2121-11-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thomas J. E., Smith M., Tonkinson J. L., Rubinfeld B., Polakis P. Induction of phosphorylation on BRCA1 during the cell cycle and after DNA damage. Cell Growth. Differ. (1997);8:801–809. [PubMed] [Google Scholar]
- 15.Sukhatme V. P., Cao X. M., Chang L. C., Tsai-Morris C. H., Stamenkovich D., Ferreira P. C., Cohen D. R., Edwards S. A., Shows T. B., Curran T., Le Beau M. M., Adamson E. D. A zinc finger-encoding gene coregulated with c-fos during growth and differentiation, and after cellular depolarization. Cell. (1988);53:37–43. doi: 10.1016/0092-8674(88)90485-0. [DOI] [PubMed] [Google Scholar]
- 16.Liu C., Rangnekar V. M., Adamson E., Mercola D. Suppression of growth and transformation and induction of apoptosis by EGR-1. Cancer Gene. Ther. (1998);5:3–28. [PubMed] [Google Scholar]
- 17.Kim J. H., Jeong I. Y., Lim Y., Lee Y. H., Shin S. Y. Estrogen receptor beta stimulates Egr-1 transcription via MEK1/Erk/Elk-1 cascade in C6 glioma cells. BMB Rep. (2011);44:452–457. doi: 10.5483/BMBRep.2011.44.7.452. [DOI] [PubMed] [Google Scholar]
- 18.Virolle T., Adamson E. D., Baron V., Birle D., Mercola D., Mustelin T., de Belle I. The Egr-1 transcription factor directly activates PTEN during irradiation- induced signalling. Nat. Cell. Biol. (2001);3:1124–1128. doi: 10.1038/ncb1201-1124. [DOI] [PubMed] [Google Scholar]
- 19.Krones-Herzig A., Adamson E., Mercola D. Early growth response 1 protein, an upstream gatekeeper of the p53 tumor suppressor, controls replicative senescence, controls replicative senescence. Proc. Natl. Acad. Sci. U.S.A. (2003);100:3233–3238. doi: 10.1073/pnas.2628034100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Choi B. H., Kim C. G., Bae Y. S., Lim Y., Lee Y. H., Shin S. Y. p21 Waf1/Cip1 expression by curcumin in U-87MG human glioma cells: role of early growth response- 1 expression. Cancer Res. (2008);68:1369–1377. doi: 10.1158/0008-5472.CAN-07-5222. [DOI] [PubMed] [Google Scholar]
- 21.Thyss R., Virolle V., Imbert V., Peyron J. F., Aberdam D., Virolle T. NF-kappaB/Egr-1/Gadd45 are sequentially activated upon UVB irradiation to mediate epidermal cell death. EMBO J. (2005);24:128–137. doi: 10.1038/sj.emboj.7600501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang Y., Zhou J., Ye X., Wan Y., Li Y., Mo X., Yuan W., Yan Y., Luo N., Wang Z., Fan X., Deng Y., Wu X. ZNF424, a novel human KRAB/C2H2 zinc finger protein, suppresses NFAT and p21 pathway. BMB Rep. (2010);43:212–218. doi: 10.5483/BMBRep.2010.43.3.212. [DOI] [PubMed] [Google Scholar]
- 23.Choi B. H., Kim C. G., Lim Y., Lee Y. H., Shin S. Y. Transcriptional activation of the human Klotho gene by epidermal growth factor in HEK293 cells; role of Egr-1. Gene. 450:121–127. doi: 10.1016/j.gene.2009.11.004. [DOI] [PubMed] [Google Scholar]


