Abstract
BACKGROUND
Genetically engineered mouse models play important roles in analyses of prostate development and pathobiology. While constitutive genetic gain-and loss-of-function models have contributed significantly to our understanding of molecular events driving these processes, the availability of a tightly regulated inducible expression system could extend the utility of transgenic approaches. Here, we describe the development of a Tet-regulatory system that employs Hoxb13 transcriptional control elements to direct reverse tetracycline transactivator (rtTA) expression in the prostate.
METHODS
Using recombineering technology, the rtTA gene was placed under Hoxb13 cis-regulatory transcriptional control in the context of a 218-kb bacterial artificial chromosome. F1 offspring carrying the Hoxb13-rtTA transgene were bred to a Tetracycline operator–Histone 2B-Green Fluorescent Protein (TetO-H2BGFP) responder line. Detailed reporter gene expression analyses, including doxycycline (Dox) induction and withdrawal kinetics, were performed in Hoxb13-rtTA|TetO-H2BGFP double transgenic adult mice and embryos.
RESULTS
Dox-dependent GFP expression was observed exclusively in the prostate and distal colon epithelia of double transgenic mice. Reporter gene mRNA was detected in the prostate within 6 hr of Dox exposure, and was extinguished within 24 hr after Dox withdrawal. Furthermore, Dox-induced reporter gene expression persisted after castration.
CONCLUSIONS
The Hoxb13-rtTA transgenic system provides a powerful tool for conditional Tet operator-driven transgene expression in the normal prostate and during disease progression. Used in conjunction with other prostate pathology models, these mice will enable precise, temporally controlled analyses of gene function and can provide opportunities for detailed analyses of molecular events underlying prostate diseases.
INTRODUCTION
The extensive morbidity and mortality associated with the prostate gland is a significant health concern (1–4). Preclinical model systems play a critical role in dissecting aspects of prostatic disease progression and in designing new therapeutic modalities (5, 6). Both gain- and loss-of-function mouse models of prostate pathology have contributed significantly to our understanding of disease progression at the molecular level (7–9). However, systems that permit analyses of temporal or context-dependent changes in clinically relevant proteins specifically in the prostate have not been reported. Genetically engineered mouse models, based on tightly regulated, inducible transgene expression can circumvent limitations of models that depend on constitutive genetic changes (10, 11). In cancer models, inducible transgene expression systems could enable study of the consequences of tumor suppressor restoration or diminished oncogene expression during cancer progression (12). Combining extant prostate cancer mouse models harboring constitutive genetic changes with inducible systems would increase the potential of these systems to closely recapitulate prostate cancer progression. In addition, these next-generation models could provide new platforms to study targeted therapeutic strategies. Despite these clear benefits, a robust, prostate-restricted, conditional transgene expression system has not been developed.
To extend the utility of prostate pathology animal models, we sought to develop a conditional expression system in the prostate based on the doxycycline (Dox)-inducible reverse tetracycline transactivator (rtTA). The rtTA system has emerged as a powerful tool for conditional, Dox-dependent, organ-specific gene expression 10. rtTA-based Tet-regulatory models have been successfully applied in etiological studies of several malignant diseases including lung, breast, and hematologic cancers (13–15).
The development of tissue-specific Tet-regulatory systems depends on the availability of cis-regulatory elements to direct transgene expression in the target organ (10). Multiple prostate-restricted promoter and enhancer elements have been used to study prostate biology in rodents (7). However, androgen-response elements are typical features of cis-regulatory regions used for prostate-specific transgene expression (16, 17). The androgen-signaling axis plays a central role in prostate biology and aberrant androgen and/or androgen receptor (AR) levels may underlie certain prostate pathologies (18). Alterations in AR activity can affect transgene expression in mouse models employing AR responsive cis-elements (19). A cis-regulatory element that supports robust prostrate-restricted transcriptional activity in the absence of androgen would be useful in a variety of pathological scenarios. For example, androgen-independence is a hallmark feature of late stage adenocarcinoma (20). The transcriptional regulatory regions of Hoxb13 support prostate-restricted expression of heterologous transgenes in the presence and absence of testicular androgens and can be used to drive a conditional expression system in the prostate (21).
Here, we describe the generation of a Hoxb13-rtTA mouse strain in which Hoxb13 cis-regulatory elements drive prostate and colon-restricted rtTA expression. We demonstrate tightly regulated induction and withdrawal kinetics of a reporter gene in a tetracycline responder strain crossed with our Hoxb13-rtTA strain 22. Our studies demonstrate the potential of this system to study the effect of regulating clinically relevant proteins in prostate biology and pathology.
MATERIALS AND METHODS
Transgene and Targeting Cassette Construction
A targeting cassette containing the rtTAS-M2 transgene (gift from H. Bujard (23)), and flanking Hoxb13 homology regions was placed in pEGFP-C1 (Clontech). An ampicillin resistance marker was cloned downstream of rtTA. Using recombineering (21, 24), the targeting cassette was introduced into the Hoxb13 bacterial artificial chromosome (BAC) RP23-335O22 to generate the Hoxb13-rtTA BAC in which the rtTA gene was placed downstream of the Hoxb13 start codon in the open reading frame (14).
Transgenic Mouse Generation and Genotyping
The Hoxb13-rtTA BAC was linearized with PI-SceI (New England Biolabs), purified by sucrose gradient centrifugation, dialyzed and injected into FVB single-cell embryos. Hoxb13-rtTA founders and their transgenic offspring were identified by Southern blot analyses. Genomic DNA was isolated using the Gentra Puregene Mouse Tail kit (Qiagen). Ten micrograms of extracted DNA was digested with EcoRI and EcoRV, electrophoresed on a 0.8% agarose gel, and then transferred to a Nytran-N membrane (Whatman). The vector pEGFP-rtTA-Amp 10 was digested with BamHI, and the primer 5′-GCCGAATTCGGATCCCGGGCCCCTGCGTCTCTTGCGTCAAG-3′ was used to create a probe for Southern blot with the Ambion Strip-EZ PCR kit (Applied Biosystems). Blot hybridization was performed using Ambion ULTRAhyb solution (Applied Biosystems), was washed according to the manufacturer's instructions, and exposed to X-ray film. Hoxb13-rtTA|TetO-H2BGFP mice were screened by Southern blot of XbaI digested genomic DNA. The probe for H2BGFP was generated using EcoRI-digested pEGFP-C1 (Clontech) and the primer 5′-TCCTTGAAGAAGATGGTGCG-3′.
Animal care was provided in accordance with the National Institutes of Health Guide for the care and use of laboratory animals. Animal use was approved by UMBC Institutional Animal Care and Use Committee.
Immunofluorescence
Whole mount prostate and colon samples were fixed in 4% paraformaldehyde in PBS (overnight) and embedded in Tissue-Tek OTC compound. Ten micrometer frozen sections were obtained and thaw-mounted on to Superfrost Plus slides. For immunofluorescence analysis, slides were allowed to achieve room temperature, were permeabilized with 0.1% Triton X-100 for 10 min and blocked with 10% FCS/1% BSA/PBS for 2 hr. Immunofluorescence was performed with a rabbit anti-Hoxb13 antibody 1:250 (21, 25) and chick anti-GFP (1:100, Covance) by incubating overnight at 4° and appropriate fluorescently tagged 2° antibodies (Vector). Extensive washes with PBS were performed between each step. DAPI was used for nuclear staining and fluorescent images were obtained using a Zeiss fluorescent microscope.
qRT-PCR Analysis
Quantitative PCR was performed using SYBR Green PCR Master Mix (Fermentas) on a Bio-Rad iCycler IQ Real-time PCR machine. Primers were designed using Primer3 software as follows: rtTA—5′-CTCTCACATCGCGACGGGGC-3′ and 5′-CCACGGCGGACAGAGCGTAC-3′, GFP—5′-AGCCGCTACCCCGACCACAT-3′ and 5′-TGCGCTCCTGGACGTAGCCT-3′, GAPDH—5′-GAAGGTGAAGGTCGGAGT-3′ and 5′-GAAGATGGTGATGGGATTTC-3′ qPCR amplifications were conducted in triplicate and the calculated thresholds (CT) were averaged (Avg CT), removing outliers with >1 cycle difference. The change in cycle threshold was normalized using the formula, dCT = (Avg CTrtTA − Avg CTGAPDH) in Hoxb13-rtTA mice and dCT = (Avg CTGFP − AvgCTrtTA) for Hoxb13-rtTA|TetO-H2BGFP mice. The fold enrichment was calculated using the formula, fold enrichment = 2−ΔCT
RESULTS AND DISCUSSION
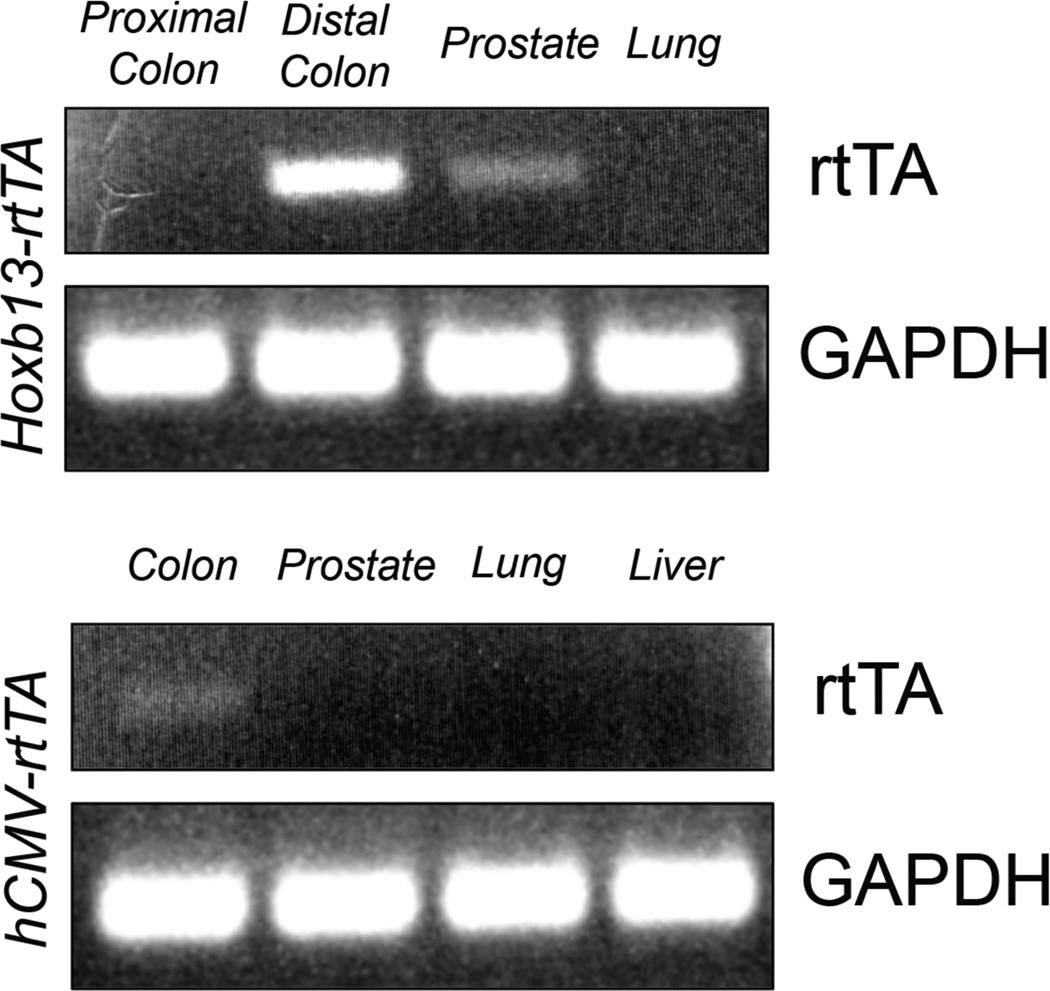
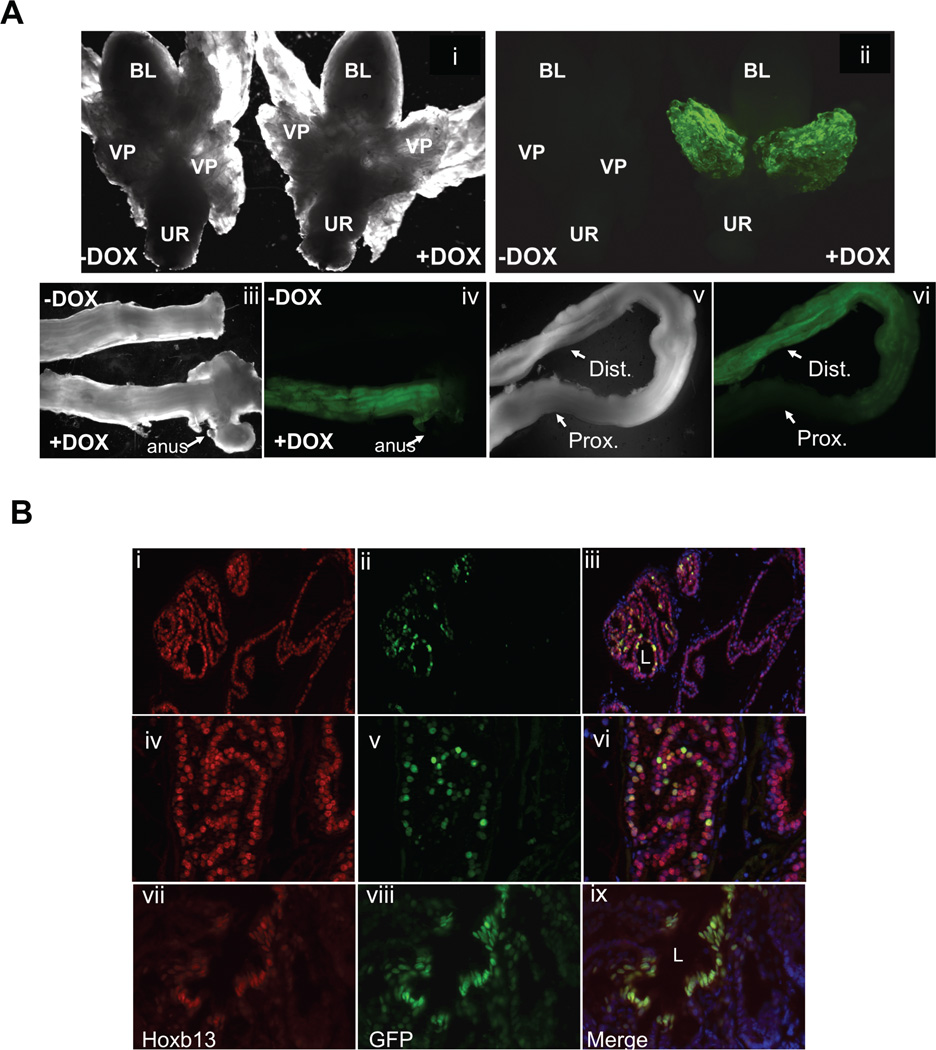
Five independent Hoxb13-rtTA founder mice indentified by Southern blot analyses were mated to the FVB/N strain. Four transmitted the Hoxb13-rtTA transgene to the F1 generation in a Mendelian manner. RT-PCR analyses of RNA obtained from tissues of F1 progeny revealed prostate- and colon-restricted rtTA expression (Fig. 1), demonstrating that the transgene is controlled by Hoxb13 cis-regulatory elements. To determine if the rtTA protein was functional, the four independent Hoxb13-rtTA strains were mated to a strain harboring a Tet operator-controlled Histone 2B-Green Fluorescent Protein (TetO-H2BGFP) responder gene to generate Hoxb13-rtTA|TetOH2BGFP double transgenic mice. Dox was administered in the drinking water to double transgenic mice for 7 days. Whole mount fluorescent microscopic analyses of tissues dissected from double transgenic mice revealed H2BGFP-mediated fluorescence restricted to the ventral prostate and distal colon exclusively in Dox treated mice (Fig. 2A). Two independent Hoxb13-rtTA strains supported Dox-inducible H2BGFP expression, albeit at varying levels (data not shown). Hoxb13-rtTA line #26 (Hoxb13-rtTA-26) displayed the most robust responder gene expression and was chosen for detailed analyses. Immunofluorescence microscopic analyses of frozen sections obtained from prostates and colons of Dox treated Hoxb13-rtTA-26|TetO-H2BGFP mice showed nuclear expression of H2BGFP in luminal epithelial cells of the ventral prostate. Expression in the distal colon was restricted to colonic epithelial cells (Fig. 2B).
Figure 1.
rtTAS-M2 is expressed in the prostate and colon of Hoxb13-rtTA transgenic mice. Tissue lysates were prepared from liver, lung, colon and prostates of Hoxb13-rtTA or hCMV-rtTA (Jackson Laboratories) transgenic mice. RNA obtained from tissues was analyzed by RTPCR using primers specific to rtTA or GAPDH (top) RNA from proximal colon, distal colon, prostate and lung of Hoxb13-rtTA-26 mice. (Bottom) RNA prepared from colon, prostate, lung, and liver of hCMV-rtTA mice. Tissue-specific expression of rtTA was seen in distal colon and prostates of Hoxb13-rtTA mice.
Figure 2.
Dox-inducible H2BGFP reporter gene expression in the ventral prostate and colon of Hoxb13-rtTA+/−|TetO-H2BGFP+/− double transgenic mice. Prostates and colons dissected from double transgenic mice treated with 2 mg/ml dox (+DOX) or water (−DOX) for 7 days were analyzed by A. Stereomicroscopy. (i, iii, v) Dark field stereomicroscopy of partial urogenital axis, colon and anus (ii, iv, vi) Fluorescent stereomicroscopic tissue in i, iii, v. Bl, Bladder; VP, ventral prostate; UR, urethra; dist., distal colon; prox., proximal colon. B: Immunofluorescence analysis. Frozen sections of prostates and colon dissected from dox-treated double transgenic mice were analyzed by immunofluorescence using chick anti-GFP and rabbit anti-Hoxb13 antibodies, fluorescein-conjugated and Texas Red-conjugated secondary antibodies were used respectively. DAPI fluorescence was used for nuclear staining. Prostate—top panel (low magnification) and middle panel (higher magnification), colon—bottom panel (i, iv, vii)— H2BGFP (ii, v, viii)—Hoxb13 (iii, vi, ix)—Merge of images demonstrating nuclear localization of GFP fluorescence in ventral prostatic and distal colonic epithelia of Dox-induced mice. L, lumen. The GFP fluorescence closely follows Hoxb13 immunofluorescence.
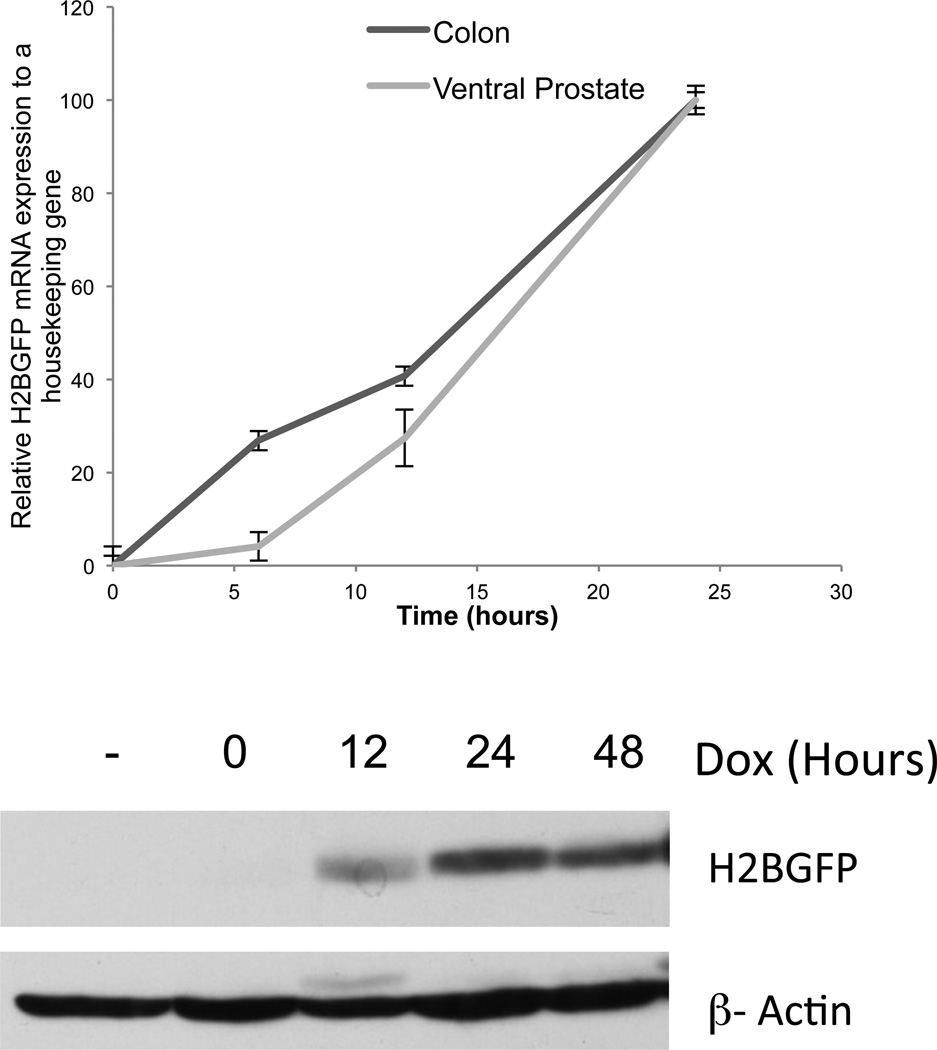
To determine responder gene induction kinetics in Hoxb13-rtTA-26, double transgenic mice were treated with Dox for up to 48 hr. Whole mount microscopic analyses and quantitative analyses of H2BGFP mRNA accumulation following Dox exposure demonstrated responder gene expression as early as 6 hr, which reached a maximum by 24 hr, showing rapid responder gene induction kinetics in the Hoxb13-rtTA-26 strain (Fig. 3).
Figure 3.
Robust H2BGFP induction kinetics of Dox-induced double transgenic mice. Double transgenic mice were treated with dox for indicated amount of time, tissue was analyzed by: Top, qRT-PCR using primers specific for H2BGFP expression which was normalized to rtTA and GAPDH mRNA level. Bottom, Western blot analyses of cell lysate obtained from treated ventral prostates using antibodies against H2BGFP and β-Actin showing longer time points.
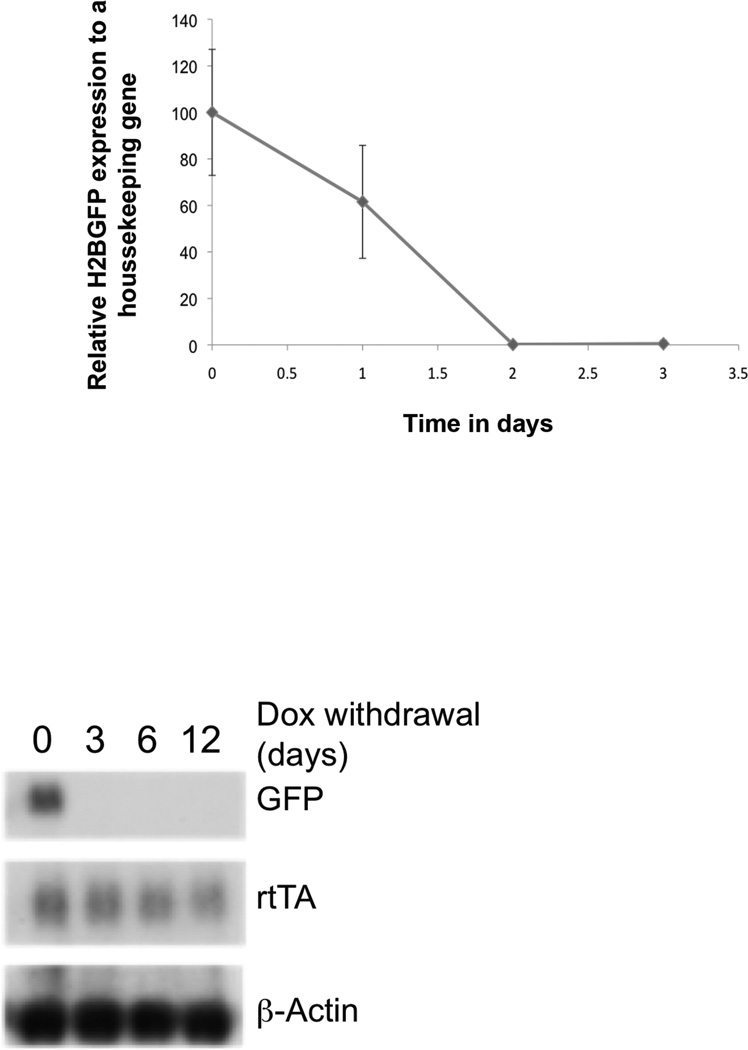
To determine the extinction kinetics of responder gene expression following Dox withdrawal, double transgenic mice were treated with Dox for 24 hr. Due to the stability of H2BGFP, whole mount microscopic analyses demonstrated perdurance of GFP fluorescence even after 6 days following Dox withdrawal (data not shown), however, qRT-PCR analyses demonstrated significant diminution of H2BGFP mRNA by 24 hr, and complete loss by 48 hr (Fig. 4). These kinetic analyses demonstrate that the Hoxb13-rtTA-26 strain enables tight regulation of responder gene expression in response to Dox availability.
Figure 4.
Hoxb13-rtTA-26 strain demonstrates rapid responder extinction kinetics following Dox-withdrawal. Double transgenic mice were treated with Dox for 24 hr followed by replacement with water for indicated period of time. Ventral prostate dissected from treated mice was analyzed by qRT-PCR (top panel). Complete loss of responder gene transcript was seen within 48 hr. Northern blot analyses of mRNA obtained from tissues of treated mice demonstrating loss of rtTA transcript at longer time points. Probes used were specific to H2BGFP or β-actin bottom panel).
Interestingly, while Northern blot analyses demonstrated rtTA expression in all prostate lobes (data not shown), whole mount fluorescent microscopy revealed GFP expression restricted to the ventral lobe of Dox-treated double transgenic mice (Fig. 2A). One possible explanation for this observation may be the stronger Hoxb13 promoter activity and corresponding rtTA accumulation in the ventral lobe in comparison to other mouse prostate lobes (21, 26). In addition, up to 70% of luminal epithelial cells demonstrated H2BGFP fluorescence at varying levels (Fig. 2B); however, this closely corresponds to Hoxb13 immunofluorescence, indicating that transgene expression is tightly regulated by Hoxb13 transcriptional activity. rtTA dimerization is required for DNA binding and transcriptional activation and consequently, subtle differences in rtTA level can significantly affect responder gene induction. In addition, the ventral-specific expression observed may be a function of H2BGFP. Analyses of the Hoxb13-rtTA-26 strain with alternative responder genes can shed light on induction kinetics in other prostate lobes.
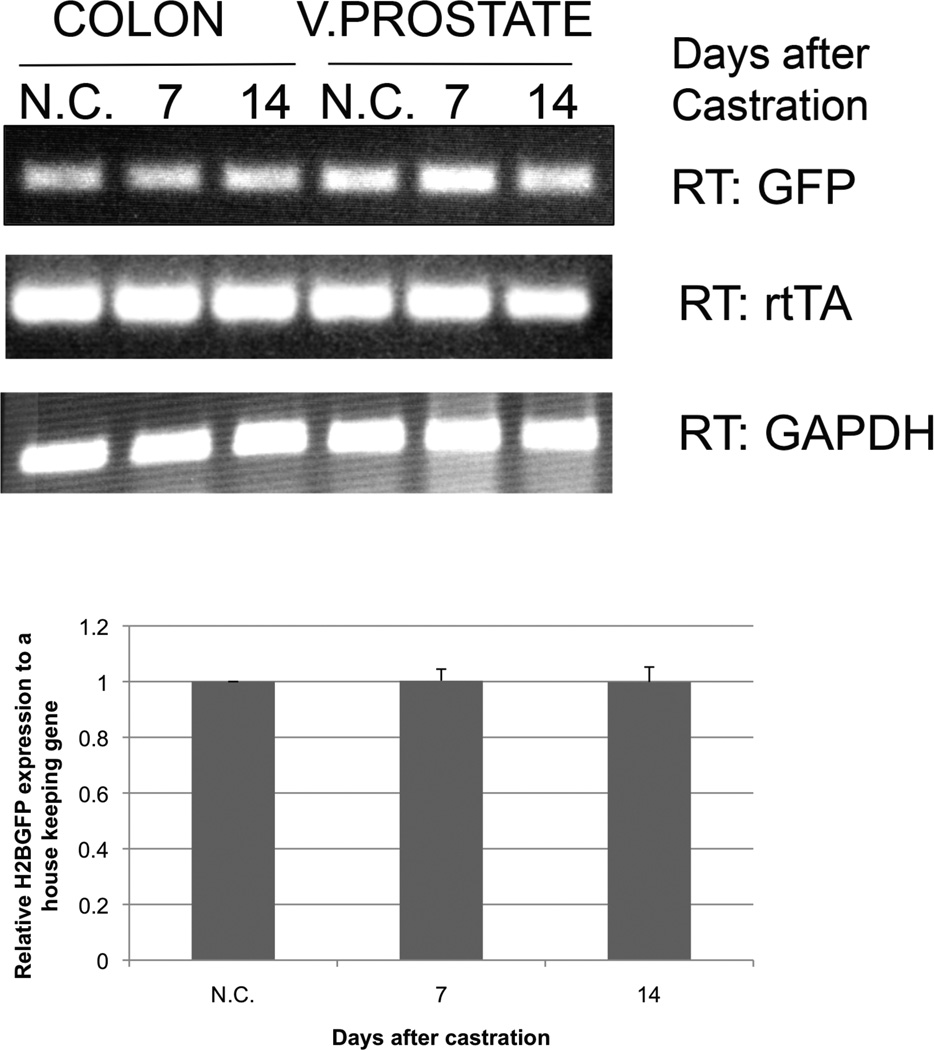
To study the effects of androgen withdrawal on transgene expression, Hoxb13-rtTA|TetO-H2BGFP double transgenic mice were surgically castrated. Dox treatment was initiated 48 hr following castration and maintained thereafter. RTPCR analyses demonstrated maintenance of rtTA and H2BGFP expression in prostates derived from castrated or intact double transgenic mice up to 14 days post-castration (Fig. 5). These analyses clearly demonstrate the utility of the Hoxb13-rtTA-26 transgenic system to regulate clinically relevant responder genes in an androgen-depleted environment.
Figure 5.
Persistent expression of H2BGP and rtTA upon surgical androgen-depletion. Testes were removed from double transgenic mice and 2-day post-castrated mice were Dox-treated for days indicated. mRNA was derived from both castated (C) and non-castrated (N.C.) mice and analyzed by RT-PCR (top) and qRT-PCR (bottom analysis). No change in rtTA or responder gene expression was detected even as late as 14 days post-castration.
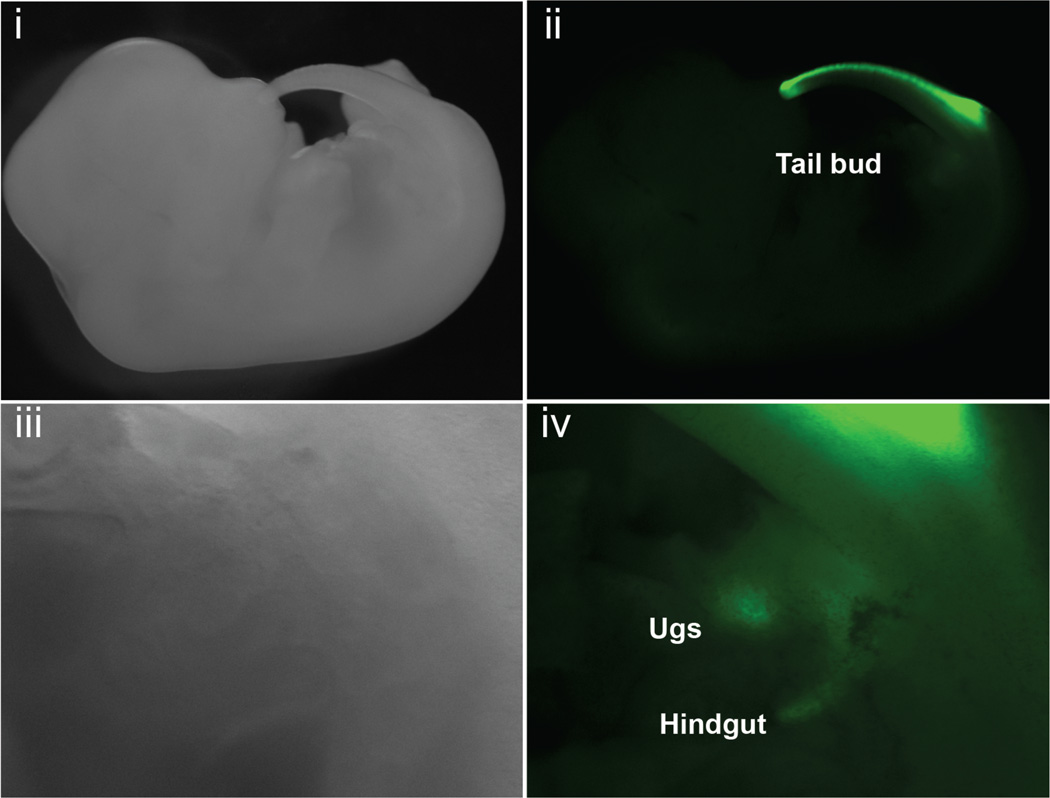
To analyze the domain of responder gene expression in embryos, Hoxb13-rtTA+/+|TetO-H2BGFP+/− males were mated to FVB/N females. Dox treatment of females was initiated upon detection of a copulation plug, and embryos were analyzed by fluorescent stereomicroscopy at (13) days post-coitum. During mouse development, Hoxb13 expression is restricted to posterior structures including the central nervous system, paraxial mesoderm, urogenital sinus and hindgut (27). Fluorescent stereomicroscopic analysis of Dox-exposed double transgenic embryos demonstrated GFP expression exclusively within the known regions of Hoxb13 transcriptional activity (Fig. 6). No responder gene expression was seen in TetO-H2BGFP single transgenic embryos.
Figure 6.
H2BGFP expression follows the domain of Hox-b13 transcriptional activity in embryos. FVB/N females were mated to double transgenic Hoxb13-rtTA+/+|TetOH2BGFP+/− male mice. Embryos were dissected at 13.5 days post-coitum. H2BGFP expression was observed by fluorescence stereomicroscopy. (i–ii) GFP fluorescence was detected in the emerging tail bud, (iii–iv) and in the urogenital sinus and hindgut (higher magnification of i and ii). Fluorescence from a portion of the tail bud is also seen in panel iv.
CONCLUSIONS
Here, we have described the development and analysis of a Hoxb13-driven transgenic mouse model for inducible gene expression in the prostate. Extensive responder gene induction analyses using a TetO-H2BGFP reporter strain demonstrated that the Hoxb13-rtTA system provides a powerful platform to achieve tightly regulated conditional gene expression under normal and androgen-depleted conditions. It is important to note that Hoxb13 cis-regulatory elements drive rtTA expression in the distal colon and rectal epithelial in addition to the prostate. Identification and inactivation of regions that support Hoxb13 transcriptional activity in the distal gastrointestinal tract would facilitate the development of a next-generation Tet-regulatory system with prostate exclusivity.
Incorporating the Hoxb13-driven rtTA system into extant mouse models of prostate cancer or other prostate disorders will enable the development of more complex disease progression models. For example, the effect of tumor suppressor restoration or oncoprotein depletion at different stages of carcinogenesis could provide new insights into the functional roles of these proteins. In addition, the cyclic inflammatory events associated with some forms of prostatitis could also be modeled. Taken together, our results demonstrate that the Hoxb13-rtTA-26 strain provides new opportunities to achieve tightly regulated gene expression in the mouse prostate. The availability of a Tet-regulated system will facilitate analyses of molecular mechanisms underlying prostate development and pathology.
ACKNOWLEDGEMENTS
This manuscript is dedicated to Jamie C. Heard 1986–2008, UMBC Undergraduate Meyerhoff Scholar (M17), valued friend and colleague. The project described was supported in part by Grant Number P20DK090921 from the National Institute of Diabetes and Digestive and Kidney Diseases. The content is solely the responsibility of the authors and does not necessarily represent the official view of the National Institute of Diabetes and Digestive and Kidney Diseases or the National Institutes of Health.
Grant sponsor: United States Army Medical Research and Materiel Command Congressionally Directed Medical Research Prostate Cancer Program; Grant number: W81XWH-07-1-0288; Grant spon- sor: National Institute of Diabetes and Digestive and Kidney Diseases; Grant number: P20DK090921.
REFERENCES
- 1.De Marzo AM, Platz EA, Sutcliffe S, Xu J, Gronberg H, Drake CG, Nakai Y, Isaacs WB, Nelson WG. Inflammation in prostate carcinogenesis. Nat Rev Cancer. 2007;7(4):256–269. doi: 10.1038/nrc2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fleshner NE, Lawrentschuk N. Risk of developing prostate cancer in the future: Overview of prognostic biomarkers. Urology. 2009;73(5 Suppl):S21–S27. doi: 10.1016/j.urology.2009.02.022. [DOI] [PubMed] [Google Scholar]
- 3.Jemal A, Siegel R, Ward E, Hao Y, Xu J, Thun MJ. Cancer statistics, 2009. CA Cancer J Clin. 2009;59(4):225–249. doi: 10.3322/caac.20006. Direct Link: [DOI] [PubMed] [Google Scholar]
- 4.Roehrborn CG. Male lower urinary tract symptoms (LUTS) and benign prostatic hyperplasia (BPH) Med Clin North Am. 95(1):87–100. doi: 10.1016/j.mcna.2010.08.013. [DOI] [PubMed] [Google Scholar]
- 5.Shappell SB, Thomas GV, Roberts RL, Herbert R, Ittmann MM, Rubin MA, Humphrey PA, Sundberg JP, Rozengurt N, Barrios R, Ward JM, Cardiff RD. Prostate pathology of genetically engineered mice: Definitions and classification. The consensus report from the Bar Harbor meeting of the Mouse Models of Human Cancer Consortium Prostate Pathology Committee. Cancer Res. 2004;64(6):2270–2305. doi: 10.1158/0008-5472.can-03-0946. [DOI] [PubMed] [Google Scholar]
- 6.Hieble JP. Animal models for benign prostatic hyperplasia. Handb Exp Pharmacol. 2011(202):69–79. doi: 10.1007/978-3-642-16499-6_4. [DOI] [PubMed] [Google Scholar]
- 7.Jeet V, Russell PJ, Khatri A. Modeling prostate cancer: A perspective on transgenic mouse models. Cancer Metastasis Rev. 29(1):123–142. doi: 10.1007/s10555-010-9212-9. [DOI] [PubMed] [Google Scholar]
- 8.Abate-Shen C, Shen MM. Mouse models of prostate carcinogenesis. Trends Genet. 2002;18(5):S1–S5. doi: 10.1016/s0168-9525(02)02683-5. [DOI] [PubMed] [Google Scholar]
- 9.Kasper S. Survey of genetically engineered mouse models for Prostate Cancer: Analyzing the molecular basis of prostate cancer development, progression and metastasis. J Cell Bio. 2005;94:279–297. doi: 10.1002/jcb.20339. Direct Link: [DOI] [PubMed] [Google Scholar]
- 10.Zhu Z, Zheng T, Lee CG, Homer RJ, Elias JA. Tetracycline-controlled transcriptional regulation systems: Advances and application in transgenic animal modeling. Semin Cell Dev Biol. 2002;13(2):121–128. doi: 10.1016/s1084-9521(02)00018-6. [DOI] [PubMed] [Google Scholar]
- 11.Belteki G, Haigh J, Kabacs N, Haigh K, Sison K, Costantini F, Whitsett J, Quaggin SE, Nagy A. Conditional and inducible transgene expression in mice through the combinatorial use of Cre-mediated recombination and tetracycline induction. Nucleic Acids Res. 2005;33(5):e51. doi: 10.1093/nar/gni051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moody SE, Sarkisian CJ, Hahn KT, Gunther EJ, Pickup S, Dugan KD, Innocent N, Cardiff RD, Schnall MD, Chodosh LA. Conditional activation of Neu in the mammary epithelium of transgenic mice results in reversible pulmonary metastasis. Cancer Cell. 2002;2(6):451–461. doi: 10.1016/s1535-6108(02)00212-x. [DOI] [PubMed] [Google Scholar]
- 13.Fechner H, Wang X, Pico AH, Wildner J, Suckau L, Pinkert S, Sipo I, Weger S, Poller W. A bidirectional Tet-dependent promotor construct regulating the expression of E1A for tight control of oncolytic adenovirus replication. J Biotechnol. 2007;127(4):560–574. doi: 10.1016/j.jbiotec.2006.09.011. [DOI] [PubMed] [Google Scholar]
- 14.Mukherjee A, Soyal SM, Fernandez-Valdivia R, DeMayo FJ, Lydon JP. Targeting reverse tetracycline-dependent transactivator to murine mammary epithelial cells that express the progesterone receptor. Genesis. 2007;45(10):639–646. doi: 10.1002/dvg.20336. Direct Link: [DOI] [PubMed] [Google Scholar]
- 15.Felsher DW, Bishop JM. Reversible tumorigenesis by MYC in hematopoietic lineages. Mol Cell. 1999;4(2):199–207. doi: 10.1016/s1097-2765(00)80367-6. [DOI] [PubMed] [Google Scholar]
- 16.Hsieh AC, Small EJ, Ryan CJ. Androgen-response elements in hormonerefractory prostate cancer: Implications for treatment development. Lancet Oncol. 2007;8(10):933–939. doi: 10.1016/S1470-2045(07)70316-9. [DOI] [PubMed] [Google Scholar]
- 17.PS Clegg N, Arnold H, Ferguson C, Bonham M, White J, Hood L, Lin B. The program of androgen-responsive genes in neoplastic prostate epithelium. Proc Natl Acad Sci USA. 2002;99(18):11890–11895. doi: 10.1073/pnas.182376299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Clegg N, Eroglu B, Ferguson C, Arnold H, Moorman A, Nelson PS. Digital expression profiles of the prostate androgen-response program. J Steroid Biochem Mol Biol. 2002;80(1):13–23. doi: 10.1016/s0960-0760(01)00167-4. [DOI] [PubMed] [Google Scholar]
- 19.Lonergan PE, Tindall DJ. Androgen receptor signaling in prostate cancer development and progression. J Carcinog. 10:20. doi: 10.4103/1477-3163.83937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lamont KR, Tindall DJ. Androgen regulation of gene expression. Adv Cancer Res. 107:137–162. doi: 10.1016/S0065-230X(10)07005-3. [DOI] [PubMed] [Google Scholar]
- 21.McMullin RP, Mutton LN, Bieberich CJ. Hoxb13 regulatory elements mediate transgene expression during prostate organogenesis and carcinogenesis. Dev Dyn. 2009;238(3):664–672. doi: 10.1002/dvdy.21870. Direct Link: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tumbar T, Guasch G, Greco V, Blanpain C, Lowry WE, Rendl M, Fuchs E. Defining the epithelial stem cell niche in skin. Science. 2004;303(5656):359–363. doi: 10.1126/science.1092436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Urlinger S, Baron U, Thellmann M, Hasan MT, Bujard H, Hillen W. Exploring the sequence space for tetracycline-dependent transcriptional activators: Novel mutations yield expanded range and sensitivity. Proc Natl Acad Sci USA. 2000;97(14):7963–7968. doi: 10.1073/pnas.130192197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Warming S, Costantino N, Court DL, Jenkins NA, Copeland NG. Simple and highly efficient BAC recombineering using galK selection. Nucleic Acids Res. 2005;33(4):e36. doi: 10.1093/nar/gni035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang L, Pu Y, Hepps D, Danielpour D, Prins GS. Posterior Hox gene expression and differential androgen regulation in the developing and adult rat prostate lobes. Endocrinology. 2007;148(3):1235–1245. doi: 10.1210/en.2006-1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sreenath T, Orosz A, Fujita K, Bieberich CJ. Androgen-independent expression of hoxb-13 in the mouse prostate. Prostate. 1999;41(3):203–207. doi: 10.1002/(sici)1097-0045(19991101)41:3<203::aid-pros8>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- 27.Zeltser L, Desplan C, Heintz N. Hoxb-13: A new Hox gene in a distant region of the HOXB cluster maintains colinearity. Development. 1996;122(8):2475–2484. doi: 10.1242/dev.122.8.2475. [DOI] [PubMed] [Google Scholar]