Abstract
The research that aims at furthering our understanding of plant primary metabolism has intensified during the last decade. The presented study validated a systems biology methodological framework for the analysis of stress-induced molecular interaction networks in the context of plant primary metabolism, as these are expressed during the first hours of the stress treatment. The framework involves the application of time-series integrated full-genome transcriptomic and polar metabolomic analyses on plant liquid cultures. The latter were selected as the model system for this type of analysis, because they provide a well-controlled growth environment, ensuring that the observed plant response is due only to the applied perturbation. An enhanced gas chromatography–mass spectrometry (GC– MS) metabolomic data correction strategy and a new algorithm for the significance analysis of time-series “omic” data are used to extract information about the plant's transcriptional and metabolic response to the applied stress from the acquired datasets; in this article, it is the first time that these are applied for the analysis of a large biological dataset from a complex eukaryotic system. The case-study involved Arabidopsis thaliana liquid cultures subjected for 30 h to elevated (1%) CO2 stress. The advantages and validity of the methodological framework are discussed in the context of the known A. thaliana or plant, in general, physiology under the particular stress. Of note, the ability of the methodology to capture dynamic aspects of the observed molecular response allowed for 9 and 24 h of treatment to be indicated as corresponding to shifts in both the transcriptional and metabolic activity; analysis of the pathways through which these activity changes are manifested provides insight to regulatory processes.
Keywords: systems biology, elevated CO2 stress, GC–MS polar metabolomic profile, functional genomics, time-series significance analysis of “omic” data
Introduction
The research, which aims at unraveling the regulatory mechanisms that govern plant primary metabolism has intensified during the last decade. This has been triggered by the Arabidopsis 2010 functional genomics initiative (Somerville and Dangl, 2000) and the new forthcoming applications of the plants in bio-fuel (Ragauskas et al., 2006), engineered bio-polymer (Slater et al., 1999), and chemical (Oksman-Caldentey and Inze, 2004) industry. The presented study validated a systems biology methodological framework for the analysis of stress-induced molecular interaction networks within the context of plant primary metabolism, as these are expressed during the first hours of the stress treatment.
This framework involves the application of integrated time-series polar metabolomic and full-genome transcriptomic analyses on plant liquid cultures. The latter were selected as the model system for this type of study, because they provide a well-controlled growth environment, ensuring that the observed response is due only to the applied perturbation. Moreover, the use of high-throughput (“omics”) techniques, and the application of the multivariate statistics and systems biology analytical toolbox, are favored for the analysis of plant primary metabolism, due to the relatively high degree of uncertainty about plant primary metabolism's pathway structure and regulation compared to other species. This level of uncertainty combined with the complexity of plant physiology usually hinder the application of the flux balance and control models (Klapa et al., 2003; Stephanopoulos et al., 1998) that are used in other biological systems and processes for the elucidation of the underlying regulatory mechanisms. High-throughput molecular analysis does not require comprehensive understanding of the interaction networks that define a particular process, while it enables the correlation between parallel occurring phenomena in a single experiment. In addition, the integrated high-throughput analyses of multiple levels of cellular function have been proven to increase the resolution of the acquired view of the cellular physiology (Hwang et al., 2005a,b; Ideker et al., 2001; Klapa and Quackenbush, 2003) compared to single level analyses. Finally, the time-series component is essential to reveal cause–effect relationships within the regulatory networks of each level, and/or between levels of cellular function. In the applied methodological framework, the analysis of the acquired datasets to extract information about the biological system's transcriptional and metabolic response to the applied stress involves the application of an enhanced gas chromatography–mass spectrometry (GC–MS) metabolomic data correction strategy (Kanani and Klapa, 2007; Kanani et al., in press), and a new algorithm for the significance analysis of time-series “omic” data (Dutta et al., 2007). In this article, it is the first time that they are applied in combination for the analysis of a large biological dataset from a complex eukaryotic system.
The particular systems biology methodological framework was applied on the A. thaliana liquid culture system to investigate its dynamic molecular response during the first 30 h of its subjection to elevated (1%) CO2 stress. The advantages and validity of the framework will be discussed in the context of the currently known regulation of the A. thaliana, in particular, and plant, in general, primary metabolism from CO2 fixation.
Materials and Methods
Experimental Design
Two sets of 20 A. thaliana liquid cultures were grown for 12 days on an orbital shaker platform (Barnstead, Melrose Park, IL) at 150 rpm, in the ambient air (350 ppm CO2) of a growth chamber (model M-40, EGC Inc., Chagrin Falls, OH), under constant white light intensity (80–100 μE m−2 s−2) and temperature (23°C) to reduce any circadian rhythm effects. Four liquid cultures from each set were then harvested to be used as markers of culture growth up to that stage (0 h). Starting on the 13th day and for the next 30 h, one set (referred to in the rest of the text as “perturbed”) grew in air of 10,000 ppm CO2 (same light and temperature conditions), while the other (referred to as “control”) kept growing under the initial conditions. The CO2 concentration increase in the air of the perturbed system's growth chamber was achieved in 5 min [WMA-4 CO2 Analyzer, PP Systems, Amesbury, MA]. The assumption of vapor-liquid equilibrium being valid (well-shaken culture system) and the Henry's law (Middleman, 1998) holding under the conditions of the experiment [i.e., constant temperature and pressure, and dissolved CO2 concentration below the maximum CO2 solubility of 1.45 g/L (Lide and Frederikse, 1995)], the concentration of the dissolved carbon dioxide in the perturbed liquid cultures was also 28-fold larger than in their control counterparts. Two liquid cultures from each set were harvested at 1, 3, 6, 9, 12, 18, 24, and 30 h of perturbation. Each harvested culture was weighed, rinsed in distilled water (the entire process of weighing and rinsing lasted ~30 s), frozen in liquid nitrogen and stored at −80°C for further analysis. One of the 6 h control cultures weighed only 9.2 g, that is 50% of the average culture weight (18.4 g) and 60% of its bio-replicates (15.2 g). Due to this macroscopic difference, supported also by the metabolomic and transcriptomic data (not shown), the culture was not considered in further analysis.
Culture Media
The seeds had been cleaned (Boyes et al., 2001) and stored overnight at 4°C prior to inoculation. The plant cultures grew in 500 mL shake flasks, each containing 200 mL B5 Gamborg media (Gamborg et al., 1976) with minimal organics (Sigma, St. Louis, MO), 2% (w/v) sucrose, and 0.1% agar, inoculated with ~100 Columbia ecotype seeds. The particular sucrose concentration, which has also been utilized in other studies involving A. thaliana liquid cultures (Liu et al., 2005; Misson et al., 2005; Osuna et al., 2007; Scheible et al., 2004; Wang et al., 2004), was selected to allow for stable growth of the A. thaliana seedlings in the liquid media by aiding root development and biomass accumulation (Hétu et al., 2005). Photosynthesis occurs in sucrose-grown liquid cultures (Cournac et al., 1991; Osuna et al., 2007), as it was also validated from the molecular data that were acquired in the present study.
Metabolomic Profiling
The polar extracts of the plant liquid cultures were obtained from 125 mg of ground culture using methanol/ water extraction (Roessner et al., 2000) and ribitol (0.2 mg/g of fresh weight) as the internal standard. The dried polar extracts were derivatized to their (Meox-) TMS-derivatives through reaction with 100 μL of 20 mg/mL methoxyamine hydrochloride solution in pyridine for 2 h, followed by reaction with 200 μL of N-methyl-trimethylsilyl-trifluoroacetamide (MSTFA) for at least 6 h (Kanani and Klapa, 2007) at room temperature. The metabolomic profiles (Fiehn et al., 2000; Roessner et al., 2000) were obtained using the Saturn 2100T (ion trap) GC–MS (Varian Inc., Palo Alto, CA). The peak identification and quantification was carried out as described in (Kanani and Klapa, 2007). The raw meta-bolomic dataset is provided in Supplementary Table I.
The relative areas (RPAs) of all detected peaks were estimated from normalization with the internal standard. The recently developed by our group data validation, normalization and correction methodology (Kanani and Klapa, 2007) was applied to account for derivatization biases (Kanani and Klapa, 2007) that are primarily due to the formation of multiple derivatives from the amine-group containing metabolites. Specifically, we first verified same GC–MS operating conditions during the acquisition of all metabolomic profiles of both the perturbed and control culture sets. Then, the derivative peak areas that corresponded to the same amine-group containing metabolite were combined into one cumulative (effective) peak area, using the weight coefficients listed in Kanani and Klapa (2007). Further, (a) one of the two Meox peaks of the known ketone-group containing metabolites (Kanani and Klapa, 2007), (b) the peaks corresponding to unknown amine-group containing metabolites (Kanani and Klapa, 2007), (c) the peaks that were identified as derivatization artifacts or with significant carry over, and (d) the metabolite peaks that were detected in less than 80% of the samples or with higher than 25% coefficient of variation between consecutive injections, were filtered out of the analysis. Subsequently, the control and perturbed RPA of each remaining metabolite at each time point was estimated as the arithmetic mean of the metabolite's RPAs in the multiple injections of all control and perturbed, respectively, biological replicates harvested at this time point. Finally, the control and perturbed RPAs of each metabolite at each time point were divided with the control and perturbed, respectively, 0 h RPA of this metabolite (the “perturbed” 0 h metabolic state refers to the four liquid cultures that were harvested at the 0 h time point from the liquid culture set that was subsequently subjected to the perturbation). These final metabolite RPA profiles over time are referred to as control and perturbed, respectively, metabolomic profiles in the text and are used to extract information about the metabolic response of the system to the applied perturbation. Any missing RPAs were imputed using the k-means neighborhood algorithm (Troyanskaya et al., 2001) as this is implemented in the TM4 MeV (V3.1) software (Saeed et al., 2003).
Transcriptomic Profiling
A. thaliana full-genome DNA amplicon microarrays (Kim et al., 2003) were used. The utilized RNA extraction, probe labeling and hybridization protocols are available in [http://atarrays.tigr.org]. The image processing and data normalization were carried out using the TIGR Spotfinder (V2.2.4) and MIDAS (V2.19), respectively, software (Saeed et al., 2003). The datafiles produced after image processing can be accessed at the public database ArrayExpress under the accession number E-MEXP-773. The normalization involved locally weighted scatterplot smoothing regression (LOWESS; smooth parameter: 0.33; reference: Cy3), variance regularization (reference: Cy3) and “flip-dye” data consistency trim (data trim option: SD cut; cross log ratio data keep range: ±2 SD). Subsequently, the control and perturbed expression of each gene at each time point was estimated as the geometric mean of its expression in all control and perturbed, respectively, biological replicates harvested at this time point. Finally, the control and perturbed expressions of each gene at each time point were divided by the control and perturbed, respectively, 0 h expression of this gene. These final gene expression profiles over time are referred to as control and perturbed, respectively, transcriptomic profiles in the text and are used to extract information about the transcriptional response of the system to the applied perturbation. Genes that were detected in less than 75% of the time points were removed from further analysis. Any missing expressions were imputed using the k-means neighborhood algorithm (Troyanskaya et al., 2001), as this is implemented in the TM4 MeV (V3.1) software (Saeed et al., 2003).
Data Analysis
The genes (or metabolites) whose expression (or concentration) was significantly higher or lower in the perturbed compared to the control culture set will be referred to as positively or negatively, respectively, significant genes (or metabolites) in the rest of the text. The significant genes and metabolites were identified using the paired significance analysis of microarrays (SAM; Tusher et al., 2001) algorithm, as this is implemented in the TM4 MeV (V3.1) software (Saeed et al., 2003). The significant genes and metabolites at each time point were identified using the recently developed in Klapa's group MiTimeS methodology (Dutta et al., 2007). MiTimeS implements a time-series significance analysis algorithm, which takes into consideration that the expression (or concentration) profiles at each time point are part of the same time-series analysis and not independent snapshot experiments. Although MiTimeS was originally developed for gene expression data, the usefulness of the algorithm for the analysis of other molecular fingerprints was validated in the presented study. In the case of the metabolomic analysis, MiTimeS was used without the Bonferroni-like correction described in Dutta et al. (2007), which was used for the transcriptomic data, due to the low number of metabolites compared to genes that are considered in the analysis. As described in Dutta et al. (2007), MiTimeS utilized the same expected gene expression and metabolite concentration, respectively, distribution as paired-SAM. MiTimeS estimates also (a) each gene's and metabolite's Significance Variability (SV) score, the latter being a measure of the gene's (or metabolite's) significance level variability over time; based on its definition, the SV score could range from 0 to 2 (zero SV score means that the gene (or metabolite) belongs to the same significance level throughout all time points, while SV score = 2 means that the gene (or metabolite) fluctuates between the positively and negatively significance level throughout the duration of the experiment), and (b) the positively or negatively gene (or metabolite) significance correlation coefficient between two time points. This is equal to the number of the common between the two time points positively or negatively, respectively, significant genes (or metabolites) normalized by the square root of the product of the total number of positively or negatively, respectively, significant genes (or metabolites) at each time point [for detailed description see Dutta et al., 2007]. The positively or negatively gene (or metabolite) significance correlation coefficients populate, respectively, the significance correlation matrix (SCM) with respect to the positively or negatively significant genes (or metabolites). The diagonal element of the positively or negatively SCM matrix that corresponds to a particular time point is defined as the number of genes (or metabolites) that are positively or negatively, respectively, significant only at the particular time point, normalized by the total number of positively or negatively, respectively, significant genes (or metabolites) at this time point (Dutta et al., 2007). Time-point correlation networks were constructed based on the corresponding SCMs. The gene ontology (GO) analysis was based on the GO terms included in the corresponding feature of the TM4 MeV(V3.1) software.
Results and Discussion
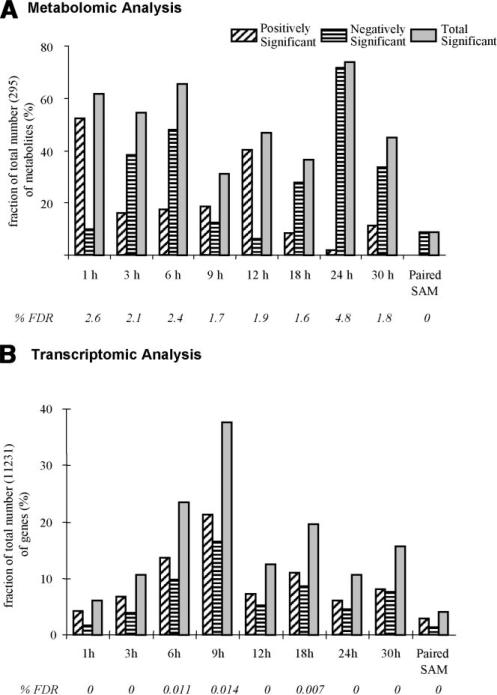
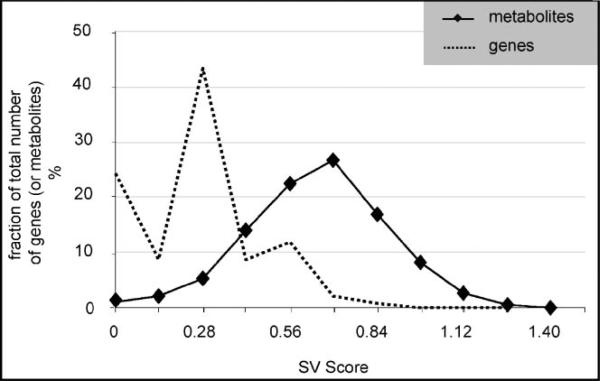
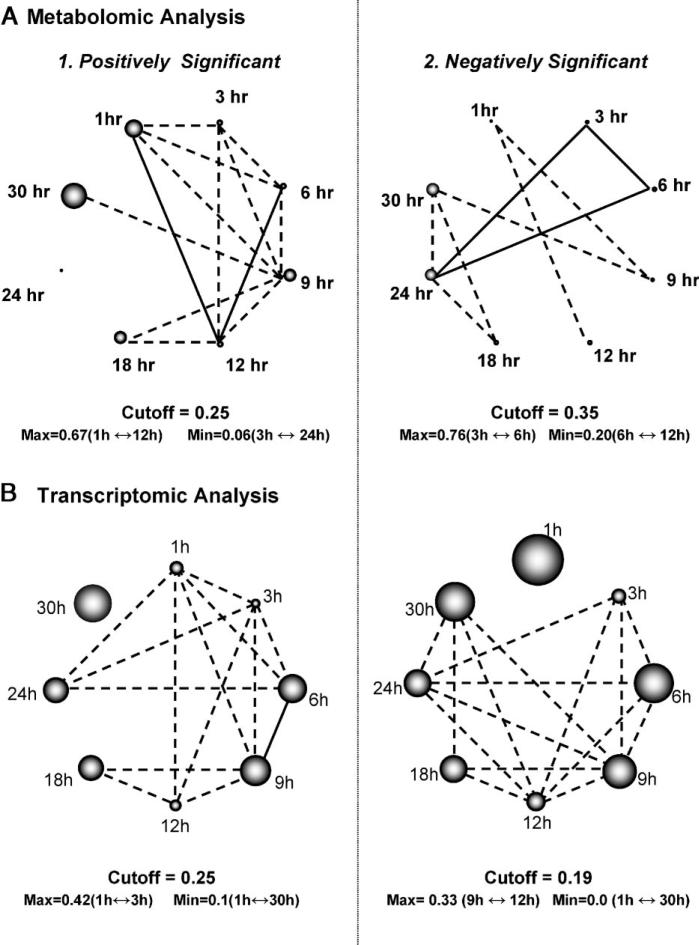
Among the 295 metabolite peaks that were used in the analysis (see Appendix A), paired-SAM [0% median false discovery rate (FDR)] identified 0 and 28 (six annotated), respectively, positively and negatively significant (Fig. 1A). This result indicates an overall decrease of the free polar metabolite pool in the perturbed compared to the control system. For the same significance threshold value, MiTimeS identified in average 66 and 91, respectively, positively and negatively significant metabolite peaks at each time point (see histogram in Fig. 1A; the significance level profiles over time and the paired-SAM classification of all annotated peaks that were identified as significant in at least one time point are shown in Supplementary Table II). Based on their SV score (Fig. 2), only three (all unidentified) metabolites were observed in the same significance level (i.e., nonsignificant) at all time points. All others were observed as positively or negatively significant at one time point at least. According to Figure 1A, the CO2 availability resulted in a drastic initial increase in the free polar metabolite pool sizes. The number of positively and negatively significant metabolite peaks at the succeeding time points up to 6 h of perturbation suggest that the net increase in the free polar metabolite pool sizes is subsequently smaller. The minimum and maximum total number of significant metabolites was observed at 9 and 24 h, respectively, after the initiation of the perturbation, indicating drastic changes in the metabolic physiology of the plant liquid cultures around these two time points. The time point correlation network (see Materials and Methods Section) based on the positively significant metabolites (Fig. 3A1; see Supplementary Table IIIA for the actual values of the time point significance correlation coefficients) indicates strong correlation between the time points up to 12 h of perturbation. During this period of treatment, among the non-negatively significant metabolites 115 were identified as positively significant at one time point at least (i.e., they were nonsignificant at the rest). The known metabolites in this group were mainly (a) amino acids and other amine-containing compounds, (b) intermediates of the one-half of the TCA cycle (citrate, iso-citrate, α-ketoglutarate), which is responsible for the production of glutamate-based amino acids and growth, (c) most of the triose- and hexose phosphates, and (d) some fatty acids (see Supplementary Table II). On the other hand, the time point correlation network based on the negatively significant metabolites indicates strong correlation between the time points of longer treatment (i.e., 18, 24, 30 h). At these time points, the number of positively significant metabolites is very low, suggesting a continuous drain of the measured free polar metabolite pools without similar increase in their production rate. Characteristically, at 24 h of perturbation, most (66%) of the free polar metabolites were identified as negatively significant. Specifically, 86 (75%) out of the 115 non-negatively significant metabolites that were identified as positively significant at one time point at least during the first 12 h of treatment belonged to the negative significance level at the late time points. It is this large decrease in the metabolite concentrations during the late time points that drives the results of paired-SAM analysis to indicate an average over time decrease in the free polar metabolite pool in the perturbed compared to the control plant system. Comparison of the time points based on the number of metabolites that were identified as positively or negatively significant at a particular time point only (i.e., the diagonal elements of the corresponding SCMs as described in the Materials and Methods section) singled out the response of the system at 30 h of continuous treatment (see Fig. 3A1–2 and Supplementary Table IIIA). This is in agreement with the previously stated observation based on the total number of significant metabolites that the 24 h of treatment coincide with a shift in the metabolic response of the system to the applied perturbation. Identification of this type of “turning points” in the response of the plants to a particular stress has not been reported before in the literature, indicating the significance of short-interval time-series analysis for this purpose. In addition, these results show the type of time-dependent information that can be extracted from the biological datasets when MiTimeS is used.
Figure 1.
Number of positively and negatively significant (A) metabolites and (B) genes at each time point (based on MiTimeS) and from paired-SAM (see Materials and Methods Section). MiTimeS and paired-SAM analyses of the metabolomic profiles were both based on the same significance threshold (delta) value of 1.2, while for the MiTimeS and paired-SAM analyses of the transcriptomic profiles the delta value was 0.9. The median% false discovery rate (FDR) of each analysis is also shown.
Figure 2.
Significance Variability (SV) score distribution of metabolomic and transcriptomic data. A gene's or metabolite's SV score (Dutta et al., 2007) is a measure of the gene's (or metabolite's) significance level variability over time. Based on its definition, the SV score could range from 0 to 2; zero SV score means that the gene (or metabolite) belongs to the same significance level throughout all time points, while SV score = 2 means that the gene (or metabolite) fluctuates between the positively and negatively significance level throughout the duration of the experiment.
Figure 3.
Time point correlation networks based on the common between time points (A1) positively significant metabolites, (A2) negatively significant metabolites, (B1) positively significant genes, and (B2) negatively significant genes. The correlation coefficient between two time points based on significant metabolites or genes is estimated as described in the Materials and Methods section [and in detail in (Dutta et al., 2007)]. In the shown networks, two time points are connected, if their correlation coefficient is larger than the mean correlation coefficient between any two time points (indicated cutoff threshold). If the correlation coefficient between two time points is larger than 0.5, then they are connected through a straight line; otherwise the line is dashed. The diameter of the sphere representing each time point in the positively or negative significant correlation networks is, respectively, proportional to the fraction of metabolites or genes that are positively or negatively significant only at that time point [see Materials and Methods section]. Thus, the wider the sphere the larger the number of (positively or negatively) significant changes at the transcriptional (or metabolic) level that are uniquely observed at the particular time point.
Among the 11,231 genes that were finally considered in the analysis after appropriate data normalization and filtering, paired-SAM (0% median FDR) identified 313 and 143, respectively, positively and negatively significant genes. For the same significance level, MiTimeS identified in average 2,000 positively and 816 negatively significant genes at each time point (Fig. 1B). It is noticeable that the number of positively and negatively significant genes exhibit similar time profiles. Specifically, they kept increasing for the first 9 h of perturbation exhibiting a maximum at the 9 h time point, which on the contrary corresponds to the minimum number of significant metabolite peaks. Despite this difference between the transcriptional and metabolic response, which could be attributed to different regulatory mechanisms governing each of these levels of cellular function, these observations indicate 9 h as an important time point for the adjustment of the plant liquid culture system to the applied stress. Another difference that was observed between the transcriptional and metabolic response referred to the 1 h of treatment that corresponded to the minimum number of significant genes in contrast to the initial drastic increase in the number of significant metabolite peaks. This indicates a faster reaction of the metabolic machinery to the applied perturbation, potentially due to the fact that this perturbation is characterized by a large change in the concentration of the main substrate of the plants. GO analysis of the negatively significant genes at the 1 h of treatment revealed the GO terms “response to fungi,” “pathogenic fungi,” “glucolipid and galactolipid biosynthesis and metabolism,” “removal and metabolism of superoxide radicals,” “stomatal movement” and “sulfur transport” as significantly underexpressed at this time point. On the other hand, the majority of the genes in the GO terms “defense response” and “response to biotic stimulus” were significantly over expressed at the same time point. At the late time points, the time point correlation network based on the positively significant genes singled out the 30 h time point (Fig. 3B1 and Supplementary Table IIIB). Thus, transcriptional analysis seem to also indicate the 24 h time point as coinciding with significant change in the molecular response of the liquid culture system to the applied stress. GO analysis at the late time points (24–30 h) identified significant decrease in the activity of the GO terms “carbon utilization” and “photosynthesis,” indicating this decrease as the potential cause of the observed re-adjustment of both the transcriptional and metabolic physiologies after 24 h of treatment. The acute and long-term physiological response to the applied stress can also be differentiated according to the GO analysis of genes based on cellular localization. Specifically, it was observed that mainly genes associated with ribosomes are negatively significant at the early time points (1–3 h), while mainly genes associated with the photosystem reaction centers I and II are negatively significant at the late time points (24–30 h).
According to the SV score distribution (Fig. 2), 2,738 (9 positively, 8 negatively, and 2,721 non-significant) genes were identified in the same significance level at all time points (i.e., SV score = 0), while only one corresponded to the highest SV score (1.14). Comparison of the metabolite and gene SV score distributions indicated that while of similar shape, the metabolite distribution was of larger mean and median. This means that the fraction of free polar metabolites that underwent multiple changes in their significance level over time was in average larger than the corresponding gene fraction. The positively significant “zero SV score” gene pool comprised eight annotated genes encoding for pyruvate decarboxylase (PDC), exportin1 (XPO1), ABC transporter family protein, oligopeptide transporter OPT family protein, sterol dehydrogenase (putative), oxidoreductase, 2OG-Fe(II) oxygenase family protein and peroxidase. PDC catalyses the conversion of pyruvate to acetaldehyde, which is subsequently reduced to ethanol through the alcohol dehydrogenase (ADH; www.kegg.com; Kursteiner et al., 2003). ADH was identified as positively significant from paired-SAM and at all but two time points from MiTimeS. Over-expression of PDC and ADH has been related to abiotic stresses in general and hypoxic stress in particular (www.kegg.com; Kursteiner et al., 2003; Seki et al., 2002; Tadege et al., 1999). In plants, the 2OG-Fe(II) oxygenase family plays an important role in the synthesis of plant hormones like ethylene, gibberellins, anthocyanidins, and flavones (Aravind and Koonin, 2001). The negatively significant “zero SV score” gene pool comprises four annotated genes encoding for xyloglucan: xyloglucosyl transferase, S-adenosyl-l-methionine: carboxyl methyltransferase family protein, NADP-dependent oxidoreductase, and F-box family protein. The enzyme xyloglucan: xyloglucosyl transferase is involved in the modification of the cell wall (Toikkanen et al., 2007; www.arabidopsis.org). GO analysis with respect to the cellular component indicated that the particular S-adenosyl-l-methionine: carboxyl methyltransferase is associated with the endomembrane, but the biological process with which it is associated remains unknown (www.arabidopsis.org). The highest SV score gene is annotated as related to a harpin-induced protein. In A. thaliana, harpin regulates the accumulation of salicylic acid (Clarke et al., 2005), which plays an important role in plant defense. Of the second highest SV score (=1) were identified genes encoding for universal stress protein (USP) family protein, dehydration responsive element (DRE)-binding transcription factor, potassium transporter (HAK5), phosphorylase family protein, and cyclin family protein. Most of these genes are negatively significant at 3, 9, 18, 30 h, and nonsignificant at the rest exhibiting an alternate significantnon-significant pattern. It has been reported (Chen et al., 2007; Sakuma et al., 2002) that the genes encoding the DRE-binding proteins are involved in gene expression regulation under conditions of drought, salt or cold stress.
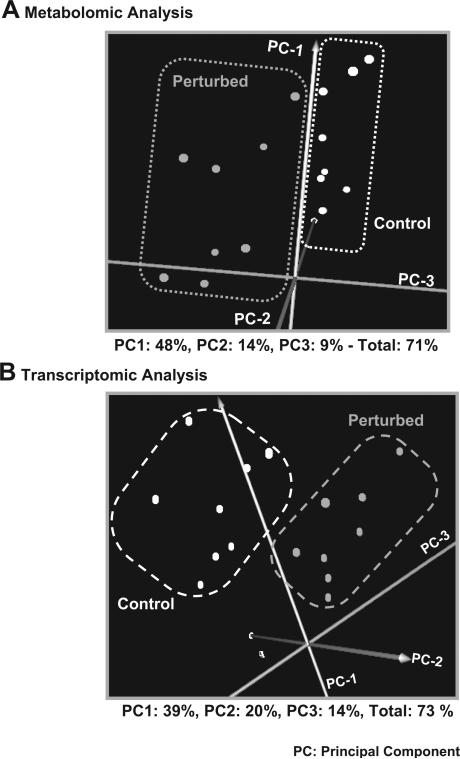
It is thus apparent that the A. thaliana plant liquid culture physiology is affected by the applied perturbation at both the metabolic and transcriptional levels even during the first 30 h of treatment. In agreement, principal component analysis (PCA) indicates that both the control metabolomic and transcriptomic profiles can be clearly differentiated from their perturbed counterparts (Fig. 4; Supplementary Table IV includes the list of all principal components and their contribution for both the metabolomic and the transcriptomic data). Both analyses indicated also a readjustment in the transcriptional and metabolic response of the liquid culture system to the applied stress at about 9 and 24 h of treatment. However, there are also differences between the transcriptional and metabolic responses, the study of which could be quite useful for the identification of interaction mechanisms between these two levels of cellular function. To isolate though the real biological differences when comparing the two cellular levels, it is necessary for the current technical differences between metabolomic and transcriptional profiling analyses to be seriously taken into consideration. According to the transcriptomics protocol, the total mRNA amount is the same in all samples. Thus, transcriptional profiling analysis quantifies the change in the mRNA composition with respect to a particular gene from one physiological state to the other. However, the total free metabolite pool weight cannot be considered the same among biological samples. Metabolomic analysis measures the concentration of a particular metabolite in certain weight of the fresh biological sample. Thus, even when the composition of the total free metabolite pool in the particular metabolite remains the same between two physiological states, the metabolite's concentration may still be different due to change in the total free metabolite pool size from one state to the other. The following sections discuss the most prominent observed changes in the physiology of the A. thaliana liquid cultures due to the applied perturbation in the context of specific pathways that are directly or indirectly related to carbon fixation. Agreement with previous knowledge regarding particular biological processes provides validation checkpoints for the applied methodological framework, while also supports any novel information that was acquired.
Figure 4.
Principal component analysis (PCA) of the (A) metabolomic and (B) transcriptomic data. Each point on the graphs represents the metabolomic (A) or transcriptomic (B) profile of the control or perturbed plant liquid cultures sets at a particular time point. The control and perturbed culture sets are separated based on their metabolomic (A) and transcriptomic (B) profiles, indicating that both the transcriptional and metabolic activity are affected by the applied perturbation even during the first 30 h of the applied treatment.
Photorespiration/Carbon Fixation and Carbohydrate Production
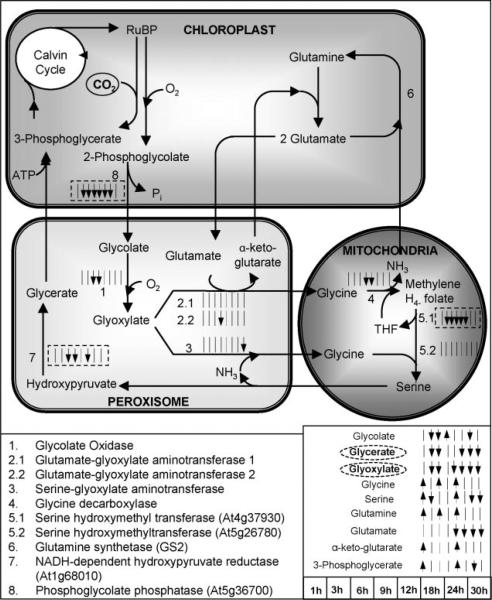
Carbon fixation and photorespiration “compete” for the Rubisco activity (Fig. 5). Thus, changes in the CO2/O2 ratio are expected to affect the flux distribution between the two pathways (see e.g., Coschigano et al., 1998; Dey and Harborne, 1997; Siedow and Day, 2001). The observed decrease in all three organic acids (glycerate, glyoxylate, glycolate) and serine in the photorespiration pathway is in agreement with the expectation that the photorespiration rate should be significantly reduced in the plant liquid cultures that experienced 25-fold increase in the CO2/O2 ratio in their growth environment. In addition, the transcriptomic data provided consistent to the metabolomic data information at the transcriptional level. Specifically, the photorespiratory pathway genes coding for serine hydroxymethyltransferase (At4g37930), NADH-dependent hydroxypyruvate reductase, and phosphoglycolate phosphatase, were identified as negatively significant based on paired-SAM and at most of the time points (Fig. 5). These observations provided a positive validation control for the presented study both with respect to the applied perturbation and the model system. It is significant that an effect that is de facto expected in soil grown plants under elevated CO2 stress, i.e., the inhibition of photorespiration, is also conclusively observed in the plant liquid culture system, at both the metabolic and transcriptional levels.
Figure 5.
The significance level profiles over time and the significance classification based on paired-SAM of the genes and metabolites in the photorespiration pathway that were included in the analysis. The transcriptional information is compartmentalized as shown in the figure, i.e., all shown genes are expressed in the particular compartments. The significance level profiles of metabolites that participate in this pathway are shown outside of the compartments, because they refer to the net over the entire liquid culture (i.e., over all compartments, tissue and cell types) metabolite pool size. If the gene encoding a particular reaction's enzyme was identified as positively or negatively significant at a time point (see time point sequence in the lower right side of the figure), the corresponding small time point arrow next to the reaction is facing up or facing down, respectively. If the gene was identified as non-significant at a time point, then a small straight line is shown in the respective location. A solid or dashed, respectively, box around the time point arrows signifies that the gene was identified as positively or negatively significant by paired-SAM. If a metabolite was identified as positively or negatively significant at a particular time point, the corresponding time point arrow next to this metabolite's name is facing up or down, respectively. If a metabolite was identified as positively or negatively significant by paired-SAM then its name is circled by a straight or dashed, respectively, line. Information about the photorespiration pathway structure was obtained from the KEGG database (www.kegg.com) and Buchanan et al. (2001). Dotted lines depict positive or negative regulation depending on the sign. (This figure can be viewed online in a color version as Supplementary Figure 1.)
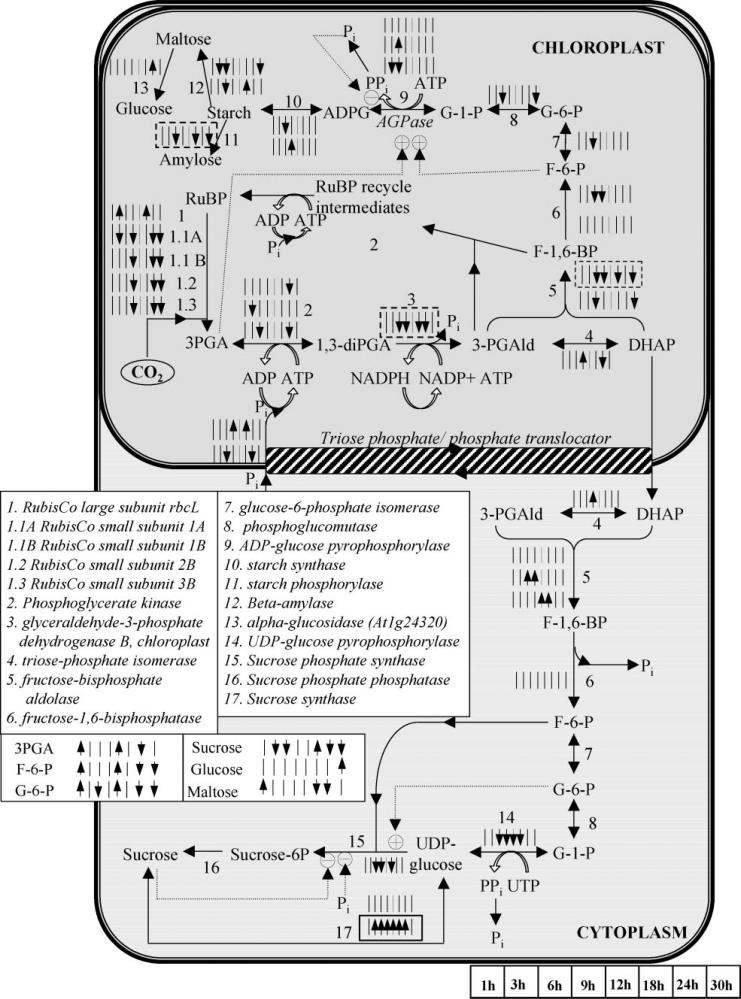
Rubisco comprises two subunits, namely the small (rbcS) and the large (rbcL), which are encoded by nuclear and chloroplast genes, respectively. The rbcL gene was positively significant at the 3 and 18 h of perturbation (Fig. 6), agreeing with the expectations for high rate of CO2 fixation in the chloroplasts during the initial period of treatment. The Arabidopsis rbcS gene family comprises four members, namely 1A, 1B, 2B, and 3B (Makino et al., 2000). All four were identified as negatively significant at the 9, 24 and 30 h of perturbation. 1B–3B are all located in the chromosome 5 and exhibit identical significance profiles over time. 1A, which is located in the chromosome 1, was identified as negatively significant at the 3 h time point too. There is no previous knowledge about the response of the two Rubisco subunits to the elevated CO2 stress. Thus, their observed differential expression during the first 30 h of the elevated CO2 treatment provides novel insight to their regulation by carbon fixation.
Figure 6.
The significance level profiles over time and the significance classification based on paired-SAM of the genes and metabolites in the Calvin cycle, starch and sucrose biosynthesis pathways that were included in the analysis. The transcriptional information is compartmentalized as shown in the figure, i.e., all shown genes are expressed in the particular compartments. The significance level profiles of metabolites that participate in these pathways are shown outside of the compartments, because they refer to the net over the entire liquid culture (i.e., over all compartments, tissue and cell types) metabolite pool size. Positively and negatively significant genes and metabolites are designated as described in the caption of Figure 5. Information about the depicted pathways’ structure was obtained from the KEGG database (www.kegg.com) and Buchanan et al. (2001). Dotted lines depict positive or negative regulation depending on the sign. (This figure can be viewed online in a color version as Supplementary Figure 2.)
Transcriptional profiling analysis of the sucrose bio-synthesis route (Fig. 6), which was constructed based on information from (www.kegg.com; Denis and Blakely, 2001; Smith, 1993; Stitt, 1991), indicated increase in sucrose degradation to produce UDP-glucose due to the elevated CO2 stress; notably there is no previous report associating the particular stress with sucrose depletion to UDP-glucose. Sucrose itself was identified as negatively significant at the 3, 6, 24, and 30 h and positively significant at the 18 h. The sucrose being, however, a main component of the culture media, its concentration measurement should be used with relative caution. Because the present study involves the average over the entire plant response of the system to the applied perturbation, the localization of the observed metabolic effect requires further, more specific investigations. In addition, the same experiment should be carried out with lower concentrations of sucrose in the supplied medium of the liquid cultures to investigate potential universality of the observed correlation. On the other hand, Figure 5 indicates accumulation of starch due to the applied treatment, which agrees with previous knowledge (Smith, 1993).
Nitrogen Assimilation and Amino Acid Metabolism
Among the essential pathways of plant primary metabolism, nitrogen assimilation is expected to be affected by the elevated CO2 stress due to (a) its direct association with the amino acid metabolism, and (b) the need for reductive power in the form of NADH/NADPH for the plants to convert nitrate/nitrite ions into amine groups. Within the context of the currently known A. thaliana physiology, there existed information only about the response of nitrate reductase (NR) genes to light (Cheng et al., 1991, 1992; Dey and Harborne, 1997) or external sugars (Coschigano et al., 1998). During the course of the present experiment, the gene encoding for the first form of the nitrate reductase (NR1) was identified as positively significant at the 18 and 30 h of perturbation. Even though the second nitrate reductase (NR2) did not pass the stringent significance test (it was identified as non-significant at all time points and also from paired SAM), it was up regulated at the 1, 3, 24, and 30 h time points. This result is in agreement with an elevated CO2 study on cucumber plants (Larios et al., 2001), which had indicated increase in the expression of the NR genes under the elevated CO2 stress. Importantly, the delayed and gradual induction of the NR1 compared to the immediate increase in the NR2 expression due to the applied perturbation had previously been observed in a study that monitored the expression of the NR1 and NR2 genes in response to light in leaves of adapted-in-the-dark, soil-grown A. thaliana plants (Cheng et al., 1991, 1992). Thus, the NR1 and NR2 gene expression appear to exhibit the same differential induction pattern in response to the elevated CO2 stress as to the light stimulation. This observation contributes to the current knowledge about the regulation of nitrogen assimilation in A. thaliana. The NR gene expressions were to-date considered to be induced only by light, nitrate ions and carbohydrates (Cheng et al., 1991, 1992; Corruzi and Last, 2001; Larios et al., 2001).
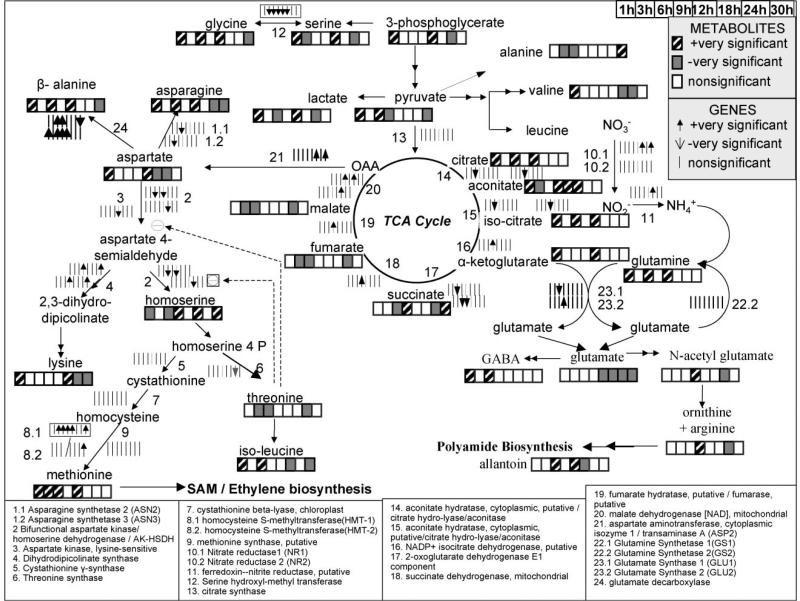
In agreement, metabolomic data indicated three of the four nitrogen-storage amino acids (Coschigano et al., 1998), glutamine, asparagine and aspartate, as positively significant at most of the time points during the first 12 h (Fig. 7). Even though the concentration of the fourth amino acid, gluta-mate, remained unaltered for the first 9 h of the perturbation, decreasing slightly at the 12 h, the amine-containing metabolites 4-aminobutyrate (GABA), ornithine/arginine and N-acetyl-glutamate, which arederived from glutamate, were identified as positively significant at most of the time points during the first 12 h (Fig. 7). In addition, aketoglutarate, required for the production of glutamate and glutamine, and its TCA cycle precursors citrate and isocitrate, were also positively significant at some/most of the time points during the first 12 h (Fig. 7). As it was also mentioned earlier within the context of all metabolomic data, the concentration of most of the known detected amino acids, their precursors and other amine compounds exhibited significant increase in response to elevated CO2 stress during the first 12 h, including valine, isoleucine, glycine, lysine, and methionine (Fig. 7).
Figure 7.
The significance level profiles over time and the significance classification based on paired-SAM of the genes and metabolites in the nitrogen assimilation and amino acid biosynthesis pathways that were included in the analysis. Positively and negatively significant genes and metabolites are designated as described in the upper right side of the figure. Information about the depicted pathways’ structure was obtained from the KEGG database (www.kegg.com) and Buchanan et al. (2001). Dashed lines depict positive or negative regulation depending on the sign.
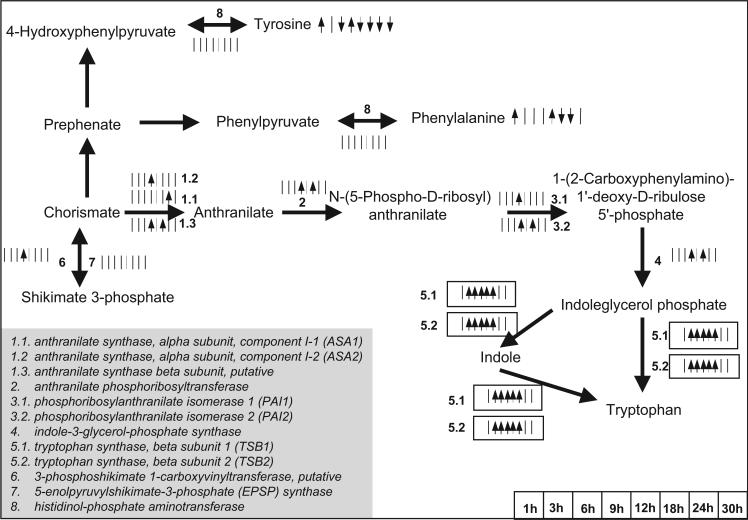
This was not, however, the case for five (out of 19) amino acid/amine compounds: serine, threonine, homoserine, tyrosine, and phenylalanine. The decrease in the tyrosine and phenylalanine concentration could be attributed to the increase in the rate of the competing pathway of tryptophan biosynthesis as this was observed at the transcriptional level (see Fig. 8). It is important to note the hereby synergy between metabolomic and transcriptomic analyses; as no free tryptophan was detected in the acquired profiles, metabolomic analysis alone would not have been conclusive about the activity of aromatic biosynthesis. Tryptophan is precursor of numerous secondary metabolites, like the hormone auxin, the indole alkanoids, the phytoalexins, and the cyclic hydroxamic acids (www.kegg.com). Increase in the rate of tryptophan biosynthesis could thus signify subsequent increase in its consumption towards secondary metabolic intermediates, a fact that could explain the negligible concentration of its free metabolite pool. This speculation is in need for further investigation through thorough study of the plant secondary metabolism's activity under the same experimental conditions.
Figure 8.
The significance level profiles over time and the significance classification based on paired-SAM of the genes and metabolites in the aromatic amino acid biosynthesis pathways that were included in the analysis. Positively and negatively significant genes and metabolites are designated as described in the caption of Figure 5. Information about the depicted pathway's structure was obtained from the KEGG database (www.kegg.com) and Buchanan et al. (2001). (This figure can be viewed online in a color version as Supplementary Figure 3.)
Beyond the first 12 h, all amino acids, except homoserine, were identified as negatively or non-significant (Fig. 7). This indicates higher rate of the amino acid pools’ depletion for the production of macromolecules without equal increase in their production rate. As described earlier, the gene expression data indicated a decrease in the transcriptional activity of the CO2 fixation after 12 h of perturbation. In the case that the transcriptional trend is indeed translated to decreased in vivo flux of CO2 fixation, then the latter could explain the observed pattern in the metabolomic data of the amino acids. Further metabolic network analysis data about the in vivo flux of the CO2 fixation under the investigated physiological conditions is needed to validate this speculation. The need for integrated “omic” studies at multiple levels of cellular function is also indicated in the context of nitrogen assimilation. Interestingly, according only to the NR gene expressions, the expected time profile of the nitrogen-storage amino acid concentrations after 12 h of treatment would have been very different. The significant decrease in the amino acid concentrations was observed even though the expression of both NR genes had increased.
Ethylene Synthesis and Signaling/Sulfur Transport and Metabolism
Ethylene is an important plant hormone regulating the plant development and morphology (Guo and Ecker, 2004), the biotic and abiotic stress response and the expression of a large family of transcription factors (Chen et al., 2005). Despite ethylene's significance in plant physiology, no previous report studying the immediate effect of elevated CO2 stress on the ethylene biosynthesis and signaling was available. In plants, the ethylene is produced through the synthesis and oxidation of the 1-aminocyclo-propane-1-carboxylate (ACC) using the ACC synthase and the ACC oxidase enzymes, respectively. In vitro studies have indicated that the ACC oxidase is activated by CO2 (Thrower et al., 2001). The A. thaliana genome comprises multiple ACC synthase and ACC oxidase copies; two of each were finally considered in the present analysis after the normalization and filtering step. One ACC synthase and one oxidase copy were identified as positively significant at five and seven time points, respectively, and from paired SAM (Supplementary Table V). The other copies were positively significant at three and one time points, respectively, but not from paired SAM (Supplementary Table V). Thus, the transcriptional activity of the ethylene biosynthesis pathway is significantly induced in response to the elevated CO2 stress, even from the first hour of perturbation (Supplementary Table V). This transcriptional observation is consistent with previous studies of the elevated CO2 stress in higher plants, which have reported a sustained ethylene release in the photosynthetic leaves (Bassi and Spencer, 1982; Dhawan et al., 1981; Wang et al., 2002). This is, however, the first time that molecular data, at any level of cellular function, that are consistent with sustained ethylene release as consequence of the elevated CO2 stress is reported in the context of the A. thaliana physiology.
The ethylene sensing and signal transduction cascade in A. thaliana and higher plants involves a large number of genes (Chen et al., 2005; Guo and Ecker, 2004; Wang et al., 2002), whose regulation and specific role in signaling are still under research. The observed transcriptional profile of the ethylene signaling pathway in response to elevated CO2 (Fig. 9) is in agreement with the reported response of soil-grown A. thaliana plants to direct treatment with ethylene or the ethylene precursor ACC (Chen et al., 2005; Hua et al., 1998). In tomatoes, the transcription of the CTR1, the second protein of the ethylene signaling cascade, is induced in response to direct treatment with ethylene (Adams-Phillips et al., 2004). This had not been, however, previously confirmed in A. thaliana (Gao et al., 2003; Kieber et al., 1992). In the present elevated CO2 study, the gene encoding for the CTR1 synthesis was identified as positively significant at three out of the eight time points (3, 9, and 24 h). Two genes encoding for subsequent proteins in the ethylene signaling cascade, the EIN2 and the EIN3, were identified as non-significant at seven out of the eight time points (see Fig. 9). These observations agree with previous studies of the ethylene stress (Chen et al., 2005), which concluded that the EIN2 and the EIN3 are not regulated by ethylene at the transcriptional level. The gene encoding for the transcription factor ERF1, which is activated by the EIN3 (Dhawan et al., 1981), was identified as negatively significant at the 9 and 18 h of perturbation (Fig. 9). These observations complement data from an ethylene stress study in soil-grown A. thaliana plants, in which it had been observed that the expression of the ERF1 was not up-regulated under conditions of high ethylene concentration (Zhong and Burns, 2003). Thus, in addition to the significant increase in the rate of ethylene synthesis at the transcriptional level, it is also the similarity between the ethylene signaling transcriptional response to elevated CO2 stress and to direct ethylene treatment that confirms the significant production of ethylene in response to elevated CO2 stress. These observations contribute to the current knowledge about the transcriptional regulation of ethylene signaling in A. thaliana and plants in general; there was only one available transcriptomic dataset of mature A. thaliana seedlings’ response to direct ethylene treatment (Zhong and Burns, 2003). Finally, among the 21 analyzed ethylene responsive element binding (EREBP) transcription factor family genes, which are regulated by the EIN3/ERF1, two were identified as positively significant, while three as negatively significant from paired-SAM (see Supplementary Table V). These observed contradictions are one additional indication of the plethora of open questions that need to be further pursued towards the full elucidation of the ethylene signaling pathway in plants.
Figure 9.
The significance level profiles over time and the significance classification based on paired-SAM of the genes and metabolites in the ethylene biosynthesis and signaling pathways that were included in the analysis. Positively and negatively significant genes and metabolites are designated as described in the caption of Figure 5. Information about the depicted pathway's structure was obtained from the KEGG database (www.kegg.com), Chen et al. (2005) and Buchanan et al. (2001). (This figure can be viewed online in a color version as Supplementary Figure 4.)
The current knowledge about the effect of elevated CO2 stress on the sulfur transport and metabolism in plants in general and A. thaliana in particular is not extensive. The acquired omic dataset contributes to the available database. The observed significance level profiles over time of the genes related to sulfur transport and metabolism are shown in Supplementary Table V.
Acknowledgments
We would like to gratefully acknowledge the financial support of the US NSF (QSB-0331312), the UMD Minta Martin Foundation and the UMD Chemical and Biomolecular Engineering Department. We would also like to thank Fenglong Liu (TIGR) for helpful discussions on the transcriptomic analysis experimental protocol; the UMD Green House facility for providing access to the growth chambers; the plant physiologist Dr. Tara Vantoai (Ohio State University) for helpful suggestions on the selection of the plant liquid culture media; and Ms. Linda Moy and Lara Linford for technical assistance with the micro-array hybridizations.
Contract grant sponsor: US NSF
Contract grant number: QSB-0331312
Contract grant sponsor: UMD Minta Martin Foundation
Contract grant sponsor: UMD Chemical and Biomolecular Engineering Department
Appendix A
GC–MS Metabolomic Data Correction and Filtering
The raw GC–MS metabolomic dataset (Supplementary Table I) comprised 612 (158 annotated) peaks, each of which was identified in at least one of the acquired meta-bolomic profiles. Among these, a total of 295 (89 annotated) were finally used in the analysis after the described in Materials and Methods Section correction, normalization and filtering. Among the 89 annotated, 61, 9, and 19, respectively, correspond to metabolites forming only one TMS-derivative (according to Kanani and Klapa [2007]) in the text they may be referred to as category-1 metabolites), to one of the two geometric isomer derivatives of ketone-group containing metabolites (according to Kanani and Klapa [2007]) in the text they may be referred to as category-2 metabolites), and to the cumulative peak areas (see relevant methodology in Kanani and Klapa [2007]) of amine-group containing metabolites (according to Kanani and Klapa [2007] in the text they may be referred to as category-3 metabolites). The average coefficient of peak area variation between injections of the same sample and between biological replicates was 6% and 17%, respectively.
It needs to be underlined that if the new GC–MS metabolomic data correction and normalization algorithm presented in Kanani and Klapa (2007) had not been used, paired-SAM analysis would have had falsely identified 11 (two annotated) additional peak areas as negatively significant (Fig. 10). This result reinforces the validity of the new algorithm and provides additional evidence in the context of a large dataset that supports the need for correcting the metabolomic profiles from the derivatization biases (Kanani and Klapa, 2007; Kanani et al., in press).
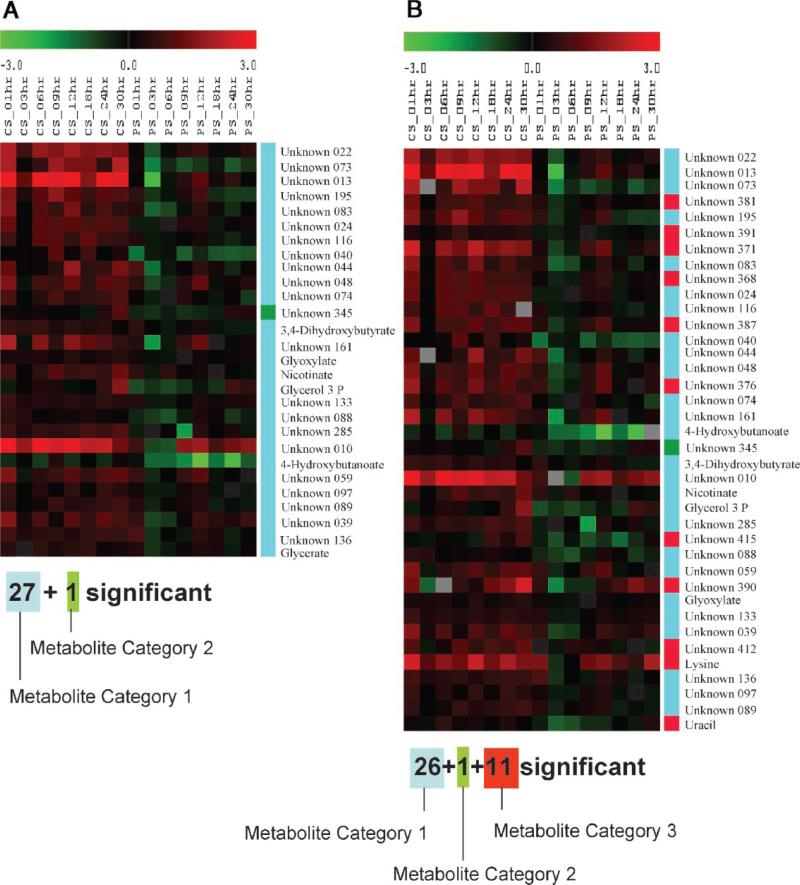
Figure 10.
The metabolite peak areas which were identified as significant by paired-SAM (0% FDR (median)) when the data correction strategy described in Kanani and Klapa (2007); (A) was, and (B) was not used. Each row of the depicted A and B tables corresponds to a metabolite identified as negatively significant in case A and B, respectively. Eleven metabolite peak areas (two annotated) would have been falsely identified as significant in case B. The color of each element of the table is proportional to the normalized with respect to 0 h relative peak area of the corresponding metabolite (row) in the sample that corresponds to the particular column (see Materials and Methods Section). CS, PS depict, respectively, the control and perturbed samples. The unknown metabolites are named based on their numbering in supplementary Table I. The tables were generated in TIGR MeV (v.3.1). Definition of metabolite category 1, 2, and 3 is provided in Appendix A.
Footnotes
Additional Supporting Information may be found in the online version of this article.
References
- Adams-Phillips L, Barry C, Kannan P, Leclercq J, Bouzayen M, Giovannoni J. Evidence that CTR1-mediated ethylene signal transduction in tomato is encoded by a multigene family whose members display distinct regulatory features. Plant Mol Biol. 2004;54:387–404. doi: 10.1023/B:PLAN.0000036371.30528.26. Arabidopsis functional genomics at TIGR http://atarrays.tigr.org/ [DOI] [PubMed] [Google Scholar]
- Aravind L, Koonin EV. The DNA-repair protein AlkB, EGL-9, and leprecan define new families of 2-oxoglutarate- and iron-dependent dioxygenases. Genome Biol. 2001;2(3):RESEARCH0007. doi: 10.1186/gb-2001-2-3-research0007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassi PK, Spencer MS. Effect of carbon dioxide and light on ethylene production in intact sunflower plants. Plant Physiol. 1982;69:1222–1225. doi: 10.1104/pp.69.5.1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyes DC, Zayed AM, Ascenzi R, McCaskill AJ, Hoffman NE, Davis KR, Gorlach J. Growth stage-based phenotypic analysis of Arabidopsis: A model for high throughput functional genomics in plants. Plant Cell. 2001;13:1499–1510. doi: 10.1105/TPC.010011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchanan B, Gruissem W, Jones R. Biochemistry & molecular biology of plants. American Society of Plant Physiologists; Rockville: 2001. [Google Scholar]
- Chen YF, Etheridge N, Schaller GE. Ethylene signal transduction. Ann Bot (Lond) 2005;95:901–915. doi: 10.1093/aob/mci100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Wang QY, Cheng XG, Xu ZS, Li LC, Ye XG, Xia LQ, Ma YZ. GmDREB2, a soybean DRE-binding transcription factor, conferred drought and high-salt tolerance in transgenic plants. Biochem Biophys Res Commun. 2007;353:299–309. doi: 10.1016/j.bbrc.2006.12.027. [DOI] [PubMed] [Google Scholar]
- Cheng CL, Acedo GN, Dewdney J, Goodman HM, Conkling MA. Differential expression of the two arabidopsis nitrate reductase genes. Plant Physiol. 1991;96:275–279. doi: 10.1104/pp.96.1.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng CL, Acedo GN, Cristinsin M, Conkling MA. Sucrose mimics the light induction of Arabidopsis nitrate reductase gene transcription. Proc Natl Acad Sci. 1992;89:1861–1864. doi: 10.1073/pnas.89.5.1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke A, Mur LA, Darby RM, Kenton P. Harpin modulates the accumulation of salicylic acid by Arabidopsis cells via apoplastic alkalization. J Exp Bot. 2005;56:3129–3136. doi: 10.1093/jxb/eri310. [DOI] [PubMed] [Google Scholar]
- Corruzi J, Last R. Amino Acids. In: Buchanan B, Gruissem W, Jones R, editors. Biochemistry & molecular biology of plants. American Society of Plant Physiologists; Rockville: 2001. pp. 358–411. [Google Scholar]
- Coschigano KT, Melo-Oliveira R, Lim J, Coruzzi GM. Arabidopsis gls mutants and distinct Fd-GOGAT genes: Implications for photorespiration and primary nitrogen assimilation. Plant Cell. 1998;10:741–752. doi: 10.1105/tpc.10.5.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cournac L, Dimon B, Carrier P, Lohou A, Chagvardieff P. Growth and photosynthetic characteristics of solanum tuberosum plantlets cultivated in vitro in different conditions of aeration, sucrose supply, and CO2 enrichment. Plant Physiol. 1991;97(1):112–117. doi: 10.1104/pp.97.1.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denis DT, Blakely SD. Carbohydrate metabolism. In: Buchanan B, Gruissem W, Jones R, editors. Biochemistry & molecular biology of plants. American Society of Plant Physiologists; Rockville: 2001. pp. 630–675. [Google Scholar]
- Dey P, Harborne J. Plant biochemistry. Academic Press; San Diego: 1997. [Google Scholar]
- Dhawan KR, Bassi PK, Spencer MS. Effects of carbon dioxide on ethylene production and action in intact sunflower plants. Plant Physiol. 1981;68:831–834. doi: 10.1104/pp.68.4.831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutta B, Snyder R, Klapa MI. Significance analysis of time-series transcriptomic data: A methodology that enables the identification and further exploration of the differentially expressed genes at each time-point. Biotechnol Bioeng. 2007;98:668–678. doi: 10.1002/bit.21432. [DOI] [PubMed] [Google Scholar]
- Fiehn O, Kopka J, Dormann P, Altmann T, Trethewey RN, Willmitzer L. Metabolite profiling for plant functional genomics. Nat Biotechnol. 2000;18:1157–1161. doi: 10.1038/81137. [DOI] [PubMed] [Google Scholar]
- Gamborg OL, Murashige T, Thorpe TA, Vasil IK. Plant tissue culture media. In Vitro. 1976;12:473–478. doi: 10.1007/BF02796489. [DOI] [PubMed] [Google Scholar]
- Gao Z, Chen YF, Randlett MD, Zhao XC, Findell JL, Kieber JJ, Schaller GE. Localization of the Raf-like kinase CTR1 to the endoplasmic reticulum of Arabidopsis through participation in ethylene receptor signaling complexes. J Biol Chem. 2003;278:34725–34732. doi: 10.1074/jbc.M305548200. [DOI] [PubMed] [Google Scholar]
- Guo H, Ecker JR. The ethylene signaling pathway: New insights. Curr Opin Plant Biol. 2004;7:40–49. doi: 10.1016/j.pbi.2003.11.011. [DOI] [PubMed] [Google Scholar]
- Hétu MF, Tremblay LJ, Lefebvre DD. High root biomass production in anchored Arabidopsis plants grown in axenic sucrose supplemented liquid culture. Biotechniques. 2005;39:245–249. doi: 10.2144/05393ST02. [DOI] [PubMed] [Google Scholar]
- Hua J, Sakai H, Nourizadeh S, Chen QG, Bleecker AB, Ecker JR, Meyerowitz EM. Ein4 and ERS2 are members of the putative ethylene receptor family in Arabidopsis. Plant Cell. 1998;10:1321–1332. doi: 10.1105/tpc.10.8.1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang D, Rust AG, Ramsey S, Smith JJ, Leslie DM, Weston AD, de Atauri P, Aitchison JD, Hood L, Siegel AF, Bolouri H. A data integration methodology for systems biology. Proc Natl Acad Sci. 2005a;102:17296–17301. doi: 10.1073/pnas.0508647102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang D, Smith JJ, Leslie DM, Weston AD, Rust AG, Ramsey S, de Atauri P, Siegel AF, Bolouri H, Aitchison JD, Hood L. A data integration methodology for systems biology: Experimental verification. Proc Natl Acad Sci. 2005b;102:17302–17307. doi: 10.1073/pnas.0508649102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ideker T, Thorsson V, Ranish JA, Christmas R, Buhler J, Eng JK, Bumgarner R, Goodlett DR, Aebersold R, Hood L. Integrated genomic and proteomic analyses of a systematically perturbed metabolic network. Science. 2001;292:929–934. doi: 10.1126/science.292.5518.929. [DOI] [PubMed] [Google Scholar]
- Kanani H, Klapa MI. Data correction strategy for metabolomics analysis using gas chromatography-mass spectrometry. Metabolic Eng. 2007;9:39–51. doi: 10.1016/j.ymben.2006.08.001. KEGG. Kyoto Encyclopedia of Genes and Genomes [ www.kegg.com] [DOI] [PubMed] [Google Scholar]
- Kanani H, Chrysanthopoulos PK, Klapa MI. Standardizing GCMS metabolomics. J Chromatogr B. In Press DOI: 10.1016/j.jchromb.2008.04.049; Available online May 21, 2008. [Google Scholar]
- Kieber JJ, Rothenberg M, Roman G, Feldmann KA, Ecker JR. CTR1, a negative regulator of the ethylene response pathway in Arabidopsis, encodes a member of the Raf family of protein kinases. Cell. 1992;72:427–441. doi: 10.1016/0092-8674(93)90119-b. [DOI] [PubMed] [Google Scholar]
- Kim H, Snesrud EC, Haas B, Cheung F, Town CD, Quackenbush J. Gene expression analyses of Arabidopsis chromosome 2 using a genomic DNA amplicon microarray. Genome Res. 2003;13:327–340. doi: 10.1101/gr.552003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klapa M, Quackenbush J. The quest for the mechanisms of life. Biotechnol Bioeng. 2003;84:739–742. doi: 10.1002/bit.10858. [DOI] [PubMed] [Google Scholar]
- Klapa MI, Aon JC, Stephanopoulos G. Systematic quantification of complex metabolic flux networks using stable isotopes and mass spectrometry. Eur J Biochem. 2003;270:3525–3542. doi: 10.1046/j.1432-1033.2003.03732.x. [DOI] [PubMed] [Google Scholar]
- Kursteiner O, Dupuis I, Kuhlemeier C. The pyruvate decarboxylase1 gene of Arabidopsis is required during anoxia but not other environmental stresses. Plant Physiol. 2003;132:968–978. doi: 10.1104/pp.102.016907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larios B, Aguera E, Haba P, Perez-Vicente R, Maldonado JM. A short-term exposure of cucumber plants to rising atmospheric CO2 increases leaf carbohydrate content and enhances nitrate reductase expression and activity. Planta. 2001;212:305–312. doi: 10.1007/s004250000395. [DOI] [PubMed] [Google Scholar]
- Lide DR, Frederikse HPR, editors. CRC handbook of chemistry and physics. 76th edition CRC Press, Inc.; Boca Raton, FL: 1995. [Google Scholar]
- Liu F, Vantoai T, Moy LP, Bock G, Linford LD, Quackenbush J. Global transcription profiling reveals comprehensive insights into hypoxic response in Arabidopsis. Plant Physiol. 2005;137(3):1115–1129. doi: 10.1104/pp.104.055475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makino A, Nakano H, Mae T, Shimada T, Yamamoto N. Photo-synthesis, plant growth and N allocation in transgenic rice plants with decreased Rubisco under CO2 enrichment. J Exp Bot. 2000;51:383–389. doi: 10.1093/jexbot/51.suppl_1.383. [DOI] [PubMed] [Google Scholar]
- Middleman S. An introduction to mass and heat transfer. John Wiley & Sons, Inc.; New York, NY: 1998. [Google Scholar]
- Misson J, Raghothama KG, Jain A, Jouhet J, Block MA, Bligny R, Ortet P, Creff A, Somerville S, Rolland N, Doumas P, Nacry P, Herrerra-Estrella L, Nussaume L, Thibaud MC. A genome-wide transcriptional analysis using Arabidopsis thaliana Affymetrix gene chips determined plant responses to phosphate deprivation. Proc Natl Acad Sci. 2005;102(33):11934–11939. doi: 10.1073/pnas.0505266102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oksman-Caldentey KM, Inze D. Plant cell factories in the post-genomic era. New ways to produce designer secondary metabolites. Trends Plant Sci. 2004;9:433–440. doi: 10.1016/j.tplants.2004.07.006. [DOI] [PubMed] [Google Scholar]
- Osuna D, Usadel B, Morcuende R, Gibon Y, Bläsing OE, Höhne M, Günter M, Kamlage B, Trethewey R, Scheible WR, Stitt M. Temporal responses of transcripts, enzyme activities and metabolites after adding sucrose to carbon-deprived Arabidopsis seedlings. Plant J. 2007;49(3):463–491. doi: 10.1111/j.1365-313X.2006.02979.x. [DOI] [PubMed] [Google Scholar]
- Ragauskas AJ, Williams CK, Davison BH, Britovsek G, Cairney J, Eckert CA, Frederick WJ, Jr., Hallett JP, Leak DJ, Liotta CL, Mielenz JR, Murphy R, Templer R, Tschaplinski T. The path forward for biofuels and biomaterials. Science. 2006;311:484–489. doi: 10.1126/science.1114736. [DOI] [PubMed] [Google Scholar]
- Roessner U, Wagner C, Kopka J, Trethewey RN, Willmitzer L. Technical advance: Simultaneous analysis of metabolites in potato tuber by gas chromatography-mass spectrometry. Plant J. 2000;23:131–142. doi: 10.1046/j.1365-313x.2000.00774.x. [DOI] [PubMed] [Google Scholar]
- Saeed AI, Sharov V, White J, Li J, Liang W, Bhagabati N, Braisted J, Klapa M, Currier T, Thiagarajan M, Sturn A, Snuffin M, Rezantsev A, Popov D, Ryltsov A, Kostukovich E, Borisovsky I, Liu Z, Vinsavich A, Trush V, Quackenbush J. TM4: A free, open-source system for microarray data management and analysis. Biotechniques. 2003;34:374–378. doi: 10.2144/03342mt01. [DOI] [PubMed] [Google Scholar]
- Sakuma Y, Liu Q, Dubouzet JG, Abe H, Shinozaki K, Yamaguchi-Shinozaki K. DNA-binding specificity of the ERF/AP2 domain of Arabidopsis DREBs, transcription factors involved in dehydrationand cold-inducible gene expression. Biochem Biophys Res Commun. 2002;290:998–1009. doi: 10.1006/bbrc.2001.6299. [DOI] [PubMed] [Google Scholar]
- Scheible WR, Morcuende R, Czechowski T, Fritz C, Osuna D, Palacios-Rojas N, Schindelasch D, Thimm O, Udvardi MK, Stitt M. Genome-wide reprogramming of primary and secondary metabolism, protein synthesis, cellular growth processes, and the regulatory infrastructure of Arabidopsis in response to nitrogen. Plant Physiol. 2004;136:2483–2499. doi: 10.1104/pp.104.047019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki M, Narusaka M, Ishida J, Nanjo T, Fujita M, Oono Y, Kamiya A, Nakajima M, Enju A, Sakurai T, Satou M, Akiyama K, Taji T, Yamaguchi-Shinozaki K, Carninci P, Kawai J, Hayashizaki Y, Shinozaki K. Monitoring the expression profiles of 7000 Arabidopsis genes under drought, cold and high-salinity stresses using a full-length cDNA microarray. Plant J. 2002;31:279–292. doi: 10.1046/j.1365-313x.2002.01359.x. [DOI] [PubMed] [Google Scholar]
- Siedow J, Day DA. Respiration and photorespiration. In: Buchanan B, Gruissem W, Jones R, editors. Biochemistry & molecular biology of plants. American Society of Plant Physiologists; Rockville: 2001. pp. 676–727. [Google Scholar]
- Slater S, Mitsky TA, Houmiel KL, Hao M, Reiser SE, Taylor NB, Tran M, Valentin HE, Rodriguez DJ, Stone DA, Padgette SR, Kishore G, Gruys KJ. Metabolic engineering of Arabidopsis and Brassica for poly(3-hydroxybutyrate-co-3-hydroxyvalerate) copolymer production. Nat Biotechnol. 1999;17:1011–1016. doi: 10.1038/13711. [DOI] [PubMed] [Google Scholar]
- Smith CJ. Carbohydrate chemistry. In: Lea PJ, Leegood RC, editors. Plant biochemistry and molecular biology. Vol. 73. John Wiley and Sons; West Sussex, England: 1993. p. 113. [Google Scholar]
- Somerville C, Dangl JL. Genomics. Plant biology in 2010. Science. 2000;290:2077–2078. doi: 10.1126/science.290.5499.2077. [DOI] [PubMed] [Google Scholar]
- Stephanopoulous G, Aristidou AA, Nielsen J. Metabolic engineering: Principals and methodologies. Academic Press; San Diego: 1998. [Google Scholar]
- Stitt M. Rising CO2 levels and their potential significance for carbon flow in photosynthetic cells. Plant Cell Environ. 1991;14:741–762. [Google Scholar]
- Tadege M, Dupuis II, Kuhlemeier C. Ethanolic fermentation: New functions for an old pathway. Trends Plant Sci. 1999;4:320–325. doi: 10.1016/s1360-1385(99)01450-8. [DOI] [PubMed] [Google Scholar]
- Thrower JS, Blalock R, 3rd, Klinman JP. Steady-state kinetics of substrate binding and iron release in tomato ACC oxidase. Biochemistry. 2001;40:9717–9724. doi: 10.1021/bi010329c. [DOI] [PubMed] [Google Scholar]
- Toikkanen JH, Niku-Paavola ML, Bailey M, Immanen J, Rintala E, Elomaa P, Helariutta Y, Teeri TH, Fagerström R. Expression of xyloglucan endotransglycosylases of Gerbera hybrida and Betula pendula in Pichia pastoris. J Biotechnol. 2007;130:161–170. doi: 10.1016/j.jbiotec.2007.03.004. [DOI] [PubMed] [Google Scholar]
- Troyanskaya O, Cantor M, Sherlock G, Brown P, Hastie T, Tibshirani R, Botstein D, Altman RB. Missing value estimation methods for DNA microarrays. Bioinformatics. 2001;17:520–525. doi: 10.1093/bioinformatics/17.6.520. [DOI] [PubMed] [Google Scholar]
- Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang KL, Li H, Ecker JR. Ethylene biosynthesis and signaling networks. Plant Cell. 2002;14:S131–S151. doi: 10.1105/tpc.001768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Tischner R, Gutierrez RA, Hoffman M, Xing X, Chen M, Coruzzi G, Crawford NM. Genomic analysis of the nitrate response using a nitrate reductase-null mutant of Arabidopsis. Plant Physiol. 2004;136(1):2512–2522. doi: 10.1104/pp.104.044610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong GV, Burns JK. Profiling ethylene-regulated gene expression in Arabidopsis thaliana by microarray analysis. Plant Mol Biol. 2003;53:117–131. doi: 10.1023/b:plan.0000009270.81977.ef. [DOI] [PubMed] [Google Scholar]