Abstract
During cap-dependent eukaryotic translation initiation, ribosomes scan mRNA from the 5′ end to the first AUG start codon with favorable sequence context1,2. For many mRNAs this AUG belongs to a short upstream open reading frame (uORF)3, and translation of the main downstream ORF requires reinitiation, an incompletely understood process1,4-6. Reinitiation is thought to involve the same factors as standard initiation1,5,7. It is unknown if any factors specifically affect translation reinitiation without affecting standard cap-dependent translation. We uncover here the non-canonical initiation factors Density Regulated Protein (DENR) and Multiple Copies in T-cell Lymphoma-1 (MCT-1) as the first selective regulators of eukaryotic reinitiation. mRNAs containing upstream Open Reading Frames with strong Kozak sequences (stuORFs) selectively require DENR•MCT-1 for their proper translation, yielding a novel class of mRNAs that can be co-regulated and that is enriched for regulatory proteins such as oncogenic kinases. Collectively, our data reveal that cells have a previously unappreciated translational control system with a key role in supporting proliferation and tissue growth.
Keywords: translational control, uORF, reinitiation, coordinated regulation, Drosophila, development, tissue growth, cell proliferation
Cellular protein abundance depends mainly on mRNA translation8. Little is known about how translation of specific sets of mRNAs can be coordinately regulated9,10. mRNAs with uORFs require reinitiation11-14, whereby ribosomes translate the uORF, terminate, and then restart translating the main ORF1,4,6,15. No metazoan trans-acting factors have yet been described that selectively affect reinitiation, enabling coordinate regulation of uORF mRNAs. Ligatin/eIF2D and the related DENR•MCT-1 complex are candidate reinitiation regulators. They associate with 40S ribosomal subunits and have domains implicated in RNA-binding and start codon recognition (Extended Data Fig. 1a). In vitro they can recycle post-termination complexes, recruit Met-tRNAiMet to mRNAs containing viral Internal Ribosome Entry Sites16,17, and affect movement of post-termination 80S complexes to nearby AUG codons6. DENR•MCT-1 was not previously implicated in reinitiation, MCT-1 is an oncogene affecting cellular mRNA translation by an unclear mechanism18-20. Collectively, these studies suggest DENR•MCT-1 and Ligatin might regulate translation of cancer relevant mRNAs through non-canonical mechanisms.
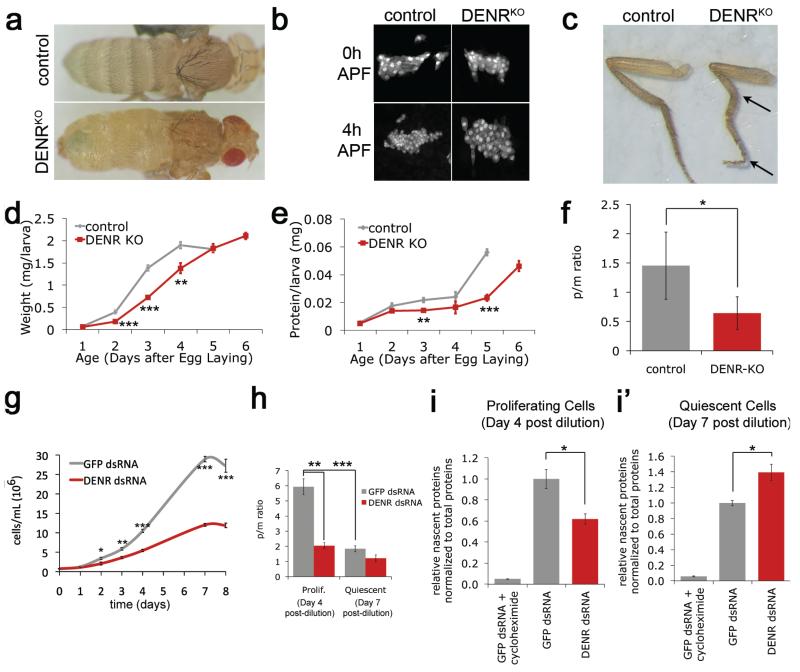
To study DENR function, we generated DENR/CG9099 knockout Drosophila lacking transcript or protein (DENRKO, ED Fig. 1b-d). DENRKO die as pharate adults with a larval-like epidermis (Fig. 1a), due to impaired proliferation of histoblast cells (Fig. 1b, ED Fig. 1e). This is rescued by expressing DENR ubiquitously (Tubulin-Gal4), or specifically in histoblast cells (escargot-GAL4) (X2–test p<0.05, ED Fig. 1f). While DENR is expressed ubiquitously (ED Fig. 1g), quickly proliferating histoblast cells appear more sensitive to DENR loss than non-proliferating tissues. DENRKO also have crooked legs and incorrectly rotated genitals (Fig. 1c, ED Fig. 1h-h’). These phenotypes are not observed in mutants with generally impaired translation (Minutes21), but are found in flies with reduced cell cycle regulators or Ecdysone Receptor signaling22,23, suggesting that DENR affects translation of a subset of mRNAs involved in cell proliferation and signaling.
Figure 1. DENR promotes cell proliferation and boosts protein synthesis in proliferating but not quiescent cells.
(a) DENRKO die as pharate adults with larval-like abdominal epidermis. (b) DENRKO anterior-dorsal histoblast nests have correct cell numbers at onset of pupation (0 h APF), but impaired proliferation the first 4 h of pupal development (4 h APF) (visualized by esgG4>GFP). (c) DENRKO have crooked legs. (d-e) DENRKO accrue mass (d) and protein (e) slowly, pupating with 1 day delay (day 6 datapoint, absent in controls). (f) Polysome profiles from DENRKO larvae have reduced polysome/monosome ratios. (g) DENR knockdown (DENR-KD) cells proliferate slowly and enter quiescence at low cell density. (h) Proliferating but not quiescent polysome profiles of DENR-KD cells have reduced p/m ratios. (i-i’) Proliferating (i) but not quiescent (i’) DENR-KD cells have reduced de novo protein synthesis rates, quantified by metabolic labeling with methionine analog. Error bars: Std dev (d-f) or SEM (g-I’). T-test (d-f) or Mann-Whitney U-Test (g-i’) *<0.05, **<0.01, ***<0.001.
Similar phenotypes were observed in flies expressing RNAi targeting MCT-1/CG5941 (ED Fig. 1i), which like human MCT-1 binds DENR (ED Fig. 1j). Reducing Ligatin gene dosage in DENRKO flies caused fewer animals to reach pupation (X2–test p<0.05, ED Fig. 1k) indicating that DENRKO phenotypes result from loss of DENR•MCT-1 with Ligatin-like activity.
DENRKO larvae and DENR knockdown (DENR-KD) S2 cells grow slowly with reduced protein accumulation rates (Fig. 1d-e, g). Mutant polysome profiles show reduced polysome/monosome (p/m) ratios (Fig. 1f, h, ED Fig. 1l-n’), suggesting defective translation initiation. Despite more ribosomes and initiator tRNA per cell (ED Fig. 1o-p), DENR-KD cells have reduced protein synthesis rates when proliferating (Fig. 1i). When quiescent, DENR-KD cells no longer display these phenotypes, and become enlarged compared to controls (Fig. 1h, i’, ED Fig. 1q-r). Thus DENR promotes translation of cellular mRNAs in proliferating but not quiescent cells.
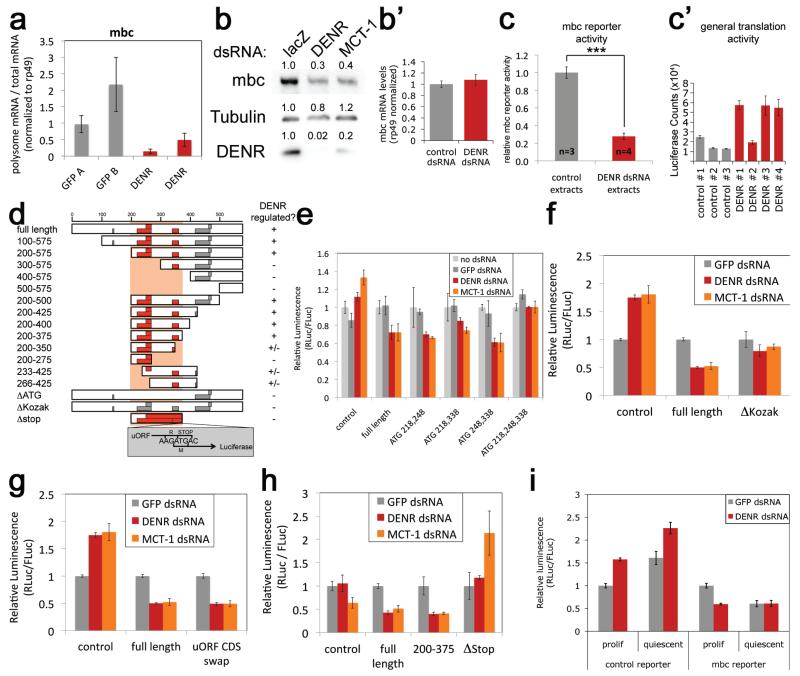
We identified ~100 mRNAs requiring DENR for efficient translation by profiling actively translated mRNAs from 80S and polysome fractions of control and DENR-KD cells and normalizing to total mRNA (Suppl. Table 1). We further analyzed myoblast city (mbc) because it was the second most under-translated mRNA and we could obtain antibody to detect it. Quantitative RT-PCR confirmed that mbc mRNA is under-represented in polysomes of DENR-KD cells (Fig. 2a), leading to reduced mbc protein but not mRNA (Fig. 2b-b’), whereas other proteins were not reduced (ED Fig. 2a). The mbc 5′UTR was sufficient to impart DENR-dependence to an RLuc reporter (ED Fig. 2b). This DENR-dependence requires the 5′ cap and is not accompanied by a drop in general translation (ED Fig. 2c-d’, 2c-c’). Combined knockdown of DENR and MCT-1 had no additive effect, as they are a functional complex (ED Fig. 2e). In sum, the DENR•MCT-1 complex selectively promotes cap-dependent translation of mbc via its 5′UTR.
Figure 2. DENR promotes reinitiation of translation downstream of uORFs in the mbc 5′UTR.
(a) qRT-PCR validation that the mbc mRNA is preferentially depleted from polysomes in DENR-KD cells (DENR A and B) compared to controls (GFP A and B). (b-b’) mbc protein (b) but not mRNA (b’) levels are reduced in DENR and MCT-1 knockdown cells. (c-c’) Translation extracts from DENR-KD cells are impaired in translating a reporter containing the mbc 5′UTR (c) but not in translating a control RLuc reporter mRNA without uORFs in the 5′UTR (c’). (d) Schematic overview of the mbc 5′UTR and the tested DNA reporter constructs, summarizing results from other panels as well as multiple (≥3) additional replicates on all the luciferase assays, not shown. Details in ED Fig. 3. (e-f) Mutating the start codons of the three mbc uORFs with strong Kozak sequences (e) or their Kozak sequences to the less functional gtgtATG (f) blunts regulation by DENR. (g) Mutating the coding sequence of mbc uORFs to poly-glutamine has no effect on DENR regulation. (h) Mutation of the stop codons of mbc uORFs 218, 248 and 338, as diagrammed in (d), causing the uORFs to extend past the RLuc ATG, leads to loss of DENR-dependent regulation. (i) DENR knockdown leads to impaired expression of the mbc 5′UTR RLuc reporter in proliferating but not quiescent S2 cells. Error bars: std dev. *t-test<0.05, ***t-test<0.001
Systematic 5′UTR truncations (ED Fig. 2f-h) identified 175-nt necessary and sufficient for DENR-dependence (Fig. 2d, ED Fig. 3a-b), containing 3 uORFs with strong Kozak sequences (stuORFs, red boxes Fig. 2d). Mutating all three stuORF ATGs, or their Kozak sequences, abolished DENR-dependence (Fig. 2e-f, ED Fig. 3c), indicating translation initiation on these stuORFs is necessary for DENR-dependence. No additional cis-acting sequences were necessary; removing sequences upstream, downstream, or between the uORFs, or mutating the uORF coding sequences, did not affect DENR-dependence (Fig 2g, ED Fig. 2f-h). Two possible explanations are: 1) DENR promotes bypass of stuORF initiation codons; 2) DENR affects reinitiation after stuORF translation. In model 1, the stuORF stop codon is irrelevant because DENR would act at the stuORF start codon. In model 2 the stuORF stop codon is crucial since translation reinitiation on the main ORF only occurs after termination on the stuORF. Two point mutations removing the stuORF stop codons completely abolished DENR-dependence (Fig. 2h, ED Fig. 3d), indicating that DENR promotes translation reinitiation.
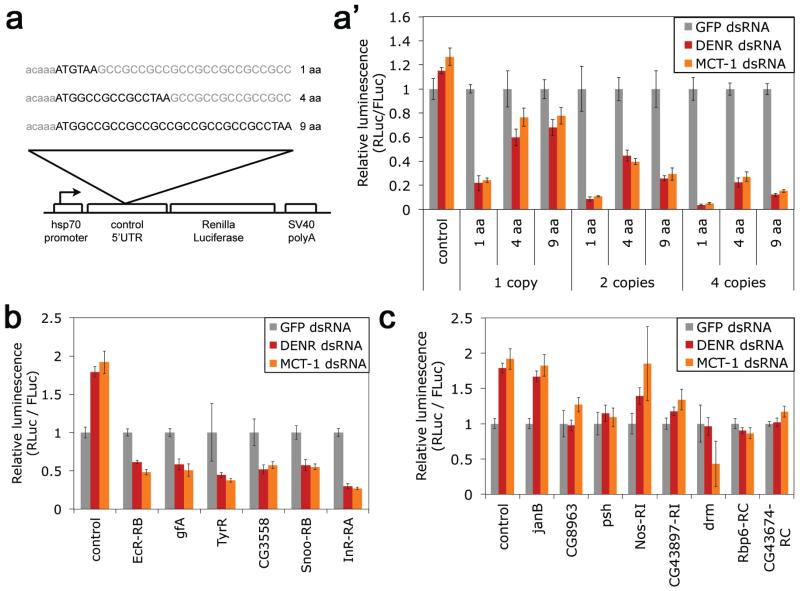
Introducing synthetic stuORFs into a control reporter was sufficient to impart DENR-dependence, with multiple stuORFs acting additively (Fig. 3a-a’). Reinitiation efficiency is reportedly inversely related to uORF length, presumably because initiation factors dissociate from ribosomes as elongation proceeds7. Consistently, the ability of DENR to promote reinitiation dropped as uORFs became longer (Fig. 3a’), reaching zero effect on a dicistronic transcript containing a long upstream ORF (not shown). In sum, mRNAs display a continuum of DENR-dependence, depending on the number and length of the uORFs and the strength of their Kozak sequences. A computational search revealed thousands of 5′UTRs containing uORFs (ED Fig. 4a). We generated a predicted “DENR-dependence score” for all transcripts based on the number of uORFs they contain and the strength of their Kozak sequences (ED Fig. 4b-b’, Suppl. Table 2). Transcripts with high “DENR-dependence scores” were significantly enriched amongst the mRNAs with reduced translation upon DENR-KD (ED Fig. 4c-d), suggesting a general mechanism. We tested 10 5′UTRs predicted to be DENR-dependent using luciferase assays. Six conferred DENR-dependence (Fig. 3b), and four inhibited reporter translation too strongly to test experimentally. Conversely, 16 5′UTRs without uORFs were not DENR-dependent (Fig. 3c and not shown). Therefore, 5′UTRs with ‘stuORFs’ are DENR-dependent, identifying a new class of transcripts whose translation can be co-regulated. Gene Ontology analysis24 revealed that these genes are enriched for transcriptional regulators and kinases (ED Fig. 4e-f).
Figure 3. uORFs with strong Kozak sequences (stuORFs) are sufficient to impart DENR-dependent regulation.
(a-a’) Introduction of synthetic uORFs bearing a ‘strong’ Kozak into a control 5′UTR imparts DENR-dependent regulation (DNA reporters). (b-c) 5′UTRs bearing stuORFs are all DENR-dependent (b) whereas 5′UTRs lacking uORFs (c) are not. (DNA reporters). Error bars: std. dev.
Immunoprecipitation of DENR showed that it binds mRNAs containing or lacking stuORFs (ED Fig. 4g), suggesting it interacts generally with initiating ribosomes, but is required on stuORF-containing mRNAs. Since only 15% of genes contain stuORFs, we were surprised to see global effects on polysomes upon DENR-KD (ED Fig. 1l). A DENR-KD timecourse revealed that stuORF-dependent translation drops prior to changes in polysome or ribosome levels (ED Fig. 5), indicating that these are likely secondary consequences.
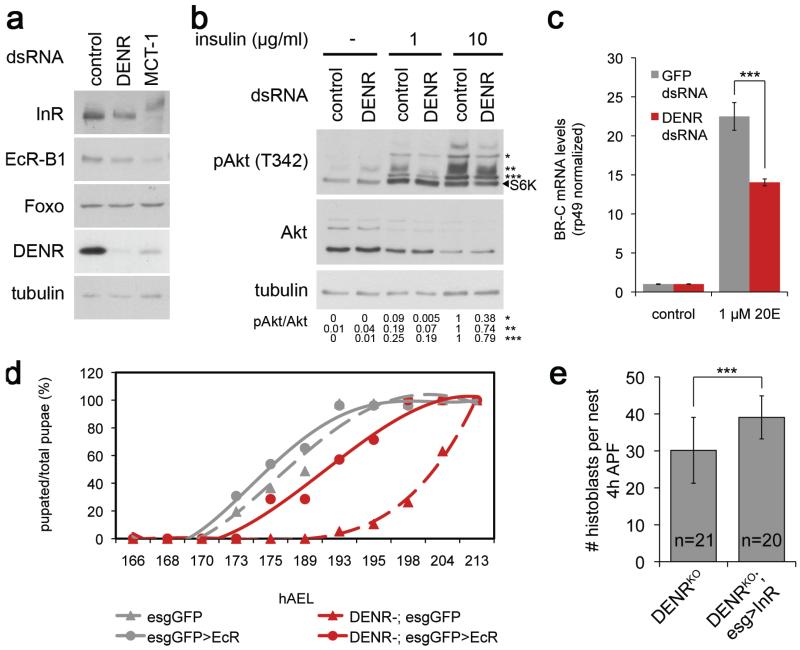
Since Insulin and Ecdysone receptors (InR and EcR) contain DENR-dependent 5′UTRs (Fig. 3b), we asked if impaired InR and EcR translation contribute to DENRKO phenotypes. Loss of DENR•MCT-1 function in S2 cells or DENRKO animals leads to reduced InR and EcR proteins, but not mRNA levels, and reduced InR and EcR signaling (Fig. 4a-c, ED Fig. 6a-c’). Reconstituting InR/EcR expression in DENRKO animals partially but significantly rescued developmental rate and histoblast proliferation (Fig. 4d-e, ED Fig. 6d). Thus, loss of DENR•MCT-1 causes reduced InR/EcR translation and signaling, and consequently impaired cell proliferation and organismal development.
Figure 4. Loss of DENR leads to reduced InR and EcR protein levels and signaling.
(a) DENR and MCT-1 knockdown cells have reduced InR and EcR protein levels. (b-c) DENR-KD cells are less sensitive to insulin stimulation (1 h) (b) and to ecdysone (1 μM, 4 h; c). (d-e) Expression of EcR (d) or InR (e) in histoblast cells and imaginal discs of DENRKO using escargot-GAL4 rescues their delayed pupation (d) and mildly but significantly their proliferation defect (e). Error bars: std dev. ***t-test<0.001
Data from DENRKO flies and DENR-KD cells suggested that proliferating cells are phenotypically more sensitive to DENR loss-of-function than quiescent cells. One explanation could be that DENR activity is low in quiescent cells, hence its removal has little effect. Using mbc and stuORF reporters, and endogenous mbc translation, as readouts for DENR activity revealed that DENR loss had a larger impact in proliferating compared to quiescent cells (Fig. 2i, ED Fig. 7). Hence DENR•MCT-1 present in quiescent cells is not very active.
To study DENR function in vivo, we generated flies carrying fluorescent reporters with or without a stuORF (ED Fig. 8a). These reporters have identical promoters, 5′UTRs and 3′UTRs, and are integrated in exactly the same genomic locus via phiC31-mediated recombination, ensuring their identical transcription. This revealed that DENR promotes stuORF reporter, but not control reporter, expression in animals (ED Fig. 8). Since stuORF-GFP reporter expression is entirely DENR-dependent, it serves as an in vivo DENR activity readout. Interestingly, the larval anterior, which contains proliferating tissues like brain and imaginal discs, shows stronger DENR activity (ED Fig. 8b). Inclusion of an RFP normalization control in trans, analogous to a dual-luciferase assay setup, revealed high DENR activity (stuORF-GFP/normalization-RFP) in proliferating tissues (brain and imaginal discs), and low activity in tissues with growing, but non-proliferating cells (salivary gland and fat body, ED Fig. 9).
We wondered how DENR•MCT-1 activity is regulated. Neither DENR protein levels nor DENR-MCT-1 binding dropped in quiescent S2 cells (ED Fig. 10ac). Phosphorylation of T82, T125 and a double-phosphorylation on T118/S119 in human and fly MCT-1 have been observed25. Using cells where endogenous MCT-1 is knocked-down via its 3′UTR and then reconstituted with MCT-1 versions lacking the endogenous 3′UTR revealed that mutations blocking T118/S119 phosphorylation abolished MCT-1 activity (ED Fig. 10d-d’). Interestingly, T118/S119 are evolutionarily conserved to humans. Although MCT-1 was observed to be phosphorylatable in vitro by Erk and Cdc226, we could not observe an effect of Erk, Cdc2, PI3K, Akt or TORC1 inhibition on stuORF reporter expression (ED Fig. 10e-g). Further work will be required to identify upstream kinases regulating DENR•MCT-1.
We have identified a new translational control system regulating an abundant class of mRNAs, featuring: 1) stuORFs as the critical cis-element, 2) DENR•MCT-1 as the trans-acting factor, and 3) proliferation as an important cellular context. This system differs fundamentally from GCN4/ATF4 paradigms both mechanistically and functionally. Unlike GCN4-type mechanisms1,2,5,27-30, DENR•MCT-1 functions in non-stressed cells, when general translation is not compromised, and independently of uORF to main-ORF distance (Fig. 2h), to promote proliferation. Importantly, DENR•MCT-1 uncouples translation reinitiation from standard initiation, since it is not required for initiation (Fig. 2c’). In contrast, GCN4-type mechanisms rely on coupling of initiation and reinitiation to antagonistically regulate GCN4/ATF4 versus all other genes (supplemental discussion). Our results suggest that reinitiation can be independently controlled via DENR•MCT-1 to modulate translation of a specific group of mRNAs.
Full Methods
Fly stocks
Escargot-GAL4, UAS-nuclear-GFP, UAS-cytoplasmic-GFP (Kyoto DGRC); UAS-MCT-1-dsRNA (Vienna Drosophila RNAi Center).
DENR knockout fly generation
DENR knockout flies were generated by homologous recombination as described 31. To generate the knockout construct, upstream and downstream genomic flanks were amplified using oligos listed below and cloned into the NotI and AscI sites of pW25 31 respectively.
Antibodies and immunoblotting
Phospho-dAkt(T342) antibody was developed in collaboration with PhosphoSolutions. Anti-mbc antibody kindly provided by Susan Abmayr32. Anti-RpS6, anti-total Akt, anti-phosphoS6K (Cell Signaling); anti-tubulin and anti-EcR (Developmental Studies Hybridoma Bank). Anti-dFOXO33 and anti-InR34. Anti-dDENR and anti-MCT-1 antibodies used in this study were either generated at DKFZ by immunizing guinea pigs with recombinant HIS-tagged full-length dDENR or MCT-1 or at Eurogentec by immunizing rabbits with two DENR-derived peptides selected to be specific and immunogenic (latter are a kind gift of Matthias Hentze). Rabbit polyclonal antibodies to Drosophila ligatin/eIF2D were raised to peptides from N- and C-terminal parts of the protein by Eurogentec (Köln, Germany), affinity purified and used 1:200 for western blotting. Mouse anti-tubulin antibody (Sigma; 1:20,000). Goat anti-rabbit-HRP and goat anti-mouse-HRP secondary antibodies from Thermo Fisher Scientific and were used at 1:4000-1:20,000 depending on the primary antibody.
Immunoblotting to nitrocellulose or PVDF was performed either with wet transfer under standard conditions35 or using an iBlot rapid transfer device (Life Technologies) according to the manufacturer’s guidelines. Blots were blocked in 5% milk/TBS-T solution and probed with antibodies diluted as indicated in this solution or in TBS-T without milk. Signals were visualized using Super Signal Dura or Femto reagent (Thermo Fisher) and imaged on a Fujifilm LAS-4000 luminescent image analyzer.
Plasmids, cloning and in vitro transcription
Sequences of all oligos used for cloning are provided in a table below. All constructs were verified by sequencing.
For most plasmid luciferase reporters, 5′UTRs were isolated by PCR using gene-specific primers as PstI-BstBI products (except the 5′UTRs of snoo and rbp6 which were isolated as PstI/ClaI products), and cloned into the respective sites of pAT1152, which contains the hsp70 basal promoter followed by a polylinker, the renilla luciferase (RLuc) ORF and an SV40 polyadenylation signal. This cloning strategy retained the identical Kozak sequence for the RLuc ORF in all constructs. RLuc reporters were cotransfected with an equivalent firefly luciferase (FLuc) reporter containing the same hsp70 basal promoter, an FLuc ORF and the same SV40 polyadenylation signal (pAT1088). Mutations of the uORF ATGs in the mbc 5′UTR were generated by mutating a single nucleotide for each ATG by point mutagenesis (oligo sequenes in table below), in a manner predicted by mfold 36 to not disrupt the secondary structure of the mbc 5′UTR. Likewise, mutagenesis of uORF Kozak sequences, uORF coding sequences and the intervening sequence between uORFs 218/248 and uORF338 were done by site-directed PCR mutagenesis using oligos described in the table below. Introduction of synthetic uORFs into an RLuc reporter containing a control 5′UTR was done as follows. The RLuc reporter containing the CG43674 5′UTR was mutagenized by site-directed PCR mutagenesis to introduce SpeI and AgeI sites into the middle of the 5′UTR. uORFs were then introduced by oligo cloning into the SpeI and AgeI sites. The cloned oligos (sequences in table below) introduce a Kpn21 site at the 5′ end of the insert, which is compatible with AgeI. This allows repeated rounds of oligo clonings using the SpeI and Kpn21 sites to introduce tandem copies of the uORFs.
For stable cell line generation, the full ORF of dMCT-1 was cloned into pMT/V5-HisA vector (Life Technologies) containing a copper-inducible metallothionein promoter, C-terminal V5 epitope tag and polyhistidine affinity tag and SV40 late polyadenylation signal using Kpn I and Xho I sites.
For inducible luciferase reporters, a complete ORF of firefly luciferase was cloned into pMT/V5-HisA (Life Technologies) using EcoR I and Xho I sites. Constructs for inducible synthetic Rluc reporters were generated by subcloning of the CG43674 5′UTR with or without 1aa uORF from corresponding original plasmids in pAT1152 into pTK122 vector (pMT-Renilla with a backbone BstB I site eliminated) using a polylinker EcoR I site and the BstB I site in RLuc.
For luciferase assays with in vitro transcribed mRNA reporters, a synthetic poly(A)72 was introduced into the basal FLuc and RLuc reporter plasmids (pAT1088 and pAT1152 respectively) by oligo cloning, followed by the actin 5′UTR, yielding pSS72 and pSS73 - the normalization control plasmid and the negative control plasmid respectively. The actin 5′UTR was replaced with the mbc 5′UTR to generate pAT1337 (oligo seqs below). The mbc ΔATG version of this construct was generated by PCR mutagenesis. Coding sequences + 5′UTR + 3′UTR were PCR amplified from the pAT1152 backbone versions using the same T7 promoter forward primer and a RLuc reverse primer as for the WT Mbc 5′UTR . This fragment was digested with BglII and BstBI and cloned into the respective sites of pMbc WT to generate pCJ1. Plasmids were linearized using a HindIII site directly downstream of the synthetic poly(A) tail for run-off transcription. In vitro transcription reactions to yield capped and A-capped mRNAs were performed as previously described 37.
Cell culture , RNAi , transfection, and reporter assays
S2 cells were grown in flasks at 25°C in either Express-Five serum-free medium (Life Technologies) or Schneider’s medium (Life Technologies or Bio&Sell) supplemented with 10% FBS (Life Technologies).
For suspension culture RNAi experiments, S2 cells grown semi-adherently at 25°C were resuspended at 2 × 106 cells/mL in Schneider’s Medium without FBS and dsRNA was added to 15μg/mL. Cells were transferred to a T-25 flask and incubated semi-adherently for 1.5 h at 25°C. Subsequently Complete Schneider’s medium was added (2 mL per 1 mL of dsRNA-treated culture), and cells were grown semi-adherently for 4 days. Cells were diluted to 0.7 × 106 cells/mL, the surfactant Pluronic F-68 (Life Technologies) was added to a final concentration of 0.1% (v/v) and cells were incubated with gentle agitation on a rocker at 25°C. DENR knockdown was efficient throughout the duration of the experiment, as quantified by immunoblotting (Extended Data Figure 7b). Cells were subsequently collected for quantification of cell density, polysome profiling, or preparation of extracts for in vitro translation assays. Cell numbers were assessed with a hemocytometer and cell viability was assessed in parallel by trypan blue staining.
For standard luciferase reporter assays with DNA or RNA reporters, S2 cells were seeded in 6-well plates and treated with dsRNAs at 12μg/mL. After 3 or 4 days of adherent culture, cells were diluted into a 96-well plate at a fixed low concentration of 90,000 cells/well. After 2 days of incubation, cells were transfected with reporters using Effectene (Qiagen) and 18 hours later they were harvested for luciferase assays.
RNAi, transfection, and reporter assays- proliferative vs. quiescent cells
To obtain proliferating versus quiescent cells in an adherent format for transfection followed by luciferase assays, 5×106 S2 cells were treated with 30μg of dsRNA in 2mL of medium in a T-25 flask. After 3 days, they were split into wells of a 24-well dish at a concentration of 0.5×106 or 2.0×106 cells per well in 1mL of medium and allowed to grow for 3 more days to obtain a state of proliferation or quiescence. For luciferase assays, cells were transfected without replacing the medium 1 day prior to lysis.
For inducible reporter assays, 1 ml of S2 cells at a concentration of 1,5×106 per mL was incubated in the presence of 15μg of dsRNA in medium lacking FCS for 1 hour. After that, 4 ml of complete medium was added and the culture was incubated in a T-25 flask for 3 days. Efficiency of the knockdown was assessed by western blotting. 1,5×106 S2 cells previously treated with dsRNA were plated onto a 6-well plate at a concentration of 1,5×106 cells per well in 2mL of medium and grown overnight. On the next day, the cells were transfected with 10 ng of inducible Fluc reporter (transfection control), 20 ng of inducible Rluc reporter with either control or 1aa uORF-containing 5′UTR and 370 ng of a non-specific plasmid using Effectene transfection reagent (Qiagen). For proliferating cells, expression of luciferase reporters was induced by addtition of CuSO4 (FC= 0.1mM) 3-6 hours post-transfection. For quiescent conditions, the transfected cells were first grown for 6 days and then induced with CuSO4 (FC= 0.1mM). In both cases induction was for 18h and the cells were then harvested and lysed with 1x Passive Lysis buffer (Promega). 10 μl of cell extracts were used for dual luciferase assays (Promega) in a multiwell plate luminometer (Perkin-Elmer).
Generation of stable S2 cell lines
For stable line generation, S2 cells grown in complete Schneider’s medium (Bio&Sell) supplemented with 10% FBS (Life Technologies) were plated onto a 6-well plate at a concentration of 1×106 per well, grown for 18h and transfected with 400 ng of pMT-MCT-1-V5His inducible construct together with 10 ng of pCoBlast plasmid (Life Technologies), which provides resistance to the antibiotic Blasticidin S. Transfection was performed using Effectene reagent (Qiagen) according to the manufacturer’s protocol and stable blasticidin resistant clones were selected by incubating of the transfected cells in the presence of decreasing concentrations of Blasticidin S (Life Technologies) from 25 μg/mL to10 μg/mL over several weeks. The resulting polyclonal stable cell lines were confirmed to enable inducible transgene expression by western blotting.
Co-immunoprecipitation from proliferating vs. quiescent cells
Stable S2 cell lines with copper-inducible V5 epitope-tagged MCT-1 were grown under proliferative or quiescent conditions and MCT-1-v5 expression was induced by addition of CuSO4 (FC= 0.5mM) for 48 hours prior to harvesting for immunoprecipitation. For the quiescent state, cells were plated at a density of 10×106 per well in a 6-well plate and grown for 4 days prior to induction. For proliferating conditions, the cells were plated at 1×106 per well in a 6-well plate and grown for 2 days prior to induction. On day 3 and 4 for quiescent and day 2 for proliferating cells, control cell counts were performed to verify the proliferation/quiescent status of the cells. The cells were harvested, washed with 1xPBS, flash-frozen in liquid nitrogen and kept at −80°C.
Frozen cells were thawed and lysed for 30 min on ice with ES2 lysis buffer (100mM KCl, 20mM Hepes pH7.5, 2.5mM EDTA, 5mM DTT, 0.05% Triton X 100 with protease and phosphatase inhibitors) and spun at 10,000 rcf for 10′ at 4 degrees. Lysate input was normalized by Bradford assay. 50 uL of Protein G Dynabeads were used for each sample. Beads were blocked by washing 5x with 3% BSA in PBS, and were incubated with 20uL of either V5 or Flag antibodies in 1mL PBS-T for 1hr at RT, rotating. Beads were then washed 2x with PBS-T, and 2x with 0.1M Sodium Borate, pH9. Antibodies were coupled to the beads during 2x 30 min incubations rotating at RT in a 1 mL 20mM DMP/0.1M Sodium Borate solution. The reaction was stopped by 2x washes of 50mM glycine, and beads were washed 3x with PBS-T. Lysate was added to the antibody-coupled beads, and samples were rotated at RT for 10 min followed by washing 3x with lysis buffer. 1X Laemmli loading buffer was added directly to the beads, and beads were boiled for 5 min at 95 degrees. Samples were run on a 15% SDS-PAGE gel, and probed using either V5 or DENR antibody.
Crosslinking and RNA-IP
S2 cells were grown to subconfluency in 10-cm dishes, two dishes were used per IP sample. Cells were washed once with PBS and cellular proteins were cross-linked with 0.5% formaldehyde in PBS for 10 min at room temperature. Further crosslinking was stopped by the addition of 0.25 M glycine (pH 7.0) for 5 min at room temperature. Cells were then scraped on ice in 1ml RNA-IP lysis buffer (50 mM Tris-HCl (pH 7.5), 150 mM NaCl, 1.5 mM MgCl2, 1 mM DTT, 1% NP-40, Mini Complete protease inhibitors EDTA-free (Roche) and 0.2 U/ml RNasin (Promega)). Lysates were cleared by centrifugation (5 min at 4°C and 2,000 rpm in a table-top centrifuge) and split into input (1:5) and IP samples (4:5). The IP samples were precleared with Protein A/G UltraLink Resin (Thermo Pierce) for 1 hr at 4°C and incubated with 2 μl anti-DENR or 3 μl anti-GFP serum for 1h at 4°C. Immuno-complexes were precipitated by incubation with protein A/G beads for 1h at 4°C. Beads were washed three times with low salt RNA IP wash buffer (50 mM Tris-HCl (pH 7.5), 150 mM NaCl, 1.5 mM MgCl2), followed by 3 washes with high salt RNA-IP wash buffer (50 mM Tris-HCl (pH 7.5), 300 mM NaCl, 1.5 mM MgCl2). RNA was eluted from the beads using 1 ml μl TriFast reagent (PeqLab) according to the manufacturer’s instructions. 15 μg GlycoBlue (Ambion) was added to the RNA prior to precipitation. 4 μl RNA were subjected to reverse transcription using random hexamer primers and Superscript III Reverse Transcriptase (Invitrogen) according to the manufacturer’s instructions, followed by qPCR analysis.
Quantitative RT-PCR for relative mRNA levels
RNA purification was performed with Trizol reagent according to the manufacturer’s recommendations. qRT-PCRs were done as previously described15. rp49 was used as a normalization control for experiments analyzing mRNAs. Primer sequences appear in the oligo table below.
Quantitative RT-PCR for rRNA and tRNA levels
To quantify 18S, 28S rRNA and initiator tRNA levels in control or DENR-KD cells, total RNA was extracted from an equal number of cells in the presence of a defined amount of spiked in RLuc RNA that had been in vitro transcribed. This RLuc spike-in was then used for normalization in qRT-PCR assays with primers and conditions described previously14,16. Primer sequences appear in the oligo table below.
Quantification of de novo protein synthesis by metabolic labeling
Nascent protein synthesis rates of DENR-depleted and control S2 cultures in either the proliferative (>= 4 independent samples) or quiescent (>= 3 independent samples) growth state were assayed using the Click-IT® L-Azidohomoalanine kit (Life Technologies) according to the manufacturer’s instructions, but adapted for use with S2 cells. 2 - 6 × 106 cells (with or without cycloheximide present as a control) were washed in 25°C PBS , resuspended in 1mL pre-warmed Schneider’s Complete Drosophila medium lacking methionine (Bio&Sell) transferred to 6-well plates and incubated at 25°C for methionine depletion. After 1 hour, the methionine analog Click-IT® L-Azidohomoalanine (final concentration= 50 μM) was added for nascent protein labeling and cells were subsequently incubated at 25°C for an additional 2 hours. Cells were harvested and lysed in lysis buffer (1% SDS in 50 mM Tris-HCl pH 8.0). Lysates were sonicated, centrifuged briefly to remove debris, and protein concentration in the resulting supernatant was measured by Bradford protein assay (BioRad). Between 0.8 and 7.8 μg of total protein were used for the chemoselective “click” reaction (20 min at RT) between the azide (l-azidohomoalanine) and alkyne (tetramethylrhodamine, TAMRA). Proteins were pelleted by methanol precipitation, solubilized in NuPAGE LDS sample buffer (Life Technologies) and incubated for 10 min at 70°C. Equal amounts of total protein were run on 4-12% Bis-Tris NuPAGE gels (Life Technologies) and TAMRA signals for nascent proteins were detected using the Fujifilm Fluorescent Image Analyzer FLA-9000 system. For quantification of total protein for normalization, gels were subsequently stained with SYPRORuby (Life Technologies) and re-scanned on the FLA-9000. TAMRA and SYPRORuby signals for each sample were quantified using FLA-9000 software and processed in Microsoft Excel.
Polysome profiling from S2 cells
Cells growing in suspension with gentle rocking were first treated with cycloheximide (CHX) to freeze polysomes prior to lysis. CHX (final conc.=100 μg/mL) was added to cultures followed by further incubation for 30 minutes at 25°C on a rocker. Cells were counted and equal cell numbers were subsequently centrifuged at 800 × g for 3 min. The supernatant was discarded and the cell pellet was washed 2 times with 1x PBS, pH=7.4. Cells were resuspended in 175 μL polysome lysis buffer (‘PLB’: 20 mM Tris, 20 mM NaCl, 3 mM MgCl2, 100 U/mL Recombinant RNasin Ribonuclease Inhibitor (Promega, Mannheim, Germany) and Roche Complete Protease Inhibitor Cocktail (Roche, Grenzach-Wyhlen, Germany). 175 μL of Polysome Extraction Buffer (‘PEB’: PLB + 1% Triton X-100, 2% Tween-20, and 1% Na-Deoxycholate; all final conc.) was added. Lysates were mixed, incubated on ice for 10 min. and then spun at maximum speed for 10 min. at 4°C in an Eppendorf microcentrifuge. Supernatants were loaded onto 14 × 95 mm Polyclear centrifuge tubes (Seton, Petaluma, CA) containing 17.5 – 50 % sucrose gradients generated using the Gradient Master 108 programmable gradient pourer (Biocomp, New Brunswick, Canada) and centrifuged for 2.5 h at 35,000 rpm in an SW40Ti rotor in a Beckman L7 ultracentrifuge (Beckman Coulter, Krefeld, Germany). After centrifugation gradients were simultaneously fractionated and measured for RNA content using a Piston Gradient Fractionator (Biocomp) attached to a UV monitor (BioRad, Hercules, CA).
For RNA and/or protein isolation after fractionation, Trizol reagent (Life Technologies, Carlsbad, CA) was added to fractions and either protein (followed by TCA precipitation) or RNA (by Pure Link RNA Mini Kit purification (Life Technologies, Calsbad, CA)) was isolated for downstream applications according to the manufacturer’s recommendations.
For quantitative analysis of individual elements (i.e. 80s monosomes, polysomes, etc.) from the polysome profiles, all traces were first exported to Microsoft Excel and adjusted as necessary to ensure matching x/y-axis scales. Traces were then exported to Adobe Photoshop and ImageJ for processing, baseline setting, cropping and pixel counting. These pixel counts were then used for determining P/M values, plot generation, and statistical analysis in Microsoft Excel.
For immunoblotting of polysome gradient fractions, proteins in fractions were precipitated with TCA, washed 2x with acetone, and resuspended in protein gel sample buffer prior to electrophoresis and immunoblotting under standard conditions.
Polysome profiling from Drosophila larvae
Polysome profiles from Drosophila larvae were performed by crushing male larvae in lysis buffer (50mM Tris pH7.4,15mM MgCl2,300mM NaCl, 1% Triton-X100, 100μg/mL cycloheximide, 0.1% β-mercaptoethanol, 1x protease inhibitor coctail from Roche, 200 U/mL RiboLock RNAse inhibitor from Fermentas) on ice. Lysates were cleared by centrifugation, equalized in protein concentration and loaded onto a 17.5-50% sucrose gradient and centrifuged for 2.5h at 35000 rpm.
In vitro translation assays with extracts from S2 cells
S2 cells extracts for in vitro translation assays were generated according to a modified version of a protocol previously used for embryo translation extracts 38, adapted for smaller cell volumes. Cells were grown in suspension culture as described above to a density of 5-6 ×106 cells/ml, harvested by low–speed centrifugation (5 min, 800×g), and cell pellets were resuspended in ice-cold PBS to wash and then respun. This PBS wash step was repeated a further 2x. After the final wash, pellet volume was carefully estimated and cells were resuspended in precisely 2 pellet volumes of DEIKM Buffer (10mM Hepes pH 7.4, 80mM K Ac 0,5 mM Mg Ac, 5mM DTT, + of Complete, Mini, EDTA-free Protease Inhibitor Cocktail (Roche,1 tablet/10ml)) and then incubated for 30-45 min on ice. Lysis was by ~20 strokes with either a 1ml or 2ml syringe. Extracts were spun for 20min at 20,000×g at 4°C. Supernatants were carefully removed and protein concentrations were checked by Bradford assay (BioRad). Extracts were adjusted to a final concentration of 10ug/ml with DEIKM buffer as necessary and adjusted to a final concentration of 10% glycerol. Extracts with concentrations below 10ug/ml were discarded, as our experience was that they displayed higher variability. Translation extracts were aliquotted, flash-frozen in liquid nitrogen, and stored at −80°C. Translation extracts were used at a final concentration of 40% for in vitro translation reactions, which were performed essentially as described (Gebauer et al 1999). The extracts were verified to be cap and poly(A) tail responsive. We also performed a titration of mRNA concentration for each reporter mRNA and used mRNA concentrations within the linear range of translation for all of the experiments shown.
Bioinformatics
All bioinformatics are based on Flybase release 5.46 39. A “DENR-dependence” score was calculated based on the number of ATG start codons in the mRNA 5′UTR and the strength of their Kozaks. To predict Kozak strength, the frequency of each nucleotide found at positions −4 to −1 relative to the ATG for ‘main ORFs’ in the genome was quantified, yielding a frequency table (Figure ED4d’) and the corresponding consensus of cAaaAUG (Figure ED4d), similar to that of other species and the ‘pregenomic’ Drosophila consensus defined with a much smaller set of genes 40. A score for the strength of any Kozak in Drosophila was then generated using a multiplicative model whereby the frequencies indicated in Figure ED4d’ for each nucleotide position from −4 to −1 relative to the ATG were multiplied together. In this manner, 4-mer sequences frequently observed upstream of main ORF ATGs in the fly genome obtain high scores (maximum of 0.0496) whereas infrequent ones obtain low scores (minimum of 0.00012). The score from all individual ATGs in a 5′UTR were summed to reach the combined score for the 5′UTR.
Oligo Sequences and number of replicates for figure data
Full oligo sequences for all oligos used, as well as information about replicate numbers for each figure panel, are in Supplementary Information.
Supplementary Material
Extended Data Figure Legends Extended Data Fig. 1: DENR knockout strategy and phenotypes
(a) Domain structures of Ligatin/eIF2D, DENR and MCT-1. (b) DENR/CG9099 genomic locus, indicating the “knockout region” that was replaced with the mini-white cassette via homology-mediated recombination. (c-d) DENRKO animals have no detectable DENR transcript by quantitative RT-PCR (c) or protein (d). (e) DENRKO anterior-dorsal histoblast nests have correct cell numbers at onset of pupation (0 h APF), but impaired proliferation the first 4 hours of pupal development (4 h APF). Quantification shown here; representative images shown in main Fig. 1b. Cells visualized by esgG4>cytoplasmic GFP+nuclear GFP. (f) DENR expression in histoblast cells of DENRKO animals rescues their viability and abdominal phenotypes (DENRKO, escargot-GAL4, UAS-DENR). (g) DENR transcript is expressed in all tissues and developmental stages tested, detected by quantitative RT-PCR, normalized to rp49. (h-h’) DENRKO animals have genitals that are not properly rotated. (i) MCT-1/CG5941 knockdown animals (Tubulin-GAL4>UAS-CG5941 RNAi) phenocopy DENRKO animals. They are pupal lethal, with abdominal epidermis that is soft and transparent, whereas head and thorax structures are comparatively better-formed. (j) HA-tagged dMCT-1 can co-immunoprecipitate endogenous dDENR from S2 cells. (k) Crossing scheme to test for genetic interaction between ligatin and DENR. DENRKO heterozygous females were crossed to males heterozygously carrying a deficiency for the ligatin locus. Resulting non-FM7 hemizygous DENR knockout male pupae were scored for presence of the TM6B balancer (using the Tubby marker) or the ligatin deficiency. If the ligatin deficiency had no effect on survival of DENR mutants, it would be present in the expected 50% mendelian ratio, in equal proportion to the FM7 balancer. Instead, only 18 out of the scored 53 animals were carrying the ligatin deficiency. (l) Polysome profiles from DENRKO larvae have high 80S and low polysomal peaks. Quantification of p/m ratio for 5 independent replicates is shown in the main Fig. 1f. (m-n’) Polysome profiles of DENR-KD cells have increased 80S peaks, reduced p/m ratios, and increased total areas under the curve at both 4 (m-m’) and 7 (n-n’) days post dilution into suspension culture. (o) DENR-KD cells have elevated levels of 18S rRNA, 28S rRNA and initiator tRNA by qRT-PCR. Experiment was performed 8 days after treatment with DENR dsRNA. (p) DENR knockdown S2 cells have elevated levels of ribosomal protein S6, quantified by western blot relative to tubulin. (q,r) Quiescent (r) but not proliferating (q) DENR-KD cells have high protein per cell. Error bars: std dev. (b-g) or SEM (o-r) *Mann-Whitney U-Test <0.05, ***t-test<0.001.
Extended Data Fig. 2: DENR promotes reinitiation of translation downstream of uORFs in the mbc 5′UTR
(a) Western blotting of extracts from control, DENR-knockdown and MCT-1 knockdown S2 cells using a panel of antibodies does not show dramatic differences in the level of most proteins. Quantification of luminescence on a LICOR Oddesey FC relative to control knockdown cells is indicated. (b) Levels of renilla luciferase (RLuc) reporters containing the mbc 5′UTR and either the mbc or hsp70 promoters are reduced in DENR and MCT-1 knockdown cells. DNA reporters were transfected into S2 cells together with a firefly luciferase (FLuc) normalization control containing an hsp70 promoter. (c) In vitro transcribed RLuc reporter mRNAs containing mbc or actin 5′UTR, co-transfected with a control FLuc reporter into DENR, MCT-1 or control knockdown cells. mRNAs were capped with the normal m7G cap (“cap”) or with an adenosine cap structure (“A-cap”) which does not bind eIF4E. Below, representative raw counts (from the control) for the three reporters shows that the signal obtained with A-capped reporters is roughly one-half to one-quarter the signal observed with a normal m7G cap, and well above background. (d-d’) Reduced translation in DENR-KD cells of an mRNA luciferase reporter containing the mbc 5′UTR is not accompanied by a reduction in the intracellular levels of the reporter mRNA. In vitro transcribed mRNA consisting of the mbc 5′UTR, the renilla luciferase ORF and a synthetic poly(A) was cotransfected with a normalization control mRNA consisting of a short 5′UTR, firefly luciferase ORF and a synthetic poly(A) into control or DENR-KD cells. Relative luminescence counts (d) and relative mRNA levels determined by quantitative RT-PCR (d’) are shown. (e) Combined knockdown of DENR and MCT-1 does not have additive effects compared to the single knockdowns on translation of a luciferase reporter containing the mbc 5′UTR, consistent with them working together in one functional complex. (f-h) High resolution deletion series of the mbc renilla luciferase reporter, summarized in Fig. 2d, identifies nt 200-375 of the mbc 5′UTR as the minimum region required to impart DENR-dependent regulation. Error bars: std dev.
Extended Data Fig. 3: Detailed sequence information for the mbc 5′UTR
(a) Full sequence of the mbc 5′UTR. ATGs are indicated in bold. Open reading frames are shown as red or grey boxes depending on whether they have a good or bad Kozak sequences, respectively. Stop codons indicated with asterisks. (b) Schematic overview of the mbc 5′UTR and the tested reporter constructs, summarizing results from other panels. Light peach-colored shading indicates the minimal region required to impart DENR-dependent regulation to a luciferase reporter. Red and grey boxes indicate upstream Open Reading Frames (uORFs) with strong and weak Kozak sequences, respectively. ΔATG construct: single point mutations were introduced to abolish the three indicated uORFs. ΔKozak construct: Kozak sequences of the indicated uORFs were mutated to gtgtATG, thereby disfavouring translation initiation at these sites. Δstop construct: stop codons of all three in-frame uORFs were mutated, resulting in a construct where the uORF stop codon is just downstream of the RLuc ATG. (c) Translation extracts from DENR-KD cells are impaired in translating a reporter containing the mbc 5′UTR. mRNA reporters containing the full-length mbc 5′UTR, or a mutated ΔATG version where the ATGs at positions 218, 248, 338 and 415 were mutated, were prepared by in vitro transcription and introduced into translation extracts prepared from S2 cells treated with either control (GFP) or DENR dsRNA. Mutating the ATGs both increases the overall translation of the reporter, and renders the reporter insensitive to DENR knockdown. Statistics performed using the two-tailed, two-sided Student’s t-Test. (d) Full sequence of the 5′UTR in the mbc Δstop construct. Mutated stop codons are shown with underlining. The same sequence (nt 200-375) without the three point mutations is regulated by DENR (Fig. 2h).
Extended Data Fig. 4: Genome-wide analysis of stuORF transcripts
(a) Genome-wide histogram of transcripts containing uORFs in their 5′UTRs. (b-b’) Frequency distribution of nucleotides found surrounding the ATG of all main ORFs annotated in the fly genome. Frequency matrix in d’ was used to generate a multiplicative score for the strength of any Kozak sequence of interest. (c) Transcripts that were translationally down-regulated in S2 polysome profiles upon DENR knockdown are enriched for stuORF containing transcripts. A translation score was measured for each transcript in the genome, consisting of the ratio of transcript levels in 80S + polysome fractions (indicating active translation) divided by transcript levels in total mRNA, yielding the percentage of mRNA being actively translated. This was calculated for both control and DENR-KD S2 cells, and compared. Transcripts were then sorted into quintiles according to this comparison, with the first quintile containing transcripts that are most down-regulated in DENR knockdown cells compared to controls, and quintile 5 containing transcripts that were up-regulated in DENR-KD cells compared to controls. For each quintile, the average stuORF score was calculated, as described in the main text. A clear correlation can be observed, with down-regulated quintiles being enriched for transcripts containing stronger stuORF score. Error bars: SEM. (d) Analysis the other way around: stuORF containing transcripts are preferentially down-regulated in the polysome analysis from DENR-KD cells compared to transcripts containing no uORFs. Two categories of transcripts were selected genome-wide: the top 210 with high stuORF scores, and the bottom 6000 transcripts containing no uORFs. For each category of transcripts, histograms of the translation score were plotted, with negative numbers representing transcripts that were translationally down-regulated in DENR-KD cells compared to controls. Transcripts containing stuORFs are preferentially down-regulated genome-wide upon DENR knockdown in a highly statistically significant manner (Mann Whitney U-test p=10−14) compared to transcripts lacking uORFs. (e) Gene ontology enrichment using DAVID on stuORF-bearing transcripts. (f) Examples of genes predicted via the “DENR-dependence score” to be DENR-dependent in Drosophila. (g) DENR binds both mRNAs containing and lacking stuORFs. Cellular proteins were cross-linked with 0.5% formaldehyde and then DENR-containing complexes were immunoprecipitated. RNA was purified from the immunoprecipitation, reverse-transcribed with random hexamers, and subjected to quantitative PCR. Both transcripts containing stuORFs (mbc, InR) and transcripts containing no or weak uORFs (RpS13, cdc2, RpL32), as well as initiator tRNA, were found to be significantly enriched, compared to RNA from a control immunoprecipitation using anti-GFP antibody. Error bars: std. dev.
Extended Data Fig. 5: Timecourse of DENR knockdown reveals that changes in stuORF translation precede global changes in polysome profiles or ribosome levels
(a) Translation of a stuORF reporter (bearing 1x ATGTAA, as in Fig. 3) progressively drops upon DENR knockdown from 1-4 days after treatment with dsRNA. A significant drop in translation can be observed already 2 and 3 days after DENR knockdown. In contrast, a control reporter is unperturbed. (b) DENR protein levels 2 and 3 days after knockdown in S2 cells, assessed by immunoblotting. (c-c’) Polysome profile of S2 cells treated for 2 (c) or 3 (c’) days with DENR dsRNA (red traces) shows no significant drop in polysome fractions compared to cells treated with control (GFP) dsRNA (grey traces), suggesting that the drop in polysome levels observed at later time points (e.g. ED Fig. 1) are in vivo secondary consequences (e.g. due to reduced translation of stuORF containing transcripts such as Insulin Receptor, leading to a global reduction in translation rates). (d-d’) Quantification of 18S RNA, 28S RNA and initiator tRNA levels in S2 cells treated with dsRNA for 2 (d) or 4 (d’) days shows little to no increase upon DENR knockdown compared to control.
Extended Data Fig. 6: Loss of DENR leads to reduced InR and EcR protein levels and signaling in vivo – support data
(a-a’) DENR and MCT-1 knockdown in S2 cells does not lead to reduced EcR-B1 and InR mRNA levels. (b) DENR-KD cells are less sensitive to ecdysone (1 μM, 4 h), assayed by qRT-PCR of two EcR target genes BR-C (Fig. 4c) and E75 (shown here), normalized to rp49. (c-c’) DENRKO pupae 4 h APF have reduced levels of InR and EcR-A protein (c) but not mRNA (c’). (d) Expression of InR in histoblast cells and imaginal discs of DENRKO animals using escargot-GAL4 rescues their delayed pupation.
Extended Data Fig. 7: DENR promotes expression of an inducible 1 aa stuORF reporter in proliferating cells more efficiently than in quiescent cells.
(a) Schematic overview of a cell-based inducible Dual Luciferase Reporter Assay to enable systematic evaluation of the effect of DENR knockdown on expression of stuORF or control reporters under different growth states. (b) DENR knockdown in S2 cells leads to efficient depletion of DENR protein for up to 18 days post-dsRNA treatment. 4 days after treatment with dsRNA, cells were diluted into either suspension culture at a density of 106/mL (for Fig. 1g-i’) on in adherent culture (for the remaining panels in this figure). Graded loading of the GFP dsRNA control sample on the blot shows that DENR knockdown yields persistent depletion of DENR protein by ≥90%. (c) Growth curve post-dilution of S2 cells treated with or without dsRNAs, indicating optimal days for reporter induction that correspond to proliferating or quiescent populations (2 d or 7 d, respectively). Under proliferative conditions both DENR-KD and control cells double within ~24 h. Conversely, under quiescent conditions no significant increase in cell number is observed, implying that the population has essentially stopped proliferating. (d) Presence of a 1 aa stuORF (ATGTAA) reduces translation of a Renilla reporter by 50% in control (GFP-KD) cells. Error bars: SEM. Data from 2 independent KDs and 4 transfections. (e) Effect of DENR knockdown on the inducible 1 aa stuORF reporter expression is significantly stronger in proliferating compared to quiescent cells. Error bars: SEM; t-test p<0.0001. (f-f’) Raw luciferase counts for the experiment shown in (e) demonstrating that upon DENR knockdown, control reporters increase in expression (consistent with what is also observed in translation extracts, Fig. 2c’) whereas the stuORF reporter decreases in expression (f). (g) Representative raw luciferase counts for main Fig. 2i. (h-h’) DENR knockdown affects mbc protein levels more strongly in proliferating than in quiescent cells. DENR knockdown leads to reduced endogenous mbc protein (h) but not mRNA (h’) in proliferating but not quiescent S2 cells. Cells were treated with dsRNA for 4 d, then seeded in parallel at two different densities to achieve proliferation or quiescence at time of assay. Error bars: Std dev.
Extended Data Fig. 8: DENR is required to express stuORF containing transcripts in vivo in Drosophila
(a) Schematic of the fluorescent reporters introduced into Drosophila. Identical reporters were generated containing a Tubulin promoter, the 5′UTR of CG43674, which does not contain any uORFs, the GFP open reading frame, and the SV40 poly-A. A 6-nt stuORF (ATGTAA) was introduced by mutagenesis into the 5′UTR of one construct, generating the “stuORF reporter”. Both constructs were inserted via phiC31-mediated recombination into the VK33 landing site at 65B2 on chromosome 3L, ensuring identical transcriptional regulation. (b) The control reporter is well expressed in both larvae containing (DENR+/+) or lacking DENR (DENRKO). Immunoblotting of larval extracts (c) shows that GFP levels of the control reporter do not drop in DENR knockouts compared to controls, indicating that DENR is not required for translation of transcripts lacking uORFs. Introduction of the 6-nt stuORF causes the reporter to be less expressed in a control DENR+/+ background, consistent with uORFs having a repressive role in translation of the main downstream ORF. In contrast to the control reporter, expression of the stuORF reporter is entirely dependent on DENR in vivo, since no GFP expression could be observed when the stuORF reporter is introduced into a DENRKO genetic background. This was confirmed by immunoblotting (c). (c) Immunoblot to detect GFP and a loading control (awd) in larval extracts of animals bearing a control or a stuORF reporter, either in a control DENR+/+ or in a DENR knockout genetic background.
Extended Data Fig. 9: DENR activity in the Drosophila larva is higher in proliferating tissues compared to non-proliferating tissues
Expression of the stuORF GFP reporter is entirely DENR-dependent (ED Fig. 8), hence it serves as a readout for DENR activity. Flies were generated bearing either a control or a stuORF-GFP reporter, combined in trans with a normalization control RFP reporter, generated by replacing the GFP of the control reporter with RFP, and inserting it into the same VK33 landing site as the GFP reporters. This setup is analogous to the dual FLuc/RLuc setup used for luciferase assays and completely controls for transcriptional effects. Various tissues were dissected and imaged by confocal microscopy, the GFP/RFP ratio was calculated, and is displayed in pseudocolor. Flies bearing a control GFP reporter and the control RFP reporter have the same ratio of GFP to RFP in all tissues (a). For instance, the strong spot in the middle of the wing disc which is due to transcriptional effects, is present in both the GFP reporter and the RFP normalization control, and is normalized out when the GFP/RFP ratio is calculated. In contrast, the stuORF reporter (b) is more strongly expressed in proliferating tissues such as the brain and associated imaginal discs, or the wing disc, compared to non-proliferating tissues such as fat body or salivary glands. This can be observed both on the overlay, which is yellow for brain and wing discs, but red for fat body and salivary gland, as well as in the pseudo-colored GFP/RFP ratio panel which is high in brain and wing disc, but low in fat body or salivary gland.
Extended Data Fig. 10: DENR activity is higher in proliferating cells compared to quiescent cells
(a-b) DENR•MCT-1 binding is not different in proliferating versus quiescent S2 cells. (b) Immunoprecipitation of endogenous DENR from proliferating or quiescent S2 cells shows similar amounts of co-immunoprecipitating endogenous MCT-1 in the two conditions. (b) A stably-transfected S2 cell line bearing a copper-inducible pMT-V5-MCT-1 construct was grown in proliferative or quiescent conditions, as in ED Fig. 7, and then induced to express MCT-1. V5-tagged MCT-1 was immunoprecipitated and endogenous DENR was detected by immunoblotting. An anti-FLAG immunoprecipitation was performed as a negative control. Near-equal levels of DENR protein co-immunoprecipitated with MCT1 in both proliferative and quiescent conditions. (c) Expression levels of DENR and eIF2D/Ligatin do not depend on each other and do not change in proliferating vs. quiescent cells. Immunoblot of S2 cells treated with DENR, eIF2D/Ligatin or control (GFP) dsRNA, in proliferative or quiescent conditions. Two different amounts of the control samples were loaded to quantitatively assess knockdown efficiencies. A non-specific band on eIF2D blot is marked with a star (*). (d-d’) Abolishing phosphorylation on T118 and S119 blunts MCT-1 activity. (d) Luciferase assay using a stuORF reporter in S2 cells depleted of endogenous MCT-1 via dsRNA targeting its 3′UTR, and re-constituted to express wild-type or mutated forms of MCT-1 (using constructs with an exogenous 3′UTR) identifies T118 and S119 as important phosphorylation sites. Four sites were tested – T118, S119, T82 and T125 – for all of which phosphorylations on endogenous MCT-1 were observed by publicly available proteome-wide mass spectrometry analyses (PhosphoPep and PhosphoSite.org). Whereas wild-type MCT-1 is able to restore expression of the stuORF reporter, a T118A/S119A mutant form of MCT-1 cannot. Testing each site individually identifies S119 as the more important of the two. In contrast, mutating T82 and T125 to alanine does not impair their ability to promote stuORF translation. ** t-test<0.01. Error bars: std. dev. (d’) MCT-1[T118A/S119A] is expressed and stable. Immunoblot of S2 cells transfected with control plasmid or plasmid expressing wild-type FLAG-MCT-1 or FLAG-MCT-1[T118A/S119A] shows that the mutant MCT-1 is expressed and roughly equivalent levels as the wild-type protein. (e-f) Inhibition of Erk, PI3K, Akt or TORC1 does not have an effect on stuORF translation. Luciferase assay with a stuORF reporter in S2 cells treated with inhibitors for Erk (U0126, ), TOR-Complex 1 (rapamycin, e), Akt (Akt Inhibitor VIII, e) or PI3K (Wortmannin, e) shows that inhibition of any of these kinases has little to no effect on stuORF translation. (g) Knockdown of Drosophila cdc2 (CG10498) does not reduce expression of a stuORF reporter. S2 cells were treated with either GFP dsRNA or CG10498 dsRNA, and then transfected with a stuORF reporter. GFP dsRNA was applied to 90 wells in a 98-well plate and the values displayed represent the average and standard deviation (error bars). CG10498 knockdown results for two separate well are shown.
Acknowledgements
We thank B. Bukau, R. Green, and P. Śoba for suggestions on the manuscript, V. Benes and T. Bähr-Ivacevic (EMBL Genomics Core Facility) for assistance with microarray experiments, S. Abmayr for anti-mbc antibody, T. Hsu for anti-awd antibody, C. Strein and M. Hentze for Drosophila DENR antibodies, P. Jakob for help with testing conditions for S2 in vitro translation extracts, L. Schibalski and J. Grawe for help with cloning, S. Hofmann for generous help with polysome analysis, and S. Lerch, A. Haffner, and M. Schroeder for technical assistance. K.K.M. is the recipient of an Alzheimer’s Research Scholarship from the Hans und Ilse Breuer Foundation. P.C.J. is supported in part by a grant from the Fritz-Thyssen Foundation to KD. This work was also supported in part by a Deutsche Forschungsgemeinschaft (DFG) grant and ERC Starting Grant to A.A.T.
Footnotes
Statements Polysome microarray data deposited at NCBI GEO (GSE54625):http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE54625
Competing financial interests The authors declare no competing financial interests.
References
- 1.Jackson RJ, Hellen CU, Pestova TV. Termination and post-termination events in eukaryotic translation. Adv Protein Chem Struct Biol. 2012;86:45–93. doi: 10.1016/B978-0-12-386497-0.00002-5. [DOI] [PubMed] [Google Scholar]
- 2.Sonenberg N, Hinnebusch AG. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell. 2009;136:731–745. doi: 10.1016/j.cell.2009.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Calvo SE, Pagliarini DJ, Mootha VK. Upstream open reading frames cause widespread reduction of protein expression and are polymorphic among humans. Proc Natl Acad Sci U S A. 2009;106:7507–7512. doi: 10.1073/pnas.0810916106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dever TE, Green R. The elongation, termination, and recycling phases of translation in eukaryotes. Cold Spring Harb Perspect Biol. 2012;4:a013706. doi: 10.1101/cshperspect.a013706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Valasek LS. ‘Ribozoomin’--translation initiation from the perspective of the ribosome-bound eukaryotic initiation factors (eIFs) Curr Protein Pept Sci. 2012;13:305–330. doi: 10.2174/138920312801619385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Skabkin MA, Skabkina OV, Hellen CU, Pestova TV. Reinitiation and Other Unconventional Posttermination Events during Eukaryotic Translation. Mol Cell. 2013 doi: 10.1016/j.molcel.2013.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Poyry TA, Kaminski A, Jackson RJ. What determines whether mammalian ribosomes resume scanning after translation of a short upstream open reading frame? Genes Dev. 2004;18:62–75. doi: 10.1101/gad.276504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schwanhausser B, et al. Global quantification of mammalian gene expression control. Nature. 2011;473:337–342. doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- 9.Gebauer F, Hentze MW. Molecular mechanisms of translational control. Nat Rev Mol Cell Biol. 2004;5:827–835. doi: 10.1038/nrm1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kong J. Lasko, P. Translational control in cellular and developmental processes. Nat Rev Genet. 2012;13:383–394. doi: 10.1038/nrg3184. [DOI] [PubMed] [Google Scholar]
- 11.Araujo PR, et al. Before It Gets Started: Regulating Translation at the 5′ UTR. Comp Funct Genomics. 2012;2012:475731. doi: 10.1155/2012/475731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ingolia NT, Lareau LF, Weissman JS. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell. 2011;147:789–802. doi: 10.1016/j.cell.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hinnebusch AG, Lorsch JR. The mechanism of eukaryotic translation initiation: new insights and challenges. Cold Spring Harb Perspect Biol. 2012;4 doi: 10.1101/cshperspect.a011544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jackson RJ, Hellen CU, Pestova TV. The mechanism of eukaryotic translation initiation and principles of its regulation. Nature reviews. Molecular cell biology. 2010;11:113–127. doi: 10.1038/nrm2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Somers J, Poyry T, Willis AE. A perspective on mammalian upstream open reading frame function. The international journal of biochemistry & cell biology. 2013;45:1690–1700. doi: 10.1016/j.biocel.2013.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dmitriev SE, et al. GTP-independent tRNA delivery to the ribosomal P-site by a novel eukaryotic translation factor. J. biological chemistry. 2010;285:26779–26787. doi: 10.1074/jbc.M110.119693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Skabkin MA, et al. Activities of Ligatin and MCT-1/DENR in eukaryotic translation initiation and ribosomal recycling. Genes Dev. 2010;24:1787–1801. doi: 10.1101/gad.1957510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dierov J, Prosniak M, Gallia G, Gartenhaus RB. Increased G1 cyclin/cdk activity in cells overexpressing the candidate oncogene, MCT-1. J Cell Biochem. 1999;74:544–550. [PubMed] [Google Scholar]
- 19.Mazan-Mamczarz K, et al. Targeted suppression of MCT-1 attenuates the malignant phenotype through a translational mechanism. Leuk Res. 2009;33:474–482. doi: 10.1016/j.leukres.2008.08.012. [DOI] [PubMed] [Google Scholar]
- 20.Prosniak M, et al. A novel candidate oncogene, MCT-1, is involved in cell cycle progression. Cancer Res. 1998;58:4233–4237. [PubMed] [Google Scholar]
- 21.Kongsuwan K, et al. A Drosophila Minute gene encodes a ribosomal protein. Nature. 1985;317:555–558. doi: 10.1038/317555a0. [DOI] [PubMed] [Google Scholar]
- 22.Hayashi S, Hirose S, Metcalfe T, Shirras AD. Control of imaginal cell development by the escargot gene of Drosophila. Development. 1993;118:105–115. doi: 10.1242/dev.118.1.105. [DOI] [PubMed] [Google Scholar]
- 23.Wilson TG, Yerushalmi Y, Donnell DM, Restifo LL. Interaction between hormonal signaling pathways in Drosophila melanogaster as revealed by genetic interaction between methoprene-tolerant and broad-complex. Genetics. 2006;172:253–264. doi: 10.1534/genetics.105.046631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37 doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bodenmiller B, et al. PhosphoPep--a phosphoproteome resource for systems biology research in Drosophila Kc167 cells. Mol Syst Biol. 2007;3:139. doi: 10.1038/msb4100182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nandi S, et al. Phosphorylation of MCT-1 by p44/42 MAPK is required for its stabilization in response to DNA damage. Oncogene. 2007;26:2283–2289. doi: 10.1038/sj.onc.1210030. [DOI] [PubMed] [Google Scholar]
- 27.Spriggs KA, Bushell M, Willis AE. Translational regulation of gene expression during conditions of cell stress. Mol Cell. 2010;40:228–237. doi: 10.1016/j.molcel.2010.09.028. [DOI] [PubMed] [Google Scholar]
- 28.Harding HP, et al. Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol Cell. 2000;6:1099–1108. doi: 10.1016/s1097-2765(00)00108-8. [DOI] [PubMed] [Google Scholar]
- 29.Vattem KM, Wek RC. Reinitiation involving upstream ORFs regulates ATF4 mRNA translation in mammalian cells. Proc Natl Acad Sci U S A. 2004;101:11269–11274. doi: 10.1073/pnas.0400541101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hood HM, Neafsey DE, Galagan J, Sachs MS. Evolutionary roles of upstream open reading frames in mediating gene regulation in fungi. Annu Rev Microbiol. 2009;63:385–409. doi: 10.1146/annurev.micro.62.081307.162835. [DOI] [PubMed] [Google Scholar]
References for Full Methods section
- 31.Huang J, Zhou W, Watson AM, Jan YN, Hong Y. Efficient ends-out gene targeting in Drosophila. Genetics. 2008;180:703–707. doi: 10.1534/genetics.108.090563. doi:genetics.108.090563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Erickson MR, Galletta BJ, Abmayr SM. Drosophila myoblast city encodes a conserved protein that is essential for myoblast fusion, dorsal closure, and cytoskeletal organization. The Journal of cell biology. 1997;138:589–603. doi: 10.1083/jcb.138.3.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Puig O, Marr MT, Ruhf ML, Tjian R. Control of cell number by Drosophila FOXO: downstream and feedback regulation of the insulin receptor pathway. Genes Dev. 2003;17:2006–2020. doi: 10.1101/gad.1098703. doi:10.1101/gad.1098703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Puig O, Tjian R. Transcriptional feedback control of insulin receptor by dFOXO/FOXO1. Genes Dev. 2005;19:2435–2446. doi: 10.1101/gad.1340505. doi:19/20/2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Duncan K, et al. Sex-lethal imparts a sex-specific function to UNR by recruiting it to the msl-2 mRNA 3′ UTR: translational repression for dosage compensation. Genes Dev. 2006;20:368–379. doi: 10.1101/gad.371406. doi:20/3/368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Duncan KE, Strein C, Hentze MW. The SXL-UNR corepressor complex uses a PABP-mediated mechanism to inhibit ribosome recruitment to msl-2 mRNA. Mol Cell. 2009;36:571–582. doi: 10.1016/j.molcel.2009.09.042. doi:S1097-2765(09)00702-3. [DOI] [PubMed] [Google Scholar]
- 38.Gebauer F, Corona DF, Preiss T, Becker PB, Hentze MW. Translational control of dosage compensation in Drosophila by Sex-lethal: cooperative silencing via the 5′ and 3′ UTRs of msl-2 mRNA is independent of the poly(A) tail. EMBO J. 1999;18:6146–6154. doi: 10.1093/emboj/18.21.6146. doi:10.1093/emboj/18.21.6146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Marygold SJ, et al. FlyBase: improvements to the bibliography. Nucleic Acids Res. 2013;41:D751–757. doi: 10.1093/nar/gks1024. doi:gks1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cavener DR. Comparison of the consensus sequence flanking translational start sites in Drosophila and vertebrates. Nucleic Acids Res. 1987;15:1353–1361. doi: 10.1093/nar/15.4.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Extended Data Figure Legends Extended Data Fig. 1: DENR knockout strategy and phenotypes
(a) Domain structures of Ligatin/eIF2D, DENR and MCT-1. (b) DENR/CG9099 genomic locus, indicating the “knockout region” that was replaced with the mini-white cassette via homology-mediated recombination. (c-d) DENRKO animals have no detectable DENR transcript by quantitative RT-PCR (c) or protein (d). (e) DENRKO anterior-dorsal histoblast nests have correct cell numbers at onset of pupation (0 h APF), but impaired proliferation the first 4 hours of pupal development (4 h APF). Quantification shown here; representative images shown in main Fig. 1b. Cells visualized by esgG4>cytoplasmic GFP+nuclear GFP. (f) DENR expression in histoblast cells of DENRKO animals rescues their viability and abdominal phenotypes (DENRKO, escargot-GAL4, UAS-DENR). (g) DENR transcript is expressed in all tissues and developmental stages tested, detected by quantitative RT-PCR, normalized to rp49. (h-h’) DENRKO animals have genitals that are not properly rotated. (i) MCT-1/CG5941 knockdown animals (Tubulin-GAL4>UAS-CG5941 RNAi) phenocopy DENRKO animals. They are pupal lethal, with abdominal epidermis that is soft and transparent, whereas head and thorax structures are comparatively better-formed. (j) HA-tagged dMCT-1 can co-immunoprecipitate endogenous dDENR from S2 cells. (k) Crossing scheme to test for genetic interaction between ligatin and DENR. DENRKO heterozygous females were crossed to males heterozygously carrying a deficiency for the ligatin locus. Resulting non-FM7 hemizygous DENR knockout male pupae were scored for presence of the TM6B balancer (using the Tubby marker) or the ligatin deficiency. If the ligatin deficiency had no effect on survival of DENR mutants, it would be present in the expected 50% mendelian ratio, in equal proportion to the FM7 balancer. Instead, only 18 out of the scored 53 animals were carrying the ligatin deficiency. (l) Polysome profiles from DENRKO larvae have high 80S and low polysomal peaks. Quantification of p/m ratio for 5 independent replicates is shown in the main Fig. 1f. (m-n’) Polysome profiles of DENR-KD cells have increased 80S peaks, reduced p/m ratios, and increased total areas under the curve at both 4 (m-m’) and 7 (n-n’) days post dilution into suspension culture. (o) DENR-KD cells have elevated levels of 18S rRNA, 28S rRNA and initiator tRNA by qRT-PCR. Experiment was performed 8 days after treatment with DENR dsRNA. (p) DENR knockdown S2 cells have elevated levels of ribosomal protein S6, quantified by western blot relative to tubulin. (q,r) Quiescent (r) but not proliferating (q) DENR-KD cells have high protein per cell. Error bars: std dev. (b-g) or SEM (o-r) *Mann-Whitney U-Test <0.05, ***t-test<0.001.
Extended Data Fig. 2: DENR promotes reinitiation of translation downstream of uORFs in the mbc 5′UTR
(a) Western blotting of extracts from control, DENR-knockdown and MCT-1 knockdown S2 cells using a panel of antibodies does not show dramatic differences in the level of most proteins. Quantification of luminescence on a LICOR Oddesey FC relative to control knockdown cells is indicated. (b) Levels of renilla luciferase (RLuc) reporters containing the mbc 5′UTR and either the mbc or hsp70 promoters are reduced in DENR and MCT-1 knockdown cells. DNA reporters were transfected into S2 cells together with a firefly luciferase (FLuc) normalization control containing an hsp70 promoter. (c) In vitro transcribed RLuc reporter mRNAs containing mbc or actin 5′UTR, co-transfected with a control FLuc reporter into DENR, MCT-1 or control knockdown cells. mRNAs were capped with the normal m7G cap (“cap”) or with an adenosine cap structure (“A-cap”) which does not bind eIF4E. Below, representative raw counts (from the control) for the three reporters shows that the signal obtained with A-capped reporters is roughly one-half to one-quarter the signal observed with a normal m7G cap, and well above background. (d-d’) Reduced translation in DENR-KD cells of an mRNA luciferase reporter containing the mbc 5′UTR is not accompanied by a reduction in the intracellular levels of the reporter mRNA. In vitro transcribed mRNA consisting of the mbc 5′UTR, the renilla luciferase ORF and a synthetic poly(A) was cotransfected with a normalization control mRNA consisting of a short 5′UTR, firefly luciferase ORF and a synthetic poly(A) into control or DENR-KD cells. Relative luminescence counts (d) and relative mRNA levels determined by quantitative RT-PCR (d’) are shown. (e) Combined knockdown of DENR and MCT-1 does not have additive effects compared to the single knockdowns on translation of a luciferase reporter containing the mbc 5′UTR, consistent with them working together in one functional complex. (f-h) High resolution deletion series of the mbc renilla luciferase reporter, summarized in Fig. 2d, identifies nt 200-375 of the mbc 5′UTR as the minimum region required to impart DENR-dependent regulation. Error bars: std dev.
Extended Data Fig. 3: Detailed sequence information for the mbc 5′UTR
(a) Full sequence of the mbc 5′UTR. ATGs are indicated in bold. Open reading frames are shown as red or grey boxes depending on whether they have a good or bad Kozak sequences, respectively. Stop codons indicated with asterisks. (b) Schematic overview of the mbc 5′UTR and the tested reporter constructs, summarizing results from other panels. Light peach-colored shading indicates the minimal region required to impart DENR-dependent regulation to a luciferase reporter. Red and grey boxes indicate upstream Open Reading Frames (uORFs) with strong and weak Kozak sequences, respectively. ΔATG construct: single point mutations were introduced to abolish the three indicated uORFs. ΔKozak construct: Kozak sequences of the indicated uORFs were mutated to gtgtATG, thereby disfavouring translation initiation at these sites. Δstop construct: stop codons of all three in-frame uORFs were mutated, resulting in a construct where the uORF stop codon is just downstream of the RLuc ATG. (c) Translation extracts from DENR-KD cells are impaired in translating a reporter containing the mbc 5′UTR. mRNA reporters containing the full-length mbc 5′UTR, or a mutated ΔATG version where the ATGs at positions 218, 248, 338 and 415 were mutated, were prepared by in vitro transcription and introduced into translation extracts prepared from S2 cells treated with either control (GFP) or DENR dsRNA. Mutating the ATGs both increases the overall translation of the reporter, and renders the reporter insensitive to DENR knockdown. Statistics performed using the two-tailed, two-sided Student’s t-Test. (d) Full sequence of the 5′UTR in the mbc Δstop construct. Mutated stop codons are shown with underlining. The same sequence (nt 200-375) without the three point mutations is regulated by DENR (Fig. 2h).
Extended Data Fig. 4: Genome-wide analysis of stuORF transcripts
(a) Genome-wide histogram of transcripts containing uORFs in their 5′UTRs. (b-b’) Frequency distribution of nucleotides found surrounding the ATG of all main ORFs annotated in the fly genome. Frequency matrix in d’ was used to generate a multiplicative score for the strength of any Kozak sequence of interest. (c) Transcripts that were translationally down-regulated in S2 polysome profiles upon DENR knockdown are enriched for stuORF containing transcripts. A translation score was measured for each transcript in the genome, consisting of the ratio of transcript levels in 80S + polysome fractions (indicating active translation) divided by transcript levels in total mRNA, yielding the percentage of mRNA being actively translated. This was calculated for both control and DENR-KD S2 cells, and compared. Transcripts were then sorted into quintiles according to this comparison, with the first quintile containing transcripts that are most down-regulated in DENR knockdown cells compared to controls, and quintile 5 containing transcripts that were up-regulated in DENR-KD cells compared to controls. For each quintile, the average stuORF score was calculated, as described in the main text. A clear correlation can be observed, with down-regulated quintiles being enriched for transcripts containing stronger stuORF score. Error bars: SEM. (d) Analysis the other way around: stuORF containing transcripts are preferentially down-regulated in the polysome analysis from DENR-KD cells compared to transcripts containing no uORFs. Two categories of transcripts were selected genome-wide: the top 210 with high stuORF scores, and the bottom 6000 transcripts containing no uORFs. For each category of transcripts, histograms of the translation score were plotted, with negative numbers representing transcripts that were translationally down-regulated in DENR-KD cells compared to controls. Transcripts containing stuORFs are preferentially down-regulated genome-wide upon DENR knockdown in a highly statistically significant manner (Mann Whitney U-test p=10−14) compared to transcripts lacking uORFs. (e) Gene ontology enrichment using DAVID on stuORF-bearing transcripts. (f) Examples of genes predicted via the “DENR-dependence score” to be DENR-dependent in Drosophila. (g) DENR binds both mRNAs containing and lacking stuORFs. Cellular proteins were cross-linked with 0.5% formaldehyde and then DENR-containing complexes were immunoprecipitated. RNA was purified from the immunoprecipitation, reverse-transcribed with random hexamers, and subjected to quantitative PCR. Both transcripts containing stuORFs (mbc, InR) and transcripts containing no or weak uORFs (RpS13, cdc2, RpL32), as well as initiator tRNA, were found to be significantly enriched, compared to RNA from a control immunoprecipitation using anti-GFP antibody. Error bars: std. dev.
Extended Data Fig. 5: Timecourse of DENR knockdown reveals that changes in stuORF translation precede global changes in polysome profiles or ribosome levels
(a) Translation of a stuORF reporter (bearing 1x ATGTAA, as in Fig. 3) progressively drops upon DENR knockdown from 1-4 days after treatment with dsRNA. A significant drop in translation can be observed already 2 and 3 days after DENR knockdown. In contrast, a control reporter is unperturbed. (b) DENR protein levels 2 and 3 days after knockdown in S2 cells, assessed by immunoblotting. (c-c’) Polysome profile of S2 cells treated for 2 (c) or 3 (c’) days with DENR dsRNA (red traces) shows no significant drop in polysome fractions compared to cells treated with control (GFP) dsRNA (grey traces), suggesting that the drop in polysome levels observed at later time points (e.g. ED Fig. 1) are in vivo secondary consequences (e.g. due to reduced translation of stuORF containing transcripts such as Insulin Receptor, leading to a global reduction in translation rates). (d-d’) Quantification of 18S RNA, 28S RNA and initiator tRNA levels in S2 cells treated with dsRNA for 2 (d) or 4 (d’) days shows little to no increase upon DENR knockdown compared to control.
Extended Data Fig. 6: Loss of DENR leads to reduced InR and EcR protein levels and signaling in vivo – support data
(a-a’) DENR and MCT-1 knockdown in S2 cells does not lead to reduced EcR-B1 and InR mRNA levels. (b) DENR-KD cells are less sensitive to ecdysone (1 μM, 4 h), assayed by qRT-PCR of two EcR target genes BR-C (Fig. 4c) and E75 (shown here), normalized to rp49. (c-c’) DENRKO pupae 4 h APF have reduced levels of InR and EcR-A protein (c) but not mRNA (c’). (d) Expression of InR in histoblast cells and imaginal discs of DENRKO animals using escargot-GAL4 rescues their delayed pupation.
Extended Data Fig. 7: DENR promotes expression of an inducible 1 aa stuORF reporter in proliferating cells more efficiently than in quiescent cells.
(a) Schematic overview of a cell-based inducible Dual Luciferase Reporter Assay to enable systematic evaluation of the effect of DENR knockdown on expression of stuORF or control reporters under different growth states. (b) DENR knockdown in S2 cells leads to efficient depletion of DENR protein for up to 18 days post-dsRNA treatment. 4 days after treatment with dsRNA, cells were diluted into either suspension culture at a density of 106/mL (for Fig. 1g-i’) on in adherent culture (for the remaining panels in this figure). Graded loading of the GFP dsRNA control sample on the blot shows that DENR knockdown yields persistent depletion of DENR protein by ≥90%. (c) Growth curve post-dilution of S2 cells treated with or without dsRNAs, indicating optimal days for reporter induction that correspond to proliferating or quiescent populations (2 d or 7 d, respectively). Under proliferative conditions both DENR-KD and control cells double within ~24 h. Conversely, under quiescent conditions no significant increase in cell number is observed, implying that the population has essentially stopped proliferating. (d) Presence of a 1 aa stuORF (ATGTAA) reduces translation of a Renilla reporter by 50% in control (GFP-KD) cells. Error bars: SEM. Data from 2 independent KDs and 4 transfections. (e) Effect of DENR knockdown on the inducible 1 aa stuORF reporter expression is significantly stronger in proliferating compared to quiescent cells. Error bars: SEM; t-test p<0.0001. (f-f’) Raw luciferase counts for the experiment shown in (e) demonstrating that upon DENR knockdown, control reporters increase in expression (consistent with what is also observed in translation extracts, Fig. 2c’) whereas the stuORF reporter decreases in expression (f). (g) Representative raw luciferase counts for main Fig. 2i. (h-h’) DENR knockdown affects mbc protein levels more strongly in proliferating than in quiescent cells. DENR knockdown leads to reduced endogenous mbc protein (h) but not mRNA (h’) in proliferating but not quiescent S2 cells. Cells were treated with dsRNA for 4 d, then seeded in parallel at two different densities to achieve proliferation or quiescence at time of assay. Error bars: Std dev.
Extended Data Fig. 8: DENR is required to express stuORF containing transcripts in vivo in Drosophila
(a) Schematic of the fluorescent reporters introduced into Drosophila. Identical reporters were generated containing a Tubulin promoter, the 5′UTR of CG43674, which does not contain any uORFs, the GFP open reading frame, and the SV40 poly-A. A 6-nt stuORF (ATGTAA) was introduced by mutagenesis into the 5′UTR of one construct, generating the “stuORF reporter”. Both constructs were inserted via phiC31-mediated recombination into the VK33 landing site at 65B2 on chromosome 3L, ensuring identical transcriptional regulation. (b) The control reporter is well expressed in both larvae containing (DENR+/+) or lacking DENR (DENRKO). Immunoblotting of larval extracts (c) shows that GFP levels of the control reporter do not drop in DENR knockouts compared to controls, indicating that DENR is not required for translation of transcripts lacking uORFs. Introduction of the 6-nt stuORF causes the reporter to be less expressed in a control DENR+/+ background, consistent with uORFs having a repressive role in translation of the main downstream ORF. In contrast to the control reporter, expression of the stuORF reporter is entirely dependent on DENR in vivo, since no GFP expression could be observed when the stuORF reporter is introduced into a DENRKO genetic background. This was confirmed by immunoblotting (c). (c) Immunoblot to detect GFP and a loading control (awd) in larval extracts of animals bearing a control or a stuORF reporter, either in a control DENR+/+ or in a DENR knockout genetic background.
Extended Data Fig. 9: DENR activity in the Drosophila larva is higher in proliferating tissues compared to non-proliferating tissues
Expression of the stuORF GFP reporter is entirely DENR-dependent (ED Fig. 8), hence it serves as a readout for DENR activity. Flies were generated bearing either a control or a stuORF-GFP reporter, combined in trans with a normalization control RFP reporter, generated by replacing the GFP of the control reporter with RFP, and inserting it into the same VK33 landing site as the GFP reporters. This setup is analogous to the dual FLuc/RLuc setup used for luciferase assays and completely controls for transcriptional effects. Various tissues were dissected and imaged by confocal microscopy, the GFP/RFP ratio was calculated, and is displayed in pseudocolor. Flies bearing a control GFP reporter and the control RFP reporter have the same ratio of GFP to RFP in all tissues (a). For instance, the strong spot in the middle of the wing disc which is due to transcriptional effects, is present in both the GFP reporter and the RFP normalization control, and is normalized out when the GFP/RFP ratio is calculated. In contrast, the stuORF reporter (b) is more strongly expressed in proliferating tissues such as the brain and associated imaginal discs, or the wing disc, compared to non-proliferating tissues such as fat body or salivary glands. This can be observed both on the overlay, which is yellow for brain and wing discs, but red for fat body and salivary gland, as well as in the pseudo-colored GFP/RFP ratio panel which is high in brain and wing disc, but low in fat body or salivary gland.
Extended Data Fig. 10: DENR activity is higher in proliferating cells compared to quiescent cells
(a-b) DENR•MCT-1 binding is not different in proliferating versus quiescent S2 cells. (b) Immunoprecipitation of endogenous DENR from proliferating or quiescent S2 cells shows similar amounts of co-immunoprecipitating endogenous MCT-1 in the two conditions. (b) A stably-transfected S2 cell line bearing a copper-inducible pMT-V5-MCT-1 construct was grown in proliferative or quiescent conditions, as in ED Fig. 7, and then induced to express MCT-1. V5-tagged MCT-1 was immunoprecipitated and endogenous DENR was detected by immunoblotting. An anti-FLAG immunoprecipitation was performed as a negative control. Near-equal levels of DENR protein co-immunoprecipitated with MCT1 in both proliferative and quiescent conditions. (c) Expression levels of DENR and eIF2D/Ligatin do not depend on each other and do not change in proliferating vs. quiescent cells. Immunoblot of S2 cells treated with DENR, eIF2D/Ligatin or control (GFP) dsRNA, in proliferative or quiescent conditions. Two different amounts of the control samples were loaded to quantitatively assess knockdown efficiencies. A non-specific band on eIF2D blot is marked with a star (*). (d-d’) Abolishing phosphorylation on T118 and S119 blunts MCT-1 activity. (d) Luciferase assay using a stuORF reporter in S2 cells depleted of endogenous MCT-1 via dsRNA targeting its 3′UTR, and re-constituted to express wild-type or mutated forms of MCT-1 (using constructs with an exogenous 3′UTR) identifies T118 and S119 as important phosphorylation sites. Four sites were tested – T118, S119, T82 and T125 – for all of which phosphorylations on endogenous MCT-1 were observed by publicly available proteome-wide mass spectrometry analyses (PhosphoPep and PhosphoSite.org). Whereas wild-type MCT-1 is able to restore expression of the stuORF reporter, a T118A/S119A mutant form of MCT-1 cannot. Testing each site individually identifies S119 as the more important of the two. In contrast, mutating T82 and T125 to alanine does not impair their ability to promote stuORF translation. ** t-test<0.01. Error bars: std. dev. (d’) MCT-1[T118A/S119A] is expressed and stable. Immunoblot of S2 cells transfected with control plasmid or plasmid expressing wild-type FLAG-MCT-1 or FLAG-MCT-1[T118A/S119A] shows that the mutant MCT-1 is expressed and roughly equivalent levels as the wild-type protein. (e-f) Inhibition of Erk, PI3K, Akt or TORC1 does not have an effect on stuORF translation. Luciferase assay with a stuORF reporter in S2 cells treated with inhibitors for Erk (U0126, ), TOR-Complex 1 (rapamycin, e), Akt (Akt Inhibitor VIII, e) or PI3K (Wortmannin, e) shows that inhibition of any of these kinases has little to no effect on stuORF translation. (g) Knockdown of Drosophila cdc2 (CG10498) does not reduce expression of a stuORF reporter. S2 cells were treated with either GFP dsRNA or CG10498 dsRNA, and then transfected with a stuORF reporter. GFP dsRNA was applied to 90 wells in a 98-well plate and the values displayed represent the average and standard deviation (error bars). CG10498 knockdown results for two separate well are shown.