Abstract
Natural history museum collections provide unique resources for understanding how species respond to environmental change, including the abrupt, anthropogenic climate change of the past century. Ideally, researchers would conduct genome-scale screening of museum specimens to explore the evolutionary consequences of environmental changes, but to date such analyses have been severely limited by the numerous challenges of working with the highly degraded DNA typical of historic samples. Here we circumvent these challenges by using custom, multiplexed, exon-capture to enrich and sequence ~11,000 exons (~4Mb) from early 20TH century museum skins. We used this approach to test for changes in genomic diversity accompanying a climate-related range retraction in the alpine chipmunks (Tamias alpinus) in the high Sierra Nevada area of California, USA. We developed robust bioinformatic pipelines that rigorously detect and filter-out base misincorporations in DNA derived from skins, most of which likely resulted from post-mortem damage. Furthermore, to accommodate genotyping uncertainties associated with low-medium coverage data, we applied a recently developed probabilistic method to call SNPs and estimate allele frequencies and the joint site frequency spectrum. Our results show increased genetic subdivision following range retraction, but no change in overall genetic diversity at either non-synonymous or synonymous sites. This case study showcases the advantages of integrating emerging genomic and statistical tools in museum collection-based population genomic applications. Such technical advances greatly enhance the value of museum collections, even where a pre-existing reference is lacking, and points to a broad range of potential applications in evolutionary and conservation biology.
Keywords: Exon capture, DNA damage, museum skins, natural history museum collections, non-model organisms, Tamias
Introduction
Natural history museums worldwide house a wealth of biological specimens that document dynamic changes in biodiversity and are hence invaluable for reconstructing patterns and processes of evolution across time and space (Wander et al. 2007). In particular, museum specimens are potentially an important source for genetic data, especially those specimens collected before the molecular era (Payne & Sorenson 2002). Early successes in obtaining genetic data from dried museum skins applied PCR to obtain mtDNA sequences from recently extinct populations or species (Higuchi et al. 1984; Cooper et al. 1992). These studies aimed to examine genetic changes through time in threatened species (Wayne & Jenks 1991; Glenn et al. 1999; Weber et al. 2000; Godoy et al. 2004) or to enhance various conservation, phylogenetic and phylogeographic studies (Thomas et al. 1990; Roy et al. 1994; Poulakakis et al. 2008; Wójcik et al. 2010; McDevitt et al. 2012). Other studies have obtained multi-locus data by amplifying nuclear microsatellite loci from museum specimens (Taylor et al. 1994; Bouzat et al. 1998; Harper et al. 2008; Peery et al. 2010; Rubidge et al. 2012), though not without considerable effort to avoid PCR artifacts (e.g. Taberlet et al. 1996). However, the extraordinary potential of museum collections has yet to be fully realized because the extensive DNA degradation typical of these specimens limits the scalability of PCR-based approaches.
The recent advent of massively parallel, next-generation sequencing (NGS) has fundamentally transformed traditional population genetics into a data-driven discipline, which enables the analyses of genome-wide patterns of sequence variation with much greater reliability (Pool et al. 2010). Various target enrichment methods (Briggs et al. 2009; Hodges et al. 2009; Maricic et al. 2010; Lemmon et al. 2012; McCormack et al. 2012; Tang et al. 2012), which selectively enrich sequences prior to NGS, substantially increase the coverage of selected targets with much reduced experimental cost (Avila-Arcos et al. 2011; Good 2011). Among these, sequence (exon) capture methods have been successfully applied to ancient bone templates to obtain genome-scale data in order to provide insights into human evolution (Burbano et al. 2010; Bos et al. 2011). Such methods have also been used to discover exonic SNPs in non-model species by relying on pre-existing, closely related reference genomes (Perry et al. 2010; Cosart et al. 2011; Good et al. 2013). These studies demonstrate the feasibility of conducting targeted genome-scale analyses from a broad range of DNA sources. However, all of these examples rely heavily on pre-existing, high quality, genomic resources to target regions of interest and to align the resulting sequences for variant discovery, a requirement that impedes application to the wide range of taxa still lacking a genomic reference.
To achieve broader, cost-effective applications, we previously showed that de novo assembled transcriptomes can be used to design exon-capture experiments, and combined this with pooled capture of barcoded libraries (Burbano et al. 2010) to enable cost-effective genomic sampling from modern samples spanning a reasonable taxonomic breadth (Bi et al. 2012). In parallel with such technical advances, powerful statistical methods are emerging to infer single nucleotide polymorphisms (SNPs), estimate allele frequencies, and jointly infer population demography and selection from NGS data (Yi et al. 2010; Nielsen et al. 2012). What remains is to demonstrate that these new approaches can be applied to museum specimens and yield high quality population genomic data despite the inevitable DNA damage involving fragmentation and base misincorporation, as well as potential contamination.
Here, we provide such a demonstration by comparing population samples of the alpine chipmunk (Tamias alpinus) obtained in the early 20TH century (museum skins) and from the same areas nearly a century later (fresh tissues). This species is endemic to the high Sierra Nevada of California and is of particular interest because its range has strongly contracted upwards over the past century in conjunction with increasing minimum environmental temperatures (Moritz et al. 2008; Rubidge et al. 2011). Using seven DNA microsatellite loci, Rubidge et al. (2012) found a decrease in allelic richness, but not heterozygosity, and increased population differentiation in modern compared to historic specimens of T. alpinus from Yosemite National Park (YNP), California, USA. These data indicate that range retraction was mainly caused by extirpation of low-elevation populations rather than population-wide range-shifts to track climatic niches (Tingley et al. 2009). Nevertheless, a small set of microsatellite markers may not sufficiently reflect underlying genome-wide diversity (Väli et al. 2008), and thus will not represent the full spectrum of genomic consequences of range retraction.
In this study we aimed to identify and compare variation across the genome from YNP T. alpinus that were collected in both historic (1915) and contemporary periods (2004–2008), and gain insight into how climate change in the past century has affected their genomic variability. Specifically, we first used de novo transcriptome-based exon capture (Bi et al. 2012) to successfully obtain genomic data from museum specimens. Second, we developed bioinformatic pipelines that rigorously detect and filter out artifacts from DNA damage and low-confidence variants. To call SNPs and estimate allele frequencies, we applied a probabilistic framework that takes into account a wide range of statistical uncertainties associated with low-medium coverage data (Nielsen et al. 2011; Nielsen et al. 2012). Finally, we used these high-quality variant data to compare patterns of genomic variation between the historic and modern specimens.
Materials and Methods
Samples
We used specimens collected from T. alpinus populations along the west slope of the Yosemite transect, at elevations ranging from 2,377 to 3,277 meters (m) (Fig. 1, Table S1). We chose 20 historic specimens that were sampled in 1915 at YNP by a research team led by Joseph Grinnell, the founding director of the Museum of Vertebrate Zoology (MVZ) at the University of California Berkeley. Historic specimens are preserved in the form of dried skins. Twenty modern specimens were sampled from the same area by the “Grinnell Resurvey” team initiated by MVZ researchers and collaborators between 2004 and 2008 (http://mvz.berkeley.edu/Grinnell/). Fresh tissues were stored at −80 °C or in 95% ethanol. The individual sample used for transcriptome sequencing was a male T. alpinus (MVZ224483) from Bullfrog Lake, Kings Canyon National Park, California, collected in November 2009. RNA was extracted from liver, kidney, spleen, and heart tissues that were fixed in RNAlater immediately following euthanasia.
Fig. 1. Sampling localities for the present study.
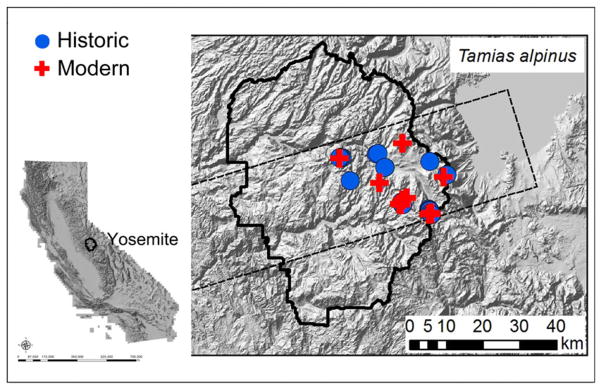
Historical sampling localities (1915) are shown in filled blue circles and modern (2004–2008) in filled red crosses. The range of Yosemite National Park (YNP) is outlined by a solid black line. The YNP transect is outlined by dashed lines. The inset shows the state of California.
DNA extractions from skins
We confined DNA extractions from museum skins to a separate lab used exclusively for historic DNA related lab work. We minimized damage to museum skins by sampling a small (2 × 2 mm) piece of toe pad tissue from each skin using a sterilized surgical blade. The skin was then kept in a 1.5mL centrifuge tube, and twice rehydrated in a 1x STE buffer (100mM NaCl, 10mM Tris-HCl pH7.5, 1mM EDTA) for 30 minutes followed by 3 minutes of rigorous vortexing. After washing, each skin was cut into small pieces inside the same tube using a straight and narrow-headed surgical blade. We used reagents provided in Qiagen DNeasy Blood & Tissue kits but purified the DNA using Qiagen PCR purification columns. PCR purification columns are more efficient at collecting the small fragments (Rowe et al. 2011), which predominate in DNA samples derived from historic specimens. DNA extractions from modern specimens were carried out using DNeasy Blood & Tissue kits following the manufacturer’s protocol.
Array design and exon capture experiments
We recently outlined an approach for using de novo assembled transcriptomes to enable cost-effective exon capture experiments across a reasonable taxonomic breadth (Bi et al. 2012). Briefly, we generated a cDNA library from combined mRNA extracted from multiple tissues of a modern T. alpinus specimen following standard procedures. The resulting cDNA library was then sequenced using one lane of Illumina GAIIx (100bp paired-end), assembled using ABySS (Birol et al. 2009), and then annotated based on BLASTX (Altschul et al. 1990) comparisons to a database of human, mouse, rat, and thirteen-lined ground squirrel proteins. Detailed information for the bioinformatic pipelines used for de novo transcriptome data processing can be found in Singhal (2013). We used the Agilent SureSelect custom 1M-feature microarrays to target 11,975 exons that were each at least 201bp long, which we identified from the annotated transcripts. Targeted exons, spanning a wide range of evolutionary rates in the genome (Bi et al. 2012), represented 6,249 protein-coding genes with a target size totaling 4Mb. In addition to the T. alpinus exons, we targeted the ~16 kb T. alpinus complete mitochondrial genome and 350 bp from the consensus Spermophilus (S. fulvus, S. major, and S. pygmaeus) SRY gene to assess empirical error rates and potential sample contamination (Bi et al. 2012). We also selected seven nuclear genes (acrosin, acp5, cmyc, rag-1, anon, zan, and zp2) previously sequenced in Tamias (6,031 bp total) (Good et al. 2008; Reid et al. 2012), which were used as positive controls in post-capture qPCR assays to determine the initial enrichment quality. Array probe design followed the recommendations of Hodges et al. (2009) and are detailed in Bi et al. (2012).
Genomic libraries for 40 specimens, 20 early 20TH century museum skins and 20 modern tissues, were constructed according to the protocol outlined by Meyer & Kircher (2010) with slight modifications: First, we sheared DNA from all modern specimens and DNA from a few historic specimens in which some high molecular weight DNA fragments were still present. Shearing DNA for most of the historic skins was not needed since fragments ranged from 100–300bp, which is the optimal size range for exon capture experiments. The adapter of each genomic library was associated with a unique 7-nt barcode introduced by indexing PCR. All libraries were amplified with Phusion High-Fidelity DNA Polymerase (Thermo Scientific) during the indexing PCR. For each individual sample we performed 2–3 indexing PCR reactions in parallel and then merged the products thereafter to decrease the PCR stochastic drift. Barcoded libraries for historic and modern T. alpinus were pooled separately in equal amounts and hybridized on one array each along with Tamias Cot-1 DNA (prepared following Trifonov et al. 2009) and blocking oligos. In addition, we multiplexed individually barcoded genomic libraries from four modern specimens of red-tailed chipmunks, T. ruficaudus, and hybridized them on an independent microarray. Tamias ruficaudus served as an outgroup to T. alpinus such that the orientation of SNPs could be polarized (ancestral vs. derived) to obtain the unfolded site frequency spectrum (SFS) (see below). Assays using qPCR and the positive control loci targeted on the same arrays allowed us to determine the post-capture enrichment efficiency of different exon capture experiments. The verified, enriched libraries were sequenced using an Illumina HiSeq2000 (100 bp paired-end). The two libraries, one consisting of 20 pooled historic and the other of 20 pooled modern specimens were each sequenced on one lane. The pooled library of T. ruficaudus was sequenced using 1/3 of a lane.
Data filtering
The captured sequence reads from T. alpinus and T. ruficaudus libraries were cleaned and assembled using the same strategy described in Singhal (2013). Briefly, raw sequence reads were filtered to remove exact duplicates, adapters, bacteria and human contamination, as well as low complexity. Overlapping paired reads were also merged. The resulting cleaned reads of the 40 libraries of T. alpinus were then assembled together to generate a final set of consensus assemblies. A reciprocal BLAST approach (Altschul et al. 1990) was applied to compare the 11,975 target exons with the consensus assemblies in order to identify the set of contigs that were associated with targets (in-target assemblies). To correct potential assembly errors that might have been introduced by de novo assembly and/or by merging raw assemblies, we used Novoalign (http://www.novocraft.com) to align cleaned reads from the T. alpinus libraries to the in-target assemblies and then corrected sites where consensus bases were not concordant with identified major alleles. We also mapped sequence reads of 4 T. ruficaudus individuals to the corrected T. alpinus in-target assemblies and used “bcftools” and “vcfutils.pl vcf2fq” implemented in SAMtools (Li et al. 2009) to generate an outgroup sequence file. We mapped sequence reads from each individual T. alpinus library to the in-target assemblies of T. alpinus, using Novoalign while constraining the number of mismatched positions to 3 or less per read pair. The output alignments in SAM format were initially analyzed using SAMtools and its associated “bcftools” to produce some of the data quality control information in VCF format. These data were then further filtered using a custom filtering program, SNPcleaner (https://github.com/fgvieira/ngsClean).
We employed three levels of filtering on the datasets in a hierarchical order: individual-level, contig-level, and site-level. The filters in each step of the hierarchy were applied only to the subset of data that passed the quality control thresholds at all previous levels. The first filters applied were the individual-level filters to remove entire individuals deviating excessively from the average across-individual coverage and error rate. Contig-level filters, followed by site-level filters were then applied to remove entire contigs and sites, respectively, that appeared to be quality outliers. All individual specimens, contigs and sites were filtered on multiple aspects of quality (e.g. potential cross-sample DNA contamination, sequencing errors, paralogy, etc.). The detailed protocol is specified below:
1. Filtering at individual level
Remove individuals having extremely low or high coverage (<1/3 or > 3X the average coverage across all individuals).
Remove individuals with excessively high sequencing error rates measured as the percentage of mismatched bases out of the total number of aligned bases in the mitochondrial genome. Remove females with a high percentage of cleaned reads being mapped to the SRY gene, and males with extremely low SRY mapping percentage relative to the across-male average.
2. Filtering at contig level
Remove contigs that show extremely low or high coverage based on the empirical coverage distribution across contigs.
Remove contigs with at least one SNP having allele frequencies highly deviating from Hardy-Weinberg equilibrium expectations (p < 0.0001) based on a two-tailed exact test (Wigginton et al. 2005).
3. Filtering at site level
Remove sites that are at least 100bp outside of exons.
Remove sites with excessively low (<19.5 percentile for historic and <18.5 percentile for modern) or high (>99.3 percentile for historic and >99.4 percentile for modern) coverage based on the empirical coverage distribution.
Remove sites with biases associated with reference and alternative allele Phred quality, mapping quality, and distance of alleles from the ends of reads. Also remove sites that show a bias towards sequencing reads coming from the forward or reverse strand.
Remove sites for which there are not at least 3/4 of the individuals sequenced at 3X coverage each.
Remove sites with a root mean square mapping quality for SNPs below 10.
Due to the high base misincorporation rate present in the historic specimens, we remove sites from all 40 individuals for which C to T, and G to A SNPs are identified.
Retain only the sites that pass all filters for both historic and modern specimens so that the modern and historic data are directly comparable.
The DNA damage filter, whereby C to T and G to A SNP sites are removed, is essential for genomic studies that use museum collections. DNA preserved from archaeological (or ancient) and historic museum specimens is often characterized by various types of post-mortem nucleotide damage (e.g. Stiller et al. 2006; Briggs et al. 2007). The majority of the damage-driven errors associated with ancient DNA are caused by hydrolytic deamination of cytosine (C) to uracil (U) residues (Hofreiter et al. 2001; Briggs et al. 2007). Most DNA polymerases replace uracils with thymines (Ts) during PCR amplification of damaged templates, resulting in an apparent C to T substitution. These spurious base misincorporations tend to occur towards the ends of molecules (Briggs et al. 2007), appear to accumulate with time, and can be significantly elevated even in century-old museum specimens (Sawyer et al. 2012). Population genetic inference based on damaged, ancient DNA sequences can be severely biased due to the base misincorporations (Axelsson et al. 2008) and should be examined before all downstream analyses. In order to characterize the patterns of DNA damage likely present in our dataset, we used a similar method to that of Briggs et al. (2007) whereby we plotted the frequency of all 12 possible mismatches against the distance from the 5′-end and 3′-end of the sequencing reads in modern and historic specimens, respectively.
SNP calling and allele frequency estimation
With low-medium coverage data (<20X per individual in the present study), SNP and genotype calls based only on allele counting, even after considering quality scores and coverage, are associated with high uncertainty. This can lead to biased estimates of allele frequency distributions and population genetic parameters (Hellmann et al. 2008; Johnson & Slatkin 2008; Lynch 2008). To account for this uncertainty, we called SNPs and estimated allele frequencies using an empirical Bayesian framework implemented in the software ANGSD (http://popgen.dk/wiki/index.php/ANGSD). Briefly, for each individual we first calculated the likelihood for all 10 possible genotypic configurations at every site that passed quality filters. We then used a maximum likelihood method to estimate the site frequency spectrum (SFS) based on the genotype likelihoods, jointly for all sites and all individuals (Nielsen et al. 2012). This was performed separately for the historic and modern population samples. The resulting population-specific SFS was used as a prior to estimate the posterior probabilities for all possible derived allele frequencies at each site in each population, which directly allowed for SNP calling at any degree of certainty. We called SNPs based on the sites having a 95% probability of being variable. A detailed description of the algorithms employed by ANGSD can be found at http://popgen.dk/wiki/index.php/ANGSD and in Nielsen et al. (2012).
Population genetic analyses
Several population genetic parameters and summary statistics of interest were calculated using the SFS. Overall genetic variation was estimated using nucleotide diversity measured as the average number of pairwise differences between sequences, π. We also estimated Watterson’s theta (Θw) (Watterson 1975) for non-synonymous (Θnon-syn) and synonymous SNPs (Θsyn) respectively. We calculated the ratio Θnon-syn/Θsyn for historic and modern populations to examine if overall genetic diversity of non-synonymous relative to synonymous sites has changed over time. We used Tajima’s D (Tajima 1989) to examine whether there was a genomic signal of past population expansion or recent decline. All of these summary statistics π, Θw, and Tajima’s D were calculated by summing over all possible allele frequencies weighted by their posterior probabilities in order to account for sequencing uncertainties.
Differentiation between the modern and historic population was measured using an estimation of FST appropriate for low coverage NGS data, while genetic relationships among the individuals in the two time periods was analyzed using principal components analysis (PCA) based on a covariance matrix of posterior genotype probabilities at every site with a minor allele frequency of at least 1.25% among all 40 individuals (Fumagalli et al. 2013). These methods are implemented in the ngsTools software package (https://github.com/mfumagalli/ngsTools). For higher resolution in the differentiation analysis we also examined the 2D-SFS between the modern and the historic population to infer shifts in allele frequencies over time. We further examined population structure using the Bayesian population clustering method implemented in Structure 2.3.4 (Pritchard et al. 2000). We used an admixture model with correlated allele frequencies and five independent runs, with each run having a burn-in of 100,000 followed by 1,000,000 MCMC (Markov Chain Monte Carlo) steps from K (number of clusters) = 1 to 10. Results across runs were summarized to determine the best K using the method of Evanno et al. (2005), implemented in Structure Harvester (Earl & vonHoldt 2012). Structure results were further analyzed and plotted using CLUMPP1.1.2 (Jakobsson & Rosenberg 2007) and DISTRUCT (Rosenberg 2004).
Results and Discussion
Transcriptome based exon capture on museum specimens
Although there has been a long history of utilizing genetic data from museum specimens, most studies have been restricted to PCR or capillary sequencing of short fragments of mitochondrial DNA, individual nuclear loci, and microsatellite loci (reviewed in Wandeler et al. 2007). The considerably high sensitivity and high throughput of NGS provides unprecedented opportunities for phylogenomic and population genomic applications based on museum collections. Rowe et al. (2011) used Illumina technology to sequence the genome from the skin and bone tissues of one historic mammal specimen. More recently, cross-species targeted hybridization with NGS was used to sequence complete mitochondrial DNA genomes from museum specimens (Mason et al. 2011; Guschanski et al. 2013). For our study, we aimed to obtain genome-wide nuclear markers for population-level comparisons between museum historic and contemporary specimens. We first generated genomic resources by de novo sequencing and assembling a multi-tissue transcriptome from a modern chipmunk, and then designed exon capture microarrays to enrich selected targets from individually-indexed, pooled, DNA derived from both historic skins and fresh tissues. Here we demonstrate that this is a fast and cost-effective approach for enriching genome-wide nuclear markers from low quality DNA typical of historic skins with sufficient sequencing coverage to characterize population genomic diversity. Our method is especially valuable for non-model organisms including those comprising the vast majority of museum collections for which a pre-existing reference genome is not usually available.
We used a multi-tissue transcriptome sequencing approach to increase the transcript diversity and ultimately the robustness of the marker development. We suggest that future projects should also consider exploring sequence resources from various other tissue types since the organs chosen in this study do not necessarily represent the best combination for maximizing the transcript diversity. For example, the testis transcriptome is typically more complex than other tissues (Soumillon et al. 2013).
De novo assembly of the Tamias transcriptome yielded 37,563 contigs (36.5 Mb), 21,262 (28.1 Mb) of which were annotated, with a mean length of 1,297 bp and an average fold coverage of 54X (Bi et al. 2012). Over 120,000 exons were identified from the annotated transcripts. 962,438 60bp probes were printed on the microarrays to target 11,975 exons that were greater than 200bp each in length (4 Mb total), the Tamias mitochondrial genome (16 Kb), the ground squirrel (Spermophilus) SRY gene (350 bp), and seven control nuclear genes (6 kb). Further details on probe and array design can be found in Bi et al. (2012).
The exon capture experiments were successful, as detailed in Table S2. For historic and modern T. alpinus specimens, 32.2 and 43.6 Gb (NCBI SRA ID: SRR847500) raw data were generated, and 14.9 and 21.8 Gb high-quality data were retained after quality filtering, respectively. Exogenous DNA contamination derived from bacteria and/or human was trivial (<0.3% for both libraries) and removed from the dataset (Table S2). De novo assembly of cleaned sequences produced 10,583 contigs (total assembly size: 7.6 Mb, N50: 722bp, mean contig length: 715bp); the identity of contigs from the in-target assemblies was determined by reciprocal BLAST to the selected exons. For all individuals, an average of 99% of the target exon bases were covered by at least one sequence read.
We found that, on average, 46.7% and 33.8% of the cleaned reads aligned to the targets for historic and modern individuals, respectively (Fig. S1). The mitochondrial genome only made up 0.4% of the total target (16Kb out of 4Mb) but among all of the reads that were mapped to targets, 6–7% were derived from the mitochondrial genome. This effect is not surprising given that there are typically several hundred copies of the mitochondrial genome per diploid cell (Robin & Wong 1988). Indeed, this inherent difference in copy number is one of the primary reasons that studies on ancient DNA have traditionally relied upon sequencing of mitochondrial DNA (e.g. Krings et al. 2000). Given this bias, including both mitochondrial and nuclear targets in the same capture experiment will inevitably result in some loss in overall sequencing efficiency. Here we included mitochondrial DNA probes specifically haploid targets that can be used as an internal control against for cross-sample contamination and recombinant PCR, and to provide an empirical estimate of sequencing and postmortem error rates (see Bi et al. 2012). We intentionally used a five-fold lower tiling density for mitochondrial relative to nuclear targets (20 versus 4-bp tiling) to help counteract the copy number bias, though our sequencing results indicate that a much lower tiling density is warranted. We recommend that future studies carefully consider the balance between quality control insights afforded by including mitochondrial targets and the potential loss in experimental efficiency. Unless targeting a large portion of the mitochondrial genome is of particular interest, it is likely that targeting a smaller subset of the mitochondrial genome and/or other haploid genetic markers (e.g., X and Y-linked loci in males) could provide sufficient internal controls while avoiding loss in overall experimental efficiency.
The higher specificity (proportion of cleaned reads that were mapped to targets) for historic individuals is noteworthy, given that degraded samples are notorious for performing poorly in PCR-based experiments. This might be because the shorter average DNA library sizes of the historic specimens (~150bp versus ~200bp for the modern samples) resulted in more efficient enrichment by the 60-bp probes of the Agilent SureSelect capture arrays. Furthermore, the specificity was highly uniform in each population, indicating that multiplexing did not bias the capture efficiency towards particular libraries. The average base fold coverage within exons was 18.9X in historic and 19.7X in modern individuals (Fig. S2). For all individuals, on average, 99% of the exons had coverage of at least 5X, and 90% had coverage of at least 10X.
The average fold coverage of each captured exon in historic and modern specimens was highly correlated (Fig. S3), suggesting consistent performance between independent capture experiments despite half the data being derived from century-old museum skins. The total length of contigs in the in-target assemblies was much greater than that of exons originally targeted due to the capture of flanking intronic sequence. However, there was an abrupt decrease in the average fold coverage near the edges of each contig making these regions more prone to assembly errors. To ensure high confidence in downstream variant calling, we only considered SNPs identified in flanking regions covered by reads that overlapped with a contiguous exon by at least 1 bp.
Data filtering and pattern of base misincorporation in historic DNA
We first applied individual-level, quality-control filters to the datasets to remove any data introducing severe bias among individual specimens. The Spermophilus SRY gene was effectively captured in all male Tamias samples. In contrast, the capture of SRY was negative in females considering that only five samples had only one to two sequence reads that were mapped to the SRY gene, indicating a negligible degree of cross-sample contamination (Table S2). We did find that the error rate was relatively greater in historic specimens (see below). However, there were no strong deviations in empirical error rate or sequencing coverage for any individual compared to the respective population average. Therefore, all 40 T. alpinus and 4 T. ruficaudus specimens passed the quality control filtering at the individual level.
The contig-level filters were subsequently employed to filter entire contigs from specimens that passed individual-level filters. The in-target assemblies of T. alpinus and T. ruficaudus were examined for potential assembly errors, resulting in 1,193 and 552 sites being corrected, respectively. After filtering, 1,334 contigs (12.6% of the total) were removed because they either showed extreme coverage or possessed SNPs that deviated significantly from HWE, which is indicative of paralogs.
Various site-level filters were lastly carried out to remove unreliable sites belonging to the contigs that passed contig-level filters. The empirical error rate was almost fivefold higher in historic (0.19%) versus modern (0.04%) samples and might be caused by the presence of excessive miscoding lesions that caused incorrect bases to be incorporated during PCR amplification (Sawyer et al. 2012). To examine this further, we plotted the frequency of all 12 possible mismatches against the distance from the 5′-end and 3′-end of the sequencing reads in modern and historic specimens, respectively (Fig. 2). For the modern specimens, the frequency of all changes remained constant and similar along the sequences, while there were excessive C to T substitutions at the 5′ ends of the historic sequences and complementary guanine (G) to adenine (A) substitutions at the 3′ ends of the respective molecules. The frequencies of C to T and G to A substitutions were ~4 fold above other possible mismatches at the 5′- and 3′-most positions, respectively. The occurrence of both misincoporation-type substitutions rapidly decreased over the first 5–10 bp from the ends of the read and then steadily decreased toward the opposite end of the sequence. However, compared to other changes, their frequencies remained elevated throughout the sequence.
Fig. 2. Patterns of mismatches in historic and modern Tamias alpinus sequences.
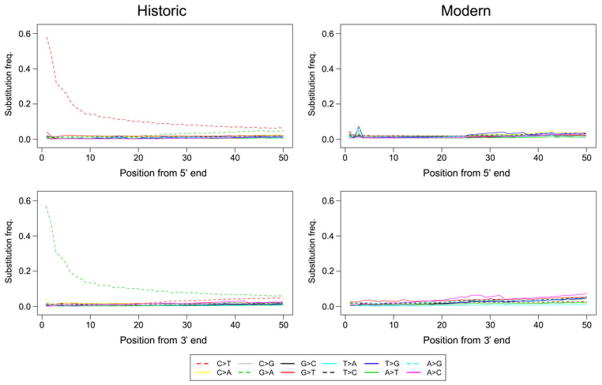
The frequencies of the 12 types of mismatches (y axis) are plotted as a function of distance from the 5′ and 3′-ends of the sequence reads (x axis). The first 50bp of the sequence reads are shown. The frequency of each particular mismatch type is calculated as the proportion of a particular alternative (non-reference) base type at a given site along the read, and is coded in different colors and line patterns explained at the bottom of the plots: “X > Y” indicates a change from reference base type X to alternative base type Y.
Overall, we observed a pattern of base misincorporation in our historic samples that is in strong agreement with patterns of damage accumulation characteristic of ancient DNA (Briggs et al. 2007). We prepared our historic DNA libraries with a Pyrococcus-like polymerase (Phusion High-Fidelity DNA Polymerase) that stalls amplification of templates containing uracil (Greagg et al. 1999; Heyn et al. 2010) and thus should reduce (but not eliminate) misincorporation errors in our sequence data (Ginolhac et al. 2011). Consistent with this, the frequency of C to T misincorporation at the end of molecules is nearly an order of magnitude lower in our data (~0.58%; Fig. 2) when compared to sequences of DNA libraries from similarly aged museum samples that were prepared with a non-proofreading enzyme (2–4%; Sawyer et al. 2012). This apparent improvement in sequence fidelity comes with two important caveats. First, proofreading enzymes that are stalled by the presence of uracil may be poorly suited for use on highly damage samples, where up to 60% of DNA fragments may contain at least one uracil (Briggs et al. 2010). Second, substantial C to T and G to A misincorporations still persist in our data (Fig. 2) despite the use of a proofreading enzyme that should not amplify molecules containing uracil. One possible source of error is that library amplification occurs after adaptor ligation and therefore will not prevent G to A errors created during blunt end repair of 5′ single stranded fragments. Another likely explanation is that not all instances of cytosine deamination result in uracil. For example, deamination of a methylated cytosine causes a direct transition to a thymine that will not be detected by proofreading polymerases and may account for up to 10% of misincorporations in ancient DNA (Briggs et al. 2010). Regardless of the source(s) of error, population genetic and demographic analyses based on damaged, ancient DNA sequences can be severely biased by base misincorporation (Axelsson et al. 2008). To accommodate this, we removed all C to T and G to A changes from both historic and modern datasets for subsequent analyses.
After the site-level filtering, we retained 3 Mb data out of the total 4.4 Mb of pre-filtered data which included exonic regions and partial flanking sequences (within a +/−99 bp window).
SNP calling and population genetic analyses
As the sequencing coverage of our data was not adequate to naively call genotypes for each individual with high confidence by counting alleles, we used a SNP calling and allele frequency estimation method based on genotype likelihoods that uses information from all individuals at all sites simultaneously (Yi et al. 2010; Nielsen et al. 2012). This approach greatly increased the statistical power to detect SNPs and improved allele frequency estimations compared to individual-based SNP calling algorithms. Using ANGSD, we were able to identify 1,578 high quality sites with at least a 95% posterior probability of being variable. Among these SNPs, 1,250 were found in historic specimens and 1,277 in modern specimens, meaning that the number of segregating sites has remained fairly constant in YNP T. alpinus over the past century.
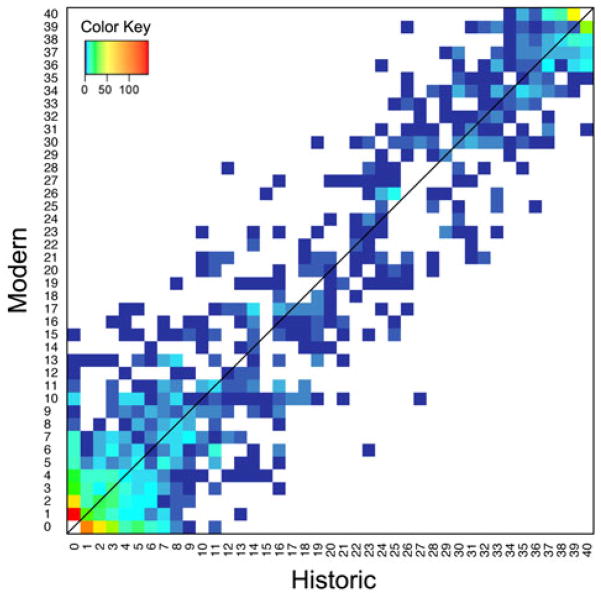
Comparison of the SFS for the temporal T. alpinus populations suggests minimal changes in allele frequencies over the period of climate change. Accordingly, the 2D-SFS showed a strong correlation between the modern and historic population allele frequency distributions (Fig. 3). This is reflected in the low global Fst (0.023) between the two time period populations. The genome wide per site nucleotide diversity (π) (Fig. 4) and Tajima’s D for the historic population were 8.179e-05 and −0.026, respectively; while for the modern population, per site π and Tajima’s D were 8.738e-05 and 0.0487. This result clearly indicates that there was minimal change in genomic diversity for these sampled populations from two time points spanning 90 years (~90 generations). In general, observable changes in allele frequencies and reduced genetic diversity requires that the number of generations during which a population experiences a reduced population size (t), should not be much smaller than the effective size of the reduced population. The expected ratio of heterozygosity for a population of size Nb before a bottleneck to a population of size Na t generations after a bottleneck, is e−t/Na((et/Na −1)Na +Nb)/Nb. So if t/Na is small (say <0.1), the bottleneck has very little effect on genetic diversity.
Fig. 3. Two-dimensional unfolded site frequency spectrum (2D-SFS) for SNPs between historic (x axis) and modern (y axis) Tamias alpinus specimens.
The color of each data point represents the number of SNPs belonging to the particular category in the 2D-SFS, which is depicted in the color key inset.
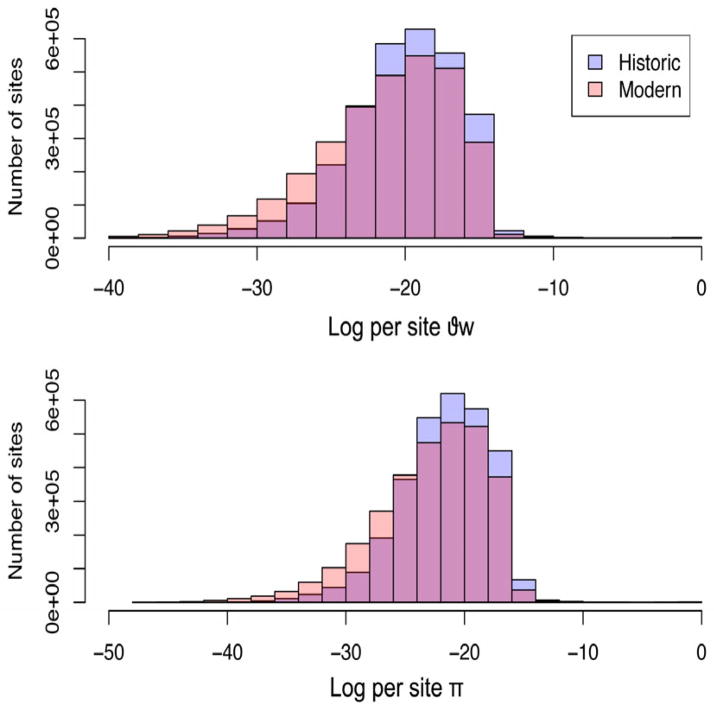
Fig. 4. Distribution of genome wide per site average pairwise nucleotide diversity (π) and Watterson’s theta (Θw) for historic and modern Tamias alpinus populations.
Histograms showing the frequencies of the log-transformed per site values of Θw (top) and π (bottom) calculated using allele frequencies weighted by their posterior probabilities for each of the 2,997,745 sites that passed quality control filters. The historic population is depicted in lavender and the modern population is in light pink as shown in the legend. The magenta portion of bars represents the overlap between the historic and modern population theta values.
Using seven microsatellite loci, Rubidge et al. (2012) surveyed 88 historic and 146 modern T. alpinus specimens and also found no change in heterozygosity in YNP temporal populations over the past century. However, they did find a significant decrease in allelic richness in modern samples. This result may not be surprising since rare alleles are lost at a greater rate than that at which heterozygosity declines after a bottleneck (e.g. Nei et al. 1975; Roderick & Navajas 2003). In this study we targeted a much smaller sample size (20 individuals in each era) and found approximately the same distribution of allele frequencies in historic and modern samples (similar values of Tajima’s D). The discrepancy between the two studies may be explained in terms of differences in sampling as we may lack power to identify rare alleles with our relatively smaller sample sizes.
We annotated and classified SNPs based on whether they are coding (synonymous and nonsynonymous) or noncoding (untranslated regions (UTRs), introns). Assuming that non-synonymous mutations tend to be more deleterious than synonymous substitutions, it is expected that Θnon-syn/Θsyn would increase in populations that undergo a size reduction because the effect of drift would overcome that of purifying selection. Thus, we might expect to see a greater Θnon-syn/Θsyn ratio in the modern population compared to the historic population of T. alpinus. However, we failed to detect such a signal; results showed that Θnon-syn/Θsyn was nearly identical for historic (0.976) and modern populations (0.986). This consistency also indicates that our data filtering pipelines worked effectively in terms of eliminating potential errors from the datasets. Our results suggest that if the T. alpinus population size is declining, the decline has not lasted long enough to leave a discernible genetic signal.
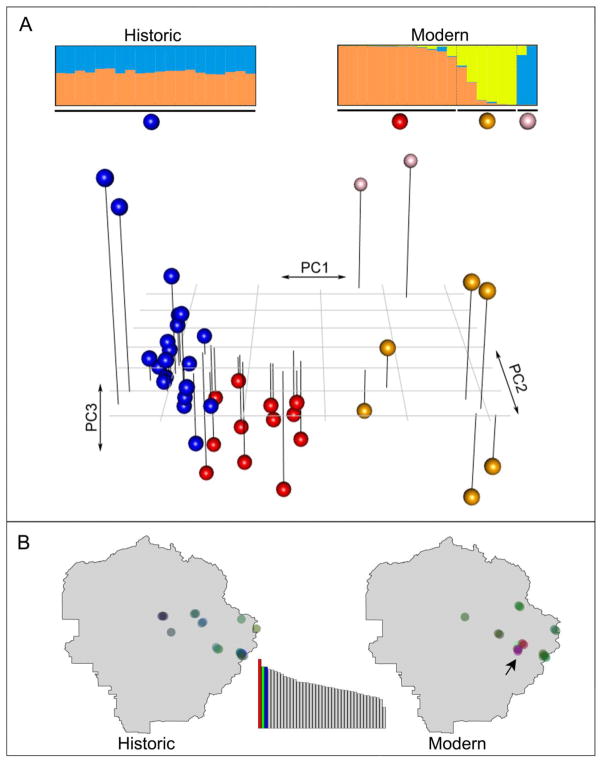
The PCA analysis (Fig. 5A) based on genotypic covariance showed that historic individuals were more genetically similar to each other than modern individuals, indicating increased genetic structure within the modern population. This result may be due to reduced gene flow among increasingly isolated modern T. alpinus subpopulations, in contrast to more widespread historic gene flow. The signal of genetic heterogeneity in modern specimens was mostly driven by individuals collected from a few geographically close localities near the Vogelsang, Evelyn, and Fletcher lakes (Fig. 5B). Individuals sampled from the remaining localities formed one genetic group. This pattern was concordant with Structure results: there was no population subdivision detected in historic specimens, while three genetic clusters (K=3; Evanno et al. 2005) were identified in modern samples. Targeting a much larger sample size, Rubidge et al. (2012) used seven microsatellite loci and detected more (K=4) genetic structure in modern T. alpinus. Thus, our results generally agree with Rubidge et al. (2012) in terms of finding increased population genetic structure in modern T. alpinus. The lack of resolution in the clustering analysis is most likely caused by our relatively modest population-level sampling since our current study is focused primarily on demonstrating the feasibility of generating high quality population genomic data from historic museum specimens. It was believed that the extinction of individuals inhabiting lower elevations could have caused the disruption of genetic connectivity among higher elevation localities, and was therefore responsible for increased genetic structure in modern T. alpinus (Rubidge et al. 2012).
Fig. 5. Genetic structure of historic and modern Tamias alpinus specimens.
(A) Principal Components Analysis (PCA) plot based on genetic covariance among all the 40 T. alpinus individuals. The first 3 principle components (PCs) are shown and cumulatively explain 10.58% of the total genetic variance (3.72%, 3.50%, and 3.36% for PCs 1, 2 and 3, respectively): The proportion of the genetic variance explained by the PCs is shown in the inset barplot at the bottom, where each bar is a PC and the y-axis is the proportion of variance explained (the first 3 PCs are colored). Each sphere in the PCA plot represents an individual specimen. Non-blue spheres are modern specimens and blue spheres are historic specimens. The corresponding 2D-PCA plots (PC1 vs PC2, PC1 vs PC3 and PC2 vs PC3) are shown in Fig. S4. The barplots at the top are Bayesian clustering results for historic (K=2, left) and modern (K=3, right) T. alpinus generated by the program STRUCTURE. Each individual specimen is represented by a horizontal line partitioned into colored segments that indicate the cluster membership. Spheres labeled at the bottom of the plots correspond to those from the PCA plot. (B) Each point on the Yosemite map corresponds to an individual T. alpinus sample and the color represents the relative contributions from the first three PCs (red, green, blue) shown in the inset barplot. Specimens collected from Vogelsang, Evelyn, and Fletcher lakes (shown as yellow and blue members in the STRUCTURE plot above for modern specimens) are indicated with an arrow.
Conclusions
For this study we designed a robust experimental, bioinformatic, and statistical framework for population genomic studies of museum specimens. Specifically, we demonstrated the feasibility of enriching high-quality, genome-wide nuclear markers and SNP calling from sequence data derived from museum historic skins. Moreover, this proof-of-concept experiment was conducted on a species group for which no independent high-quality genomic resources exist.
Our approach is likely to be useful across a very broad range of evolutionary and ecological questions. A large number of museum specimens represent the past and recent genetic diversity of small, declining or extinct populations, making them especially valuable because collecting new material is consequently difficult or impossible. Paradoxically, the extensive, random fragmentation that makes DNA from museum skins a poor template for PCR is a boon for short-read parallel sequencing. That said, there remains significant bioinformatic challenges for the analyses of historic DNA. We observed substantial base misincorporations in DNA derived from century old museum skins and developed pipelines for data filtration at different levels (individuals, contigs, sites), which addresses post mortem damages. Low-medium coverage sequence data will be common in cost-effective, population genomic, study designs that use NGS. To account for the genotyping uncertainties associated with such data, we integrated a recently developed Bayesian SNP calling framework based on joint information from all individuals in a sample, which is expected to be more powerful than variant calling based on single individuals (Nielsen et al. 2012).
Transcriptome-based, multiplexed, exon capture is a fast and cost-effective approach for gathering thousands of nuclear loci derived from museum specimens that lack a pre-existing reference genome. This opens the door to population level, genomic comparisons for diverse taxa in museum collections. Furthermore, the initial development of genome resources does not rely on historic specimens. Rather, this approach produces a high quality transcriptome reference by using only one modern specimen and then uses exon capture to enrich homologous genes from historic specimens by DNA hybridization. Moreover, because this method works at modest phylogenetic scales (Bi et al. 2012; Jin et al. 2012), if the species of interest is extinct or cannot be sampled in modern times, genetically close relatives can be used to design the capture array.
There are a few alternative approaches that might suffice to generate an initial reference when transcriptome sequencing is not feasible and moderately divergent genomic references are not available. Though it remains unclear how robust hybridization-based captures are across deeper evolutionary divergences, it is likely that more divergent genome references could be used to generate an initial capture experiment where one or a few individuals are captured and sequenced to high coverage. A second, species-specific capture could then be designed from the subset of regions that are successfully recovered. Alternatively, probes could also be designed using shotgun genomic NGS data from one individual, which would rely on assembled genome scaffolds and direct alignment of the sequence reads to the exome of the most closely related species with an available genome reference. The experiments described in the present study were carried out using array-based exon capture. It is worth noting that in-solution based methods utilizing longer probes will likely be more cost-effective for targeting large genomic regions and large sample sizes, and are more likely to provide sufficient enrichment from divergent references or poorly preserved DNA typical of museum skins.
All of these capture-based approaches are likely to cost more and require greater initial experimental and bioinformatic investment than alternative reduced representation approaches on non-model organisms that rely upon restriction digests to enrich anonymous regions of the genome (i.e., RAD-seq and related approaches; Baird et al. 2008). However, RAD-seq is poorly suited for use on limited or highly degraded DNA sources (Etter et al. 2011), precluding its broad application to historic museum samples. Moreover, transcriptome-based exon capture offers a number of other decisive advantages over RAD-seq including greater experimental control, lower experimental variance among loci and samples, some a priori knowledge of target function, and the ability to explicitly partition different site classes in population genetic analyses (e.g., synonymous and non-synonymous positions).
One of the keys to museum population genomic applications is to ensure similar sequencing coverage among historic and modern specimens. Uneven coverage among specimens adds difficulty to the statistical analyses that could lead to biases if the variance in coverage and errors are not modeled accurately. Many current methods are designed to accommodate this issue (e.g., ANGSD) but could fail if the error structure is not modeled accurately. In this regard, we recommend that researchers always aim for a balanced design including managing enrichment PCR and equimolar pooling of pre-capture libraries to ensure similar coverage among historic and modern specimens.
We demonstrated that we were able to use the methods herein described to obtain biologically meaningful results. The population genomic analyses of early 20TH century museum historic skins and contemporary specimens of Alpine chipmunks (T. alpinus) showed that there was no major change in overall genetic diversity over the time period examined. However, we observed increased population genetic structure within the modern period, which is likely a consequence of reduced gene flow among patchy subpopulations due to climate-change induced range contraction. Future research will focus on larger specimen sample and genetic target sizes for T. alpinus, and will compare subsequent findings to those from populations that have remained stable during climate change to understand its demographic and evolutionary consequences for wildlife populations.
Acknowledgments
The authors would like to thank Chris Conroy, Eileen Lacey, James Patton, Karen Rowe, Kevin Rowe and Jack Sullivan for providing access to museum specimens and tissues, Hernán Burbano for sharing scripts for array design, Maria Santos for helping with maps, and Brice Sarver for providing Tamias mitochondrial sequence. We also thank the Texas Advanced Computing Center (TACC) at the University of Texas at Austin for providing computational support. We would also like to acknowledge Roberta Damasceno, Sean Maher, and Sonal Singhal for their insightful comments on this manuscript and Johannes Krause and Adrian Briggs for helpful conversations on damage patterns in ancient DNA. This work was supported by an NSERC postdoctoral fellowship (KB), University of Montana start-up funds (JG), and University of California Berkeley VCR-BiGCB and the Gordon and Betty Moore Foundation (RN & CM). TL was in-part supported by the NIH Genomics Training Grant (Grant T32HG000047-13).
Footnotes
Data Accessibility
The T. alpinus RNAseq and exon capture sequencing data are available in the NCBI sequence read archive (SRA) (IDs: SRR504595 & SRR847500). The bioinformatic pipelines used in this study are available at https://github.com/MVZSEQ/Exon-capture. The T. alpinus annotated transcripts and sequences of short DNA baits for exon capture are available at the DRYAD, entry doi:10.5061/dryad.s296n
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. Journal of Molecular Biology. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Avila-Arcos MC, Cappellini E, Romero-Navarro JA, Wales N, Moreno-Mayar JV, Rasmussen M, Fordyce SL, Montiel R, Vielle-Calzada JP, Willerslev E, Gilbert MT. Application and comparison of large-scale solution-based DNA capture-enrichment methods on ancient DNA. Scientific reports. 2011;1:74. doi: 10.1038/srep00074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axelsson E, Willerslev E, Gilbert MTP, Nielsen R. The effect of ancient DNA damage on inferences of demographic histories. Molecular Biology and Evolution. 2008;25:2181–2187. doi: 10.1093/molbev/msn163. [DOI] [PubMed] [Google Scholar]
- Baird NA, Etter PD, Atwood TS, Currey MC, Shiver AL, Lewis ZA, Selker EU, Cresko WA, Johnson EA. Rapid SNP discovery and genetic mapping using sequenced RAD markers. PLoS ONE. 2008;3:e3376. doi: 10.1371/journal.pone.0003376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi K, Vanderpool D, Singhal S, Linderoth T, Moritz C, Good JM. Transcriptome-based exon capture enables highly cost-effective comparative genomic data collection at moderate evolutionary scales. BMC Genomics. 2012;13:403. doi: 10.1186/1471-2164-13-403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birol İ, Jackman SD, Nielsen CB, Qian JQ, Varhol R, Stazyk G, Morin RD, Zhao Y, Hirst M, Schein JE, Horsman DE, Connors JM, Gascoyne RD, Marra MA, Jones SJ. De novo transcriptome assembly with ABySS. Bioinformatics. 2009;25:2872–2877. doi: 10.1093/bioinformatics/btp367. [DOI] [PubMed] [Google Scholar]
- Bos KI, Schuenemann VJ, Golding GB, Burbano HA, Waglechner N, Coombes BK, McPhee JB, DeWitte SN, Meyer M, Schmedes S, Wood J, Earn DJ, Herring DA, Bauer P, Poinar HN, Krause J. A draft genome of Yersinia pestis from victims of the Black Death. Nature. 2011;478:506–510. doi: 10.1038/nature10549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouzat JL, Lewin HA, Paige KN. The ghost of genetic diversity past: historical DNA analysis of the greater prairie chicken. The American Naturalist. 1998;152:1–6. doi: 10.1086/286145. [DOI] [PubMed] [Google Scholar]
- Briggs AW, Good JM, Green RE, Krause J, Maricic T, Stenzel U, Lalueza-Fox C, Rudan P, Brajkovic D, Kucan Z, Gusic I, Schmitz R, Doronichev VB, Golovanova LV, de la Rasilla M, Fortea J, Rosas A, Pääbo S. Targeted retrieval and analysis of five Neandertal genomes. Science. 2009;325:318–321. doi: 10.1126/science.1174462. [DOI] [PubMed] [Google Scholar]
- Briggs AW, Stenzel U, Johnson PL, Green RE, Kelso J, Prüfer K, Meyer M, Krause J, Ronan MT, Lachmann M, Pääbo S. Patterns of damage in genomic DNA sequences from a Neandertal. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:14616–14621. doi: 10.1073/pnas.0704665104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggs AW, Stenzel U, Meyer M, Krause J, Kircher M, Pääbo S. Removal of deaminated cytosines and detection of in vivo methylation in ancient DNA. Nucleic Acids Research. 2010;38:e87. doi: 10.1093/nar/gkp1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burbano HA, Hodges E, Green RE, Briggs AW, Krause J, Meyer M, Good JM, Maricic T, Johnson PLF, Xuan Z, Rooks M, Bhattacharjee A, Brizuela L, Albert FW, de la Rasilla M, Fortea J, Rosas A, Lachmann M, Hannon GJ, Pääbo S. Targeted investigation of the Neandertal genome by array-based sequence capture. Science. 2010;328:723–725. doi: 10.1126/science.1188046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper A, Mourer-Chauvire C, Chambers GK, et al. Independent origins of New Zealand moas and kiwis. Proceedings of the National Academy of Sciences of the United States of America. 1992;89:8741–8744. doi: 10.1073/pnas.89.18.8741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosart T, Beja-Pereira A, Chen S, Ng SB, Shendure J, Luikart G. Exome-wide DNA capture and next generation sequencing in domestic and wild species. BMC Genomics. 2011;12:347. doi: 10.1186/1471-2164-12-347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earl DA, vonHoldt BM. STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conservation Genetics Resources. 2012;4:359–361. [Google Scholar]
- Etter P, Bassham S, Hohenlohe PA, Johnson E, Cresko WA. SNP discovery and genotyping for evolutionary genetics using RAD sequencing. In: Orgogozo V, Rockman MV, editors. Molecular Methods for Evolutionary Genetics, Methods in Molecular Biology. Vol. 772. Humana Press; New York, USA: 2011. pp. 157–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evanno G, Regnaut S, Goudet J. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Molecular Ecology. 2005;14:2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x. [DOI] [PubMed] [Google Scholar]
- Fumagalli M, Vieira FG, Korneliussen TS, Linderoth T, Huerta-Sanchez E, Anders Albrechtsen A, Nielsen R. Quantifying population genetic differentiation from Next-Generation Sequencing data. Genetics. 2013 doi: 10.1534/genetics.113.154740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greagg MA, Fogg MJ, Panayotou G, Evans SJ, Connolly BA, Pearl LH. A read-ahead function in archaeal DNA polymerases detects promutagenic template-strand uracil. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:9045–9050. doi: 10.1073/pnas.96.16.9045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginolhac A, Rasmussen M, Gilbert MTP, Willerslev E, Orlando L. mapDamage: testing for damage patterns in ancient DNA sequences. Bioinformatics. 2011;27:2153–2155. doi: 10.1093/bioinformatics/btr347. [DOI] [PubMed] [Google Scholar]
- Glenn TC, Stephan W, Braun MJ. Effects of a population bottleneck on whooping crane mitochondrial DNA variation. Conservation Biology. 1999;13:1097–1107. [Google Scholar]
- Godoy JA, Negro JJ, Hiraldo F, Donázar JA. Phylogeography, genetic structure and diversity in the endangered bearded vulture (Gypaetus barbatus, L) as revealed by mitochondrial DNA. Molecular Ecology. 2004;13:371–390. doi: 10.1046/j.1365-294x.2003.02075.x. [DOI] [PubMed] [Google Scholar]
- Good JM, Hird S, Reid N, Demboski JR, Steppan SJ, Martin-Nims TR, Sullivan J. Ancient hybridization and mitochondrial capture between two species of chipmunks. Molecular Ecology. 2008;17:1313–1327. doi: 10.1111/j.1365-294X.2007.03640.x. [DOI] [PubMed] [Google Scholar]
- Good JM. Reduced representation methods for subgenomic enrichment and next-generation sequencing. In: Orgogozo V, Rockman MV, editors. Molecular Methods for Evolutionary Genetics, Methods in Molecular Biology. Vol. 772. Humana Press; New York, USA: 2011. pp. 85–103. [DOI] [PubMed] [Google Scholar]
- Good JM, Wiebe V, Albert FW, Burbano HA, Kircher M, Green RE, Halbwax M, André C, Atencia R, Fischer A, Pääbo S. Comparative population genomics of the ejaculate in humans and the great apes. Molecular Biology and Evolution. 2013;30:964–976. doi: 10.1093/molbev/mst005. [DOI] [PubMed] [Google Scholar]
- Guschanski K, Krause J, Sawyer S, Valente LM, Bailey S, Finstermeier K, Sabin R, Gilissen E, Sonet G, Nagy ZT, Lenglet G, Mayer F, Savolainen V. Next-Generation Museomics Disentangles One of the Largest Primate Radiations. Systematic Biology. 2013 doi: 10.1093/sysbio/syt018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper GL, McClean N, Goulson D. Analysis of museum specimens suggests extreme genetic drift in the adonis blue butterfly (Polyommatus bellargus) Biological Journal of the Linnean Society. 2008;88:447–452. [Google Scholar]
- Hellmann I, Mang Y, Gu Z, Li P, de la Vega FM, Clark AG, Nielsen R. Population genetic analysis of shotgun assemblies of genomic sequences from multiple individuals. Genome Research. 2008;18:1020–1029. doi: 10.1101/gr.074187.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyn P, Stenzel U, Briggs AW, Kircher M, Hofreiter M, Meyer M. Road blocks on paleogenomes--polymerase extension profiling reveals the frequency of blocking lesions in ancient DNA. Nucleic Acids Research. 2010;38:e161. doi: 10.1093/nar/gkq572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higuchi R, Bowman B, Freiberger M, Ryder OA, Wilson AC. DNA sequences from the quagga, and extinct member of the horse family. Nature. 1984;312:282–284. doi: 10.1038/312282a0. [DOI] [PubMed] [Google Scholar]
- Hodges E, Rooks M, Xuan Z, Bhattacharjee A, Benjamin Gordon D, Brizuela L, Richard McCombie W, Hannon GJ. Hybrid selection of discrete genomic intervals on custom-designed microarrays for massively parallel sequencing. Nature Protocol. 2009;4:960–974. doi: 10.1038/nprot.2009.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofreiter M, Jaenicke V, Serre D, von Haeseler A, Paabo S. DNA sequences from multiple amplifications reveal artifacts induced by cytosine deamination in ancient DNA. Nucleic Acids Research. 2001;29:4793–4799. doi: 10.1093/nar/29.23.4793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakobsson M, Rosenberg N. CLUMPP: a cluster matching and permutation program for dealing with multimodality in analysis of population structure. Bioinformatics. 2007;23:1801–1806. doi: 10.1093/bioinformatics/btm233. [DOI] [PubMed] [Google Scholar]
- Jin X, He M, Ferguson B, Meng Y, Ouyang L, Ren J, Mailund T, Sun F, Sun L, Shen J, Zhuo M, Song L, Wang J, Ling F, Zhu Y, Hvilsom C, Siegismund H, Liu X, Gong Z, Ji F, Wang X, Liu B, Zhang Y, Hou J, Wang J, Zhao H, Wang Y, Fang X, Zhang G, Wang J, Zhang X, Schierup MH, Du H, Wang J, Wang X. An effort to use human-based exome capture methods to analyze chimpanzee and macaque exomes. PLoS ONE. 2012;7:e40637. doi: 10.1371/journal.pone.0040637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson PLF, Slatkin M. Accounting for bias from sequencing error in population genetic estimates. Molecular Biology and Evolution. 2008;25:199–206. doi: 10.1093/molbev/msm239. [DOI] [PubMed] [Google Scholar]
- Krings M, Capelli C, Tschentscher F, Geisert H, Meyer S, von Haeseler A, Grossschmidt K, Possnert G, Paunovic M, Paabo S. A view of Neandertal genetic diversity. Nature Genetics. 2000;26:144–146. doi: 10.1038/79855. [DOI] [PubMed] [Google Scholar]
- Lemmon A, Emme S, Lemmon E. Anchored hybrid enrichment for massively high-throughput phylogenomics. Systematic Biology. 2012;61:727–744. doi: 10.1093/sysbio/sys049. [DOI] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. Genome Project Data Processing Subgroup (2009) The Sequence alignment/map (SAM) format and SAMtools. Bioinformatics. 1000;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M. Estimation of Nucleotide Diversity, Disequilibrium Coefficients, and Mutation Rates from High-Coverage Genome-Sequencing Projects. Molecular Biology and Evolution. 2008;25:2409–2419. doi: 10.1093/molbev/msn185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maricic T, Whitten M, Pääbo S. Multiplexed DNA Sequence Capture of Mitochondrial Genomes Using PCR Products. PLoS ONE. 2010;5:e14004. doi: 10.1371/journal.pone.0014004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason VC, Li G, Helgen KM, Murphy WJ. Efficient cross-species capture hybridization and next-generation sequencing of mitochondrial genomes from noninvasively sampled museum specimens. Genome Research. 2011;21:1695–1704. doi: 10.1101/gr.120196.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormack JE, Faircloth BC, Crawford NG, Gowaty PA, Brumfield RT, Glenn TC. Ultraconserved elements are novel phylogenomic markers that resolve placental mammal phylogeny when combined with species-tree analysis. Genome Research. 2012;22:746–754. doi: 10.1101/gr.125864.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDevitt AD, Zub K, Kawałko A, Oliver MK, Herman JS, Wójcik JM. Climate and refugial origin influence the mitochondrial lineage distribution of weasels (Mustela nivalis) in a phylogeographic suture zone. Biological Journal of the Linnean Society. 2012;106:57–69. [Google Scholar]
- Meyer M, Kircher M. Illumina sequencing library preparation for highly multiplexed target capture and sequencing. Cold Spring Harbor Protocols. 2010;2010 doi: 10.1101/pdb.prot5448. pdb.prot5448. [DOI] [PubMed] [Google Scholar]
- Moritz C, Patton JL, Conroy CJ, Parra JL, White GC, Beissinger SR. Impact of a century of climate change on small-mammal communities in Yosemite National Park, USA. Science. 2008;322:261–264. doi: 10.1126/science.1163428. [DOI] [PubMed] [Google Scholar]
- Nei M, Maruyama T, Chakraborty R. The bottleneck effect and genetic variability in populations. Evolution. 1975;29:1–10. doi: 10.1111/j.1558-5646.1975.tb00807.x. [DOI] [PubMed] [Google Scholar]
- Nielsen R, Korneliussen T, Albrechtsen A, Li Y, Wang J. SNP calling, genotype calling, and sample allele frequency estimation from New-Generation Sequencing data. PLoS One. 2012;7:e37558. doi: 10.1371/journal.pone.0037558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen R, Paul JS, Albrechtsen A, Song YS. Genotype and SNP calling from next-generation sequencing data. Nature Review Genetics. 2011;12:443–451. doi: 10.1038/nrg2986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne RB, Sorenson MD. Museum collections as sources of genetic data. Bonn zoological Bulletin. 2002;51:97–104. [Google Scholar]
- Peery MZ, Hall LA, Sellas A, Beissinger SR, Moritz C, Bérubé M, Raphael MG, Nelson SK, Golightly RT, McFarlane-Tranquilla L, Newman S, Palsbøll PJ. Genetic analyses of historic and modern marbled murrelets suggest decoupling of migration and gene flow after habitat fragmentation. Proceedings of the Royal Society B-Biological Sciences. 2010;277:697–706. doi: 10.1098/rspb.2009.1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry GH, Marioni JC, Melsted P, Gilad Y. Genomic-scale capture and sequencing of endogenous DNA from feces. Molecular Ecology. 2010;19:5332–5344. doi: 10.1111/j.1365-294X.2010.04888.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pool JE, Hellmann I, Jensen JD, Nielsen R. Population genetic inference from genomic sequence variation. Genome Research. 2010;29:291–300. doi: 10.1101/gr.079509.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulakakis N, Glaberman S, Russello M, Beheregaray LB, Ciofi C, Powell JR, Caccone A. Historical DNA analysis reveals living descendants of an extinct species of Galápagos tortoise. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:15464–15469. doi: 10.1073/pnas.0805340105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid N, Demboski JR, Sullivan J. Phylogeny estimation of the radiation western American chipmunk (Tamias) in the face of introgression using reproductive protein genes. Systematic Biology. 2012;61:44–62. doi: 10.1093/sysbio/syr094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robin ED, Wong R. Mitochondrial DNA molecules and virtual number of mitochondria per cell in mammalian cells. Journal of Cellular Physiology. 1988;136:507–513. doi: 10.1002/jcp.1041360316. [DOI] [PubMed] [Google Scholar]
- Roderick GK, Navajas M. Genes in new environments: genetics and evolution in biological control. Nature Reviews Genetics. 2003;4:889–899. doi: 10.1038/nrg1201. [DOI] [PubMed] [Google Scholar]
- Rosenberg N. DISTRUCT: a program for the graphical display of population structure. Molecular Ecology Resources. 2004;4:137–138. [Google Scholar]
- Rowe KC, Singhal S, Macmanes MD, Ayroles JF, Morelli TL, Rubidge EM, Bi K, Moritz CC. Museum genomics: low-cost and high-accuracy genetic data from historical specimens. Molecular Ecology Resources. 2011;11:1082–1092. doi: 10.1111/j.1755-0998.2011.03052.x. [DOI] [PubMed] [Google Scholar]
- Roy MS, Girman DJ, Taylor AC, Wayne RK. The use of museum specimens to reconstruct the genetic-variability and relationships of extinct populations. Experientia. 1994;50:551–557. doi: 10.1007/BF01921724. [DOI] [PubMed] [Google Scholar]
- Rubidge EM, Monahan WB, Parra JL, Cameron SE, Brashares JS. The role of climate, habitat, and species co-occurrence as drivers of change in small mammal distributions over the past century. Global Change Biology. 2011;17:696–708. [Google Scholar]
- Rubidge EM, Patton JL, Lim M, Burton AC, Brashares JS, Moritz C. Climate-induced range contraction drives genetic erosion in an alpine mammal. Nature Climate Change. 2012;2:285–288. [Google Scholar]
- Sawyer S, Krause J, Guschanski K, Savolainen V, Paabo S. Temporal patterns of nucleotide misincorporations and DNA fragmentation in ancient DNA. PLoS ONE. 2012;7:e34131. doi: 10.1371/journal.pone.0034131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soumillon M, Necsulea A, Weier M, Brawand D, Zhang X, Gu H, Barthes P, Kokkinaki M, Nef S, Gnirke A, Dym M, de Massy B, Mikkelsen TS, Kaessmann H. Cellular source and mechanisms of high transcriptome complexity in the mammalian Testis. Cell Reports. 2013;3:2179–2190. doi: 10.1016/j.celrep.2013.05.031. [DOI] [PubMed] [Google Scholar]
- Singhal S. De novo transcriptomic analyses for non-model organisms: an evaluation of methods across a multi-species data set. Molecular Ecology Resources. 2013;13:403–416. doi: 10.1111/1755-0998.12077. [DOI] [PubMed] [Google Scholar]
- Stiller M, Green RE, Ronan M, Simons JF, Du L, He W, Egholm M, Rothberg JM, Keates SG, Ovodov ND, Antipina EE, Baryshnikov GF, Kuzmin YV, Vasilevski AA, Wuenschell GE, Termini J, Hofreiter M, Jaenicke-Després V, Pääbo S. Patterns of nucleotide misincorporations during enzymatic amplification and direct large-scale sequencing of ancient DNA. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:13578–13584. doi: 10.1073/pnas.0605327103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taberlet P, Griffin S, Goossens B, Questiau S, Manceau V, Escaravage N, Waits LP, Bouvet J. Reliable genotyping of samples with very low DNA quantities using PCR. Nucleic Acids Research. 1996;24:3189–3194. doi: 10.1093/nar/24.16.3189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989;123:585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W, Qian D, Ahmad S, Mattox D, Todd NW, et al. A low-cost exon capture method suitable for large-scale screening of genetic deafness by the massively-parallel sequencing approach. Genetic testing and molecular biomarkers. 2012;16:536–542. doi: 10.1089/gtmb.2011.0187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor AC, Sherwin WB, Wayne RK. Genetic variation of microsatellite loci in a bottlenecked species: the northern hairy-nosed wombat Lasiorhinus krefftii. Molecular Ecology. 1994;3:277–290. doi: 10.1111/j.1365-294x.1994.tb00068.x. [DOI] [PubMed] [Google Scholar]
- Thomas WK, Pääbo S, Villablanca FX, Wilson AC. Spatial and temporal continuity of kangaroo rat populations shown by sequencing mitochondrial DNA from museum specimens. Journal of Molecular Evolution. 1990;31:101–112. doi: 10.1007/BF02109479. [DOI] [PubMed] [Google Scholar]
- Tingley MW, Monahan WB, Beissinger SR, Moritz C. Birds track their Grinnellian niche through a century of climate change. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:19637–19643. doi: 10.1073/pnas.0901562106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trifonov VA, Vorobieva NN, Rens W. FISH With and without COT1 DNA. In: Liehr T, editor. Fluorescence In Situ Hybridization (FISH): Application Guide. Springer-Verlag; Berlin, Heidelberg: 2009. pp. 99–109. [Google Scholar]
- Väli U, Einarsson A, Waits L, Ellegren H. To what extent do microsatellite markers reflect genome-wide genetic diversity in natural populations? Molecular Ecology. 2008;17:3808–3817. doi: 10.1111/j.1365-294X.2008.03876.x. [DOI] [PubMed] [Google Scholar]
- Wandeler P, Hoeck PE, Keller LF. Back to the future: museum specimens in population genetics. Trends in Ecology & Evolution. 2007;22:634–642. doi: 10.1016/j.tree.2007.08.017. [DOI] [PubMed] [Google Scholar]
- Watterson GA. On the number of segregating sites in genetical models without recombination. Theoretical Population Biology. 1975;7:256–276. doi: 10.1016/0040-5809(75)90020-9. [DOI] [PubMed] [Google Scholar]
- Wayne RK, Jenks SM. Mitochondrial DNA analysis implying extensive hybridization of the endangered red wolf Canis rufus. Nature. 1991;351:565–568. [Google Scholar]
- Weber DS, Stewart BS, Garza JC, Lehman N. An empirical genetic assessment of the severity of the northern elephant seal population bottleneck. Current Biolology. 2000;10:1287–1290. doi: 10.1016/s0960-9822(00)00759-4. [DOI] [PubMed] [Google Scholar]
- Wigginton JE, Cutler DJ, Abecasis GR. A note on exact tests of Hardy-Weinberg equilibrium. The American Journal of Human Genetics. 2005;76:887–893. doi: 10.1086/429864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wójcik JM, Kawałko A, Marková S, Searle JB, Kotlík P. Phylogeographic signatures of northward post-glacial colonization from high-latitude refugia: a case study of bank voles using museum specimens. Journal of Zoology. 2010;281:249–262. [Google Scholar]
- Yi X, Liang Y, Huerta-Sanchez E, Jin X, Cuo ZX, Pool JE, Xu X, Jiang H, Vinckenbosch N, Korneliussen TS, Zheng H, Liu T, He W, Li K, Luo R, Nie X, Wu H, Zhao M, Cao H, Zou J, Shan Y, Li S, Yang Q, Asan, Ni P, Tian G, Xu J, Liu X, Jiang T, Wu R, Zhou G, Tang M, Qin J, Wang T, Feng S, Li G, Huasang, Luosang J, Wang W, Chen F, Wang Y, Zheng X, Li Z, Bianba Z, Yang G, Wang X, Tang S, Gao G, Chen Y, Luo Z, Gusang L, Cao Z, Zhang Q, Ouyang W, Ren X, Liang H, Zheng H, Huang Y, Li J, Bolund L, Kristiansen K, Li Y, Zhang Y, Zhang X, Li R, Li S, Yang H, Nielsen R, Wang J, Wang J. Sequencing of 50 human exomes reveals adaptation to high altitude. Science. 2010;329:75–78. doi: 10.1126/science.1190371. [DOI] [PMC free article] [PubMed] [Google Scholar]