Abstract
Under conditions of genotoxic stress, human p53 activates the apoptotic effectors BAX or BAK, resulting in mitochondrial outer membrane permeabilization and apoptosis. Anti-apoptotic BCL-2 family member BCL-xL opposes this activity by sequestering cytosolic p53 via association with its DNA-binding domain, an interaction that is enhanced by p53 tetramerization. Here we characterized the BCL-xL – p53 complex using NMR spectroscopy and modulated it through mutagenesis to determine the relative contributions of BCL-xL’s interactions with p53, or with other BCL-2 family proteins, to BCL-xL-dependent inhibition of UV irradiation-induced apoptosis. Under our experimental conditions, one third of the anti-apoptotic activity of BCL-xL was mediated by p53 sequestration and the remaining two thirds through sequestration of pro-apoptotic BCL-2 family members. Our studies define the contributions of cytosolic p53 to UV irradiation-induced apoptosis and provide opportunities to explore its contributions to other, p53-dependent apoptotic signaling pathways.
INTRODUCTION
The tumor suppressor p53 exhibits a subset of cytosolic functions, independent of nuclear transactivation, including activation of apoptosis through interactions with the apoptotic effector proteins BAX and BAK1. Activated effectors form oligomers within and permeabilize the outer mitochondrial membrane (OMM), resulting in the release of cytochrome c and other signaling molecules and induction of apoptosis2,3.
Effector activation is antagonized through sequestration of cytosolic p53 by the anti-apoptotic BCL-xL1,4. BCL-xL and homolog proteins also sequester “BH3-only” pro-apoptotic proteins (BID, BIM) that can also activate BAX and BAK. These proteins are down-regulated in inhibitory complexes when their BH3 domains fold into α-helices and bind a hydrophobic groove within BCL-xL5–8. Unlike other interaction partners of BCL-xL, p53 does not exhibit a BH3 domain9, suggesting that the BCL-xL – p53 complex differs from those between BCL-xL and BH3-only proteins or between p53 and Mdm210.
Full-length p53 is a multi-domain, tetrameric protein11 (Fig. 1). Studies with individual p53 domains indicate that the DNA-binding domain (p53 DBD) contributes predominantly to BCL-xL binding12–15 without engaging the hydrophobic groove that is the binding site for BH3-only proteins12–15. While lacking a BH3 domain, p53 contains two transactivation domains (TAD1, residues 15–30; TAD2, residues 50–65) within its N-terminus (p53 NTD) that fold into amphipathic α–helices upon complex formation10,16,17 and bind weakly to the BH3 groove of BCL-xL18–20. However, how NTD and DBD cooperate when full-length p53 binds to BCL-xL is not understood. Here, we endeavored to characterize the interaction of near full-length, tetrameric p53 with BCL-xL and elucidate the thermodynamic interplay between p53 domains upon binding BCL-xL. Using NMR spectroscopy, we determined the solution structure of the BCL-xL – p53 DBD complex and characterized the same interactions in the context of tetrameric p53. Our results confirmed that the BCL-xL-binding surface of p53 DBD largely overlaps with its DNA-binding surface12 and explained BCL-xL binding specificity. Using mutagenesis, we identified sites within p53 DBD that contribute primarily to BCL-xL binding or to both BCL-xL and DNA binding, providing opportunities to dissect p53’s nuclear and cytosolic functions. The diverse regulatory functions of p53, ranging from cell cycle arrest21 to senescence22 and apoptosis23, are mediated by promiscuous interactions of its individual domains with other proteins and DNA. The p53 DBD contributes to this hub-like behavior, by binding to double-stranded DNA, BCL-xL, and other proteins24,25. Our studies reveal the mechanism by which BCL-xL binds the same surface of p53 DBD that has evolved to also bind a diverse family of DNA gene regulatory sites.
Figure 1.
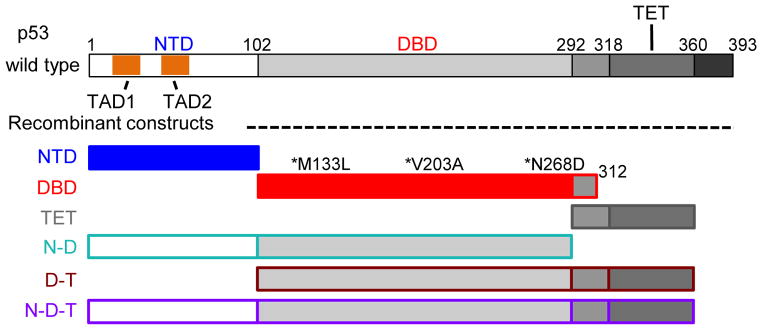
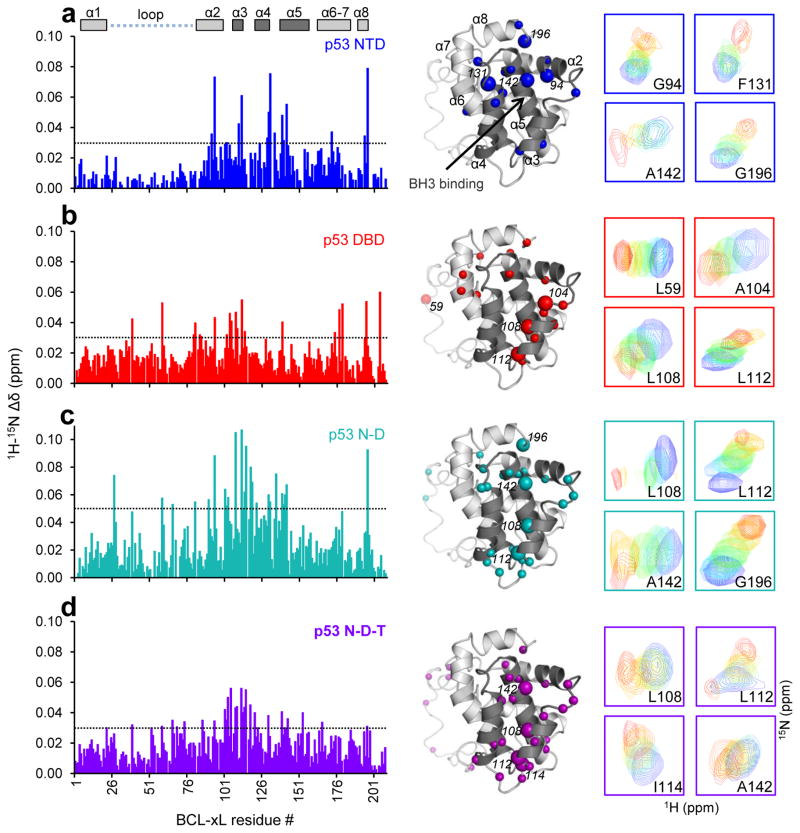
Schematic representation of p53 constructs employed in this study: N terminal transcriptional activation domain (NTD); DNA-binding domain (DBD); tetramerization domain (TET) and multi-domain constructs containing NTD and DBD (N-D); DBD and TET (D-T) or NTD, DBD, TET (N-D-T) respectively. The first and second transcriptional activation regions (TAD1 and 2) within the NTD are highlighted in the schematic of the full-length protein. All the constructs that encompassed the DBD contained three buried, core-stabilizing mutations (marked with asterisks)28. The color coding for various p53 domain constructs used in this figure will be employed throughout the following figures.
RESULTS
Tetramerization of p53 promotes binding to BCL-xL
To establish a baseline for the p53 constructs used here, we first analyzed binding of individual p53 domains to BCL-xL. We monitored chemical shift perturbations (CSPs) of 15N-labeled BCL-xL upon titration with unlabeled, single-domain p53 constructs26: the N-terminal transcriptional activation domain (p53 NTD, residues 1–102), the DNA-binding domain (p53 DBD, residues 102–312), and the tetramerization domain (p53 TET, residues 310–360; Fig. 1). We also measured CSPs using 15N-labeled p53 constructs upon titration of unlabeled BCL-xL. For these experiments we used a BCL-xL construct lacking 22 C-terminal residues (termed BCL-xLΔC27) and a stabilized, triple mutant form of p53 DBD (hereafter referred to as p53 DBD28). These p53 mutations affect buried residues and do not alter DNA binding28.
Titration of unlabeled p53 NTD into 15N-BCL-xLΔC caused CSPs for residues within the BCL-xL BH3-binding groove between α-helices 3 and 4 (α3 and α4) and within α2, α5 and α8 (Fig. 2a; Supplementary Fig. 1). When we titrated unlabeled BCL-xLΔC into 15N-p53 NTD, we observed CSPs for residues within TAD1 and, to a lesser extent, TAD2 of p53 (Fig. 3a; Supplementary Fig. 2), confirming past reports18–20.
Figure 2.
Titrations of various unlabeled p53 domain constructs into 15N-labeled BCL-xLΔC monitored by 1H-15N-TROSY. The panels display sequence (left) and structure mapping (right) of 15N-BCL-xLΔC chemical shift perturbations (CSPs) induced by incremental addition of unlabeled p53 NTD (panels a); p53 DBD (b); p53 N-D (c); and p53 N-D-T (d). The threshold value for CSPs displayed on the BCL-xL structure for each dataset is indicated as a dashed line on the corresponding bar plot (p53 NTD, DBD, N-D-T: 0.03 ppm; p53 N-D: 0.05 ppm). The perturbation trend of selected resonances, highlighted with larger spheres in the respective structure mapping of CSPs, is displayed at the far right of each panel. Perturbation of resonances within the BH3 binding groove of BCL-xL(α helices 4 and 5 – α4-5) is observed upon addition of p53 NTD, in agreement with previous reports18,19, while p53 constructs that contain the DBD induce noticeable perturbation of resonances within α3 of BCL-xL.
Figure 3.
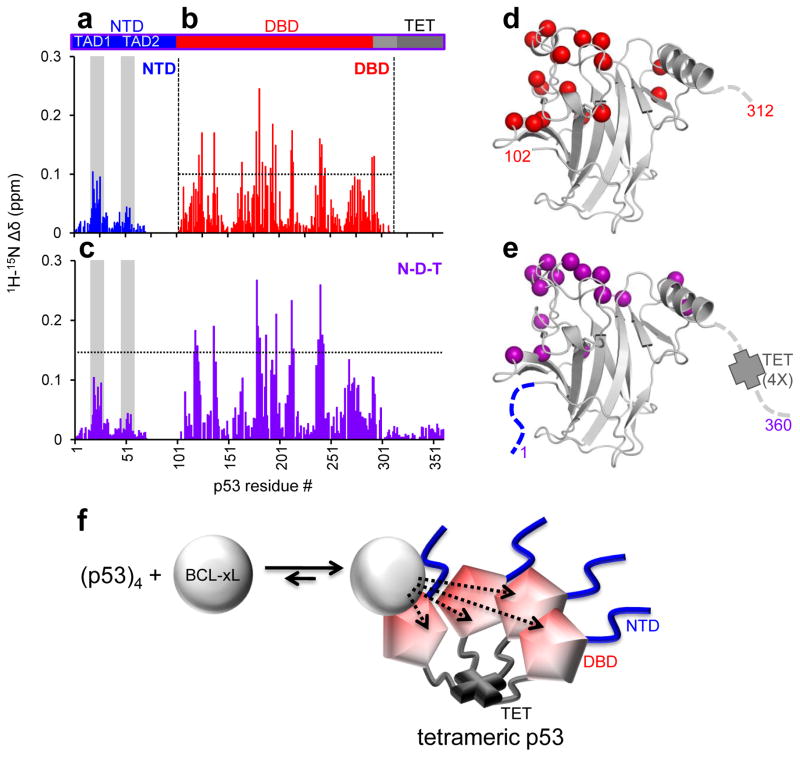
Titrations of unlabeled BCL-xLΔC into different 15N-labeled p53 constructs monitored by 1H-15N-TROSY. a. Sequence mapping of average 15N-p53 NTD chemical shift perturbations (CSPs). b, c. Similar mapping of CSPs within p53 DBD and N-D-T respectively. d, e. Mapping of the data illustrated in panels b, c, respectively, on the structure of p53 DBD. The residues highlighted on the p53 DBD structure showed CSPs above the thresholds marked by a dotted line in b, c. The perturbed resonances cluster in both cases on the basic DNA-interface region of this domain. f. Explanatory model for the observed increase in binding affinity between BCL-xL and tetramerization-competent p53 in the absence of a direct interaction between BCL-xL and p53 TET. When bound to p53 at sub-stoichiometric concentrations, BCL-xL will have increased chances to re-associate to p53, which exhibits high local concentration in the tetrameric state, rather than diffuse away and fully dissociate from the complex.
Titration of p53 DBD caused extensive but quantitatively more subtle CSPs in 15N-BCL-xLΔC (Fig. 2b), with effects on residues within α2 and α3, the BH3-binding groove, the hinge between α6 and α7, and the N-terminus of the flexible loop between α1 and α2 (residues 21–84). Widespread CSPs suggested that conformational changes may accompany BCL-xL binding to p53 DBD, as reported previously14. Titration of unlabeled BCL-xLΔC into 15N-p53 DBD defined a broad interface that, as shown by others12,14, coincided with the DNA-binding surface of p53 (Fig. 3b, d). Finally, unlabeled p53 TET did not induce CSPs in 15N-BCL-xLΔC, indicating that these constructs did not interact (Supplementary Fig. 1).
NMR titrations with multi-domain p53 constructs (bearing the DBD stabilizing mutations described above), including p53 N-D (residues 1–292) and p53 N-D-T (residues 1–360; Fig 1) revealed CSP patterns similar to those observed with individual domains. Titration of 15N-BCL-xLΔC with p53 N-D caused CSPs similar to but larger than those observed with p53 NTD or p53 DBD (Fig. 2c), while p53 N-D-T caused marked resonance broadening at high concentrations (Fig. 2d). When we titrated unlabeled BCL-xLΔC into 2H/15N-p53 N-D or N-D-T, CSPs within the NTD and DBD regions closely matched the perturbations observed in individual domains (Fig. 3c, e).
We then used isothermal titration calorimetry (ITC) to determine the affinities of these interactions. Titrations between BCL-xLΔC and p53 constructs that contained the DBD exhibited weakly endothermic binding which approached the signal-to-noise limits of the calorimeter and precluded quantitative analysis. Titrations performed using a BCL-xL construct lacking the disordered α1-α2 loop (BCL-xLΔLΔC), however exhibited exothermic binding suitable for quantitative analysis (Supplementary Table 1; Supplementary Note). The change in binding enthalpy (ΔH) upon deletion of the disordered BCL-xL loop suggested that this segment somehow participated in binding; however qualitative comparison of titrations performed with BCL-xLΔC or BCL-xLΔLΔC indicated that it did not affect binding free energies (ΔG). The comparable CSP patterns observed upon titration of p53 NTD, p53 DBD and (reported previously29) p53 N-D-T into 15N-BCL-xLΔC or 15N-BCL-xLΔLΔC further confirmed that the two BCL-xL constructs bound p53 similarly and that the disordered loop does not influence the structural features of BCL-xL – p53 interactions (Supplementary Note).
ITC titrations showed that p53 DBD binds BCL-xL one order of magnitude more tightly (Kd = 17 μM; Table 1) than p53 NTD (Kd = 250 μM), both with 1:1 stoichiometry and p53 TET did not bind BCL-xL. Linkage of p53 NTD and DBD (p53 N-D) did not dramatically enhance binding affinity compared with p53 DBD alone, indicating that, while p53 TAD1 and TAD2 transiently bound the BH3-binding groove of BCL-xL, they did not cooperate with the DBD to enhance affinity for BCL-xL. We next investigated the role of p53 TET within multi-domain p53 constructs in BCL-xL binding. The limited solubility of p53 D-T precluded its use in NMR experiments, however ITC titrations showed that linkage of DBD and TET enhanced binding affinity for BCL-xLΔLΔC approximately one order of magnitude over that of p53 DBD (Kd = 1.9 μM); p53 N-D-T showed a further increase in affinity for BCL-xLΔLΔC (Kd = 0.6 μM). Both titrations exhibited 1:1 stoichiometry (BCL-xLΔLΔC:p53 construct monomer). NMR data confirmed the tetrameric state of p53 N-D-T30, and its concentration in ITC experiments (30 μM) exceeded the Kd for p53 tetramers (< 1 μM)31. These results supported that p53 tetramerization enhanced BCL-xL binding but also suggested that tetramerization did not affect the general mode of interaction, with BCL-xL binding 1:1 to individual N-D domains within tetrameric p53 N-D-T.
Table 1.
Dissociation constants between BCL-xL and p53 constructs or between BCL-xL, pre-incubated with p53 constructs, and BID BH3 peptide*
| BCL-xL – p53 Kd (μM) | BID – BCL-xL Kd (nM)** | ||
|---|---|---|---|
|
| |||
| p53 construct | ITC | CSP | (free BCL-xL) 23±7 |
|
| |||
| NTD | 250±80 | 230±90 | 37±6 |
| DBD | 17±2*** | 30±20 | 7±2 |
| TET | no binding | no binding | N.D. |
| N-D | 30±20*** | 40±10 | 63±8 |
| D-T | 1.9±0.1*** | N.D. | 7±4 |
| N-D-T | 0.6±0.3*** | 1.2±0.6 | 35±4 |
Affinities were determined by isothermal titration calorimetry (ITC – average of two independent titrations; error range: s.e.m.; N.D.: not determined) or fitting of NMR chemical shift perturbation data (CSP - for BCL-xL – p53 binding only; error ranges indicate uncertainties of fit for curves obtained from average CSP of 10–15 well resolved resonances).
Measured by ITC, titration of BID BH3 into BCL-xL pre-incubated with the indicated p53 construct
Titration of BCL-xLΔLΔC
Fitting of CSP curves for the titration of 15N-BCL-xLΔC with p53 N-D-T revealed an apparent Kd value (1.2 μM) lower than that obtained with p53 N-D (40 μM), consistent with ITC data showing that tetramerization enhanced binding of p53 to BCL-xL (Supplementary Note). However, the similarity of CSPs caused by p53 N-D and p53 N-D-T within 15N-BCL-xLΔC demonstrated that the two p53 constructs bound BCL-xL through a similar structural mechanism, involving primarily p53 DBD and secondarily p53 NTD interactions. CSPs were not present within the tetramerization domain of 15N-p53 N-D-T, indicating that this domain plays an indirect role in enhancing the affinity of p53 for BCL-xL. We reasoned that this enhancement of affinity is caused by increased local concentration of p53 DBD in the context of tetrameric p53 (Fig. 3f).
Charge interactions drive BCL-xL – p53 complex formation
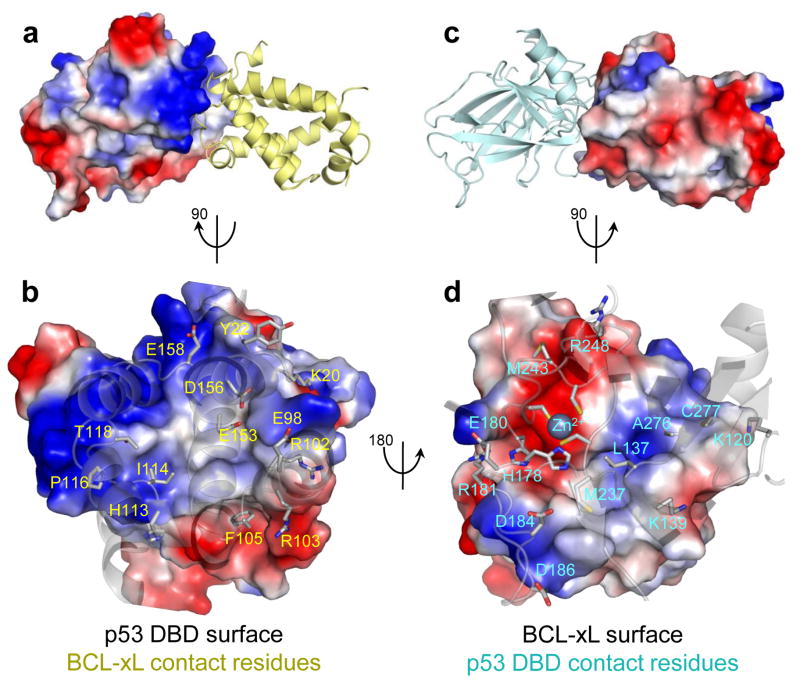
After establishing that p53 DBD primarily mediates binding to BCL-xL, we endeavored to determine the solution structure of the BCL-xL – p53 DBD complex. CSPs identified general contact interfaces, but were insufficient to define the relative orientations of the two proteins. We therefore analyzed paramagnetic relaxation enhancement (PRE) effects on 15N-p53 DBD upon addition of BCL-xLΔC constructs labeled with the nitroxide MTSL. We probed three labeling sites: wild-type Cys151 (Supplementary Fig. 3), Cys2, or Cys122 (for the latter two, the wild-type residue was Ser and we mutated wild-type Cys151 to Ser; Supplementary Note). PRE measurements yielded distance restraints between the MTSL-labeled sites in BCL-xLΔC and amide moieties in 15N-p53 DBD. We also introduced paramagnetic Co2+ into the Zn2+ coordination site of p53 DBD and measured PREs on backbone amide, carbonyl resonances and Isoleucine, Leucine, Valine side-chains in selectively labeled (1H/13C-ILV-methyl)-2H/13C/15N-BCL-xLΔC (Supplementary Fig. 3; Supplementary Note). We further detected a small number of intermolecular 1H-1H NOEs between amide protons of 2H/15N-p53 DBD and side-chains of unlabeled BCL-xLΔC (Supplementary Fig. 4). Using distance restraints spanning the intermolecular interface, we determined the structure of the complex in two stages. First, recognizing, based on our own and others’ data14, that binding of p53 DBD alters the conformation of BCL-xL, we determined the structure of BCL-xLΔC, selectively labeled as above, when bound to unlabeled p53 DBD. We validated this structure determination protocol29 by determining the structure of apo-BCLxL ΔC (2.1 Å r.m.s.d. for backbone atoms with respect to the crystal structure, 1R2D; Supplementary Fig. 5; Table 2). Second, we used the p53 DBD-bound BCL-xL structure and the crystal structure of p53 DBD (PDB: 2AC0)32 to determine the solution structure of the BCL-xLΔC – p53 DBD complex (Fig. 4; Table 3).
Table 2.
NMR and refinement statistics for structures of apo- and p53 DBD-bound BCL-xL
| Apo-BCL-xLΔC | p53 DBD-bound BCL-xLΔC | |
|---|---|---|
| NMR distance and dihedral constraints | ||
| Distance constraints | ||
| Total NOE | 607 | 766 |
| Intra-residue | 59 | 54 |
| Inter-residue | 548 | 712 |
| Sequential (|i − j| = 1) | 225 | 283 |
| Medium-range (|i − j| < 4) | 164 | 266 |
| Long-range (|i − j| > 5) | 159 | 163 |
| Intermolecular | 0 | 0 |
| Hydrogen bonds | 48 | 70 |
| Total dihedral angle restraints | 216 | 208 |
| ϕ | 108 | 104 |
| ψ | 108 | 104 |
| Structure statistics | ||
| Violations (mean ± s.d.) | ||
| Distance constraints (Å) | 0.019 (0.002) | 0.016 (0.003) |
| Dihedral angle constraints (°) | 0.73 (0.16) | 0.73 (0.17) |
| Max. dihedral angle violation (°) | 5.25 | 4.69 |
| Max. distance constraint violation (Å) | 0.33 | 0.35 |
| Deviations from idealized geometry | ||
| Bond lengths(Å) | 0.013 | 0.013 |
| Bond angles (°) | 1.7 | 1.7 |
| Impropers(°) | ||
| Average pairwise r.m.s. deviation** (Å) | ||
| Heavy | 2.2 | 1.8 |
| Backbone | 1.6 | 1.1 |
r.m.s.d. calculated for the 20 lowest energy structures of each model and folded core residues 3–20, 87–193.
Figure 4.
Structure of the complex between p53 DBD and BCL-xL a. Side view of the complex with surface and electrostatic representation of p53 DBD and cartoon representation of BCL-xL. b. Top view of the interaction surface on p53 DBD showing the key interacting residues on BCL-xL. c. Side view of the complex with surface and electrostatic representation of BCL-xL and cartoon representation of p53 DBD. d. Top view of the interaction surface on BCL-xL showing the key interacting residues on p53 DBD, highlighting an electrostatic interaction between the positively charged Zn2+ coordination site on p53 DBD and a pair of acidic residues at the C terminus of BCL-xL α5. Residues within α3 of BCL-xL (R103, F105) also establish interactions with p53 DBD.
Table 3.
NMR and refinement statistics for the BCL-xLΔC – p53 DBD complex
| BCL-xLΔC – p53 DBD | |
|---|---|
| NMR distance constraints | |
| Distance constraints (intermolecular) | |
| Total PRE | 223 |
| 1H PREs (BCL-xL Cys151-MTSL) | |
| Attractive (<24 Å) | 25 |
| Repulsive (>20 Å) | 34 |
| 1H PREs (BCL-xL Cys122-MTSL) | |
| Attractive | 15 |
| Repulsive | 33 |
| 1H PREs (BCL-xL Cys2-MTSL) | |
| Attractive | 0 |
| Repulsive | 58 |
| 1H PREs (p53 Co2+) | |
| Attractive | 33 |
| Repulsive | 5 |
| 13C PREs (p53 Co2+) | |
| Attractive (<17 Å) | 20 |
| NOEs | 13 |
| Structure statistics | |
| Violations (mean and s.d.) | |
| Distance constraints (Å) | 0.13 (0.06) |
| Max. distance constraint violation (Å) | 1.39 |
| Deviations from idealized geometry | |
| Bond lengths (Å) | 0.004 |
| Bond angles (°) | 0.6 |
| Impropers (°) | |
| Average pairwise r.m.s. deviation** (Å) | |
| Heavy | 0.4 |
| Backbone | 0.2 |
r.m.s.d. calculated for the 20 lowest energy structures
The BCL-xLΔC – p53 DBD complex revealed highly charged interfaces, in agreement with the ionic strength dependence of this interaction14. Interfacial residues on BCL-xL are found within the N terminus of the α1-α2 loop, the α3-α4 and α5-α6 loops, as previously reported12, and within the α2-α3 loop and α-helix 3. The interfacial residues within p53 DBD are clustered in two major patches: one overlaps with the DNA-binding region, the other, encompassing α-helix 1 and the Zn2+ coordination site, is not directly involved in DNA binding, however it forms intermolecular DBD-DBD contacts when p53 binds to DNA33. At the complex interface, basic residues on p53 DBD interact with acidic residues on BCL-xL, including the Zn2+ coordination site on p53 DBD and Glu153, Asp156 on BCL-xL; Arg248 on p53 and Asp158 on BCL-xL. Interactions also occur between acidic residues on p53 DBD and basic residues on BCL-xL (Glu180 on p53 and Lys20 on BCL-xL, and Arg103 on BCL-xL and Asp186 on BCL-xL). These contacts, with opposite charge complementarity compared to the overall interface, likely determine the orientation of the two proteins. A few hydrophobic interactions engage residues within α-helix 3 of BCL-xL, and Leu137 and Met237 within p53 DBD (Supplementary Note).
The p53 DBD-bound conformation of BCL-xLΔC differed from its apo form in the N terminal region of the α1-α2 loop; α2 was extended and α3 repositioned, becoming nearly perpendicular to α4 (Supplementary Fig. 5). These rearrangements affect a region of BCL-xL that is involved in binding to BH3-only proteins; consequently, we investigated whether the binding of p53 to BCL-xL modulated its interactions with BH3 domains. We measured by ITC the binding of a BID BH3 domain peptide to BCL-xLΔC that was simultaneously bound to different p53 constructs (Table 1). Preliminary binding of p53 DBD to BCL-xL did not noticeably alter binding of BID to the BH3-groove of BCL-xL, which mediates most known anti-apoptotic functions of BCL-xL34. In summary, the structure of the BCL-xLΔC – p53 complex illustrates how BCL-xL is able to sequester cytosolic p53 without altering its anti-apoptotic functions involving interactions with BH3-only pro-apoptotic proteins.
Tuning p53 binding to DNA versus BCL-xL through mutagenesis
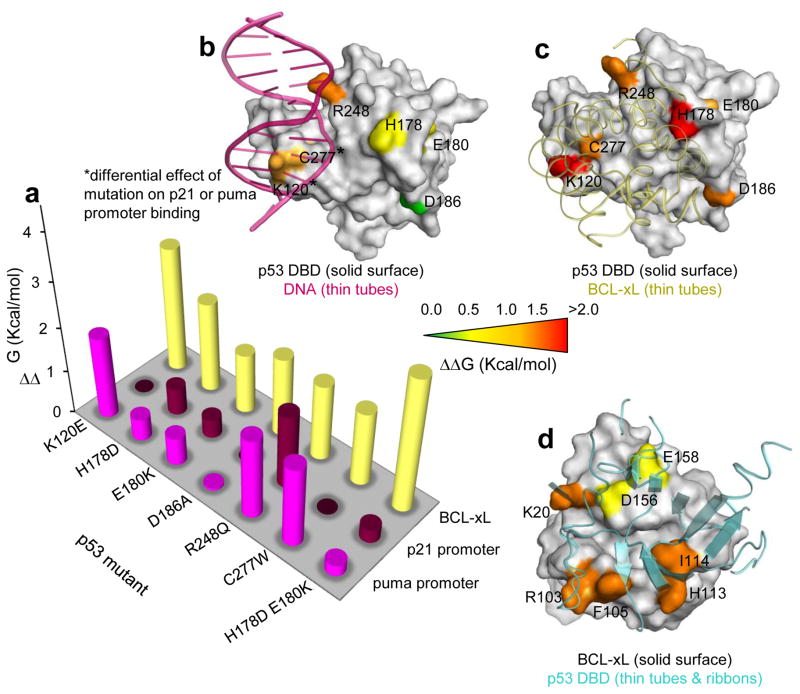
The structure of BCL-xLΔC – p53 DBD revealed that only a portion of the DBD surface engaged in BCL-xL binding is also utilized for DNA binding (Fig. 5a–c), suggesting the possibility to selectively disrupt the binding of p53 DBD to BCL-xL through mutagenesis. We mutated DBD residues within p53 N-D-T predicted35,36 or reported37–39 to not disrupt domain folding and/or known to maintain transactivation function40; some of these mutations had been observed in human cancers35,36. We also mutated corresponding interacting residues on BCL-xLΔC (Fig. 5d). We used fluorescence polarization to monitor binding of p53 mutants to fluorescein-labeled BCL-xLΔC (F-BCL-xLΔC); we measured binding of unlabeled BCL-xLΔC mutants to wild-type p53 N-D-T using this assay in a competition setting (Supplementary Fig. 6).
Figure 5.
Mutagenesis analysis of individual p53 N-D-T residue contributions to DNA or BCL-xL binding, and BCL-xLΔC residue contributions to p53 binding. a. Plot of measured ΔΔG values for various p53 mutants binding to BCL-xL or DNA consensus sequences. b. Surface representation of p53 DBD bound to DNA (purple ribbon; PDB: 2AC0)32 displaying mutated residues. Residues K120 and W277, whose mutation resulted in differential effects on p21 or puma promoter binding, are marked with an asterisk. c. Equivalent representation of p53 DBD bound to BCL-xL. d. Surface representation of BCL-xL bound to p53 displaying the de-stabilizing effect of BCL-xL mutations on p53 binding.
Individual mutations within the DBD of p53 N-D-T reduced affinities for BCL-xL by one-two orders of magnitude, supporting our structural model. We observed the largest effects for mutation of K120E and of H178D. Lys120 is located in a region of p53 DBD where BCL-xL and DNA interfaces overlap, while His178 occupies a position that principally contributes to BCL-xL binding. Combining of a second mutation (E180K) with H178D, resulted in over two orders of magnitude reduction in BCL-xL binding affinity. Individual mutations within BCL-xL reduced affinity for p53 N-D-T about one order of magnitude; binding could not be detected for two dual mutants (I114A combined with either K20A or E158K). These results further support our structural model of the BCL-xLΔC – p53 DBD complex (Fig. 5; Table 4).
Table 4.
Variations in binding affinities (Kd) and free energies (ΔΔG) between BCL-xL and p53 upon mutagenesis of interfacial residues*
| p53 N-D-T mutant | Wild Type | K120E | H178D | E180K | D186A | R248Q | C277W | H178D E180K | Bac-p53 |
|---|---|---|---|---|---|---|---|---|---|
| Kd (μM) | 1.3±0.1 | 150±20 | 41±2 | 13±1 | 27±3 | 24±1 | 22±2 | 210±20 | 1.5±0.4 |
| ΔΔG Wild Type** | 0 | +2.81 | +2.04 | +1.36 | +1.80 | +1.73 | +1.67 | +3.01 | n.d. |
| BCL-xL mutant | Wild Type | K20A | R103E | F105A | H113A | I114A | D156K | E158K | K20A I114A | E158K I114A |
|---|---|---|---|---|---|---|---|---|---|---|
| Kd (μM) | 1.9±0.4 | 36±2 | 46±7 | 26±3 | 35±5 | 42±7 | 8±1 | 7±1 | >100 | >100 |
| ΔΔG Wild Type** | 0 | +1.74 | +1.89 | +1.55 | +1.72 | +1.83 | +0.85 | +0.77 | >+2.3 | >+2.3 |
Binding between wild-type or mutant p53 N-D-T and wild-type fluorescent BCL-xL (F-BCL-xL) was measured by fluorescence polarization titrations (top rows). Binding between wild-type or mutant BCL-xL and wild-type p53 N-D-T was measured by competition titrations against F-BCl-xL (bottom rows). Error ranges indicate uncertainties of fit for curves obtained from the average of three independent titrations.
Determined as follows: ΔΔG = −RT*ln(Kd mut/Kd wild type); values in Kcal/mol
We also measured binding of wild-type and mutated p53 N-D-T to fluorescently-labeled DNA oligonucleotides encoding the promoter sequences from the p21 and puma genes (Fig. 5; Table 5; Supplementary Fig. 7). We fit binding curves to a cooperative binding model assuming equal affinities of p53 DBD for the two, suboptimal consensus sites within each promoter. This basic analysis indicated that wild-type p53 N-D-T bound both oligonucleotides with high affinity (Kd app < 6.4 nM and Hill coefficient ~ 2, indicating cooperative binding of p53 tetramers), in agreement with previous reports41. The R248Q cancer mutation42 impaired binding to both DNA sequences; K120E or C277W mutations primarily impaired binding to the puma promoter. Acetylation of Lys12043,44 and mutation of Cys277 38 are known to alter p53 DNA binding specificities, consistent with this selective effect. Mutations outside the DNA interface, including the H178D E180K double mutant, that bound BCL-xL very weakly, bound DNA tightly but with low cooperativity, likely because the mutated residues are involved in DBD-DBD contacts that stabilize interactions between dimeric or tetrameric p53 DBD units and DNA33.
Table 5.
Apparent dissociation constants (Kdapp), Hill coefficients and variations in binding free energy (ΔΔG) for wild-type and mutant p53 N-D-T binding to fluorescently labeled double stranded oligonucleotides*
| p53 N-D-T mutant | Wild Type | K120E | H178D | E180K | D186A | R248Q | C277W | H178D E180K | Bac-p53 |
|---|---|---|---|---|---|---|---|---|---|
| p21 promoter | |||||||||
| Kdapp. (nM) | ≤6.4 | ≤6.4 | 18±8 | 13±3 | 6±3 | 110±10 | ≤6.4 | 11±10 | 6.2±0.6 |
| Hill Coefficient | 2.05 | 2.04 | 1.05 | 1.01 | 1.49 | 0.59 | 1.65 | 1.14 | 1.59 |
| ΔΔG Wild Type** | 0 | N.D. | +0.61 | +0.42 | N.D. | +1.68 | N.D. | +0.30 | n.d. |
|
| |||||||||
| puma promoter | |||||||||
| Kdapp. (nM) | ≤6.4 | 150±20 | 15±6 | 19±5 | 7±3 | 140±40 | 130±10 | 10±4 | 7.0±0.4 |
| Hill Coefficient | 1.78 | 0.69 | 1.13 | 1.08 | 1.55 | 0.66 | 0.63 | 1.27 | 1.50 |
| ΔΔG Wild Type** | 0 | +1.86 | +0.50 | +0.63 | +0.05 | +1.81 | +1.80 | +0.25 | n.d. |
Error ranges indicate uncertainties of fit for curves obtained from the average of three independent titrations.
Determined as follows: ΔΔG = −RT*ln(Kdapp.mut/Kdapp.wild type); values in Kcal/mol
To confirm the overlapping and mutually exclusive nature of interaction sites for DNA and BCL-xL on the p53 surface, we verified that a non-fluorescent p21 promoter oligonucleotide competitively inhibited formation of the complex between F-BCL-xLΔC and wild-type p53 N-D-T. Importantly, p53 N-D-T and full length, wild-type human p53 (Bac-p53) exhibited equivalent affinities for F-BCL-xL ΔC and DNA oligonucleotides, ruling out potential interferences of the core stabilizing mutations on NMR experiments and binding assays (Supplementary Figs. 6,7).
Collectively these experiments demonstrate that mutation of p53 residues that are involved in binding to both DNA and BCL-xL disrupted both interactions, while mutation of residues that principally contact BCL-xL predominantly disrupted interactions with BCL-xL with limited effects on DNA binding. The DNA binding affinities of p53 mutants also correlated with transactivation activities measured in a yeast reporter assay40 (Supplementary Fig. 8), indicating that mutagenesis may help discriminate between the nuclear and cytosolic functions of p53.
Modulation of cytosolic p53 function through mutagenesis
The knowledge gained from in vitro mutagenesis experiments allowed us to selectively manipulate the apoptotic signaling axis of cytosolic p53. First, we evaluated the cellular functionality of p53 mutants that showed low affinity for BCL-xL but retained partial DNA binding in vitro. However, these experiments indicated that in vitro binding results predicted some but not all cellular behaviors of the p53 mutants. This outcome could be due to several factors, including the potential for p53 mutants to exhibit altered aggregation propensity in cells (possibly due the lack of enhanced stability conferred by the stabilizing mutations) and experience altered interactions with other protein partners24,25, including activating interactions with BAX-BAK.
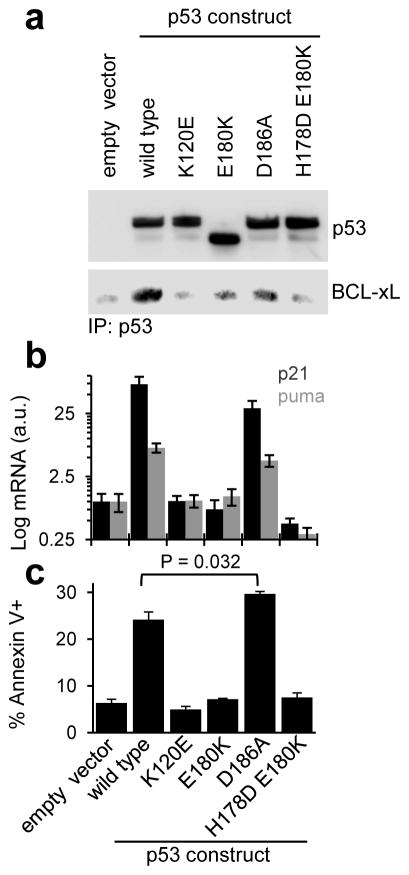
In agreement with in vitro BCL-xL binding assays, binding deficient p53 mutants expressed in p53-null H1299 cells showed little co-immunoprecipitation with endogenous BCL-xL (Fig. 6a). Most of these poorly BCL-xL-sequestered p53 mutants, however, did not activate p21 or puma transcription, despite exhibiting near wild-type-like DNA binding in vitro (Fig. 6b). As partial exception, p53 D186A significantly activated transcription of both genes (P = 0.0055 and 0.0036 for p21 and puma, respectively), although at lower levels than wild-type p53. Additional experiments provided insights into these transactivation deficiencies; the K120E mutant was defective in nuclear localization (Supplementary Fig. 9) and the E180K mutant exhibited altered post translational modification or proteolytic processing (Fig. 6a). The double mutant, H178D - E180K, exhibited weak DNA binding cooperativity in vitro (Table 5). Consistent with transactivation data, wild-type p53 and the D186A mutant were able to induce cell death (Fig. 6c). Interestingly, the latter mutant was associated with slightly higher levels of apoptosis than wild-type p53 (~30% vs ~24% Annexin V+, P = 0.032), despite its lower transcriptional activity. Asp186 within p53 DBD is not known to be involved in other established protein-protein interactions43–45; therefore we attribute the increased apoptotic function of the D186A mutant to decreased sequestration of p53 by BCL-xL and concomitant enhanced access to BAX. Conversely, the failure of other mutants to induce apoptosis cannot be ascribed solely to their lack of nuclear transactivation, as the same mutations may also alter other functions of p53, including activation of BAX or BAK in the cytosol. To evaluate this possibility, we determined the ability of wild-type and mutant p53 N-D-T to activate BAX and permeabilize model membranes that mimic the outer mitochondrial membrane [large unilamellar vesicles (LUVs)]46,47. All of the p53 mutants (in the context of p53 N-D-T) caused reduced BAX activation and membrane permeabilization compared to wild-type p53, indicating a possible overlap between the contact regions of BCL-xL and BAX on p53 (Supplementary Fig. 8). These findings underscored the challenge of using mutagenesis to dissect the various nuclear and cytosolic functions of p53 that are mediated by largely overlapping regions of the protein surface.
Figure 6.
Functional effects of mutations within p53 that destabilize its interaction with BCL-xL. a. Co-immunoprecipitation of endogenous BCL-xL and wild-type or mutant p53, transiently transfected in p53-null H1299 cells for 24 hours. The screened mutants displayed decreased levels of BCL-xL binding compared to wild-type p53. Full images of western blots are displayed in Supplementary Fig. 9. b. Quantitative polymerase chain reaction (qPCR) measurement of p21 and puma mRNA levels after transient transfection of the same p53 constructs. c. Fluorescence activated cell sorting (FACS) analysis measuring apoptosis in H1299 cells after transient transfection of the same p53 constructs. For panels b and c, error bars indicate s.e.m (n = 3 cell cultures); P values were determined by two-tailed Student’s t test.
To circumvent the complexities associated with mutating p53, we evaluated the effects of BCL-xL mutations on its ability to sequester p53 and inhibit BAX activation. While BCL-xL is also a multi-functional protein, most of its activities are mediated by interactions of BH3 domain-containing proteins with its BH3-binding groove29. The fact that p53 binds to BCL-xL via an alternative surface (vide supra) enabled interrogation of the contributions of these two independent binding surfaces to anti-apoptotic BCL-xL function. We monitored LUV permeabilization upon BAX activation by p53 N-D-T in the absence or presence of wild-type or mutant BCL-xLΔC (Fig. 7a,b). To prevent BAX inhibition through sequestration of its BH3 domain by BCL-xL48, we pre-incubated BCL-xL with either the BH3 domain of the apoptotic de-repressor BAD or the small molecule inhibitor ABT-73749. As ABT-737 also binds to the BH3-binding groove of BCL-xL, we verified using FP that this interaction only marginally affected the binding of BCL-xL to p53 [F-BCL-xLΔC – p53 N-D-T, Kd = 1.3 μM; F-BCL-xL ΔC·ABT-737 – p53 N-D-T, Kd = 3.0 μM (Supplementary Fig. 6)]. We also verified that all the BCL-xL mutants bound a BAX BH3 domain peptide with affinities comparable to that of wild-type BCL-xL (Kd ~ 500 nM; Supplementary Note), confirming that the mutations selectively altered binding of BCL-xL to p53 without compromising binding to BH3 domains.
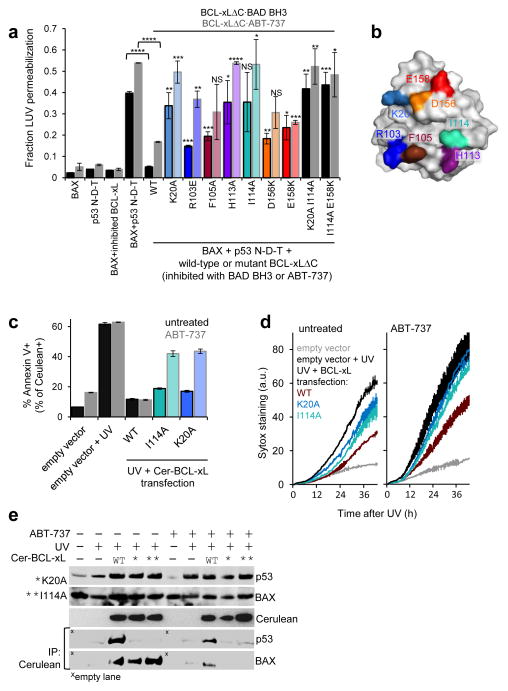
Figure 7.
Functional effects of mutations within BCL-xL that destabilize its interaction with p53. a. Fractional permeabilization of Large Unilamellar Vesicles (LUV) relative to a CHAPS detergent positive control in the presence of the indicated proteins; error bars indicate s.e.m (n = 4 experiments). *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001; NS: not statistically significant. The color coding for BCL-xLΔC mutations corresponds to that used in the surface representation of BCL-xL shown in panel b. c. FACS analysis measuring protective effects against UV-induced apoptosis in HCT116 cells, expressing wild-type p53, previously treated by transient transfection with wild-type or mutant BCL-xL fused to the fluorescent protein Cerulean. The levels of apoptosis shown in the plot are measured for Cerulean positive cell populations. Error bars, s.e.m. (n = 3 cell cultures). P values were determined by two-tailed Student’s t test. d. Differential kinetics of HCT116 cell membrane permeabilization, measured by Sytox green staining, upon transient transfection with wild-type or mutant BCL-xL followed by UV treatment, as described for panel c. Error bars, values range (n = 2 cell cultures). e. Western blot analysis of total protein levels and Cerulean co-immunoprecipitation for p53, BAX and Cerulean BCL-xL in HCT116 cells upon UV irradiation, transient transfection of wild-type or mutant Cerulean BCL-xL, and ABT-737 treatment. Full images of western blots are displayed in Supplementary Fig. 9.
The p53 N-D-T construct caused BAX activation-dependent LUV permeabilization that was inhibited by wild-type BCL-xLΔC·BAD BH3 or by wild-type BCL-xLΔC·ABT-737. Inhibition of BAX activation was dramatically diminished by mutation of Lys20, His113, and Ile114 of BCL-xLΔC and less so by mutation of Arg103, Phe105, Asp156, and Glu158, in approximate agreement with the effects of these mutations on p53 N-D-T binding. Mutation of Ile114 together with either Lys20 or Glu158 within BCL-xLΔC abrogated p53 binding and inhibition of BAX activation. In the absence of BAD BH3 or ABT-737, however, BCL-xLΔC mutants that displayed impaired protection when bound to these molecules, were able to inhibit LUV permeabilization, due to their unaltered ability to bind the BH3 domain of partially active BAX48 (Supplementary Fig. 8). These results demonstrate that BCL-xL mutations that disrupted binding to p53 N-D-T also impaired the ability of BCL-xL to inhibit p53-stimulated BAX activation.
To confirm these findings, we transiently transfected HCT116 cells, expressing wild-type p5350, with Cerulean-tagged, wild-type or mutant BCL-xL (Cer-BCL-xL) followed by UV irradiation to activate and stabilize p53. This treatment induced apoptosis in the absence and presence of ABT-737 in mocktransfected cells (~62% Annexin V+, Fig. 7c), which was inhibited in Cerulean positive cells expressing wild-type BCL-xL (~11% Annexin V+ regardless of ABT-737 treatment). Wild-type BCL-xL·ABT-737 complexes cannot sequester proapoptotic BH3 regions, but can still bind p53 and inhibit p53 activation of BAX-BAK; strikingly, ABT-737 was insufficient to abrogate BCL-xL’s anti-apoptotic function. However, cells expressing mutant BCL-xL defective in p53 binding exhibited enhanced apoptosis compared to those expressing wild-type BCL-xL in the absence of ABT-737 and significantly higher levels of apoptosis when treated with ABT-737 (19% and 17% Annexin V+ w/o ABT-737 versus 42% and 44% Annexin V+ with ABT-737 for the Ile114 and Lys20 mutants, respectively; P = 0.0003 and P <0.0001, respectively). We further used fluorescence imaging to monitor the viability of HCT116 cells transfected with the same BCL-xL reagents. This experiment, performed with longer incubation times than the previous assay, revealed differential cell death between samples transfected with wild-type or mutant BCL-xL both in the absence and presence of ABT-737 (Fig. 7d). Co-immunoprecipitation demonstrated the dependence of these results on differential sequestration of p53 by wild-type or mutant BCL-xL constructs (Fig. 7e). Treatment with ABT-737 decreased, but did not abrogate, coimmunoprecipitation of p53 with wild-type Cer-BCL-xL. In support of BCL-xL-dependent inhibition of BAX activation, BAX co-immunoprecipitated with all of the Cer-BCL-xL constructs, wild-type and mutant, and this interaction was nearly abrogated in the presence of ABT-737. Furthermore, the two BCL-xL mutants exhibited wild-type-like affinity for the BH3 domains of BIM and BID, and were equivalent to wild-type BCL-xL in inhibiting BIM or BID-dependent BAX activation in LUV assays (Supplementary Note). Mutations designed to selectively disrupt p53 binding therefore did not alter the BH3 domain binding properties of BCL-xL. In ABT-737-treated cells, not only were BH3 domains released from the BCL-xL hydrophobic groove, but binding and sequestration of cytosolic p53 by mutant BCL-xL was also impaired, providing multiple mechanisms for BAX activation. These results support the role of the surface of BCL-xL shown in our solution structure of the BCL-xLΔC – p53 DBD complex in inhibiting p53-dependent BAX activation and induction of apoptosis in human cells.
DISCUSSION
Past studies of BCL-xL – p53 interactions have dealt with individual p53 domains12–15,18–20 without addressing how they synergize within the full-length protein to engage BCL-xL. Our results confirm that p53 DBD is thermodynamically dominant in mediating binding to BCL-xL, and show that, while p53 NTD interacts weakly with the BH3 groove of BCL-xL, as previously reported18–20, it does not synergize with other domains to enhance BCL-xL binding. Tetramerization of p53, however, indirectly enhances binding to BCL-xL by increasing the local concentration of DBDs. Our determination of the solution structure of the BCL-xLΔC – p53 DBD complex reveals the molecular basis for sequestration of p53 by BCL-xL to inhibit BAX-BAK activation. This knowledge empowered the elucidation of differential roles of BCL-xL in regulating apoptosis through protein-protein interactions mediated by its BH3-binding groove and its p53-binding surface. The interface between the two proteins buries 1821 Å2 of surface area, dominated by charged and hydrophilic (1076 Å2) versus hydrophobic (745 Å2) amino acids. This situation arises because the BCL-xL surface recognizes the same region of p53 DBD utilized for binding DNA33. While BCL-xL does not structurally mimic duplex DNA, charged residues on its surface engage p53 DBD residues that make contacts with bases or phosphate groups of promoter DNA. The electrostatic nature of the interaction patch that overlaps with the p53 DNA-binding surface, as well as contacts involving the Zn2+ coordination site in p53 and acidic residues in BCL-xL, result in a low interface hydrophobicity (−0.43 Kcal/residue calculated solvation energy, versus an average value of −0.26 Kcal/mol for heterodimeric complexes)51. These results define the structural and functional roles of different p53 domains in binding to BCL-xL.
The p53 DBD surface that engages BCL-xL is larger than that utilized to bind DNA, suggesting that it was possible to modulate this interaction through p53 DBD mutagenesis without abrogating DNA binding. Efforts to selectively eliminate BCL-xL binding through mutagenesis, however, were confounded by additional effects on protein stability and/or other interactions mediated by the same region of p5324,25. Additional functions affected may include the activation of BAX or BAK, previously reported to engage the DNA-binding domain of p5313. Despite this caveat, the slightly higher levels of apoptosis induced by the D186A mutant, which is released from BCL-xL, reinforces the role of BCL-xL as an inhibitor of p53’s cytosolic, pro-apoptotic activity. This result suggests that decreased BCL-xL sequestration of this mutant compensates for its partial impairment in activation of both transcription and BAX, leading to efficient induction of apoptosis.
In contrast to the complexities associated with selectively disrupting p53 binding to BCL-xL, structure-based mutagenesis of BCL-xL selectively disrupted BCL-xL – p53 binding in vitro and in cells, and enhanced p53-dependent apoptosis without disrupting BCL-xL’s functions associated with sequestration of BH3 domains33. In the absence of ABT-737, the equivalent inhibitory activities of wild-type and mutant BCL-xL constructs against BH3 domains overrode their differential ability to bind and sequester UV-activated p53. In the presence of ABT-737 these anti-apoptotic activities were inhibited, to reveal the differential p53 binding-related, anti-apoptotic functions of wild-type versus mutant BCL-xL. In cells expressing wild-type BCL-xL, capable of sequestering cytosolic p53, addition of ABT-737 was insufficient to abrogate its protective effects against UV-induced apoptosis. However, BCL-xL mutants with impaired p53 binding displayed markedly impaired anti-apoptotic effects. These results underscore the activity of cytosolic p53 to mediate UV-induced apoptosis and the ability of BCL-xL to oppose this activity.
The BCL-xL – p53 DBD structure expands our understanding of p53 DBD’s binding and functional versatility: this domain exploits the same surfaces to bind diverse protein partners (e.g., BCL-xL, 53BP143, 53BP244, and sv4045) or target DNA in different regulatory contexts. It is fascinating that the DBD, long thought to dominate p53’s tumor suppressor functions through its nuclear gene regulatory activity21,22,23,52, is also an instrument of apoptotic regulation in the cytosol. We have characterized the structural details of p53’s interaction with BCL-xL and the role this interaction plays in restraining UV- and p53-dependent BAX activation and apoptosis. Our data show that p53 functionally interacts with BAX, as shown previously2, but molecular details of these interactions are poorly understood. Future studies into the structural basis of this interaction, as well as that with the other apoptotic effector, BAK, will provide deeper understanding of p53’s cytosolic, apoptotic regulatory network and its contributions to tumor suppression.
ONLINE METHODS
Reagents
Detailed information about reagents employed in this study can be found in the accompanying Supplementary Note.
Protein expression and purification
Detailed descriptions of recombinant protein expression and purification procedures can be found in the accompanying Supplementary Note. Briefly, all proteins were expressed in E. coli BL21(DE3) expression strain. Unlabeled proteins were expressed in LB media, isotopically labeled ones in MOPS minimal media supplemented with the appropriate labeling reagents dissolved in H2O or D2O. Selective protonation of Isoleucine, Leucine, Valine methyl moieties was performed according to established procedures53. To maximize the yield, the insoluble fraction of isotopically labeled 2H/13C/15N-p53 N-D-T was recovered from inclusion bodies and refolded according to an established procedure54, then purified as described in the Supplementary Note.
NMR spectroscopy
All samples for NMR experiments were prepared in 10 mM sodium phosphate, 40 mM NaCl, 2 mM DTT (or 5 mM DTT for 3D p53 DBD samples), 0.01% NaN3, pH 7.0 unless otherwise specified. Sample concentrations were 100 μM for 15N-TROSY chemical shift titrations, 200 μM for 13C-HSQC chemical shift titrations, 0.5 mM (p53 DBD), 0.6–1 mM (free BCL-xLΔC), 0.6 mM (BCL-xLΔC bound to unlabeled p53 DBD) for samples employed for 3D experiments. Unlabeled sample components were added in 1.1 molar excess over the labeled one unless otherwise specified. All experiments were performed at 25 °C on 600 MHz or 800 MHz Bruker Avance spectrometers both equipped with cryogenically cooled triple resonance z-gradient probes. Data were processed using Bruker Topspin software and analyzed with the program CARA55. Resonance assignments were derived from published assignments of each studied construct, confirmed and integrated with triple-resonance experiments as described in detail in the Supplementary Note. For Three-dimensional NOESY experiments, A NOE mixing time of 120 ms was employed, with the exception of 15N-edited TROSY-NOESY spectra of 2H/15N-p53 DBD (450 μM) in the absence and presence of 2:1 molar excess unlabeled BCL-xLΔC, collected over 24 scans, 2048 × 40 × 192 increments with a NOE mixing time of 160 ms. To perform paramagnetic relaxation enhancement (PRE) experiments a S-(2,2,5,5-tetramethyl-2,5-dihydro-1H-pyrrol-3-yl)methyl methanesulfonothioate) (MTSL) spin label was introduced on BCL-xL Cys151 or, after mutating the latter to Ser, S2C and S122C mutants by adding a 10:1 molar excess of the label dissolved in acetonitrile to BCL-xL samples in 25 mM tris, 100 mM NaCl, pH 7.5. Reaction mixtures were incubated at room temperature in the dark for two hours followed by overnight incubation at 4 °C. Excess unreacted spin label was removed by extensive buffer exchange over a Centricon centrifugal device. NMR samples containing 100 μM 2H/15N-labeled p53 DBD and the various MTSL labeled BCL-xL constructs (150 μM) were prepared in the standard NMR sample buffer (see above) lacking DTT. TROSY spectra relevant to the oxidized state of the spin label were acquired swiftly in ~1.5 hours, followed by immediate addition of excess solid DTT (to a final concentration ~10 mM) which allowed to reduce the spin label and stabilize the cystein-rich p53 DBD. A reference TROSY spectrum was then acquired for each sample after the spin label had been reduced. For PRE experiments involving isotopically labeled BCL-xL (100 μM) and paramagnetic Co2+ labeled p53 DBD (150 μM), the TROSY spectra of these samples were referenced against identically prepared samples containing Zn2+ expressed p53 DBD. For 13C-detected and methyl region13C-HSQC PRE experiments, samples were employed containing 0.6 mM 2H/13C/15N-labeled BCL-xLΔC with selective protonation of I,L,V side-chain methyl groups in the presence of 1.1 molar excess Zn2+ or Co2+ expressed p53 DBD.
Structure calculation
Solution structure of BCL-xL free and p53-bound conformation
Calculations were performed using the program Cyana56 for 100 structures with 10,000 annealing steps, the 20 top ranking structures being selected as output. Distance restraints were generated from lists of NOESY cross peaks imported from CARA depositories using the program Caliba for restraint calibration and merging. The output restraint lists were inspected by comparison with actual NOESY spectra to check for incorrect integrations due to peak overlaps. Dihedral restraints were generated using the program Talos57. Hydrogen bond restraints were generated manually after inspection of the presence or absence of amide-water cross peaks in NOESY spectra. The Ramachandran statistics for the two models, assessed with the Procheck58 program were as follows. Apo BCL-xL; Most favored regions: 68.1% of all residues (84.6% of structured residues: 1–27, 86–196); Additionally allowed regions: 25.5% (12.1%); Generously allowed regions: 4.9% (2.2%); Disallowed regions 1.5% (1.1%). For p53-bound BCL-xL: Most favored regions: 69.2% (86.3%); Additionally allowed regions: 23.4% (9.8%); Generously allowed regions: 6.4 (3.5%); Disallowed regions: 1.1% (0.3%).
Structure of BCL-xL – p53 DBD complex
Calculations were performed using the web-server version of the program Haddock59 with default settings and unambiguous distance restraints. PRE distance restraints were defined according to the formula of PRE distance dependence60 with error ranges accounting for the undefined distance between the paramagnetic center and the restraint location (Cα of the labeled residue) for MTSL-derived restraints. NOE restraints were assigned to the heavy atom attached to the involved protons and, based on the cross-peak intensity, defined as strong (1–7 Å) or weak (3–9 Å). Input structures were ‘chain A’ of the crystal structure of free p53 DBD (pdb 2OCJ)61 or DNA bound p53 DBD (pdb 2AC0)32 and the top ranking NMR solution structure of p53-bound BCL-xL. No noticeable difference was observed between calculations performed free or DNA-bound p53 DBD input structures, however the latter produced a slightly better energy score (−220.5 ± 2.1 versus −203.6 ± 7.5) and was therefore chosen for further refinement performed with the Haddock refinement function. This step, which reduced the output cluster size from 200 to 20 structures, was repeated for three iterations until no further improvement of the structure energy function was observed (final score = −257.6±4.5). Interface properties and hydrophobicity were analyzed using the ProtorP62, ProFace63 and MolSurfer64 web servers. The Ramachandran statistics for the model were as follows: Most favored regions: 85.0%; Additionally allowed regions: 13.8%; Generously allowed regions: 0.7%; Disallowed regions: 0.5%.
Isothermal titration calorimetry
All ITC experiments were performed over 19 injections of 2 μL each at 25 °C in 20 mM Tris, 40 mM NaCl, 2 mM TCEP, pH 7.0 using a MicroCal ITC200 instrument. Data were analyzed using the MicroCal ITC plugins within the Origin software suite (OriginLab). All titrations were performed in duplicate. For detailed descriptions of conditions for each set of titrations refer to the accompanying Supplementary Note.
Fluorescence polarization
The BCL-xL C151S-S2C double mutant was labeled with 5- Iodoacetamidofluorescein (5-IAF, Thermo Scientific) in its stock buffer (25 mM Tris, 200 mM NaCl, 1mM EDTA, 5 mM DTT, pH 7.0) according to the manufacturer’s reaction protocol in presence of DTT. Excess unreacted dye was removed by extensive buffer exchange over a centrifugal device. Fluorescence polarization measurements were performed at 25°C in 96-well plates on a Perkin Elmer EnVision plate reader equipped with FITC excitation and emission filters. Instrument settings were optimized automatically. The buffer employed was the same used for ITC experiments. The concentration of fluorescent probe (F-BCL-xL ΔC or DNA oligos) was 20 nM in all cases. For direct binding experiments 1:1 serial dilutions of WT or mutant p53 1–360 were performed over the range of concentrations indicated in the relevant figure, for competition experiments between F-BCL-xLΔC and unlabeled BCL-xL mutants or p21 promoter oligonucleotide, the latter were serially diluted (1:1) into a mix of 20 nM F-BCL-xL ΔC and 1.5 μM p53 1–360. All samples were incubated at room temperature in the dark for 2 hours before fluorescence readings. Titrations were fit to a Langmuir isotherm equation, for DNA binding titrations by p53 a Hill coefficient (h) was further introduced to account for the cooperative effect of binding by p53 tetramers: Fraction bound = 1/[1+(Kd/conc)h], where Kd is the fitted dissociation constant and conc the variable concentration of the titrated species. Competition titrations were fit as described previously29. Fitting of each titration was performed on the average of three independent experiments.
Large unilamellar vesicle assay
The assay was performed according to established procedures47. Fluorescence readings were carried out at 37°C in a Horiba Fluorolog 3 spectrofluorimeter using a 3 mM path-length, 45 μL volume quartz cuvette. Details about specific settings and sample concentrations are provided in the accompanying Supplementary Note.
Cell death analysis
NCI-H1299 or HCT116 cells (American Type Culture Collection – ATCC), cultured under prophylactic Plasmocin treatment (InvivoGen), were plated at 105 cells/mL (1 mL in 12-well plates) in Dulbecco modified Eagle medium supplemented with 10% fetal calf serum supplemented with 2mM glutamine, penicillin and streptomycin, transiently transfected for 24 hours with 1 μg of the indicated p53 (H1299 cells) or Cerulean BCL-xL (HCT116) constructs (all cloned in pcDNA3.1 vector, Invitrogen) using lipofectamine. BCL-xL transfected HCT116 cells were then UV irradiated (10 mJ/cm2) and incubated for additional 16 hours. Supernatant medium and trypsinized cells were collected, stained with APC-labeled Annexin V (eBiosciences) and cell death was analyzed by flow cytometry. A total of 104 cells were analyzed for each sample, each condition was tested in three independent samples. Equivalent sets of samples were then pooled, harvested and immediately lysed and analyzed by coimmunoprecipitation as described below.
Cell death kinetics analysis
HCT116 cells were manipulated and transiently transfected BCL-xL constructs for 24 hours as described above (105 cells/mL; 0.5 mL seeding in 24-well plates). After UV irradiation (10 mJ/cm2) and delivery of ABT-737 for samples that required this treatment, cultures were imaged for a total of 42 hours for Sytox Green (Invitrogen) fluorescence and phase contrast imaging in an IncuCyte (Essen BioScience) instrument. Four images were collected for each well at 30′ intervals, duplicate samples were analyzed for each condition. Average object count figures were normalized to an approximate 0–100 scale.
Co-immunoprecipitation
Cells were lysed in 0.4 mL ice-cold buffer containing 50 mM tris, 150 mM NaCl, 1 mM EDTA, 0.5 % NP40, pH 7.4 supplemented with protease inhibitor tablets (Roche). Cell lysates were incubated with 20 μL Protein A/G beads (Santa Cruz Biotechnology) supplemented with 1 μg anti-p53 antibody (clone DO-7, Santa Cruz Biotechnology at 1:1000 dilution – for H1299 cells transfected with p53 constructs), or 1 μg anti GFP antibody (mixed 7.1 and 13.1 clones, Roche at 1:2000 dilution – for HCT116 cells transfected with Cerulean BCL-xL constructs) for 2 hours at 4 degrees, washed 4 times with cold lysis buffer and eluted by boiling in 50 μL 2X XT sample buffer (BioRad). The protein concentration from total lysate aliquots was determined by BCA assay and normalized amounts of immunoprecipitated samples were analyzed by SDS-PAGE and western blot. H1299 cell samples were analyzed for p53 (antibody clone DO-7) and endogenous BCL-xL (antibody clone S18, Santa Cruz Biotechnology, at 1:2000 dilution). HCT116 cell samples were analyzed for Cerulean BCL-xL (GFP antibody mixed clones 7.1, 13.1), p53 (antibody clone DO-7), BAX (antibody clone N20, Santa Cruz Biotechnology). Validation of antibodies employed in these assays can be found on the respective manufacturers’ websites.
Quantitative polymerase chain reaction
NCI-H1299 cells were manipulated and transiently transfected with p53 constructs as described above (105 cells/mL; 2 mL seeding in 6-well plates). RNA was isolated using a NucleoSpin RNAII kit (Macherey-Nagel) and 1 μg aliquots were employed for first-strand cDNA synthesis (SuperScript kit, Invitrogen). Equivalent aliquots of cDNA were analyzed by quantitative polymerase chain reaction using Sybr green staining (Applied Biosystems) in an Applied Biosystems 7900HT instrument. Expression of p53 target genes p21 and puma was normalized to h18S levels. Triplicate samples were analyzed for each condition.
Subcellular fractionation
NCI-H1299 cells were manipulated and transfected as described above (105 cells/mL; 2 mL seeding in 6-well plates). Cells were trypsinized, harvested and resuspended in 0.1 mL fractionation buffer containing 250 mM sucrose, 20 mM HEPES (pH: 7.4). 10 mM KCl, 1.5 mM MgCl2, 1 mM EDTA, 1 mM EGTA, 1 mM DTT, protease inhibitor cocktail (Roche). After 20 min. incubation on ice, lysates were centrifuged at 720 G for 5′. The obtained nuclear pellets were further washed with 0.2 mL fractionation buffer. Supernatants were centrifuged at 10000 G for 5′ to separate heavy membrane (pellet) from cytosolic (supernatant) fractions. Nuclear and heavy membrane pellets were resuspended in fractionation buffer supplemented with 10% glycerol and 0.1% SDS. The concentration of all fractions was determined by absorbance at 280 nm, normalized amounts were analyzed by SDS page and western blotting for p53 (antibody clone DO-7), GAPDH (antibody clone 6C5, Chemicon, at 1:2000 dilution), and Lamin B (polyclonal antibody AbCam ab16048, at 1:2000 dilution). Validation of antibodies employed in these assays can be found on the respective manufacturers’ websites.
Supplementary Material
Acknowledgments
The authors acknowledge Christy R. Grace (at St. Jude Children’s Research Hospital) for assistance with NMR experiments, Cheon-Gil Park (at St. Jude Children’s Research Hospital) for assistance with protein preparation and isotope labeling, and Jerry Chipuk (at Mt. Sinai Medical Center, New York, NY) for helpful discussion and advice. This work was supported by US National Institutes of Health (NIH) grants R01CA082491 and 1R01GM083159 (to R.W.K.), NIH R01GM52735 and R01GM96208 (to D.R.G.), a US National Cancer Institute Cancer Center Support Grant P30CA21765 (at St. Jude Children’s Research Hospital), and ALSAC. A. V. F. is the recipient of the Neoma Boadway Fellowship from St. Jude Children’s Research Hospital.
Footnotes
AUTHOR CONTRIBUTIONS
A.V.F. and L.O. contributed to NMR experiments and data analysis; A.V.F. contributed to structural calculations, biophysical and biochemical assays; A.V.F, F.L. and K.B contributed to cellular assays; A.V.F., D.R.G. and R.K. wrote the manuscript. All authors contributed to different aspects of the design and conception of this study.
ACCESSION CODES
NMR data and structures of BCL-xL calculated with selective isotope labeling in its free and p53 DBD-bound conformation have been deposited to the Biological Magnetic Resonance Bank (BMRB) and Protein Data Bank (PDB) under the following accession codes. Free BCL-xL: 19521 (BMRB), 2me9 (PDB); p53 DBD-bound BCL-xL conformation: 19520 (BMRB), 2me8 (PDB). The NMR structure of the BCL-xL – p53 DBD complex and related NMR data have been deposited under accession codes 19522 (BMRB) and 2mej (PDB).
References
- 1.Green DR, Kroemer G. Cytoplasmic functions of the tumour suppressor p53. Nature. 2009;458:1127–1130. doi: 10.1038/nature07986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chipuk JE, et al. Direct activation of Bax by p53 mediates mitochondrial membrane permeabilization and apoptosis. Science. 2004;303:1010–1014. doi: 10.1126/science.1092734. [DOI] [PubMed] [Google Scholar]
- 3.Leu JI, Dumont P, Hafey M, Murphy ME, George DL. Mitochondrial p53 activates Bak and causes disruption of a Bak-Mcl1 complex. Nat Cell Biol. 2004;6:443–450. doi: 10.1038/ncb1123. [DOI] [PubMed] [Google Scholar]
- 4.Chipuk JE, Bouchier-Hayes L, Kuwana T, Newmeyer DD, Green DR. PUMA couples the nuclear and cytoplasmic proapoptotic function of p53. Science. 2005;309:1732–1735. doi: 10.1126/science.1114297. [DOI] [PubMed] [Google Scholar]
- 5.Sattler M, et al. Structure of Bcl-xL-Bak peptide complex: recognition between regulators of apoptosis. Science. 1997;275:983–986. doi: 10.1126/science.275.5302.983. [DOI] [PubMed] [Google Scholar]
- 6.Petros AM, et al. Rationale for Bcl-xL/Bad peptide complex formation from structure, mutagenesis, and biophysical studies. Protein Sci. 2000;9:2528–2534. doi: 10.1110/ps.9.12.2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Feng W, Huang S, Wu H, Zhang M. Molecular basis of Bcl-xL’s target recognition versatility revealed by the structure of Bcl-xL in complex with the BH3 domain of Beclin-1. J Mol Biol. 2007;372:223–235. doi: 10.1016/j.jmb.2007.06.069. [DOI] [PubMed] [Google Scholar]
- 8.Liu X, Dai S, Zhu Y, Marrack P, Kappler JW. The structure of a Bcl-xL/Bim fragment complex: implications for Bim function. Immunity. 2003;19:341–352. doi: 10.1016/s1074-7613(03)00234-6. [DOI] [PubMed] [Google Scholar]
- 9.Cheng EH, et al. BCL-2, BCL-X(L) sequester BH3 domain-only molecules preventing BAX- and BAK-mediated mitochondrial apoptosis. Molecular Cell. 2001;8:705–711. doi: 10.1016/s1097-2765(01)00320-3. [DOI] [PubMed] [Google Scholar]
- 10.Kussie PH, et al. Structure of the MDM2 oncoprotein bound to the p53 tumor suppressor transactivation domain. Science. 1996;274:948–953. doi: 10.1126/science.274.5289.948. [DOI] [PubMed] [Google Scholar]
- 11.Wang P, et al. p53 domains: structure, oligomerization, and transformation. Mol Cell Biol. 1994;14:5182–5191. doi: 10.1128/mcb.14.8.5182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Petros AM, Gunasekera A, Xu N, Olejniczak ET, Fesik SW. Defining the p53 DNA-binding domain/Bcl-x(L)-binding interface using NMR. FEBS Lett. 2004;559:171–174. doi: 10.1016/S0014-5793(04)00059-6. [DOI] [PubMed] [Google Scholar]
- 13.Sot B, Freund SM, Fersht AR. Comparative biophysical characterization of p53 with the pro-apoptotic BAK and the anti-apoptotic BCL-xL. J Biol Chem. 2007;282:29193–29200. doi: 10.1074/jbc.M705544200. [DOI] [PubMed] [Google Scholar]
- 14.Hagn F, et al. BclxL changes conformation upon binding to wild-type but not mutant p53 DNA binding domain. J Biol Chem. 2010;285:3439–3450. doi: 10.1074/jbc.M109.065391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yao H, et al. Anti-apoptosis Proteins Mcl-1 and Bcl-xL Have Different p53-Binding Profiles. Biochemistry. 2013:6324–6334. doi: 10.1021/bi400690m. [DOI] [PubMed] [Google Scholar]
- 16.Di Lello P, et al. Structure of the Tfb1/p53 complex: Insights into the interaction between the p62/Tfb1 subunit of TFIIH and the activation domain of p53. Mol Cell. 2006;22:731–740. doi: 10.1016/j.molcel.2006.05.007. [DOI] [PubMed] [Google Scholar]
- 17.Lee CW, Martinez-Yamout MA, Dyson HJ, Wright PE. Structure of the p53 transactivation domain in complex with the nuclear receptor coactivator binding domain of CREB binding protein. Biochemistry. 2010;49:9964–9971. doi: 10.1021/bi1012996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu H, Tai J, Ye H, Kang CB, Yoon HS. The N-terminal domain of tumor suppressor p53 is involved in the molecular interaction with the anti-apoptotic protein Bcl-Xl. Biochem Biophys Res Commun. 2006;341:938–944. doi: 10.1016/j.bbrc.2005.12.227. [DOI] [PubMed] [Google Scholar]
- 19.Xu H, et al. The MDM2-binding region in the transactivation domain of p53 also acts as a Bcl-X(L)-binding motif. Biochemistry. 2009;48:12159–12168. doi: 10.1021/bi901188s. [DOI] [PubMed] [Google Scholar]
- 20.Ha JH, et al. Dual-site interactions of p53 protein transactivation domain with anti-apoptotic Bcl–2 family proteins reveal a highly convergent mechanism of divergent p53 pathways. J Biol Chem. 2013;288:7387–7398. doi: 10.1074/jbc.M112.400754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Livingstone LR, et al. Altered cell cycle arrest and gene amplification potential accompany loss of wild-type p53. Cell. 1992;70:923–935. doi: 10.1016/0092-8674(92)90243-6. [DOI] [PubMed] [Google Scholar]
- 22.Serrano M, Lin AW, McCurrach ME, Beach D, Lowe SW. Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell. 1997;88:593–602. doi: 10.1016/s0092-8674(00)81902-9. [DOI] [PubMed] [Google Scholar]
- 23.Yonish-Rouach E, et al. Wild-type p53 induces apoptosis of myeloid leukaemic cells that is inhibited by interleukin-6. Nature. 1991;352:345–347. doi: 10.1038/352345a0. [DOI] [PubMed] [Google Scholar]
- 24.Mietz JA, Unger T, Huibregtse JM, Howley PM. The transcriptional transactivation function of wild-type p53 is inhibited by SV40 large T-antigen and by HPV-16 E6 oncoprotein. EMBO J. 1992;11:5013–5020. doi: 10.1002/j.1460-2075.1992.tb05608.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schultz LB, Chehab NH, Malikzay A, Halazonetis TD. p53 binding protein 1 (53BP1) is an early participant in the cellular response to DNA double-strand breaks. J Cell Biol. 2000;151:1381–1390. doi: 10.1083/jcb.151.7.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pervushin K, Riek R, Wider G, Wuthrich K. Attenuated T-2 relaxation by mutual cancellation of dipole-dipole coupling and chemical shift anisotropy indicates an avenue to NMR structures of very large biological macromolecules in solution. Proc Natl Acad Sci U S A. 1997;94:12366–12371. doi: 10.1073/pnas.94.23.12366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Muchmore SW, et al. X-ray and NMR structure of human Bcl-xL, an inhibitor of programmed cell death. Nature. 1996;381:335–341. doi: 10.1038/381335a0. [DOI] [PubMed] [Google Scholar]
- 28.Joerger AC, Allen MD, Fersht AR. Crystal structure of a superstable mutant of human p53 core domain. Insights into the mechanism of rescuing oncogenic mutations. J Biol Chem. 2004;279:1291–1296. doi: 10.1074/jbc.M309732200. [DOI] [PubMed] [Google Scholar]
- 29.Follis AV, et al. PUMA binding induces partial unfolding within BCL-xL to disrupt p53 binding and promote apoptosis. Nat Chem Biol. 2013;9:163–168. doi: 10.1038/nchembio.1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Galea C, Bowman P, Kriwacki RW. Disruption of an intermonomer salt bridge in the p53 tetramerization domain results in an increased propensity to form amyloid fibrils. Protein Sci. 2005;14:2993–3003. doi: 10.1110/ps.051622005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Johnson CR, Morin PE, Arrowsmith CH, Freire E. Thermodynamic Analysis of the Structural Stability of the Tetrameric Oligomerization Domain of P53 Tumor-Suppressor. Biochemistry. 1995;34:5309–5316. doi: 10.1021/bi00016a002. [DOI] [PubMed] [Google Scholar]
- 32.Kitayner M, et al. Structural basis of DNA recognition by p53 tetramers. Mol Cell. 2006;22:741–753. doi: 10.1016/j.molcel.2006.05.015. [DOI] [PubMed] [Google Scholar]
- 33.Cho Y, Gorina S, Jeffrey PD, Pavletich NP. Crystal structure of a p53 tumor suppressor-DNA complex: understanding tumorigenic mutations. Science. 1994;265:346–355. doi: 10.1126/science.8023157. [DOI] [PubMed] [Google Scholar]
- 34.Chipuk JE, Moldoveanu T, Llambi F, Parsons MJ, Green DR. The BCL-2 family reunion. Mol Cell. 2010;37:299–310. doi: 10.1016/j.molcel.2010.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hollstein M, et al. Database of p53 gene somatic mutations in human tumors and cell lines. Nucleic Acids Res. 1994;22:3551–3555. [PMC free article] [PubMed] [Google Scholar]
- 36.Hernandez-Boussard T, Rodriguez-Tome P, Montesano R, Hainaut P. IARC p53 mutation database: a relational database to compile and analyze p53 mutations in human tumors and cell lines. International Agency for Research on Cancer. Hum Mutat. 1999;14:1–8. doi: 10.1002/(SICI)1098-1004(1999)14:1<1::AID-HUMU1>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 37.Bullock AN, et al. Thermodynamic stability of wild-type and mutant p53 core domain. Proc Natl Acad Sci U S A. 1997;94:14338–14342. doi: 10.1073/pnas.94.26.14338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Inga A, Resnick MA. Novel human p53 mutations that are toxic to yeast can enhance transactivation of specific promoters and reactivate tumor p53 mutants. Oncogene. 2001;20:3409–3419. doi: 10.1038/sj.onc.1204457. [DOI] [PubMed] [Google Scholar]
- 39.Bullock AN, Henckel J, Fersht AR. Quantitative analysis of residual folding and DNA binding in mutant p53 core domain: definition of mutant states for rescue in cancer therapy. Oncogene. 2000;19:1245–1256. doi: 10.1038/sj.onc.1203434. [DOI] [PubMed] [Google Scholar]
- 40.Kato S, et al. Understanding the function-structure and function-mutation relationships of p53 tumor suppressor protein by high-resolution missense mutation analysis. Proc Natl Acad Sci U S A. 2003;100:8424–8429. doi: 10.1073/pnas.1431692100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Weinberg RL, Veprintsev DB, Fersht AR. Cooperative binding of tetrameric p53 to DNA. J Mol Biol. 2004;341:1145–1159. doi: 10.1016/j.jmb.2004.06.071. [DOI] [PubMed] [Google Scholar]
- 42.Hollstein M, Sidransky D, Vogelstein B, Harris CC. p53 mutations in human cancers. Science. 1991;253:49–53. doi: 10.1126/science.1905840. [DOI] [PubMed] [Google Scholar]
- 43.Joo WS, et al. Structure of the 53BP1 BRCT region bound to p53 and its comparison to the Brca1 BRCT structure. Genes Dev. 2002;16:583–593. doi: 10.1101/gad.959202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gorina S, Pavletich NP. Structure of the p53 tumor suppressor bound to the ankyrin and SH3 domains of 53BP2. Science. 1996;274:1001–1005. doi: 10.1126/science.274.5289.1001. [DOI] [PubMed] [Google Scholar]
- 45.Lilyestrom W, Klein MG, Zhang R, Joachimiak A, Chen XS. Crystal structure of SV40 large T-antigen bound to p53: interplay between a viral oncoprotein and a cellular tumor suppressor. Genes Dev. 2006;20:2373–2382. doi: 10.1101/gad.1456306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kuwana T, et al. Bid, Bax, and lipids cooperate to form supramolecular openings in the outer mitochondrial membrane. Cell. 2002;111:331–342. doi: 10.1016/s0092-8674(02)01036-x. [DOI] [PubMed] [Google Scholar]
- 47.Asciolla JJ, Renault TT, Chipuk JE. Examining BCL-2 family function with large unilamellar vesicles. J Vis Exp. 2012 doi: 10.3791/4291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Llambi F, et al. A unified model of mammalian BCL-2 protein family interactions at the mitochondria. Mol Cell. 2011;44:517–531. doi: 10.1016/j.molcel.2011.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Oltersdorf T, et al. An inhibitor of Bcl-2 family proteins induces regression of solid tumours. Nature. 2005;435:677–681. doi: 10.1038/nature03579. [DOI] [PubMed] [Google Scholar]
- 50.Liu Y, Bodmer WF. Analysis of P53 mutations and their expression in 56 colorectal cancer cell lines. Proc Natl Acad Sci U S A. 2006;103:976–981. doi: 10.1073/pnas.0510146103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jones S, Thornton JM. Principles of protein-protein interactions. Proceedings of the National Academy of Sciences of the United States of America. 1996;93:13–20. doi: 10.1073/pnas.93.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Miyashita T, Reed JC. Tumor suppressor p53 is a direct transcriptional activator of the human bax gene. Cell. 1995;80:293–299. doi: 10.1016/0092-8674(95)90412-3. [DOI] [PubMed] [Google Scholar]
- 53.Wang Y, Filippov I, Richter C, Luo R, Kriwacki RW. Solution NMR studies of an intrinsically unstructured protein within a dilute, 75 kDa eukaryotic protein assembly; probing the practical limits for efficiently assigning polypeptide backbone resonances. Chembiochem. 2005;6:2242–2246. doi: 10.1002/cbic.200500260. [DOI] [PubMed] [Google Scholar]
- 54.Bell S, Hansen S, Buchner J. Refolding and structural characterization of the human p53 tumor suppressor protein. Biophys Chem. 2002;96:243–257. doi: 10.1016/s0301-4622(02)00011-x. [DOI] [PubMed] [Google Scholar]
- 55.Keller RJJ. PhD thesis. Eidgenossische Technishe Hochschule; 2005. Optimizing the process of nuclear magnetic resonance spectrum analysis and computer aided resonance assingment. [Google Scholar]
- 56.Guntert P. Automated NMR structure calculation with CYANA. Methods Mol Biol. 2004;278:353–378. doi: 10.1385/1-59259-809-9:353. [DOI] [PubMed] [Google Scholar]
- 57.Cornilescu G, Delaglio F, Bax A. Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J Biomol Nmr. 1999;13:289–302. doi: 10.1023/a:1008392405740. [DOI] [PubMed] [Google Scholar]
- 58.Laskowski RA, Rullmannn JA, MacArthur MW, Kaptein R, Thornton JM. AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J Biomol NMR. 1996;8:477–486. doi: 10.1007/BF00228148. [DOI] [PubMed] [Google Scholar]
- 59.de Vries SJ, van Dijk M, Bonvin AM. The HADDOCK web server for data-driven biomolecular docking. Nat Protoc. 2010;5:883–897. doi: 10.1038/nprot.2010.32. [DOI] [PubMed] [Google Scholar]
- 60.Simon B, Madl T, Mackereth CD, Nilges M, Sattler M. An efficient protocol for NMR-spectroscopy-based structure determination of protein complexes in solution. Angewandte Chemie. International Ed in English. 2010;49:1967–1970. doi: 10.1002/anie.200906147. [DOI] [PubMed] [Google Scholar]
- 61.Wang Y, Rosengarth A, Luecke H. Structure of the human p53 core domain in the absence of DNA. Acta Crystallogr D Biol Crystallogr. 2007;63:276–281. doi: 10.1107/S0907444906048499. [DOI] [PubMed] [Google Scholar]
- 62.Reynolds C, Damerell D, Jones S. ProtorP: a protein-protein interaction analysis server. Bioinformatics. 2009;25:413–414. doi: 10.1093/bioinformatics/btn584. [DOI] [PubMed] [Google Scholar]
- 63.Saha RP, Bahadur RP, Pal A, Mandal S, Chakrabarti P. ProFace: a server for the analysis of the physicochemical features of protein-protein interfaces. BMC Struct Biol. 2006;6:11. doi: 10.1186/1472-6807-6-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gabdoulline RR, Wade RC, Walther D. MolSurfer: A macromolecular interface navigator. Nucleic Acids Res. 2003;31:3349–3351. doi: 10.1093/nar/gkg588. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.