Abstract
The regions of transcriptional enhancer factor-1 (TEF-1) required for its activation function and sequence-specific DNA binding have been determined. Deletion analysis of a chimera between TEF-1 and the GAL4 DNA binding domain (DBD) indicated that at least three regions of TEF-1 were involved in transactivation. However, none of these regions functioned as independent activating domains. Moreover, none of the GAL4 chimeras containing individual TEF-1 regions interfered with the activity of endogenous HeLa cell TEF-1, while interference was observed with the GAL4-TEF-1 chimeras which functioned as transactivators. These results indicate that there is a general correlation between the abilities of a given GAL4-TEF-1 chimera to function in transcriptional activation and interference, thus supporting the idea that transactivation by TEF-1 is mediated by a limiting transcriptional intermediary factor. In addition, we show experimentally that the TEA/ATTS domain is a novel class of DBD involved in the sequence-specific DNA binding of TEF-1 and its Drosophila homologue scalloped. Two other regions of TEF-1 are also required for DNA binding. These regions are not part of the minimum DBD, but may function by antagonizing the effect of sequences which negatively regulate DNA binding mediated by both the TEF-1 TEA/ATTS domain and the GAL4 DBD. In addition, analysis of TEF-1 and scalloped derivatives in which their TEA/ATTS domains have been interchanged further indicates that the TEA/ATTS domain is not the only determinant of DNA binding specificity.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Andrianopoulos A., Timberlake W. E. ATTS, a new and conserved DNA binding domain. Plant Cell. 1991 Aug;3(8):747–748. doi: 10.1105/tpc.3.8.747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg J. M. Zinc fingers and other metal-binding domains. Elements for interactions between macromolecules. J Biol Chem. 1990 Apr 25;265(12):6513–6516. [PubMed] [Google Scholar]
- Berger S. L., Cress W. D., Cress A., Triezenberg S. J., Guarente L. Selective inhibition of activated but not basal transcription by the acidic activation domain of VP16: evidence for transcriptional adaptors. Cell. 1990 Jun 29;61(7):1199–1208. doi: 10.1016/0092-8674(90)90684-7. [DOI] [PubMed] [Google Scholar]
- Blank V., Kourilsky P., Israël A. NF-kappa B and related proteins: Rel/dorsal homologies meet ankyrin-like repeats. Trends Biochem Sci. 1992 Apr;17(4):135–140. doi: 10.1016/0968-0004(92)90321-y. [DOI] [PubMed] [Google Scholar]
- Brou C., Chaudhary S., Davidson I., Lutz Y., Wu J., Egly J. M., Tora L., Chambon P. Distinct TFIID complexes mediate the effect of different transcriptional activators. EMBO J. 1993 Feb;12(2):489–499. doi: 10.1002/j.1460-2075.1993.tb05681.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brou C., Wu J., Ali S., Scheer E., Lang C., Davidson I., Chambon P., Tora L. Different TBP-associated factors are required for mediating the stimulation of transcription in vitro by the acidic transactivator GAL-VP16 and the two nonacidic activation functions of the estrogen receptor. Nucleic Acids Res. 1993 Jan 11;21(1):5–12. doi: 10.1093/nar/21.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushman F. D., Shang C., Ptashne M. A single glutamic acid residue plays a key role in the transcriptional activation function of lambda repressor. Cell. 1989 Sep 22;58(6):1163–1171. doi: 10.1016/0092-8674(89)90514-x. [DOI] [PubMed] [Google Scholar]
- Bürglin T. R. The TEA domain: a novel, highly conserved DNA-binding motif. Cell. 1991 Jul 12;66(1):11–12. doi: 10.1016/0092-8674(91)90132-i. [DOI] [PubMed] [Google Scholar]
- Campbell S., Inamdar M., Rodrigues V., Raghavan V., Palazzolo M., Chovnick A. The scalloped gene encodes a novel, evolutionarily conserved transcription factor required for sensory organ differentiation in Drosophila. Genes Dev. 1992 Mar;6(3):367–379. doi: 10.1101/gad.6.3.367. [DOI] [PubMed] [Google Scholar]
- Carey M., Leatherwood J., Ptashne M. A potent GAL4 derivative activates transcription at a distance in vitro. Science. 1990 Feb 9;247(4943):710–712. doi: 10.1126/science.2405489. [DOI] [PubMed] [Google Scholar]
- Carey M. Mechanistic advances in eukaryotic gene activation. Curr Opin Cell Biol. 1991 Jun;3(3):452–460. doi: 10.1016/0955-0674(91)90073-8. [DOI] [PubMed] [Google Scholar]
- Casaz P., Sundseth R., Hansen U. trans activation of the simian virus 40 late promoter by large T antigen requires binding sites for the cellular transcription factor TEF-1. J Virol. 1991 Dec;65(12):6535–6543. doi: 10.1128/jvi.65.12.6535-6543.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee V. K., Madison L. D., Mayo S., Jameson J. L. Repression of the human glycoprotein hormone alpha-subunit gene by glucocorticoids: evidence for receptor interactions with limiting transcriptional activators. Mol Endocrinol. 1991 Jan;5(1):100–110. doi: 10.1210/mend-5-1-100. [DOI] [PubMed] [Google Scholar]
- Courey A. J., Tjian R. Analysis of Sp1 in vivo reveals multiple transcriptional domains, including a novel glutamine-rich activation motif. Cell. 1988 Dec 2;55(5):887–898. doi: 10.1016/0092-8674(88)90144-4. [DOI] [PubMed] [Google Scholar]
- Cousens D. J., Greaves R., Goding C. R., O'Hare P. The C-terminal 79 amino acids of the herpes simplex virus regulatory protein, Vmw65, efficiently activate transcription in yeast and mammalian cells in chimeric DNA-binding proteins. EMBO J. 1989 Aug;8(8):2337–2342. doi: 10.1002/j.1460-2075.1989.tb08361.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cress W. D., Triezenberg S. J. Critical structural elements of the VP16 transcriptional activation domain. Science. 1991 Jan 4;251(4989):87–90. doi: 10.1126/science.1846049. [DOI] [PubMed] [Google Scholar]
- Danielsen M., Hinck L., Ringold G. M. Two amino acids within the knuckle of the first zinc finger specify DNA response element activation by the glucocorticoid receptor. Cell. 1989 Jun 30;57(7):1131–1138. doi: 10.1016/0092-8674(89)90050-0. [DOI] [PubMed] [Google Scholar]
- Davidson I., Fromental C., Augereau P., Wildeman A., Zenke M., Chambon P. Cell-type specific protein binding to the enhancer of simian virus 40 in nuclear extracts. Nature. 1986 Oct 9;323(6088):544–548. doi: 10.1038/323544a0. [DOI] [PubMed] [Google Scholar]
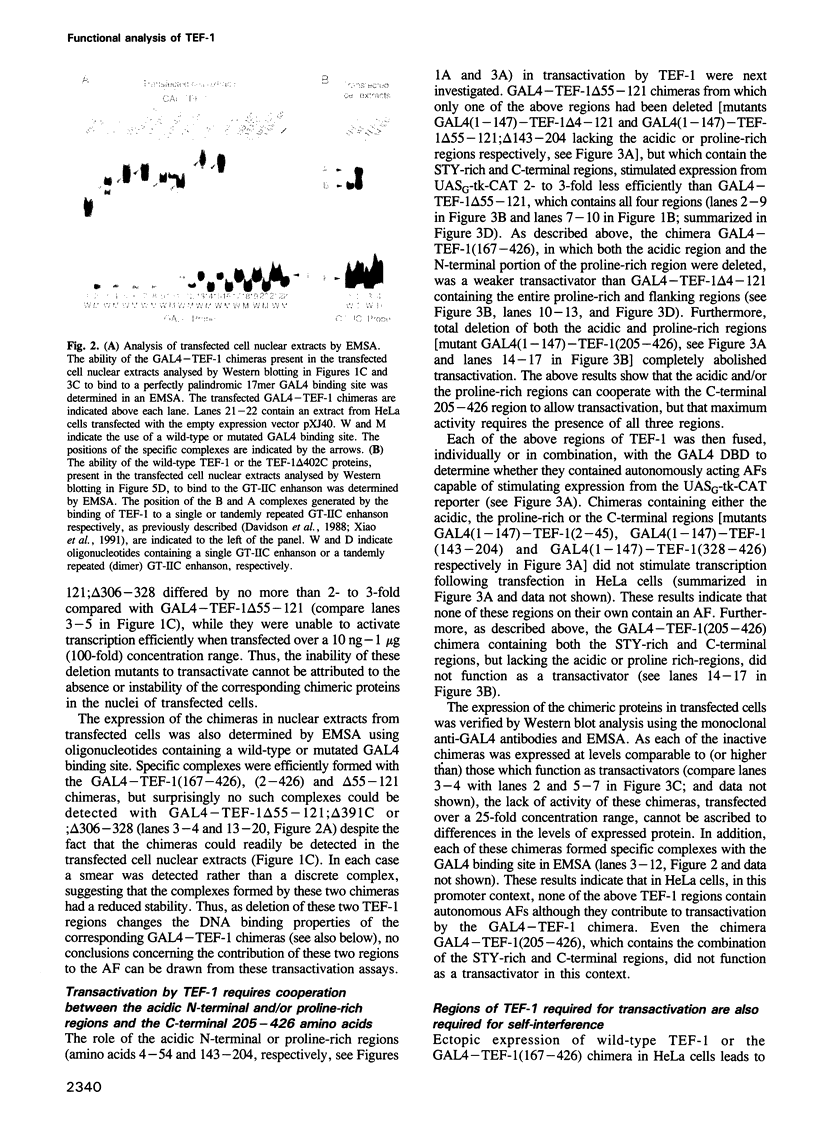
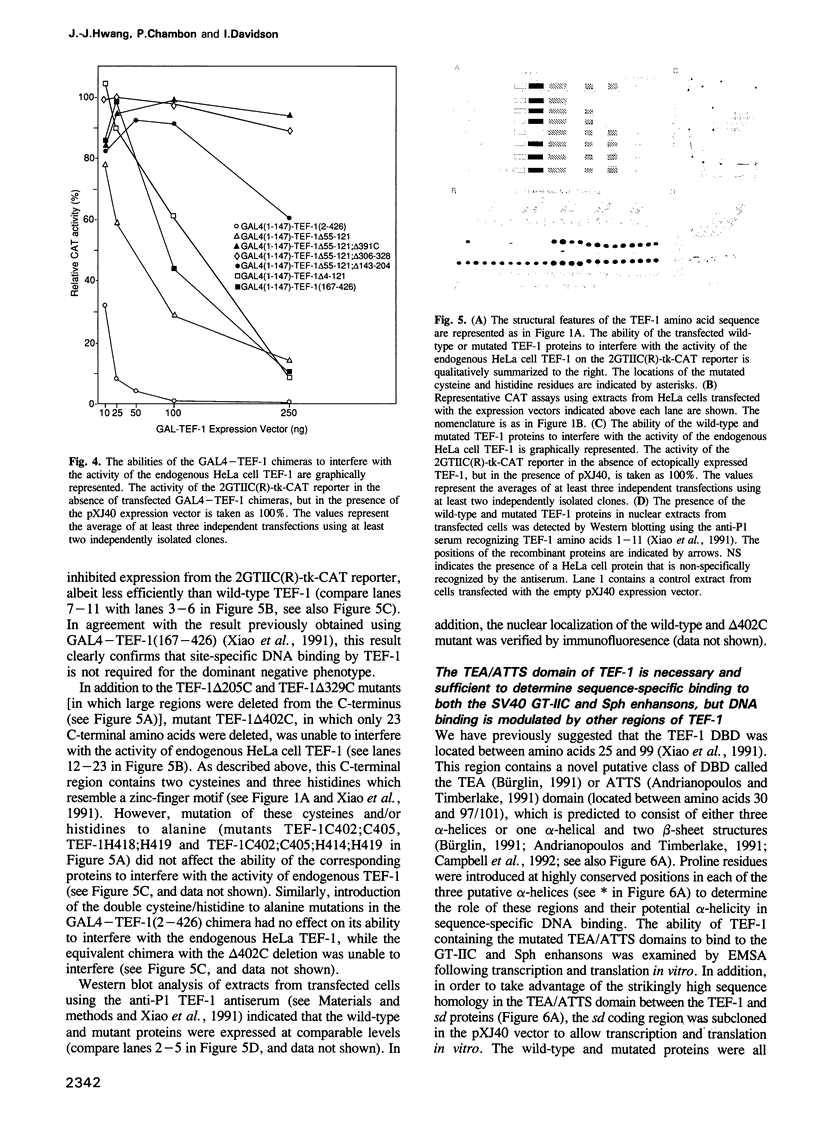
- Davidson I., Xiao J. H., Rosales R., Staub A., Chambon P. The HeLa cell protein TEF-1 binds specifically and cooperatively to two SV40 enhancer motifs of unrelated sequence. Cell. 1988 Sep 23;54(7):931–942. doi: 10.1016/0092-8674(88)90108-0. [DOI] [PubMed] [Google Scholar]
- Davis R. L., Weintraub H. Acquisition of myogenic specificity by replacement of three amino acid residues from MyoD into E12. Science. 1992 May 15;256(5059):1027–1030. doi: 10.1126/science.1317057. [DOI] [PubMed] [Google Scholar]
- Dynlacht B. D., Hoey T., Tjian R. Isolation of coactivators associated with the TATA-binding protein that mediate transcriptional activation. Cell. 1991 Aug 9;66(3):563–576. doi: 10.1016/0092-8674(81)90019-2. [DOI] [PubMed] [Google Scholar]
- Farrance I. K., Mar J. H., Ordahl C. P. M-CAT binding factor is related to the SV40 enhancer binding factor, TEF-1. J Biol Chem. 1992 Aug 25;267(24):17234–17240. [PubMed] [Google Scholar]
- Flanagan P. M., Kelleher R. J., 3rd, Sayre M. H., Tschochner H., Kornberg R. D. A mediator required for activation of RNA polymerase II transcription in vitro. Nature. 1991 Apr 4;350(6317):436–438. doi: 10.1038/350436a0. [DOI] [PubMed] [Google Scholar]
- Fromental C., Kanno M., Nomiyama H., Chambon P. Cooperativity and hierarchical levels of functional organization in the SV40 enhancer. Cell. 1988 Sep 23;54(7):943–953. doi: 10.1016/0092-8674(88)90109-2. [DOI] [PubMed] [Google Scholar]
- Gerster T., Balmaceda C. G., Roeder R. G. The cell type-specific octamer transcription factor OTF-2 has two domains required for the activation of transcription. EMBO J. 1990 May;9(5):1635–1643. doi: 10.1002/j.1460-2075.1990.tb08283.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill G., Ptashne M. Negative effect of the transcriptional activator GAL4. Nature. 1988 Aug 25;334(6184):721–724. doi: 10.1038/334721a0. [DOI] [PubMed] [Google Scholar]
- Gill G., Tjian R. Eukaryotic coactivators associated with the TATA box binding protein. Curr Opin Genet Dev. 1992 Apr;2(2):236–242. doi: 10.1016/s0959-437x(05)80279-5. [DOI] [PubMed] [Google Scholar]
- Gilmore T. D. NF-kappa B, KBF1, dorsal, and related matters. Cell. 1990 Sep 7;62(5):841–843. doi: 10.1016/0092-8674(90)90257-f. [DOI] [PubMed] [Google Scholar]
- Giniger E., Ptashne M. Transcription in yeast activated by a putative amphipathic alpha helix linked to a DNA binding unit. Nature. 1987 Dec 17;330(6149):670–672. doi: 10.1038/330670a0. [DOI] [PubMed] [Google Scholar]
- Green S., Chambon P. Nuclear receptors enhance our understanding of transcription regulation. Trends Genet. 1988 Nov;4(11):309–314. doi: 10.1016/0168-9525(88)90108-4. [DOI] [PubMed] [Google Scholar]
- Gronemeyer H. Transcription activation by estrogen and progesterone receptors. Annu Rev Genet. 1991;25:89–123. doi: 10.1146/annurev.ge.25.120191.000513. [DOI] [PubMed] [Google Scholar]
- Gruda M. C., Alwine J. C. Simian virus 40 (SV40) T-antigen transcriptional activation mediated through the Oct/SPH region of the SV40 late promoter. J Virol. 1991 Jul;65(7):3553–3558. doi: 10.1128/jvi.65.7.3553-3558.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruda M. C., Zabolotny J. M., Xiao J. H., Davidson I., Alwine J. C. Transcriptional activation by simian virus 40 large T antigen: interactions with multiple components of the transcription complex. Mol Cell Biol. 1993 Feb;13(2):961–969. doi: 10.1128/mcb.13.2.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutman A., Wasylyk B. Nuclear targets for transcription regulation by oncogenes. Trends Genet. 1991 Feb;7(2):49–54. doi: 10.1016/0168-9525(91)90231-E. [DOI] [PubMed] [Google Scholar]
- Harrison S. C. A structural taxonomy of DNA-binding domains. Nature. 1991 Oct 24;353(6346):715–719. doi: 10.1038/353715a0. [DOI] [PubMed] [Google Scholar]
- Hayashi S., Scott M. P. What determines the specificity of action of Drosophila homeodomain proteins? Cell. 1990 Nov 30;63(5):883–894. doi: 10.1016/0092-8674(90)90492-w. [DOI] [PubMed] [Google Scholar]
- Henthorn P., Kiledjian M., Kadesch T. Two distinct transcription factors that bind the immunoglobulin enhancer microE5/kappa 2 motif. Science. 1990 Jan 26;247(4941):467–470. doi: 10.1126/science.2105528. [DOI] [PubMed] [Google Scholar]
- Herbomel P., Bourachot B., Yaniv M. Two distinct enhancers with different cell specificities coexist in the regulatory region of polyoma. Cell. 1984 Dec;39(3 Pt 2):653–662. doi: 10.1016/0092-8674(84)90472-0. [DOI] [PubMed] [Google Scholar]
- Herr W., Clarke J. The SV40 enhancer is composed of multiple functional elements that can compensate for one another. Cell. 1986 May 9;45(3):461–470. doi: 10.1016/0092-8674(86)90332-6. [DOI] [PubMed] [Google Scholar]
- Hollenberg S. M., Evans R. M. Multiple and cooperative trans-activation domains of the human glucocorticoid receptor. Cell. 1988 Dec 2;55(5):899–906. doi: 10.1016/0092-8674(88)90145-6. [DOI] [PubMed] [Google Scholar]
- Hope I. A., Struhl K. Functional dissection of a eukaryotic transcriptional activator protein, GCN4 of yeast. Cell. 1986 Sep 12;46(6):885–894. doi: 10.1016/0092-8674(86)90070-x. [DOI] [PubMed] [Google Scholar]
- Hoppe-Seyler F., Butz K., Rittmüller C., von Knebel Doeberitz M. A rapid microscale procedure for the simultaneous preparation of cytoplasmic RNA, nuclear DNA binding proteins and enzymatically active luciferase extracts. Nucleic Acids Res. 1991 Sep 25;19(18):5080–5080. doi: 10.1093/nar/19.18.5080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter T., Karin M. The regulation of transcription by phosphorylation. Cell. 1992 Aug 7;70(3):375–387. doi: 10.1016/0092-8674(92)90162-6. [DOI] [PubMed] [Google Scholar]
- Hwang J. J., Curthoys N. P. Effect of acute alterations in acid-base balance on rat renal glutaminase and phosphoenolpyruvate carboxykinase gene expression. J Biol Chem. 1991 May 25;266(15):9392–9396. [PubMed] [Google Scholar]
- Ingles C. J., Shales M., Cress W. D., Triezenberg S. J., Greenblatt J. Reduced binding of TFIID to transcriptionally compromised mutants of VP16. Nature. 1991 Jun 13;351(6327):588–590. doi: 10.1038/351588a0. [DOI] [PubMed] [Google Scholar]
- Ishiji T., Lace M. J., Parkkinen S., Anderson R. D., Haugen T. H., Cripe T. P., Xiao J. H., Davidson I., Chambon P., Turek L. P. Transcriptional enhancer factor (TEF)-1 and its cell-specific co-activator activate human papillomavirus-16 E6 and E7 oncogene transcription in keratinocytes and cervical carcinoma cells. EMBO J. 1992 Jun;11(6):2271–2281. doi: 10.1002/j.1460-2075.1992.tb05286.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson S. P. Regulating transcription factor activity by phosphorylation. Trends Cell Biol. 1992 Apr;2(4):104–108. doi: 10.1016/0962-8924(92)90014-e. [DOI] [PubMed] [Google Scholar]
- Johnson P. F., McKnight S. L. Eukaryotic transcriptional regulatory proteins. Annu Rev Biochem. 1989;58:799–839. doi: 10.1146/annurev.bi.58.070189.004055. [DOI] [PubMed] [Google Scholar]
- Jones N. Transcriptional regulation by dimerization: two sides to an incestuous relationship. Cell. 1990 Apr 6;61(1):9–11. doi: 10.1016/0092-8674(90)90207-u. [DOI] [PubMed] [Google Scholar]
- Karim F. D., Urness L. D., Thummel C. S., Klemsz M. J., McKercher S. R., Celada A., Van Beveren C., Maki R. A., Gunther C. V., Nye J. A. The ETS-domain: a new DNA-binding motif that recognizes a purine-rich core DNA sequence. Genes Dev. 1990 Sep;4(9):1451–1453. doi: 10.1101/gad.4.9.1451. [DOI] [PubMed] [Google Scholar]
- Kelleher R. J., 3rd, Flanagan P. M., Kornberg R. D. A novel mediator between activator proteins and the RNA polymerase II transcription apparatus. Cell. 1990 Jun 29;61(7):1209–1215. doi: 10.1016/0092-8674(90)90685-8. [DOI] [PubMed] [Google Scholar]
- Kelly J. J., Wildeman A. G. Role of the SV40 enhancer in the early to late shift in viral transcription. Nucleic Acids Res. 1991 Dec 25;19(24):6799–6804. doi: 10.1093/nar/19.24.6799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerr L. D., Inoue J., Verma I. M. Signal transduction: the nuclear target. Curr Opin Cell Biol. 1992 Jun;4(3):496–501. doi: 10.1016/0955-0674(92)90017-7. [DOI] [PubMed] [Google Scholar]
- Kissinger C. R., Liu B. S., Martin-Blanco E., Kornberg T. B., Pabo C. O. Crystal structure of an engrailed homeodomain-DNA complex at 2.8 A resolution: a framework for understanding homeodomain-DNA interactions. Cell. 1990 Nov 2;63(3):579–590. doi: 10.1016/0092-8674(90)90453-l. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lai J. S., Cleary M. A., Herr W. A single amino acid exchange transfers VP16-induced positive control from the Oct-1 to the Oct-2 homeo domain. Genes Dev. 1992 Nov;6(11):2058–2065. doi: 10.1101/gad.6.11.2058. [DOI] [PubMed] [Google Scholar]
- Laloux I., Dubois E., Dewerchin M., Jacobs E. TEC1, a gene involved in the activation of Ty1 and Ty1-mediated gene expression in Saccharomyces cerevisiae: cloning and molecular analysis. Mol Cell Biol. 1990 Jul;10(7):3541–3550. doi: 10.1128/mcb.10.7.3541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leid M., Kastner P., Chambon P. Multiplicity generates diversity in the retinoic acid signalling pathways. Trends Biochem Sci. 1992 Oct;17(10):427–433. doi: 10.1016/0968-0004(92)90014-z. [DOI] [PubMed] [Google Scholar]
- Lillie J. W., Green M. R. Transcription activation by the adenovirus E1a protein. Nature. 1989 Mar 2;338(6210):39–44. doi: 10.1038/338039a0. [DOI] [PubMed] [Google Scholar]
- Lin A., Frost J., Deng T., Smeal T., al-Alawi N., Kikkawa U., Hunter T., Brenner D., Karin M. Casein kinase II is a negative regulator of c-Jun DNA binding and AP-1 activity. Cell. 1992 Sep 4;70(5):777–789. doi: 10.1016/0092-8674(92)90311-y. [DOI] [PubMed] [Google Scholar]
- Lin Y. S., Green M. R. Mechanism of action of an acidic transcriptional activator in vitro. Cell. 1991 Mar 8;64(5):971–981. doi: 10.1016/0092-8674(91)90321-o. [DOI] [PubMed] [Google Scholar]
- Ma J., Ptashne M. A new class of yeast transcriptional activators. Cell. 1987 Oct 9;51(1):113–119. doi: 10.1016/0092-8674(87)90015-8. [DOI] [PubMed] [Google Scholar]
- Mader S., Kumar V., de Verneuil H., Chambon P. Three amino acids of the oestrogen receptor are essential to its ability to distinguish an oestrogen from a glucocorticoid-responsive element. Nature. 1989 Mar 16;338(6212):271–274. doi: 10.1038/338271a0. [DOI] [PubMed] [Google Scholar]
- Martin K. J., Lillie J. W., Green M. R. Evidence for interaction of different eukaryotic transcriptional activators with distinct cellular targets. Nature. 1990 Jul 12;346(6280):147–152. doi: 10.1038/346147a0. [DOI] [PubMed] [Google Scholar]
- Martinez E., Dusserre Y., Wahli W., Mermod N. Synergistic transcriptional activation by CTF/NF-I and the estrogen receptor involves stabilized interactions with a limiting target factor. Mol Cell Biol. 1991 Jun;11(6):2937–2945. doi: 10.1128/mcb.11.6.2937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- May E., Omilli F., Ernoult-Lange M., Zenke M., Chambon P. The sequence motifs that are involved in SV40 enhancer function also control SV40 late promoter activity. Nucleic Acids Res. 1987 Mar 25;15(6):2445–2461. doi: 10.1093/nar/15.6.2445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meisterernst M., Roy A. L., Lieu H. M., Roeder R. G. Activation of class II gene transcription by regulatory factors is potentiated by a novel activity. Cell. 1991 Sep 6;66(5):981–993. doi: 10.1016/0092-8674(91)90443-3. [DOI] [PubMed] [Google Scholar]
- Mermod N., O'Neill E. A., Kelly T. J., Tjian R. The proline-rich transcriptional activator of CTF/NF-I is distinct from the replication and DNA binding domain. Cell. 1989 Aug 25;58(4):741–753. doi: 10.1016/0092-8674(89)90108-6. [DOI] [PubMed] [Google Scholar]
- Meyer M. E., Gronemeyer H., Turcotte B., Bocquel M. T., Tasset D., Chambon P. Steroid hormone receptors compete for factors that mediate their enhancer function. Cell. 1989 May 5;57(3):433–442. doi: 10.1016/0092-8674(89)90918-5. [DOI] [PubMed] [Google Scholar]
- Mirabito P. M., Adams T. H., Timberlake W. E. Interactions of three sequentially expressed genes control temporal and spatial specificity in Aspergillus development. Cell. 1989 Jun 2;57(5):859–868. doi: 10.1016/0092-8674(89)90800-3. [DOI] [PubMed] [Google Scholar]
- Mitchell P. J., Tjian R. Transcriptional regulation in mammalian cells by sequence-specific DNA binding proteins. Science. 1989 Jul 28;245(4916):371–378. doi: 10.1126/science.2667136. [DOI] [PubMed] [Google Scholar]
- Morimoto R. I. Transcription factors: positive and negative regulators of cell growth and disease. Curr Opin Cell Biol. 1992 Jun;4(3):480–487. doi: 10.1016/0955-0674(92)90015-5. [DOI] [PubMed] [Google Scholar]
- Müller-Immerglück M. M., Schaffner W., Matthias P. Transcription factor Oct-2A contains functionally redundant activating domains and works selectively from a promoter but not from a remote enhancer position in non-lymphoid (HeLa) cells. EMBO J. 1990 May;9(5):1625–1634. doi: 10.1002/j.1460-2075.1990.tb08282.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolov D. B., Hu S. H., Lin J., Gasch A., Hoffmann A., Horikoshi M., Chua N. H., Roeder R. G., Burley S. K. Crystal structure of TFIID TATA-box binding protein. Nature. 1992 Nov 5;360(6399):40–46. doi: 10.1038/360040a0. [DOI] [PubMed] [Google Scholar]
- Nolan G. P., Baltimore D. The inhibitory ankyrin and activator Rel proteins. Curr Opin Genet Dev. 1992 Apr;2(2):211–220. doi: 10.1016/s0959-437x(05)80276-x. [DOI] [PubMed] [Google Scholar]
- Nomiyama H., Fromental C., Xiao J. H., Chambon P. Cell-specific activity of the constituent elements of the simian virus 40 enhancer. Proc Natl Acad Sci U S A. 1987 Nov;84(22):7881–7885. doi: 10.1073/pnas.84.22.7881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ondek B., Gloss L., Herr W. The SV40 enhancer contains two distinct levels of organization. Nature. 1988 May 5;333(6168):40–45. doi: 10.1038/333040a0. [DOI] [PubMed] [Google Scholar]
- Ondek B., Shepard A., Herr W. Discrete elements within the SV40 enhancer region display different cell-specific enhancer activities. EMBO J. 1987 Apr;6(4):1017–1025. doi: 10.1002/j.1460-2075.1987.tb04854.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pabo C. O., Sauer R. T. Transcription factors: structural families and principles of DNA recognition. Annu Rev Biochem. 1992;61:1053–1095. doi: 10.1146/annurev.bi.61.070192.005201. [DOI] [PubMed] [Google Scholar]
- Peterson M. G., Tanese N., Pugh B. F., Tjian R. Functional domains and upstream activation properties of cloned human TATA binding protein. Science. 1990 Jun 29;248(4963):1625–1630. doi: 10.1126/science.2363050. [DOI] [PubMed] [Google Scholar]
- Pomerantz J. L., Kristie T. M., Sharp P. A. Recognition of the surface of a homeo domain protein. Genes Dev. 1992 Nov;6(11):2047–2057. doi: 10.1101/gad.6.11.2047. [DOI] [PubMed] [Google Scholar]
- Ptashne M., Gann A. A. Activators and targets. Nature. 1990 Jul 26;346(6282):329–331. doi: 10.1038/346329a0. [DOI] [PubMed] [Google Scholar]
- Ptashne M. How eukaryotic transcriptional activators work. Nature. 1988 Oct 20;335(6192):683–689. doi: 10.1038/335683a0. [DOI] [PubMed] [Google Scholar]
- Pugh B. F., Tjian R. Diverse transcriptional functions of the multisubunit eukaryotic TFIID complex. J Biol Chem. 1992 Jan 15;267(2):679–682. [PubMed] [Google Scholar]
- Sadowski I., Ma J., Triezenberg S., Ptashne M. GAL4-VP16 is an unusually potent transcriptional activator. Nature. 1988 Oct 6;335(6190):563–564. doi: 10.1038/335563a0. [DOI] [PubMed] [Google Scholar]
- Sauer F., Jäckle H. Concentration-dependent transcriptional activation or repression by Krüppel from a single binding site. Nature. 1991 Oct 10;353(6344):563–566. doi: 10.1038/353563a0. [DOI] [PubMed] [Google Scholar]
- Schena M., Freedman L. P., Yamamoto K. R. Mutations in the glucocorticoid receptor zinc finger region that distinguish interdigitated DNA binding and transcriptional enhancement activities. Genes Dev. 1989 Oct;3(10):1590–1601. doi: 10.1101/gad.3.10.1590. [DOI] [PubMed] [Google Scholar]
- Schirm S., Jiricny J., Schaffner W. The SV40 enhancer can be dissected into multiple segments, each with a different cell type specificity. Genes Dev. 1987 Mar;1(1):65–74. doi: 10.1101/gad.1.1.65. [DOI] [PubMed] [Google Scholar]
- Schmitz M. L., Henkel T., Baeuerle P. A. Proteins controlling the nuclear uptake of NF-kappa B, Rel and dorsal. Trends Cell Biol. 1991 Nov;1(5):130–137. doi: 10.1016/0962-8924(91)90118-s. [DOI] [PubMed] [Google Scholar]
- Schwarz J. J., Chakraborty T., Martin J., Zhou J. M., Olson E. N. The basic region of myogenin cooperates with two transcription activation domains to induce muscle-specific transcription. Mol Cell Biol. 1992 Jan;12(1):266–275. doi: 10.1128/mcb.12.1.266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott M. P., Tamkun J. W., Hartzell G. W., 3rd The structure and function of the homeodomain. Biochim Biophys Acta. 1989 Jul 28;989(1):25–48. doi: 10.1016/0304-419x(89)90033-4. [DOI] [PubMed] [Google Scholar]
- Seipel K., Georgiev O., Schaffner W. Different activation domains stimulate transcription from remote ('enhancer') and proximal ('promoter') positions. EMBO J. 1992 Dec;11(13):4961–4968. doi: 10.1002/j.1460-2075.1992.tb05603.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith D. B., Johnson K. S. Single-step purification of polypeptides expressed in Escherichia coli as fusions with glutathione S-transferase. Gene. 1988 Jul 15;67(1):31–40. doi: 10.1016/0378-1119(88)90005-4. [DOI] [PubMed] [Google Scholar]
- Stringer K. F., Ingles C. J., Greenblatt J. Direct and selective binding of an acidic transcriptional activation domain to the TATA-box factor TFIID. Nature. 1990 Jun 28;345(6278):783–786. doi: 10.1038/345783a0. [DOI] [PubMed] [Google Scholar]
- Struhl G., Struhl K., Macdonald P. M. The gradient morphogen bicoid is a concentration-dependent transcriptional activator. Cell. 1989 Jun 30;57(7):1259–1273. doi: 10.1016/0092-8674(89)90062-7. [DOI] [PubMed] [Google Scholar]
- Takada R., Nakatani Y., Hoffmann A., Kokubo T., Hasegawa S., Roeder R. G., Horikoshi M. Identification of human TFIID components and direct interaction between a 250-kDa polypeptide and the TATA box-binding protein (TFIID tau). Proc Natl Acad Sci U S A. 1992 Dec 15;89(24):11809–11813. doi: 10.1073/pnas.89.24.11809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka M., Herr W. Differential transcriptional activation by Oct-1 and Oct-2: interdependent activation domains induce Oct-2 phosphorylation. Cell. 1990 Feb 9;60(3):375–386. doi: 10.1016/0092-8674(90)90589-7. [DOI] [PubMed] [Google Scholar]
- Tanese N., Pugh B. F., Tjian R. Coactivators for a proline-rich activator purified from the multisubunit human TFIID complex. Genes Dev. 1991 Dec;5(12A):2212–2224. doi: 10.1101/gad.5.12a.2212. [DOI] [PubMed] [Google Scholar]
- Tasset D., Tora L., Fromental C., Scheer E., Chambon P. Distinct classes of transcriptional activating domains function by different mechanisms. Cell. 1990 Sep 21;62(6):1177–1187. doi: 10.1016/0092-8674(90)90394-t. [DOI] [PubMed] [Google Scholar]
- Theill L. E., Castrillo J. L., Wu D., Karin M. Dissection of functional domains of the pituitary-specific transcription factor GHF-1. Nature. 1989 Dec 21;342(6252):945–948. doi: 10.1038/342945a0. [DOI] [PubMed] [Google Scholar]
- Thompson C. C., Brown T. A., McKnight S. L. Convergence of Ets- and notch-related structural motifs in a heteromeric DNA binding complex. Science. 1991 Aug 16;253(5021):762–768. doi: 10.1126/science.1876833. [DOI] [PubMed] [Google Scholar]
- Treisman J., Gönczy P., Vashishtha M., Harris E., Desplan C. A single amino acid can determine the DNA binding specificity of homeodomain proteins. Cell. 1989 Nov 3;59(3):553–562. doi: 10.1016/0092-8674(89)90038-x. [DOI] [PubMed] [Google Scholar]
- Treisman R., Ammerer G. The SRF and MCM1 transcription factors. Curr Opin Genet Dev. 1992 Apr;2(2):221–226. doi: 10.1016/s0959-437x(05)80277-1. [DOI] [PubMed] [Google Scholar]
- Turcotte B., Guarente L. HAP1 positive control mutants specific for one of two binding sites. Genes Dev. 1992 Oct;6(10):2001–2009. doi: 10.1101/gad.6.10.2001. [DOI] [PubMed] [Google Scholar]
- Voit R., Schnapp A., Kuhn A., Rosenbauer H., Hirschmann P., Stunnenberg H. G., Grummt I. The nucleolar transcription factor mUBF is phosphorylated by casein kinase II in the C-terminal hyperacidic tail which is essential for transactivation. EMBO J. 1992 Jun;11(6):2211–2218. doi: 10.1002/j.1460-2075.1992.tb05280.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Gralla J. D., Carey M. The acidic activator GAL4-AH can stimulate polymerase II transcription by promoting assembly of a closed complex requiring TFIID and TFIIA. Genes Dev. 1992 Sep;6(9):1716–1727. doi: 10.1101/gad.6.9.1716. [DOI] [PubMed] [Google Scholar]
- Wasylyk C., Kerckaert J. P., Wasylyk B. A novel modulator domain of Ets transcription factors. Genes Dev. 1992 Jun;6(6):965–974. doi: 10.1101/gad.6.6.965. [DOI] [PubMed] [Google Scholar]
- Webster L. C., Ricciardi R. P. trans-dominant mutants of E1A provide genetic evidence that the zinc finger of the trans-activating domain binds a transcription factor. Mol Cell Biol. 1991 Sep;11(9):4287–4296. doi: 10.1128/mcb.11.9.4287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster N., Jin J. R., Green S., Hollis M., Chambon P. The yeast UASG is a transcriptional enhancer in human HeLa cells in the presence of the GAL4 trans-activator. Cell. 1988 Jan 29;52(2):169–178. doi: 10.1016/0092-8674(88)90505-3. [DOI] [PubMed] [Google Scholar]
- Weintraub H., Dwarki V. J., Verma I., Davis R., Hollenberg S., Snider L., Lassar A., Tapscott S. J. Muscle-specific transcriptional activation by MyoD. Genes Dev. 1991 Aug;5(8):1377–1386. doi: 10.1101/gad.5.8.1377. [DOI] [PubMed] [Google Scholar]
- White J. H., Brou C., Wu J., Burton N., Egly J. M., Chambon P. Evidence for a factor required for transcriptional stimulation by the chimeric acidic activator GAL-VP16 in HeLa cell extracts. Proc Natl Acad Sci U S A. 1991 Sep 1;88(17):7674–7678. doi: 10.1073/pnas.88.17.7674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White J., Brou C., Wu J., Lutz Y., Moncollin V., Chambon P. The acidic transcriptional activator GAL-VP16 acts on preformed template-committed complexes. EMBO J. 1992 Jun;11(6):2229–2240. doi: 10.1002/j.1460-2075.1992.tb05282.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wildeman A. G., Zenke M., Schatz C., Wintzerith M., Grundström T., Matthes H., Takahashi K., Chambon P. Specific protein binding to the simian virus 40 enhancer in vitro. Mol Cell Biol. 1986 Jun;6(6):2098–2105. doi: 10.1128/mcb.6.6.2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams T., Tjian R. Analysis of the DNA-binding and activation properties of the human transcription factor AP-2. Genes Dev. 1991 Apr;5(4):670–682. doi: 10.1101/gad.5.4.670. [DOI] [PubMed] [Google Scholar]
- Winter B., Braun T., Arnold H. H. Co-operativity of functional domains in the muscle-specific transcription factor Myf-5. EMBO J. 1992 May;11(5):1843–1855. doi: 10.1002/j.1460-2075.1992.tb05236.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolberger C., Vershon A. K., Liu B., Johnson A. D., Pabo C. O. Crystal structure of a MAT alpha 2 homeodomain-operator complex suggests a general model for homeodomain-DNA interactions. Cell. 1991 Nov 1;67(3):517–528. doi: 10.1016/0092-8674(91)90526-5. [DOI] [PubMed] [Google Scholar]
- Xiao J. H., Davidson I., Ferrandon D., Rosales R., Vigneron M., Macchi M., Ruffenach F., Chambon P. One cell-specific and three ubiquitous nuclear proteins bind in vitro to overlapping motifs in the domain B1 of the SV40 enhancer. EMBO J. 1987 Oct;6(10):3005–3013. doi: 10.1002/j.1460-2075.1987.tb02606.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
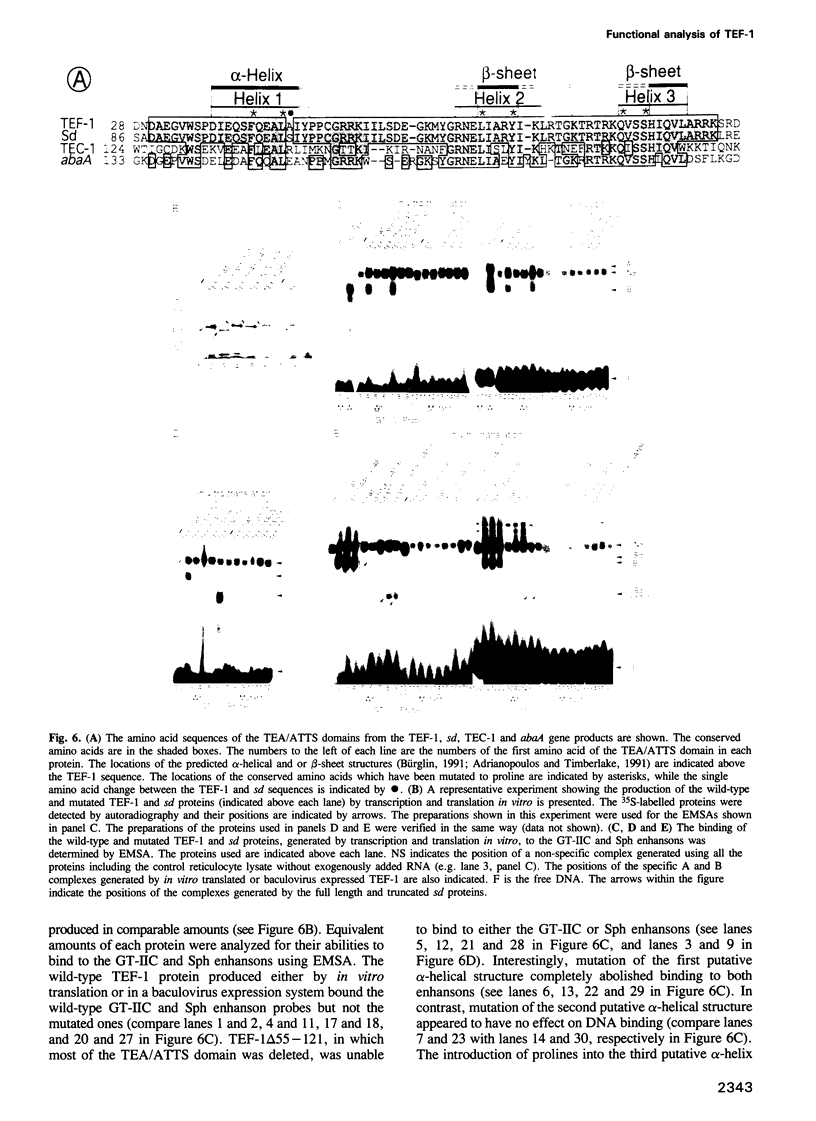
- Xiao J. H., Davidson I., Matthes H., Garnier J. M., Chambon P. Cloning, expression, and transcriptional properties of the human enhancer factor TEF-1. Cell. 1991 May 17;65(4):551–568. doi: 10.1016/0092-8674(91)90088-g. [DOI] [PubMed] [Google Scholar]
- Zenke M., Grundström T., Matthes H., Wintzerith M., Schatz C., Wildeman A., Chambon P. Multiple sequence motifs are involved in SV40 enhancer function. EMBO J. 1986 Feb;5(2):387–397. doi: 10.1002/j.1460-2075.1986.tb04224.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q., Lieberman P. M., Boyer T. G., Berk A. J. Holo-TFIID supports transcriptional stimulation by diverse activators and from a TATA-less promoter. Genes Dev. 1992 Oct;6(10):1964–1974. doi: 10.1101/gad.6.10.1964. [DOI] [PubMed] [Google Scholar]
- Zhu H., Pyrwes R. Identification of a coactivator that increases activation of transcription by serum response factor and GAL4-VP16 in vitro. Proc Natl Acad Sci U S A. 1992 Jun 15;89(12):5291–5295. doi: 10.1073/pnas.89.12.5291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Groot R. P., Sassone-Corsi P. Activation of Jun/AP-1 by protein kinase A. Oncogene. 1992 Nov;7(11):2281–2286. [PubMed] [Google Scholar]