Abstract
Despite the pressing need to noninvasively monitor transplanted cells in vivo with fluorescence imaging, desirable fluorescent agents with rapid labeling capability, durable brightness, and ideal biocompatibility remain lacking. Herein we report phosphorylcholine-coated near-infrared (NIR) fluorescent semiconducting polymer nanoparticles (SPNs) as a new class of rapid, efficient and cytocompatible labeling nanoagents for in vivo cell tracking. The phosphorylcholine coating results in efficient and rapid endocytosis and allows the SPN to enter cells within 0.5 h in complete culture medium apparently independent of the cell type, while its NIR fluorescence leads to a tissue penetration depth of 0.5 cm. In comparison to quantum dots and Cy5.5, the SPN is tolerant to physiologically ubiquitous reactive oxygen species ROS, resulting in durable fluorescence both in vitro and in vivo. These desirable physical and physiological properties of the SPN permit cell tracking of human renal cell carcinoma (RCC) cells in living mice at a lower limit of detection of 10,000 cells with no obvious alteration of cell phenotype after 12 days. SPNs thus could provide unique opportunities for optimizing cellular therapy and deciphering pathological processes as a cell tracking label.
Keywords: Semiconducting polymer nanoparticles, fluorescence imaging, cell labelling
1. Introduction
The ability to noninvasively monitor transplanted cells in vivo is a pressing need not only for optimizing cell-based therapeutics but also for understanding many life-threatening pathological processes such as cancer metastasis.[1] Fluorescence imaging as a powerful non-ionizing technique to visualize biology and pathology can provide a sensitive and safe way to track cells in living animals.[2] Fluorescent nanoparticles usually have prolonged intracellular retention as compared with small-molecule dyes due to their larger size, making them suited for long-term cell tracking.[3] Although semiconductor quantum dots (QDs) have been shown for cell tracking and QD-based labelling agents are commercially available,[4] they could be readily degraded in the presence of reactive oxygen species (ROS).[5] This characteristic could not only cause the loss of fluorescence but also trigger the release of toxic heavy metal ions, potentially impairing transplanted cell function, reducing therapeutic effect, and preventing the long-term localization of cells.[6] As ROS are integral chemical mediators ubiquitous in living animals, and their concentrations can be at micromolar level in phagocytic cells (e.g., neutrophils and monocytes),[7], alternative fluorescent nanoparticles with higher ROS stability would be more preferred for in vivo cell tracking.
Semiconducting polymer nanoparticles (SPNs) represent a new class of fluorescent nanomaterials with high brightness and controllable dimensions.[8] With completely organic and biologically benign components, SPNs circumvent the issue of heavy metal ion-induced toxicity to living organisms, and display good biocompatibility.[8c] In addition to excellent photostability, SPNs are highly tolerant to ROS and thus are stably fluorescent under physiological conditions.[8c, 8f] These attractive features have generated intense interest in developing SPN probes for molecular imaging.[8f, 9] Recently, we developed self-luminescing SPNs by the attachment of a Renilla luciferase mutant as the bioluminescence source to enhance imaging depth, resulting in improved tumor imaging in living animals.[10] SPNs have also been demonstrated as a new class of contrast nanomatreials for photoacoustic molecular imaging.[11] Despite the great potential of SPNs in biomedical applications, its suitability for in vivo cell tracking has not been fully tested yet.[12]
The key challenges to accomplish cell tracking with SPNs lie in nanoparticle engineering to confer rapid and efficient cellular uptake, as well as sufficient in vivo imaging depth. As existing SPNs usually possess passivated surfaces covered with poly(ethylene glycol) (PEG),[13] silica,[14] or carboxyl groups,[9a] they show very slow and limited cell internalization, requiring at least overnight incubation prior to imaging acquisition.[10–11] Although bioconjugation with specific antibodies or small molecular ligands promotes receptor-mediated endocytosis, the ability to label different cell lines with a single nanoparticle formulation is compromised. Owing to their short-wavelength absorption and fluorescence,[15] conventional SPNs also suffer from the interference of tissue autofluorescence and light scattering, making them less ideal for optical imaging in living animals.
Herein, we report the development of phosphorylcholine-coated near-infrared (NIR) SPNs as a new class of rapid and efficient cell labelling nanoagents that are applicable to in vivo tracking of primary human cancer cells. Phosphorylcholine, a zwitterionic molecular segment abundant on the extracellular face of the cell membrane, was utilized to decorate the SPN surface. As phosphorylcholine-containing polymers and nanoparticles have been report to have high affinity to the cell membrane,[16] this characteristic allowed the SPN to undergo efficient and rapid endocytosis. In conjunction, a far-red absorbing and NIR-emitting semiconducting polymer was employed as the nanoparticle core to enhance tissue penetration depth. We found that the NIR SPN was able to label cells rapidly within 30 min, track cultured cells for more than five days, and be clearly visualized at the tissue penetration depth of 0.5 cm. With these advantages, we demonstrated that the phosphorylcholine-coated NIR SPN permitted effective long-term tracking of as few as 10,000 primary human renal cell carcinoma (RCC) cells in living mice.
2. Results and Discussion
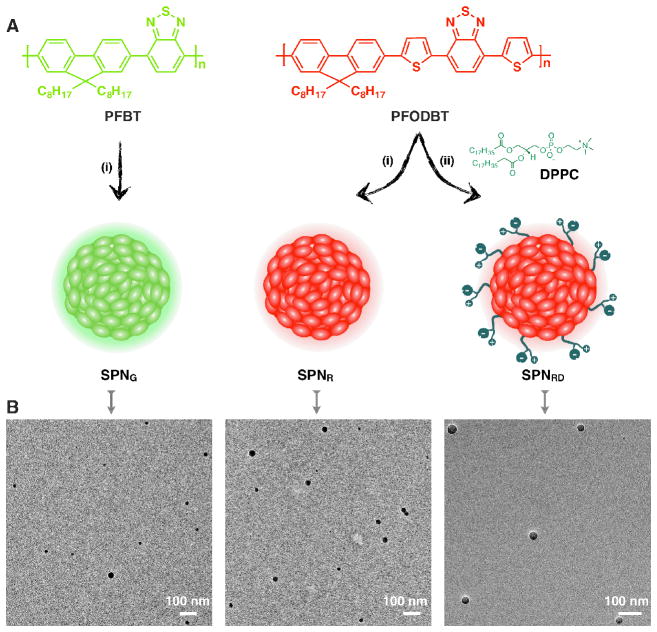
A nanoprecipitation method was used to synthesize the SPNs as shown in Figure 1A. Direct precipitation of poly[2,7-(9,9-dioctylfluorene)-alt-4,7-(2,1,3-benzothiadiazole)] (PFBT) and poly[2,7-(9,9-dioctylfluorene)-alt-4,7-bis(thiophen-2-yl)benzothiadiazole] (PFODBT) in water yielded two bare SPNs, SPNG and SPNR, respectively. Coprecipitation of PFODBT and 1,2-dipalmitoyl-sn-glycero-3-phosphocholine (DPPC) yielded the zwitterionic SPN (SPNRD). During nanoparticle formation, the hydrophobic lipid tails of DPPC, driven by their strong hydrophobic interaction, should most likely be embedded inside the nanoparticles, while the zwitterionic phosphorylcholine head should extend outside into the aqueous environment, constituting a lipophilic surface with high cell-membrane affinity to facilitate cellular internalization.[16a,16b,16d] The bare SPNs (SPNR and SPNG) served as respective controls to demonstrate the importance of the phosphorylcholine shell in nanoparticle internalization, and to determine the relative importance of absorption, emission and fluorescence quantum yield properties for enhanced imaging depth.
Figure 1. Synthesis and characterization of SPNs.
(A) The molecular structures of PFBT and PFOBT, and the schematic illustration of synthesis of SPNG, SPNR and SPNRD. (i) Direct precipitation of polymer in water; (ii) coprecipitation of polymer with DPPC. (B) TEM images of SPNG, SPNR, and SPNRD.
Transmission electron microscopy (TEM) (Figure 1B) showed that SPNG, SPNR and SPNRD had similar spherical morphological with average diameters of 29, 31, and 35 nm, respectively. Dynamic light scattering (DLS) confirmed their narrow size distribution with polydispersity indexes of 0.16 ± 0.05, 0.15 ± 0.03 and 0.18 ± 0.03 for SPNG, SPNR and SPNRD, respectively. The sizes of these SPNs are larger than QDs (<20 nm) which might be beneficial for their cellular retention. The zeta potentials of SPNG, SPNR and SPNRD were measured to be −22 ± 6, −25 ± 5 and −36 ± 7 mV in phosphate-buffered saline (PBS) (50 mM, pH = 7.4), respectively. These data reveal that the presence of DPPC slightly affects the size of SPNs but increases the zeta potential, potentially leading to improved aqueous stability for SPNRD.
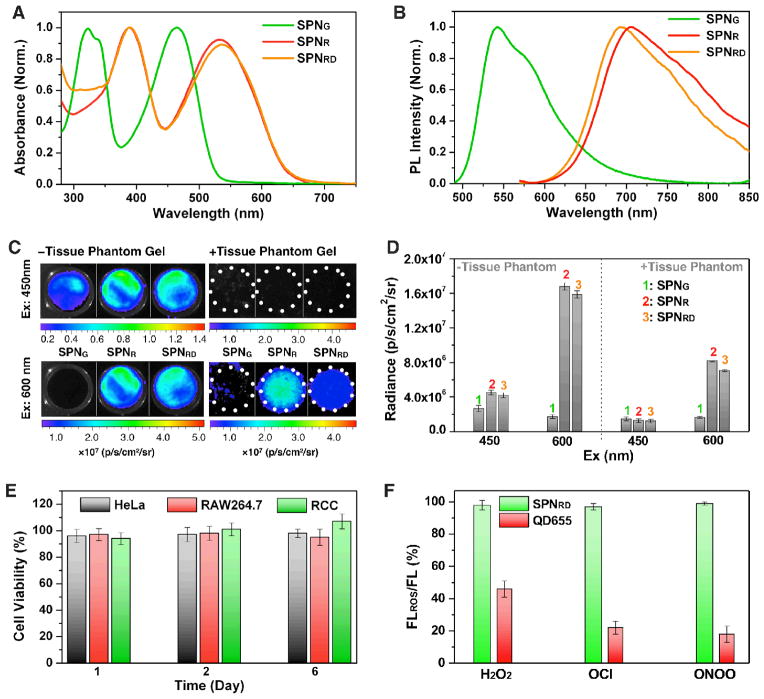
The UV-vis absorption and photoluminescence (PL) spectra of synthesized SPNs are shown in Figures 2A and 2B, respectively. Similar to organic dyes, SPNs have relatively broad spectra in comparison with QDs. SPNR and SPNRD have absorption maximum at 535 nm, which was red-shifted as compared to SPNG (465 nm). Different from green fluorescent SPNG (542 nm), SPNR and SPNRD emitted NIR fluorescence with maximum at 705 and 695 nm, respectively. The PL quantum yields of SPNG, SPNR and SPNRD in 1×PBS (pH = 7.4) were determined to be 22%, 2.8%, and 2.5%, respectively. The lower quantum yield of SPNR and SPNRD relative to SPNG should be attributed to the strong intramolecular charged transfer feature of PFODBT that quenched polymer fluorescence in a highly polar aqueous solution.[17]
Figure 2. Optical characterization of SPNs.
UV-vis (A) and (B) PL spectra of SPNG, SPNR and SPNRD. (C) Fluorescence/bright-field overlaid images of the nanoparticle solutions with (right) and (left) without tissue phantom gel (0.5 cm) covering. Excitation wavelength was at 450 (top) or 600 nm (bottom), with emission wavelength fixed at 700 ± 10 nm. (D) Quantification of fluorescence intensities of the corresponding nanoparticle solutions in C. (E) In vitro viability of SPNRD-labeled HeLa, RAW264.7 and human RCC cells after growing for 1, 2 or 6 days. [SPNRD] = 100 μg/mL. The percentage of viable treated cells is calculated relative to that of untreated cells with the control viability defined as 100%. Error bars are standard deviation. (F) Changes in the fluorescence intensities of SPNRD and QD655 in the presence of different ROS (20 μM). [SPNRG] = 1 μg/mL; [QD655] = 20 nM. Excitation wavelength was 465 nm.
To examine fluorescence imaging depth of these SPNs, gel phantoms with a thickness of 0.5 cm were made from gelatin, hemoglobin, sodium azide, and intralipid to mimic the absorbance and scattering characteristics of living tissues. Fluorescence images of nanoparticle solutions at the same mass concentrations (20 μg/mL) were acquired at 700 nm with and without the phantom gel covering (Figure 2C). Despite the much lower quantum yield of SPNR and SPNRD relative to SPNG, their fluorescent intensities at 700 nm upon excitation at 450 and 600 nm were respectively ~2- and 10-times those of SPNG, attributable to their high absorption and brightness at the NIR region (Figure 2D). In the presence of the gel phantom, there was no detectable fluorescence upon excitation at 450 nm for any nanoparticle solution; however, after excitation at 600 nm, strong fluorescence was observed for SPNR and SPNRD. While the fluorescent signal with the gel phantom decreased 50% relative to the signal without the phantom for SPNR and SPNRD, no fluorescence signal was observed above background for SPNG. These data illustrate that long-wavelength absorption and NIR emission rather than PL brightness more strongly constrain the imaging depth of SPNs, and clearly the NIR SPNs are more suitable than the green SPN for cell tracking in living animals.
Cytocompatibility and stability under physiologically relevant conditions in addition to rapid and efficient cell labelling is essential for long-term in vivo cell tracking. With completely organic and biologically benign components, SPNRD was found to be non-toxic to both immortal cells (HeLa and RAW264.7) and primary human RCC cells (Figure 2E). Additionally, the fluorescence of SPNRD remained nearly unchanged in the presence of ROS such as peroxynitrite (ONOO−), hypochlorite (ClO−) and hydrogen peroxide (H2O2), while significant fluorescence drops (20–40%) were observed for PEG-coated CdSe-ZnS QD (QD655) under the same conditions (Figure 2F). This result illustrates the key attributes of SPNRG over QDs for cell tracking in living animals.
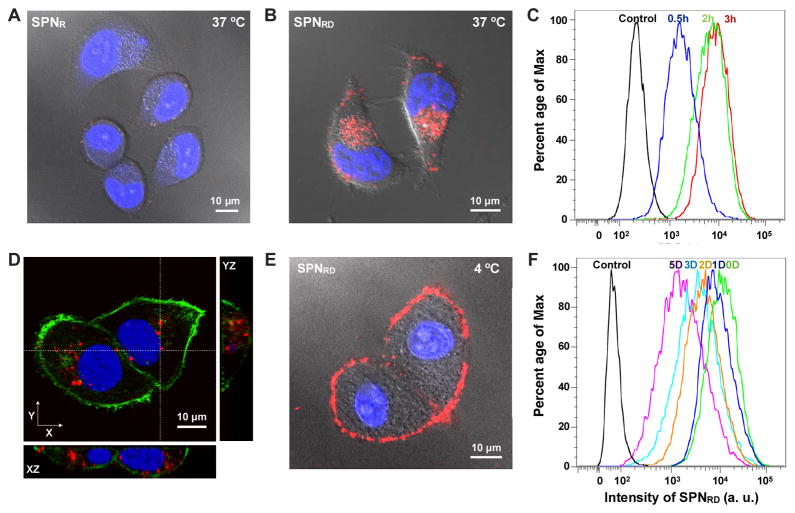
To test the in vitro cell-labelling efficiency of SPNR and SPNRD, HeLa cells were used as a model cell line. After incubation with the SPNs (50 μg/mL) at 37 °C for 3h, cellular nuclei were stained with Hoechst 33342 and fluorescence images of living cells were then acquired with confocal laser scanning microscopy (CLSM). While little fluorescence was detected for SPNR-treated cells (Figure 3A), bright NIR fluorescence was observed for SPNRD-treated cells (Figure 3B). Flow cytometry analysis confirmed that almost every cell was labelled with SPNRD within 30 min in complete cell culture media, with fluorescence intensity reaching a plateau at 3 h (Figure 3C). CLSM Z-stack images with orthogonal views were acquired from fixed HeLa cells treated with SPNRD under the same conditions. Co-staining of cellular nuclei with DAPI (blue) and filamentous actins with an Alexa488-phalloidin conjugate (green) revealed that the nanoparticle clusters of SPNRD were located in the cytoplasm rather than limited to the cell membrane (Figure 3D).
Figure 3. In vitro tracking of HeLa cells.
CLSM images of live HeLa cells after incubation with SPNR (A) and SPNRD (B) at 37 °C for 3 h. The cell nuclei were stained with Hoechst 33342 (blue). (C) Flow cytometry profiles of HeLa cells after incubation with SPNRD at 37 °C for 0.5, 2 or 3h. (D) CLSM orthogonal images of fixed SPNRD-labelled HeLa cells. Cellular nuclei and filamentous actins were stained by DAPI (blue) and Alexa488-phalloidin conjugate (green), respectively. (E) CLSM image of live HeLa cells after incubation with SPNRD at 4 °C for 3h. (F) Flow cytometry profiles of SPNRD-labelled HeLa cells after growth for indicated period of time. [SPN] = 50 μg/mL.
The rapid and efficient internalization capability of SPNRD was clearly associated with its surface coating with phosphorylcholine groups, as the bare nanoparticle, SPNR, was not efficiently internalized. Similar cellular uptake behaviour was also observed for phosphorylcholine-functionalized inorganic nanoparticles (e.g., silver and gold nanoparticles), but the mechanism was not reported.[18] To understand the internalization of SPNRD, HeLa cells were incubated with nanoparticles at 4 °C for 3 h and washed with PBS prior to CLSM imaging. Little fluorescence was detectable within the cells, but was rather confined to the cell periphery (Figure 3E). This suggested that SPNRD were internalized through an energy-dependent pathway, consistent with previous reports that SPNs entered cells through endocytosis.[19] As the pH of the cell culture medium was neutral, SPNRD should mainly bind to but not fuse into the cell membrane according to a previous study on the fusogenic capability of DPPC.[20]
While further investigation is required to gain more insights into the uptake mechanism of SPNRD, it is clear that the phosphorylcholine groups on the nanoparticle surface plays an indispensable role in inducing the rapid and efficient endocytosis of SPNRD. Cell uptake of SPNRD may not depend on specific cell surface receptors since both the murine stromal MS-5 cell line and the primary human RCC cells were also rapidly and efficiently labelled with SPNRD (Supporting Information, Figures S1 and S2). In comparison, the QDs are reported to usually require overnight incubation for sufficient cell labelling.[4a]
The capability of SPNRD for long-term in vitro cell tracking was then examined with HeLa cells. After 3 h incubation with nanoparticles and washing with PBS, HeLa cells were trypsinized and inoculated into fresh medium at low cell numbers. Flow cytometry was used to track the fluorescence intensities of cells over a 5-day period. Due to cell proliferation, nanoparticles were distributed into daughter cells through cytokinesis, inevitably decreasing nanoparticle concentration per cell and leading to a progressive decrease in average fluorescence intensities per cell (Figure 3F). The efficient cytokinetic redistribution of nanoparticles to daughter cells and the low exocytosis of SPNRD was suggested as almost all cells maintained fluorescent even after cell growth for 5 days, indicating the applicability of SPNRD for long-term cell tracking.
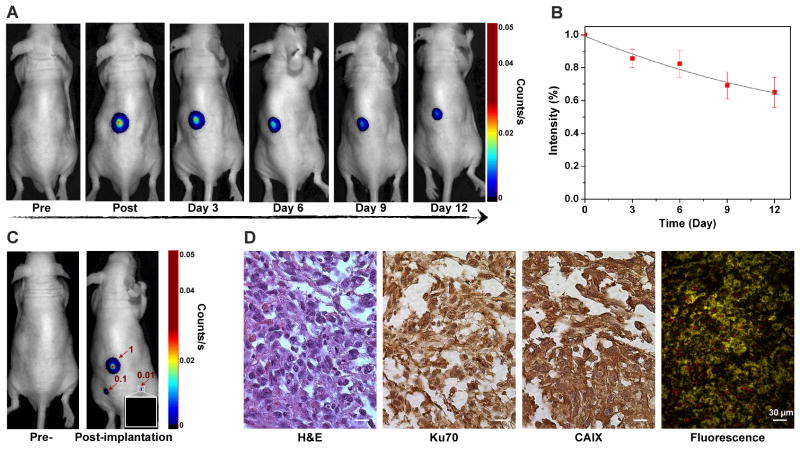
The practicability of using SPNRD for in vivo cell tracking was tested by establishing xenografts with nanoparticle-loaded primary human RCC cells. As realistic preclinical models for RCC, the deadliest urological cancer, are limited,[21] tracking primary RCC could provide an efficient and more authentic model compared to established cell lines to image the progression of the disease. SPNRD-labelled cells (1×106) were subcutaneously implanted into mice and fluorescence images were acquired from 10 min to 12 days post-injection. Fluorescence was detectable throughout this long period with just a 35±9.5% decrease after 12 days of growth (Figure 4A and 4B). The limit of detecting SPNRD-labeled cells in living mice was found to be 10,000 cells (Figure 4C). Histological analysis was performed on RCC tumours recovered after 12 days of growth (Figure 4D). RCC tumours retained expression of carbonic anhydrase IX (CAIX), a specific phenotypic marker of RCC.[22] Nuclear staining with anti-Ku70 confirmed the human origin of the RCC tumour (Figure 4D). Thus, the presence of SPNRD had little influence on the phenotypes and genotypes of RCC. Additionally, as a result of the stability of SPNRD, NIR fluorescence was still detectable ex vivo after tissue processing for histological analysis, including formalin fixation and paraffin embedding. These data demonstrate that SPNRD is well suited for in vivo cell tracking of primary human cancer cells with minimal perturbation to the transplanted cell phenotype.
Figure 4. In vivo tracking of primary human RCC.
(A) Fluorescence images of a representative mouse (n= 3) subcutaneously implanted with primary human RCC cells (1×106) pre-labelled for 30 min with SPNRG. (B) Quantification of fluorescence intensities of the implanted cells as a function of post-implantation time. Intensity (%) stands for the percentage of the intensity at the indicated day relative to the initial value. (C) Fluorescence images of a mouse before (left) and 10 min after (right) subcutaneous implantation with 1×106 (1), 1×105 (0.1), and 1×104 (0.01) primary human RCC cells pre-labeled with SPNRD. (D) Histological examination of human RCC xenograft 12 days after implantation. Sections were stained by hematoxylin & eosin (H&E), for the human cell nuclear marker Ku70, and for the RCC phenotype marker CAIX. An unstained section showed nanoparticle retention (red fluorescence).
3. Conclusion
In conclusion, we have developed phosphorylcholine-coated NIR SPNs as a new class of efficient, rapid, and durable cell labelling nanoagents for long-term cell tracking in living animals. SPNRD can enter cells within 30 min of incubation in complete culture medium apparently independent of the cell type, demonstrating its potential as a general cell tracker. Although it possesses a relatively low quantum yield, the far-red absorbing and NIR-emitting features of SPNRD allow it to be clearly visualized at a minimum tissue penetration depth of 0.5 cm, which was otherwise unachievable for the highly fluorescent green SPN (SPNG). In comparison to QDs, SPNRD has a broader absorption and emission spectrum, and its size is generally larger than QDs. However, SPNRD is tolerant to ubiquitous ROS, resulting in durable in vivo fluorescence. Additionally, the completely organic and biologically benign components make SPNRD highly cytocompatible to both immortal and more fragile primary cells, which minimizes the probability of perturbing the phenotype of transplanted cells in living animals and maximizes the bioorthogonal nature of SPNRD as cell tracking material.
With these desirable physical and physiological properties, SPN-based in vivo cell tracking has been demonstrated with primary human RCC cells, with its effectiveness witnessed by durable fluorescence and a lower limit of detection of 10,000 cells with no obvious alteration of phenotype after 12 days. Considering the synthetic and structural flexibility of SPNs, they can easily be tailored into multimodality cell tracking agents by, for instance, incorporating magnetic resonance imaging agents (e.g., iron oxide nanoparticles or gadolinium-lipids).[23] Thus, we believe that SPNs could provide unique opportunities for optimizing cellular therapy and deciphering pathological processes as a cell tracking label.
4. Experimental Section
Chemicals
All chemicals were obtained from Sigma-Aldrich unless otherwise stated. 1,2-Dipalmitoyl-sn-glycero-3-phosphocholinewere was purchased from Avanti Lipids. Poly[(9,9-di-n-octylfluorenyl-2,7-diyl)-alt-(benzo[2,1,3]thiadiazol-4,8-diyl)] (MW = 10,000–20,000) and poly[2,7-(9,9-dioctylfluorene)-alt-4,7-bis(thiophen-2-yl)benzo-2,1,3-thiadiazole] (PFODBT) (MW = 10,000–50,000) were purchased from Sigma-Aldrich. Hoechst 33342, 4′,6-diamidino-2-Phenylindole, dihydrochloride (DAPI), Alexa488 phalloidin conjugate and MTS assay were purchased from Invitrogen. QD655 was also purchased from Invitrogen with the commercial description as Qdot® 655 ITK™ carboxyl quantum dots (CdSe-ZnS). Anti-Ku-70 and anti-CAIX antibody was purchased from Abcam and Novus Biologicals, respectively.
Materials characterization
TEM images were obtained on a JEM 1230 transmission electron microscope with an accelerating voltage of 200 kV. Dynamic light scattering (DLS) was performed on a 90 plus particle size analyzer (Brookhaven Instruments Corporation). Zeta potential measurements were conducted on the Malvern ZetaSizer Nano S. UV-vis spectra were recorded on an Agilent spectrophotometer. Fluorescence measurements were carried out on a wavelength-calibrated FluoroMax-3 fluorometer (Horiba Jobin Yvon). Confocal laser scanning microscopy (CLSM) was conducted on LSM 510 Meta Confocal Scope. Flow cytometry analysis of cells was run on the BD FACScan analyzer. Fluorescence animal imaging was acquired with a CRi Maestro spectral fluorescence imager. The quantum yields were measured using fluorescein as the standard with known quantum yield of 95% in 0.1 M NaOH.
Preparation of SPNs
A THF solution of PFBT or PFODBT (0.25 mg/mL, 1 mL) was rapidly injected into distilled-deionized water (9 mL) under continuous sonication with a microtip-equipped probe sonicator (Branson, W-150) at a power output of 6 watts RMS for 30 S. Then, DPPC (0.25 mg/mL, 1 mL) in THF/water (4: 6) was added into the mixture. After sonication for additional 1 min, THF was evaporated at 50 °C above the transition temperature of DPPC (41 °C) under nitrogen atmosphere. The aqueous solution was filtered through a polyvinylidene fluoride (PVDF) syringe driven filter (0.22 μm) (Millipore), and washed three times using 30 K centrifugal filter units (Millipore) under centrifugation at 4,500 rpm for 5 min at 4 °C. The nanoparticle solution was finally concentrated to 0.08 mg/mL (based on the mass of SP) by ultrafiltration and stored in the dark at 8 °C.
Measurement of imaging depth
The tissue phantom gel was prepared according to a previous report.[24] Tris-buffered saline was poured on top of a known amount of sodium azide and gelatin to make a final concentration of 15 mM and 10 w/v%, respectively. The mixture was heated to 50 °C using a water bath under constant stirring. After the gelatin had dissolved, the mixture was cooled to 37 °C with constant stirring. Hemoglobin and intralipid were added to reach concentrations of 170 μM and 1 v/v%, respectively. A gel cassette intended for electrophoresis was used as a mold to form a 1 cm thick tissue phantom. The fluorescence intensities of the nanoparticle solutions (150 μL, 100 μg/mL) in 96-plate wells were recorded with IVIS spectrum imaging system before and after covering with the tissue phantom gel. Excitation wavelength was at 450 or 600 nm with emission wavelength fixed at 700 nm.
Culture of immortal cell lines
The human cervical cancer HeLa cells and the murine macrophage RAW 264.7 cells were purchased from the American Type Culture Collection (ATCC). Both HeLa cells and RAW264.7 were cultured in Dulbecco’s Modified Eagle Medium (DMEM) (GIBCO) supplemented with 10% fetal bovine serum (FBS) (GIBCO). The murine stromal MS-5 cells were cultured in Iscove’s modified Dulbecco’s medium (IMDM) supplemented with 10% FBS. The cell lines were maintained in an atmosphere of 5% CO2 and 95% humidified air at 37 °C.
Culture of primary human RCC cells
Primary cultures of RCC cells were established according to the method of Valente et al. from a tumorgraft of clear cell RCC of Fuhrman grade III/IV and stage pT2a, NxMx.[25] Briefly, tissue was minced and digested in 200 U/ml collagenase type I (Sigma-Aldrich) until single cells were released. The digested tissue was then incubated with Red Cell Lysis Buffer (eBioscience) followed by passage through 70-μm and 40-μm cell strainers (BD Biosciences). The predominantly single cells passing through the 40-μm filter were centrifuged, resuspended in DMEM/F12 + GlutaMAX-I™ (GIBCO) supplemented with 10% FBS and holo-transferrin (5 μg/ml, Sigma-Aldrich) and placed in collagen-coated cell culture plates. Cell cultures were negative for mycoplasma. After serial passage, retention of the immuno and genetic phenotype of the primary tumour was confirmed in the cultured cells.
Cell imaging
Cells were seeded into 8-well chamber slides and allowed to adhere for 24 h in a humidified atmosphere of 5% CO2 and 95% air at 37 °C. The cells were treated with SPNs (final 50 μg/mL) at 37 or 4 °C in culture medium for 3 or 0.5 h, and then were washed three times with 1 × PBS. The cell nuclei were stained with Hoechst 33342 prior to live cell imaging. Excitation at 405 nm and emission at 420 nm were used for Hoechst 33342, and excitation at 567 nm and emission above 650 nm were used for SPNs. For fixed cell imaging, cells were grown on sterile glass slides, treated with SPNs under the same conditions, and then fixed with 4% paraformaldehyde (PFA). Cellular nuclei and filamentous actins were stained by DAPI and an Alexa488 phalloidin conjugate, respectively. Excitation at 405 nm (Argon laser) and emission at 450 ± 30 nm were used for DAPI, excitation at 458 nm (Argon laser) and emission at 527 ± 22 nm were used for Alexa488, and excitation at 567 nm (diode-pumped solid state laser) and emission above 650 nm (long-pass filter) were used for SPNs.
Flow cytometry
HeLa cells were seeded in a culture dish and allowed to adhere for 24 h. After incubation with SPNRD (50 μg/mL) for 0.5, 1, or 3 h, the cells were washed with 1×PBS, trypsinized, and centrifuged. The cell pellets were collected and suspended in 1×PBS for flow cytometry analysis. For long-term cell tracking, the SPNRD-labelled cells were inoculated into new culture dishes, allowed to grow for 1, 2, 3, 4, or 5 days, and then trypsinized and centrifuged. The cell pellets were collected and suspended in cold 1×PBS for flow cytometry analysis. Cy5.5 channel was used to detect signals.
Cytotoxicity test
Cells were seeded in a culture dish and allowed to adhere for 24 h. The cells were incubated with or without (control) SPNRD (100 μg/mL) for 12 h, and then the cells were washed with 1×PBS, trypsinized, and inoculated into new culture dishes. After growing for 1, 2 and 6 days, cell proliferation was measured with an MTS assay of both SPNRD-labeled and control cells. The absorbance of MTS at 490 nm was measured by using a microplate reader. Cell viability was expressed by the ratio of the absorbance of SPNRD-labeled cells to that of cells incubated with culture medium only.
Animal Models
All animal experiments were performed in compliance with the Guidelines for the Care and Use of Research Animals established by the Stanford University Animal Studies Committee. Male nude mice (12 weeks-old) were implanted subcutaneously with the specified number of RCC cells, pre-labeled with SPNRD in culture medium overnight. Cells were mixed 1:1 with Matrigel prior to inoculation into the animal. Mice were anesthetized with isoflurane, and then imaged using the CRi Maestro imaging system with excitation 595±25 nm and emission from 630–800 nm. Spectra of the SPN and autofluorescence were recorded and images were spectrally deconvolved using CRi software. On day 12, mice were euthanized by CO2 asphyxiation and tumor tissue was excised and placed in formalin for processing by immunohistochemistry.
Immunohistochemistry
Immunohistochemistry was performed according to our previous report.[26] Resected tumor tissue was fixed in 10% buffered formalin overnight and embedded in paraffin. Five-micron tissue sections were deparaffinized with xylene and rehydrated with ethanol, followed by antigen retrieval by heating in citrate buffer (pH 6.0). Endogenous peroxidase activity was blocked by incubation in a methanolic solution of 0.3% hydrogen peroxide. 10% horse serum was used to block nonspecific binding of antibodies, followed by incubation with primary Anti-Ku-70 (Abcam) and anti-CAIX antibodies (Novus Biologicals) at 1:200 dilutions. Following incubation with biotinylated secondary antibody, then peroxidase-conjugated streptavidin, color was developed with 3,3-diaminoben-zidine (DakoCytomation California, Inc., Carpinteria, CA). Counter staining was performed with H&E.
Supplementary Material
Acknowledgments
We acknowledge the use of the SCi3 Core Facility. This work was supported by the NIH National Cancer Institute (NCI) grants R01CA135294, R21CA138353A2, the Stanford University National Cancer Institute (NCI) CCNE-T grant (U54CA119367) and ICMIC (P50CA114747). AJS also thanks the Susan G. Komen For The Cure for fellowship support. MPV thanks the Instrumentarium Science Foundation, Finland, the Finnish Medical Foundation. MS appreciates the support of the Ferdinand Eisenberger Grant of the German Society of Urology ID SaM1/FE-11. Supporting Information is available online from Wiley InterScience or from the author.
Footnotes
Supporting Information is available online from the Wiley Online Library or from the authors.
Contributor Information
Dr. Kanyi Pu, Molecular Imaging Program at Stanford, Department of Radiology, School of Medicine, Stanford University, USA.
Dr. Adam J. Shuhendler, Molecular Imaging Program at Stanford, Department of Radiology, School of Medicine, Stanford University, USA.
Dr. Maija P. Valta, Department of Urology, School of Medicine, Stanford University, USA. Division of Medicine, Turku University Hospital and University of Turku, Finland.
Dr. Lina Cui, Molecular Imaging Program at Stanford, Department of Radiology, School of Medicine, Stanford University, USA
Dr. Matthias Saar, Department of Urology School of Medicine, Stanford University, USA. Department of Urology and Pediatric Urology, University of Saarland, Homburg/Saar, Germany
Prof. Donna M. Peehl, Department of Urology School of Medicine, Stanford University, USA
Prof. Jianghong Rao, Email: jrao@stanford.edu, Molecular Imaging Program at Stanford, Department of Radiology, School of Medicine, Stanford University, USA
References
- 1.a) de Vries IJ, Lesterhuis WJ, Barentsz JO, Verdijk P, van Krieken JH, Boerman OC, Oyen WJ, Bonenkamp JJ, Boezeman JB, Adema GJ, Bulte JW, Scheenen TW, Punt CJ, Heerschap A, Figdor CG. Nat Biotechnol. 2005;23:1407–1413. doi: 10.1038/nbt1154. [DOI] [PubMed] [Google Scholar]; b) Lewin M, Carlesso N, Tung CH, Tang XW, Cory D, Scadden DT, Weissleder R. Nat Biotechnol. 2000;18:410–414. doi: 10.1038/74464. [DOI] [PubMed] [Google Scholar]; c) Dudley ME, Rosenberg SA. Nat Rev Cancer. 2003;3:666–675. doi: 10.1038/nrc1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.a) Rao J, Dragulescu-Andrasi A, Yao H. Curr Opin Biotech. 2007;18:17–25. doi: 10.1016/j.copbio.2007.01.003. [DOI] [PubMed] [Google Scholar]; b) Ntziachristos V, Ripoll J, Wang LV, Weissleder R. Nat Biotechnol. 2005;23:313–320. doi: 10.1038/nbt1074. [DOI] [PubMed] [Google Scholar]; c) Frangioni JV. Curr Opin Chem Biol. 2003;7:626–634. doi: 10.1016/j.cbpa.2003.08.007. [DOI] [PubMed] [Google Scholar]
- 3.a) Pittet MJ, Swirski FK, Reynolds F, Josephson L, Weissleder R. Nat Protoc. 2006;1:73–79. doi: 10.1038/nprot.2006.11. [DOI] [PubMed] [Google Scholar]; b) Xu C, Miranda-Nieves D, Ankrum JA, Matthiesen ME, Phillips JA, Roes I, Wojtkiewicz GR, Juneja V, Kultima JR, Zhao W, Vemula PK, Lin CP, Nahrendorf M, Karp JM. Nano Lett. 2012;12:4131–4139. doi: 10.1021/nl301658q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.a) Shah BS, Clark PA, Moioli EK, Stroscio MA, Mao JJ. Nano Lett. 2007;7:3071–3079. doi: 10.1021/nl071547f. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Voura EB, Jaiswal JK, Mattoussi H, Simon SM. Nat, Med. 2004;10:993–998. doi: 10.1038/nm1096. [DOI] [PubMed] [Google Scholar]; c) Dubertret B, Skourides P, Norris DJ, Noireaux V, Brivanlou AH, Libchaber A. Science. 2002;298:1759–1762. doi: 10.1126/science.1077194. [DOI] [PubMed] [Google Scholar]; d) Rivera_Gil P, Yang F, Thomas H, Li L, Terfort A, Parak WJ. Nano Today. 2011;6:20–27. [Google Scholar]
- 5.a) Mancini MC, Kairdolf BA, Smith AM, Nie SM. J Am Chem Soc. 2008;130:10836–10837. doi: 10.1021/ja8040477. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Winnik FM, Maysinger D. Acc Chem Res. 2013;46:672–680. doi: 10.1021/ar3000585. [DOI] [PubMed] [Google Scholar]; c) Zhang Y, He J, Wang PN, Chen JY, Lu ZJ, Lu DR, Guo J, Wang CC, Yang WL. J Am Chem Soc. 2006;128:13396–13401. doi: 10.1021/ja061225y. [DOI] [PubMed] [Google Scholar]; d) Yan Y, Wang S, Liu Z, Wang H, Huang D. Anal Chem. 2010;82:9775–9781. doi: 10.1021/ac101929q. [DOI] [PubMed] [Google Scholar]
- 6.Dupont KM, Sharma K, Stevens HY, Boerckel JD, Garcia AJ, Guldberg RE. Proc Natl Acad Sci U S A. 2010;107:3305–3310. doi: 10.1073/pnas.0905444107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Klebanoff SJ. J Leukoc Biol. 2005;77:598–625. doi: 10.1189/jlb.1204697. [DOI] [PubMed] [Google Scholar]
- 8.a) Zhu C, Liu L, Yang Q, Lv F, Wang S. Chem Rev. 2012;112:4687–4735. doi: 10.1021/cr200263w. [DOI] [PubMed] [Google Scholar]; b) Pecher J, Mecking S. Chem Rev. 2010;110:6260–6279. doi: 10.1021/cr100132y. [DOI] [PubMed] [Google Scholar]; c) Wu C, Chiu DT. Angew Chem Int Ed Engl. 2013;52:3086–3109. doi: 10.1002/anie.201205133. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Pu KY, Liu B. Adv Funct Mater. 2011;21:3408–3423. [Google Scholar]; e) Moon JH, McDaniel W, Maclean P, Hancock LF. Angew Chem Int Ed Engl. 2007;46:8223–8225. doi: 10.1002/anie.200701991. [DOI] [PubMed] [Google Scholar]; f) Pu K, Shuhendler AJ, Rao J. Angew Chem Int Ed Engl. 2013;52:10325–10329. doi: 10.1002/anie.201303420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.a) Wu C, Hansen SJ, Hou Q, Yu J, Zeigler M, Jin Y, Burnham DR, McNeill JD, Olson JM, Chiu DT. Angew Chem Int Ed Engl. 2011;50:3430–3434. doi: 10.1002/anie.201007461. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Kim S, Lim CK, Na J, Lee YD, Kim K, Choi K, Leary JF, Kwon IC. Chem Commun. 2010;46:1617–1619. doi: 10.1039/b923309a. [DOI] [PubMed] [Google Scholar]; c) Ding D, Liu J, Feng G, Li K, Hu Y, Liu B. Small. 2013;9:3093–3102. doi: 10.1002/smll.201300171. [DOI] [PubMed] [Google Scholar]; d) Ding D, Li K, Qin W, Zhan R, Hu Y, Liu J, Tang BZ, Liu B. Adv Healthc Mater. 2013;2:500–507. doi: 10.1002/adhm.201200243. [DOI] [PubMed] [Google Scholar]
- 10.Xiong L, Shuhendler AJ, Rao J. Nat Commun. 2012;3:1193. doi: 10.1038/ncomms2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pu K, Shuhendler AJ, Jokerst JV, Mei J, Gambhir SS, Bao Z, Rao J. Nat Nanotech. 2014 doi: 10.1038/NNANO.2013.302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Feng G, Li K, Liu J, Ding D, Liu B. Small. 2013 doi: 10.1002/smll.201302161. [DOI] [Google Scholar]
- 13.a) Wu C, Schneider T, Zeigler M, Yu J, Schiro PG, Burnham DR, McNeill JD, Chiu DT. J Am Chem Soc. 2010;132:15410–15417. doi: 10.1021/ja107196s. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Howes P, Green M, Levitt J, Suhling K, Hughes M. J Am Chem Soc. 2010;132:3989–3996. doi: 10.1021/ja1002179. [DOI] [PubMed] [Google Scholar]; c) Koner AL, Krndija D, Hou Q, Sherratt DJ, Howarth M. ACS Nano. 2013;7:1137–1144. doi: 10.1021/nn3042122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu C, Szymanski C, McNeill J. Langmuir. 2006;22:2956–2960. doi: 10.1021/la060188l. [DOI] [PubMed] [Google Scholar]
- 15.a) Fischer CS, Baier MC, Mecking S. J Am Chem Soc. 2013;135:1148–1154. doi: 10.1021/ja311497e. [DOI] [PubMed] [Google Scholar]; b) Chan YH, Ye F, Gallina ME, Zhang X, Jin Y, Wu IC, Chiu DT. J Am Chem Soc. 2012;134:7309–7312. doi: 10.1021/ja3022973. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Petkau K, Kaeser A, Fischer I, Brunsveld L, Schenning AP. J Am Chem Soc. 2011;133:17063–17071. doi: 10.1021/ja2075345. [DOI] [PubMed] [Google Scholar]
- 16.a) Hafez IM, Cullis PR. Adv Drug Deliv Rev. 2001;47:139–148. doi: 10.1016/s0169-409x(01)00103-x. [DOI] [PubMed] [Google Scholar]; b) Lonez C, Lensink MF, Kleiren E, Vanderwinden JM, Ruysschaert JM, Vandenbranden M. Cell Mol Life Sci. 2010;67:483–494. doi: 10.1007/s00018-009-0197-x. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Ishihara K, Goto Y, Takai M, Matsuno R, Inoue Y, Konno T. BBA-Gen Sub. 2011;1810:268–275. doi: 10.1016/j.bbagen.2010.04.008. [DOI] [PubMed] [Google Scholar]; d) Son S, Kim G, Singha K, Park S, Ree M, Kim WJ. Small. 2011;7:2991–2997. doi: 10.1002/smll.201100232. [DOI] [PubMed] [Google Scholar]
- 17.Thomas KRJ, Lin JT, Velusamy M, Tao YT, Chuen CH. Adv Func Mater. 2004;14:83–90. [Google Scholar]
- 18.a) Zhou WB, Shao JY, Qiao J, Wei QS, Tang JG, Jian J. Chem Commun. 2010;46:1479–1481. doi: 10.1039/b915125g. [DOI] [PubMed] [Google Scholar]; b) Chung YC, Chen IH, Chen CJ. Biomaterials. 2008;29:1807–1816. doi: 10.1016/j.biomaterials.2007.12.032. [DOI] [PubMed] [Google Scholar]
- 19.a) Fernando LP, Kandel PK, Yu JB, McNeill J, Ackroyd PC, Christensen KA. Biomacromolecules. 2010;11:2675–2682. doi: 10.1021/bm1007103. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Lee J, Twomey M, Machado C, Gomez G, Doshi M, Gesquiere AJ, Moon JH. Macromol Biosci. 2013;13:913–920. doi: 10.1002/mabi.201300030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Knoll G, Burger KNJ, Bron R, Vanmeer G, Verkleij AJ. J Cell Biol. 1988;107:2511–2521. doi: 10.1083/jcb.107.6.2511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Siegel R, Naishadham D, Jemal A. CA Cancer J Clin. 2012;62:10–29. doi: 10.3322/caac.20138. [DOI] [PubMed] [Google Scholar]
- 22.Al-Ahmadie HA, Alden D, Fine SW, Gopalan A, Touijer KA, Russo P, Reuter VE, Tickoo SK. Am J Surg Pathol. 2011;35:949–961. doi: 10.1097/PAS.0b013e31821e25cd. [DOI] [PubMed] [Google Scholar]
- 23.a) Vuu K, Xie J, McDonald MA, Bernardo M, Hunter F, Zhang Y, Li K, Bednarski M, Guccione S. Bioconjug Chem. 2005;16:995–999. doi: 10.1021/bc050085z. [DOI] [PubMed] [Google Scholar]; b) Nasongkla N, Bey E, Ren JM, Ai H, Khemtong C, Guthi JS, Chin SF, Sherry AD, Boothman DA, Gao JM. Nano Lett. 2006;6:2427–2430. doi: 10.1021/nl061412u. [DOI] [PubMed] [Google Scholar]
- 24.Shuhendler AJ, Prasad P, Chan HK, Gordijo CR, Soroushian B, Kolios M, Yu K, O’Brien PJ, Rauth AM, Wu XY. ACS Nano. 2011;5:1958–1966. doi: 10.1021/nn103024b. [DOI] [PubMed] [Google Scholar]
- 25.Valente MJ, Henrique R, Costa VL, Jeronimo C, Carvalho F, Bastos ML, de Pinho PG, Carvalho M. PLoS One. 2011;6:e19337. doi: 10.1371/journal.pone.0019337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhao HJ, Nolley R, Chen ZX, Peehl DM. Am J Pathol. 2010;177:229–239. doi: 10.2353/ajpath.2010.090821. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.