Abstract
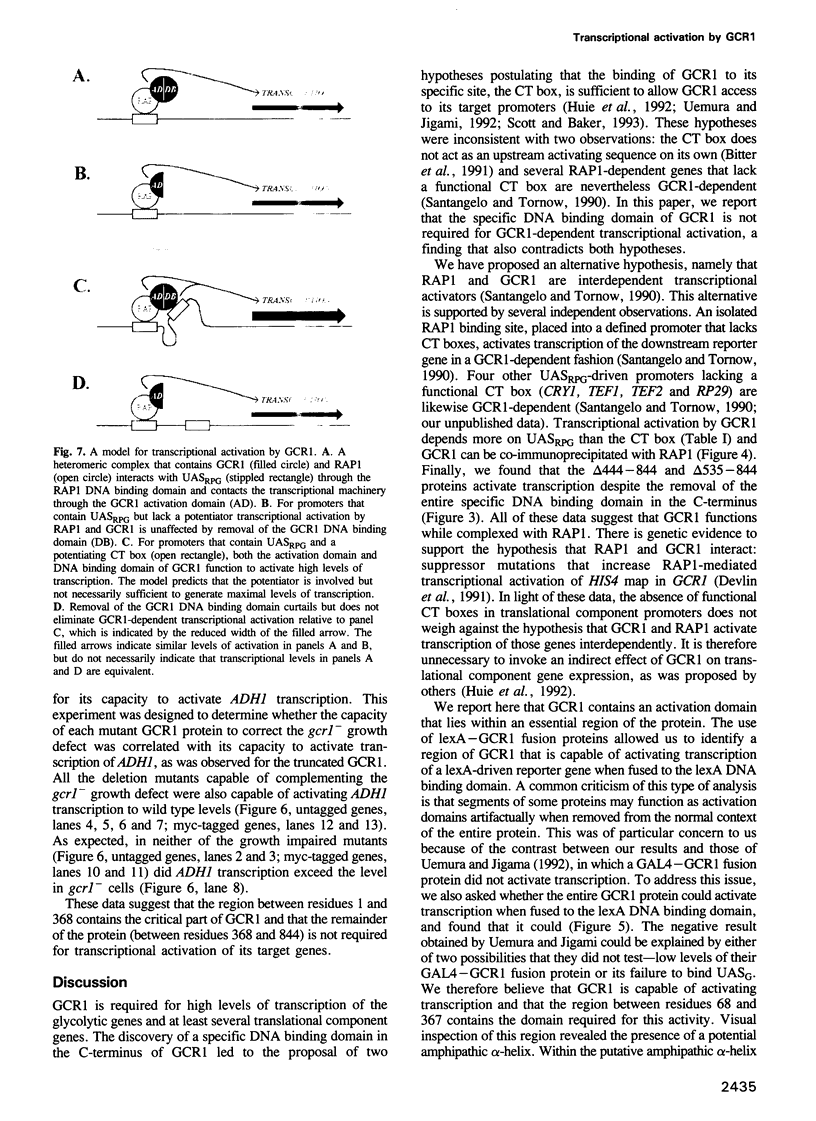
In Saccharomyces cerevisiae, efficient expression of glycolytic and translational component genes requires two DNA binding proteins, RAP1 (which binds to UASRPG) and GCR1 (which binds to the CT box). We generated deletions in GCR1 to test the validity of several different models for GCR1 function. We report here that the C-terminal half of GCR1, which includes the domain required for DNA binding to the CT box in vitro, can be removed without affecting GCR1-dependent transcription of either the glycolytic gene ADH1 or the translational component genes TEF1 and TEF2. We have also identified an activation domain within a segment of the GCR1 protein (the N-terminal third) that is essential for in vivo function. RAP1 and GCR1 can be co-immunoprecipitated from whole cell extracts, suggesting that they form a complex in vivo. The data are most consistent with a model in which GCR1 is attracted to DNA through contact with RAP1.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baker H. V. GCR1 of Saccharomyces cerevisiae encodes a DNA binding protein whose binding is abolished by mutations in the CTTCC sequence motif. Proc Natl Acad Sci U S A. 1991 Nov 1;88(21):9443–9447. doi: 10.1073/pnas.88.21.9443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker H. V. Glycolytic gene expression in Saccharomyces cerevisiae: nucleotide sequence of GCR1, null mutants, and evidence for expression. Mol Cell Biol. 1986 Nov;6(11):3774–3784. doi: 10.1128/mcb.6.11.3774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bitter G. A., Chang K. K., Egan K. M. A multi-component upstream activation sequence of the Saccharomyces cerevisiae glyceraldehyde-3-phosphate dehydrogenase gene promoter. Mol Gen Genet. 1991 Dec;231(1):22–32. doi: 10.1007/BF00293817. [DOI] [PubMed] [Google Scholar]
- Broach J. R., Strathern J. N., Hicks J. B. Transformation in yeast: development of a hybrid cloning vector and isolation of the CAN1 gene. Gene. 1979 Dec;8(1):121–133. doi: 10.1016/0378-1119(79)90012-x. [DOI] [PubMed] [Google Scholar]
- Buchman A. R., Lue N. F., Kornberg R. D. Connections between transcriptional activators, silencers, and telomeres as revealed by functional analysis of a yeast DNA-binding protein. Mol Cell Biol. 1988 Dec;8(12):5086–5099. doi: 10.1128/mcb.8.12.5086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnette W. N. "Western blotting": electrophoretic transfer of proteins from sodium dodecyl sulfate--polyacrylamide gels to unmodified nitrocellulose and radiographic detection with antibody and radioiodinated protein A. Anal Biochem. 1981 Apr;112(2):195–203. doi: 10.1016/0003-2697(81)90281-5. [DOI] [PubMed] [Google Scholar]
- Chambers A., Stanway C., Kingsman A. J., Kingsman S. M. The UAS of the yeast PGK gene is composed of multiple functional elements. Nucleic Acids Res. 1988 Sep 12;16(17):8245–8260. doi: 10.1093/nar/16.17.8245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clifton D., Fraenkel D. G. The gcr (glycolysis regulation) mutation of Saccharomyces cerevisiae. J Biol Chem. 1981 Dec 25;256(24):13074–13078. [PubMed] [Google Scholar]
- Clifton D., Weinstock S. B., Fraenkel D. G. Glycolysis mutants in Saccharomyces cerevisiae. Genetics. 1978 Jan;88(1):1–11. doi: 10.1093/genetics/88.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cress W. D., Triezenberg S. J. Critical structural elements of the VP16 transcriptional activation domain. Science. 1991 Jan 4;251(4989):87–90. doi: 10.1126/science.1846049. [DOI] [PubMed] [Google Scholar]
- Devlin C., Tice-Baldwin K., Shore D., Arndt K. T. RAP1 is required for BAS1/BAS2- and GCN4-dependent transcription of the yeast HIS4 gene. Mol Cell Biol. 1991 Jul;11(7):3642–3651. doi: 10.1128/mcb.11.7.3642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evan G. I., Lewis G. K., Ramsay G., Bishop J. M. Isolation of monoclonal antibodies specific for human c-myc proto-oncogene product. Mol Cell Biol. 1985 Dec;5(12):3610–3616. doi: 10.1128/mcb.5.12.3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerster T., Roeder R. G. A herpesvirus trans-activating protein interacts with transcription factor OTF-1 and other cellular proteins. Proc Natl Acad Sci U S A. 1988 Sep;85(17):6347–6351. doi: 10.1073/pnas.85.17.6347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy C. F., Sussel L., Shore D. A RAP1-interacting protein involved in transcriptional silencing and telomere length regulation. Genes Dev. 1992 May;6(5):801–814. doi: 10.1101/gad.6.5.801. [DOI] [PubMed] [Google Scholar]
- Himmelfarb H. J., Pearlberg J., Last D. H., Ptashne M. GAL11P: a yeast mutation that potentiates the effect of weak GAL4-derived activators. Cell. 1990 Dec 21;63(6):1299–1309. doi: 10.1016/0092-8674(90)90425-e. [DOI] [PubMed] [Google Scholar]
- Holland M. J., Yokoi T., Holland J. P., Myambo K., Innis M. A. The GCR1 gene encodes a positive transcriptional regulator of the enolase and glyceraldehyde-3-phosphate dehydrogenase gene families in Saccharomyces cerevisiae. Mol Cell Biol. 1987 Feb;7(2):813–820. doi: 10.1128/mcb.7.2.813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huet J., Cottrelle P., Cool M., Vignais M. L., Thiele D., Marck C., Buhler J. M., Sentenac A., Fromageot P. A general upstream binding factor for genes of the yeast translational apparatus. EMBO J. 1985 Dec 16;4(13A):3539–3547. doi: 10.1002/j.1460-2075.1985.tb04114.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huet J., Sentenac A. TUF, the yeast DNA-binding factor specific for UASrpg upstream activating sequences: identification of the protein and its DNA-binding domain. Proc Natl Acad Sci U S A. 1987 Jun;84(11):3648–3652. doi: 10.1073/pnas.84.11.3648. [DOI] [PMC free article] [PubMed] [Google Scholar]
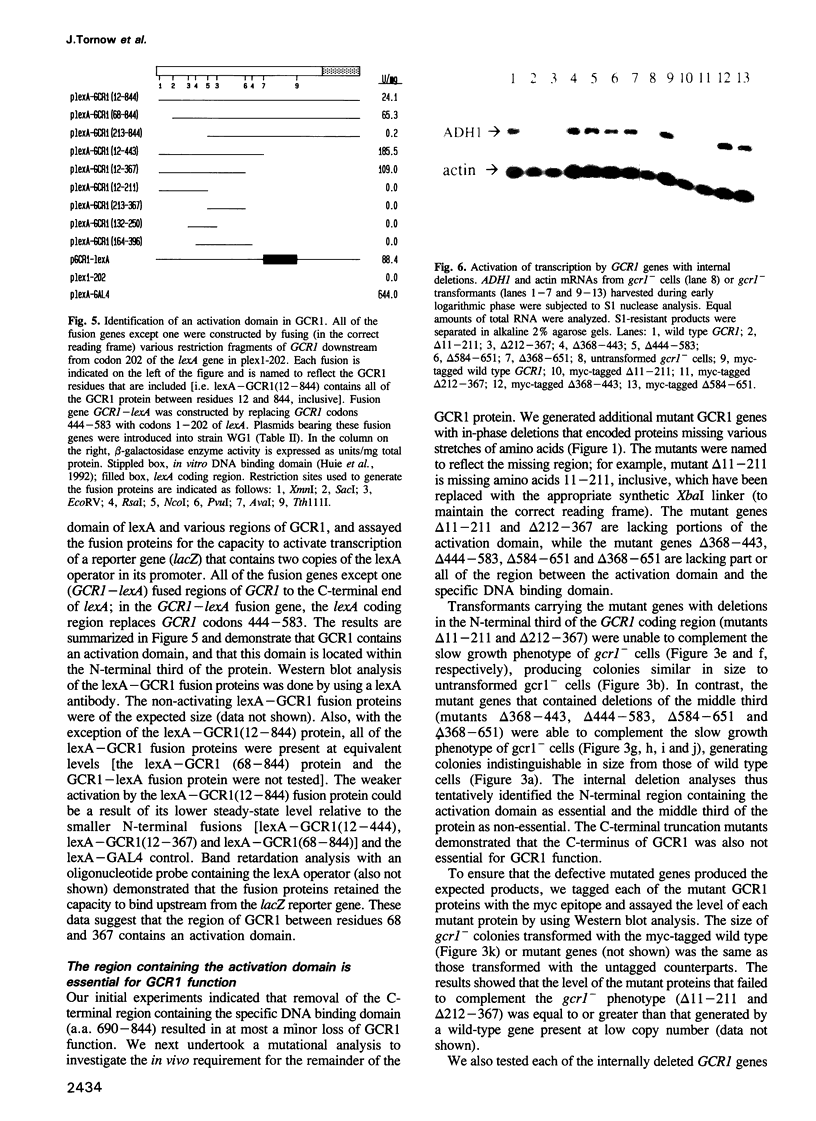
- Huie M. A., Scott E. W., Drazinic C. M., Lopez M. C., Hornstra I. K., Yang T. P., Baker H. V. Characterization of the DNA-binding activity of GCR1: in vivo evidence for two GCR1-binding sites in the upstream activating sequence of TPI of Saccharomyces cerevisiae. Mol Cell Biol. 1992 Jun;12(6):2690–2700. doi: 10.1128/mcb.12.6.2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito H., Fukuda Y., Murata K., Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983 Jan;153(1):163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lai J. S., Herr W. Ethidium bromide provides a simple tool for identifying genuine DNA-independent protein associations. Proc Natl Acad Sci U S A. 1992 Aug 1;89(15):6958–6962. doi: 10.1073/pnas.89.15.6958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lustig A. J., Kurtz S., Shore D. Involvement of the silencer and UAS binding protein RAP1 in regulation of telomere length. Science. 1990 Oct 26;250(4980):549–553. doi: 10.1126/science.2237406. [DOI] [PubMed] [Google Scholar]
- Machida M., Uemura H., Jigami Y., Tanaka H. The protein factor which binds to the upstream activating sequence of Saccharomyces cerevisiae ENO1 gene. Nucleic Acids Res. 1988 Feb 25;16(4):1407–1422. doi: 10.1093/nar/16.4.1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieuwint R. T., Mager W. H., Maurer K. C., Planta R. J. Mutational analysis of the upstream activation site of yeast ribosomal protein genes. Curr Genet. 1989 Apr;15(4):247–251. doi: 10.1007/BF00447039. [DOI] [PubMed] [Google Scholar]
- Runge K. W., Zakian V. A. Introduction of extra telomeric DNA sequences into Saccharomyces cerevisiae results in telomere elongation. Mol Cell Biol. 1989 Apr;9(4):1488–1497. doi: 10.1128/mcb.9.4.1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santangelo G. M., Tornow J. Efficient transcription of the glycolytic gene ADH1 and three translational component genes requires the GCR1 product, which can act through TUF/GRF/RAP binding sites. Mol Cell Biol. 1990 Feb;10(2):859–862. doi: 10.1128/mcb.10.2.859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santangelo G. M., Tornow J., McLaughlin C. S., Moldave K. Properties of promoters cloned randomly from the Saccharomyces cerevisiae genome. Mol Cell Biol. 1988 Oct;8(10):4217–4224. doi: 10.1128/mcb.8.10.4217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott E. W., Allison H. E., Baker H. V. Characterization of TPI gene expression in isogeneic wild-type and gcr1-deletion mutant strains of Saccharomyces cerevisiae. Nucleic Acids Res. 1990 Dec 11;18(23):7099–7107. doi: 10.1093/nar/18.23.7099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott E. W., Baker H. V. Concerted action of the transcriptional activators REB1, RAP1, and GCR1 in the high-level expression of the glycolytic gene TPI. Mol Cell Biol. 1993 Jan;13(1):543–550. doi: 10.1128/mcb.13.1.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shore D., Nasmyth K. Purification and cloning of a DNA binding protein from yeast that binds to both silencer and activator elements. Cell. 1987 Dec 4;51(5):721–732. doi: 10.1016/0092-8674(87)90095-x. [DOI] [PubMed] [Google Scholar]
- Shore D., Stillman D. J., Brand A. H., Nasmyth K. A. Identification of silencer binding proteins from yeast: possible roles in SIR control and DNA replication. EMBO J. 1987 Feb;6(2):461–467. doi: 10.1002/j.1460-2075.1987.tb04776.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tornow J., Santangelo G. M. Efficient expression of the Saccharomyces cerevisiae glycolytic gene ADH1 is dependent upon a cis-acting regulatory element (UASRPG) found initially in genes encoding ribosomal proteins. Gene. 1990 May 31;90(1):79–85. doi: 10.1016/0378-1119(90)90441-s. [DOI] [PubMed] [Google Scholar]
- Uemura H., Jigami Y. Role of GCR2 in transcriptional activation of yeast glycolytic genes. Mol Cell Biol. 1992 Sep;12(9):3834–3842. doi: 10.1128/mcb.12.9.3834. [DOI] [PMC free article] [PubMed] [Google Scholar]