Abstract
The Src-homology 3 (SH3) region is a protein domain consisting of approximately 60 residues. It occurs in a large number of eukaryotic proteins involved in signal transduction, cell polarization and membrane--cytoskeleton interactions. The function is unknown, but it is probably involved in specific protein--protein interactions. Here we report the crystal structure of the SH3 domain of Fyn (a Src family tyrosine kinase) at 1.9 A resolution. The crystals have two SH3 molecules per asymmetric unit. These two Fyn SH3 domains are not related by a local twofold axis. The crystal structures of spectrin and Fyn SH3 domains as well as the solution structure of the Src SH3 domain show that these all have the same basic fold. A protein domain which has the same topology as SH3 is present in the prokaryotic regulatory enzyme BirA. The comparison between the crystal structures of Fyn and spectrin SH3 domains shows that a conserved surface patch, consisting mainly of aromatic residues, is flanked by two hairpin-like loops (residues 94-104 and 114-118 in Fyn). These loops are different in tyrosine kinase and spectrin SH3 domains. They could modulate the binding properties of the aromatic surface.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Benner S. A., Cohen M. A., Gerloff D. Correct structure prediction? Nature. 1992 Oct 29;359(6398):781–781. doi: 10.1038/359781a0. [DOI] [PubMed] [Google Scholar]
- Benner S. A., Cohen M. A., Gerloff D. Predicted secondary structure for the Src homology 3 domain. J Mol Biol. 1993 Jan 20;229(2):295–305. doi: 10.1006/jmbi.1993.1035. [DOI] [PubMed] [Google Scholar]
- Booker G. W., Breeze A. L., Downing A. K., Panayotou G., Gout I., Waterfield M. D., Campbell I. D. Structure of an SH2 domain of the p85 alpha subunit of phosphatidylinositol-3-OH kinase. Nature. 1992 Aug 20;358(6388):684–687. doi: 10.1038/358684a0. [DOI] [PubMed] [Google Scholar]
- Broek D., Toda T., Michaeli T., Levin L., Birchmeier C., Zoller M., Powers S., Wigler M. The S. cerevisiae CDC25 gene product regulates the RAS/adenylate cyclase pathway. Cell. 1987 Mar 13;48(5):789–799. doi: 10.1016/0092-8674(87)90076-6. [DOI] [PubMed] [Google Scholar]
- Brünger A. T., Kuriyan J., Karplus M. Crystallographic R factor refinement by molecular dynamics. Science. 1987 Jan 23;235(4787):458–460. doi: 10.1126/science.235.4787.458. [DOI] [PubMed] [Google Scholar]
- Chant J., Corrado K., Pringle J. R., Herskowitz I. Yeast BUD5, encoding a putative GDP-GTP exchange factor, is necessary for bud site selection and interacts with bud formation gene BEM1. Cell. 1991 Jun 28;65(7):1213–1224. doi: 10.1016/0092-8674(91)90016-r. [DOI] [PubMed] [Google Scholar]
- Chenevert J., Corrado K., Bender A., Pringle J., Herskowitz I. A yeast gene (BEM1) necessary for cell polarization whose product contains two SH3 domains. Nature. 1992 Mar 5;356(6364):77–79. doi: 10.1038/356077a0. [DOI] [PubMed] [Google Scholar]
- Cicchetti P., Mayer B. J., Thiel G., Baltimore D. Identification of a protein that binds to the SH3 region of Abl and is similar to Bcr and GAP-rho. Science. 1992 Aug 7;257(5071):803–806. doi: 10.1126/science.1379745. [DOI] [PubMed] [Google Scholar]
- Clark S. G., Stern M. J., Horvitz H. R. C. elegans cell-signalling gene sem-5 encodes a protein with SH2 and SH3 domains. Nature. 1992 Mar 26;356(6367):340–344. doi: 10.1038/356340a0. [DOI] [PubMed] [Google Scholar]
- Drubin D. G., Miller K. G., Botstein D. Yeast actin-binding proteins: evidence for a role in morphogenesis. J Cell Biol. 1988 Dec;107(6 Pt 2):2551–2561. doi: 10.1083/jcb.107.6.2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drubin D. G., Mulholland J., Zhu Z. M., Botstein D. Homology of a yeast actin-binding protein to signal transduction proteins and myosin-I. Nature. 1990 Jan 18;343(6255):288–290. doi: 10.1038/343288a0. [DOI] [PubMed] [Google Scholar]
- Eck M. J., Shoelson S. E., Harrison S. C. Recognition of a high-affinity phosphotyrosyl peptide by the Src homology-2 domain of p56lck. Nature. 1993 Mar 4;362(6415):87–91. doi: 10.1038/362087a0. [DOI] [PubMed] [Google Scholar]
- Hughes D. A., Fukui Y., Yamamoto M. Homologous activators of ras in fission and budding yeast. Nature. 1990 Mar 22;344(6264):355–357. doi: 10.1038/344355a0. [DOI] [PubMed] [Google Scholar]
- Jackson P., Baltimore D. N-terminal mutations activate the leukemogenic potential of the myristoylated form of c-abl. EMBO J. 1989 Feb;8(2):449–456. doi: 10.1002/j.1460-2075.1989.tb03397.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janin J., Miller S., Chothia C. Surface, subunit interfaces and interior of oligomeric proteins. J Mol Biol. 1988 Nov 5;204(1):155–164. doi: 10.1016/0022-2836(88)90606-7. [DOI] [PubMed] [Google Scholar]
- Jones T. A., Zou J. Y., Cowan S. W., Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A. 1991 Mar 1;47(Pt 2):110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- Kabsch W., Sander C. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983 Dec;22(12):2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- Kato J. Y., Takeya T., Grandori C., Iba H., Levy J. B., Hanafusa H. Amino acid substitutions sufficient to convert the nontransforming p60c-src protein to a transforming protein. Mol Cell Biol. 1986 Dec;6(12):4155–4160. doi: 10.1128/mcb.6.12.4155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakami T., Pennington C. Y., Robbins K. C. Isolation and oncogenic potential of a novel human src-like gene. Mol Cell Biol. 1986 Dec;6(12):4195–4201. doi: 10.1128/mcb.6.12.4195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitamura D., Kaneko H., Miyagoe Y., Ariyasu T., Watanabe T. Isolation and characterization of a novel human gene expressed specifically in the cells of hematopoietic lineage. Nucleic Acids Res. 1989 Nov 25;17(22):9367–9379. [PMC free article] [PubMed] [Google Scholar]
- Koch C. A., Anderson D., Moran M. F., Ellis C., Pawson T. SH2 and SH3 domains: elements that control interactions of cytoplasmic signaling proteins. Science. 1991 May 3;252(5006):668–674. doi: 10.1126/science.1708916. [DOI] [PubMed] [Google Scholar]
- Leto T. L., Lomax K. J., Volpp B. D., Nunoi H., Sechler J. M., Nauseef W. M., Clark R. A., Gallin J. I., Malech H. L. Cloning of a 67-kD neutrophil oxidase factor with similarity to a noncatalytic region of p60c-src. Science. 1990 May 11;248(4956):727–730. doi: 10.1126/science.1692159. [DOI] [PubMed] [Google Scholar]
- Martinez R., Mathey-Prevot B., Bernards A., Baltimore D. Neuronal pp60c-src contains a six-amino acid insertion relative to its non-neuronal counterpart. Science. 1987 Jul 24;237(4813):411–415. doi: 10.1126/science.2440106. [DOI] [PubMed] [Google Scholar]
- Matthews D. A., Smith S. L., Baccanari D. P., Burchall J. J., Oatley S. J., Kraut J. Crystal structure of a novel trimethoprim-resistant dihydrofolate reductase specified in Escherichia coli by R-plasmid R67. Biochemistry. 1986 Jul 29;25(15):4194–4204. doi: 10.1021/bi00363a005. [DOI] [PubMed] [Google Scholar]
- Mayer B. J., Baltimore D. Signalling through SH2 and SH3 domains. Trends Cell Biol. 1993 Jan;3(1):8–13. doi: 10.1016/0962-8924(93)90194-6. [DOI] [PubMed] [Google Scholar]
- Mayer B. J., Jackson P. K., Baltimore D. The noncatalytic src homology region 2 segment of abl tyrosine kinase binds to tyrosine-phosphorylated cellular proteins with high affinity. Proc Natl Acad Sci U S A. 1991 Jan 15;88(2):627–631. doi: 10.1073/pnas.88.2.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCaffrey G., Clay F. J., Kelsay K., Sprague G. F., Jr Identification and regulation of a gene required for cell fusion during mating of the yeast Saccharomyces cerevisiae. Mol Cell Biol. 1987 Aug;7(8):2680–2690. doi: 10.1128/mcb.7.8.2680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mian I. S., Bradwell A. R., Olson A. J. Structure, function and properties of antibody binding sites. J Mol Biol. 1991 Jan 5;217(1):133–151. doi: 10.1016/0022-2836(91)90617-f. [DOI] [PubMed] [Google Scholar]
- Morris A. L., MacArthur M. W., Hutchinson E. G., Thornton J. M. Stereochemical quality of protein structure coordinates. Proteins. 1992 Apr;12(4):345–364. doi: 10.1002/prot.340120407. [DOI] [PubMed] [Google Scholar]
- Murzin A. G. Familiar strangers. Nature. 1992 Dec 17;360(6405):635–635. doi: 10.1038/360635a0. [DOI] [PubMed] [Google Scholar]
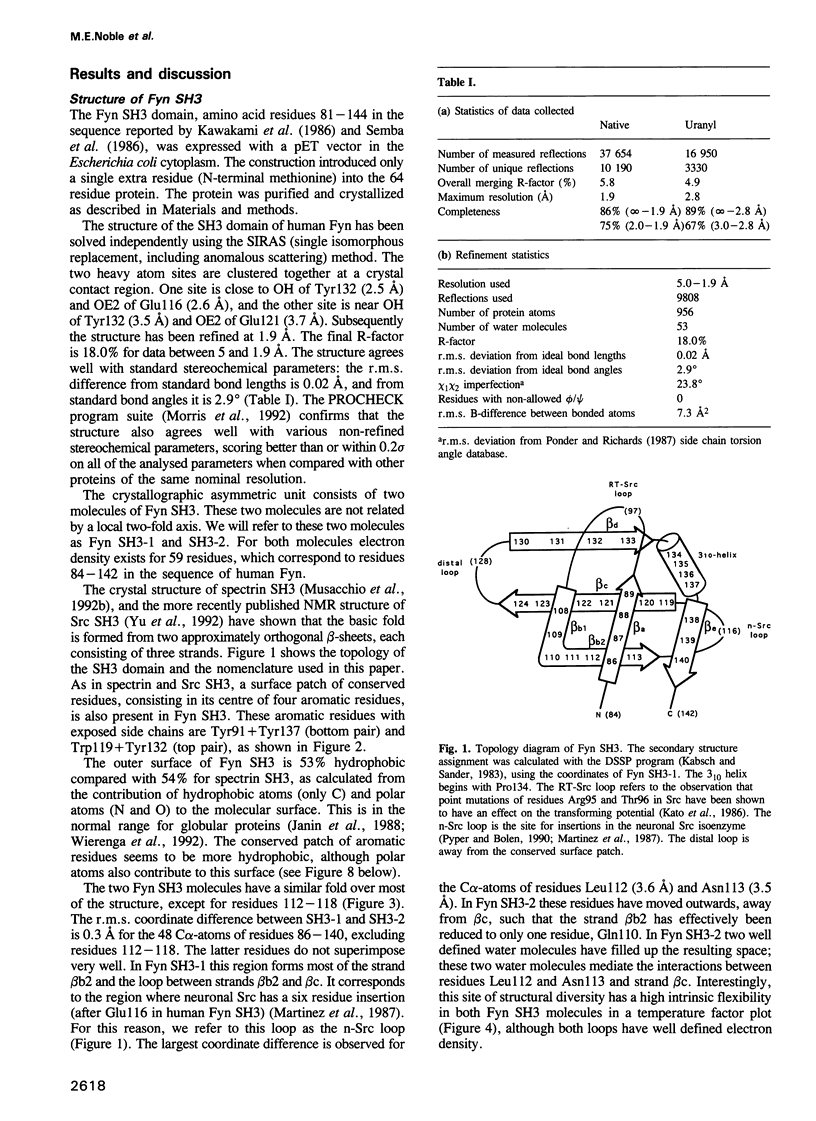
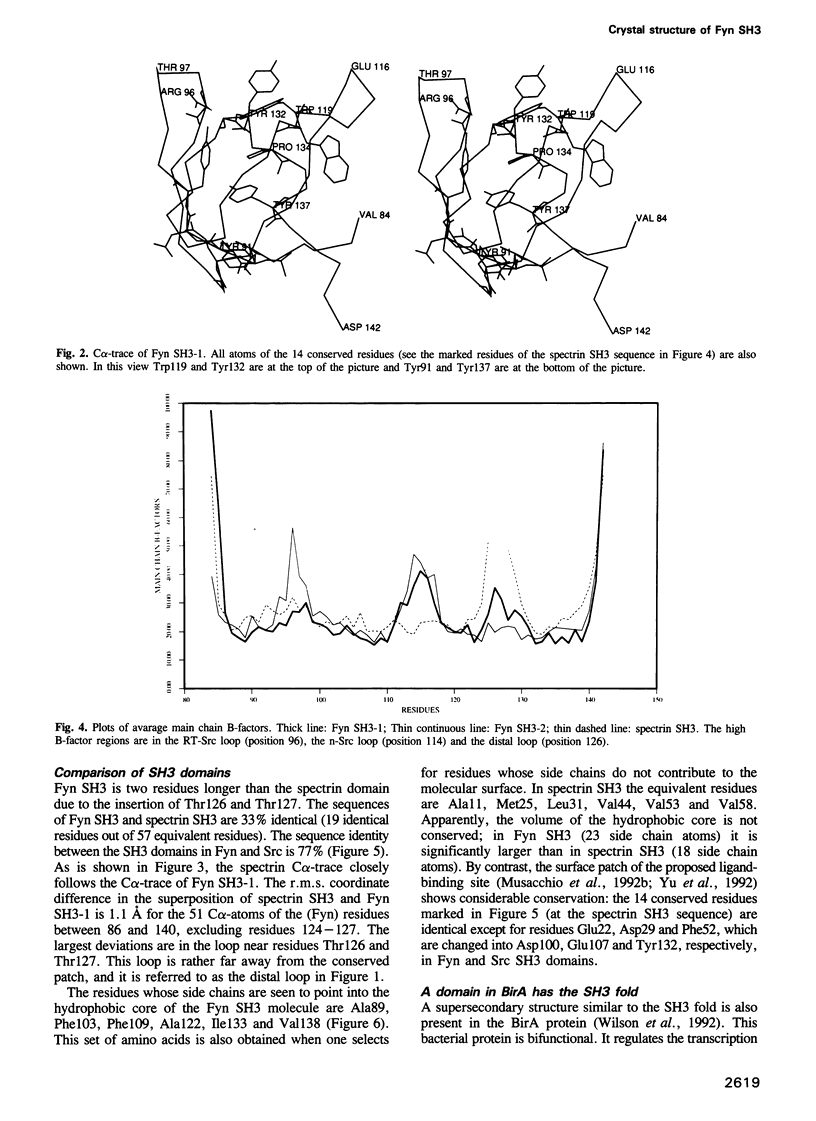
- Musacchio A., Gibson T., Lehto V. P., Saraste M. SH3--an abundant protein domain in search of a function. FEBS Lett. 1992 Jul 27;307(1):55–61. doi: 10.1016/0014-5793(92)80901-r. [DOI] [PubMed] [Google Scholar]
- Musacchio A., Noble M., Pauptit R., Wierenga R., Saraste M. Crystal structure of a Src-homology 3 (SH3) domain. Nature. 1992 Oct 29;359(6398):851–855. doi: 10.1038/359851a0. [DOI] [PubMed] [Google Scholar]
- Otsu M., Hiles I., Gout I., Fry M. J., Ruiz-Larrea F., Panayotou G., Thompson A., Dhand R., Hsuan J., Totty N. Characterization of two 85 kd proteins that associate with receptor tyrosine kinases, middle-T/pp60c-src complexes, and PI3-kinase. Cell. 1991 Apr 5;65(1):91–104. doi: 10.1016/0092-8674(91)90411-q. [DOI] [PubMed] [Google Scholar]
- Overduin M., Rios C. B., Mayer B. J., Baltimore D., Cowburn D. Three-dimensional solution structure of the src homology 2 domain of c-abl. Cell. 1992 Aug 21;70(4):697–704. doi: 10.1016/0092-8674(92)90437-h. [DOI] [PubMed] [Google Scholar]
- Padlan E. A. On the nature of antibody combining sites: unusual structural features that may confer on these sites an enhanced capacity for binding ligands. Proteins. 1990;7(2):112–124. doi: 10.1002/prot.340070203. [DOI] [PubMed] [Google Scholar]
- Ponder J. W., Richards F. M. Tertiary templates for proteins. Use of packing criteria in the enumeration of allowed sequences for different structural classes. J Mol Biol. 1987 Feb 20;193(4):775–791. doi: 10.1016/0022-2836(87)90358-5. [DOI] [PubMed] [Google Scholar]
- Pyper J. M., Bolen J. B. Identification of a novel neuronal C-SRC exon expressed in human brain. Mol Cell Biol. 1990 May;10(5):2035–2040. doi: 10.1128/mcb.10.5.2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren R., Mayer B. J., Cicchetti P., Baltimore D. Identification of a ten-amino acid proline-rich SH3 binding site. Science. 1993 Feb 19;259(5098):1157–1161. doi: 10.1126/science.8438166. [DOI] [PubMed] [Google Scholar]
- Rost B., Sander C. Jury returns on structure prediction. Nature. 1992 Dec 10;360(6404):540–540. doi: 10.1038/360540b0. [DOI] [PubMed] [Google Scholar]
- Sahr K. E., Laurila P., Kotula L., Scarpa A. L., Coupal E., Leto T. L., Linnenbach A. J., Winkelmann J. C., Speicher D. W., Marchesi V. T. The complete cDNA and polypeptide sequences of human erythroid alpha-spectrin. J Biol Chem. 1990 Mar 15;265(8):4434–4443. [PubMed] [Google Scholar]
- Seidel-Dugan C., Meyer B. E., Thomas S. M., Brugge J. S. Effects of SH2 and SH3 deletions on the functional activities of wild-type and transforming variants of c-Src. Mol Cell Biol. 1992 Apr;12(4):1835–1845. doi: 10.1128/mcb.12.4.1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semba K., Nishizawa M., Miyajima N., Yoshida M. C., Sukegawa J., Yamanashi Y., Sasaki M., Yamamoto T., Toyoshima K. yes-related protooncogene, syn, belongs to the protein-tyrosine kinase family. Proc Natl Acad Sci U S A. 1986 Aug;83(15):5459–5463. doi: 10.1073/pnas.83.15.5459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl M. L., Ferenz C. R., Kelleher K. L., Kriz R. W., Knopf J. L. Sequence similarity of phospholipase C with the non-catalytic region of src. Nature. 1988 Mar 17;332(6161):269–272. doi: 10.1038/332269a0. [DOI] [PubMed] [Google Scholar]
- Trahey M., Wong G., Halenbeck R., Rubinfeld B., Martin G. A., Ladner M., Long C. M., Crosier W. J., Watt K., Koths K. Molecular cloning of two types of GAP complementary DNA from human placenta. Science. 1988 Dec 23;242(4886):1697–1700. doi: 10.1126/science.3201259. [DOI] [PubMed] [Google Scholar]
- Trueheart J., Boeke J. D., Fink G. R. Two genes required for cell fusion during yeast conjugation: evidence for a pheromone-induced surface protein. Mol Cell Biol. 1987 Jul;7(7):2316–2328. doi: 10.1128/mcb.7.7.2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volpp B. D., Nauseef W. M., Donelson J. E., Moser D. R., Clark R. A. Cloning of the cDNA and functional expression of the 47-kilodalton cytosolic component of human neutrophil respiratory burst oxidase. Proc Natl Acad Sci U S A. 1989 Sep;86(18):7195–7199. doi: 10.1073/pnas.86.18.7195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vriend G. WHAT IF: a molecular modeling and drug design program. J Mol Graph. 1990 Mar;8(1):52-6, 29. doi: 10.1016/0263-7855(90)80070-v. [DOI] [PubMed] [Google Scholar]
- Waksman G., Kominos D., Robertson S. C., Pant N., Baltimore D., Birge R. B., Cowburn D., Hanafusa H., Mayer B. J., Overduin M. Crystal structure of the phosphotyrosine recognition domain SH2 of v-src complexed with tyrosine-phosphorylated peptides. Nature. 1992 Aug 20;358(6388):646–653. doi: 10.1038/358646a0. [DOI] [PubMed] [Google Scholar]
- Waksman G., Shoelson S. E., Pant N., Cowburn D., Kuriyan J. Binding of a high affinity phosphotyrosyl peptide to the Src SH2 domain: crystal structures of the complexed and peptide-free forms. Cell. 1993 Mar 12;72(5):779–790. doi: 10.1016/0092-8674(93)90405-f. [DOI] [PubMed] [Google Scholar]
- Wasenius V. M., Saraste M., Salvén P., Erämaa M., Holm L., Lehto V. P. Primary structure of the brain alpha-spectrin. J Cell Biol. 1989 Jan;108(1):79–93. doi: 10.1083/jcb.108.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wierenga R. K., Noble M. E., Davenport R. C. Comparison of the refined crystal structures of liganded and unliganded chicken, yeast and trypanosomal triosephosphate isomerase. J Mol Biol. 1992 Apr 20;224(4):1115–1126. doi: 10.1016/0022-2836(92)90473-w. [DOI] [PubMed] [Google Scholar]
- Wilson K. P., Shewchuk L. M., Brennan R. G., Otsuka A. J., Matthews B. W. Escherichia coli biotin holoenzyme synthetase/bio repressor crystal structure delineates the biotin- and DNA-binding domains. Proc Natl Acad Sci U S A. 1992 Oct 1;89(19):9257–9261. doi: 10.1073/pnas.89.19.9257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H., Rosen M. K., Shin T. B., Seidel-Dugan C., Brugge J. S., Schreiber S. L. Solution structure of the SH3 domain of Src and identification of its ligand-binding site. Science. 1992 Dec 4;258(5088):1665–1668. doi: 10.1126/science.1280858. [DOI] [PubMed] [Google Scholar]
- van der Oost J., Lappalainen P., Musacchio A., Warne A., Lemieux L., Rumbley J., Gennis R. B., Aasa R., Pascher T., Malmström B. G. Restoration of a lost metal-binding site: construction of two different copper sites into a subunit of the E. coli cytochrome o quinol oxidase complex. EMBO J. 1992 Sep;11(9):3209–3217. doi: 10.1002/j.1460-2075.1992.tb05398.x. [DOI] [PMC free article] [PubMed] [Google Scholar]