Abstract
Aphelids are a poorly known group of parasitoids of algae that have raised considerable interest due to their pivotal phylogenetic position. Together with Cryptomycota and the highly derived Microsporidia, they have been recently re-classified as Opisthosporidia, being the sister group to fungi. Despite their huge diversity, as revealed by molecular environmental studies, and their phylogenetic interest, only three genera have been described (Aphelidium, Amoeboaphelidium, and Pseudaphelidium), from which 18S rRNA gene sequences exist only for Amoeboaphelidium species. Here, we describe the life cycle and ultrastructure of Aphelidium aff. melosirae, and provide the first 18S rRNA gene sequence obtained for this genus. Molecular phylogeny analysis indicates that Aphelidium is very distantly related to Amoebaphelidium, highlighting the wide genetic diversity of the aphelids. The parasitoid encysts and penetrates the host alga, Tribonema gayanum through an infection tube. Cyst germination leads to a young trophont that phagocytes the algal cell content and progressively develops a plasmodium, which becomes a zoospore-producing sporangium. Aphelidium aff. melosirae has amoeboflagellate zoospores, tubular/lamellar mitochondrial cristae, a metazoan type of centrosome, and closed orthomitosis with intranuclear spindle. These features together with trophont phagocytosis distinguish Aphelidium from fungi and support the erection of the new superphylum Opisthosporidia as sister to fungi.
Keywords: Rozella, parasitoids, ultrastucture, molecular phylogeny, life cycle, ecology
Introduction
The aphelids are a phylogenetically divergent group of intracellular parasitoids of common species of eukaryotic phytoplankton. There are three known genera: Aphelidium, Amoeboaphelidium, and Pseudaphelidium, but the group is very diverse, including many related environmental sequences (Karpov et al. 2013; 2014 in press). The phyla Microsporidia and Cryptomycota (Rozellida), and the class Aphelidea have recently been shown to be the deepest branches of the ‘Holomycota’ lineage forming the so-called ARM-clade (Aphelidea-Rozellida-Microsporidia), which is sister to the classical fungi (Karpov et al. 2013). Consequently, the taxonomy of ARM clade has been reorganized, and a new superphylum Opisthosporidia with three phyla: Aphelida, Cryptomycota and Microsporidia, has been proposed (Karpov et al. 2014 in press). Opisthosporidia are not considered true fungi. Not only their phylogenetic position place them as sister to true fungi, but also several of their biological peculiarities do not conform the classical definition of fungi. The most remarkable of these is the fact that the trophonts of Aphelida and Cryptomycota (but not Microsporidia, which are extremely specialized and derived parasites) engulf the host cytoplasm by phagocytosis, like amoebae (Gromov 2000).
Even if the aphelids seem to encompass a huge genetic diversity (Karpov et al. 2013), only two strains of Amoeboaphelidium protococcarum have been investigated using molecular phylogeny: X-5 CALU (Karpov et al. 2013) and FD01 (Letcher et al. 2013). Despite both have similar morphology, they are very divergent genetically (Karpov et al. 2014 in press; Letcher et al. 2013). Here, we propose the morphological and molecular phylogenetic study of another genus - Aphelidium (strain P-1 CALU) - which is supported in culture on the alga Tribonema gayanum Pasch. (strain 20 CALU) as host. By light microscopy, this strain corresponds to the description of Aphelidium melosirae Scherffel 1925 (Gromov 1972, 2000; Karpov et al. 2014 in press), although its host is the yellow-green alga Tribonema instead of the diatom Melosira.
Results
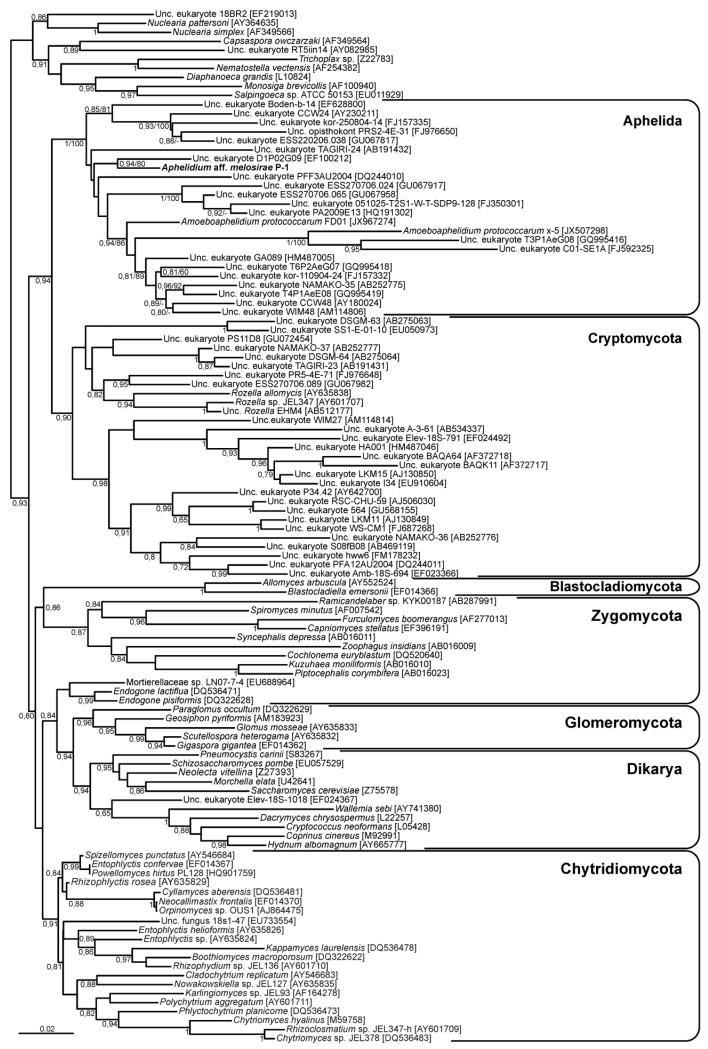
Molecular phylogeny
We reconstructed maximum likelihood (ML) and Bayesian inference (BI) phylogenetic trees including the new 18S rDNA sequence of strain P-1 and a selection of fungal sequences similar to the one previously used by Karpov et al. (2013). In these trees, strain P-1 branched within a strongly supported group (ML bootstrap proportion 100% and Bayesian posterior probability 1) that contained the two available Amoebaphelidium protococcarum sequences (strain x-5 from Karpov et al. 2013 and strain FD01 from Letcher et al. 2013) as well as a large number of environmental 18S rDNA sequences (Fig. 1). The closest relative to our strain P-1 was the unpublished environmental clone D1P02G09, retrieved from oxygen-depleted intertidal marine sediment (accession number EF100212), although both 18S rDNA sequences shared only 93% sequence identity. Strain P-1 was very distantly related to the A. protococcarum sequences (88% and 92% sequence identity with A. protococcarum strains x-5 and FD01, respectively).
Figure 1. 18S rDNA-based Bayesian phylogenetic tree showing the position of Aphelidium aff. melosirae.
The tree contains a sampling of sequences from fungi, cryptomycota and aphelids, rooted on nucleariid amoebal and animal sequences. The tree was constructed using 1414 conserved positions. Numbers at branches are Bayesian posterior probabilities. For the Aphelida, maximum likelihood bootstrap values are also shown after the posterior probabilities.
Life cycle
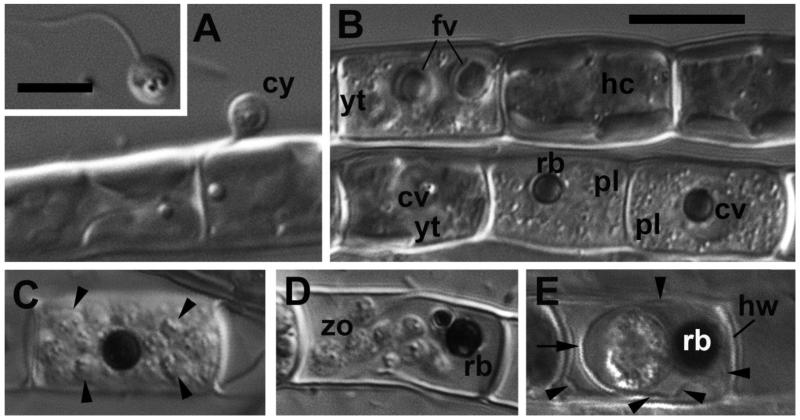
The observed life cycle of the strain P-1 CALU was typical for the genus Aphelidium, and encompassed several phases as follows (Fig. 2). The zoospore attaches to the host alga, involves its flagellum and encysts (Fig. 2 A). The cyst germinates and penetrates the host cell wall with an infection tube. A conspicuous enlarging vacuole pushes the contents of the cyst towards the interior of the host cell through the infection tube. The parasitoid becomes an intracellular phagotrophic amoeba which engulfs the host cytoplasm forming food vacuoles. The parasitoid continues to grow and forms an endobiotic plasmodium with a residual body while it totally consumes the cytoplasm of the host cell (Fig. 2 B, C). A multinucleate plasmodium is formed with a large central vacuole and a residual excretion body. The mature plasmodium then divides into a number of uninucleated cells (Fig. 2 C, D). After maturation, the uniflagellated zoospores of Aphelidium are released from the empty host cell after splitting the two parts of the Tribonema cell wall, and then infect other host algal cells (Fig. 2 E, insert in A). In the following sections, we describe in more detail each phase of the cell cycle.
Figure 2. Main stages of the life cycle of Aphelidium aff. melosirae observed in living material by differential interference contrast (DIC) microscopy.
A – cyst (cy) on the Tribonema filament. Insert – swimming zoospore. B – healthy cells of the host (hc), young trophont (yt) of parasitoid with food vacuoles (fv), a central vacuole (cv), and two plasmodia (pl) containing residual bodies (rb) in the central vacuole. C – mature sporangium with divided cells (arrowheads). D – mature zoospores (zo) and residual body (rb) inside the host cell wall. Note flexible ridges of the wall permitting the zoospores to come out. E – resting spore (sporocyst), surrounded by cyst wall (arrow) and spore wall (arrowheads) adpressed to the host cell wall (hw). The residual body lies in between of the cyst and spore walls.
Scale bars: insert in A – 5 μm, B – 10 μm for A-E.
Cyst
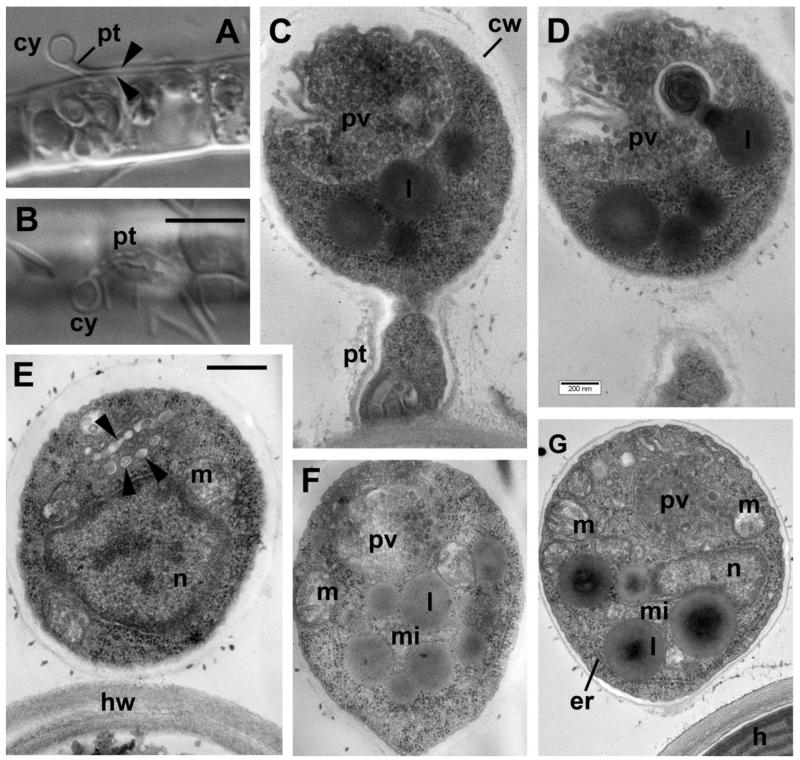
Before encystment, zoospores can settle down on the Tribonema filament at any place, but they are able to penetrate the cell wall only through the gap occurring between the two halves of the algal cell wall overlapping each other. Thus, after settlement the zoospore becomes amoeboflagellate and moves with immotile flagellum along the algal filament using short lobopodia. It attaches near to the end of the host cell. Just before attachment, the zoospore involves its flagellum: the cell rotates quickly along the lateral axis while the flagellum twists like a belt around the cell submerging into the cell; only an acroneme remains sticking out for some time. After encystment, the parasitoid germinates into the gap between the inner and outer halves of the host cell wall and reaches the plasma membrane (Fig. 3 A, B). The length of the infection tube may reach up to 5 times the cyst diameter, around 3-3.5 μm (Fig. 3 B). Empty cysts remain attached to the host cells through their infection tubes for a long time.
Figure 3. Cyst structure of Aphelidium aff. melosirae under light microscopy (LM) (A, B) and transmission electron microscopy (TEM) sections (C-G).
A – cyst (cy) with penetration tube (pt) between two halves of the host cell wall (arrowheads). B – empty cyst with penetrative tube on the surface of Tribonema filament. C-D and E-F – section pairs of cysts from two different series. Arrowheads on E show vesicles derived of the Golgi apparatus connected with posterior vacuole (pv) on F. Other abbreviations: cw – cyst wall, er-endoplasmic reticulum, h-host, hw-host cell wall, l-lipid globule, m-mitochondrion, mi-microbody, n-nucleus, pv-posterior vacuole. Scale bars: A, B – 10 μm, C, D – 200 nm, E-G – 500 nm.
The cyst is covered with a rather thick polysaccharide wall, which normally looks electron translucent, as the carbohydrates are not fixed with glutaraldehyde and osmium tetroxide (Fig. 3 C-G). After encystment, the cell contains one centrally located nucleus associated with a few lipid globules and a microbody (Fig. 3 E, G). Heterochromatin is visible in the center and periphery of the nucleus. The nucleolus is not perceptible. Mitochondria have flat, rarely tubular cristae. Cisterns of endoplasmic reticulum (ER) are located at the cell periphery, normally parallel to the plasma membrane. In some cysts the remnants of degraded axonemes are present in the peripheral cytoplasm.
The most prominent organelle in the mature cyst is a large posterior vacuole containing a large number of membranous structures, and rather dark contents (Fig. 3 C-G). The vacuole contents may be formed by activity of the Golgi apparatus (Fig. 3 D, arrowheads), the endoplasmic reticulum (ER), and even the lipid globules losing their contents in the process of vacuole formation (Fig. 3 D). In some of our thin sections, the process of the vacuole content extrusion was fixed (Fig. 3 C, D). The posterior vacuole may occupy more than half of the cyst volume, as seen on serial sections (Fig. 3 C, D). On some sections it looks electron translucent, probably after cell excretion. The contents of the cyst move through the penetrative tube into the space between the host plasma membrane and the cell wall to form a young uninucleate trophont (Fig. 4 A).
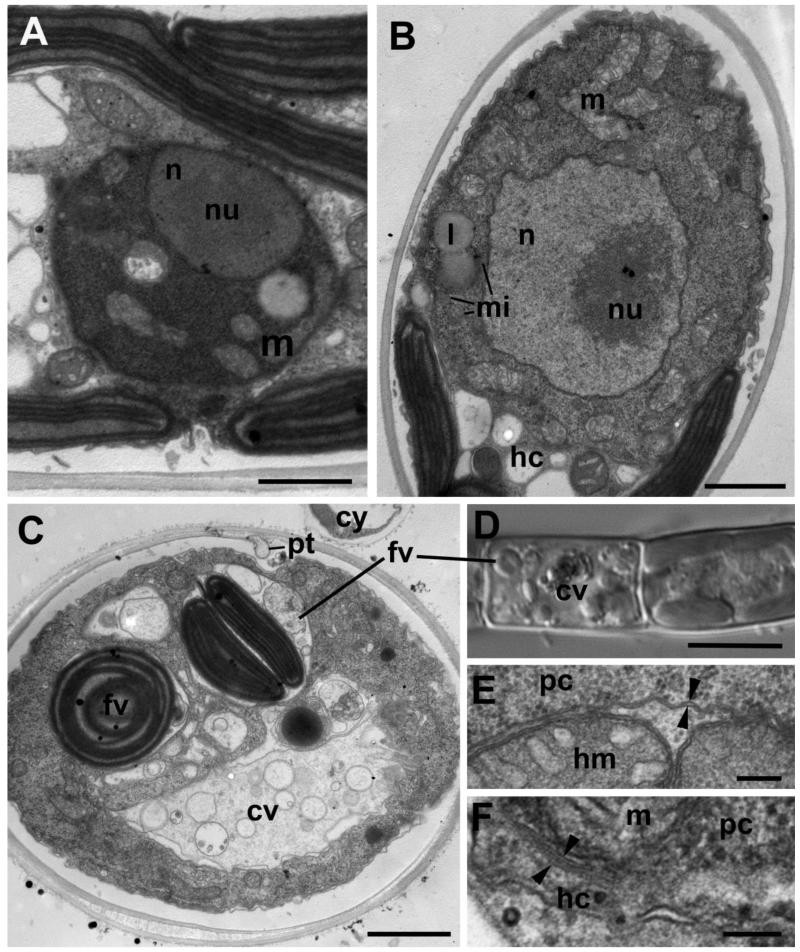
Figure 4. Structure of the Aphelidium aff. melosirae young trophont seen on ultrathin sections under TEM (A-C, E-F) and LM (D).
A – one of the first stages of trophont development after invasion, B – bigger uninuclear trophont adpresses the host cytoplasm, C – trophont with two food vacuoles and central vacuole, D – LM of the same stage as in C. E – host plasma membrane and food vacuole membrane (arrowheads) delimiting food vacuole. F – two membranes (arrowheads), bordering the parasitoid cell in the host. Abbreviations: cv-central vacuole, cw-cyst wall, cy-cyst, er-endoplasmic reticulum, fv-food vacuoles, hc-host cytoplasm, hm-host mitochondrion, hw-host cell wall, l-lipid globule, m-mitochondrion, mi-microbody, n-nucleus, nu-nucleolus, pc-parasitoid cytoplasm, pt-penetration tube. Scale bars: A-C – 2 μm, D – 10 μm, E – 400 nm, F – 100 nm.
Young trophont
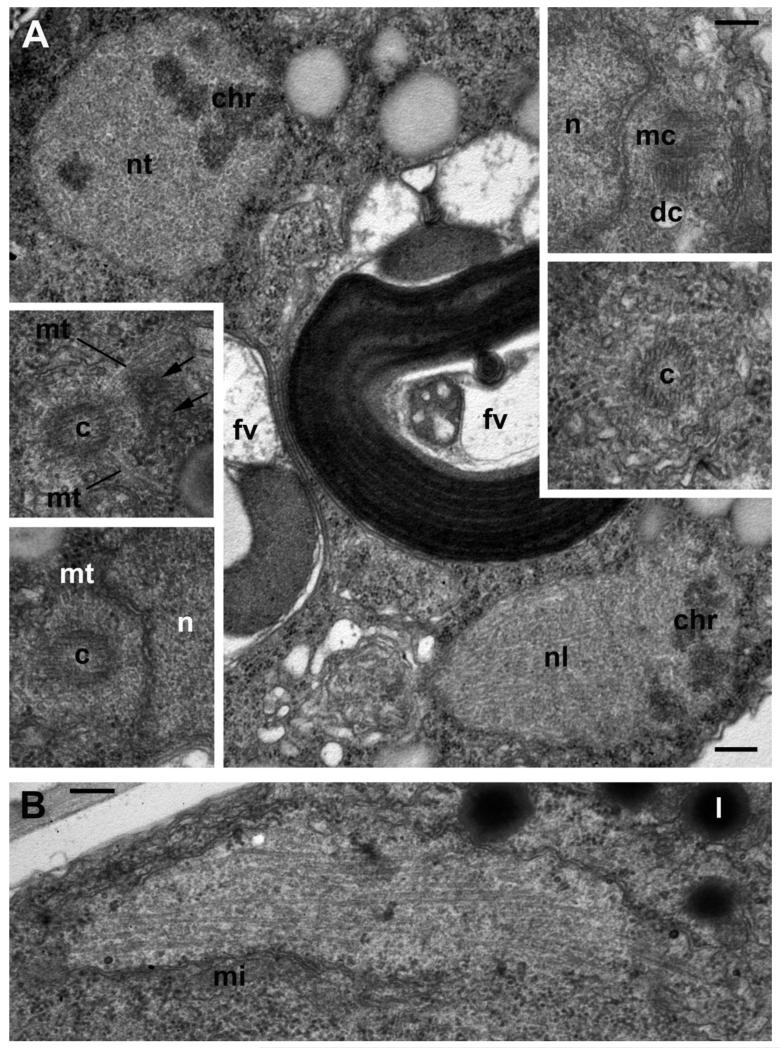
The trophic stage is highly proliferative, as can be deduced from the overall cell structure: a nucleus that does not contain heterochromatin but a huge slightly eccentric nucleolus; many mitochondrial profiles with densely packed tubular/lamellar cristae. Lipid globules are not visible at the earliest stages (Fig. 4). The parasitoid feeds by phagocytosis, what is confirmed by conspicuous food vacuole formation (Fig. 4 C, D). Such vacuoles contain parts of the host organelles (chloroplasts and mitochondria) covered with host plasma membrane (Fig. 4 E). These food vacuoles have been just formed and they are not yet digestive as even the host plasma membrane remains intact. Later on, the host plasma membrane disintegrates and only the food vacuole membrane separates food from the parasitoid cytoplasm (Fig. 5 A).
Figure 5. Centrosome structure and nuclear division in the young trophont of Aphelidium aff. melosirae.
A – prometaphase in two dividing nuclei with transverse (nt) and nearly longitudinal (nl) sections of mitotic spindle. Inserts show centrioles (c) with radiating microtubules (mt), centrosome composed of two orthogonal centrioles: mother (mc) and daughter (dc). Arrows show nuclear pores. B – elongated nucleus at late anaphase/early telophase. Other abbreviations as in previous Figures. Scale bars: A – 500 nm, A inserts – 200 nm, B – 250 nm.
At later stages, the trophont can contain several food vacuoles, more than one nucleus, and a big central vacuole (Fig. 6). Each nucleus has a centrosome (MTOC) composed of two orthogonal centrioles with microtubules radiating from the mother’s one (Fig. 5 A). Some microtubules pass along the nuclear surface. In the young trophont the nucleus divides with a totally internal microtubular spindle and seems to keep an intact nuclear envelope, i.e. by a closed mitosis mechanism (Fig. 5). Nevertheless, the possibility of semiopen mitosis can also be envisaged, as a pair of centrioles with radiating microtubules is present at each pole of the nucleus in nuclear invaginations (Fig. 5 A inserts). This means that centriolar microtubules may take part in chromosome separation during anaphase, but in the metaphase figures this microtubule penetration in the nucleus was not noticed (Fig. 5 A). Further nuclear divisions occur in different planes: spindles are orthogonal in the neighbor nuclei (Fig. 5 A). Many small lipid droplets, associated with small microbody profiles also appear at the stage with several nuclei (Fig. 5). At this stage, the parasitoid is still enclosed by its own membrane and the host cell membrane, both membranes are tightly close to each other and therefore difficult to distinguish (Fig. 4 F).
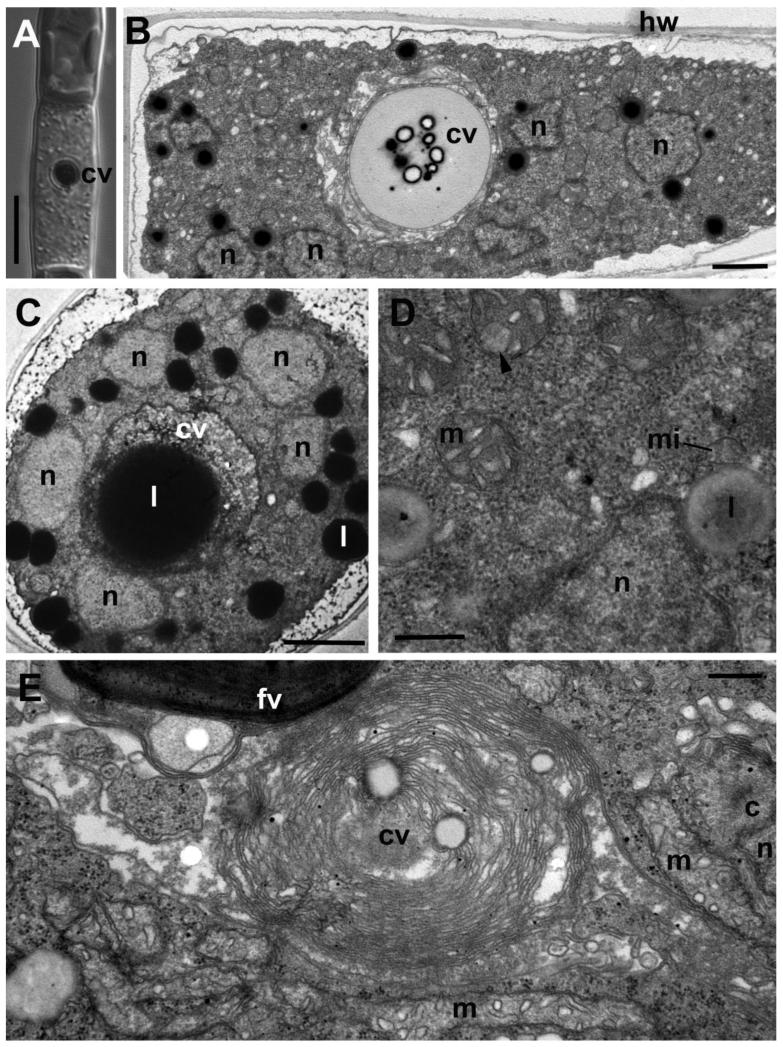
Figure 6. Plasmodium structure of Aphelidium aff. melosirae. A-B – plasmodium seen on longitudinal sections observed under LM (A) and TEM (B).
C – plasmodium on transverse section. D – portion of plasmodium at higher magnification, E – central vacuole formation. Abbreviations as in previous Figures. Scale bars: A – 10 μm, B – 2 μm, C – 2 μm, D – 500 nm, E – 300 nm.
Plasmodium
The plasmodium stage with a large central vacuole containing a red residual body is the most commonly observed in the infected culture, as it is the longest in the cycle. The plasmodium occupies all the space inside the host cell wall (Fig. 6). The multiple nuclei are located at more or less equal distance from each other, and around the central vacuole (Fig. 6 B, C). The plasmodium lives inside the totally degraded host cell and seems to be enclosed by the host plasma membrane. In the mature plasmodia the nuclear heterochromatin is well visible. Nuclear divisions stop in the mature plasmodium. Numerous active mitochondria with predominantly flat cristae are present in the cytoplasm (Fig. 6 D, E). Many lipid globules associated with rare microbodies locate around the nuclei. Typical dictyosomes are absent, groups of small vesicles seems to represent the Golgi apparatus. A large central vacuole surrounded with several layers of membranes is visible (Fig. 6 B, E). A stage of the central vacuole formation can be seen in Fig. 6 E. The residual body contains predominantly lipids and some membrane remnants (Fig. 6 B, C).
Sporangium
The sporangium formation is a very short stage which is constrained by zoospore maturation. It includes a multiple division of the plasmodium into 10-30 cells (Fig. 7 A), and zoospore formation. Recently divided cells are densely packed inside the host wall. Mature zoospores develop a flagellum and can move out of host cell wall by two ways: breaking the wall into two halves (Fig. 7 B), or penetrating the space in between the two halves of the cell wall from inside (note the flexible tips of the internal parts of host cell wall - Fig. 2 E) and leave the host one by one.
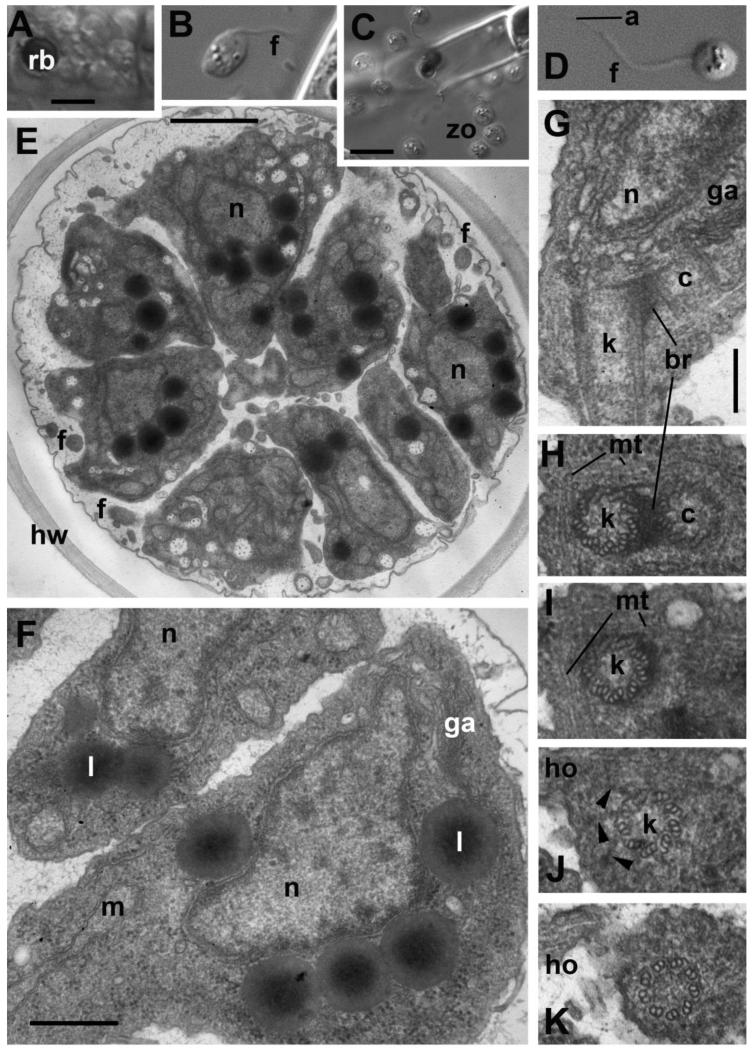
Figure 7. Zoospore structure and development in Aphelidium aff. melosirae seen by LM o living cells(A-D) and ultrathin sections observed by TEM (E-H).
A – recently divided plasmodium, B – amoeboid appearance of zoospore in algal vicinity, C – zoospore releasing, D – mature free-swimming zoospore. E-H – zoospore ultrastructure. E – general view of maturing zoospores inside the host cell wall, F – zoospore structure at higher magnification, G – kinetid at LS, H-K – series of kinetid consecutive thin sections from kinetosome and centriole (H) to flagellar transition zone (K). View from outside the cell. Arrowheads show the kinetosome transitional fibers. Abbreviations: a-acronema, br-bridge between kinetosome and centriole, c-centriole, f-flagellum, ga-Golgi apparatus, ho-horn, hw-host cell wall, k-kinetosome, l-lipid globule, m-mitochondrion, mi-microbody, mt-microtubules, n-nucleus, nu-nucleolus, rb-residual body, zo-zoospores. Scale bars: A, B, D - 5 μm, C – 10 μm, E – 1 μm, F – 500 nm, G-K – 200 nm.
Zoospores
Each free swimming zoospore is 4-5 μm in diameter with one acronematic flagellum of ~15 μm including an acroneme of 5 μm (Fig. 7 C, D). In the vicinity of the host algal filament, the zoospore stops swimming and begins to produce broad and very short lobopodia consecutively in anterior and lateral directions (Fig. 7 B). Rarely, short filopodia are also visible in the cell posterior. In such “amoeboid” state, the zoospore has an anterior cap of hyaloplasm, which enlarges its length up to 6 μm, and an average size of the amoeboid body is 4×6 μm (Fig. 7 B).
Zoospore maturation includes the development of the flagellum, which appears at the zoospore distal end with respect to the center of the sporangium (Fig. 7 E), and the reduction of cell volume during the transformation from an amoeboid to a flagellate state. We did not observe free swimming zoospores in ultrathin sections, just inside the sporangium, where the flagellum was not still completely formed.
Many lipid globules, which probably contain storage material for the future life of the free swimming zoospore, are associated with a microbody located around the nucleus (Fig. 7 E, F). Mitochondria show rare tubular to sac-like cristae, some degraded. Perhaps the mitochondrion becomes more compact in the smaller volume available in the free swimming zoospore. The microtubular cytoskeleton is extraordinarily developed and associated with a kinetosome (Fig. 7 G-K). The flagellar apparatus does not yet have a definitive shape: the growing flagellum is like a stub with a short axoneme often having an incomplete set of microtubules, and immature transition zone (Fig. 7 G). The kinetosome and centriole are composed of microtubule triplets and lie at an angle from 0° to ~30°, being connected to each other by a broad fibrillar bridge (Fig. 7 G-H). A cone of disordered microtubules is initiated at a side of the kinetosome, nearly opposite to the centriole. Most of the microtubular singlets pass from this point deep in the cell around the nucleus and under the plasma membrane, but a bundle of a few microtubules passes in the opposite direction forming a small horn parallel to the flagellum (Fig. 7 G-K). These microtubules are probably involved in the zoospore morphogenesis from the amoeboid to the flagellate shape. No definite roots were found at this stage of cytoskeleton formation. The kinetid is underlined by the dictyosome, and the nucleus is in close vicinity to the kinetid (Fig. 7G).
Many small holes at the cell periphery are the profiles of cell invaginations; these likely occur because of the highly flexible cell ridges during zoospore morphogenesis. The interphase nucleus contains heterochromatin without microtubules.
Sporocyst
Resting spores or sporocysts are very rare in the culture of P-1. They appear under particular environmental conditions, which are not yet clearly defined. The rounded spore is 8-10 μm in diameter, being surrounded by a relatively thin spore wall. The spore lies inside the elongated and sculpted cyst wall, underlying the host cell wall (Fig. 2 E). The residual body lies in the space between spore and cyst walls.
Discussion
Species identification
Our strain P-1 (CALU) certainly belong to the genus Aphelidium by its life cycle, type of zoospore and ultrastructure (Gromov 2000; Gromov and Mamkaeva 1975). Among four species and one forma known for this genus, strain P-1 appears to be most similar to A. melosirae Scherffel 1925, the only one having such big oval zoospores (4 × 6 μm) with several refractive granules. Its zoospores are slightly amoeboid, can produce short lobopodia and move like amoebae having an immotile flagellum. Our strain differs by its longer flagellum (15 vs. 10 μm) and by the host. If Scherffel did not notice the very thin acronema of 5 μm long, which is possible using a microscope of less quality at that time, then the length of the flagellum might be the same as in P-1. A. melosirae is a parasitoid of the diatom alga Melosira varians Ag. Scherffel certainly did not check the possibility of A. melosirae to infect other algae than Melosira. Taking into account that the majority of aphelid strains can infect several different algae and do not have strong host specifity in general (Karpov et al. 2014 in press), we cannot exclude that the original A. melosirae can infect Tribonema gayanum. At the same time, the resting spore of A. melosirae, unlike of P-1, is elongated, slightly larger (12-14 × 10 μm) and lies freely in the host cell having no cyst wall. This difference has essential taxonomic significance, but the structure and dimension of parasitoid sporocyst can differ in different hosts.
Thus, to decide whether P-1 belongs to Aphelidium melosirae or not, we will need to study the morphology and molecular phylogeny of A. melosirae parasitizing Melosira varians Ag. For the moment, we have to consider our strain P-1 as Aphelidium aff. melosirae.
Molecular phylogeny
18S rDNA-based phylogenetic analyses strongly supported the monophyly of the three known aphelid strains (A. protococcarum strains x-5 and FD01 and the new Aphelidium aff. melosirae P-1) together with a variety of environmental sequences retrieved from marine, freshwater, and soil environments (Fig. 1). Within this group, the three aphelid sequences branched very distantly from known sequences, in agreement with their low sequence identity level (between 85 and 92%). Since these three sequences were well distributed within the group with the environmental sequences, we assumed that this whole group corresponded to aphelid sequences (namely, the Aphelida). Thus, aphelids appear to be extremely diverse in terms of within-group sequence divergence, with levels comparable to those of important groups such as the Chytridiomycota or the Cryptomycota.
This group most likely contains a high level of cryptic diversity, as exemplified by the two A. protococcarum sequences, which only had 85% sequence identity as already noticed by Letcher et al. (2013). Thus, despite having been classified as the same species, they are so distant that they probably correspond to different genera. In the case of strain A. protococcarum x-5 (Karpov et al. 2013), it belonged to a group of fast-evolving sequences, as shown by their very long branches. This probably reflects different ecological constraints on these species. By contrast, the sequence of Aphelidium aff. melosirae P-1 was placed as the shortest branch among all the aphelid sequences (Fig. 1). This suggested that this may be a slow-evolving aphelid species, which makes it an interesting target for future massive sequencing approaches for genomic and phylogenomic studies.
Interestingly, the large aphelid clade branched as sister group of the Cryptomycota. This relationship was moderately supported (BP 64%, PP 0.94) but in agreement with results of a combined 18S+28S rDNA phylogenetic analysis (Letcher et al. 2013). This was also in agreement with the recent proposal that Aphelida and Cryptomycota, together with the highly derived Microsporidia, form a major eukaryotic group, the Opisthosporidia, sister to the true fungi (Karpov et al. 2014). Aphelida also appeared as sister to the branch Cryptomycota+Microsporidia in a very recent analysis based on the 18S+5.8S+(partial) 28S rRNA genes (Corsaro et al. 2014). The authors erroneously labeled Amoeboaphelidium protococcarum strain X-5 as Aphelidium (Fig. 3 in Corsaro et al. 2014). The 18S rRNA gene for a representative of the genus Aphelidium has been sequenced for the first time in the present work (Fig. 1). Corsaro et al (2014) proposed a new name for the phylum Cryptomycota, the Rozellomycota (James and Berbee 2012) D. Corsaro and R. Michel 2014 with a new diagnosis, a suggestion previously made by other authors (James et Berbee 2012). Unfortunately, the authors still described this group as “unicellular fungi”. However, these protists that branch as a sister group to fungi possess their own idiosyncratic characteristics distinctive from fungi. This led to Karpov et al. (2014) to amend the diagnosis of Cryptomycota (Jones & Richards 2011) Karpov et Aleoshin, which belong now to the superphylum Opisthosporidia.
Ultrastructure
The ultrastructure of Aphelidium has been described for only two species: Aphelidium sp. (Schnepf et al. 1971), where the cyst and trophic stage were studied, and A. chlorococcarum f. majus (Gromov and Mamkaeva 1975), illustrated at all the stages of its life cycle, including zoospore structure. Unfortunately, the fixation with permanganate and osmium tetroxide did not allow good preservation of cell structures (Gromov and Mamkaeva 1975). Despite so, some comparative points can be discussed here.
The cyst wall in both species is also electron translucent and approximately of the same thickness as in A. aff. melosirae. Gromov and Mamkaeva (1975) showed the passage of the cyst contents into the host, and defined the order of steps in this process: a small portion of cytoplasm enters the host first, then the mitochondria and other organelles, and the nucleus is always the last to enter the host.
The posterior vacuole filled with dark granules and membranes is well known in the spores of microsporidia as a posteriosome, which then becomes the posterior vacuole (Vávra and Lukeš 2013). Dense dark vesicles in the growing vacuole were also found in the cyst of Rozella allomycis (Held, 1973). All these taxa are close relatives of the aphelids and belong to the same monophyletic superphylum Opisthosporidia. Thus, we can propose that the vacuole in the cyst of Aphelida and Cryptomycota grows first as a posteriosome, and that this structure is homologous to posteriosome of Microsporidia.
Like in A. aff. melosirae, the trophont of A. chlorococcarum f. majus (Gromov and Mamkaeva 1975) and Aphelidium sp. (Schnepf et al. 1971) was separated from the host cytoplasm by two membranes, and a central vacuole was surrounded with several membrane layers. A straight contact of the plasma membrane with the host cytoplasm was shown for Rozella polyphagi (Powell 1984). Also, the trophont of Paramicrosporidium (Cryptomycota) lies freely in the nucleoplasm of Vannella (Corsaro et al. 2014).
Nucleus
A typical metazoan centrosome composed of mother and daughter centrioles, and an intranuclear mitosis have been found here for the aphelids for the first time. The centriole close to the nucleus in A. chlorococcarum f. majus was also visible at some sections. The structure of the centrosome was studied by Schweikert and Schnepf (1997) in Pseudaphelidium drebesii. This centrosome is also composed of two centrioles, but lying antiparallel to each other at one axis. Both locate in a small invagination at the nuclear pole and each centriole produces many radiating microtubules passing also along the nuclear surface.
The very early prophase and late telophase for the dividing nuclei were shown in the Pseudaphelidium drebesii plasmodium (Schweikert and Schnepf 1997). At both stages intranuclear microtubules and an intact nuclear envelope were observed. Thus, the nuclear division of the aphelids seems to be a closed orthomitosis with intranuclear spindle according to the classification by Raikov (1994). The same type of nuclear division has been shown for the metaphase in Rozella polyphagi (Cryptomycota) (Powell 1984).
In the case of A. chlorococcarum f. majus, although nuclear division was not specifically observed, several nuclei closely attached to each other under the envelope of the “mother” nucleus were seen. We think that the partly broken “mother nucleus envelope” is just an ER cistern which normally occurs in peripheral cytoplasm. In contrast with A. chlorococcarum f. majus, we never observed a tight contact of the nuclei to each other in A. aff. melosirae.
Mitochondrial cristae
It is clear now that the shape of mitochondrial cristae in the aphelids is not lamellar (Cavalier-Smith 2013), as is the fungal type. Early works using fixations without glutaraldehyde (permanganate and osmium tetroxide), showed tubular cristae in most cases (Gromov and Mamkaeva 1970a,b; 1975; Schnepf et al. 1971), but these results remained to be confirmed. Using the modern fixation methods we can unambiguously say that the shape of the cristae varies in different stages of the aphelid life cycle (Karpov et al. 2014 in press). Thus, in Aphelidium aff. melosirae mitochondrial cristae are predominantly lamellar with rare tubular profiles in the cyst, lamellar but becoming progressively saccular in the young trophont, lamellar, saccular and tubular in nearly equal proportions in the plasmodium, and saccular and tubular in the maturing zoospore. Unfortunately, we could not observed them in free swimming zoospores. So far, the ultrastructure of zoospores for the genus Aphelidium has been only studied in A. chlorococcarum f. majus, which has tubular cristae (Gromov and Mamkaeva 1975). Pseudaphelidium mitochondrial cristae are lamellar in the encysting zoospore and tubular in the plasmodium (Schweikert and Schnepf 1997). Similar variations were shown earlier for two species of Cryptomycota: flat mitochondrial cristae in zoospores of Rozella allomycis (Held, 1975), tubular cristae in the plasmodium of R. polyphagi (Powell, 1984), and flat with tubular (rare) cristae in the plasmodium of Rozella allomycis (Held, 1980). Thus, the variable shape of mitochondrial cristae is common for two phyla of Opisthosporidia: Aphelida and Cryptomycota.
Phagocytosis
We did not find in the trophont of A. aff. melosirae obvious pseudopodia as in A. chlorococcarum f. majus (Gromov and Mamkaeva 1975). However, the presence of large food vacuoles containing host plastids and mitochondria unambiguously confirm the capacity of phagocytosis in Aphelidium. The central vacuole seems to be digestive, as food vacuoles normally contain intact organelles. Food vacuoles are in close contact with the central one during its formation (Fig. 6 E), being likely involved in the transport of food to the central vacuole for digestion (Gromov 2000). Multimembrane structures in and around the central vacuole were described in Aphelidium sp. (Schnepf et al. 1971) and A. chlorococcarum f. majus (Gromov and Mamkaeva 1975), but they were not observed for Amoeboaphelidium chlorellavorum and A. protococcarum (Gromov and Mamkaeva 1970 a,b).
The amoeboflagellate zoospore
Amoeboid activity is common for Aphelidium zoospores (Gromov 2000; Karpov et al. 2014 in press). Two types of pseudopodia can be produced: filopodia (A. chlorococcarum f. majus, A. tribonemae) and short lobopodia (A. melosirae). In A. chlorococcarum f. majus filopodia appear at the anterior end of the zoospore (Gromov and Mamkaeva 1975), whereas the zoospore of A. tribonemae produces filopodia in all directions (Gromov 1972; Karpov et al. in preparation). Up to now this character looks discriminant for the species description. Mature zoospores in the sporangium were also shown in ultrathin sections of Aphelidium sp. (Schnepf et al. 1971) at low magnification. In spite of flagella presence, zoospore shape was fairly amoeboid. Unlike in Aphelidium aff. melosirae, the residual body in this Aphelidium sp. was in the centre of the mature sporangium and also inside the resting spore (Schnepf et al. 1971).
Material and methods
Isolation and cultivation of Aphelidium aff. melosirae
The strain P-1 (CALU) was isolated by M. Mamkaeva in May 2013 from the roadside ditch in the park Sergievka, Stary Petershof, Leningrad region, Russia. It was maintained in culture on Tribonema gayanum Pasch. (strain 20 CALU) as the host. The culture of the host was grown on mineral medium (KNO3, 2 g L−1; KH2PO4, 0.3 g L−1; MgSO4, 0.15 g L−1; EDTA, 10 mg L−1; FeSO4, 5 mg L−1; NaBO3, 1.4 mg L−1; (NH4)6Mo7O2, 4.1 mg L−1; CaCl2, 0.6 mg L−1; ZnSO4, 0.1 mg L−1; CuSO4, 50 g L−1, Co(NO3)2, 20 g L−1) at room temperature in the presence of white light. After inoculation with strain of Aphelidium, the cultures were incubated for 1-2 weeks in order to reach the maximum infection of host cells. The cells were then harvested and used directly for DNA extraction.
Light and transmission electron microscopy
Light and DIC microscopy observations of living cultures were carried out on a Zeiss Axioplan microscope equipped with black and white MRm Axiocam. For electron microscopy, the algal filaments were fixed with 2% glutaraldehyde on 0.1 M cacodylate buffer for two hours on ice, rinsed in the same buffer, and fixed with 1% OsO4 during one hour on ice. After rinsing in 0.1 M cacodylate buffer for 5 min, the filaments were dehydrated in alcohol series (30°-50°-70°-90°-100°) and in propylen oxid, then embedded in Spurr resin (Sigma Aldrich, St Louis, USA). Ultrathin sections were prepared using Leica Ultracut ultratome with diamond knife. After double staining, the sections were observed at a JEM 1400 (Jeol) microscope equipped with digital camera Olympus Veleta.
DNA purification and sequencing
DNA from infected cultures was extracted using the kit PowerSoil (MoBio) following the manufacturer’s instructions. To avoid amplifying an excess of host genes, the aphelid 18S rRNA gene was amplified by polymerase chain reaction using the ‘fungi’-specific primers UF1 (5′-CGAATCGCATGGCCTTG) and AU4 (5′-RTCTCACTAAGCCATTC). Each PCR reaction was carried out in a volume of 25 μl of reaction buffer, containing 1 μl of the eluted DNA, 1.5mM MgCl2, dNTPs (10 nmol each), 20 pmol of each primer, and 0.2 U TaqPlatinum DNApolymerase (Invitrogen). PCR reactions were performed under the following conditions: 2 min denaturation at 94 °C; 35 cycles consisting of a denaturation step at 94 °C for 15 s, a 30 s annealing step at 55 °C and an extension step at 72 °C for 2 min;; and a final elongation step of 7 min at 72 °C. Negative controls without template DNA were used at all amplification steps. Since the cultures were not axenic, the amplified 18S rRNA gene fragments were cloned using the Topo TA Cloning System (Invitrogen, Carlsbad, CA, USA) following the manufacturer’s instructions. Clone inserts were PCR-amplified using flanking vector primers, and inserts of expected size (1,400 bp) were then sequenced bidirectionally using vector primers ((Beckman Coulter Genomics, Takeley, UK). Retrieved sequences were compared by BLAST to sequences in the database and non-aphelid sequences discarded. All the aphelid sequences retrieved from the isolate P1 were identical.
Molecular phylogenetic analyses
The Aphelidium aff. melosirae 18S rRNA gene sequences were aligned together with the sequences previously used by Karpov et al. (2013) using MUSCLE 3.7 (Edgar 2004). The resulting alignment was manually inspected using the program ED of the MUST package (Philippe 1993) to remove ambiguously aligned regions and gaps. Phylogenetic trees were reconstructed by Maximum Likelihood (ML) and Bayesian Inference (BI) analyses. ML analyses were done with the program TREEFINDER (Jobb et al. 2004) applying a GTR+G+I model of nucleotide substitution, taking into account a proportion of invariable sites, and a Gamma-shaped distribution of substitution rates with four rate categories. Bootstrap values were calculated using 1,000 pseudoreplicates with the same substitution model. The BI analyses were carried out with the program MrBayes (Ronquist et al. 2012) applying the GTR+G+I model with four chains and 10,000,000 generations per run. After checking for convergence and eliminating the first 15,000 trees (burn-in), a consensus tree was constructed sampling every 100 trees. The sequence was deposited in GenBank under accession number KJ566931.
Acknowlegements
The research leading to these results received funding from the Russian Foundation for Basic Research (projects No 12-04-01486), the program “Problems of life origin and biosphere development” launched by the Presidium of the Russian Academy of Sciences, and the European Research Council under the European Union’s Seventh Framework Program ERC Grant Agreement 322669 “ProtistWorld”. S. K. was recipient of a “Research in Paris” grant from the Mairie de Paris. We also thank the Research Resource Center for Molecular and Cell Technologies (RRC MCT) at St. Petersburg State University (SPbSU) for access to the EM facilities.
References
- Cavalier-Smith T. Early evolution of eukaryote feeding modes, cell structural diversity, and classification of the protozoan phyla Loukozoa, Sulcozoa, and Choanozoa. Eur J Protistol. 2013;49:115–178. doi: 10.1016/j.ejop.2012.06.001. [DOI] [PubMed] [Google Scholar]
- Corsaro D, Walochnik J, Venditti D, Steinmann J, Müller KD, Michel R. Microsporidia-like parasites of amoebae belong to the early fungal lineage Rozellomycota. Parasitol Res. 2014 doi: 10.1007/s00436-014-3838-4. doi:10.1007/s00436-014-3838-4. [DOI] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gromov BV. Aphelidium tribonemae Scherffel parasitizing yellow green algae. Mikol Fitopatol. 1972;6:443–445. In Russian. [Google Scholar]
- Gromov BV. Algal parasites of the genera Aphelidium, Amoeboaphelidium and Pseudoaphelidium from the Cienkovski’s “Monadea” group as representatives of new class. Zool Zhurnal. 2000;79:517–525. In Russian. [Google Scholar]
- Gromov BV, Mamkaeva KA. Sensitivity of different Scenedesmus strains to the endoparasitic microorganism Amoeboaphelidium. Phycol. 1968;7:19–23. [Google Scholar]
- Gromov BV, Mamkaeva KA. Electron-microscopic investigations of development cycle and feeding behaviour of intercellular parasite of Chlorella, Amoebaphelidium chlorellavorum. Tsilogiya. 1970a;12:1191–1196. In Russian. [Google Scholar]
- Gromov BV, Mamkaeva KA. The fine structure of Amoeboaphilidium protococcarum - an endoparasite of green alga Scenedesmus. Arch Hydrobiol. 1970b;67:452–459. [Google Scholar]
- Gromov BV, Mamkaeva KA. Zoospore ultrastructure of Aphelidium chlorococcarum Fott. Mikol Fitopatol. 1975;9:190–193. In Russian. [Google Scholar]
- James TY, Berbee ML. No jacket required--new fungal lineage defies dress code: recently described zoosporic fungi lack a cell wall during trophic phase. Bioessays. 2012;34:94–102. doi: 10.1002/bies.201100110. [DOI] [PubMed] [Google Scholar]
- Jobb G, von Haeseler A, Strimmer K. TREEFINDER: a powerful graphical analysis environment for molecular phylogenetics. BMC Evol Biol. 2004;4:18. doi: 10.1186/1471-2148-4-18. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Karpov SA, Mikhailov KV, Mirzaeva GS, Mirabdullaev IM, Mamkaeva KA, Titova NN, Aeoshin VV. Obligately phagotrophic aphelids turned out to branch with the earliest-diverging fungi. Protist. 2013;164:195–205. doi: 10.1016/j.protis.2012.08.001. [DOI] [PubMed] [Google Scholar]
- Karpov SA, Mamkaeva MA, Aleoshin VV, Lilje O, Gleason FH. Morphology, phylogeny and ecology of the aphelids (Aphelidea, Opisthokonta) with proposal of new superphylum Opisthosporidia. Front Microbiol. 2014;5:1–11. doi: 10.3389/fmicb.2014.00112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lara E, Moreira D, Lopez-Garcia P. The environmental clade LKM11 and Rozella form the deepest branching clade of fungi. Protist. 2010;161:116–121. doi: 10.1016/j.protis.2009.06.005. [DOI] [PubMed] [Google Scholar]
- Letcher PM, Lopez S, Schmieder R, Lee PA, Behnke C, Powell MJ, McBride RC. Characterization of Amoeboaphelidium protococcarum, an algal parasite new to the cryptomycota isolated from an outdoor algal pond used for the production of biofuel. PLoS One. 2013;8:2. doi: 10.1371/journal.pone.0056232. doi: 10.1371/journal.pone.0056232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paps J, Medina-Chacón LA, Marshall W, Suga H, Ruiz-Trillo I. Molecular phylogeny of unikonts: new insights into the position of apusomonads and ancyromonads and the internal relationships of opisthokonts. Protist. 2013;164:2–12. doi: 10.1016/j.protis.2012.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippe H. MUST, a computer package of management utilities for sequences and trees. Nucleic Acids Res. 1993;21:5264–5272. doi: 10.1093/nar/21.22.5264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raikov IB. The diversity of forms of mitosis in Protozoa: a comparative review. Europ J Protistol. 1994;30:253–269. [Google Scholar]
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 2012;61:539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherffel A. Endophytische Phycomyceten-Parasiten der bacilleriaceen und einige nene Mondinen. Arch Protistenkd. 1925;52:1–141. [Google Scholar]
- Schnepf E, Hegewald E, Soeder CJ. Electronen-mikroskopische Beobachtungen an Parasiten aus Scenedesmus-Massenkulturen. 2. Uber Entwicklung und Parasit-Wirt-Kontakt von Aphelidium und virusartige Partikel im Cytoplasma infizierter Scenedesmus-Zellen. Acta Mikrobiol. 1971;75:209–229. [Google Scholar]
- Schweikert M, Schnepf E. Electron microscopical observations on Pseudoaphelidium drebesii Schweikert and Schnepf, a parasite of the centric diatom Thalassiosira punctigera. Protoplasma. 1997;199:113–123. [Google Scholar]
- Torruella G, Derelle R, Paps J, Lang BF, Roger AJ, Shalchian-Tabrizi K, Ruiz-Trillo I. Phylogenetic relationships within the Opisthokonta based on phylogenomic analyses of conserved single-copy protein domains. Mol Biol Evol. 2012;29:531–544. doi: 10.1093/molbev/msr185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zopf W. Zur Morphologic und Biologic der niederen Pilztiere (Monadinen) Leipzig: 1885. [Google Scholar]