Abstract
A normal-phase HPLC-MS method was established to analyze mitochondrial phospholipids quantitatively as well as qualitatively. An efficient extraction procedure and chromatographic conditions were developed using twelve standardized phospholipids and lysophospholipids. The chromatographic conditions provided physical separation of phospholipids by class, and efficient ionization allowed detection of low abundance phospholipids such as phosphatidylglycerol and monolysocardiolipin. The chromatographic separation of each class of phospholipid permitted qualitative identification of molecular species without interference from other classes. This is advantageous for mitochondrial lipidomics because the composition of mitochondrial phospholipids varies depending on tissue source, pathological condition, and nutrition. Using the method, seven classes of phospholipids (phosphatidylethanolamine, phosphatidylcholine, phosphatidylglycerol, phosphatidylinositol, phosphatidylserine, cardiolipin, and monolysocardiolipin) were detected in rat heart and skeletal muscle mitochondria and all but phosphatidylserine were quantified. The concentration was calculated using standard curves with an internal standard generated for each class of phospholipid. The method was validated for intraday and interday variation and showed excellent reproducibility and accuracy. This new method, with each step documented, provides a powerful tool for accurate quantitation of phospholipids, a basic structural component of mitochondrial membranes.
Keywords: normal-phase HPLC, mass spectrometry, solid phase extraction, internal standard, standard curve, ion suppression, cardiolipin, monolysocardiolipin
1. Introduction
Phospholipids containing a hydrophilic head group and hydrophobic acyl chains are an important component of mitochondria in maintenance of membrane structure and proper enzyme activity [1,2]. The polar head group determines the class of phospholipid and each class consists of numerous species depending on the acyl chains, which vary in chain length and degree of unsaturation. With a combination of different head groups and acyl chains, hundreds of different species of phospholipids exist in biological samples.
Although mass spectrometry has been widely used to analyze phospholipids [3-5], the large number of species makes analysis a challenge; because of ion suppression, low abundance phospholipids are often not detected when analyzed in the presence of high abundance phospholipids [3,5]. Therefore, chromatographic separation of each phospholipid class before mass analysis should increase detection of low abundance phospholipids.
Chromatographic separation also provides a mass spectrum that shows the species of only a specific class of phospholipid, which makes it simple to identify compositional changes or the appearance of new species within a class of phospholipids. In particular, this profile of phospholipids is important for analysis of mitochondrial phospholipids, especially of cardiolipin (CL).
CL, an unusual phospholipid highly localized to the mitochondrial inner membrane, is essential for the proper function of mitochondrial enzymes and for membrane integrity. Molecular species of CL contain multiple polyunsaturated fatty acids, which make CL highly susceptible to oxidative modification [6-9]. The removal of these oxidized acyl chains of CL has been suggested to be the major mechanism of CL remodeling under oxidative stress, which often results in changes in composition or content of CL species.
CL affects mitochondrial function. For example, Barth syndrome is a genetic disorder with mutations in the tafazzin gene resulting in mitochondrial dysfunction caused by a decrease in the content and/or a change in the composition of CL. These changes also have been observed in various pathological conditions or with different fat diets [10-11]. However, the detailed mechanisms how CL is synthesized or how mitochondrial pathology is related to the structure of CL are not well understood.
In biological systems, CL is made by de-novo synthesis from phosphatidylglycerol (PG) and by remodeling through monolysocardiolipin (MLCL). Therefore, analysis of CL, PG, and MLCL will provide mechanistic information concerning the biosynthesis of CL. Recently, MALDI-TOF analysis of phospholipids in intact mitochondria was reported [12]. The method provides a quick and simple procedure and detection of various lipids classes, including these low abundance phospholipids. Although content of MLCL is enhanced in certain pathological conditions such as Barth syndrome [13], PG and MLCL usually are below the detection limit of common analytical methods and the detailed compositions of these phospholipids compared to CL have not been studied. Reasons for their inconsistent detection may be ion suppression caused by more abundant analytes. Therefore, separation of phospholipids into their classes before mass analysis should increase the sensitivity and reproducibility, especially of low abundance phospholipids.
Since the pioneering work of W.W. Christie, traditionally normal phase chromatography has been used to separate phospholipids into their classes [14,15]. Previous studies successfully applied HPLC-MS for phospholipids analysis [15-19]. The class separation enabled sensitive detection of phospholipids and extended the application of mass spectrometry to various biological sources. We have developed and validated a normal-phase HPLC-MS method for quantitative and qualitative analysis of mitochondrial phospholipids. The chromatographic conditions provide separation of 12 classes of standardized phospholipids. The sample simplification procedure involves organic extraction followed by harvesting phospholipids by means of a pre-packed silica gel solid phase extraction (SPE) column optimized for recovery. We separated and detected seven classes of phospholipids from mitochondria and six of these phospholipids, including MLCL, were quantified. We examined rat heart and mouse heart mitochondria for the composition of MLCL compared to the composition of CL. The new method provides optimized extraction and simplification and, in addition, reproducible HPLC-ESI-MS analysis of mitochondrial phospholipids for quantitative and qualitative analysis.
2. Materials and methods
2.1 Chemicals and materials
HPLC-grade solvents, chloroform (CHCl3), acetonitrile (ACN), isopropanol (IPA), methanol (MeOH), isooctane (IOA), ethylacetate (EA), acetic acid (AcOH), and formic acid were obtained from Fisher Scientific (Pittsburgh, PA, USA). tert-Butylmethylether (TBME) was purchased from Alfa Aesar (Ward Hill, MA, USA). Standard phospholipids of biological origin, PG from chicken egg, phosphatidylinositol (PI) from bovine liver, phosphatidylethanolamine (PE) from bovine heart, phosphatidylcholine (PC) from bovine heart, phosphatidylserine (PS) from porcine brain, sphingomyelin (SM) from porcine brain, lyso-PE (LPE) from chicken egg, lyso-PI (LPI) from bovine liver, and lyso-PC (LPC) from chicken egg, and synthetic standard phospholipids, CL(14:0)4, CL(18:1)4, PC(16:0)2, PC(22:0)2, and PC(18:0)(22:6), were obtained from Avanti Polar Lipids (Alabaster, AL, USA). CL from bovine heart, 1,2-dipalmitoyl-sn-glycero-3-phospho-N-methylethanolamine (PME used as the internal standard), butylated hydroxytoluene, and ammonium formate were purchased from Sigma Aldrich (St. Louis, MO, USA). 1,2-dipalmitoyl-sn-glycero-3-phospho-N,N-dimethylethanolamine was purchased from 3B Scientific Corp. (Libertyville, IL, USA). Bovine Albumin (BSA) Fraction V Solution was purchased from Invitrogen (Carlsbad, CA, USA) and diluted in 100 mM (pH 7.4) potassium phosphate buffer (KPi). Pre-packed silica gel SPE columns were purchased from Biotage (Charlotte, NC, USA). MLCL and dilysocardiolipin (DLCL) were prepared as previously reported [20]. Heart (subsarcolemmal) mitochondria were isolated from Sprague-Dawley rat or C57BL/6 mouse, and skeletal muscle mitochondria from Sprague-Dawley rat by differential centrifugation [21,22]. Reagent grade water was prepared using a Milli-Q reagent water system (Millipore, Inc., Bedford, MA, USA).
2.2 Standardization of phospholipids
Stock standard phospholipids and PME solutions were prepared by dissolving phospholipids in CHCl3:MeOH, 4:1; their concentrations were determined by organic phosphorus analysis as described by Bartlett [22,23]. The concentration of the stock solution was expressed as molarity of phospholipid (2 M phosphate is equivalent to 1 M phospholipid for CL and MLCL). The purity and masses of species in each class of phospholipid were examined by HPLC-MS and were in accord with the manufacturer's label.
A working solution of standardized phospholipid mixture in CHCl3 was prepared by mixing standardized phospholipids to yield final concentrations in μM of 80 (PE), 3.2 (PG), 16 (PI), 24 (CL), 0.8 (MLCL), and 120 (PC). Five additional solutions with different concentrations were prepared by consecutive 1:1 dilution starting with the original working solution in CHCl3. A total of six working solutions were used to generate standard curves for each class of phospholipid. The masses of each of these standardized phospholipids are shown in Figure S1 (supplementary data).
2.3 Extraction of mitochondrial phospholipids
Isolated mitochondria were diluted in 50 μL of 100 mM KPi buffer and extracted by the method of Christiansen [24]. Mitochondria (30 μg or 15 μg) in 50 μL of 100 mM KPi buffer was mixed with 72 pmol of PME (internal standard) and extracted with 1 mL of CHCl3:MeOH, 2:1 containing 2 mM butylated hydroxytoluene as an antioxidant. The mixture was vortexed for 2 min and centrifuged for 10 min at 2000×g. The supernate was decanted and evaporated to dryness under N2.
2.4 Sample simplification
Phospholipids were separated from the lipid mixture extracted from mitochondria using a pre-packed silica gel SPE column (25 mg, Biotage). To prepare the column for SPE, the column was washed with 1 mL of 10 mM ammonium acetate in MeOH and equilibrated with 0.5 mL of IOA:EA:AcOH (75:25:2)
The mitochondrial lipid residue was reconstituted in 0.2 mL of IOA:EA:AcOH (75:25:2) and applied to the column. An additional 0.2 mL of IOA:EA:AcOH (75:25:2) was used to rinse the tube and applied to the column. The lipid-loaded column was further washed with 0.5 mL of IOA:EA:AcOH (75:25:2) followed by 0.5 mL of IOA:EA (75:25). Finally, phospholipids were eluted with 1.5 mL of MeOH and evaporated to dryness under N2.
2.5 Normal-phase HPLC-MS
The HPLC system (Agilent 1100 Series) to separate each class of phospholipid was developed by modification of a previously reported method [18]. A Nucleosil diol column (5 μm, 3×250 mm) from Macherey-Nagel (Duren, Germany) was used. Eluent A contained IPA:TBME:ammonium formate (340:170:50) and eluent B contained MeOH. Aqueous ammonium formate (pH ∼2.5) was prepared by dissolving 295 mg of ammonium formate and 2 mL of formic acid in 50 mL of water. The gradients used for the 45 min chromatogram were as follows: 100 % A for 20 min, 100% A to 20% A over 10 min, 20% A for 6 min, 20% A to 100% A over 1 min, and hold 100% A for 6 min. The flow rate was 0.3 mL/min and the column temperature was 30°C. At the end of each batch of analysis, which includes both standardized and mitochondrial phospholipids, the HPLC column was exhaustively washed with MeOH for 20 min. For a new set of analysis, the HPLC column was equilibrated with eluent A until the retention times of standardized phospholipids stabilized (up to 5 h). After equilibration, the phospholipid fraction from the SPE was evaporated and reconstituted in 100 μL of eluent A and 25 μL of this solution was injected in to the HPLC. MS data were obtained with a Thermo LCQ Deca mass spectrometer operated in the negative ion mode, collecting full scan MS data from 400 – 2000 m/z. The source parameters were: sheath gas flow (80 units), auxiliary gas flow (30 units), spray voltage (-5 kV), capillary temperature (300 °C), and capillary voltage (-25 V).
2.6 Generation of standard curves
Standard curves were generated using standardized phospholipids, which had been mixed and extracted from BSA. Briefly, each working solution of standardized phospholipids was mixed with PME (internal standard) and evaporated under N2. The residue was mixed with 50 μL of 0.2% BSA in 100 mM KPi and extracted by the method of Christiansen. The extract was subjected to SPE and analyzed by HPLC-MS as described above. The intensity of each class of phospholipid with six different concentrations was compared to that of the same amount of PME to generate standard curves.
2.7 Influence of acyl chain on ionization efficiency
The influence of acyl chain on the ionization efficiency of species of PC and CL was tested using two species of CL, CL(14:0)4 and CL(18:1)4, and three species of PC, PC(16:0)2, PC(22:0)2, and PC(18:0)(22:6). Each of these phospholipids was analyzed by HPLC-MS at the concentrations used to generate the standard curves.
2.8 Efficiency of extraction solution
To assess the efficiency of extraction, 2 nmol each of 12 standardized phospholipids were mixed with 50 μL of 0.2% BSA in 100 mM KPi (pH 7.4) and phospholipids were extracted with 1 mL of ACN:IPA, 3:1, 1 mL of ACN:MeOH, 3:1, the method of Folch [25], or of Christiansen (1 mL of CHCl3:MeOH, 2:1) [24]. The phospholipids extracted were subjected to pre-packed silica gel SPE column (see below for details), mixed with PME (external standard), and injected for HPLC-MS analysis. The same amounts of standardized phospholipids and PME without extraction were used as the control. Efficiency was determined by comparing the peak area ratios of all species in each class of phospholipid recovered from the extractions to those of control phospholipids. The recovery of phospholipids from pre-packed silica gel SPE column alone also was determined in a similar manner. The methods of Folch and of Christiansen also were compared for extraction of phospholipids from 30 μg of mitochondria using the same procedure from BSA.
2.9 Recovery of standardized phospholipids from mitochondria
The Christiansen method was tested for phospholipid extraction efficiency from mitochondria by adding back the standardized phospholipids. Standardized phospholipids in different concentrations were mixed with 10 μg (for PE and PC) or 30 μg (for PG, PI, CL, and MLCL) of rat skeletal muscle mitochondria in the presence of PME. Both exogenous and endogenous phospholipids were extracted from mitochondria by the method of Christiansen and simplified by SPE as described above. The phospholipid fraction was analyzed by HPLC-MS and the amount of each class of phospholipid was calculated using the standard curve generated with the same standardized phospholipids.
2.10 Precision of the method
To assess the precision of the method, quantitative analysis of mitochondrial phospholipids was performed for intraday and interday variations. Three aliquots of rat heart and skeletal muscle mitochondria suspension were kept at -60°C and one aliquot of each type of mitochondrion was thawed each day just before use. Heart and muscle mitochondria were diluted to 1 mg/mL concentration in 100 mM KPi buffer and 30 μg each was distributed into five or six tubes for skeletal mitochondria and 30 μg or 15 μg for heart mitochondria. Mitochondria in each tube were mixed with PME and extracted by the procedure of Christiansen. Phospholipids were separated by SPE and analyzed by HPLC-MS as described above.
The same procedure was repeated on three different days; concentrations of mitochondrial phospholipids were calculated with a different standard curve for each day.
3. Results
3.1 Phospholipids separation by chromatography
The HPLC system based on the method of Sommer et al. [18] was significantly modified to detect and quantify all classes of mitochondrial phospholipids, especially CL and MLCL. Different polarities and charges on the head groups of phospholipids provided obstacles in identification of an eluting system for both chromatographic separation and efficient ionization.
We tried multiple HPLC eluting systems, varying ratios of solvents and pH, and identified optimal conditions for MS analysis of all 12 standardized phospholipids of biological origin in a single ion mode by reducing the pH of ammonium formate to ∼2.5. This low optimal pH also was essential to detect PS, because at higher pH, PS gave no detectable chromatographic peak. This phenomenon may be based on an intramolecular hydrogen bond between adjacent carboxyl group and amine group in the serine moiety; formation of this bond will be prevented by protonation on carboxylic group at pH 2.5. Alternatively, the increased amount of proton has a similar effect as did EDTA used by Myint et al. [26] by reducing the interaction of metal ions with PS. Representative ion chromatograms of 12 standardized phospholipids are shown in Figure S2 (supplementary data); ion chromatograms were generated using masses of the most abundant species in each class. The retention times and species of each class of phospholipid were determined by individual HPLC-MS analysis of the standardized phospholipids of biological origin.
Additionally, the eluting conditions, especially pH, increased the sensitivity of negatively-charged phospholipids in the negative ion mode. For example, the lower limit of quantitation of PG, PI, CL, and MLCL is 4 pmol injected, whereas the limit of detection of these phospholipids is 1 or 2 pmol injected. This high sensitivity for negatively-charged phospholipids is a great advantage for analysis of mitochondrial phospholipids, because mitochondria have a lower content of negatively-charged phospholipids compared to positively-charged phospholipids.
Using the extraction method, SPE, and HPLC-MS, seven classes of phospholipids, PE, PG, PI, PS, CL, MLCL, and PC were detected in isolated heart and skeletal muscle mitochondria. The content of PS was below the limit of quantitation and we excluded it from analysis.
In the analysis of mitochondrial phospholipids, individual phospholipid peaks were identified either by comparing the masses to those of the standardized phospholipids of biological origin or by MS/MS. One example of the MS/MS spectrum of a PE species in rat skeletal muscle mitochondria is shown in the supplement data (Figure S3). For each new sample, this procedure was followed to establish an appropriate range for each phospholipid class. Inspection of ions recorded provided a range from lowest to highest mass; the range was used to generate the ion chromatograms used for both quantitation and identification of that particular phospholipid. The ranges for rat heart and skeletal muscle mitochondria in m/z were 737-800 (PE), 704-708 (PME, internal standard), 744-775 (PG), 860-915 (PI), 1420-1520 (CL), 1185-1188 (MLCL), and 740-885 (PC), which were similar to the ranges of standardized phospholipids of biological origin. The mass spectra of these six phospholipids extracted from skeletal muscle mitochondria are shown in Figure 1 and the chromatograms are in Figure 2.
Figure 1.
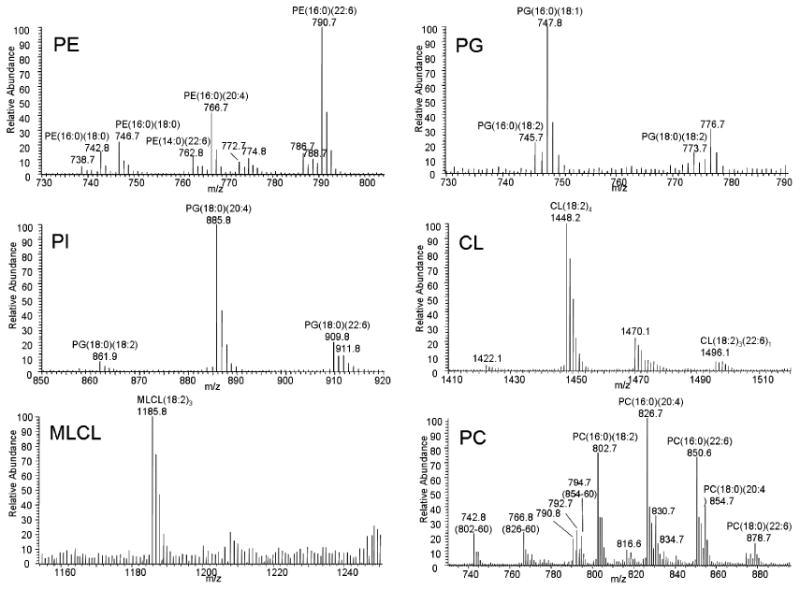
Mass spectra of phospholipids from skeletal muscle mitochondria. Mass spectra ranges are derived from the ion chromatogram eluting at the same retention time window as standardized phospholipids.
Figure 2.
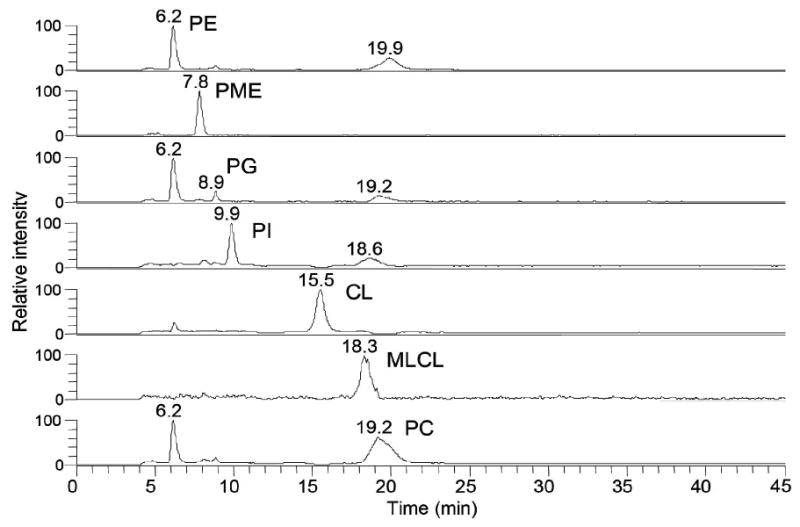
Representative ion chromatogram of skeletal muscle mitochondrial phospholipids. Mass ranges for each chromatographic peak are PE, 737-800, PME, 704-708; PG, 744-775; PI, 860-915; CL,1420-1520; MLCL, 1185-1189; PC, 740-885.
The HPLC method showed good long term reproducibility. The retention times of standardized phospholipids used to generate standard curves were evaluated over a 7 month period. A sample from each day that contains the same concentrations was chosen to compare the retention times. As shown in Table 1, coefficients of variation of the retention time are less than 7% for all class of phospholipids. All data were generated using the same column that had been used to develop chromatographic conditions.
Table 1. Long-term reproducibility of retention time of standardized phospholipids (N=17, 7 month period).
| PE | PME | PG | PI | CL | MLCL | PC | |
|---|---|---|---|---|---|---|---|
| Avg. | 6.1 | 7.9 | 8.8 | 9.9 | 15.9 | 18.9 | 21 |
| STDV | 0.1 | 0.2 | 0.1 | 0.1 | 0.3 | 0.4 | 1.4 |
| C.V. (%) | 1.6 | 2.5 | 1.1 | 1.0 | 1.9 | 2.1 | 6.7 |
3.2 Internal standard
Lysyl phosphatidylglycerol, 1,2-dipalmitoyl-sn-glycero-3-phospho-N,N-dimethylethanolamine, and PME were tested as internal standards. However, the former two were not chromatographically well separated from the other phospholipids and lysyl phosphatidylglycerol yielded broad split chromatographic peaks under the final chromatographic conditions. PME eluted as a well-defined peak between PE and LPE. Although PME eluted in the early part of the chromatogram, its use as an internal standard for late eluting phospholipids, such as CL and PC, provided a linear mass response over a wide range of concentrations. Thus, for quantitation of mitochondrial phospholipids, PME was a good internal standard. The mass range for PME was 704-708
3.3 Standard curves
There were large variations in the concentration of each different class of mitochondrial phospholipid. For example, the concentrations of PE and PC were about 40 times greater than that of PG and over 100 times greater than that of MLCL. Thus, it was incumbent on us to find the ranges of standard curves that cover the concentration of each phospholipid in mitochondria. Using chromatographic separation of each class of phospholipid, the final method generated standard curves with excellent R2 that covered the required ranges. Figure 3 shows typical standard curves with the ranges generated during one of the days for validation of the method.
Figure 3.
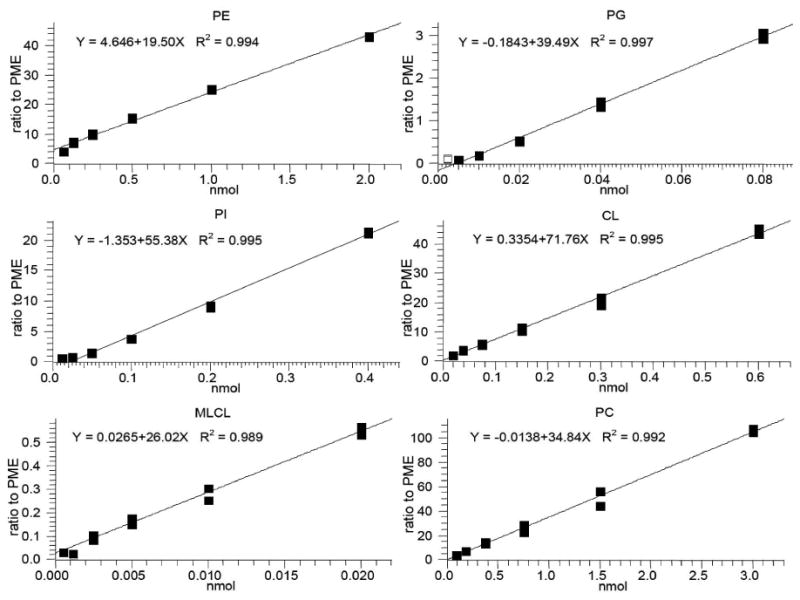
Representative standard curves of six classes of phospholipids with PME as internal standard. Standardized phospholipids with six different concentrations were extracted and analyzed by HPLC-MS. Each data point represents the relative areas of chromatographic peaks of phospholipid compared to that of PME at a given concentration.
We optimized the amount of mitochondria for extraction based on the concentration of mitochondrial phospholipids in relationship to the standard curve for each individual phospholipid. Phospholipids extracted from 30 μg of heart and skeletal muscle mitochondria provided an adequate signal for all the phospholipids to fit within their standard curves. At this amount of mitochondria, the concentration of MLCL injected was at the low end of its standard curve, and thus just above the limit of quantitation. However, the amount of mitochondria extracted could not be increased to obtain a larger signal for MLCL because the PE signal was at the high end of its standard curve (2 nmol) and the linearity of PE fell off quickly above this amount.
3.4 Influence of acyl chain on ionization efficiency
The ionization efficiency of phospholipids mainly depends on the head group that determines the class of phospholipid [27]. Additionally, the chain length and the number of double bonds of the acyl chains also affect the ionization efficiency and the response over a range of concentrations [28,29]. However, the influence of acyl chain becomes insignificant in dilute concentrations [29,30]. Han et al. stated that the acyl chain should not affect the quantitation of phospholipids at lower than 10 pmol/μl [30]. Because we used chromatography, whereas Han's statement is based on infusion studies, we tested the influence of acyl chain on ionization efficiency under the conditions used for our assay.
We examined the ionization efficiency of two CL species, CL(14:0)4 and CL(18:1)4, and three PC species, PC(16:0)2, PC(18:0)(22:6), and PC(22:0)2 at the highest concentrations used for these phospholipids to generate the standard curves. The result showed that two species of CL at 0.6 nmol total had almost identical response (Figure 4A) indicating that the concentration of CL was below the concentration in which the influence of acyl chain is significant. For PC species at the concentration of 3 nmol total, PC(16:0)2 and PC(18:0)(22:6) had essentially identical responses but the response of PC(22:0)2 was about 60% of the other two species (Figure 4B). The difference in the response of PC(22:0)2 was gradually reduced with consecutive dilutions (Figure 4B). We considered that the lower ionization efficiency of PC(22:0)2 was derived from a combination of long chain length and complete saturation of the acyl chain as shown previously [29].
Figure 4.
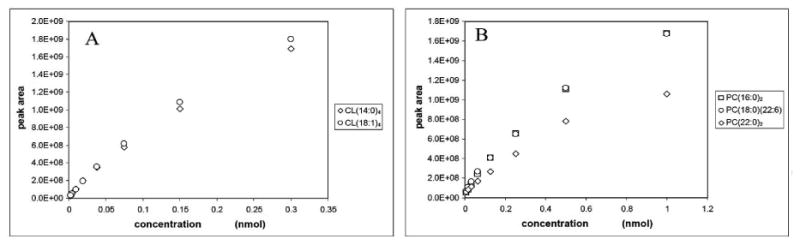
The comparison of mass responses of two species in CL and three species in PC over a range of concentrations.
However, PC(22:0)2 is a synthetic, non-biological species and, if found, would be at a level much below where acyl chains have an influence on the ionization efficiency. PC(16:0)2 and PC(18:0)(22:6) well represent PC species of biological origin and have the same ionization efficiency. This indicates that the influence of acyl chains is unlikely to be significant in the quantitation of actual biological samples. We examined PC species in various biological origins and the molecular profiles are shown in the supplement data (Figures S5 – S10). The data clearly show that the majority of PC species are within a mass window between PC(16:0)2 and PC(18:0)(22:6). Therefore, we concluded that the influence of acyl chain on the quantitation of phospholipid is negligible under the conditions used with biological samples. Moreover, the use of standardized phospholipids, whose molecular patterns are similar to those of analytes, further reduces the variation caused by acyl chain as discussed previously [31].
3.5 Sample preparation
We previously reported sample simplification using a silica gel column to harvest phospholipids from lipid extracts [25,32]. The direct application of this sampling procedure was unsuccessful because the HPLC separation of phospholipids was altered by unknown impurities, particularly in the PI and CL area accompanied by a high background. The method developed herein uses significantly smaller amounts of mitochondria than previously used [25,32]. We suspected that the impurities were from the silica gel column and were noticeable because of the smaller mitochondria sample. For this reason, we evaluated commercially pre-packed smaller silica gel SPE columns. The impurities eluting from the Biotage (25 mg) column were greatly reduced. We additionally examined washing procedures and found that washing the column with 1 mL of 10 mM ammonium acetate in MeOH markedly reduced any further eluting impurities.
The combination of the Biotage column and the washing procedure stabilized the chromatography of the extract. Because we used a different silica gel column with a different washing and eluting procedure from our previous work [25,32], we needed to document the recovery of phospholipids from the SPE step. Using the procedure given in the Methods Section, all 12 standardized phospholipids were recovered in a quantitative fashion (Figure S11, supplementary data).
Four extraction systems were evaluated to determine extraction efficiency for standardized phospholipids from BSA, which was used as a surrogate biological system. In general, Folch extraction provided good extraction efficiency for most phospholipids except PS (60%), DLCL (66%), LPC (40%) and LPI (8%). The extraction procedure described by Christiansen provided equal extraction efficiency for phospholipids and improved extraction efficiency with LPC (68%) and LPI (47%). We also tested two other extraction systems used in our lab, ACN:IPA (3:1, 1 mL) [33] and ACN:MeOH (3:1, 1 mL) [34], and both were not as efficient as the method of Christiansen (Figure 5).
Figure 5.
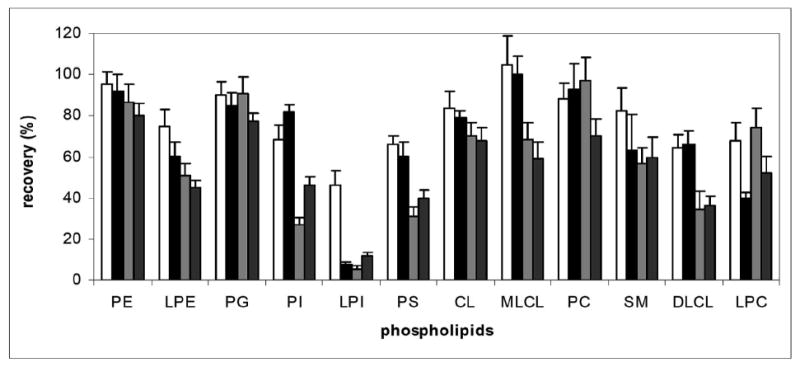
Recovery of standardized phospholipids extracted from BSA and from pre-packed silica gel SPE column (n=6). Standardized phospholipids were mixed with 0.2% BSA and extracted with ACN:MeOH (dark grey), ACN:IPA (light grey), method of Folch (black) or method of Christiansen (white). The extracts were subjected to the silica gel column and recovery was determined by comparing the amounts of standardized phospholipids recovered from extraction and silica gel column to the same amount of control standardized phospholipids using PME as an external standard.
We next compared the methods of Folch [25] and Christiansen [24] for phospholipid extraction from mitochondria. Although not significant, there was a consistent trend in greater recovery with Christiansen's method (Figure 6). With similar, or slightly improved, extraction, but with the more convenient monophasic system, we selected Christiansen's method [24] for phospholipid extraction.
Figure 6.
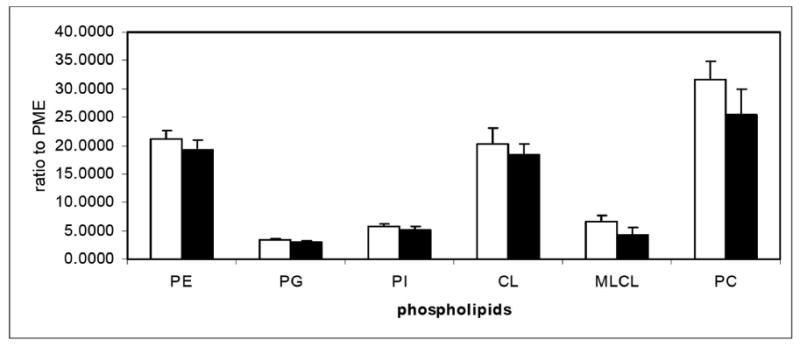
Relative recovery of mitochondrial phospholipids (n=6). Mitochondria were extracted by the method of Folch (black) or by the method of Christiansen (white) and the amount of phospholipids extracted were compared using PME as an external standard.
Finally, the extraction efficiency of the Christiansen method of phospholipids over a range of concentration was tested. Standardized phospholipids in different concentrations were added to mitochondria and both endogenous and exogenous phospholipids were extracted and analyzed. Because of the high concentrations of endogenous PC and PE in mitochondria, 10 μg of mitochondria was used for PE and PC, whereas 30 μg of mitochondria was used for the remaining phospholipids. The amounts of each class of phospholipid were quantified using standard curves generated by the same standardized phospholipids mixed and extracted from BSA. The amounts of phospholipids added were plotted against amount detected. Figure 7 shows that for all phospholipids, the extraction was almost equally efficient from BSA and mitochondria and the responses were linear in the ranges examined.
Figure 7.
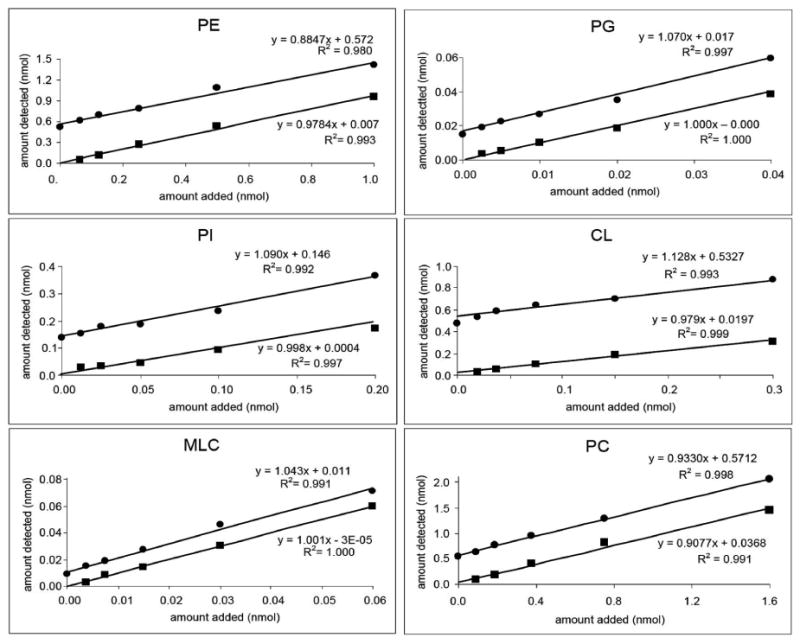
Standardized phospholipids were added to mitochondria (circle) or BSA (square), extracted, and analyzed by HPLC-MS. The amount of each class of phospholipid was calculated using the standard curve and plotted against the amount added to mitochondria (10 μg for PE and PC, 30 μg for PG, PI, CL, and MLCL).
3.6 Validation and quantitation
To assess the precision of the method, the concentration of each class of phospholipid was determined on three different days with standard curves on each day. Phospholipids were extracted from 30 μg of heart or skeletal muscle mitochondria and 15 μg of heart mitochondria for low concentration. The amount of each class of phospholipid detected is shown in Table 2. The interday and the intraday variations were within 10%, except for PG and MLCL. The accuracy of the concentration of MLCL was limited by the fact that the amount of MLCL detected by HPLC-MS was just above the quantitation limit where noise became significant.
Table 2.
Validation-Precision and Reproducibility. Amount (nmol) detected in 25 μL injected of 100 μL final extract.
| rat mitochondria | PE | PG | PI | CL | MLCL | PC | total | |
|---|---|---|---|---|---|---|---|---|
| A - 15 μg heart | ||||||||
| Trial 1 (N=6) | Avg. | 0.718 | 0.016 | 0.052 | 0.177 | NQ | 0.673 | 1.640 |
| STDV | 0.084 | 0.002 | 0.006 | 0.023 | 0.021 | 0.111 | ||
| CV (%) | 11 | 13 | 12 | 13 | 3.1 | 6.8 | ||
| Trial 2 (N=6) | Avg. | 0.694 | 0.018 | 0.051 | 0.183 | NQ | 0.679 | 1.629 |
| STDV | 0.039 | 0.001 | 0.002 | 0.011 | 0.079 | 0.121 | ||
| CV (%) | 5.6 | 5.6 | 3.9 | 6.0 | 12 | 7.4 | ||
| Trial 3 (N=6) | Avg. | 0.722 | 0.016 | 0.058 | 0.179 | NQ | 0.614 | 1.592 |
| STDV | 0.040 | 0.000 | 0.000 | 0.010 | 0.060 | 0.100 | ||
| CV (%) | 5.5 | 0.0 | 0.0 | 5.6 | 9.8 | 6.3 | ||
|
|
||||||||
| Total (N=18) | Avg. | 0.711 | 0.017 | 0.054 | 0.180 | NQ | 0.655 | 1.616 |
| STDV | 0.056 | 0.002 | 0.005 | 0.016 | 0.063 | 0.106 | ||
| CV (%) | 7.8 | 9.9 | 9.0 | 8.6 | 9.5 | 6.5 | ||
| B - 30 μg heart | ||||||||
| Trial 1 (N=5) | Avg. | 1.267 | 0.032 | 0.119 | 0.314 | 0.006 | 1.212 | 2.950 |
| STDV | 0.132 | 0.003 | 0.011 | 0.029 | 0.002 | 0.073 | 0.218 | |
| CV (%) | 10 | 9.4 | 9.2 | 9.2 | 33 | 6.0 | 7.4 | |
| Trial 2 (N=5) | Avg. | 1.275 | 0.035 | 0.102 | 0.291 | 0.004 | 1.152 | 2.860 |
| STDV | 0.138 | 0.011 | 0.004 | 0.025 | 0.000 | 0.144 | 0.089 | |
| CV (%) | 10.8 | 31.4 | 3.9 | 8.6 | 10 | 12.5 | 3.1 | |
| Trial 3 (N=6) | Avg. | 1.277 | 0.029 | 0.103 | 0.304 | 0.005 | 1.164 | 2.882 |
| STDV | 0.064 | 0.002 | 0.006 | 0.015 | 0.001 | 0.064 | 0.133 | |
| CV (%) | 5.0 | 6.9 | 5.8 | 4.9 | 20 | 5.5 | 4.6 | |
|
|
||||||||
| Total (N=16) | Avg. | 1.241 | 0.031 | 0.107 | 0.296 | 0.005 | 1.142 | 2.887 |
| STDV | 0.105 | 0.007 | 0.010 | 0.024 | 0.001 | 0.095 | 0.149 | |
| CV (%) | 8.3 | 20.8 | 9.6 | 7.8 | 29.8 | 8.1 | 5.1 | |
| C - 30 μg skeletal muscle | ||||||||
| Trial 1 (N=6) | Avg. | 1.401 | 0.018 | 0.155 | 0.356 | 0.010 | 1.491 | 3.431 |
| STDV | 0.066 | 0.001 | 0.008 | 0.024 | 0.002 | 0.087 | 0.143 | |
| CV (%) | 4.7 | 5.6 | 5.2 | 6.7 | 20 | 5.8 | 4.2 | |
| Trial 2 (N=6) | Avg. | 1.316 | 0.021 | 0.143 | 0.349 | 0.006 | 1.474 | 3.308 |
| STDV | 0.069 | 0.001 | 0.011 | 0.017 | 0.000 | 0.086 | 0.170 | |
| CV (%) | 5.2 | 4.8 | 7.7 | 4.9 | 0.0 | 5.8 | 5.1 | |
| Trial 3 (N=6) | Avg. | 1.415 | 0.020 | 0.160 | 0.369 | 0.008 | 1.434 | 3.407 |
| STDV | 0.087 | 0.001 | 0.010 | 0.021 | 0.001 | 0.063 | 0.153 | |
| CV (%) | 6.1 | 5.0 | 6.3 | 5.7 | 13 | 4.4 | 4.5 | |
|
|
||||||||
| Total (N=18) | Avg. | 1.377 | 0.020 | 0.153 | 0.358 | 0.008 | 1.466 | 3.372 |
| STDV | 0.083 | 0.002 | 0.011 | 0.022 | 0.002 | 0.079 | 0.156 | |
| CV (%) | 6.1 | 8.6 | 7.5 | 6.0 | 25.9 | 5.4 | 4.6 | |
In a preliminary experiment, the concentration of MLCL in frozen mitochondria appeared to be ∼2 or 3 times higher than in fresh mitochondria, but no changes in the amount of other phospholipids were observed. We believe that the level of MLCL is artificially increased in frozen mitochondria and that the increase is due to CL degradation. However, a two to three fold increase in the content of MLCL (0.6 nmol in freshly isolated mitochondria) will decrease CL only by ∼ 1 nmol from 40 nmol of CL. The change is less than 3%. The difference between 39 mmol and 40 nmol of CL is not easily observed.
Using the developed method, the concentration of each class of phospholipid in rat heart mitochondria and rat skeletal muscle mitochondria was determined. Table 3 shows the concentrations of each class of phospholipid per mg mitochondrial protein. Generally, the concentrations of phospholipids were higher in skeletal muscle mitochondria than in heart mitochondria, except for PG. The concentration MLCL was the lowest of the mitochondrial phospholipids observed and accounted for only about 0.2% of total phospholipid content.
Table 3. Phospholipid content in rat heart mitochondria (N=16) and rat skeletal muscle (N=18) mitochondria (nmol/mg mitochondria protein).
| PE | PG | PI | CL | MLCL | PC | total | |
|---|---|---|---|---|---|---|---|
| Heart | 165.5 | 4.1 | 14.3 | 39.5 | 0.7 | 152.3 | 384.9 |
| STDV | 7.1 | 0.3 | 1.5 | 3.1 | 0.2 | 11.2 | 6.3 |
| CV (%) | 4.3 | 7.3 | 10.5 | 7.8 | 29 | 7.4 | 1.6 |
| muscle | 183.7 | 2.7 | 20.4 | 47.7 | 1.1 | 195.5 | 449.6 |
| STDV | 7.2 | 0.3 | 1.2 | 1.3 | 0.3 | 3.9 | 8.7 |
| CV (%) | 3.9 | 11 | 5.9 | 2.7 | 27 | 2.0 | 1.9 |
The concentrations of mitochondrial phospholipids were higher than those previously reported using the organic phosphorous assay [22,32]. We believe these differences are the result of the method we used employs an internal standard added before extraction, thus compensating for any losses during extraction or in the silica gel column. The phosphate assay starts with the organic PO4- content of the phospholipid fraction obtained from the silica gel column [32], and thus does not account for any losses during the Folch extraction or the silica gel step. Additionally, using the external standard recovery method, we demonstrate that the Christiansen method has a slightly higher efficiency than does the Folch extraction, a fact that may contribute to the observed difference.
3.7 Qualitative analysis of CL and MLCL
CL, a phospholipid mainly found in the inner membrane of mitochondria, has been extensively studied for its role in the activities of electron transport chain complexes and translocase enzymes [35,36]. The composition of CL varies depending on the species, organ [37], and pathological conditions [38,39]. The composition of CL depends mainly on the remodeling process. MLCL is suggested to be the key intermediate for CL remodeling, but the composition of MLCL, unlike that of CL, is not well characterized.
We compared the mass spectra of CL and MLCL between rat heart and mouse heart mitochondria. A significant amount of the CL species containing docosahexaenoic acid (22:6) or oleic acid (18:1) was found in mouse heart mitochondria; the major species of CL in mouse heart mitochondria were CL(18:2)4, CL(18:2)3(22:6)1, and CL(18:2)2(22:6)2, whereas CL(18:2)4 was the only major species in rat heart mitochondria (Figure 8), in agreement with previous results from our laboratory based on analysis of CL by reverse-phase HPLC-MS/MS [37].
Figure 8.
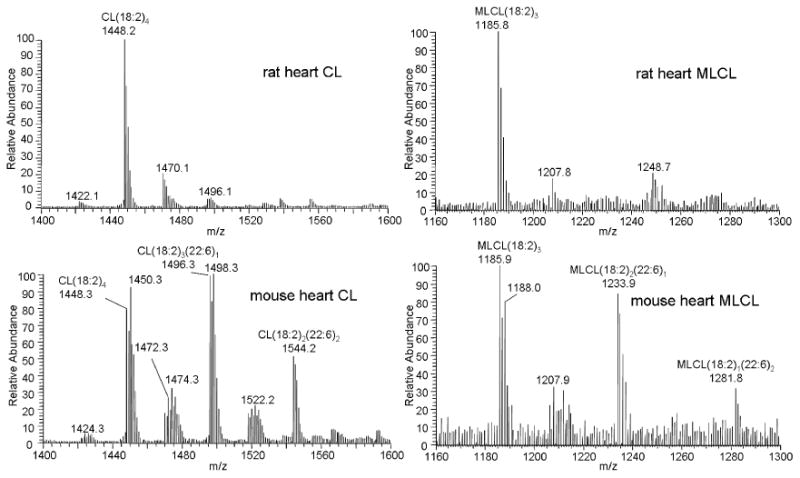
Mass spectra of CL and MLCL extracted from rat heart and mouse heart mitochondria. The mass spectra show increased amounts of CL and MLCL species containing docosahexaenoic acid (22:6) and oleic acid (18:1) in mouse heart. The structure of the peaks was assigned based on their molecular weights.
Interestingly, these same molecular patterns were observed with MLCL (Figure 8). The major species in MLCL from rat heart was MLCL(18:2)3, indicated by the nominal peak at m/z 1186. The same peak in the mass spectrum of MLCL in mouse heart indicated that MLCL(18:2)3 was still the most abundant MLCL species. Consistent with CL, the amounts of docosahexaenoic acid and oleic acid were significantly increased in MLCL in mouse heart. This finding is clearly shown in Figure 8 by the increase in the nominal peaks at m/z 1188, 1234, and 1282 corresponding to MLCL(18:2)2(18:1)1, MLCL(18:2)2(22:6)1, and MLCL(18:2)1(22:6)2, respectively. Thus the composition of MLCL correlates with the composition of CL.
In rat heart mitochondria, Bird et al. reported a change in the content of MLCL(18:2)3 compared to the total content of CL following different fat diets [40]. However, the interpretation of the change using a single species of MLCL as reflecting the change in the total content of MLCL needs to be more carefully addressed, because different fat diets may have changed both the composition of MLCL, as well as of CL, as shown in Figure 8. Our data show the advantage of chromatographic separation before MS analysis especially of low abundance phospholipids. To the best of our knowledge, we provide the first demonstration showing the species-dependent composition of MLCL compared to CL. Further analysis of CL and MLCL will help to understand the mechanisms of CL remodeling with different fatty acyl chains.
4. Discussion
Each class of phospholipid is composed of various amounts of individual species. The composition and content of these individual species in each class of phospholipid vary depending on the tissue and the species [11]. These variations in content and composition and the large number of phospholipids limit the accuracy of MS quantitative analysis because of ion suppression [41]. Moreover, ion suppression will be even more significant in the presence of other types of lipids, such as triacylglycerol and cholesterols.
Ion suppression can be significantly reduced by simplification of the sample and chromatographic separation of the phospholipids into their respective classes before MS analysis. Using chromatographic separation, we can readily monitor the appearance of new species in a class of phospholipid and quantitate them without interference from other classes. Therefore, sample simplification and normal-phase HPLC-MS provide more accurate quantitative analysis of phospholipids.
The chromatographic separation also increased sensitivity, especially of CL. And our procedure provide the most sensitive method to detect CL. Bird et al. showed in their recent paper, 5 pmol was close to the limit of detection of CL using Orbitrap MS [40]. This sensitivity was comparable to the triple quadrupole LC-MS [42]. Herein, we report a limit of quantitation of 4 pmol and a limit of detection of 1 pmol for CL using our new method.
5. Conclusions
A new method to analyze mitochondrial phospholipids was developed using an efficient extraction procedure and normal-phase HPLC-MS. The convenient Christiansen extraction provided a better extraction efficiency than Folch extraction for most phospholipids tested. The inclusion of PME as an internal standard and standardization of phospholipids were used to generate the standard curves to accurately quantify phospholipids in mitochondria. Chromatographic separation and high sensitivity for each class of phospholipid enabled quantitative as well as qualitative analysis of mitochondrial phospholipids, including MLCL. Application of this method to rat heart and skeletal muscle mitochondria showed that the HPLC-MS method is a powerful tool for reliable and reproducible quantitation of mitochondrial phospholipids.
Supplementary Material
Highlights.
We developed a comprehensive analytical method based on HPLC-MS to analyze mitochondrial phospholipids.
We optimized extraction and simplification procedures.
We developed normal phase HPLC conditions for a rigorous quantitation with an internal standard and standard curves.
We detected and quantitated low abundance phospholipids such as monolysocardiolipin and phosphatidylglycerol.
Acknowledgments
We thank Paul Minkler for his advice and assistance for this study, Dr. Mariana Rosca for providing isolated mitochondria, and Lori Hezel for performing the phosphorus analysis to standardize phospholipids. We appreciate Dr. Bernard Tandler and the Hoppel lab “Writing with Style” group for editorial assistance. This work was supported by NIH/NIA grant (5P01 AG015885).
Abbreviations
- CL
cardiolipin
- MLCL
monolysocardiolipin
- PG
Phosphatidylglycerol
- PI
phosphatidylinositol
- PE
phosphatidylethanolamine
- PC
phosphatidylcholine
- PS
phosphatidylserine
- SM
sphingomyelin
- DLCL
dilysocardiolipin
- SPE
solid phase extraction
- KPi
potassium phosphate buffer
- PME
1,2-dipalmitoyl-sn-glycero-3-phospho-N-methylethanolamine
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Paradies G, Ruggiero FM, Petrosillo G, Quagliariello E. Arch Gerontol Geriatr. 1993;16:263. doi: 10.1016/0167-4943(93)90037-i. [DOI] [PubMed] [Google Scholar]
- 2.Kiebish MA, Han X, Seyfried TN. Methods Mol Biol. 2009;579:3. doi: 10.1007/978-1-60761-322-0_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sato Y, Nakamura T, Aoshima K, Oda Y. Anal Chem. 2010;82:9858. doi: 10.1021/ac102211r. [DOI] [PubMed] [Google Scholar]
- 4.Han X, Yang J, Cheng H, Yang K, Abendschein DR, Gross RW. Biochemistry. 2005;44:16684. doi: 10.1021/bi051908a. [DOI] [PubMed] [Google Scholar]
- 5.Houjou T, Yamatani K, Imagawa M, Shimizu T, Taguchi R. Rapid Commun Mass Spectrom. 2005;19:654. doi: 10.1002/rcm.1836. [DOI] [PubMed] [Google Scholar]
- 6.Kim J, Minkler PE, Salomon RG, Anderson VE, Hoppel CL. J Lipid Res. 2011;52:125. doi: 10.1194/jlr.M010520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim J, Rodriguez ME, Oleinick NL, Anderson VE. Free Radic Biol Med. 2010;49:718. doi: 10.1016/j.freeradbiomed.2010.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim J, Fujioka H, Oleinick NL, Anderson VE. Free Radic Biol Med. 2010;49:726. doi: 10.1016/j.freeradbiomed.2010.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bayir H, Tyurin VA, Tyurina YY, Viner R, Ritov V, Amoscato AA, Zhao Q, Zhang XJ, Janesko-Feldman KL, Alexander H, Basova LV, Clark RS, Kochanek PM, Kagan VE. Ann Neurol. 2007;62:154. doi: 10.1002/ana.21168. [DOI] [PubMed] [Google Scholar]
- 10.Han X, Yang J, Yang K, Zhao Z, Abendschein DR, Gross RW. Biochemistry. 2007;46:6417. doi: 10.1021/bi7004015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hong MY, Chapkin RS, Barhoumi R, Burghardt RC, Turner ND, Henderson CE, Sanders LM, Fan YY, Davidson LA, Murphy ME, Spinka CM, Carroll RJ, Lupton JR. Carcinogenesis. 2002;23:1919. doi: 10.1093/carcin/23.11.1919. [DOI] [PubMed] [Google Scholar]
- 12.Angelini R, Vitale R, Patil VA, Cocco T, Ludwig B, Greenberg ML, Corcelli A. J Lipid Res. 2012;53:1417. doi: 10.1194/jlr.D026203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van Werkhoven MA, Thorburn DR, Gedeon AK, Pitt JJ. J Lipid Res. 2006;47:2346. doi: 10.1194/jlr.D600024-JLR200. [DOI] [PubMed] [Google Scholar]
- 14.McLaren DG, Miller PL, Lassman ME, Castro-Perez JM, Hubbard BK, Roddy TP. Anal Biochem. 2011;414:266. doi: 10.1016/j.ab.2011.03.009. [DOI] [PubMed] [Google Scholar]
- 15.Uran S, Larsen A, Jacobsen PB, Skotland T. J Chromatogr B Biomed Sci Appl. 2001;758:265. doi: 10.1016/s0378-4347(01)00188-8. [DOI] [PubMed] [Google Scholar]
- 16.Malavolta M, Bocci F, Boselli E, Frega NG. J Chromatogr B Analyt Technol Biomed Life Sci. 2004;810:173. doi: 10.1016/j.jchromb.2004.07.001. [DOI] [PubMed] [Google Scholar]
- 17.Hayakawa J, Okabayashi Y. J Pharm Biomed Anal. 2004;35:583. doi: 10.1016/j.jpba.2004.02.007. [DOI] [PubMed] [Google Scholar]
- 18.Sommer U, Herscovitz H, Welty FK, Costello CE. J Lipid Res. 2006;47:804. doi: 10.1194/jlr.M500506-JLR200. [DOI] [PubMed] [Google Scholar]
- 19.Narvaez-Rivas M, Gallardo E, Rios JJ, Leon-Camacho M. J Chromatogr A. 2011;1218:3453. doi: 10.1016/j.chroma.2011.03.067. [DOI] [PubMed] [Google Scholar]
- 20.Kim J, Hoppel CL. J Lipid Res. 2011;52:389. doi: 10.1194/jlr.D010587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Puchowicz MA, Varnes ME, Cohen BH, Friedman NR, Kerr DS, Hoppel CL. Mitochondrion. 2004;4:377. doi: 10.1016/j.mito.2004.07.004. [DOI] [PubMed] [Google Scholar]
- 22.Moghaddas S, Stoll MS, Minkler PE, Salomon RG, Hoppel CL, Lesnefsky EJ. J Gerontol A Biol Sci Med Sci. 2002;57:B22. doi: 10.1093/gerona/57.1.b22. [DOI] [PubMed] [Google Scholar]
- 23.Bartlett GR. J Biol Chem. 1959;234:466. [PubMed] [Google Scholar]
- 24.Christiansen K. Anal Biochem. 1975;66:93. doi: 10.1016/0003-2697(75)90728-9. [DOI] [PubMed] [Google Scholar]
- 25.Ingalls ST, Kriaris MS, Xu Y, DeWulf DW, Tserng KY, Hoppel CL. J Chromatogr. 1993;619:9. doi: 10.1016/0378-4347(93)80441-6. [DOI] [PubMed] [Google Scholar]
- 26.Myint KT, Uehara T, Aoshima K, Oda Y. Anal Chem. 2009;81:7766. doi: 10.1021/ac901269h. [DOI] [PubMed] [Google Scholar]
- 27.Kim HY, Wang TC, Ma YC. Anal Chem. 1994;66:3977. doi: 10.1021/ac00094a020. [DOI] [PubMed] [Google Scholar]
- 28.Brugger B, Erben G, Sandhoff R, Wieland FT, Lehmann WD. Proc Natl Acad Sci U S A. 1997;94:2339. doi: 10.1073/pnas.94.6.2339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Koivusalo M, Haimi P, Heikinheimo L, Kostiainen R, Somerharju P. J Lipid Res. 2001;42:663. [PubMed] [Google Scholar]
- 30.Han X, Gross RW. Mass Spectrom Rev. 2005;24:367. doi: 10.1002/mas.20023. [DOI] [PubMed] [Google Scholar]
- 31.Norris C, Fong B, MacGibbon A, McJarrow P. Lipids. 2009;44:1047. doi: 10.1007/s11745-009-3357-8. [DOI] [PubMed] [Google Scholar]
- 32.Lesnefsky EJ, Stoll MS, Minkler PE, Hoppel CL. Anal Biochem. 2000;285:246. doi: 10.1006/abio.2000.4783. [DOI] [PubMed] [Google Scholar]
- 33.Minkler PE, Kerner J, Ingalls ST, Hoppel CL. Anal Biochem. 2008;376:275. doi: 10.1016/j.ab.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Minkler PE, Stoll MS, Ingalls ST, Yang S, Kerner J, Hoppel CL. Clinical Chemistry. 2008;54:1451. doi: 10.1373/clinchem.2007.099226. [DOI] [PubMed] [Google Scholar]
- 35.Beyer K, Klingenberg M. Biochemistry. 1985;24:3821. doi: 10.1021/bi00336a001. [DOI] [PubMed] [Google Scholar]
- 36.Paradies G, Petrosillo G, Pistolese M, Di Venosa N, Federici A, Ruggiero FM. Circ Res. 2004;94:53. doi: 10.1161/01.RES.0000109416.56608.64. [DOI] [PubMed] [Google Scholar]
- 37.Minkler PE, Hoppel CL. J Lipid Res. 2010;51:856. doi: 10.1194/jlr.D002857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sparagna GC, Johnson CA, McCune SA, Moore RL, Murphy RC. J Lipid Res. 2005;46:1196. doi: 10.1194/jlr.M500031-JLR200. [DOI] [PubMed] [Google Scholar]
- 39.Sparagna GC, Chicco AJ, Murphy RC, Bristow MR, Johnson CA, Rees ML, Maxey ML, McCune SA, Moore RL. J Lipid Res. 2007;48:1559. doi: 10.1194/jlr.M600551-JLR200. [DOI] [PubMed] [Google Scholar]
- 40.Bird SS, Marur VR, Sniatynski MJ, Greenberg HK, Kristal BS. Anal Chem. 2011;83:940. doi: 10.1021/ac102598u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gross RW, Han X. Chemistry and Biology. 2011;18:284. doi: 10.1016/j.chembiol.2011.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Houtkooper RH, Rodenburg RJ, Thiels C, van Lenthe H, Stet F, Poll-The BT, Stone JE, Steward CG, Wanders RJ, Smeitink J, Kulik W, Vaz FM. Anal Biochem. 2009;387:230. doi: 10.1016/j.ab.2009.01.032. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


